Proteolysis of Cortactin by Calpain Regulates Membrane Protrusion during Cell Migration (original) (raw)
Abstract
Calpain 2 regulates membrane protrusion during cell migration. However, relevant substrates that mediate the effects of calpain on protrusion have not been identified. One potential candidate substrate is the actin binding protein cortactin. Cortactin is a Src substrate that drives actin polymerization by activating the Arp2/3 complex and also stabilizes the cortical actin network. We now provide evidence that proteolysis of cortactin by calpain 2 regulates membrane protrusion dynamics during cell migration. We show that cortactin is a calpain 2 substrate in fibroblasts and that the preferred cleavage site occurs in a region between the actin binding repeats and the α-helical domain. We have generated a mutant cortactin that is resistant to calpain proteolysis but retains other biochemical properties of cortactin. Expression of the calpain-resistant cortactin, but not wild-type cortactin, impairs cell migration and increases transient membrane protrusion, suggesting that calpain proteolysis of cortactin limits membrane protrusions and regulates migration in fibroblasts. Furthermore, the enhanced protrusion observed with the calpain-resistant cortactin requires both the Arp2/3 binding site and the Src homology 3 domain of cortactin. Together, these findings suggest a novel role for calpain-mediated proteolysis of cortactin in regulating membrane protrusion dynamics during cell migration.
INTRODUCTION
Regulated cell migration is critical to both normal and pathological processes, including development, immune function, and tumor metastasis. During migration, actin polymerization drives the extension of membrane projections at the leading edge, including filopodia and lamellipodia, which are required for directional migration. Our recent studies implicate a central role for the intracellular protease calpain 2 in the regulation of membrane protrusion during cell migration. Fibroblasts deficient in calpain 2 display increased transient protrusion activity and impaired efficiency of lamellipodial extension at the leading edge (Franco et al., 2004a). Calpain activity is also required for neuronal growth cone turning and stabilization of filopodia during growth cone guidance (Robles et al., 2003).
Calpain 2 is a ubiquitous, intracellular, calcium-dependent protease that has previously been implicated in cell migration through its capacity to regulate integrin-mediated adhesion complexes (Huttenlocher et al., 1997; Dourdin et al., 2001; Bhatt et al., 2002). Calpain is thought to regulate adhesions through proteolysis of key cytoskeletal and regulatory proteins such as talin, FAK, and paxillin. Previous studies have demonstrated that calpain proteolysis of talin occurs between the head and rod domain and functions to promote adhesion complex disassembly (Franco et al., 2004b). However, key substrates that mediate the effects of calpain 2 on membrane protrusion dynamics have not been identified.
Protrusion depends on actin polymerization at the barbed ends of actin filaments and their formation into a highly branched dendritic network that drives membrane extension at the leading edge of lamellipodia. The Arp2/3 complex has been identified as a key regulator of the actin network that acts by mediating actin nucleation at branch points on F-actin filaments. Arp2/3 is activated by several proteins, including the Rac and Cdc42 effector proteins of the WASp/SCAR/WAVE family, ActA, Abp1, and cortactin (reviewed in Pollard and Borisy, 2003). Cortactin has the additional function of stabilizing actin branch points, suggesting cortactin may play a key role in the dynamic assembly and disassembly of actin polymerization at the cell periphery (Weaver et al., 2001). Studies have shown that overexpression of cortactin may be associated with increased cell migration under some conditions (Patel et al., 1998) as well as enhanced membrane protrusion, most notably in the presence of the cortactin binding protein WIP (Kinley et al., 2003).
Cortactin contains an N-terminal acidic domain that mediates binding to Arp2/3 and binds actin through an adjacent set of six and a half tandem repeats of 37 amino acids (Weed et al., 2000). Following the tandem repeats is an α-helical domain of unknown function and a proline-rich domain, which is phosphorylated by Src and extracellular signal-regulated kinase (ERK) family kinases (reviewed in Weed and Parsons, 2001). Cortactin further regulates protrusion and migration through a C-terminal Src homology (SH)3 domain that binds to adaptor and signaling proteins, including WIP, WASp, dynamin, and the Cdc42 guanine nucleotide exchange factor Fgd1 (McNiven et al., 2000; Hou et al., 2003; Kinley et al., 2003; Martinez-Quiles et al., 2004). In addition to regulation by both Src and ERK-mediated phosphorylation (Huang et al., 1997a; Martinez-Quiles et al., 2004), there is evidence that proteolysis by calpain may regulate cortactin function in platelets (Huang et al., 1997b).
In this study, we sought to determine whether calpain cleavage of cortactin regulates protrusion in fibroblasts. We found that calpain 2 proteolyzes cortactin in fibroblasts and that the preferred calpain cleavage site is in a region between the actin binding repeats and the α-helical domain. We generated a mutant version of cortactin that is resistant to calpain proteolysis but retains other biochemical properties of cortactin, including SH3 domain function and Arp2/3 activation. Expression of calpain-resistant cortactin, but not wild-type cortactin, impairs cell migration and increases transient membrane protrusion, suggesting that calpain proteolysis of cortactin limits membrane protrusions in fibroblasts. Furthermore, the enhanced protrusion dynamics observed with the calpain-resistant cortactin requires both the Arp2/3 binding site and the SH3 domain of cortactin. Together, these findings suggest a novel role for calpain-mediated proteolysis of cortactin in regulating membrane protrusion dynamics during cell migration.
MATERIALS AND METHODS
Cell Lines
Chinese hamster ovary (CHO)-K1 and human embryonic kidney (HEK)-293 cells were cultured as described previously (Cox et al., 2001; Franco et al., 2004b). CHO-K1 cells expressing enhanced green fluorescent protein (EGFP)-epidermal growth factor receptor (EGFR) were kindly provided by Doug Lauffenburger (Massachusetts Institute of Technology, Cambridge, MA) and maintained as described previously (Harms et al., 2005). CHO-K1 cells were transfected with DNA plasmids or small interfering RNA (siRNA) oligonucleotides (oligos) using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) according to manufacturer's instructions. Knockdown cells were assessed 72 h after transfection. HEK-293 cells were transfected by standard calcium phosphate precipitation. Stable cell lines were selected and maintained in 1 mg/ml Geneticin (G418) (Invitrogen) and were sorted for green fluorescent protein (GFP) expression by flow cytometry.
Antibodies and Reagents
Fibronectin was purified from human plasma by affinity chromatography as described previously (Ruoslahti et al., 1982). Anti-calpain 2 antibody was obtained from Triple Point Biologics (Forest Grove, OR); anti-talin (clone 8d4), anti-FLAG antibodies were purchased from Sigma-Aldrich (St. Louis, MO), and anti-phospho-tyrosine antibody (4G10) was obtained from Upstate Biotechnology (Lake Placid, NY). Anti-cortactin (clone 4F11) was a kind gift from A. Reynolds (Vanderbilt University, Nashville, TN). Anti-GFP and Alexa Fluor-680 goat-anti-mouse IgG secondary antibody were purchased from Invitrogen. IRDye 800CW goat-anti-rabbit IgG secondary antibody was obtained from Rockland (Gilbertsville, PA). Glutathione-Sepharose was purchased from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). Calpain inhibitor 1 (ALLN) was purchased from Calbiochem (San Diego, CA) and used at a concentration of 50 μg/ml. Control siRNA oligo against the target sequence 5′-TTCTCCGAACGTGTCACGT-3′ and calpain 2-specific siRNA oligo against the mouse target sequence 5′-AAGGATGGCGATTTCTGCATC-3′ were purchased from QIAGEN (Valencia, CA). Cortactin siRNA oligo against the mouse target sequence 5′-GGAACACATCAACATTCAC 3′ was purchased from Ambion (Austin, TX).
Constructs and siRNA
FLAG-cortactin was a generous gift from A. Weaver (Vanderbilt University). FLAG-cortactin-D6 was generated by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. FLAG-cortactin-D28 was generated by separately amplifying the regions upstream and downstream of the deletion with the following primers from FLAG-cortactin template: upstream forward 5′-_CAGTGTGGTGGAATT_CATGGACTACAAGGACGACG-3′, reverse 5′-GGCAGATGGCACCTGGACC-3′; downstream forward 5′-_CAGGTGCCATCTGCC_GAGCAGGAGGACAGGCGG-3′, reverse 5′-_GATATCTGCAGAATT_CCTACTGCCGCAGCTCCACATAG-3′. The PCR fragments were assembled in a reaction with pcDNA3.1 digested with _Eco_RI using the PCR In-Fusion cloning kit (Clontech, Mountain View, CA). The In-Fusion enzyme-mediated recombination between complementary regions (15 base pairs, in italics) in the upstream forward primer and the downstream reverse primer to the ends of pcDNA3.1 digested with EcoR1, and in the downstream forward primer to the upstream reverse primer. GFP-tagged constructs were generated by amplifying the respective cortactin constructs by PCR, and cloning into pEGFP-C1 (Clontech). Cortactin bearing a C-terminal glutathione _S_-transferase (GST) fusion was generated by amplifying cortactin (forward primer 5′-AGGAGATATACCATGTGGAAAGCCTCTGCAGGC-3′, reverse primer 5′-TAGTATAGGGGACATCTCGAGCTGCCGCAGCTCCACATAG-3′) and GST (forward primer 5′-ATGTCCCCTATACTAGGTTATT-3′, reverse primer 5′-GTTAGCAGCCGGATCCCTAATCCGATTTTGGAGGATGGTCG-3′), and assembling the fragments in pET 16b (Novagen, Madison, WI) digested with _Nco_1 and _Xho_1 using the PCR In-Fusion cloning kit (Clontech). FLAG-cortactin and FLAG-cortactin-D28 were amplified from the respective pcDNA constructs by PCR (forward primer 5′-TAATAGGATCCACCATGGACTACAAGGACGACG-3′ and reverse primer 5′-GATATCTGCAGAATTCCTACTGCCGCAGCTCCACATAG-3′), digested with _Bam_HI and _Eco_RI, and ligated into similarly digested pFastBac (Invitrogen). To express cortactin after siRNA treatment, five silent mutations were introduced into GFP-cortactin and GFP-cortactin-D28 by site-directed mutagenesis (Stratagene). Cortactin or cortactin-D28 containing the silent mutations as well as the cortactin N-terminal fragment, were cloned to pCS2-mRFP (Benink and Bement, 2005) to generate monomeric red fluorescent protein (mRFP) fusions. GST-verprolin-cofilin-acidic motif (VCA) was kindly provided by J. T. Parsons (University of Virginia, Charlottesville, VA). All constructs were sequenced before use.
Immunoblot Analysis
Cells were plated in DMEM with 0.2% bovine serum albumin (BSA) on dishes coated with 10 μg/ml fibronectin and incubated at 37°C under 5% CO2 for 1 h. Cells were scraped into lysis buffer (50 mM Tris, pH 7.6, 500 mM NaCl, 0.1% SDS, 0.5% deoxycholate, 1% Triton X-100, 0.5 mM MgCl2, and 0.2 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml pepstatin, 2 μg/ml aprotinin and 1 μg/ml leupeptin) on ice and clarified by centrifugation. Protein concentrations were determined using a BCA protein assay kit (Pierce Chemical, Rockford, IL) according to manufacturer's instructions. Equal amounts of total protein were denatured in SDS sample buffer, run on 4–20% gradient SDS-polyacrylamide gels, and transferred to nitrocellulose. Western blots were imaged and quantified with an Odyssey Infrared Imaging System (LI-COR Biosciences, Omaha, NE).
Immunoprecipitation
HEK-293 cells were transfected and lysed 24–48 h later in immunoprecipitation buffer (20 mM HEPES, pH 7.5, 50 mM KCl, 1 mM EDTA, 1% NP-40, 0.2 mM PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, and 1 μg/ml leupeptin). CHO-K1 cells were lysed in modified radioimmunoprecipitation assay buffer (50 mM HEPES, pH 7.2, 150 mM NaCl, 2 mM EDTA, 0.5% deoxycholate, 1% NP-40, 0.2 mM PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 mM vanadate) (Head et al., 2003). Lysates were incubated with 3 μg of anti-GFP antibody or 3 μg of rabbit IgG. Immune complexes were captured on GammaBind G-Sepharose beads (GE Healthcare), washed in lysis buffer, and analyzed by immunoblotting with anti-FLAG or anti-GFP antibody.
Protein Purification
Cortactin-GST and GST-VCA were purified from Escherichia coli (BL21) using glutathione-Sepharose (GE Healthcare). Briefly, overnight cultures were diluted 1:10 to fresh LB containing 100 μg/ml ampicillin and 30 μg/ml chloramphenicol and grown at 37°C for 1 h. Expression was induced with 0.4 mM isopropyl β-d-thiogalactoside (IPTG) for 4 h at 37°C. Cells were lysed in TS (10 mM Tris-HCl, pH 7.5, 100 mM NaCl, and 0.2 mM PMSF) by sonication, 1% Triton X-100 was added, the lysate was clarified by centrifugation, and the supernatant was incubated with glutathione-Sepharose beads for 30 min, and washed in TS. GST-VCA was eluted by incubation with 20 mM reduced glutathione, dialyzed into buffer D (10 mM Tris-HCl, pH 7.5, 150 mM KCl, 2 mM MgCl2, and 1 mM dithiothreitol [DTT]) and the concentration was determined by A280 measurement using a calculated extinction coefficient of 30,600 M–1 cm–1.
FLAG-cortactin and FLAG-cortactin-D28 pFastBac constructs were transformed into DH10BAC E. coli cells (Invitrogen) for recombination into bacmid DNA. High-titer viral stocks were used to infect SF9 insect cells. Seventy-two hours after infection cells were lysed in phosphate-buffered saline (PBS) (10 mM sodium phosphate, 138 mM NaCl, and 2.7 mM KCl, pH 7.4) with 1% Triton X-100, 0.2 mM PMSF, 1 μg/ml pepstatin, 2 μg/ml aprotinin, 1 μg/ml leupeptin, and 1 μM E-64 (Sigma-Aldrich). Lysate was circulated over anti-FLAG M2-agarose (Sigma-Aldrich), washed with PBS with 0.1% Triton X-100 and 0.2 mM PMSF, and eluted in PBS with 100 μg/ml FLAG peptide (Sigma-Aldrich). Proteins were dialyzed into buffer D, and the concentration was determined by _A_280 measurements and calculated extinction coefficients of 70,340 M–1 cm–1 for FLAG-cortactin and 69,060 M–1 cm–1 for FLAG-cortactin-D28.
Actin was purified from rabbit skeletal muscle powder (Spudich and Watt, 1971) and gel filtered on Sephacryl S-300 in Ca-G buffer (2 mM imidazole, pH 7.0, 0.2 mM ATP, 0.1 mM CaCl2, and 0.5 mM DTT). Pyrenyl-actin was produced by labeling Cys374 with pyrene iodoacetamide (Kouyama and Mihashi, 1981; Pollard, 1984).
In Vitro Actin Polymerization
Immediately before polymerization, Ca-ATP-actin was converted to Mg-ATP-actin by incubation with 1/10 volume of 10× exchange buffer (10 mM EGTA and 1 mM MgCl2) for 1 min. Proteins were mixed at 22°C in Mg-G buffer with 1× KMEI (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole). Actin polymerization reactions were started by mixing with 3 μM actin (5% pyrene labeled), and fluorescence was monitored continuously with 365-nm excitation and 407-nm emission on an ISS PC1 photon counting spectrofluorometer (ISS, Champaign, IL).
Protein Localization
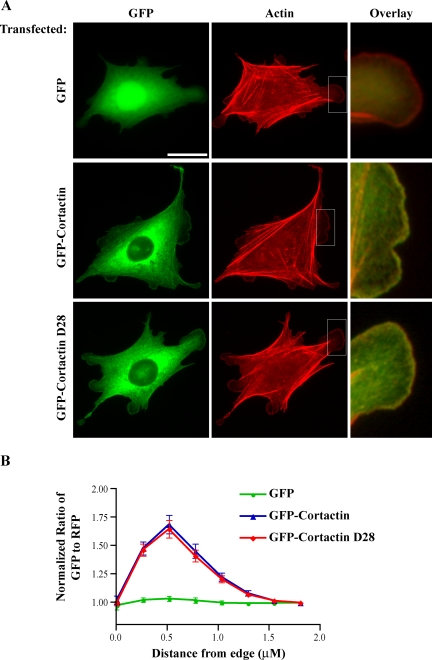
CHO-K1 cells expressing GFP-cortactin constructs were plated for 12 h in DMEM with 0.2% BSA on glass coverslips coated with 10 μg/ml fibronectin for 1 h at 37°C. The media were exchanged for DMEM with 1% fetal bovine serum (FBS) for 30 min, cells were fixed with 3% formaldehyde, and stained for actin with rhodamine-phalloidin (Invitrogen). For quantification of cortactin localization, CHO-K1 cells that stably express GFP, GFP-cortactin, or GFP-cortactin-D28 were transiently transfected with dsRed (Clontech) to serve as a volumetric control, plated, and fixed as described above. To correct for potential volume differences at the cell periphery, MetaMorph software was used to determine the ratio of GFP fluorescent intensity to dsRed fluorescent intensity along a 1.8-μm line extending from the cell edge toward the cytoplasm. Each ratio along the line was normalized to a cytoplasmic value to allow for comparison between cells with different expression levels of GFP or dsRed.
Protrusion Analysis
Protrusion analysis was performed using a 40× phase contrast objective on an Olympus IX-70 inverted microscope housed in a closed system to maintain temperature at 37°C and CO2 levels at 5%. Dishes (35 mm) were coated with 10 μg/ml fibronectin for 1 h at 37°C. Cells were serum starved for 16 h in DMEM with 1% FBS or in DMEM with 0.2% BSA for experiments using epidermal growth factor (EGF). For microscopy experiments, cells were plated in DMEM plus 0.2% fatty acid-free BSA, with or without 25 nM EGF (Sigma-Aldrich) as indicated, at subconfluence and allowed to adhere for 1.5 h before imaging. MetaVue Imaging software (Molecular Devices, Sunnyvale, CA) was used to take phase contrast images at 6-s intervals for 10 min. MetaVue software was used to quantify protrusion with modifications of previously described methods (Cox et al., 2001). In brief, from each frame in the stack of images, the subsequent image was digitally subtracted, leaving only regions where the pixel intensity changed. A mask was applied around the periphery of the cell to quantify changes in peripheral protrusions. The resulting image was then subjected to a threshold to quantify pixel changes with intensity greater than average background intensity. The integrated intensity of those pixels was determined for each frame of the movie and averaged to determine a protrusion index for each cell. Statistical significance was calculated in GraphPad Prism using analysis of variance with a Bonferroni posttest.
Cleavage Site Mapping
Cortactin-GST on glutathione beads were washed and resuspended in proteolysis buffer (50 mM Tris-HCl, pH 7.5, 134 mM KCl, 1 mM MgCl2, and 1 mM CaCl2). Purified calpain 2 (Sigma-Aldrich) was added to the beads (40–200 μg/ml), and the reaction was incubated at 37°C for 1 h. To analyze the digest, bead volumes were boiled in SDS-PAGE sample buffer and separated on SDS-PAGE gels and stained with Coomassie blue. For N-terminal sequencing, beads were washed extensively with TS after digestion with calpain before electrophoresis. Protein was transferred to polyvinylidene difluoride membrane (Bio-Rad, Hercules, CA), stained with Coomassie blue, and an individual band was subjected to sequencing (performed at the Baylor College of Medicine [Houston, TX] protein services core).
Random Migration
CHO-K1 cell lines were serum starved in DMEM with 2% fatty acid-free BSA and nonessential amino acids for 20 h, lifted off the plate with 0.04% EDTA in PBS, and held in suspension for 30 min in DMEM with 2% BSA. Glass-bottomed dishes were coated with 10 μg/ml fibronectin in PBS overnight at 4°C, washed with PBS, blocked with 2% BSA for 30 min at 37°C, and washed with PBS. Cells were plated in Opti-MEM (Invitrogen) with 1% FBS, nonessential amino acids, and Pen/Strep and allowed to adhere for 90 min before imaging every 10 min for 4 h with a 10× objective. Cell speeds were obtained from the resulting movie by using MetaMorph software to track cells that did not divide, touch other cells, or migrate out of the field of view for the duration of observation.
Transwell Migration Assay
Transwell filters (Corning Life Sciences, Acton, MA) with 8-μm pores were coated on the top and bottom with 10 μg/ml fibronectin for 1 h at 37°C and blocked with 2% BSA for 30 min at 37°C. Filters were dried after coating. Serum-starved CHO-K1 cell lines were lifted with 0.02% EDTA, and 1 × 105 cells were plated in the top chamber in DMEM with 0.2% BSA. DMEM with 10% FBS was added to the bottom chamber, and cells were allowed to migrate for 3 h. Cells were wiped from the top surface of the filter, and cells on the bottom surface were fixed and stained as described previously (Huttenlocher et al., 1996).
RESULTS
Proteolysis of Cortactin by Calpain 2 in Fibroblasts
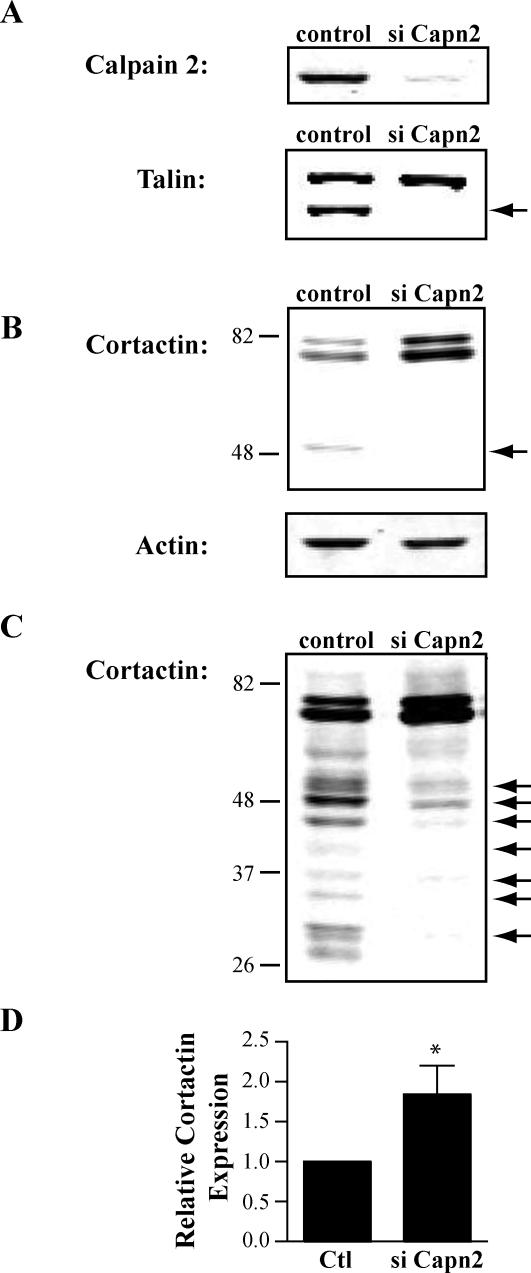
Previous studies have demonstrated that calpain 2 regulates membrane protrusion dynamics in fibroblasts (Franco et al., 2004a). Cortactin is an attractive candidate substrate that may mediate the effects of calpain 2 on protrusion dynamics. To determine whether cortactin is a calpain 2 substrate in fibroblasts, we generated calpain 2-deficient CHO-K1 and NIH-3T3 lines using calpain 2-specific siRNA. In both CHO-K1 cells (Figure 1A) and NIH-3T3 cells (Franco et al., 2004b), calpain 2 siRNA reduced calpain 2 protein expression compared with nonsilencing control oligos. Accordingly, reduced proteolysis of talin was observed in the presence of calpain 2-specific oligos (Figure 1A). In both CHO-K1 cells and NIH-3T3 cells, cortactin proteolysis was observed in control cells but not in the calpain 2-deficient cells (Figure 1B; our unpublished data). Interestingly, there was a substantial increase (2-fold) in intact cortactin in the calpain 2-deficient fibroblasts compared with control cells (Figure 1D), suggesting that calpain 2 regulates the levels of intact cortactin in cells. This observation is in contrast to talin, where the amount of intact talin is not significantly altered by calpain 2 siRNA (Figure 1A; Franco et al., 2004b). These findings suggest that calpain mediates cortactin degradation, rather than limited proteolysis. In agreement with this possibility, longer exposures of cortactin blots demonstrate multiple cleavage fragments. These degradative fragments were significantly reduced in the calpain 2-deficient CHO-K1 cells (Figure 1C). Together, these results suggest that calpain 2 proteolyzes cortactin, which leads to cortactin degradation and may provide a mechanism to regulate cortactin levels locally within the cell.
Figure 1.
Calpain mediates cortactin proteolysis in fibroblasts. CHO-K1 cells were transfected with nonsilencing control siRNA oligos (control) or siRNA oligos targeting calpain 2 (siCapn2). Equal amounts of lysate (15 μg) were separated by SDS-PAGE, transferred to nitrocellulose membrane, and probed with specific antibodies. (A) Immunoblot analysis of calpain 2 and talin. The talin cleavage product is indicated by an arrow. (B) Short exposure of cortactin immunoblot. Arrow indicates calpain-dependent cleavage fragment. Actin was probed as a loading control. (C) Long exposure of the cortactin immunoblot. Arrows indicate calpain-dependent cleavage fragments. (D) Quantification of full-length cortactin protein level. Intact cortactin in either control or siCapn2 lysate from three separate experiments was quantified (*p < 0.05).
Calpain Cleaves Cortactin between the Actin Binding Repeats and the α-Helical Domain
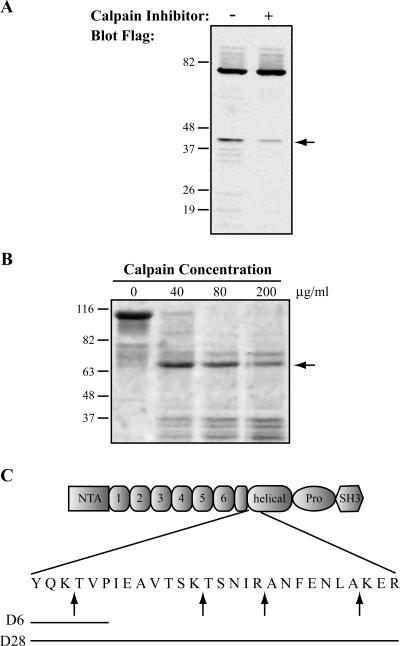
Previous studies have demonstrated that there is no primary amino acid consensus sequence in proteins that defines the calpain cleavage site; rather, calpain tends to preferentially cleave substrates between protein domains in relatively unstructured regions of proteins (Tompa et al., 2004). One predominant calpain cleavage fragment was observed both endogenously (Figure 1B) and with ectopic expression of FLAG-tagged cortactin in HEK-293 cells (Figure 2A). These findings suggest that this region of cortactin is particularly susceptible to calpain proteolysis. To map this proteolytic site, purified cortactin-GST was treated with increasing amounts of purified calpain 2 (Figure 2B). In accordance with the findings in cultured cells, calpain-mediated proteolysis of cortactin in vitro generated one predominant cleavage fragment that was analyzed by N-terminal sequencing. Interestingly, the sequenced band contained fragments produced from four different cleavage sites. All four sites are located in a 22-amino acid region without predicted secondary structure (nn-predict; our unpublished data), located immediately following the last half repeat of the actin binding domain and before the α-helical domain (Figure 2C).
Figure 2.
Calpain cleaves cortactin between the actin binding repeats and the α-helical domain. (A) Immunoblot analysis of exogenously expressed FLAG-cortactin. HEK-293 cells were transfected with FLAG-cortactin, treated with vehicle control or with calpain inhibitor (ALLN), and blotted as in Figure 1. The blot was probed with FLAG antibody. The arrow indicates the calpain-dependent cleavage product. (B) In vitro calpain cleavage of cortactin-GST. Cortactin-GST bound to glutathione-Sepharose beads was incubated with increasing concentrations of purified calpain 2. The reaction mixture was separated by SDS-PAGE, and the gel was stained with Coomassie blue. The arrow indicates the band analyzed by N-terminal sequencing. (C) Schematic of cortactin. Arrows indicate sites of calpain cleavage identified by N-terminal sequencing within cortactin. Solid lines represent the regions deleted in cortactin-D6 or cortactin-D28.
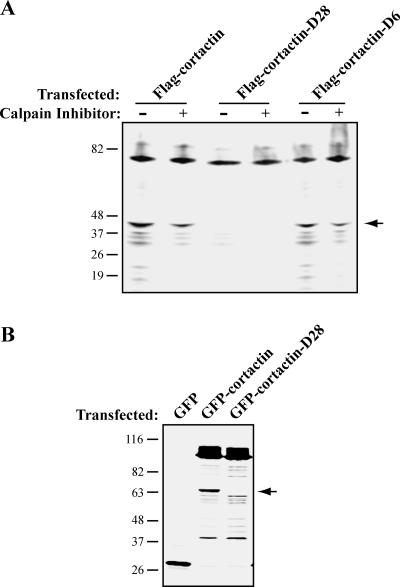
To generate a mutant form of cortactin that is resistant to calpain proteolysis, we made two deletion constructs that encompass the cleavage sites. The first deletion removed six amino acids (YQKTVP) centered on the first cleavage site (cortactin-D6), and the second deletion removed 28 amino acids encompassing the four cleavage sites (cortactin-D28) (Figure 2C). When expressed in HEK-293 cells, cortactin-D6 remained susceptible to calpain proteolysis (Figure 3A), suggesting that there is flexibility within the calpain cleavage region without a clear requirement for a specific site. However, cortactin-D28 was significantly less susceptible to calpain proteolysis (Figure 3, A and B). These findings demonstrate that the mapped cleavage sites in cortactin are important for calpain proteolysis, and deletion of those sites renders cortactin resistant to calpain proteolysis.
Figure 3.
A deletion mutation renders cortactin resistant to calpain proteolysis. (A) Immunoblot analysis of mutant cortactin in HEK-293 cells. HEK-293 cells were transfected with FLAG-cortactin, FLAG-cortactin-D6, or FLAG-cortactin-D28, treated with vehicle control or with calpain inhibitor (ALLN), and blotted with FLAG antibody. The arrow indicates the calpain-dependent cleavage product. (B) Immunoblot analysis of GFP-cortactin in CHO-K1 cells. CHO-K1 cells were transfected with cortactin siRNA and GFP, GFP-cortactin, or GFP-cortactin-D28 and blotted for GFP. The arrow indicates the predominant calpain-dependent cleavage fragment.
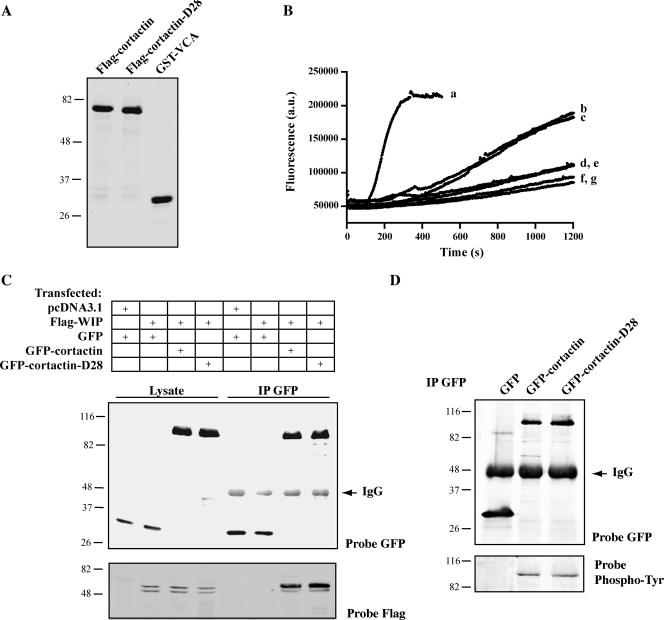
To determine whether the calpain-resistant cortactin is functional, we examined the biochemical properties and localization of wild-type cortactin and cortactin-D28. In vitro actin polymerization assays demonstrated that both purified wild-type FLAG-cortactin and FLAG-cortactin-D28 (Figure 4A) increased actin polymerization to a similar extent compared with actin alone, actin with Arp2/3, or actin with cortactin (Figure 4B). These findings demonstrate that cortactin-D28 retains the ability to activate Arp 2/3-mediated actin polymerization in vitro. We next examined its interaction with WIP, a protein known to bind the SH3 domain of cortactin. Both GFP-cortactin and GFP-cortactin-D28 interacted with WIP in coimmunoprecipitation experiments (Figure 4C), suggesting that the C-terminal SH3 domain of cortactin is functional. To further characterize mutant cortactin, we checked the amount of tyrosine phosphorylation of GFP-cortactin and GFP-cortactin-D28 in CHO-K1 cells (Figure 4D). Both proteins displayed equivalent phosphorylation, suggesting that GFP-cortactin and GFP-cortactin-D28 are not differentially phosphorylated by Src family kinases. We also examined the localization of GFP-cortactin and GFP-cortactin-D28 in CHO-K1 cells. Both proteins displayed similar subcellular localization to the perinuclear region and to the membrane as reported previously for cortactin (Figure 5, A and B) (Cao et al., 2005). This suggests that the deletion mutation does not affect the intracellular distribution of cortactin-D28. Together, these results indicate that the principal calpain cleavage region in cortactin resides between the actin binding repeats and the α-helical domain of cortactin and that this region is not required for cortactin function.
Figure 4.
Cortactin-D28 retains biochemical activity. (A) Purified cortactin and GST-VCA. Purified FLAG-cortactin, FLAG-cortactin-D28, or GST-VCA were separated by SDS-PAGE and stained with Coomassie blue. (B) I vitro actin polymerization assay. FLAG-cortactin, FLAG-cortactin-D28, or GST-VCA was incubated with Arp2/3 complex, upon the addition of 3 μM actin (5% pyrene labeled) polymerization was monitored by continuous measurement of fluorescence at 407 nm. Reactions included 3 μM actin and 20 nM Arp2/3 complex with the following additions: 200 nM GST-VCA (a), 500 nM FLAG-cortactin (b), or 500 nM FLAG-cortactin-D28 (c). Controls included 3 μM actin with the following additions: 500 nM FLAG-cortactin (d), 500 nM FLAG-cortactin-D28 (e), 20 nM Arp2/3 complex (f), or buffer (g). (C) Immunoblot analysis of GFP-cortactin and WIP coimmunoprecipitation. HEK-293 cells were transfected with GFP, GFP-cortactin, or GFP-cortactin-D28 in combination with Flag-WIP. Protein was immunoprecipitated with anti-GFP antibody, and samples of lysate and immunoprecipitate were blotted as described in Figure 1. Blots were probed with antibody against either GFP to detect expressed cortactin or FLAG to detect WIP. (D) Immunoblot analysis of cortactin tyrosine phosphorylation. CHO-K1 cells transfected with GFP, GFP-cortactin, GFP-cortactin-D28, and protein was immunoprecipitated with anti-GFP antibody. The immunoprecipitate was probed with antibody against GFP or phosphotyrosine.
Figure 5.
Calpain-resistant cortactin demonstrates normal subcellular localization. CHO-K1 cells stably expressing GFP, GFP-cortactin, or GFP-cortactin-D28 were plated on coverslips as described in Materials and Methods. (A) Localization of GFP-cortactins. The actin cytoskeleton was stained with rhodamine-phalloidin, and GFP was visualized to localize cortactin. Boxed regions were enlarged to show colocalization of GFP-cortactin or GFP-cortactin-D28 with actin at the cell periphery. Bar, 20 μm. (B) Quantification of cortactin localization. Cells were transfected with dsRed to serve as a volumetric control. The normalized ratio of GFP-to-dsRed intensity was determined at the edge of cells (n = 25).
Calpain-resistant Cortactin Impairs Cell Migration
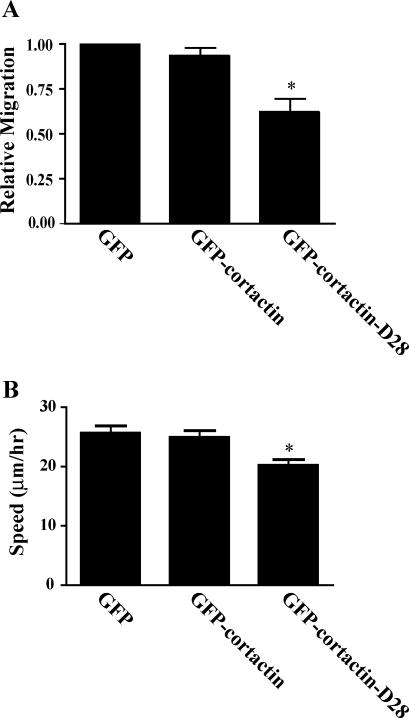
To determine whether calpain-mediated proteolysis of cortactin is important for cell migration, we characterized the migration of CHO-K1 cells that express calpain-resistant cortactin or comparable levels of wild-type cortactin using a transwell assay and by time-lapse microscopy. CHO-K1 cell lines were generated that express GFP, GFP-cortactin, or GFP-cortactin-D28 and were sorted for similar levels of GFP expression by flow cytometry (our unpublished data). Serum-starved CHO-K1 cells were assayed for their ability to migrate toward serum in a transwell assay (Figure 6A) or to undergo random migration on a fibronectin-coated surface (Figure 6B). We found that overexpression of GFP-cortactin did not significantly alter cell migration rates compared with control cells that express GFP. In contrast, overexpression of GFP-cortactin-D28 led to a reduction in cell migration both by transwell assay and live imaging compared with control cells. Interestingly, the difference in migration was somewhat more dramatic by transwell assay, which may indicate differences in actin cytoskeletal stability mediated by the calpain-resistant cortactin. Together, these findings suggest that calpain-mediated proteolysis of cortactin is required for efficient cell migration.
Figure 6.
Cortactin-mediated proteolysis of cortactin modulates cell migration. CHO-K1 cells were transfected with GFP, GFP-cortactin, or GFP-cortactin-D28, selected with G418, and sorted for GFP expression to generate stable lines. (A) Migration by transwell assay. Cells were serum starved for 16 h, plated in the top of a Boyden chamber that had been coated with 10 μg/ml fibronectin, and allowed to migrate toward serum in the bottom chamber for 4 h. The cells that migrated through the membrane were counted. The results from three separate experiments were normalized to the GFP control cell line. The error bars represent SEM (*p < 0.01). (B) Cell speeds during random migration. Serum-starved cells were plated in glass-bottomed dishes coated with 10 μg/ml fibronectin, and migration was recorded by time-lapse microscopy. At least 50 GFP-positive cells were tracked to determine cell speeds. Error bars represent SEM (*p < 0.01).
Proteolysis of Cortactin Regulates Membrane Protrusion
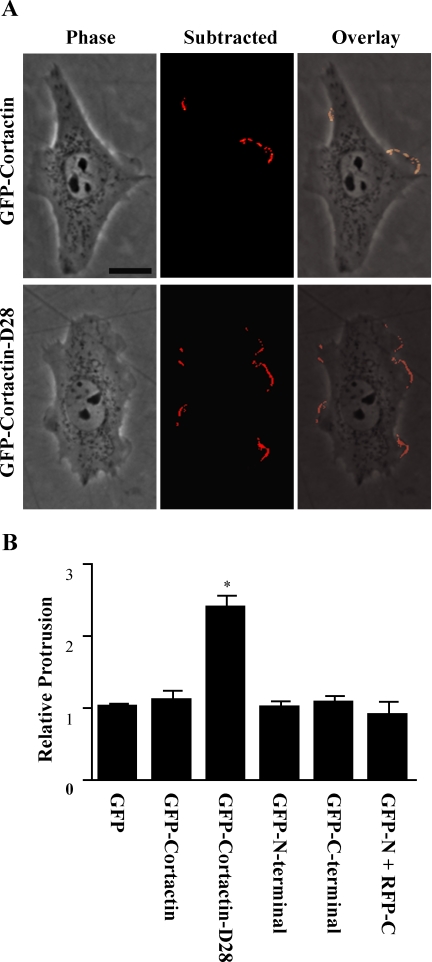
Because calpain 2 has been implicated in membrane protrusion (Franco et al., 2004a), we next sought to determine whether calpain proteolysis of cortactin regulates membrane protrusion. To this end, we compared transient protrusion in CHO-K1 cells that express GFP, GFP-cortactin, or GFP-cortactin-D28. Additionally, we also compared protrusion in cells that express the N-terminal or C-terminal proteolytic fragments of cortactin (GFP-N-terminal and GFP-C-terminal) as well as cells that express both fragments (mRFP-N-terminal and GFP-C-terminal). The constructs correspond to the fragments that are produced upon proteolysis by calpain, based on the mapped calpain-sensitive region in cortactin. Compared with control cells expressing GFP, expression of GFP-cortactin, GFP-N-terminal or GFP-C-terminal did not alter average protrusion rates or cell morphology (Figure 7). However, cells expressing GFP-cortactin-D28 displayed more than a twofold increase in protrusion activity compared with control cells (Figure 7B). Additionally, in contrast to control cells that demonstrate more polarized protrusion activity, cells that express cortactin-D28 displayed protrusion activity through out the cell periphery that was not limited to specific regions of the cell (Figure 7A).
Figure 7.
Calpain-mediated proteolysis of cortactin regulates membrane protrusion kinetics. Stable cells lines were generated as in Figure 6 that express GFP, GFP-cortactin, GFP-cortactin D28, GFP-cortactin N-terminal, or GFP-cortactin C-terminal. Cells were serum starved for 16 h and plated on 10 μg/ml fibronectin in serum-free media. Phase contrast images were acquired every 6 s for 10 min. (A) Image and subtracted image from time-lapse movie. For each image in the movie, the subsequent image was digitally subtracted, and the cell was outlined to isolate intensity changes at the cell periphery as described in Materials and Methods. The overlay highlights the protruding regions of the cells. Bar, 15 μm. (B) Quantification of protrusion dynamics. From each cell, the average integrated intensity change for each frame of the movie was calculated. Each cell was normalized to the average protrusion value of the GFP cell line. The mean and SE of the mean (SEM) were graphed (*p < 0.001).
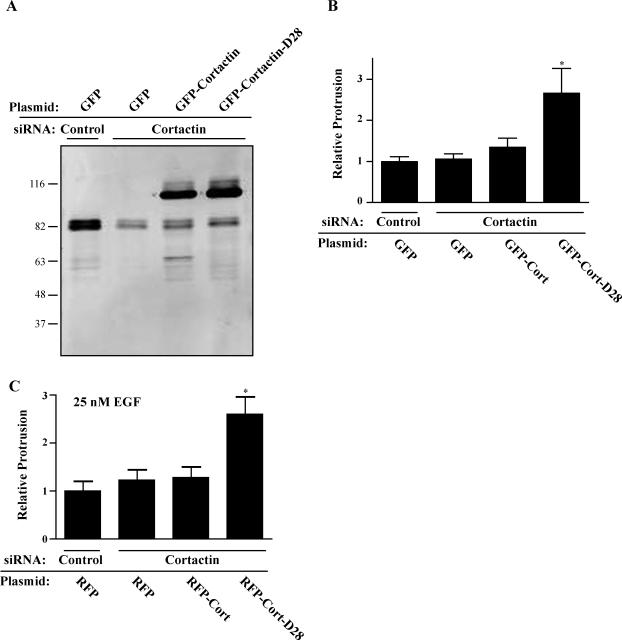
To determine whether endogenous cortactin is required for membrane protrusion in CHO-K1 cells, we generated cortactin-deficient CHO-K1 cells by knocking down expression of endogenous cortactin using siRNA. A recent study suggests that cell lines that are deficient in cortactin may show enhanced membrane protrusion under some conditions, suggesting that cortactin may be inhibitory to membrane protrusion (Kempiak et al., 2005). Under the conditions of our assay, we found no significant difference in membrane protrusion dynamics in cells that expressed control or cortactin siRNA, despite a reduction in cortactin expression of ∼80% (Figure 8). To determine the effects of the calpain-resistant cortactin on membrane protrusion dynamics in the absence of endogenous cortactin, we generated stable cell lines that express GFP, GFP-cortactin, or GFP-cortactin-D28 with silent mutations that produced mismatches in the region homologous to the siRNA sequence. Cell lines were sorted by flow cytometry to ensure equal GFP expression (our unpublished data). Treatment with cortactin siRNA successfully reduced expression of endogenous cortactin without affecting the expression of GFP-cortactin or GFP-cortactin-D28 in these cell lines. In the absence of significant endogenous cortactin, we found that expression of GFP-cortactin-D28 increased membrane protrusion dynamics by more than twofold as compared with GFP-cortactin (Figure 8B). To determine the effect of cortactin-D28 on protrusion in the presence of growth factors, we transfected CHO-K1 cells that stably express EGFP-EGFR (Harms et al., 2005) with cortactin siRNA and mRFP, mRFP-cortactin, or mRFP-cortactin-D28. We observed that EGF-treated cells were more protrusive than untreated cells (our unpublished data) and that cortactin siRNA alone had no effect on membrane protrusion. However, expression of mRFP-cortactin-D28 increased EGF-stimulated membrane protrusion by more than twofold compared with control cells or cells that express mRFP-cortactin (Figure 8C). These findings demonstrate that calpain-resistant cortactin enhances membrane protrusion dynamics in both the presence and absence of growth factors. Together, the results indicate that calpain-mediated proteolysis of cortactin functions to limit membrane protrusion, thereby allowing for efficient cell migration.
Figure 8.
Calpain-resistant cortactin promotes protrusion independently of endogenous cortactin in the presence and absence of EGF. GFP-cortactin and GFP-cortactin-D28 containing silent mutations in siRNA targeting sequences were constructed, and stable cells lines were generated as in Figure 6. Cells were then transfected with control siRNA or siRNA targeting cortactin. (A) Immunoblot analysis of cortactin in CHO-K1 cell lines transfected with siRNA oligos. Endogenous cortactin, running at ∼80 KDa, is significantly reduced with siRNA treatment, whereas GFP-Cortactin and GFP-cortactin-D28, running at ∼110 KDa, remain. (B) Quantification of protrusion. Protrusion of CHO-K1 cell lines transfected with control of cortactin-targeting siRNA oligos was analyzed as in Figure 7. The mean and SEM are plotted (*p < 0.01). (C) Quantification of protrusion in the presence of EGF. CHO-K1 cells stably expressing GFP-EGFR were transiently transfected with cortactin siRNA and mRFP, mRFP-cortactin, or mRFP-cortactin-D28, and protrusion was analyzed in media containing 25 nM EGF (*p < 0.01).
Cortactin Requires a Functional SH3 Domain and Arp2/3 Binding to Modulate Protrusion
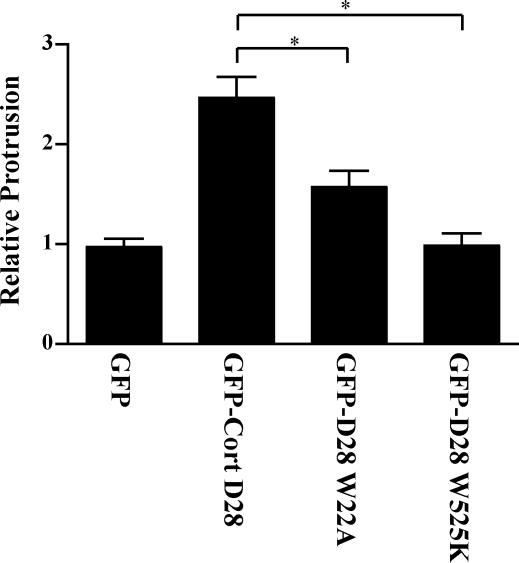
Previous studies have demonstrated that cortactin SH3 domain binding interactions with WIP, N-WASP, and dynamin as well as the cortactin N-terminal binding interactions with Arp2/3 are important for cortactin-mediated effects on the actin cytoskeleton, membrane protrusion, and cell migration (Schafer et al., 2002; Kinley et al., 2003; Kowalski et al., 2005). Mutations in cortactin that abrogate Arp2/3 activation (W22A) or SH3 domain binding function (W525K) have been described previously (Weaver et al., 2001; Kinley et al., 2003). To determine whether these sites are critical for the effects of calpain-resistant cortactin on membrane protrusion, we generated cortactin-D28 constructs with either the W525K or W22A mutation. We then assayed membrane protrusion in CHO-K1 cell lines that express GFP-cortactin-D28, GFP-cortactin-D28-W22A and GFP-cortactin-D28-W525K. We found that the enhanced protrusion mediated by GFP-cortactin-D28 required a functional SH3 domain because cells expressing GFP-cortactin-D28-W525K displayed protrusion rates comparable with control cells (Figure 9). Dependence on the SH3 domain is consistent with previous studies demonstrating that the cortactin SH3 domain recruits WIP, resulting in enhanced membrane protrusion at the cell periphery (Kinley et al., 2003). The Arp2/3 binding site was also important for the effects of cortactin-D28 on protrusion, although, cortactin-D28-W22A displayed an intermediate phenotype compared with cortactin-D28 and control cells (Figure 9). Together, the findings suggest that calpain-resistant cortactin mediates its effects on membrane protrusion through its interaction with both Arp2/3 and SH3 domain binding proteins, such as N-WASP, WIP, or dynamin.
Figure 9.
The Arp2/3 binding site and SH3 domain of cortactin are required for cortactin effects on membrane protrusion. CHO-K1 cells were transfected with GFP, GFP-cortactin-D28, GFP-cortactin-D28-W22A, or GFP-cortactin-D28-W525K and analyzed for protrusion as in Figure 7. The mean and SEM were plotted as in Figure 7 (*p < 0.01).
DISCUSSION
Recent studies have demonstrated that calpain 2 plays a crucial role in regulating the actin cytoskeleton and membrane protrusion events at the cell periphery; however, the mechanisms by which this is accomplished have not previously been defined. We now provide direct evidence to suggest that calpain proteolysis of an actin binding protein, cortactin, regulates membrane protrusion dynamics. This is the first study to show that cortactin is cleaved and degraded by calpain 2 in fibroblasts. We have identified the putative preferred cortactin cleavage site and have successfully generated a cortactin mutant that is resistant to calpain proteolysis but retains the other biochemical properties of intact cortactin. The mutant cortactin localizes normally and enhances membrane protrusion activity in CHO-K1 cells. The effects of calpain-resistant cortactin on membrane protrusion require a functional SH3 domain and Arp2/3 binding site. Together, the data suggest that calpain proteolysis of cortactin regulates cortactin function to limit membrane protrusion and allow for efficient cell migration.
Previous studies have demonstrated that calpain mediates the limited proteolysis of many of its substrates, thereby generating proteolytic fragments that may be long lived and have distinct intracellular distributions and functions. This is most well defined with the proteolysis of talin into a head and rod domain, with the talin head domain binding to integrin with high affinity and modulating its ligand binding affinity (Yan et al., 2001). In contrast, calpain-mediated proteolysis of cortactin seems to perform a degradative function, cleaving cortactin at multiple sites and regulating its expression levels. Decreasing the level of calpain 2 by siRNA leads to a twofold increase in the amount of cortactin, thereby providing a potential mechanism by which cortactin levels may be regulated both temporally and spatially in migrating cells. In this study, we mapped and mutated the region of cortactin that contains the preferred sites of calpain-mediated cleavage. Interestingly, despite residual cleavage sites, cortactin-D28 significantly increased membrane protrusion in CHO-K1 cells, suggesting that proteolysis in the region between the actin binding repeats and α-helical domain may be important for regulating cortactin function. Expression of the predominant cleavage fragments did not have an apparent effect on membrane protrusion, suggesting that these fragments may not have independent functions. Together, the findings suggest that calpain cleaves cortactin at multiple sites but that calpain proteolysis of the region between the actin repeats and the proline-rich domain may play a key regulatory role in modulating membrane protrusion.
The cortical localization of cortactin, particularly in response to growth factors, suggests a role in protrusion. However, the specific effects of cortactin remain to be fully determined. Under some conditions, overexpression of cortactin enhances protrusion and migration (Patel et al., 1998; Bryce et al., 2005), but in other studies it has no effect (Kinley et al., 2003; Lua and Low, 2004). Under our conditions, cortactin was not sufficient to increase protrusion or migration (Figures 6 and 7), consistent with a previous study also in CHO-K1 cells under similar conditions (Kinley et al., 2003). Knockdown of cortactin has also produced varied results. In one study, siRNA knockdown of cortactin resulted in increased actin polymerization and protrusion in response to EGF-coated beads as well as increased cellular protrusion in general (Kempiak et al., 2005). A separate study demonstrated that cells lacking cortactin produce less stable lamellipodia and are less migratory (Bryce et al., 2005). We observed no change in protrusion in cells lacking endogenous cortactin under our experimental conditions. It is likely that cell type and experimental conditions contribute to the varied effect of cortactin overexpression or knockdown on protrusion and migration.
Proteolysis of cortactin by calpain seems to be critical for properly regulated protrusion because calpain-resistant cortactin acts as a dominant active protein that promotes protrusion in the presence or absence of endogenous cortactin. It is interesting to speculate that calpain-mediated proteolysis of cortactin may provide a mechanism to limit membrane protrusion to specific regions of the cell, thus allowing for efficient cell migration. Cortactin is poised to serve as a switch between assembly and disassembly of the actin network because cortactin not only activates Arp2/3 and stimulates actin polymerization but also stabilizes actin branch points (Weed et al., 2000; Weaver et al., 2001). It is therefore conceivable that calpain-mediated cleavage of cortactin may regulate membrane protrusion by negatively regulating the polymerization or stability of the cortical actin network. Additionally, a recent study demonstrated that calpain proteolyzes WAVE1, 2, and 3 in platelets, suggesting that calpain may degrade several proteins critical for actin-based protrusion (Oda et al., 2005). Alternatively, calpain-mediated cleavage of cortactin may have a role in the turnover of focal adhesions, a process in which calpains have been implicated. Recently it was reported that the cortactin SH3 domain binding protein dynamin is required for microtubule targeted focal adhesion disassembly (Ezratty et al., 2005). Although cortactin has not yet been localized to focal adhesions, it is possible that dynamin and cortactin regulate focal adhesion turnover through endocytosis (Burridge, 2005). In future studies, it will be interesting to study the role of calpain-mediated proteolysis of cortactin and the relationship between focal adhesion dynamics and protrusion.
Our results also support a critical role for both the Arp2/3 binding site and the SH3 domain of cortactin for protrusion. Interestingly, the SH3 domain is required for the enhancing effects of calpain-resistant cortactin on membrane protrusion. Many cortactin-SH3 domain binding proteins, notably N-WASP, WIP, and dynamin, promote motility, protrusion, or rearrangements of the actin cytoskeleton (Schafer et al., 2002; Kinley et al., 2003; Krueger et al., 2003; Kowalski et al., 2005). The Arp2/3 binding site is also important, but not absolutely required, for the enhancing effects of the calpainresistant cortactin on protrusion activity. It is possible that cortactin can promote protrusion independently of Arp2/3 through the activities of proteins bound to the SH3 domain. Regardless, the results suggest that both the SH3 domain and Arp2/3 binding sites are important for cortactin regulation of membrane protrusion.
Cortactin and calpain have both been implicated in cancer. Cortactin is expressed at high levels in some metastatic cancers and is a Src substrate (Patel et al., 1996; Li et al., 2001; Chuma et al., 2004). Calpain may be important for metastasis of some cancers (Mamoune et al., 2003) and has recently been implicated in Src-mediated transformation (Carragher et al., 2001, 2004). Interestingly, Src-mediated phosphorylation of cortactin increases its susceptibility to proteolysis in vitro (Huang et al., 1997b), suggesting that calpain proteolysis of cortactin may be an important regulatory mechanism in invasive cancer cells. Although our study suggests that calpain functions to degrade endogenous cortactin, it is likely that increased expression of both proteins can combine to contribute to a migratory phenotype. For example, increased levels of cortactin may enhance protrusion at the leading edge of a cell, whereas calpain 2 may function to increase migration efficiency by suppressing inappropriate protrusion. Further understanding of how calpain proteolysis of cortactin is spatially regulated within the cell will clarify how these proteins impact migration and tumor metastasis.
In summary, this study demonstrates a novel role for calpain 2-mediated proteolysis of cortactin in regulating membrane protrusion dynamics and cell migration. Calpain proteolysis of cortactin leads to degradation of cortactin and expression of calpain-resistant cortactin increases membrane protrusion and reduces cell migration rates, an effect that requires an intact SH3 domain and Arp2/3 binding site in cortactin. Together, our results suggest a novel mechanism whereby calpain-mediated proteolysis of cortactin may suppress membrane protrusion, thereby allowing for efficient cell migration.
Supplementary Material
[Supplemental Material]
Acknowledgments
We thank Santos Franco and Jonathan Mathias for useful discussions and for critical reading of the manuscript. This research was supported by the National Institutes of Health Grants R01CA85862-01 (to A. H.) and T32GM07215-25 (to B.J.P.).
References
- Benink, H. A., and Bement, W. M. (2005). Concentric zones of active RhoA and Cdc42 around single cell wounds. J. Cell Biol. 168, 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A., Kaverina, I., Otey, C., and Huttenlocher, A. (2002). Regulation of focal complex composition and disassembly by the calcium-dependent protease calpain. J. Cell Sci. 115, 3415–3425. [DOI] [PubMed] [Google Scholar]
- Bryce, N. S., Clark, E. S., Leysath, J. L., Currie, J. D., Webb, D. J., and Weaver, A. M. (2005). Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15, 1276–1285. [DOI] [PubMed] [Google Scholar]
- Burridge, K. (2005). Foot in mouth: do focal adhesions disassemble by endocytosis? Nat. Cell Biol. 7, 545–547. [DOI] [PubMed] [Google Scholar]
- Cao, H., Weller, S., Orth, J. D., Chen, J., Huang, B., Chen, J. L., Stamnes, M., and McNiven, M. A. (2005). Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat. Cell. Biol. 7, 483–492. [DOI] [PubMed] [Google Scholar]
- Carragher, N. O., Fincham, V. J., Riley, D., and Frame, M. C. (2001). Cleavage of focal adhesion kinase by different proteases during SRC-regulated transformation and apoptosis. Distinct roles for calpain and caspases. J. Biol. Chem. 276, 4270–4275. [DOI] [PubMed] [Google Scholar]
- Carragher, N. O., Fonseca, B. D., and Frame, M. C. (2004). Calpain activity is generally elevated during transformation but has oncogene-specific biological functions. Neoplasia 6, 53–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma, M., Sakamoto, M., Yasuda, J., Fujii, G., Nakanishi, K., Tsuchiya, A., Ohta, T., Asaka, M., and Hirohashi, S. (2004). Overexpression of cortactin is involved in motility and metastasis of hepatocellular carcinoma. J. Hepatol. 41, 629–636. [DOI] [PubMed] [Google Scholar]
- Cox, E. A., Sastry, S. K., and Huttenlocher, A. (2001). Integrin-mediated adhesion regulates cell polarity and membrane protrusion through the Rho family of GTPases. Mol. Biol. Cell 12, 265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourdin, N., Bhatt, A. K., Dutt, P., Greer, P. A., Arthur, J. S., Elce, J. S., and Huttenlocher, A. (2001). Reduced cell migration and disruption of the actin cytoskeleton in calpain-deficient embryonic fibroblasts. J. Biol. Chem. 276, 48382–48388. [DOI] [PubMed] [Google Scholar]
- Ezratty, E. J., Partridge, M. A., and Gundersen, G. G. (2005). Microtubule-induced focal adhesion disassembly is mediated by dynamin and focal adhesion kinase. Nat. Cell Biol. 7, 581–590. [DOI] [PubMed] [Google Scholar]
- Franco, S., Perrin, B., and Huttenlocher, A. (2004a). Isoform specific function of calpain 2 in regulating membrane protrusion. Exp. Cell. Res. 299, 179–187. [DOI] [PubMed] [Google Scholar]
- Franco, S. J., Rodgers, M. A., Perrin, B. J., Han, J., Bennin, D. A., Critchley, D. R., and Huttenlocher, A. (2004b). Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell. Biol. 6, 977–983. [DOI] [PubMed] [Google Scholar]
- Harms, B. D., Bassi, G. M., Horwitz, A. R., and Lauffenburger, D. A. (2005). Directional persistence of EGF-induced cell migration is associated with stabilization of lamellipodial protrusions. Biophys. J. 88, 1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head, J. A., Jiang, D., Li, M., Zorn, L. J., Schaefer, E. M., Parsons, J. T., and Weed, S. A. (2003). Cortactin tyrosine phosphorylation requires Rac1 activity and association with the cortical actin cytoskeleton. Mol. Biol. Cell 14, 3216–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, P., Estrada, L., Kinley, A. W., Parsons, J. T., Vojtek, A. B., and Gorski, J. L. (2003). Fgd1, the Cdc42 GEF responsible for Faciogenital Dysplasia, directly interacts with cortactin and mAbp1 to modulate cell shape. Hum. Mol. Genet. 12, 1981–1993. [DOI] [PubMed] [Google Scholar]
- Huang, C., Ni, Y., Wang, T., Gao, Y., Haudenschild, C. C., and Zhan, X. (1997a). Down-regulation of the filamentous actin cross-linking activity of cortactin by Src-mediated tyrosine phosphorylation. J. Biol. Chem. 272, 13911–13915. [DOI] [PubMed] [Google Scholar]
- Huang, C., Tandon, N. N., Greco, N. J., Ni, Y., Wang, T., and Zhan, X. (1997b). Proteolysis of platelet cortactin by calpain. J. Biol. Chem. 272, 19248–19252. [DOI] [PubMed] [Google Scholar]
- Huttenlocher, A., Ginsberg, M. H., and Horwitz, A. F. (1996). Modulation of cell migration by integrin-mediated cytoskeletal linkages and ligand-binding affinity. J. Cell Biol. 134, 1551–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttenlocher, A., Palecek, S. P., Lu, Q., Zhang, W., Mellgren, R. L., Lauffenburger, D. A., Ginsberg, M. H., and Horwitz, A. F. (1997). Regulation of cell migration by the calcium-dependent protease calpain. J. Biol. Chem. 272, 32719–32722. [DOI] [PubMed] [Google Scholar]
- Kempiak, S. J., Yamaguchi, H., Sarmiento, C., Sidani, M., Ghosh, M., Eddy, R. J., Desmarais, V., Way, M., Condeelis, J., and Segall, J. E. (2005). A neural Wiskott-Aldrich Syndrome protein-mediated pathway for localized activation of actin polymerization that is regulated by cortactin. J. Biol. Chem. 280, 5836–5842. [DOI] [PubMed] [Google Scholar]
- Kinley, A. W., Weed, S. A., Weaver, A. M., Karginov, A. V., Bissonette, E., Cooper, J. A., and Parsons, J. T. (2003). Cortactin interacts with WIP in regulating Arp2/3 activation and membrane protrusion. Curr. Biol. 13, 384–393. [DOI] [PubMed] [Google Scholar]
- Kouyama, T., and Mihashi, K. (1981). Fluorimetry study of N-(1-pyrenyl) iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur. J. Biochem. 114, 33–38. [PubMed] [Google Scholar]
- Kowalski, J. R., Egile, C., Gil, S., Snapper, S. B., Li, R., and Thomas, S. M. (2005). Cortactin regulates cell migration through activation of N-WASP. J. Cell Sci. 118, 79–87. [DOI] [PubMed] [Google Scholar]
- Krueger, E. W., Orth, J. D., Cao, H., and McNiven, M. A. (2003). A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factorstimulated cells. Mol. Biol. Cell 14, 1085–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y., Tondravi, M., Liu, J., Smith, E., Haudenschild, C. C., Kaczmarek, M., and Zhan, X. (2001). Cortactin potentiates bone metastasis of breast cancer cells. Cancer Res. 61, 6906–6911. [PubMed] [Google Scholar]
- Lua, B. L., and Low, B. C. (2004). BPGAP1 interacts with cortactin and facilitates its translocation to cell periphery for enhanced cell migration. Mol. Biol. Cell 15, 2873–2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoune, A., Luo, J. H., Lauffenburger, D. A., and Wells, A. (2003). Calpain-2 as a target for limiting prostate cancer invasion. Cancer Res. 63, 4632–4640. [PubMed] [Google Scholar]
- Martinez-Quiles, N., Ho, H. Y., Kirschner, M. W., Ramesh, N., and Geha, R. S. (2004). Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol. Cell. Biol. 24, 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNiven, M. A., Kim, L., Krueger, E. W., Orth, J. D., Cao, H., and Wong, T. W. (2000). Regulated interactions between dynamin and the actin-binding protein cortactin modulate cell shape. J. Cell Biol. 151, 187–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, A., et al. (2005). WAVE/Scars in platelets. Blood 105, 3141–3148. [DOI] [PubMed] [Google Scholar]
- Patel, A. M., Incognito, L. S., Schechter, G. L., Wasilenko, W. J., and Somers, K. D. (1996). Amplification and expression of EMS-1 (cortactin) in head and neck squamous cell carcinoma cell lines. Oncogene 12, 31–35. [PubMed] [Google Scholar]
- Patel, A. S., Schechter, G. L., Wasilenko, W. J., and Somers, K. D. (1998). Overexpression of EMS1/cortactin in NIH3T3 fibroblasts causes increased cell motility and invasion in vitro. Oncogene 16, 3227–3232. [DOI] [PubMed] [Google Scholar]
- Pollard, T. D. (1984). Purification of a high molecular weight actin filament gelation protein from Acanthamoeba that shares antigenic determinants with vertebrate spectrins. J. Cell Biol. 99, 1970–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard, T. D., and Borisy, G. G. (2003). Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- Robles, E., Huttenlocher, A., and Gomez, T. M. (2003). Filopodial calcium transients regulate growth cone motility and guidance through local activation of calpain. Neuron 38, 597–609. [DOI] [PubMed] [Google Scholar]
- Ruoslahti, E., Hayman, E. G., Pierschbacher, M., and Engvall, E. (1982). Fibronectin: purification, immunochemical properties, and biological activities. Methods Enzymol. 82, 803–831. [DOI] [PubMed] [Google Scholar]
- Schafer, D. A., Weed, S. A., Binns, D., Karginov, A. V., Parsons, J. T., and Cooper, J. A. (2002). Dynamin2 and cortactin regulate actin assembly and filament organization. Curr. Biol. 12, 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J. A., and Watt, S. (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosintroponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871. [PubMed] [Google Scholar]
- Tompa, P., Buzder-Lantos, P., Tantos, A., Farkas, A., Szilagyi, A., Banoczi, Z., Hudecz, F., and Friedrich, P. (2004). On the sequential determinants of calpain cleavage. J. Biol. Chem. 279, 20775–20785. [DOI] [PubMed] [Google Scholar]
- Weaver, A. M., Karginov, A. V., Kinley, A. W., Weed, S. A., Li, Y., Parsons, J. T., and Cooper, J. A. (2001). Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374. [DOI] [PubMed] [Google Scholar]
- Weed, S. A., Karginov, A. V., Schafer, D. A., Weaver, A. M., Kinley, A. W., Cooper, J. A., and Parsons, J. T. (2000). Cortactin localization to sites of actin assembly in lamellipodia requires interactions with F-actin and the Arp2/3 complex. J. Cell Biol. 151, 29–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weed, S. A., and Parsons, J. T. (2001). Cortactin: coupling membrane dynamics to cortical actin assembly. Oncogene 20, 6418–6434. [DOI] [PubMed] [Google Scholar]
- Yan, B., Calderwood, D. A., Yaspan, B., and Ginsberg, M. H. (2001). Calpain cleavage promotes talin binding to the beta 3 integrin cytoplasmic domain. J. Biol. Chem. 276, 28164–28170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]