Evolution of alternative splicing after gene duplication (original) (raw)
Abstract
Alternative splicing and gene duplication are two major sources of proteomic function diversity. Here, we study the evolutionary trend of alternative splicing after gene duplication by analyzing the alternative splicing differences between duplicate genes. We observed that duplicate genes have fewer alternative splice (AS) forms than single-copy genes, and that a negative correlation exists between the mean number of AS forms and the gene family size. Interestingly, we found that the loss of alternative splicing in duplicate genes may occur shortly after the gene duplication. These results support the subfunctionization model of alternative splicing in the early stage after gene duplication. Further analysis of the alternative splicing distribution in human duplicate pairs showed the asymmetric evolution of alternative splicing after gene duplications; i.e., the AS forms between duplicates may differ dramatically. We therefore conclude that alternative splicing and gene duplication may not evolve independently. In the early stage after gene duplication, young duplicates may take over a certain amount of protein function diversity that previously was carried out by the alternative splicing mechanism. In the late stage, the gain and loss of alternative splicing seem to be independent between duplicates.
Alternative splicing, which was discovered decades ago, is a common post-transcriptional process in eukaryotic organisms to produce multiple transcript isoforms from a single gene (Black 2003). Although substantial evidence has shown the functional importance of alternative splicing in development, differentiation, and cancer (Lopez 1998; Jiang et al. 2000; Venables 2002), alternative splicing was conventionally thought of as an exceptional event occurring in only 5% of human genes (Sharp 1994). However, this view has been shown incorrect by genomic data. Indeed, many studies have revealed a very different picture—that >50% of human or mouse genes are alternatively spliced (Mironov et al. 1999; Brett et al. 2000, 2002; Kan et al. 2001; Kim et al. 2004). Though the estimation of alternative splice (AS) forms has been rough, and may vary among different approaches and EST data sets, it has been generally accepted recently that alternative splicing may serve as one major mechanism for generating proteomic complexity in higher eukaryotes (Graveley 2001; Maniatis and Tasic 2002; Kriventseva et al. 2003).
The finding of a high percentage of alternatively spliced genes in humans and mice raises several interesting evolutionary questions. For instance, gene duplications have been widely proposed as the major resource for the origin of new genes to increase the proteomic complexity by the follow-up functional divergence (Ohno 1970; Hughes 1994; Li 1997). So, what would happen when an alternatively spliced gene is duplicated? Apparently, each duplicate copy could lose some AS isoforms due to the functional redundancy, or they could acquire new isoforms. Yu et al. (2003) found that two duplicates of Fugu synapsin-2 genes, SYN2a and SYN2b, corresponded to each of the AS isoforms of the single human SYN2 gene. Using the model of subfunctionization (Force et al. 1999), the investigators suggested that the ancestral Fugu gene prior to duplication may have had two AS isoforms, each of which was kept by one duplicate gene, respectively; each duplicate gene lost the potential to produce the other AS isoform. Another example is the teleost mitf duplicate genes (mitfa and mitfb). Altschmied et al. (2002) found that long indels in the 5′ terminal might be the result of complementary degeneration of alternative 5′ exons of the ancestral gene after gene duplication.
In spite of these interesting case studies that indicate gene duplications may reduce the level of alternative splicing, it remains largely unclear at the genome level how alternative splicing evolves after gene duplications. Since both mechanisms are important for the evolution of functional diversity, some intriguing questions arise. For instance, how long does it take to generate alternative splicing difference between duplicates? Is alternative splicing evolution asymmetric? Do alternative splicing and gene duplication evolve independently? To address these issues, we developed a computational pipeline to predict the number of AS isoforms for each human gene, which was used as the proxy of the alternative splicing functional divergence. We then conducted a comprehensive analysis to investigate the evolutionary pattern of alternative splicing after gene duplications.
Results
Detecting human alternative splice forms
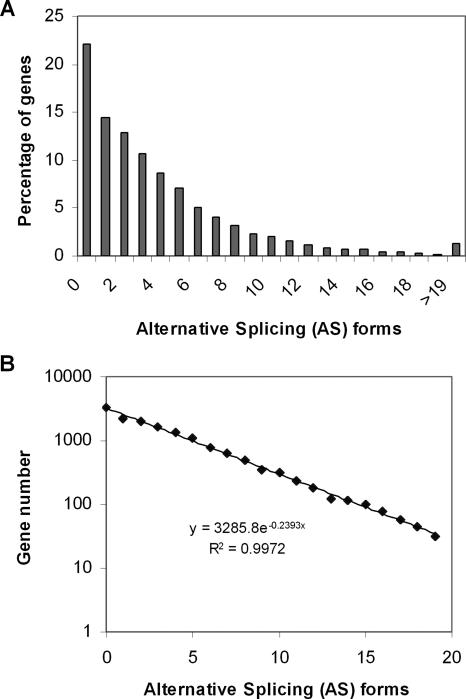
Our interest here is to identify all potential AS forms for well annotated human genes. To this end, we developed a computational pipeline to predict the number of AS isoforms for each human gene by comparing human ESTs and mRNAs to the fine-assembled human genome sequence (see Methods). We examined 15,422 non-redundant human genes (RefSeq) and found that 12,014 of them (77.9%) are possibly alternatively spliced; that is, they may have at least two forms of messenger RNA, or at least one AS form. The average number of AS forms is 3.9 per human gene. However, the range of AS forms is unexpectedly broad among genes. For instance, there are 1167 genes (7.6%) that have >10 AS forms. We found that the number of genes decays with the number of AS forms, following an exponential law perfectly (Fig. 1, R = 0.99). That is, the frequency (f)of k - AS forms can be characterized by a geometric distribution, f(k) = P(1 - P)k, where k = 0, 1, 2,... the parameter P was estimated as P ≈ 0.26.
Figure 1.
The distribution of alternative splice (AS) forms in human genes. (A) Each bar represents the percentage of human genes with the given number of AS forms. (B) The number of genes decays with the number of AS forms, following an exponential law (>19 AS forms excluded for simplicity).
Duplicates may have fewer alternative splice forms than single-copy genes
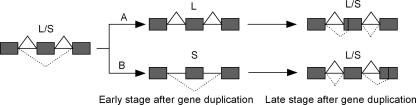
Gene duplications are widely believed to be the major source of genetic novelties (Ohno 1970). Meanwhile, numerical examples have shown that combinations of alternative splicing in specific genes can significantly expand the coding capacity of genome, such as for cell adhesion molecules or ion channels (Lopez 1998; Copley 2004). The “independent model” claims that alternative splicing and gene duplication are two independent mechanisms for increasing the proteomic complexity. Alternatively, the “function-sharing model” claims that some proteomic components can be performed either by alternatively spliced genes or duplicate genes. We have noticed that these two competing models have distinct predictions about the level of AS forms. For instance, the independent model predicts a similar level of AS forms between single-copy and duplicate genes, while the function-sharing model predicts a higher level of alternative splicing in single-copy genes than in duplicate genes, because AS forms can be fixed in each copy, respectively, after gene duplication (Fig. 2).
Figure 2.
Schematic illustration for the evolution of alternative splicing after gene duplication. The ancestral gene has two alternative splice forms, L (long) and S (short). In the early stage after gene duplication, the L and S forms may become dominantly expressed in one of the duplicate copies A and B, respectively. In the late stage, some novel alternative splice forms may be generated.
Yu et al. (2003) conducted a few case studies to support the function-sharing model. To further test whether it is the general case, we used a BLAST search to classify all well annotated human genes under study into 8819 single-copy genes and 6603 duplicate genes (see Methods). The percentage of alternative splicing of single-copy genes is 7090/8819 ≈ 80.4%, while that of duplication genes is given by 4924/6603 ≈ 74.6%. The difference between single-copy genes and duplicates is statistically highly significant (χ2 = 74.3, P < 10-5). Similarly, the mean number of alternative splicing duplicate genes is 3.52 ± 0.05, which is significantly lower than that of single-copy genes (4.11 ± 0.05 [_t_-test, _P_ < 0.001]). Besides, we found a higher proportion of single-copy genes with many AS forms (8.3% for >10 forms) than duplicates (6.6%). In short, our analysis suggests that, at the genome level, functional divergence among duplicate genes may reduce the number of AS forms, which is more consistent with the prediction of the function-sharing model than the independent model. Roughly speaking, one duplicate gene may take over a certain amount of proteomic diversity that previously was carried out by, on average, 0.59 (=4.11-3.52) AS forms. In other words, in the human genome, there are ∼6603 × 0.59 ≈ 3896 losses of AS forms due to gene duplications.
Large gene families tend to have fewer alternative splice forms
Another prediction from the function-sharing model is that large gene families tend to have fewer AS forms, because multiple rounds of gene duplication may result in more loss of AS forms. Except for 1486 genes that cannot be mapped to Ensembl gene ID or were not assigned an Ensembl gene family ID, we further classified the remaining 13,936 human genes into 8211 gene families (see Methods). For each gene family size, we estimated the mean of AS forms and the percentage of genes that are not alternatively spliced (no-AS). Figure 3A shows the mean of AS forms plotted against the gene family size; a single-copy gene is regarded as a gene family with size 1. It appears that the mean of AS forms remains roughly constant for gene families with small to moderate sizes, e.g., size 1 to 4, but decreases for larger gene families (R = -0.85, P < 0.0037). Similarly, Figure 3B shows that the proportion of no-AS forms increases with increasing gene family size (R = 0.93, P < 0.0004). In short, the negative correlation between the number of AS forms and the gene family size is likely to be caused by the effect of function-sharing after gene duplications.
Figure 3.
Fewer AS forms in larger gene families. Error bar, standard error. (A) Mean number of AS forms plotted against human gene family size (size = 1 means single-copy gene). (B) Proportion of genes that are not alternatively spliced plotted against the human gene family size.
Loss of alternative splicing may occur only shortly after gene duplication
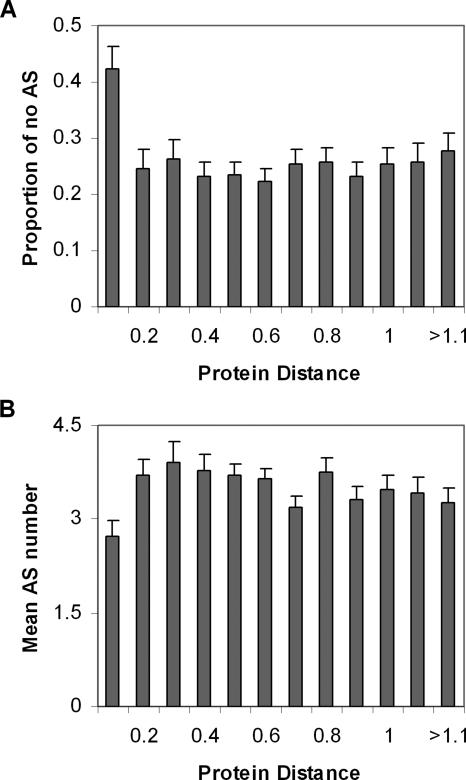
To further explore the evolutionary pattern of alternative splicing after gene duplications, we compiled independent 2875 human duplicate pairs and calculated the protein sequence distance (d) between duplicates (see Methods). As shown by a histogram (Supplemental Fig. 1), there is a peak of duplicate pairs around d = 0.7, roughly corresponding to 500-600 million years ago when the duplication time was estimated by the method of molecular clock (Gu et al. 2002b).
Using the protein sequence distance as a proxy of the age of the duplicate, we first grouped duplicate gene pairs with similar protein sequence distance (with a bin of 0.1 distance unit, ∼80 million years ago, or the time of mammalian radiation), and calculated the percentage of no-AS. Interestingly, the percentage of no-AS in the most recent duplicate group (d < 0.1) is 42%, which is almost two times higher than that in the other more ancient groups (22% on average, Fig. 4A); a χ2 test showed the difference is statistically significant (P < 0.001). Similarly, the mean number of AS forms (2.7) in the most recent duplicate genes is smaller than that of more ancient duplicate genes (3.5; Fig. 4B). Hence, both measures, the percentage of no-AS and the mean number of AS forms, indicated a rapid reduction of AS forms in young duplicates.
Figure 4.
The proportion of no alternatively spliced (no-AS) genes (A) and the mean of AS forms (B) plotted against the protein distance between duplicates (with a bin of 0.1 distance unit). Error bar, standard error.
Duplication versus alternative splicing in other model organisms
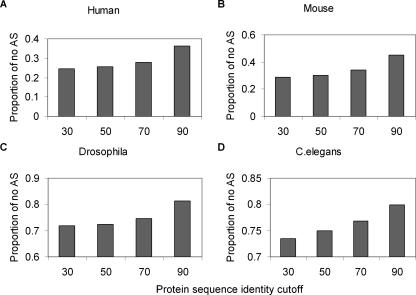
The above analysis was based on human AS isoforms. It would be interesting to test whether duplicates in other organisms also tend to have a lower number of AS isoforms. We developed a simple approach to compare the evolutionary patterns of alternative splicing in several model animals, and found that it might be the case (Fig. 5). First, we used the amino acid identity percentage (I) and the BLAST search E-value as the criteria to define duplicate genes. For example, the identity I = 30 and E-value 1E-10 mean genes have identity ≥30% and E-value is ≤1E-10. Second, we changed the criterion I from 30% to 50%, 70%, and 90%, respectively, and calculated the corresponding proportion of no-AS genes. Since the amino acid identity (I) is a proxy for young and ancient duplicate genes, the proportion of no-AS genes is expected to increase with the increasing of I, if the loss of AS isoforms in duplicates may occur shortly after the gene duplication. For comparison, we also included the human genome. As shown in Figure 5, we found that in all organisms we examined (human, mouse, Drosophila, and Caenorhabditis elegans), the proportion of no-AS genes tends to increase with the sequence identity criterion. Our analysis indicated that the loss of AS forms in young duplicates may be a general pattern in animals.
Figure 5.
Recent duplicated genes are unlikely to be alternatively spliced (A-D). Each bar represents the proportion of genes that is not alternatively spliced in four model organisms. Genes having the sequence identity >30% and E-value <1E-10 were classified to the group 30. Similarly, the groups 50, 70, and 90 were under the identity cutoff 50%, 70%, and 90% respectively.
Alternative splicing of duplicates after the human-mouse split
To further test the hypothesis that loss of alternative splicing may occur only shortly after gene duplication, we classified all human genes into two groups using mouse orthologs as the reference. The A group (H1) includes 9640 one-to-one and 282 one-to-many human-mouse orthologous genes, while the B group (Hx) includes 419 human duplicate genes that were duplicated after the human-mouse split. We analyzed the alternative splicing evolution for the B group, i.e., human recent duplicates after the human-mouse split. We found that the mean AS forms of the Hx group (2.83 ± 0.2) is significantly lower than that of the H1 group (3.66 ± 0.04), as well as that of whole human duplicate genes (3.52 ± 0.05). Similarly, the percentage of no-AS forms in Hx (0.369 ± 0.024) is significantly higher than in the H1 group (0.221 ± 0.0042). These results indicate that the reduction of AS forms in duplicate genes mainly happened in the early stage after the gene duplication.
Put together, we propose a scenario for the evolution of alternative splicing after the gene duplication (Fig. 2). For simplicity, consider a gene that has two AS forms, L (long) and S (short), for distinct physiological roles. After the gene/genome duplication, two duplicate copies (A and B) inherit both AS forms, but may start to have differential expression profiles. Because of the functional redundancy, the transcription of the L form in gene A becomes more dominant, while that of the S form in gene B becomes dominant. This evolutionary transition from alternative splicing diversity to functional divergence of duplicates may occur in the early stage after gene duplication. In the late stage, novel alternative splicing may be added, increasing the overall number of AS forms.
Testing asymmetric evolution of alternative splicing after gene duplication
Although there are 571 (19.8%) and 575 (20%) duplicate pairs with no or only one difference, respectively (Fig. 6), we indeed observed a significant portion of duplicate pairs showing dramatic differences in their number of AS forms. For instance, 206 duplicate pairs (7.2%) were found to have >10 differences of AS forms, indicating the possibility of an asymmetric pattern of alternative splicing evolution after gene duplication. It implies that not all duplicate genes have a similar number of AS forms. To address this issue vigorously, we used the binomial test to obtain the _P_-value (type-one error) for each duplicate pair under the null hypothesis of no difference in the number of AS forms. As a result, 418 pairs (14.5%) have significantly different AS forms (P < 0.05), and 181 pairs (6.3%) have _P_-values <0.01 (Fig. 7). Table 1 shows the 36 duplicate pairs that have the largest AS form difference (P < 0.0001). (For all 181 pairs, see Supplemental Table.) However, since it involves 2875 simultaneous statistical tests, the multiple-test problem should not be neglected. For instance, at the significance level of 0.05, there are ∼2875 × 0.05 ≈ 144 significant cases by pure chance. In other words, at this 0.05 significance level, we observed 418 significant cases; only 418 - 144 = 274 cases are likely to be truly significant, whereas others are false-positive. Statistically, it may be evaluated by the false-positive discovery rate (FDR). At the 0.05 significance level, we calculated FDR ≈ 34%. For 181 significant cases at the level of 0.01, the FDR is ∼16%. At any rate, this analysis indicates that a significant portion of human duplicate genes evolve asymmetrically in the AS forms.
Figure 6.
The distribution of the number of AS form differences in all duplicate pairs. _P_-values for asymmetric AS evolution between duplicates <0.01 (black bars) and >0.01 (white bars) are shown.
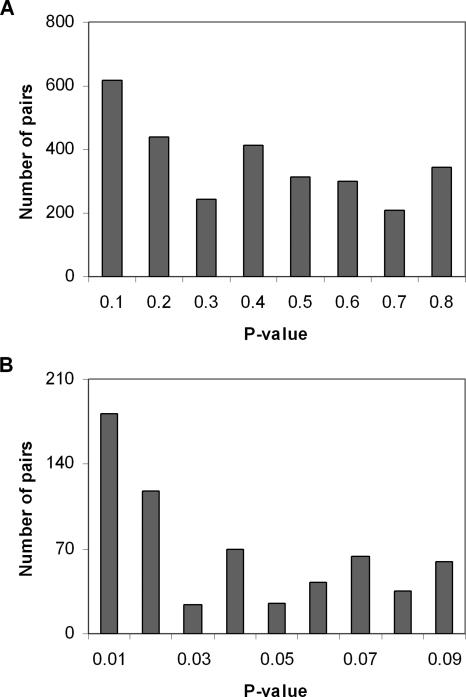
Figure 7.
The _P_-value distribution of all duplicate pairs for asymmetric AS evolution after gene duplication, which was calculated by the binomial test under the null hypothesis of no difference in the number of AS forms (see Methods). (A) represents all duplicate pairs; (B) represents duplicate pairs having _P_-value <0.1.
Table 1.
Duplicate pairs that have the highest alternative splicing divergence
| Gene 1 | AS forms of gene 1 | Gene 2 | AS forms of gene 2 | Function of gene 1 | Function of gene 2 | _P_-value |
|---|---|---|---|---|---|---|
| NM_014364 | 2 | NM_002046 | 63 | GAPDH-2 | Glyceraldehyde-3-phosphate dehydrogenase, GAPDH | 0 |
| NM_000090 | 57 | NM_000393 | 2 | Collagen, type III, alpha 1 | Collagen, type V, alpha 2 | 1.5E – 14 |
| NM_080426 | 46 | NM_002073 | 0 | GNAS complex locus | G protein, alpha z polypeptide | 1.74E – 13 |
| NM_001658 | 39 | NM_001659 | 3 | ADP-ribosylation factor 1 | ADP-ribosylation factor 3 | 8.53E – 09 |
| NM_003970 | 38 | NM_003803 | 3 | Myomesin (M-protein) 2 | Myomesin 1 (skelemin) | 1.55E – 08 |
| NM_184041 | 43 | NM_005165 | 6 | Aldolase A | Aldolase C | 6.06E – 08 |
| NM_003127 | 30 | NM_003126 | 1 | α-spectrin, non-erythrocytic 1 | α-spectrin, erythrocytic 1 | 6.54E – 08 |
| NM_003971 | 3 | NM_033392 | 35 | Sperm associated antigen 9 | Protein kinase binding | 9.29E – 08 |
| NM_002473 | 30 | NM_005964 | 2 | Myosin, heavy polypeptide 9 | Myosin, heavy polypeptide 10 | 3.83E – 07 |
| NM_000188 | 23 | NM_000189 | 0 | Hexokinase 1 | Hexokinase 2 | 7.75E – 07 |
| NM_006087 | 1 | NM_178014 | 25 | Tubulin, beta 4 | Tubulin, beta polypeptide | 1.52E – 06 |
| NM_012268 | 25 | NM_138790 | 1 | Phospholipase D3 | Hydrolase activity | 1.52E – 06 |
| NM_016521 | 0 | NM_007111 | 21 | Transcription factor Dp-3 | Transcription factor Dp-1 | 2.86E – 06 |
| NM_018677 | 25 | NM_032501 | 2 | ACAS2 (ADP forming) | ACAS2 (AMP forming)-like | 7.62E – 06 |
| NM_002293 | 22 | NM_006059 | 1 | Laminin, gamma 1 | Laminin, gamma 3 | 9.72E – 06 |
| NM_006366 | 1 | NM_006367 | 22 | CAP, adenylate cyclase-associated protein, 2 | CAP, adenylate cyclase-associated protein 1 | 9.72E – 06 |
| NM_005061 | 0 | NM_000967 | 19 | Ribosomal protein L3-like | Ribosomal protein L3 | 1.05E – 05 |
| NM_015528 | 19 | NM_181710 | 0 | Ring finger protein 167 | Zinc and ring finger 4 | 1.05E – 05 |
| NM_001416 | 29 | NM_014740 | 4 | Eukaryotic translation initiation factor 4A, isoform 1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 48 | 1.12E – 05 |
| NM_004739 | 3 | NM_004689 | 26 | MTA2 | Metastasis associated 1 | 1.7E – 05 |
| NM_017772 | 1 | NM_014346 | 21 | TBC1, member 22B | TBC1, member 22A | 1.79E – 05 |
| NM_002209 | 21 | NM_000887 | 1 | Integrin, alpha L | Integrin, alpha X | 1.79E – 05 |
| NM_000224 | 21 | NM_019010 | 1 | Keratin 18 | Keratin 20 | 1.79E – 05 |
| NM_004953 | 37 | NM_003760 | 9 | Eukaryotic translation initiation factor 4 gamma, 1 | Eukaryotic translation initiation factor 4 gamma, 3 | 3.08E – 05 |
| NM_182743 | 27 | NM_000637 | 4 | Thioredoxin reductase 1 | Glutathione reductase | 3.31E – 05 |
| NM_003380 | 31 | NM_001927 | 6 | Vimentin | Desmin | 3.51E – 05 |
| NM_021804 | 0 | NM_152831 | 17 | Angiotensin 1 converting enzyme (peptidyl-dipeptidase A) 2 | Angiotensin 1 converting enzyme (peptidyl-dipeptidase A) 1 | 3.81E – 05 |
| NM_198597 | 24 | NM_014822 | 3 | SEC24 related gene family, member C | SEC24 related gene family, member D | 5.19E – 05 |
| NM_152856 | 32 | NM_005778 | 7 | RNA binding motif protein 10 | RNA binding motif protein 5 | 5.61E – 05 |
| NM_019854 | 4 | NM_198319 | 26 | HMT1 hnRNP methyltransferase-like 4 | HMT1 hnRNP methyltransferase-like 2 | 5.65E – 05 |
| NM_006431 | 4 | NM_006429 | 26 | Chaperonin containing TCP1, subunit 2 (beta) | Chaperonin containing TCP1, subunit 7 (eta) | 5.65E – 05 |
| NM_006472 | 19 | NM_183376 | 1 | Thioredoxin interacting protein | Arrestin domain containing 4 | 6.06E – 05 |
| NM_021971 | 2 | NM_205847 | 21 | GDP-mannose pyrophosphorylase B | GDP-mannose pyrophosphorylase A | 7.83E – 05 |
| NM_012302 | 21 | NM_014921 | 2 | Latrophilin 2 | Latrophilin 1 | 7.83E – 05 |
| NM_054013 | 21 | NM_012214 | 2 | MGAT4B, transferase activity | MGAT4A, transferase activity | 7.83E – 05 |
| NM_000477 | 29 | NM_001134 | 6 | Albumin | α-fetoprotein | 9.55E – 05 |
The effects of EST coverage and expression level
Clearly, AS form detecting is affected by EST coverage, because the more ESTs found for a given gene, the more likely AS form(s) can be detected (Hide et al. 2001; Kan et al. 2002). When the size of the library is sufficiently large, the EST coverage of genes is mainly determined by the gene expression levels. Indeed, we observed a positive correlation between the detected AS form number and the EST hit number in our data set (P < 0.0001, Supplemental Fig. 2A). In addition, the number of genes decays exponentially with the number of AS forms (Fig. 1), as well as the number of EST hits (R = 0.93, Supplemental Fig. 2B).
We have run several tests to examine whether our duplicate AS analysis is affected by the EST coverage, measured by the number of ESTs aligned to a given gene (EST hits; see Methods). We found a very similar EST hit distribution between single-copy and duplicate genes; the mean number of EST hits in single-copy genes is 196 ± 3.7, with no significant difference from that in duplicate genes (206.9 ± 6.0). Moreover, we grouped duplicate genes and single-copy genes with similar EST coverage (with a bin of 40 EST hits) and found duplicate genes always have a larger percentage of no-AS genes and a smaller mean of AS forms in any EST coverage span (Supplemental Fig. 3).
In the AS-gene family size and AS-duplicate distance analyses, we classified all human genes into two categories: the H category for 7713 highly expressed genes with >112 EST hits, and the L category for 7709 other weakly expressed genes (<112 EST hits). We found a positive correlation between the proportion of no-AS genes and gene family size in both H and L categories (Supplemental Fig. 4). For the L category, the proportion of no-AS decreases with increasing duplicate protein distance. However, in the H category, the proportion of no-AS in single-copy genes and duplicate genes decreases with increasing of the duplicate protein distance when d < 0.3, but seems to increase in more ancient duplicate pairs (Supplemental Fig. 4). Though this pattern needs to be elaborated further, it may imply that highly expressed duplicate genes may lose some AS forms in the late stage after gene duplication.
Finally, we compared the duplicate pairs that have significant asymmetric evolution of AS forms (Table 2). For 181 duplicate pairs with highly significant asymmetric AS forms (P < 0.01), there are 98 pairs (54%) that have similar EST coverage (both belong to the _H_ or _L_ category). Though it is reasonably (but not much) lower than that for the rest of 2694 duplicate pairs with _P_ > 0.01 (67%), it indicates that the asymmetric evolution of alternative splicing after gene duplication may not merely be because of the gene expression divergence difference.
Table 2.
Summary of expression and alternative splicing divergence of duplicates
| P < 0.01 (181) | P > 0.01 (2694) | |||
|---|---|---|---|---|
| Duplicate pair categories | Number of pairs | Proportion | Number of pairs | Proportion |
| HHa | 89 | 0.49 | 798 | 0.3 |
| LLb | 9 | 0.05 | 1018 | 0.37 |
| LHc | 83 | 0.46 | 878 | 0.33 |
Discussion
We conducted a large-scale alternative splicing analysis of the human genome, using human AS forms predicted from the EST databases. Many methods or programs for identification of alternative splicing have been developed recently, such as mapping ESTs onto mRNA sequences (Brett et al. 2002); aligning ESTs with genomic sequences (Mironov et al. 1999); multiply aligning ESTs, mRNAs, and genomic sequences (Modrek et al. 2001; Modrek and Lee 2003); and aligning transcribed consensus sequences to genomic sequences (Gupta et al. 2004). A number of studies have relied on EST self-clustering to assemble alternatively spliced transcripts (Mironov et al. 1999; Quackenbush et al. 2001). Some studies have combined genome-based EST clustering and transcripts' assembly approaches to reconstruct the alternatively spliced isoforms (Kan et al. 2001; Xing et al. 2004; Kim et al. 2005b). All these efforts attempt to address the “garbage EST” issue. The analysis pipeline we developed has adopted several techniques to reduce the potential garbage EST effects (also see Methods). First, a simple method was implemented to verify splice forms based on the pairwise alignments between EST/RefSeq and genomic sequences to alleviate the error-prone nature of EST consensus sequences caused by high sequencing error rates in ESTs, experimental contaminations, chimeric clones, redundant copies, paralogous genes, or pseudogenes. Second, any EST that results in an extremely short exon or intron was removed because extremely short exons and introns are error-prone. Third, splice-site motifs (GT-AG/GC-AG) were used to validate our detected AS forms. And finally, to avoid the high false-positive rate of the transcript assembly procedure (Bouck et al. 1999; Xing et al. 2004), we did not assemble the ESTs to full-length transcripts. Instead, we simply detected the number of mutually exclusive splice, using this as a proxy for the level of AS-related functional diversity.
We found that the percentage of alternatively spliced genes and the average number of AS forms per gene we estimated in the human genome are somewhat higher than previous prediction (e.g., Kim et al. 2004). The difference may be from the different sampling strategies adopted. To obtain a more precise evaluation of alternative splicing, we selected the well annotated human genes in the RefSeq database (Pruitt et al. 2005). These well annotated genes obviously have higher EST coverage than other genes.
Many nonfunctional alternatively spliced transcripts may be produced during pre-mRNA splicing (for reviews, see Modrek and Lee 2002; Lareau et al. 2004), such as aberrant splicing in some tumors, genome contamination, unspliced mRNA, splicing errors that arose from various reasons, or background splicing without any effect on the cellular function. Some of these artifacts can be detected by identifying the premature termination codon (PTC) in alternative splicing isoforms to find the putative targets of nonsense-mediated decay (NMD) (Lewis et al. 2003; Xing and Lee 2004). In this current study, we were not able to use such an approach because we did not assemble the ESTs to full-length transcripts. Instead, we excluded these garbage ESTs by requiring pairs of mutually exclusive splices in different ESTs. Since observing a given splice form in one EST may be insufficient, we tested a stricter criterion by requiring that the two ESTs share one splice site but differ at RefSeq. As expected, we detected about half the number of AS forms than before, but our analysis showed it did not affect our main conclusions (Supplemental Fig. 5).
In spite of these precautions, inevitably, the AS forms detected still had a certain level of error. To test whether our main results were not sensitive to the inherent bias in our analysis pipeline, we repeated our analysis using independent predicted alternative splicing data derived from the ECgene database (Version 1.2) (http://genome.ewha.ac.kr/ECgene/) (Kim et al. 2005a,b). As shown in Supplemental Figure 6, we obtained virtually similar results.
Conclusions
In this study, we discovered that the level of alternative splicing in duplicate genes is usually lower than that of single-copy genes. Further analyses indicated that the number of AS forms is negatively associated with the gene family size, and the loss of AS forms may happen shortly after the gene duplication. Moreover, we demonstrated asymmetric AS evolution; i.e., the AS numbers between some duplicates may differ dramatically. Some of these results have been confirmed in other organisms, including mouse, Drosophila, and C. elegans. We therefore conclude that AS and gene duplication, two mechanisms for proteomic function diversity, may not evolve independently, supporting the model of function-sharing. That is, in the early stage after gene duplication, the young duplicates may take over a certain amount of protein function diversity that previously was carried out by the alternative splicing mechanism. After that, the evolution of AS forms may be independent between duplicates.
Several studies have shown that the evolutionary rate of coding sequences may accelerate shortly after gene duplication (Lynch and Force 2000; Kondrashov et al. 2002; Conant and Wagner 2003; Zhang et al. 2003). Gu et al. (2005) found that the initial rate for either expression or regulatory network evolution after yeast gene duplications is much higher than that of the late stage. The current study for the loss of AS forms between duplicates presents another example to support the viewpoint of rapid evolution in the early stage after gene duplication. Moreover, the pattern of rapid AS form loss in young duplicates is consistent with the model of subfunctionalization (Lister et al. 2001; Altschmied et al. 2002; Yu et al. 2003). We suggested that AS subfunctionalization of duplicate genes may be a general phenomenon that happened in the early stage after gene duplication. On the other hand, alternative splicing may contribute to the neofunctionalization by acquiring new functional AS variants, resulting in the increase of AS isoforms for ancient duplicate genes (Fig. 2).
Methods
Sequence data
The reference sequences (RefSeq) of human, mouse, Drosophila, and C. elegans were downloaded from NCBI (ftp://ftp.ncbi.nih.gov/refseq/). Only the mRNA sequences of IDs starting with the prefix “NM_” were extracted, and the corresponding protein sequences were also extracted. There were 21,267, 16,863, 18,648, and 20,785 mRNA reference sequences for human, mouse, Drosophila, and C. elegans, respectively. The alignments between mRNA reference sequences and genome sequence were downloaded from the UCSC genome center (http://genome.ucsc.edu/), and they were generated by the BLAT program (Kent et al. 2002). The alignment information was used to remove redundant sequences. In the case of multiple mRNA sequences being mapped to one genomic region, only the sequence resulting in the longest protein product was accepted.
Identification of duplicate genes
We used the method of Gu et al. (2002a) to identify duplicate genes. Briefly, every protein was used as the query to search against all other proteins by using BLASTP (E = 10). Two proteins are scored as forming a link if (1) the BLASTP alignable region between them is >80% of the longer protein, and (2) the identity (I) between them is ≥30% if the alignable region is longer than 150 amino acids; I ≥ 0.01_n_ + 4.8_L_-0.32[1+exp(-L/1000)] (Rost 1999) for all other protein pairs, in which n = 6 and L is the alignable length between the two proteins. Duplicate pairs were seeded with a two-way best pairwise match. The protein sequence distance (d) between duplicates was calculated by d = -ln(I/100), the Poisson correction.
Identifying gene family size and genes duplicated after the human-mouse split
All human gene family IDs were extracted using the EnsMart tool (Kasprzyk et al. 2004). The size of each gene family was obtained from counting the frequency of this gene's family ID in whole genome. Orthologs and in-paralogs between human and mouse were extracted from the Inparanoid database (version 4.0) (O'Brien et al. 2005). Human genes having one-to-one and one-to-many relationships with mouse were classified into the H1 category. Similarly, human genes having many-to-one and many-to-many relationships with mouse were classified into the Hx category. RefSeq/Ensembl gene mapping was extracted using EnsMart. Only reciprocally unique pairs were further analyzed.
Identification of alternative splice forms
Alternative splice forms were identified by using alignments of human EST/RefSeq and genomic sequences as follows: The EST/RefSeq sequences that were highly similar to only one genomic region were selected. The remaining EST/RefSeq sequences were discarded. The selected EST/RefSeq sequences were grouped together by genomic regions, one group per genomic region. If there were multiple RefSeq sequences in a group, then the RefSeq sequence with the longest protein sequence was the leader sequence for the group. For each EST/RefSeq sequence, splice sites in the genomic region were identified by the GeneSplicer program (Pertea et al. 2001), and exons and introns of sufficient lengths in the genomic region were identified based on the splice sites and the alignment of the EST/RefSeq sequence and the genomic region. Any EST/RefSeq sequence without any exons and introns was discarded. The pattern of splicing for each EST/RefSeq sequence was indicated by a list of all exons in order of increasing coordinate. For each group of EST/RefSeq sequences, alternative splice forms were identified by comparing the exon list of each EST/RefSeq sequence with the exon list of the leader sequence and finding differences in exon coordinates. Redundant alternative splice forms were removed.
The method given above finds and removes garbage ESTs from aberrant transcripts or abnormal cell lines by three measures: First, any EST that is not highly similar to a genomic region over a majority of its length is removed. Second, any EST that results in an extremely short exon or intron is removed because extremely short exons and introns are error-prone. Third, any EST that results in a weak splice site in a genomic region is removed.
Additional methods for finding garbage ESTs have been suggested by several previous studies (Modrek and Lee 2002; Lareau et al. 2004; Xing and Lee 2004). One of them is to use only AS forms that are confirmed by two independent libraries. We are currently experimenting with this method and other methods to see if they affect the main observations in the paper.
Acknowledgments
This work was supported by NIH grants (X.G.).
Article published online ahead of print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.4197006.
Footnotes
[Supplemental material is available online at www.genome.org.\]
References
- Altschmied, J., Delfgaauw, J., Wilde, B., Duschl, J., Bouneau, L., Volff, J.N., and Schartl, M. 2002. Subfunctionalization of duplicate mitf genes associated with differential degeneration of alternative exons in fish. Genetics 161 259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black, D.L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72 291-336. [DOI] [PubMed] [Google Scholar]
- Bouck, J., Yu, W., Gibbs, R., and Worley, K. 1999. Comparison of gene indexing databases. Trends Genet. 15 159-162. [DOI] [PubMed] [Google Scholar]
- Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbruck, S., Krueger, S., Reich, J., and Bork, P. 2000. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474 83-86. [DOI] [PubMed] [Google Scholar]
- Brett, D., Pospisil, H., Valcarcel, J., Reich, J., and Bork, P. 2002. Alternative splicing and genome complexity. Nat. Genet. 30 29-30. [DOI] [PubMed] [Google Scholar]
- Conant, G.C. and Wagner, A. 2003. Asymmetric sequence divergence of duplicate genes. Genome Res. 13 2052-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copley, R.R. 2004. Evolutionary convergence of alternative splicing in ion channels. Trends Genet. 20 171-176. [DOI] [PubMed] [Google Scholar]
- Force, A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.L., and Postlethwait, J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 17 100-107. [DOI] [PubMed] [Google Scholar]
- Gu, Z., Cavalcanti, A., Chen, F.C., Bouman, P., and Li, W.H. 2002a. Extent of gene duplication in the genomes of Drosophila, nematode, and yeast. Mol. Biol. Evol. 19 256-262. [DOI] [PubMed] [Google Scholar]
- Gu, X., Wang, Y., and Gu, J. 2002b. Age distribution of human gene families shows significant roles of both large- and small-scale duplications in vertebrate evolution. Nat. Genet. 31 205-209. [DOI] [PubMed] [Google Scholar]
- Gu, X., Zhang, Z., and Huang, W. 2005. Rapid evolution of expression and regulatory divergences after yeast gene duplication. Proc. Natl. Acad. Sci. 102 707-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, S., Zink, D., Korn, B., Vingron, M., and Haas, S.A. 2004. Genome wide identification and classification of alternative splicing based on EST data. Bioinformatics 20 2579-2585. [DOI] [PubMed] [Google Scholar]
- Hide, W.A., Babenko, V.N., van Heusden, P.A., Seoighe, C., and Kelso, J.F. 2001. The contribution of exon-skipping events on chromosome 22 to protein coding diversity. Genome Res. 11 1848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. 1994. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256 119-124. [DOI] [PubMed] [Google Scholar]
- Jiang, Z., Cote, J., Kwon, J.M., Goate, A.M., and Wu, J.Y. 2000. Aberrant splicing of τ pre-mRNA caused by intronic mutations associated with the inherited dementia frontotemporal dementia with parkinsonism linked to chromosome 17. Mol. Cell. Biol. 20 4036-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, Z., Rouchka, E.C., Gish, W.R., and States, D.J. 2001. Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res. 11 889-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan, Z., States, D., and Gish, W. 2002. Selecting for functional alternative splices in ESTs. Genome Res. 12 1837-1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzyk, A., Keefe, D., Smedley, D., London, D., Spooner, W., Melsopp, C., Hammond, M., Rocca-Serra, P., Cox, T., and Birney, E. 2004. EnsMart: A generic system for fast and flexible access to biological data. Genome Res. 14 160-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W.J., Sugnet, C.W., Furey, T.S., Roskin, K.M., Pringle, T.H., Zahler, A.M., and Haussler, D. 2002. The human genome browser at UCSC. Genome Res. 12 996-1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H., Klein, R., Majewski, J., and Ott, J. 2004. Estimating rates of alternative splicing in mammals and invertebrates. Nat. Genet. 36 915-916; author reply 916-917. [DOI] [PubMed] [Google Scholar]
- Kim, P., Kim, N., Lee, Y., Kim, B., Shin, Y., and Lee, S. 2005a. ECgene: Genome annotation for alternative splicing. Nucleic Acids Res. 33 D75-D79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, N., Shin, S., and Lee, S. 2005b. ECgene: Genome-based EST clustering and gene modeling for alternative splicing. Genome Res. 15 566-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, F.A., Rogozin, I.B., Wolf, Y.I., and Koonin, E.V. 2002. Selection in the evolution of gene duplications. Genome Biol. 3 research0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriventseva, E.V., Koch, I., Apweiler, R., Vingron, M., Bork, P., Gelfand, M.S., and Sunyaev, S. 2003. Increase of functional diversity by alternative splicing. Trends Genet. 19 124-128. [DOI] [PubMed] [Google Scholar]
- Lareau, L.F., Green, R.E., Bhatnagar, R.S., and Brenner, S.E. 2004. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 14 273-282. [DOI] [PubMed] [Google Scholar]
- Lewis, B.P., Green, R.E., and Brenner, S.E. 2003. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 100 189-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.-H. 1997. Molecular evolution. Sinauer Associates, Sunderland, MA.
- Lister, J.A., Close, J., and Raible, D.W. 2001. Duplicate mitf genes in zebrafish: Complementary expression and conservation of melanogenic potential. Dev. Biol. 237 333-344. [DOI] [PubMed] [Google Scholar]
- Lopez, A.J. 1998. Alternative splicing of pre-mRNA: Developmental consequences and mechanisms of regulation. Annu. Rev. Genet. 32 279-305. [DOI] [PubMed] [Google Scholar]
- Lynch, M. and Force, A. 2000. The probability of duplicate gene preservation by subfunctionalization. Genetics 154 459-473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T. and Tasic, B. 2002. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature 418 236-243. [DOI] [PubMed] [Google Scholar]
- Mironov, A.A., Fickett, J.W., and Gelfand, M.S. 1999. Frequent alternative splicing of human genes. Genome Res. 9 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek, B. and Lee, C. 2002. A genomic view of alternative splicing. Nat. Genet. 30 13-19. [DOI] [PubMed] [Google Scholar]
- ———. 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat. Genet. 34 177-180. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., and Lee, C. 2001. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 29 2850-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, K.P., Remm, M., and Sonnhammer, E.L. 2005. Inparanoid: A comprehensive database of eukaryotic orthologs. Nucleic Acids Res. 33 D476-D480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S. 1970. Evolution by gene and genome duplication. Springer, Berlin.
- Pertea, M., Lin, X., and Salzberg, S.L. 2001. GeneSplicer: A new computational method for splice site prediction. Nucleic Acids Res. 29 1185-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K.D., Tatusova, T., and Maglott, D.R. 2005. NCBI reference sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 33 D501-D504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush, J., Cho, J., Lee, D., Liang, F., Holt, I., Karamycheva, S., Parvizi, B., Pertea, G., Sultana, R., and White, J. 2001. The TIGR gene indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 29 159-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rost, B. 1999. Twilight zone of protein sequence alignments. Protein Eng. 12 85-94. [DOI] [PubMed] [Google Scholar]
- Sharp, P.A. 1994. Split genes and RNA splicing. Cell 77 805-815. [DOI] [PubMed] [Google Scholar]
- Venables, J.P. 2002. Alternative splicing in the testes. Curr. Opin. Genet. Dev. 12 615-619. [DOI] [PubMed] [Google Scholar]
- Xing, Y. and Lee, C.J. 2004. Negative selection pressure against premature protein truncation is reduced by alternative splicing and diploidy. Trends Genet. 20 472-475. [DOI] [PubMed] [Google Scholar]
- Xing, Y., Resch, A., and Lee, C. 2004. The multiassembly problem: Reconstructing multiple transcript isoforms from EST fragment mixtures. Genome Res. 14 426-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W.P., Brenner, S., and Venkatesh, B. 2003. Duplication, degeneration and subfunctionalization of the nested synapsin-Timp genes in Fugu. Trends Genet. 19 180-183. [DOI] [PubMed] [Google Scholar]
- Zhang, P., Gu, Z., and Li, W.H. 2003. Different evolutionary patterns between young duplicate genes in the human genome. Genome Biol. 4 R56. [DOI] [PMC free article] [PubMed] [Google Scholar]