Differential Distribution of Exosome Subunits at the Nuclear Lamina and in Cytoplasmic Foci (original) (raw)
Abstract
The exosome complex plays important roles in RNA processing and turnover. Despite significant mechanistic insight into exosome function, we still lack a basic understanding of the subcellular locales where exosome complex biogenesis and function occurs. Here, we employ a panel of Drosophila S2 stable cell lines expressing epitope-tagged exosome subunits to examine the subcellular distribution of exosome complex components. We show that tagged Drosophila exosome subunits incorporate into complexes that recover endogenous nuclear and cytoplasmic exosome subunits. Immunolocalization analyses demonstrate that subsets of both epitope-tagged and endogenous exosome subunits are enriched in discrete subcellular compartments. In particular, dRrp4, dRrp42, dRrp46, and dCsl4 are enriched in cytoplasmic foci. Although dRrp4 and dRrp42 sometimes colocalize with dCsl4, these subunits are predominantly found in distinct cytoplasmic compartments. Strikingly, dRrp44/dDis3 and dRrp41/dSki6 colocalize with the nuclear lamina and often exhibit a restricted and asymmetric distribution at the nuclear periphery. Taken together, these observations indicate that individual exosome subunits have distinct localizations in vivo. These different distribution patterns presumably reflect distinct exosome subunit subcomplexes with correspondingly specialized functions.
INTRODUCTION
The exosome performs important roles in surveillance, 3′ processing, and turnover of distinct RNA substrates (Mitchell and Tollervey, 2000; Butler, 2002; Raijmakers et al., 2004). Consistent with its myriad functions, exosome subunits reside in the nucleus and the cytoplasm (Kadowaki et al., 1995; Allmang et al., 1999; Brouwer et al., 2001; Raijmakers et al., 2002b) and exosome complexes can be recovered from extracts representing these subcellular domains (Allmang et al., 1999; Andrulis et al., 2002; Forler et al., 2003). Exosome complexes have been isolated from eukaryotes such as Saccharomyces cerevisiae (Mitchell et al., 1997; Allmang et al., 1999), Drosophila melanogaster (Andrulis et al., 2002; Forler et al., 2003), Arabidopsis thaliana (Chekanova et al., 2000), Homo sapiens (Chen et al., 2001), and Trypanosoma brucei (Estevez et al., 2001; Estevez et al., 2003) as well as from the archaeon Sulfobactus solftaricus (Evguenieva-Hackenburg et al., 2003). The exosome complex is structurally and functionally conserved, as several yeast exosome mutant alleles can be complemented by their cognate human genes (Mitchell et al., 1997; Allmang et al., 1999).
In yeast, the cytoplasmic extract-derived exosome complex (“core”) contains 10 subunits in apparent stoichiometry, whereas the “nuclear” complex has one additional subunit, Rrp6 (Ribosomal RNA processing; Allmang et al., 1999). The majority of these subunits are predicted to be 3′ to 5′ exoribonucleases (Mitchell and Tollervey, 2000). In this regard, purified recombinant Rrp6 and three additional recombinant exosome polypeptides, Rrp4, Rrp44/Dis3, and Rrp41/Ski6, have 3′ to 5′ exoribonucleolytic activity in vitro (Mitchell et al., 1997; Burkard and Butler, 2000; Chekanova et al., 2000, 2002; Estevez et al., 2001). Several yeast exosome subunits, namely Rrp44/Dis3, Rrp4, Rrp40, and Csl4 contain S1 RNA-binding domains. The reason for grouping so many RNA binding and degradative activities in one protein complex is unclear, although it has been hypothesized that they are important for distinct RNA substrate recognition and regulation (Mitchell and Tollervey, 2000).
Several studies have addressed this hypothesis by examining exosome complex structure. Both high-throughput (Uetz et al., 2000; Ito et al., 2001) and directed (Oliveira et al., 2002; Raijmakers et al., 2002a; Estevez et al., 2003) two-hybrid analyses have revealed specific interactions among and between exosome subunits. These interactions and a three-dimensional (3D) reconstruction of electron microscopic images of the purified yeast exosome complex resulted in a hypothetical structure for exosome core complex (Aloy et al., 2002; Estevez et al., 2003). Recently, the Sulfobactus exosome core was defined to 2.8 Å resolution as a hexameric ring structure consisting of a trimer of Rrp41-Rrp42 heterodimers; the coordination of the exoribonuclease active sites appears to facilitate the recognition and processing of the RNA substrate (Lorentzen and Conti, 2005; Lorentzen et al., 2005). Eukaryotic exosome subunits are thought to assemble in a similar manner in vitro (Lorentzen et al., 2005). In addition, exosome complexes assembled either from the recombinant, purified Archaeoglobus fulgidus (Afu) exosome subunits _Afu_Rrp4, _Afu_Rrp41, and _Afu_Rrp42 or from _Afu_Csl4, _Afu_Rrp41, and _Afu_Rrp42 are stoichiometric (Buttner et al., 2005).
Despite the fact that exosome subunits can form subcomplexes in vitro, it is generally assumed that all exosome subunits function together in a single complex in vivo. However, whether exosome subunits form subcomplexes in vivo has not been formally addressed. This question requires consideration, because the core and “nuclear” exosome complexes have been exclusively implicated in numerous distinct RNA metabolic pathways. In this regard, although most exosome genes were identified in screens for rRNA processing mutants (Mitchell et al., 1996, 1997), a select few exosome subunits have been shown to localize along transcriptionally active genes (Rrp6, Rrp4, Csl4; Andrulis et al., 2002; Hieronymus et al., 2004). Furthermore, only a few subunits have been implicated in transcription-site retention of aberrant transcripts (Rrp6; Hilleren et al., 2001; Libri et al., 2002), processing or degradation of terminator read-through transcripts (Rrp6 and Rrp41; Torchet et al., 2002), nonstop mRNA decay (Csl4; van Hoof et al., 2002), RNA interference (RNAi)-mediated mRNA turnover (Rrp4, Csl4; Orban and Izaurralde, 2005), NMD-mediated mRNA decay (Rrp4, Rrp41, and Rrp6/PM-Scl100 Lejeune et al., 2003; Mitchell and Tollervey, 2003; Gatfield and Izaurralde, 2004), tRNA surveillance (Rrp44/Dis3; Kadaba et al., 2004), mRNA export (Mtr3 and Rrp6; Kadowaki et al., 1995; Hieronymus et al., 2004), and RNA damage response (Rrp6; Hieronymus et al., 2004). Given that each of these studies focused on specific, rather than all exosome subunits, it is difficult to determine whether functions that are ascribed to the exosome complex are actually mediated by the exosome complex, or are mediated by individual exosome subunits or by subunits in distinct complexes.
To refine our understanding of exosome complex structure and function in vivo, we are studying interactions and localizations of exosome subunits in D. melanogaster S2 cells. Our findings suggest that several distinct exosome subunit subcomplexes exist in vivo.
MATERIALS AND METHODS
Cell Culture
D. melanogaster embryonic Schneider (S2) cell lines were grown in HyQ-CCM3 or HyQ-SFX media (Hyclone, Logan, UT) at 27°C. Stable cell lines of tagged Drosophila exosome subunits were prepared as described (Andrulis et al., 2002) and grown in media supplemented with 300 μg/ml hygromycin B (Invitrogen, Carlsbad, CA).
Cloning, Expression, and Bioinformatic Analysis of Drosophila Exosome Subunits
All plasmids were made using basic molecular cloning techniques. Exosome genes were PCR amplified from full-length cDNAs (Invitrogen) or available laboratory clones using primers with unique restriction enzyme sites and with an in-frame 3′ FLAG (DYKDDDK) tag or both 3′ FLAG and 6xHis epitope. After digestion with the appropriate enzymes, the tagged exosome genes were ligated into pRmHa3 to obtain tagged exosome genes downstream of the metallothionein (Mtn) promoter. Mtn-exosome constructs were transiently transfected using CELLFectin (Invitrogen) and tested for copper-inducible expression. Maltose-binding protein (MBP) fusions to exosome genes were made similarly, except the genes were cloned into the pMAL-c2 vector (New England Biolabs, Beverly, MA) and the 3′ primer lacked the epitope tags. Recombinant MBP-exosome subunit fusions were purified from Escherichia coli using single-step amylose resin affinity purification and maltose elution of MBP fusion proteins, followed by extensive dialysis. Protein domains were identified and sequence alignments were performed using BLAST: http://www.ncbi.nlm.nih.gov/BLAST/. Hypothetical nuclear localization signals were identified by PSORT: http://psort.nibb.ac.jp/.
Antibodies
Polyclonal antibodies were raised against the recombinant polypeptides MBP-dDis3, -dRrp46, and -dRrp40. Recombinant proteins were injected into animals and sera recovered (Pocono Rabbit Farm and Lab, Canadensis, PA). Rabbit anti-dDis3, guinea pig anti-dRrp46, and rabbit anti-dRrp40 bleeds were all compared against preimmune sera from the respective animals to determine specificity. Guinea pig anti-dCsl4, anti-dRrp42, and anti-dRrp4 were affinity-purified against the cognate purified recombinant protein.
Cell Fractionation
Cells were grown to a density of 1 × 107 cells/ml on a 100-mm Petri dish and recovered, centrifuged, washed with wash buffer (10 mM Tris, pH 7.4, 140 mM NaCl), centrifuged, and then lysed, with vigorous pipetting, in the presence of lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 3 mM MgCl2, 0.5 mM EDTA, pH 8.0, 0.5 mM DTT, 1% Triton X-100, 10% glycerol, and protease inhibitor cocktail; Invitrogen) on ice for 5 min. After centrifugation, supernatant was removed (∼150 μl), the resulting pellet was resuspended in an equivalent volume to the reserved supernatant, and both supernatant and pellet were mixed with 1× SDS loading dye and analyzed by Western blotting.
Immunoprecipitation and Immunoaffinity Purification
For exosome complex immunoaffinity purifications, whole cell extracts were prepared from each of the stable cell lines as described above for the cell fractionation extracts, except cell lysis was for 30 min, and all steps were performed in the presence of 450 NaCl. About 150 μl of soluble extract was obtained from ∼1 × 108 cells, and ∼90% of this was placed into an Eppendorf tube with 800 μl wash buffer (lysis buffer but 0.1% nonidet P-40 rather than TX-100), and 20 μl of a 50% slurry of M2 anti-FLAG affinity resin (Sigma, St. Louis, MO) that had been pre-equilibrated in wash buffer. After gentle end-over-end mixing for 1–2 h, beads were spun down, and washed exhaustively, tagged subunits were eluted with FLAG peptide (300 μg/ml), and eluates were resuspended in 1× SDS loading dye for analysis by SDS-PAGE and Western blotting.
Double-stranded RNA Preparation and RNAi
In vitro transcription templates were generated by PCR using exosome subunit-specific primers with T7 overhangs and transcribed using a MEGAscript T7 kit (Ambion, Austin, TX) according to the manufacturer's instructions. The resulting double-stranded RNA (dsRNA) was purified and annealed and its concentration was determined by UV spectrophotometry. S2 cells in a 24-well dish were treated with 5 μg of dsRNA three times over a period of 4 d. The cells were then either fixed and stained for immunofluorescence (see below) or pelleted, lysed in 1× SDS dye, and assayed by Western blotting.
Indirect Immunofluorescence
Stably transfected or untransfected S2 cells were plated in 100-mm Petri dishes and cultured for 1–2 d at early to midlog phase. The stably transfected S2 were induced with the appropriate CuSO4 concentration for 16 h. Cells were collected by centrifugation, washed once in phosphate-buffered saline (PBS), fixed in freshly prepared 3.7% formaldehyde in PBS for 10 min, and then washed once in PBS. All antibodies were diluted in PBS with 0.1% saponin and 1–2% normal donkey serum (PSN; Jackson ImmunoResearch, West Grove, PA). One-fifth of the cells were placed in amber Eppendorf tubes, stained for 1 h in primary antibodies (mouse anti-FLAG [1:500 dilution; Sigma], rabbit anti-dRrp6 [1:500], rabbit anti-dRrp40 [1:100], or affinity-purified guinea pig anti-dCsl4, anti-dRrp42, or anti-dRrp4 [1:5]), washed three times in PBS/0.05% saponin, stained for 45 min in specific secondary antibody (anti-mouse TR, anti-rabbit Cy2, or anti-guinea pig Cy2 at 1:100 dilutions; Jackson ImmunoResearch) and then washed three times with PBS before mounting (in 90% glycerol, 10% 100 mm Tris, pH 8.0, and 0.5% _n_-propyl gallate). For double immunofluorescence, cells were blocked overnight in PSN and probed with rabbit anti-Drosophila lamin Dm0 (kind gift of Paul A. Fisher, State University of New York, Stony Brook, NY) at 1:2000 and then washed and probed with secondary antibody as described above. DAPI (4′-6-diamidino-2-phenylindole; 1 μg/ml) was added during the second wash of the secondary antibody.
Imaging Analysis
Nondeconvolved images (see Figures 1 and 4) were obtained by mounting cells on 12-well slides (Carlson Scientific, Peotone, IL) and capturing data with a Zeiss Axioplan 2 microscope (Thornwood, NY)/Hamamatsu Digital CCD camera (Bridgewater, NJ). Z-series color images (see Figures 5 and 7) were obtained by mounting cells on cover slips in GelMount (Biomeda) and capturing data with a DeltaVision Restoration microscope system using an Olympus IX70 microscope (Melville, NY) fitted with an automated stage (Applied Precision) and a CCD digital camera. Optical sections (0.2–0.25 μm) were acquired using a 100× (Nikon, PlanApo, NA 0.45 lens) objective lens. The Softworks deconvolution software (Applied Precision, Issaquah, WA) was used to remove out-of-focus light for deconvolved images and to obtain the Pearson coefficient of correlation. For experiment colocalizing lamin and the FLAG-tagged exosome subunits, the nucleus of each cell evaluated was defined as a region of interest through the _z_-series and then compared for staining intensity of both the FLAG and the lamin stain. All images were arranged using Adobe Photoshop (San Jose, CA).
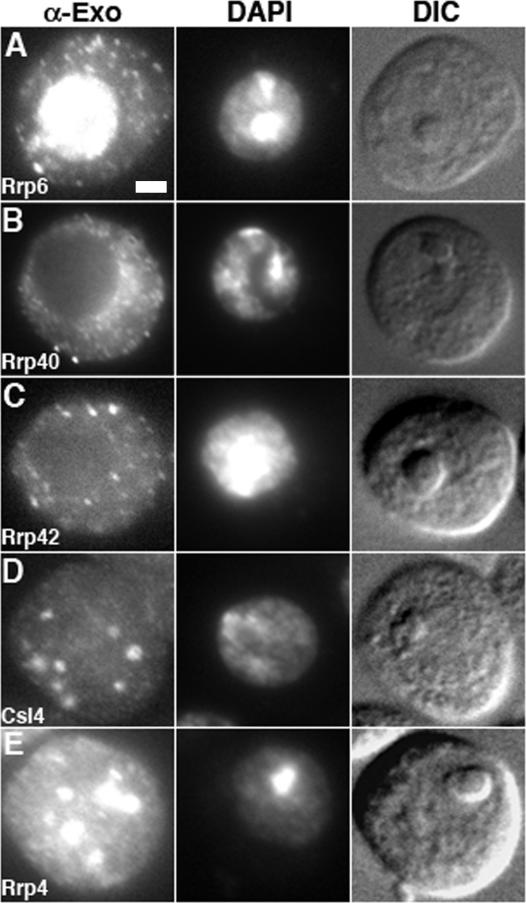
Figure 1.
Immunolocalization of endogenous exosome subunits. (A) dRrp6 is enriched in the nucleus yet also localizes to numerous small cytoplasmic foci. Bar, 2 μm. (B and C) dRrp40 and dRrp42 localize throughout the cytoplasm and in small foci. (D and E) dCsl4 and dRrp4 are enriched in discrete large cytoplasmic structures that, in some cases, are on or within the nucleus. α-Exo, anti-exosome subunit antibody. DAPI, 4′-6-diamidino-2-phenylindole. DIC, differential interference contrast.
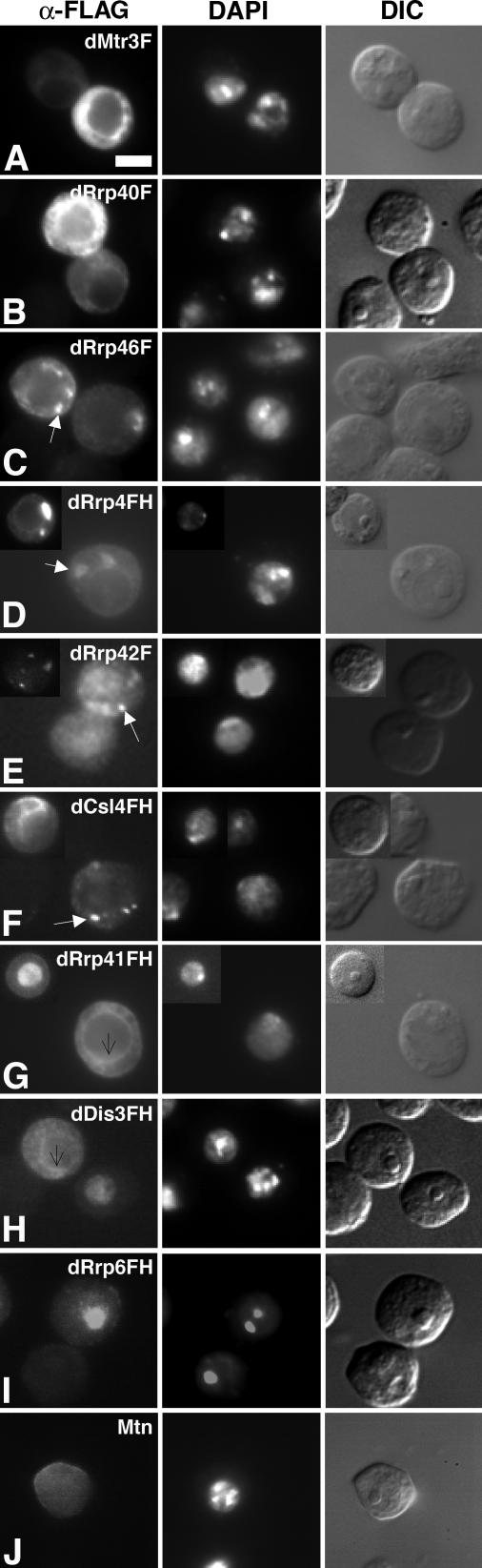
Figure 4.
Epitope-tagged exosome subunits are enriched in distinct subcellular locations. (A and B) dMtr3F and dRrp40F are localized predominantly to the cytoplasm and are differentially enriched around the nucleus and/or excluded from the nucleus. Bar, 5 μm. (C–F) Epitope-tagged dRrp46F, dRrp4FH, dRrp42F, and dCsl4FH localize to foci that appear cytoplasmic (white arrows). dCsl4FH is also found in cytoplasmic structures that may represent extensions of foci (inset; see Supplementary Movies 4–6). (G and H) Both dRrp41FH and dDis3FH localize exclusively to the nucleus or to the cytoplasm in individual cells. Note that these polypeptides localize to what appears to be the nuclear rim (thin black arrows). (I) dRrp6FH is enriched in the nucleus and nucleolus. (J) Background staining is observed in cells expressing the Mtn vector.
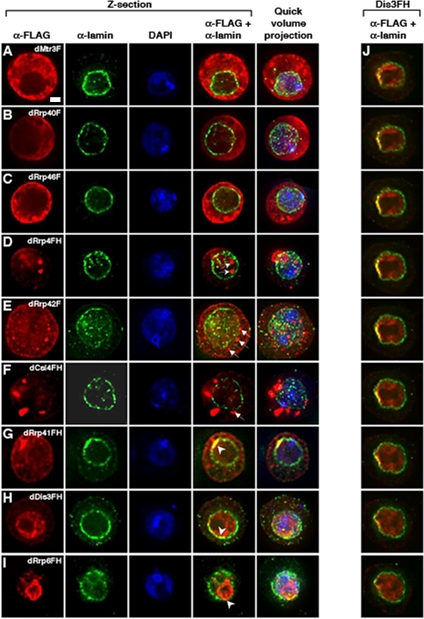
Figure 5.
Colocalization of exosome subunits with the nuclear lamina. Stable cell lines expressing individual epitope-tagged exosome subunits were fixed and costained with antibodies to the FLAG epitope (red) and to the nuclear lamina (green); colocalization of these two antibodies is observed (yellow); DNA is stained with DAPI (blue). Deconvolved images of a _Z_-series section are presented alongside a quick volume projection of the particular cell. (A–C) In the _Z_-section, dMtr3F appears to colocalize with the outer edges of the lamin stain, whereas dRrp40F and dRrp46F do not show significant overlap with the lamina. Bar, 2 μm. (D and E) Some tagged dRrp4 and dRrp42 foci localize directly on the nuclear lamina, whereas others are within the nucleoplasm or cytoplasm (white arrows). (F) dCsl4FH localizes to cytoplasmic foci that are proximal to, but not on the nuclear lamina (e.g., white arrow). (G and H) Both dRrp41FH and dDis3FH show strong and asymmetric colocalization with the lamina (white arrowhead). (I) Deconvolution of dRrp6FH nucleolar and nuclear staining shows localization to the nucleolar periphery with colocalization with the nuclear lamina (white arrowhead). (J) dDis3 nucleoperipheral localization is highly restricted along one face of the nuclear lamina. The initial panel represents the first _Z_-section in a series of nine sections from the top to the bottom of area manifesting dDis3FH-lamina colocalization. The 3D reconstruction of this cell can be viewed in Supplementary Movie 1.
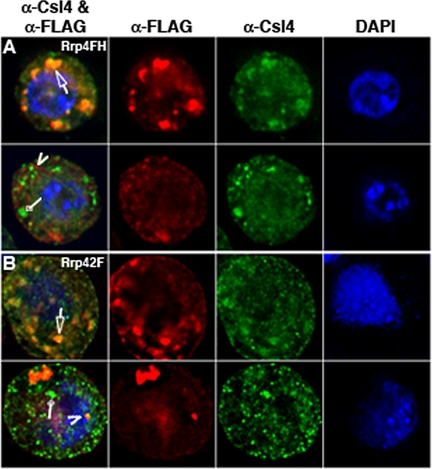
Figure 7.
Colocalization of dRrp4FH and dRrp42F with Endogenous dCsl4. (A) Endogenous dCsl4 shows significant overlap with dRrp4FH in foci in ∼20% of cells (top cell, arrow). Note how the dCsl4 staining appears within the dRrp4FH staining pattern. In most cells, there is little or no signal overlap between dCsl4 (diamond arrow) and dRrp4F (arrow-head). (B) Endogenous dCsl4 colocalizes with dRrp42F in cytoplasmic foci (top cell, arrow) in ∼25% of cells. The majority of cells show little or no colocalization (bottom cell). dCsl4 staining alone is marked with diamond arrow and dRrp42F staining alone with an arrowhead. Images are deconvolved and represent one section through a _Z_-series.
RESULTS
Endogenous Drosophila Exosome Subunits Localize Differentially
Given the numerous and varied roles of exosome subunits in RNA metabolism, we suspected that they might exhibit distinct localization patterns in vivo. To directly test this idea, we performed indirect immunofluorescence on D. melanogaster S2 tissue culture cells using antibodies to endogenous exosome subunits. Endogenous dRrp6 was found predominantly in the nucleus and nucleolus, coincident with the DNA stain (Figure 1A) as seen in yeast (Allmang et al., 1999; Burkard and Butler, 2000), and also in the cytoplasm, as has been observed in mammalian cells (Lejeune et al., 2003). In contrast, endogenous dRrp40 and dRrp42 exhibited a diffuse cytoplasmic localization in small cytoplasmic foci and were apparently excluded from the nucleus (Figures 1, B and C). Appropriate preimmune sera showed nonspecific background staining (Supplementary Figure 1). Affinity-purified antibodies to dCsl4 and dRrp4 detected these subunits not only diffuse throughout the cytoplasm but also concentrated in foci that appeared within the cytoplasm and on or around the nucleus (Figure 1, D and E). These data indicate that individual endogenous Drosophila exosome subunits show at least three discrete subcellular localization patterns. A more comprehensive investigation into exosome subunit distribution is therefore of interest.
Expression of Epitope-tagged Exosome Subunits in S2 Cells
Given these distinct immunolocalization patterns of exosome subunits in S2 cells, we next wanted to inquire whether other exosome subunits have similar or different distribution profiles. As a first step toward addressing this issue, we developed a set of stable cell lines expressing individual epitope-tagged exosome subunits (Figure 2A).
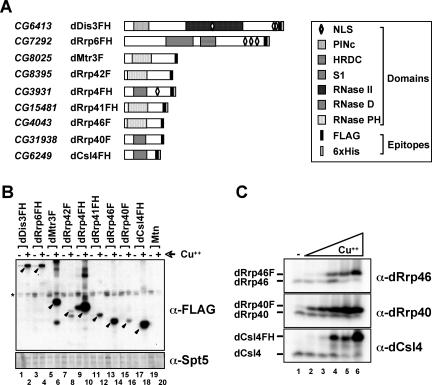
Figure 2.
Establishment and characterization of Drosophila S2 stable cell lines expressing individual exosome subunits. (A) Schematic representation of exosome subunits expressed stably in S2 embryonic tissue culture cells. The Drosophila exosome subunits have domain structures that resemble their respective homologues in yeast and humans (Chen et al., 2001; Aloy et al., 2002). For example, dRrp4, dRrp40, and dCsl4 all have an RNA-binding S1 domain (Bycroft et al., 1997). Both dDis3 and dRrp6 contain conserved domains (PINc and HRDC, respectively) that are predicted to be involved in nucleic acid binding (Clissold and Ponting, 2000; Phillips and Butler, 2003). Only dDis3, dRrp6, and dRrp4 have predicted nuclear localization signals. Exosome subunits are tagged with FLAG alone (F) or both FLAG and 6xHis (FH) epitopes. CG numbers represent Drosophila gene numbers. (B) Analysis of stable S2 cell lines containing copper-inducible epitope-tagged exosome subunits. Full-length subunits (closed arrowheads) were detected by Western blotting using an antibody directed against the FLAG epitope. The bottom panel is a Western blot of endogenous dSpt5 as a loading control; the band of faster mobility is a dSpt5 degradation product. (C) Optimizing expression conditions of epitope-tagged exosome subunits. Stably transfected S2 cells were either grown in the presence or absence of copper (Cu2+). Cu2+ is in a fourfold serial dilution range from 5.4 μM to 1.4 mM.
Each exosome subunit was epitope tagged with either FLAG (F) or FLAG and 6xHis (FH) and cloned downstream of the metallothionein (Mtn) promoter to allow for copper-inducible expression. After establishment of stable D. melanogaster S2 cell lines, we used Western blot analysis to show that all exosome subunits were up-regulated after copper addition to media (Figure 2B). In several of these cell lines, the tagged exosome subunits were significantly overexpressed relative to the cognate endogenous polypeptides. Tagged exosome subunit overexpression led to the reduction of the cognate endogenous exosome polypeptide (Figures 2C and 3A; as observed in Estevez et al., 2003). Thus, the copper concentration in media was reduced to allow for approximately equal amounts of tagged protein relative to the endogenous protein, as judged by Western blotting (Figure 2C). All subsequent experiments were performed under induction conditions that have been optimized for each epitope-tagged exosome subunit.
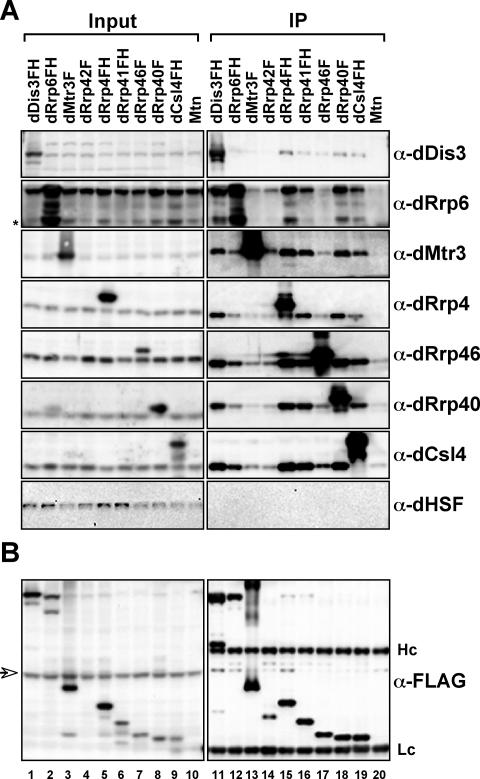
Figure 3.
Purification and analysis of exosome complexes. (A) Epitope-tagged exosome subunits incorporate into and coprecipitate endogenous exosome complexes. Whole cell extracts prepared from stable cell lines expressing tagged exosome subunits or vector control (Mtn) were incubated with anti-FLAG affinity resin. The resin was washed and complexes eluted with FLAG peptide. The amount of immunoprecipitated material (IP) and a known amount of reserved material (input) were run on SDS-PAGE, transferred to nitrocellulose, and then probed with antibodies to endogenous exosome subunits or with a control antibody (HSF). Input, 2.5%; IP, 10%. Asterisk, dRrp6 degradation product. (B) Recovery of epitopetagged exosome subunits. Samples were isolated and analyzed as above but were probed with an antibody to the FLAG epitope in order to determine efficiency of tagged protein recovery. Hc and Lc, IgG heavy and light chains, respectively. Arrow, anti-FLAG antibody cross-reactive band.
Epitope-tagged Exosome Subunits Incorporate into Complexes
To assess the functionality of the tagged exosome subunits, we tested whether they incorporated into complexes with other subunits in vivo by using immunoaffinity chromatography. Our previous study (Andrulis et al., 2002) indicated that the Drosophila exosome complex is salt-stable. Thus, S2 whole cell extracts were prepared and immunoprecipitations were performed in the presence of 0.45 M NaCl to both extract nucleoplasmic complexes and to assess the stability of the interactions among the Drosophila exosome subunits.
Tagged exosome subunits were purified from whole cell extracts by binding to an anti-FLAG affinity resin. After extensive washing of the resin, complexes were eluted with FLAG peptide. Eluates were analyzed by SDS-PAGE and compared with a loading control (input). Three generally different exosome complex recovery efficiencies are observed. First, dDis3FH, dRrp4FH, dRrp41FH, dRrp40F, and dCsl4FH (Figure 3A, lanes 11, 15, 16, 18, and 19) were most effective at recovering endogenous exosome subunits, with similar amounts of subunits recovered as judged by Western blotting. Second, dRrp6FH and dRrp46F (Figure 3A, lanes 12 and 17) recovered less of each subunit examined. Surprisingly, epitope-tagged dRrp46 recovers endogenous dDis3 better than dRrp6, which is deficient in dDis3 recovery. This would suggest that dDis3 is not stably associated with all exosome subunits, as has been shown for yeast Dis3 (Allmang et al., 1999). Finally, the recovery of endogenous exosome subunits with dMtr3F and dRrp42F was consistently poor (Figure 3A, lanes 13 and 14). Because dRrp42F is expressed rather poorly, this provides an explanation for the low recovery of associated exosome components. Nonetheless, all exosome subunits were coprecipitated specifically, as judged by an inability to recover an unrelated protein, HSF. Moreover, complexes were coprecipitated at levels above background binding to the affinity resin (Figure 3A, lane 20). Importantly, the tagged subunits themselves were recovered with similar efficiencies (Figure 3B), suggesting that these different exosome complex recovery efficiencies are not attributable to an inability to precipitate the tagged polypeptide from whole cell extracts. Thus, tagged exosome subunits incorporate into complexes composed of the endogenous exosome subunits found in whole cell extracts.
To determine the efficiency of endogenous exosome subunit extraction from these cytoplasmic foci and the nucleus, we examined the distribution of endogenous exosome subunits in fractionated cell extracts. Standard cell lysis and fractionation, in buffer containing physiological salt (0.15 M NaCl) and nonionic detergent, elicits a supernatant fraction (S), which includes soluble and cytoplasmic proteins, and a pellet fraction (P), which includes insoluble and nuclear proteins. Under these conditions, endogenous dRrp6 is enriched in the insoluble fraction, and dRrp4, dRrp41, and dCsl4 are predominantly soluble (Supplementary Figure 2A). This solubility profile extends across a range of salt conditions (Supplementary Figure 2B). These data demonstrate that the whole cell extracts prepared for immunoprecipitations in Figure 3 contain the majority of endogenous nuclear and cytoplasmic exosome subunits.
Tagged Exosome Subunits Localize to and Are Enriched in Distinct Subcellular Compartments
Because tagged exosome subunits incorporate into nuclear- and cytoplasmically derived complexes (Figure 3), we surmised that they would localize properly in vivo. To test this idea, we performed indirect immunofluorescence using an antibody to the FLAG epitope to detect each subunit. There was extremely good correlation between the localization of tagged and endogenous exosome polypeptides (cf. Figures 1 and 4). Moreover, consistent with our immunolocalization analysis of endogenous exosome subunits, several distinct tagged subunit localization patterns were observed.
dMtr3F and dRrp40F were observed throughout the cytoplasm and around the nucleus (Figure 4, A and B). An additional set of exosome subunits, dRrp46F, dRrp4FH, dRrp42F, and dCsl4FH were likewise found in the cytoplasm yet specifically enriched in one, two, or more large foci or structures (0.5–3 μm) and/or multiple small foci (0.1–0.4 μm; Figures 4, C–F). These foci vary in size and number depending upon the growth state of the cells and the level of exosome subunit overexpression (Table 1; A. C. Graham and E. D. Andrulis, unpublished observations). Both dDis3FH and dRrp41FH showed restricted localization patterns to either the cytoplasm or the nucleus in individual cells, with dDis3FH predominantly nuclear and dRrp41FH predominantly cytoplasmic (Figure 4, G and H; Table 1). dDis3FH and dRrp41FH also appeared to be enriched around the nucleus. By comparison, dRrp6FH was enriched in the nucleolus (compare immunofluorescence with differential interference contrast panels) and in the nucleus but not in the cytoplasm (Figure 4I). Importantly, indirect immunofluorescence with the anti-FLAG antibody in cells expressing the Mtn vector had background signal that was observed only upon increasing the exposure time (Figure 4J). These images thus reflect the localization of FLAG-tagged exosome subunits and are not due to nonspecific signal from the anti-FLAG antibody.
Table 1.
Localization pattern of epitope-tagged Drosophila exosome subunits
| % Cytoplasm/focia | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Exosome subunit | % Nucleolus | % Nucleus | [a] | [b] | [c] | [d] | [e] | [f] | No. of cells scoredb |
| dRrp6FH | 93 | 7 | N/A | N/A | N/A | N/A | N/A | N/A | 102 |
| dDis3FH | N/A | 83 | 10 | N/A | 7 | N/A | N/A | N/A | 100 |
| dRrp41FH | N/A | 13 | 76 | N/A | 10 | 1 | N/A | N/A | 104 |
| dMtr3F | N/A | N/A | 32 | 37 | 30 | 2 | N/A | N/A | 105 |
| dRrp40F | N/A | N/A | 9 | 17 | 60 | 14 | N/A | N/A | 89 |
| dRrp46F | N/A | N/A | 20 | 16 | 9 | 16 | 22 | 17 | 96 |
| dRrp42F | N/A | N/A | 4 | N/A | 3 | 14 | 44 | 35 | 103 |
| dCsl4FH | N/A | N/A | 8 | N/A | 9 | 22 | 19 | 42 | 103 |
| dRrp4FH | N/A | N/A | N/A | N/A | 7 | 16 | 21 | 56 | 102 |
Because these stable cell lines are not clonal and individual subunits showed subtle changes in distribution from cell to cell, we examined ∼100 cells for the localization of each tagged exosome subunit (Table 1). The most obvious difference uncovered was variation in the number and size of cytoplasmic foci for dRrp42F, dRrp4FH, dRrp46F, and dCsl4FH. In addition, dMtr3F, dRrp40F, and dRrp46F were infrequently found in cytoplasmic regions proximal to the plasma cell membrane. This immunofluorescence analysis shows that individual Drosophila exosome subunits reside in different subcellular locations, with some variation in these localization patterns from cell to cell.
Exosome Subunits Localize to the Nuclear Lamina
Several endogenous and tagged exosome subunits appeared to localize on or around the nuclear rim. Because this localization could correspond to the outer nuclear membrane, inner nuclear membrane, or nuclear lamina, we needed to examine the distribution of exosome subunits relative to an established marker protein. We chose lamin Dm0, a Drosophila B-type lamin that defines the most peripheral part of the nucleoplasm. To obtain higher-resolution images, we used deconvolution microscopy.
Double immunofluorescence detected both lamin Dm0 and FLAG epitopes in the stable S2 cell lines. A portion of dMtr3F localized along parts of the “outside” of the nuclear lamina (Figure 5A). Most strikingly, dDis3FH and dRrp41FH showed appreciable colocalization with lamin that was often restricted to one side of the nucleus, regardless of whether these exosome subunits were nuclear or cytoplasmically localized (Figures 5, G and H; Supplementary Videos 1–3). This is particularly clear in a series of _Z_-sections taken from a cell expressing dDis3FH (Figure 5J). Although dRrp4FH, dRrp46F, dRrp42F, and dCsl4FH foci often appear proximal to the nuclear rim, modest or negligible costaining with the lamina is observed (Figures 5, C–F; Supplementary Videos 4–6). Unexpectedly, we also observe nuclear foci in cells expressing either dRrp4FH and dRrp42F (Figure 5, D and E; Supplementary Video 5). Lastly, analysis of dRrp6FH localization shows distribution along the outer edges of what is predicted to be the nucleolus and partial overlap with the lamina (Figure 5I).
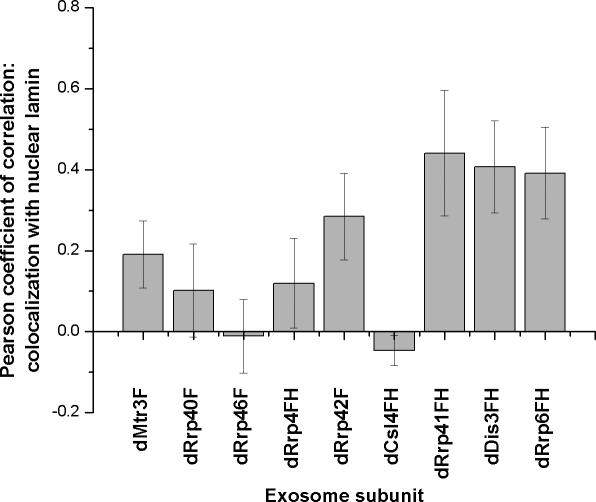
To determine whether the exosome-lamin colocalization was statistically significant, we calculated the Pearson coefficient of correlation in ∼10 cell nuclei for each tagged exosome subunit (Figure 6). dRrp6FH, dDis3FH, and dRrp41FH colocalization with lamina scored Pearson coefficients of ∼0.4 (closest to a score of 1 indicating direct correlation). dMtr3F, dRrp42F, dRrp40F and dRrp4FH had intermediate Pearson coefficients of ∼0.2, whereas dRrp46F and dCsl4FH scored close to or below zero. These data suggest that several exosome subunits having overlapping localization with lamin in fixed S2 cells.
Figure 6.
Quantitative analysis of colocalization of exosome subunits and lamin. A Pearson coefficient of +1.0 is complete coincidence of two signals in the area examined, whereas –1.0 is no overlap of two signals. Several, but not all exosome subunits are enriched at the nuclear periphery.
Exosome Subunits in Distinct Cytoplasmic Compartments
Several exosome subunits localize to cytoplasmic foci (Figures 1, 4, and 5) that vary in terms of size and number (Table 1). We next inquired whether these foci are similar or represent different cytoplasmic compartments. This was accomplished by first localizing endogenous dCsl4 with dRrp4FH. Although we observed cells in which there was a significant degree of dCsl4 and dRrp4FH signal overlap (Figure 7A, top panels), these proteins are in separate cytoplasmic compartments in ∼80% of the cells examined (Figure 7A, bottom panels). A similar finding was made with dRrp42F and dCsl4. In this case, however, ∼75% of the cells exhibited little or no overlap between dRrp42F and dCsl4 (Figure 7B, bottom panel), whereas the remainder of the cells show a high degree of subunit coincidence (Figure 7B, top panel).
Based on these observations, we tested whether Csl4 localization to these foci required Rrp4. Treating S2 cells with dsRNA to dRrp4 reduced endogenous dRrp4 levels to ∼10% without affecting dCsl4 protein levels (Supplementary Figure 3). Nonetheless, there was no change in dCsl4 subcellular distribution under these conditions (unpublished data). We conclude that dRrp4 is not required for the targeting of dCsl4 to, or for the stability of dCsl4 within, cytoplasmic foci.
DISCUSSION
Exosome Subunit Subcomplexes Distinct from the Core Exosome Complex
If all exosome subunits were found exclusively and stoichiometrically in the core exosome complex, one might expect that purification of each exosome subunit from cell extracts should yield equivalent amounts of other exosome complex subunits. However, this is not the case observed here, as several individual exosome subunits (e.g., dRrp42, dMtr3, dRrp6, dRrp46) fail to efficiently recover the exosome complex and another subunit (dRrp6) is deficient at recovering one subunit. The latter class suggests that an individual subunit may not associate with (or may be dissociated from) the complex being purified. The former suggests that certain exosome subunits, although stoichiometric with the exosome complex, comprise additional complexes. Indeed, a high-throughput proteomic analysis of yeast multiprotein particles has demonstrated that individual exosome subunits associate with protein complexes that are distinct from the exosome complex (Gavin et al., 2002). In addition to this proteomic study, another group (Krogan et al., 2004) showed that purified exosome complexes are often missing one or more subunits and contain additional factors that are unique to the precipitated subunit. In this regard, our previous work indicates that the exosome exists in a nuclear complex containing the transcription elongation factor dSpt6 yet lacking the exosome subunit dRrp45 (Andrulis et al., 2002). Finally, consistent with the idea that exosome subunits assemble into distinct complexes, the initial exosome complex comprised 5 subunits in apparent stoichiometry (Mitchell et al., 1997), but the subsequent core complex contained 10 stoichiometric subunits (Allmang et al., 1999).
Recent in vitro studies provide a framework on which to examine our findings. For example, that dRrp41, dRrp42, and dMtr3 are in similar subcellular compartments is consistent with the ability of these subunits to associate in complexes. Indeed, homologues of Rrp42 complex with Rrp41 (Buttner et al., 2005; Lorentzen and Conti, 2005; Lorentzen et al., 2005) and with Mtr3 (Lorentzen et al., 2005). In addition, that dRrp4 and dCsl4 do not always colocalize is consistent with the model that these proteins can assemble either into distinct complexes or into the same complex. Indeed, this is the case with homologues of these subunits in vitro (Buttner et al., 2005). These data indicate an unanticipated diversity of exosome complexes.
Exosome Subunits and Cytoplasmic Foci
That several, but not all, exosome subunits localize to different cytoplasmic foci provides support for cytoplasmic exosome subcomplexes. Two different cytoplasmic structures have been implicated in RNA metabolism: stress granules and processing bodies (P-bodies; also called GW bodies), distinct compartments for mRNA sequestration, storage, or decay (Sheth and Parker, 2003; Cougot et al., 2004; Kedersha et al., 2005). Although the yeast exosome is not known to localize to P-bodies, the entire set of subunits have not been exhaustively examined (Brengues et al., 2005). This is particularly important to explore in light of our findings that only a few of the exosome subunits are enriched in cytoplasmic foci. Interestingly, dRrp4 and dCsl4, both of which localize to cytoplasmic foci in this study, have been shown to be required for RNAi-mediated mRNA decay (Orban and Izaurralde, 2005). Notably, the RNA-induced silencing complex machinery, required for degrading RNAi-targeted mRNAs, localizes to P-bodies in mammalian cells (Liu et al., 2005). Thus, we favor the idea that some of these exosome subunit cytoplasmic foci correspond to P-bodies. However, this explanation is insufficient to explain the observation that dRrp4FH and dRrp42F are largely in cytoplasmic compartments distinct those containing dCsl4. Thus, the composition and functions of these cytoplasmic structures awaits identification.
Are Exosome Subunits Positioned on the Nuclear Lamina for mRNA Surveillance?
The localization of several exosome subunits to the nuclear lamina is likewise interesting. It is possible that nucleoperipheral domain represents a storage or nucleocytoplasmic transit site for exosome subunits. An alternative possibility is that exosome subunits at the lamina are critical for surveillance during mRNA export (Kadowaki et al., 1995; Hilleren et al., 2001; Jensen et al., 2001; Zenklusen et al., 2002; Thomsen et al., 2003; Galy et al., 2004; Hieronymus et al., 2004). Interestingly, two of the lamina-localizing exosome subunits were identified as yeast mutants (mtr3 and rrp6) defective for mRNA export (Kadowaki et al., 1995; Hieronymus et al., 2004). The asymmetric laminar localization of dRrp41 and dDis3 is reminiscent of the yeast myosinlike proteins (Mlp), factors critical for the nuclear retention of aberrant mRNAs (Galy et al., 2004; Vinciguerra et al., 2005). Because the yeast exosome proteins Rrp41/Ski6 and Dis3/Rrp44 are required for turnover of these aberrant mRNAs in the nucleus (Bousquet-Antonelli et al., 2000), it is appealing to speculate that these asymmetric, lamina-directed exosome subunits may function in a similar mRNA export and surveillance pathway in Drosophila cells.
Differential Localization of Exosome Subunits
Both individual endogenous and epitope-tagged exosome subunits localize differentially in Drosophila S2 cells. Although several exosome subunits in yeast and mammalian cells fractionate and localize similarly to their Drosophila homologues (Kadowaki et al., 1995; Allmang et al., 1999; Brouwer et al., 2001; Raijmakers et al., 2002b), others do not (Brouwer et al., 2001; Raijmakers et al., 2002b). Nevertheless, we find it unlikely that the epitope-tagged Drosophila exosome subunits mislocalize, as they localize similarly to the cognate endogenous protein and incorporate into exosome complexes.
We find exosome subunits specifically enriched at the nucleolus, nucleus, nuclear foci, nuclear rim, cytoplasm, cytoplasmic foci, and plasma cell membrane. Why so many distinct locales? There are several possibilities. First, perhaps exosome subunits are dynamic, and our localization analysis in fixed cells provides a limited perspective. In one particular example, the nuclear- or cytoplasmic-restricted localization patterns of dDis3FH and dRrp41FH suggest that these two proteins may shuttle between the nucleus and cytoplasm. However, treating cells with the drug leptomycin B does not change dDis3FH and dRrp41FH distribution (unpublished data), indicating that these proteins do not use a Crm1-dependent export pathway. Interestingly, Schizosaccharomyces pombe and human Dis3 interact directly with Ran (Noguchi et al., 1996; Shiomi et al., 1998), a protein essential for nucleocytoplasmic transport of protein and ribonucleoprotein cargoes (Macara, 2001). Second, exosome subunits may localize to different subcellular domains depending on changes in the cell cycle or growth conditions. Third, individual exosome subunits, singly or in combination with other subunits, may occupy specific subcellular domains crucial for specialized roles in mRNA processing, turnover, transport, or surveillance or in the metabolism of other RNA species. Finally, exosome subunits may reside in complexes distinct from the exosome complex, where they perform functions that are unrelated to RNA regulation. In this regard, Dis3 has a mitotic role in fission yeast (Kinoshita et al., 1991) and Rrp6 is implicated in DNA damage surveillance and recombination in budding yeast (Hieronymus et al., 2004; Luna et al., 2005).
To date, most studies on exosome function have utilized either mutants or RNAi-depletions of one or two subunits to suggest a role for the core or nuclear exosome complex in a particular pathway. Our results provide compelling evidence for the existence of several distinct, subcellularly compartmentalized exosome subcomplexes. In so doing, this study promotes the idea that exosome subunits, singly or in exosome subunit subcomplexes, function in several of the RNA metabolic pathways that have been ascribed to the exosome complex.
Supplementary Material
[Supplemental Material]
Acknowledgments
We thank Drs. Piet de Boer and David McDonald for microscope use; Drs. Piet de Boer, Jonathan Karn, Jo Ann Wise, Patrick Viollier, Elisa Izaurralde, and Alan Tartakoff for comments on the manuscript; and Dr. Paul Fisher for antibodies. E.D.A. is especially grateful to Dr. John Lis for guidance and support during the initial phases of this project. E.D.A. is a Mount Sinai Health Care Foundation Scholar.
References
- Allmang, C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., and Mitchell, P. (1999). The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev. 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aloy, P., Ciccarelli, F. D., Leutwein, C., Gavin, A. C., Superti-Furga, G., Bork, P., Bottcher, B., and Russell, R. B. (2002). A complex prediction: three-dimensional model of the yeast exosome. EMBO Rep. 3, 628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis, E. D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J. T. (2002). The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420, 837–841. [DOI] [PubMed] [Google Scholar]
- Bousquet-Antonelli, C., Presutti, C., and Tollervey, D. (2000). Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 102, 765–775. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., and Parker, R. (2005). Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 310, 486–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer, R., Allmang, C., Raijmakers, R., van Aarssen, Y., Egberts, W. V., Petfalski, E., van Venrooij, W. J., Tollervey, D., and Pruijn, G. J. (2001). Three novel components of the human exosome. J. Biol. Chem. 276, 6177–6184. [DOI] [PubMed] [Google Scholar]
- Burkard, K. T., and Butler, J. S. (2000). A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20, 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, J. S. (2002). The yin and yang of the exosome. Trends Cell Biol. 12, 90–96. [DOI] [PubMed] [Google Scholar]
- Buttner, K., Wenig, K., and Hopfner, K. P. (2005). Structural framework for the mechanism of archaeal exosomes in RNA processing. Mol. Cell 20, 461–471. [DOI] [PubMed] [Google Scholar]
- Bycroft, M., Hubbard, T. J., Proctor, M., Freund, S. M., and Murzin, A. G. (1997). The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell 88, 235–242. [DOI] [PubMed] [Google Scholar]
- Chekanova, J. A., Dutko, J. A., Mian, I. S., and Belostotsky, D. A. (2002). Arabidopsis thaliana exosome subunit AtRrp4p is a hydrolytic 3′→5′ exonuclease containing S1 and KH RNA-binding domains. Nucleic Acids Res. 30, 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova, J. A., Shaw, R. J., Wills, M. A., and Belostotsky, D. A. (2000). Poly(A) tail-dependent exonuclease AtRrp41p from Arabidopsis thaliana rescues 5.8 S rRNA processing and mRNA decay defects of the yeast ski6 mutant and is found in an exosome-sized complex in plant and yeast cells. J. Biol. Chem. 275, 33158–33166. [DOI] [PubMed] [Google Scholar]
- Chen, C. Y., Gherzi, R., Ong, S. E., Chan, E. L., Raijmakers, R., Pruijn, G. J., Stoecklin, G., Moroni, C., Mann, M., and Karin, M. (2001). AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell 107, 451–464. [DOI] [PubMed] [Google Scholar]
- Clissold, P. M., and Ponting, C. P. (2000). PIN domains in nonsense-mediated mRNA decay and RNAi. Curr. Biol. 10, R888–R890. [DOI] [PubMed] [Google Scholar]
- Cougat, N., Babajko, S., and Seraphin, B. (2004). Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165, 31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez, A. M., Kempf, T., and Clayton, C. (2001). The exosome of Trypanosoma brucei. EMBO J. 20, 3831–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez, A. M., Lehner, B., Sanderson, C. M., Ruppert, T., and Clayton, C. (2003). The roles of intersubunit interactions in exosome stability. J. Biol. Chem. 278, 34943–34951. [DOI] [PubMed] [Google Scholar]
- Evguenieva-Hackenburg, E., Walter, P., Hochleitner, E., Lottspeich, F., and Klug, G. (2003). An exosome-like complex in Sulfolobus solfataricus. EMBO Rep. 4, 889–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler, D., Kocher, T., Rode, M., Gentzel, M., Izaurralde, E., and Wilm, M. (2003). An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat. Biotechnol. 21, 89–92. [DOI] [PubMed] [Google Scholar]
- Galy, V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. (2004). Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116, 63–73. [DOI] [PubMed] [Google Scholar]
- Gatfield, D., and Izaurralde, E. (2004). Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature 429, 575–578. [DOI] [PubMed] [Google Scholar]
- Gavin, A. C. et al. (2002). Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Hieronymus, H., Yu, M. C., and Silver, P. A. (2004). Genome-wide mRNA surveillance is coupled to mRNA export. Genes Dev. 18, 2652–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren, P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T. H. (2001). Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413, 538–542. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, T. H., Boulay, J., Rosbash, M., and Libri, D. (2001). The DECD box putative ATPase Sub2p is an early mRNA export factor. Curr. Biol. 11, 1711–1715. [DOI] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A. M., Hinnebusch, A. G., and Anderson, J. (2004). Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev. 18, 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki, T., Schneiter, R., Hitomi, M., and Tartakoff, A. M. (1995). Mutations in nucleolar proteins lead to nucleolar accumulation of polyA+ RNA in Saccharomyces cerevisiae. Mol. Biol. Cell 6, 1103–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha et al. (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita, N., Goebl, M., and Yanagida, M. (1991). The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol. 11, 5839–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J. et al. (2004). High-definition macromolecular composition of yeast RNA-processing complexes. Mol. Cell 13, 225–239. [DOI] [PubMed] [Google Scholar]
- Lejeune, F., Li, X., and Maquat, L. E. (2003). Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol. Cell 12, 675–687. [DOI] [PubMed] [Google Scholar]
- Libri, D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T. H. (2002). Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22, 8254–8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M. A., Hannon, G. J., and Parker, R. (2005). MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7, 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzen, E., and Conti, E. (2005). Structural basis of 3′ end RNA recognition and exoribonucleolytic cleavage by an exosome RNase PH core. Mol. Cell 20, 473–481. [DOI] [PubMed] [Google Scholar]
- Lorentzen, E., Walter, P., Fribourg, S., Evguenieva-Hackenberg, E., Klug, G., and Conti, E. (2005). The archaeal exosome core is a hexameric ring structure with three catalytic subunits. Nat. Struct. Mol. Biol. 12, 575–581. [DOI] [PubMed] [Google Scholar]
- Luna, R., Jimeno, S., Marin, M., Huertas, P., Garcia-Rubio, M., and Aguilera, A. (2005). Interdependence between transcription and mRNP processing and export, and its impact on genetic stability. Mol. Cell 18, 711–722. [DOI] [PubMed] [Google Scholar]
- Macara, I. G. (2001). Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65, 570–594, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., Shevchenko, A., Mann, M., and Tollervey, D. (1997). The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell 91, 457–466. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Petfalski, E., and Tollervey, D. (1996). The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev. 10, 502–513. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., and Tollervey, D. (2000). Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 7, 843–846. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., and Tollervey, D. (2003). An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′→5′ degradation. Mol. Cell 11, 1405–1413. [DOI] [PubMed] [Google Scholar]
- Noguchi, E. et al. (1996). Dis3, implicated in mitotic control, binds directly to Ran and enhances the GEF activity of RCC1. EMBO J. 15, 5595–5605. [PMC free article] [PubMed] [Google Scholar]
- Oliveira, C. C., Gonzales, F. A., and Zanchin, N. I. (2002). Temperature-sensitive mutants of the exosome subunit Rrp43p show a deficiency in mRNA degradation and no longer interact with the exosome. Nucleic Acids Res. 30, 4186–4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orban, T. I., and Izaurralde, E. (2005). Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA 11, 459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, S., and Butler, J. S. (2003). Contribution of domain structure to the RNA 3′ end processing and degradation functions of the nuclear exosome subunit Rrp6p. RNA 9, 1098–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raijmakers, R., Egberts, W. V., van Venrooij, W. J., and Pruijn, G. J. (2002a). Protein-protein interactions between human exosome components support the assembly of RNase PH-type subunits into a six-membered PNPase-like ring. J. Mol. Biol. 323, 653–663. [DOI] [PubMed] [Google Scholar]
- Raijmakers, R., Noordman, Y. E., van Venrooij, W. J., and Pruijn, G. J. (2002b). Protein-protein interactions of hCsl4p with other human exosome subunits. J. Mol. Biol. 315, 809–818. [DOI] [PubMed] [Google Scholar]
- Raijmakers, R., Schilders, G., and Pruijn, G. J. (2004). The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 83, 175–183. [DOI] [PubMed] [Google Scholar]
- Sheth, U., and Parker, R. (2003). Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi, T., Fukushima, K., Suzuki, N., Nakashima, N., Noguchi, E., and Nishimoto, T. (1998). Human dis3p, which binds to either GTP- or GDP-Ran, complements Saccharomyces cerevisiae dis3. J. Biochem. (Tokyo) 123, 883–890. [DOI] [PubMed] [Google Scholar]
- Thomsen, R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T. H. (2003). Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9, 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchet, C., Bousquet-Antonelli, C., Milligan, L., Thompson, E., Kufel, J., and Tollervey, D. (2002). Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9, 1285–1296. [DOI] [PubMed] [Google Scholar]
- Uetz, P. et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., Frischmeyer, P. A., Dietz, H. C., and Parker, R. (2002). Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295, 2262–2264. [DOI] [PubMed] [Google Scholar]
- Vinciguerra, P., Iglesias, N., Camblong, J., Zenklusen, D., and Stutz, F. (2005). Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export. EMBO J. 24, 813–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen, D., Vinciguerra, P., Wyss, J. C., and Stutz, F. (2002). Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell. Biol. 22, 8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material]