Comparative Mapping in the Pinaceae (original) (raw)
Abstract
A comparative genetic map was constructed between two important genera of the family Pinaceae. Ten homologous linkage groups in loblolly pine (Pinus taeda L.) and Douglas fir (Pseudotsuga menziesii [Mirb.] Franco) were identified using orthologous expressed sequence tag polymorphism (ESTP) and restriction fragment length polymorphism (RFLP) markers. The comparative mapping revealed extensive synteny and colinearity between genomes of the Pinaceae, consistent with the hypothesis of conservative chromosomal evolution in this important plant family. This study reports the first comparative map in forest trees at the family taxonomic level and establishes a framework for comparative genomics in Pinaceae.
THE essence of comparative genome analysis is the extrapolation of information from one organism to another. Comparative mapping and comparative sequence analysis are the key components of comparative genomics. Comparative mapping establishes the syntenic relationships between genomes of different species, assisting in genetic map consolidation, verification of quantitative trait loci (QTL), identification of candidate genes underlying QTL, and a better understanding of genome evolution (Sankoff and Nadeau 2000; Kliebenstein et al. 2001; Murphy et al. 2001; Zhang et al. 2001; Schmidt 2002).
Comparative genome analysis is often performed between model and nonmodel species (for reviews, see Paterson et al. 2000; Hall et al. 2002; Schmidt 2002). For example, thale cress (Arabidopsis thaliana) and rice (Oryza sativa) are model species for dicots and monocots, respectively. Putative syntenic regions have been identified for dicots in comparisons between soybean (Glycine max), barrel medic (Medicago truncatula), cabbage (Brassica oleracea), potato (Solanum tuberosum), and A. thaliana (Grant et al. 2000; Babula et al. 2003; Gebhardt et al. 2003; Lukens et al. 2003; Zhu et al. 2003). Similar comparisons have been done for monocots between sorghum (Sorghum bicolor), barley (Hordeum vulgare), wheat (Triticum aestivum), maize (Zea mays), and O. sativa (Klein et al. 2003; Ware and Stein 2003). Comparative maps have been constructed among several species within a few important families of plants, notably Brassicaceae (Paterson et al. 2000; Barnes 2002; Hall et al. 2002), Solanaceae (Doganlar et al. 2002), Fabaceae (Boutin et al. 1995; Yan et al. 2003), and Poaceae (Feuillet and Keller 2002; Laurie and Devos 2002; Ware et al. 2002; Ware and Stein 2003). A comparative map framework among these taxa facilitates the transfer of information across species and enables a taxonomic family to be viewed as a single genetic system (Freeling 2001).
Pinaceae is the most important among eight families of the order Coniferales (conifers). This family comprises 11 genera and 232 species distributed throughout the world (Frankis 1989), primarily in the temperate region of the northern hemisphere. Members of the Pinaceae have large economic importance as a source of timber, pulp, and resins. They also play a very significant ecological role by producing large biomass and creating habitat for many other organisms. Forest trees of the Pinaceae are essential for carbon sequestration that may affect global climate.
Pinaceae genomes are very large compared to nearly all other plant species and are unlikely to be completely sequenced in the near future. Pinaceae DNA contents vary from 5.8 to 32.2 pg per haploid genome (1C) with 22 pg on average for 83 species studied (Murray 1998; Leitch et al. 2001; Bennett and Leitch 2003). Pinaceae genomes are 6-fold larger than the human genome (3.5 pg; Morton 1991) and 100-fold larger than that of A. thaliana (0.18 pg; Bennett and Smith 1991). In the absence of a genome sequence for a member of the Pinaceae, comparative mapping becomes even more important as the primary tool for integrating information across species.
Loblolly pine (Pinus taeda L., 2_n_ = 2_x_ = 24) is the most genetically studied conifer species and was chosen as the reference species for comparative mapping in Pinaceae. Although still far from being a true model species, loblolly pine has rich genetic resources, well-developed genetic and QTL maps (Sewell et al. 1999; Brown et al. 2001, 2003; Temesgen et al. 2001), and expressed sequence tag (EST) databases (Allona et al. 1998; Whetten et al. 2001; Kirst et al. 2003). Douglas fir (Pseudotsuga menziesii [Mirb.] Franco, 2_n_ = 2_x_ = 26) is the most important species of the genus Pseudotsuga with well-studied genetic and QTL maps (Krutovskii et al. 1998; Jermstad et al. 1998, 2001a,b, 2003). Together, they are the most commercially important forest tree species in the United States, and a comparative map between species would have significant practical value. It might also help to establish the origin of the thirteenth chromosome pair in Douglas fir, the only species not having 12 chromosome pairs in the Pinaceae.
Orthologous markers are essential for constructing comparative maps. Fitch (1970)(2000) defined orthologous genes as homologous genes whose divergence follows a speciation event, while paralogs are defined as genes whose divergence follows a duplication event within a species. Orthologs are expected to have similar function, expression, amino acid and nucleotide sequence, and genome location in closely related species (e.g., Mirny and Gelfand 2002). Two criteria, high sequence similarity and genome location, were used as evidence for orthology in this study.
Restriction fragment length polymorphism (RFLP), based on Southern hybridization with single-copy genomic clones to ensure orthology, has been used broadly for comparative mapping in plants (Ahn et al. 1993; Sherman et al. 1995; Gale and Devos 1998; Yan et al. 2003). Comparative mapping using single- or low-copy cDNA as hybridization probes has been also successful in the Brassicaceae (Lan et al. 2000; Barnes 2002; Babula et al. 2003), Solanaceae (Doganlar et al. 2002), Poaceae (Smilde et al. 2001; Feuillet and Keller 2002; Laurie and Devos 2002), and across different families (Davis et al. 1999; Gebhardt et al. 2003). However, RFLP methods have had limited application in conifers due to genome complexity and numerous multigene families (Kinlaw and Neale 1997). Although comparative mapping using RFLP markers has been successful in pine species (Devey et al. 1999), it is difficult to apply RFLPs across different genera in conifers. Many loblolly pine probes produced a complex multiband pattern in hybridization with Douglas fir genomic DNA (Jermstad et al. 1994, 1998). PCR-amplified EST polymorphisms (ESTPs) have emerged recently as an alternative to RFLP markers for comparative mapping (Brown et al. 2001). A set of orthologous ESTP markers developed in loblolly pine has been established and successfully used in comparative mapping in the genus Pinus (Brown et al. 2001; Chagné et al. 2003; Komulainen et al. 2003). However, only 22% of these markers were amplified in Douglas fir (Brown et al. 2001). Mutations in primer binding sites have made comparative mapping via a common set of PCR primers practically impossible between conifer genera. To overcome this problem a computational approach was used to identify Douglas fir ESTs with high homology, and putative orthology, to ESTPs mapped in loblolly pine. A similar approach was used in recent comparative mapping studies between tomato, potato, and Arabidopsis (Fulton et al. 2002; Gebhardt et al. 2003). The selected Douglas fir ESTs were used to design Douglas fir-specific PCR primers to amplify loblolly pine orthologs in Douglas fir for subsequent genetic mapping. This approach allowed comparative mapping to be extended to the family level and established a framework for comparative genomics in Pinaceae. This study is a part of the Conifer Comparative Genomics Project (CCGP) formed as an international collaboration at the Institute of Forest Genetics (U.S. Department of Agriculture Forest Service) to develop the orthologous genetic markers and publicly available reference mapping populations that can be shared among different laboratories to facilitate comparative mapping (http://dendrome.ucdavis.edu/ccgp).
MATERIALS AND METHODS
Mapping populations and reference maps:
The loblolly pine and Douglas fir mapping populations were three-generation outbred pedigrees consisting of four grandparents, two F1 parents, and several hundred progeny (Jermstad et al. 1998, 2003; Sewell et al. 1999). The loblolly pine reference map was based on RFLP and ESTP markers as reported in Sewell et al. (1999), Temesgen et al. (2001), and Brown et al. (2001). This map is a consensus map between two pedigrees. Syntenic relationships with other Pinus species were established previously for most of the 12 linkage groups (Devey et al. 1999; Brown et al. 2001; Chagné et al. 2003; Komulainen et al. 2003). These groups included 302 markers (166 RFLP, 5 isozyme, and 131 EST markers), with a total map length of 1274 cM. The Douglas fir reference map was based on 376 markers [172 RFLP, 77 randomly amplified polymorphic DNA (RAPD), and 2 isozyme markers (Jermstad et al. 1998) with 20 simple sequence repeat (SSR), 4 sequence-tagged site (STS), and 101 ESTP markers added in this study (see supplemental Table S1 at http://www.genetics.org/supplemental/)].
Markers analyzed:
Three types of markers were used to develop the loblolly pine and Douglas fir comparative map: (1) single- or low-copy RFLP markers developed from loblolly cDNA clones, (2) ESTP markers developed in several pine and spruce (Picea) species that amplified a single locus in previous studies, and (3) ESTP and STS markers developed in Douglas fir. RFLP markers were mapped and sequenced previously in both species (Jermstad et al. 1998; Sewell et al. 1999). ESTP markers developed in pines and spruces were mapped in both species also according to methods described in Brown et al. (2001). ESTP and STS markers from Douglas fir are described in detail below. Briefly, EST and STS sequences were selected initially for evaluation as putative homologs to mapped loblolly pine markers on the basis of sequence similarity. One PCR primer of each pair was situated preferentially in an untranslated region to favor the selective amplification in Douglas fir of a single member of a gene family. At the intergeneric level, this strategy precluded the mapping of Douglas fir markers in loblolly pine. However, the orthologous relationships of these loci in both species were assessed by sequence similarity of amplified fragments and their conserved mapped location.
Douglas fir EST and STS markers:
A cDNA library was obtained from 1-month-old Douglas fir seedlings in collaboration with Integrated Genomics (Chicago). Total RNA was extracted from ∼5 g of ground tissue following the protocol of Chang et al. (1993). Double-stranded cDNA was prepared using the Universal RiboClone cDNA Synthesis System (Promega, Madison, WI), filtered through a Sephacryl S-400 column, ligated into the _Eco_ICR I-cut dephosphorylated pGEM-3Z sequencing vector, and electroporated into Escherichia coli DH5α. A total of 5031 EST sequences were obtained and assembled into contigs. These ESTs and contigs and four Douglas fir STSs available from GenBank were used to query the December 2002 assembly of loblolly pine ESTs, which is accessible at http://pine.ccgb.umn.edu, using BLASTn and tBLASTx. The assembly contained 20,456 contigs and singletons derived from 59,430 sequences from six xylem libraries. Douglas fir sequences homologous to mapped loblolly pine loci were selected for further study, if they showed nucleotide similarity >80% and expected values of ≤_E_-15 over a minimum of 100 bp.
PCR and detection of polymorphisms:
PCR primers were designed using the Douglas fir EST and STS sequences homologous to mapped loblolly pine loci. Primers were designed using the computer program GeneRunner v3.04 (Hastings Software, Hudson, NY) to yield products of 300–500 bp. A typical reaction volume was 25 μl and included 10 mm TRIS-HCl pH 8.3, 50 mm KCl, 1.5 mm MgCl2, 200 μm of each dNTP, 1 μm of each primer, 12 ng of DNA template, and 0.5 units of HotStart Taq DNA Polymerase from QIAGEN (Valencia, CA). Following HotStart Taq activation (94° for 15 min), PCR amplification involved denaturation at 94° for 1 min, annealing for 0.5 min, and extension for 2 min. The annealing temperature during the initial cycles was lowered from X to Y by 1° every second cycle. Standard PCR conditions (X = 65° and Y = 60° or 55°) were used for most ESTP markers, although for primer pairs that failed to amplify, the stringency was reduced (X = 60° and Y = 50°). An additional 30 cycles of amplification were performed upon reaching the final annealing temperature (Y) followed by a final extension at 72° for 10 min. Amplification products from the parents of the Douglas fir pedigree were screened for polymorphism in 2% agarose gels and by denaturing gradient gel electrophoresis (DGGE) using a 15–45% denaturing gradient and a DCode apparatus (Bio-Rad, Hercules, CA) according to Temesgen et al. (2000). Finally, ESTP segregation data were collected for the 94 progeny of the Douglas fir pedigree.
DNA sequencing:
To support the possibility that PCR markers amplified in Douglas fir were orthologs to the mapped loblolly pine markers, amplifications products were directly sequenced and compared to the original Douglas fir and loblolly pine sequences. To use direct sequencing without cloning 33 markers were selected that amplified a single locus and had no amplification background, such as light additional bands that may interfere with sequencing. The selected markers represented 26 putative orthologous markers (5 loblolly pine-based ESTP, 18 Douglas fir-based ESTP, and 3 STS markers) and 7 nonorthologous Douglas fir-based ESTP markers. Four alleles at each locus were sequenced using DNA extracted from the haploid megagametophyte tissue of four Douglas fir seeds. DNA sequences were obtained from both strands with the primers used for PCR amplification and the ABI PRISM BigDye Primer Cycle Sequencing Kit v.3.1 (Applied Biosystems, Foster City, CA). Fragments were detected on an ABI 3730 DNA Analyzer at the Genomics Facility Center at the University of California, Davis. Raw sequences were base called by the PHRED program (Ewing and Green 1998; Ewing et al. 1998), assembled using the PHRAP program, and viewed through CONSED (Gordon et al. 1998, 2001).
Linkage analysis:
Genotypic data were scored visually and tested for Mendelian segregation. Markers showing only slight segregation distortion (0.01 < P < 0.05) were not excluded from linkage analysis because recombination estimators are still valid when distortion is observed at only one locus of a linked pair of loci (Bailey 1961; Ott 1991). ESTP markers were added to the existing segregation data (Jermstad et al. 1998) and the linkage analysis was repeated. A sex-averaged consensus map was produced using JOINMAP versions 1.4 and 2.0 (Stam 1993; Stam and Van Ooijen 1995). Linkage groups were assigned at the LOD thresholds of 4 and 5. Grouping was almost identical at both thresholds, except a few loci unmapped at LOD = 5 and a spurious merging of two apparently independent linkage groups at LOD = 4. Therefore, we used mainly LOD = 4 for grouping, except two linkage groups that were assembled at LOD = 5. The procedure for ordering markers was the same as described in Jermstad et al. (1998). The Kosambi function was used to estimate map distances.
Nomenclature and informatics:
Mapped loci were named according to guidelines for submitting data to the TreeGenes database (http://dendrome.ucdavis.edu/Tree_Page.htm). A mapped marker is defined by its experiment, source, accession number, and locus identifier fields. For example, an ESTP derived from the loblolly pine cDNA clone PtIFG_8732 and mapped in both loblolly pine and Douglas fir in this study is referenced as IFGREF_ estPtIFG_8732_a and IFGLXD_ estPtIFG_8732_a in the loblolly pine and Douglas fir maps, respectively. For brevity, however, experiment fields have been omitted (Figure 1).
Figure 1.—
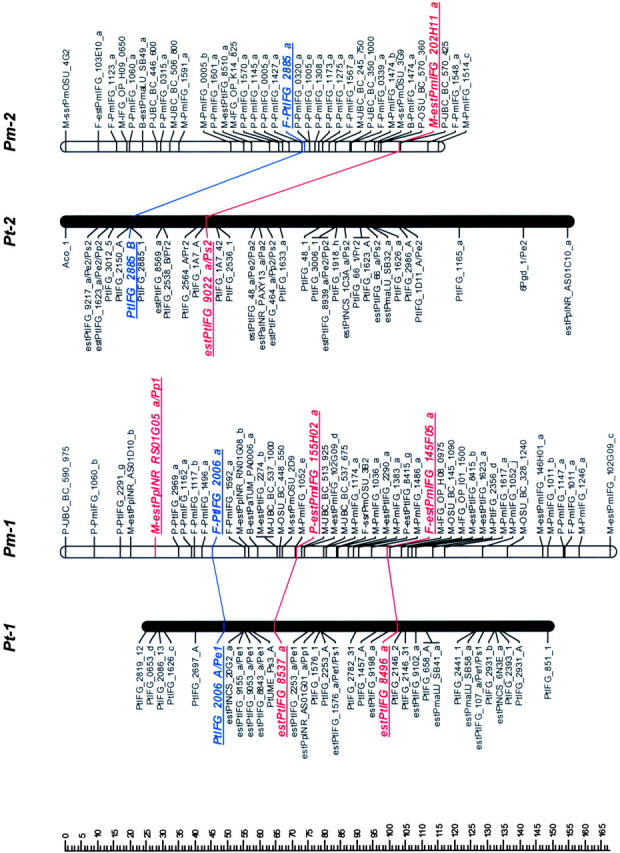
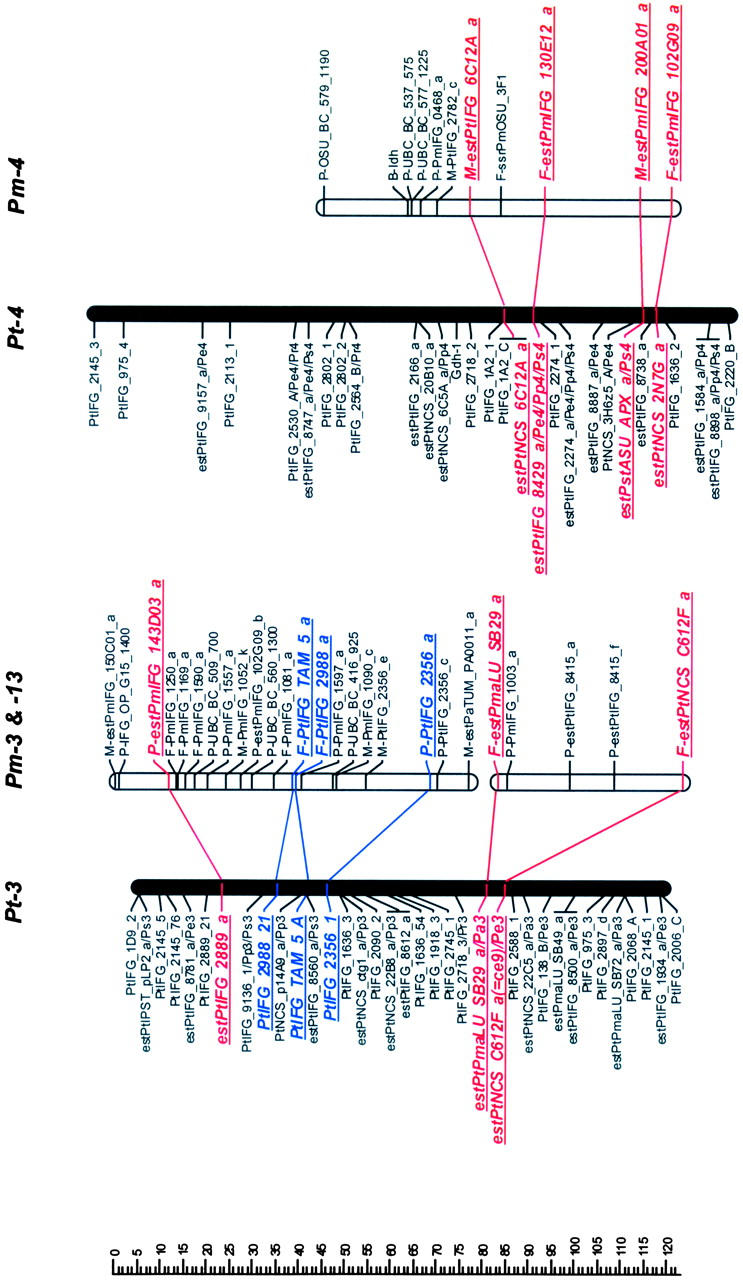
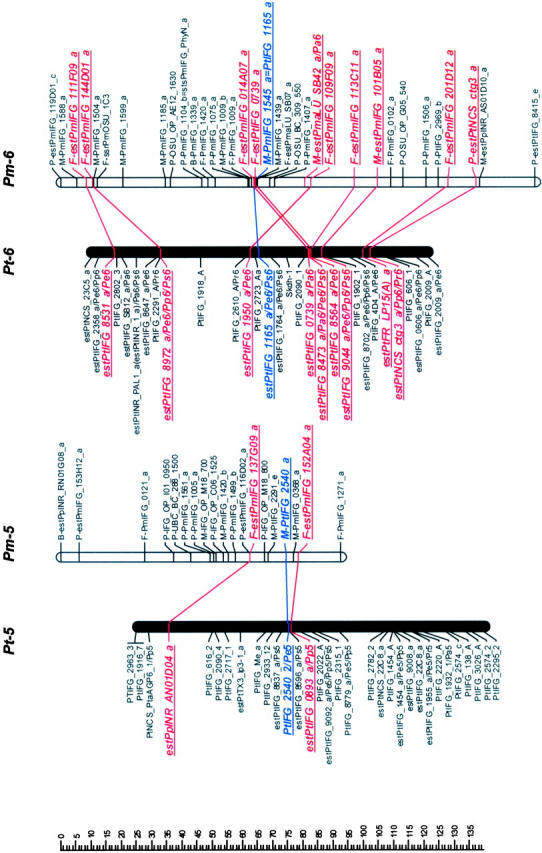
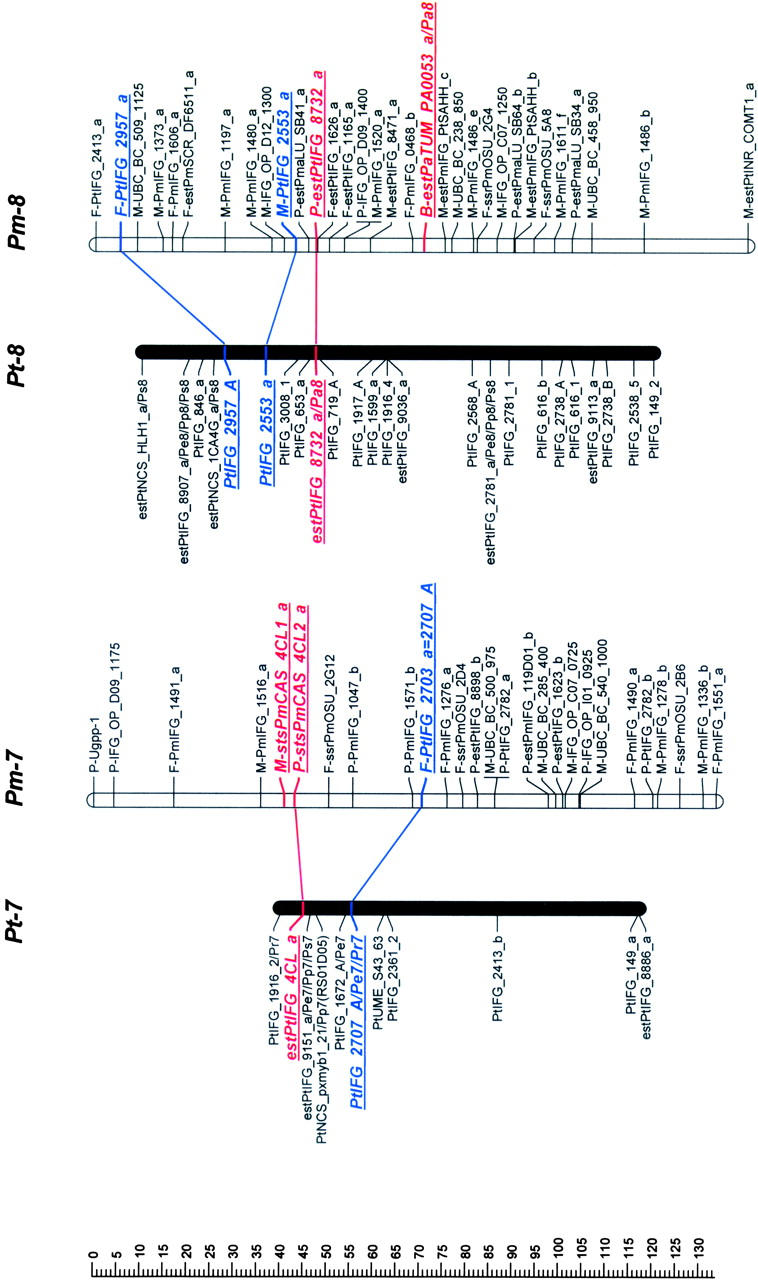
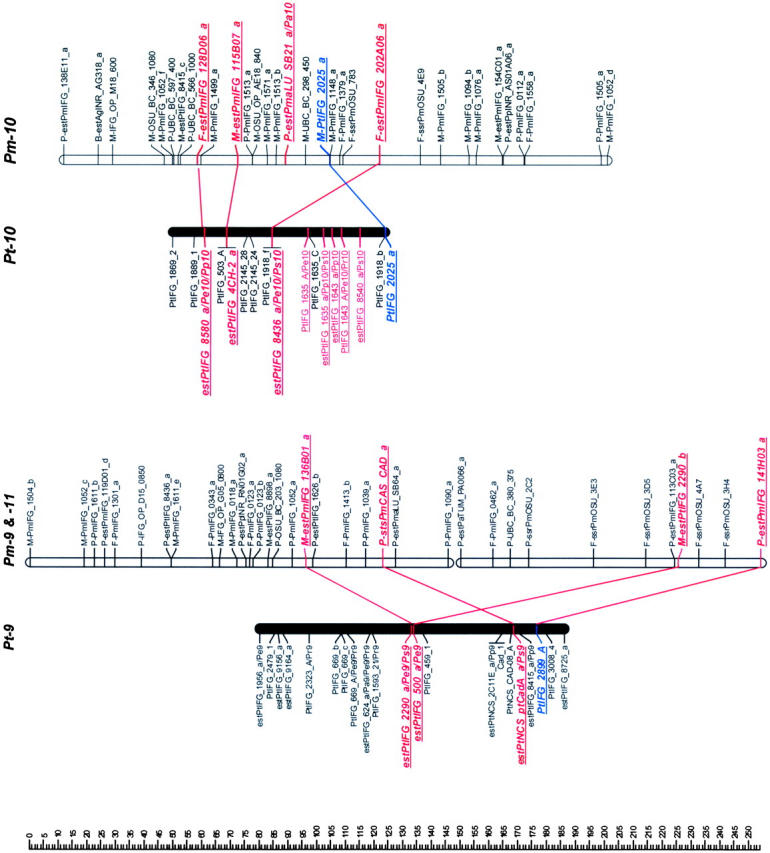
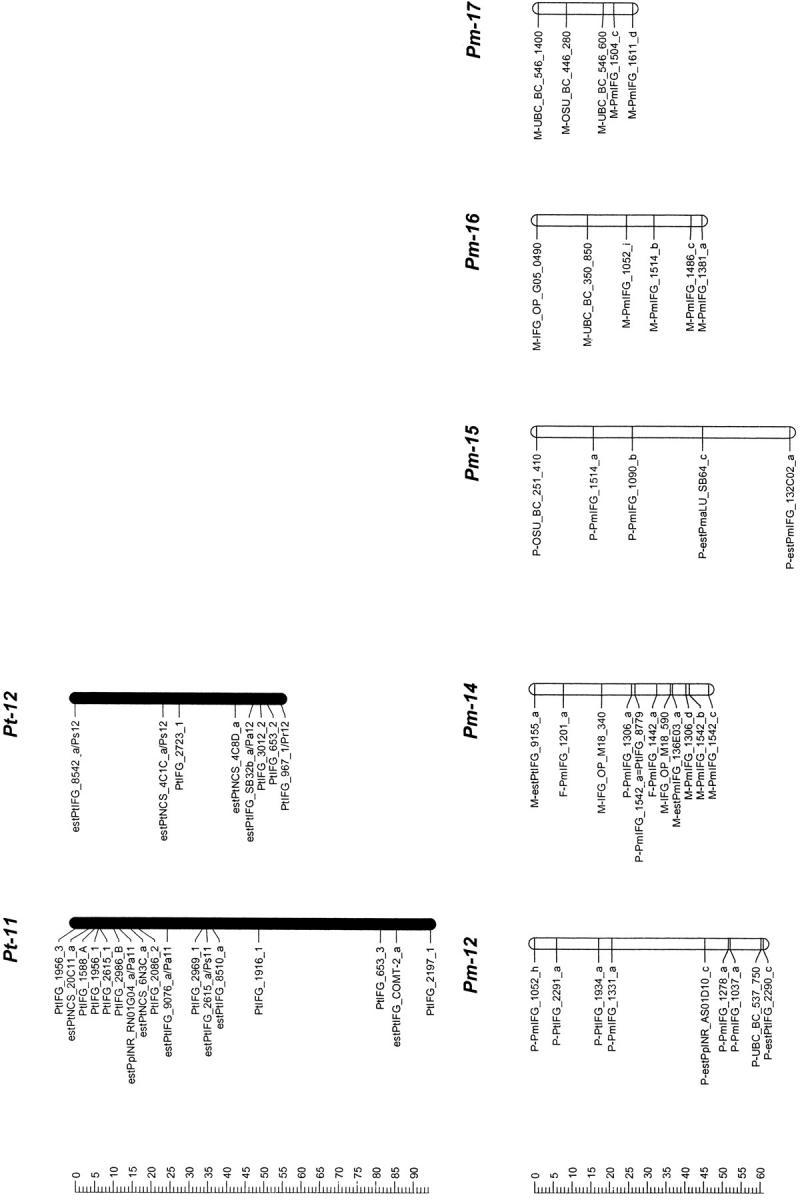
Genetic maps of loblolly pine (Pinus taeda L.), Pt, and Douglas fir (Pseudotsuga menziesii [Mirb.] Franco), Pm. Orthologous markers are highlighted by larger, boldface, italicized, and underlined type and are connected by lines, except four of them that were mapped in pine species other than loblolly, but in the same syntenic linkage group (LG) [_estPpINR_RS01G05_a_ was mapped in LG1 in _Pinus pinaster_ (Chagné et al. 2003), and estPmaLU_SB42_a, estPaTUM_PA0053_a, and estPmaLU_SB21_a were mapped in LG6, LG8, and LG10 in Picea abies, respectively (our unpublished data)]. The loci were named following guidelines for the TreeGenes genome database (http://dendrome.ucdavis.edu/Tree_Page.htm; see also materials and methods). Abbreviations placed after the underscore in some loblolly pine marker names show other conifer species and syntenic linkage groups in which these markers were also mapped. For instance, Pe1 in the PtIFG_2006_A/Pe1 marker name in the linkage group Pt-1 means that the PtIFG_2006_A marker was also mapped in the Pinus elliottii syntenic linkage group 1. Similarly, Pp stands for P. pinaster, Pr for P. radiata, Ps for P. sylvestris, and Pa for Picea abies.
RESULTS
Orthologous RFLP and ESTP markers derived from pine and spruce species:
Twenty-six RFLP markers were mapped in both loblolly pine and Douglas fir (Jermstad et al. 1998; Sewell et al. 1999). Seven markers met criteria of orthologous markers. Four markers (PtIFG_2006_a, PtIFG_2356_a, PtIFG_2988_a, and PtIFG_2540_a) revealed more than a single locus, but were mapped into syntenic regions and can be also conditionally considered as orthologous markers.
The ESTP primer pairs developed from pine and spruce species amplified Douglas fir templates with variable success, ranging from 24% amplification success with primers derived from loblolly pine to 93% success with primers derived from Norway spruce, Picea abies (L.) Karst. (Table 1). In total, 55 ESTPs from the four species were mapped in Douglas fir, but only 11 markers met the criteria of orthologous markers.
TABLE 1.
RFLP, ESTP, and STS markers screened in Douglas fir
| Marker | Species | Screened | Amplified(%)a | Mapped(%)a | Orthologous(%)b |
|---|---|---|---|---|---|
| RFLP | Loblolly pine (Pinus taeda)c | 171 | 122 (71)h | 26 (15) | 11 (42) |
| ESTP | Loblolly pined | 156 | 37 (24) | 30 (19) | 6 (20) |
| Maritime pine (P. pinaster)e | 50 | 38 (76) | 9 (18) | 1 (11) | |
| Black spruce (Picea mariana)f | 50 | 42 (84) | 11 (22) | 3 (27) | |
| Norway spruce (P. abies)g | 15 | 14 (93) | 5 (33) | 1 (20) | |
| Total pine and spruce ESTPs | 271 | 131 (48) | 55 (20) | 11 (22) | |
| Douglas fir (Pseudotsuga menziesii) | 75 | 73 (97) | 39 (52) | 21 (54) | |
| STS | Douglas fir | 4 | 4 (100) | 4 (100) | 3 (75) |
| Total | 521 | 330 (63) | 124 (24) | 46 (37) |
Orthologous ESTP and STS markers derived from Douglas fir:
Of 5031 ESTs analyzed, 1992 sequences were assembled into 621 contigs. There were an additional 55 singletons and 2984 singlets. All ESTs and contigs were compared to the genetically mapped loblolly pine ESTs, and Douglas fir sequences with >80% nucleotide identity were used for PCR primer design. Most primers (97%) designed in this manner amplified a single Douglas fir product of expected size, and 39 markers were mapped. Twenty-one of the 39 markers met the criteria of orthologous markers (Table 1). Three Douglas fir genomic sequences available from GenBank had orthologs among mapped loblolly pine ESTs. They were mapped and met the criteria of orthologous markers (Table 1).
Douglas fir linkage map:
In total, 376 markers (172 RFLPs, 77 RAPDs, 20 SSRs, 2 isozymes, 4 STSs, and 101 ESTPs) were mapped to 22 linkage groups consisting of 3 or more markers (Table 1). There were 17 major linkage groups that consisted of 5 or more markers (Figure 1). The total length of the linkage map was 1664 cM for the 17 major linkage groups and 1859 cM for all 22 linkage groups.
Sequence of PCR-amplified ESTP and STS markers:
PCR products for 26 putative orthologs (5 ESTPs amplified using primers based on loblolly pine ESTs, 18 ESTPs amplified using primers based on Douglas fir ESTs, and 3 STSs) and 7 nonorthologs, which were amplified and mapped in Douglas fir, were sequenced and compared to the original Douglas fir and loblolly pine EST or genomic sequences. All sequences confirmed the identity and origin of amplified markers. The orthology of all 26 putative orthologs, from which PCR amplification products were sequenced, was also confirmed (Table 2). The identity between Douglas fir and loblolly pine sequences was 89 ± 4% for 26 orthologous and 83 ± 2% for 7 nonorthologous markers on average (_t_-test P = 0.012).
TABLE 2.
Orthologous markers mapped in both Douglas fir and loblolly pine
| Comparison between Douglas firand loblolly pine sequences | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Type | Marker mappedin Douglas fir | Douglas firlinkagegroup | Position(cM) | GenBankaccessionno. | Orthologous markermapped in loblolly pine | Syntenic linkagegroup inloblolly pine | GenBankaccessionno. | Nucleotide identity (%) | BLASTN _e_-value |
| ESTP | estPpINR_RS01G05_aa | 1 | 27.5 | AL750905 | estPpINR_RS01G05_a in Pinus pinaster | 1 | AL750905 | NAa | NAa |
| RFLP | PtIFG_2006_ab | 1 | 45.1 | H75041H75042 | PtIFG_2006_A | 1 | H75041H75042 | NAb | NAb |
| ESTP | estPmIFG_155H02_ac | 1 | 71.4 | CN637754 | estPtIFG_8537_a | 1 | AA739563 | 89 | 1.E-107 |
| ESTP | estPmIFG_145F05_ac | 1 | 99.2 | CN637175 | estPtIFG_8496_a | 1 | AA739536 | 82 | 9.E-17 |
| ESTP | estPmIFG_202H11_ac | 2 | 11.8 | CN638460 | estPtIFG_9022_a | 2 | AI725138 | 93 | 1.E-160 |
| RFLP | PtIFG_2885_ab | 2 | 41.3 | NA | PtIFG_2885_B | 2 | NA | NAb | NAb |
| RFLP | PtIFG_2356_ab | 3 | 8.3 | H75085 | PtIFG_2356_a | 3 | H75085 | NAb | NAb |
| RFLP | PtIFG_2988_ab | 3 | 37.6 | NA | PtIFG_2988_a | 3 | NA | NAb | NAb |
| RFLP | PtIFG_TAM_5_ab | 3 | 38.2 | NA | PtIFG_TAM_5_a | 3 | NA | NAb | N/Ab |
| ESTP | estPmIFG_143D03_ac | 3 | 65.1 | CN637040 | estPtIFG_2889_a | 3 | H75234 | 85 | 7.E-19 |
| ESTP | estPmaLU_SB29_ad | 13 | 0.0 | AF051222 | estPmaLU_SB29_a | 3 | AF051222 | NSd | NSd |
| ESTP | estPtNCS_6C12F_ac | 13 | 39.9 | AA556811 | estPtNCS_6C12F_a | 3 | AA556811 | 85e | 3.E-30e |
| ESTP | estPmIFG_102G09_aa | 4 | 0.0 | CN634675 | estPtNCS_2N7G_a | 4 | AA556198 | 84 | 3.E-50 |
| ESTP | estPmIFG_200A01_ac | 4 | 6.8 | CN638230 | estPtIFG_APX | 4 | AF326783 | 93 | 2.E-86 |
| ESTP | estPmIFG_130E12_ac | 4 | 27.9 | CN636293 | estPtIFG_8429_a | 4 | AA739505 | 91 | 1.E-57 |
| ESTP | estPtIFG_6C12A_ac | 4 | 44.2 | AA556806 | estPtIFG_6C12A_a | 4 | AA556806 | 93e | 1.E-103e |
| ESTP | estPmIFG_137G09_ad | 5 | 62.6 | CN636731 | estPpINR_AN01D04_a | 5 | AL749558 | 92f | 0f |
| RFLP | PtIFG_2540_ab | 5 | 74.4 | H75131 | PtIFG_2540_a | 5 | H75131 | NAb | NAb |
| ESTP | estPmIFG_152A04_ac | 5 | 78.6 | CN637533 | estPtIFG_0893_a | 5 | H75118 | 87 | 4.E-08g |
| ESTP | estPmIFG_111F09_ac | 6 | 8.4 | CN635180 | estPtIFG_8531_a | 6 | AA739558 | 82 | 3.E-57 |
| ESTP | estPmIFG_144D01_ac | 6 | 10.5 | CN637093 | estPt_8647_a | 6 | AA739625 | 95 | 1.E-22 |
| ESTP | estPmIFG_014A07_aa | 6 | 63.8 | CN634509 | estPtIFG_8473_a | 6 | AA739526 | 84 | 7.E-16 |
| ESTP | estPtIFG_0739_ac | 6 | 64.2 | H75167H75168 | estPtIFG_0739_a | 6 | H75167H75168 | 92e | 1.E-22e |
| RFLP | PmIFG_1545_a | 6 | 64.4 | AA701802H75180 | PtIFG_1165_a | 6 | AA701802H75180 | 86h | 1.E-10h |
| ESTP | estPmaLU_SB42_aa | 6 | 80.3 | AF051232 | estPmaLU_SB42_a in Picea abies | 6 | AF051232 | NAa | NAa |
| ESTP | estPmIFG_109F09_ac | 6 | 82.5 | CN635076 | estPtIFG_1950_a | 6 | H75126 | 84 | 3.E-11 |
| ESTP | estPmIFG_113C11_ac | 6 | 96.8 | CN635266 | estPtIFG_8564_a | 6 | AA739580 | 95 | 5.E-55 |
| ESTP | estPmIFG_101B05_ac | 6 | 104.3 | CN634590 | estPtIFG_9044_a | 6 | AA739876 | 94 | 1.E-115 |
| ESTP | estPmIFG_201D12_ac | 6 | 127.8 | CN638348 | estPtFR_LP15_a | 6 | AF013803 | 82 | 9.E-25 |
| ESTP | estPtNCS_ctg3_ac | 6 | 137.0 | AF036095 | estPtNCS_CCoAOMT_a= estPtNCS_ctg3_a | 6 | AF036095 | 83e | 2.E-26e |
| RFLP | PtIFG_2703_a (=PtIFG_2707_A)b | 7 | 63.1 | H75113H75227H75228 | PtIFG_2707_A (=PtIFG_2703_a)b | 7 | H75113H75227H75228 | NAb | NAb |
| STS | stsPmCAS_4CL2_ac | 7 | 90.5 | AF144507 | estPtIFG_4CL_a | 7 | U12012U39405 | 88 | 1.E-173 |
| STS | stsPmCAS_4CL1_ac | 7 | 92.7 | AF144506AF144508 | estPtIFG_4CL_a | 7 | U12012U39405 | 87 | 1.E-166 |
| RFLP | PtIFG_2957_ab | 8 | 5.3 | NA | PtIFG_2957_a | 8 | NA | NAb | NAb |
| RFLP | PtIFG_2553_ab | 8 | 42.9 | H75210 | PtIFG_2553_a | 8 | H75210 | NAb | NAb |
| ESTP | estPtIFG_8732_ac | 8 | 47.3 | AA739680 | estPtIFG_8732_a | 8 | AA739680 | 90e | 7.E-19e |
| ESTP | estPaTUM_PA0053_aa | 8 | 70.5 | AJ132535 | estPaTUM_PA0053_a in Picea abies | 8 | AJ132535 | NAa | NAa |
| STS | stsPmCAS_CAD_ac | 9 | 22.7 | AF145985AF145986 | estPtNCS_ptCadA_a | 9 | Z37991 | 90 | 2.E-92 |
| ESTP | estPmIFG_136B01_ac | 9 | 49.4 | CN636620 | estPtIFG_500_a | 9 | H75151 | 86 | 2.E-51 |
| ESTP | estPmIFG_141H03_ac | 11 | 0.0 | CN636962 | PtIFG_2899_a | 9 | H75245 | 95 | 2.E-71 |
| ESTP | estPtIFG_2290_ba | 11 | 28.8 | H75067 | estPtIFG_2290_a | 9 | H75067 | NAa | NAa |
| ESTP | estPmIFG_202A06_ac | 10 | 72.8 | CN638390 | estPtIFG_8436_a | 10 | AA739508 | 93 | 2.E-87 |
| RFLP | PtIFG_2025_ab | 10 | 88.8 | H75045H75046 | PtIFG_2025_a | 10 | H75045H75046 | NAb | NAb |
| ESTP | estPmaLU_SB21_ad | 10 | 103.0 | AF051216 | estPmaLU_SB21_a in Picea abies | 10 | AF051216 | NSd | NSd |
| ESTP | estPmIFG_115B07_ac | 10 | 118.2 | CN635370 | estPtIFG_4CH-2_a | 10 | AF096998 | 89 | 3.E-52 |
| ESTP | estPmIFG_128D06_ac | 10 | 131.1 | CN636156 | estPtIFG_8580_a | 10 | AA739590 | 87 | 1.E-22 |
Orthologous markers and homologous linkage groups:
Comparison of Douglas fir and loblolly pine maps revealed 10 linkage groups (LG1–LG10) in loblolly pine that shared 2–10 orthologous markers with 12 apparently syntenic linkage groups in Douglas fir based on 46 orthologous markers (Table 2, Figure 1). Primer sequences and PCR conditions that were used to amplify orthologous markers are presented in the supplemental Table S2, and their homology analysis and annotation in the supplemental Table S3 (see http://www.genetics.org/supplemental/). Markers mapped in pine species other than loblolly helped to strengthen the comparative mapping. For example, the estPpINR_RS01G05_a marker was mapped in maritime pine (P. pinaster Ait; Chagné et al. 2003) and in Douglas fir, but not in loblolly pine. However, in both species this marker was mapped in the linkage group that was homologous to the same LG1 in loblolly pine based on other orthologous markers and therefore corroborated syntenic relationships between these groups (Table 2; Figure 1). Similarly, the estPmaLU_SB42_a marker was mapped in Norway spruce (our unpublished data) and Douglas fir, but not in loblolly pine. The linkage group containing this marker in both Norway spruce and Douglas fir was homologous to LG6 in loblolly pine based on other markers (Table 2; Figure 1). The total lengths of the syntenic linkage groups that shared orthologous markers were 1125 cM for 10 groups in loblolly pine and 1421 cM for 12 groups in Douglas fir.
Seven of 12 linkage groups in Douglas fir had three or more orthologous markers that allow inspection of colinearity. Gene order was completely colinear in the syntenic regions of 5 of these 7 groups (LG1, LG3, LG4, LG5, and LG8) and partly colinear in LG6 (Figure 1). Local noncolinearity was observed only in two groups, LG6 and LG10 (Figure 1). There were two cases when two linkage groups in Douglas fir showed synteny with a single linkage group in loblolly pine: (1) Pm-3 and Pm-13 vs. Pt-3 (LG3) and (2) Pm-9 and Pm-11 vs. Pt-9 (LG9).
DISCUSSION
Comparative mapping in Pinaceae:
The macrosyntenic relationships between species of two genera of the family Pinaceae were established for nearly all major linkage groups. Ten homologous linkage groups were identified in loblolly pine that shared two or more orthologous markers with Douglas fir linkage groups (Figure 1). The same 10 homologous linkage groups were identified between loblolly pine and maritime pine (Chagné et al. 2003). All 12 homologous linkage groups were identified between loblolly pine and Scots pine, but only 10 linkage groups shared two and more orthologous markers (Komulainen et al. 2003). Therefore, the intergeneric maps in this study have identified practically all the same homologous linkage groups as were found in the intrageneric pine maps.
Chromosomal evolution in Pinaceae:
The high level of synteny and colinearity among species within the Pinaceae supports the general hypothesis based on cytogenetic data that major chromosomal rearrangements have not been frequent in the evolution of the Pinaceae (Prager et al. 1976). Except in a few species, there is no evidence for major chromosomal rearrangement or polyploidy in Pinaceae (Prager et al. 1976). We also observed substantial colinearity between genera at the macrosyntenic level. Local noncolinearity was observed only in LG6 and LG10 and could be easily explained by a single inversion (Figure 1). However, it would be preliminary to speculate about local rearrangements and microcolinearity from these data. Local noncolinearity might also result from mapping paralogs and/or other members of multigene families in the same syntenic regions. Approximately 40% of all ESTP, STS, and RFLP markers tested could serve as orthologous loci. Some of the remaining nonorthologous markers were assayed under less stringent conditions and may therefore have revealed paralogs or different members of multigene families. This can be easily expected due to the complexity of conifer genomes (Kinlaw and Neale 1997). This is also supported by lower levels of identity observed between Douglas fir and loblolly pine sequences for nonorthologous (83%) vs. orthologous (89%) markers. There were a few ESTP primers that amplified two products that were mapped in the same region, showing that tandem duplication might be common in Pinaceae genome evolution. Paralogs complicate construction of comparative maps in Pinaceae, but are also of great interest for studying evolution of multigene families.
Pseudotsuga and Larix are the next closest genera to Pinus after Picea on the basis of phylogenetic studies (Wang et al. 2000; Rydin et al. 2002). However, the karyotype of Douglas fir is unique in Pinaceae. It has 13 chromosome pairs (2_n_ = 26), while all other species, including closely related Pseudotsuga and Larix species, have only 12 pairs (2_n_ = 24). The karyotype of Douglas fir includes 2 telocentric chromosomes that are strikingly dissimilar to the other 11 chromosomes (5 metacentric and 6 submetacentric chromosomes). Their length is also less than one-half that of the metacentric chromosomes (Doerksen and Ching 1972), suggesting that these two chromosomes originated by centromeric fission of one of the metacentric chromosomes. We could not resolve unambiguously the question of the origin of the thirteenth chromosome pair in Douglas fir, but our data allow some speculation. Douglas fir linkage groups Pm-3 and Pm-13 were syntenic with loblolly pine LG3, as were Pm-9 and Pm-11 with LG9 (Figure 1). One of these syntenic linkage pairs could represent two different chromosomes in Douglas fir. Orthologous markers mapped in these groups would be good candidates for FISH to resolve this question.
Potential applications of comparative-mapping results:
Comparative mapping is an important tool for integrating genetic data among related taxa. It helps to consolidate genetic maps and bridge linkage gaps. For instance, comparative mapping has helped to assign several small unlinked groups to the larger homologous linkage groups in pine species (Brown et al. 2001; Komulainen et al. 2003). The loblolly pine × Douglas fir comparative map now integrates the mapping data between different genera of the family Pinaceae and also between the two most important tree species in North America. For example, the QTL mapped in loblolly pine (Sewell et al. 2000, 2002; Brown et al. 2003) and in Douglas fir (Jermstad et al. 2001a,b, 2003) can now be compared across different genera the same way as it was done across different species within genus Eucalyptus (Marques et al. 2002) and Pinus (Chagné et al. 2003). Mapped orthologous markers that consistently associated with the same QTL across different species can be used to confirm and verify QTL and to identify candidate genes for quantitative traits. The orthologous markers that have been developed and mapped in this study could also be used with species of the remaining genera of Pinaceae that also have worldwide importance, such as Picea, Larix, Abies, and Tsuga. For example, these markers have recently been used to construct a comparative map between loblolly pine and Norway spruce (our unpublished data) that will further contribute toward a deeper understanding of the evolution of conifer genomes.
Establishing Pinaceae as a genetic system:
We were able to establish basic syntenic relationships in Pinaceae. High synteny and conserved gene order found in this study for such distantly related species as Douglas fir and loblolly pine open the possibility of comparative mapping at the family level and establish a comparative genomics framework in Pinaceae that can now be viewed as a genetic system. However, more studies are needed to establish a comparative genomics system that would rival the existing model systems. ESTs represent a good alternative to complete genome sequencing for nonmodel organisms or organisms with large genomes, such as conifers. As more conifer EST and genomic sequences become available in public databases their homology can be tested and more orthologs can be established.
Acknowledgments
We thank the members of the Conifer Comparative Genomics Project for their participation and contributions; Valerie Hipkins and Randy Meyer (U.S. Department of Agriculture (USDA) Forest Service, National Forest Gel Electrophoresis Laboratory) for isozyme analysis in Douglas fir; Thomas Adams, Glenn Howe, and Gancho Slavov (Oregon State University) for SSR genotyping; Nicholas Wheeler (Weyerhaeuser) for providing the reference mapping populations; Santiago Gonzalez-Martinez (Department of Biotechnology and Breeding, Center of Forest Research, Spain) and Geoffrey Gill (ViaLactia Biosciences, Auckland, New Zealand) for help in marker development; and Jennifer Manares (University of California, Davis) for technical assistance with PCR and gel electrophoresis. Funding for this project was provided by the USDA Plant Genome National Research Initiative (USDA National Research Initiative grant no. 00-35300-9316). Trade names and commercial products or enterprises are mentioned solely for information and no endorsement by the USDA is implied.
References
- Ahn, S., J. A. Anderson, M. E. Sorrells and S. D. Tanksley, 1993. Homoeologous relationships of rice, wheat and maize chromosomes. Mol. Gen. Genet. 241**:** 483–490. [DOI] [PubMed] [Google Scholar]
- Allona, I., M. Quinn, E. Shoop, K. Swope, S. St. Cyr et al., 1998. Analysis of xylem formation in pine by cDNA sequencing. Proc. Natl. Acad. Sci. USA 95**:** 9693–9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babula, D., M. Kaczmarek, A. Barakat, M. Delseny, C. F. Quiros et al., 2003. Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: complexity of the comparative map. Mol. Genet. Genomics 268**:** 656–665. [DOI] [PubMed] [Google Scholar]
- Bailey, N. T. J., 1961 Mathematical Theory of Genetic Linkage. Clarendon Press, London.
- Barnes, S., 2002. Comparing Arabidopsis to other flowering plants. Curr. Opin. Plant Biol. 5**:** 128–134. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., and I. J. Leitch, 2003 Plant DNA _C_-values database (release 2.0, January 2003). http://www.rbgkew.org.uk/cval/homepage.html.
- Bennett, M. D., and J. B. Smith, 1991. Nuclear DNA amounts in angiosperms. Philos. Trans. R. Soc. Lond. B 334**:** 309–345. [DOI] [PubMed] [Google Scholar]
- Boutin, S. R., N. D. Young, T. Olson, Z.-H. Yu, R. C. Shoemaker et al., 1995. Genome conservation among three legume genera detected with DNA markers. Genome 38**:** 928–937. [DOI] [PubMed] [Google Scholar]
- Brown, G. R., E. E. Kadel, III, D. L. Bassoni, K. L. Kiehne, B. Temesgen et al., 2001. Anchored reference loci in loblolly pine (Pinus taeda L.) for integrating pine genomics. Genetics 159**:** 799–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, G. R., D. L. Bassoni, G. P. Gill, J. R. Fontana, N. C. Wheeler et al., 2003. Identification of quantitative trait loci influencing wood property traits in loblolly pine (Pinus taeda L.). III. QTL verification and candidate gene mapping. Genetics 164**:** 1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagné, D., G. R. Brown, C. Lalanne, D. Madur, D. Pot et al., 2003. Comparative genome and QTL mapping between maritime and loblolly pines. Mol. Breed. 12**:** 185–195. [Google Scholar]
- Chang, S., J. Puryear and J. Cairney, 1993. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11**:** 113–116. [Google Scholar]
- Davis, G. L., M. D. Mcmullen, C. Baysdorfer, T. Musket, D. Grant et al., 1999. A maize map standard with sequenced core markers, grass genome reference points, and 932 expressed sequence tagged sites (ESTs) in a 1736 locus map. Genetics 152**:** 1137–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devey, M. E., K. D. Jermstad, C. G. Tauer and D. B. Neale, 1991. Inheritance of RFLP loci in a loblolly pine three-generation pedigree. Theor. Appl. Genet. 83**:** 238–242. [DOI] [PubMed] [Google Scholar]
- Devey, M. E., T. A. Fiddler, B.-H. Liu, S. J. Knapp and D. B. Neale, 1994. An RFLP linkage map for loblolly pine based on a three-generation outbred pedigree. Theor. Appl. Genet. 88**:** 273–278. [DOI] [PubMed] [Google Scholar]
- Devey, M. E., M. M. Sewell, T. L. Uren and D. B. Neale, 1999. Comparative mapping in loblolly and radiata pine using RFLP and microsatellite markers. Theor. Appl. Genet. 99**:** 656–662. [DOI] [PubMed] [Google Scholar]
- Doerksen, A. H., and K. K. Ching, 1972. Karyotypes in the genus Pseudotsuga. For. Sci. 18**:** 66–69. [Google Scholar]
- Doganlar, S., A. Frary, M. C. Daunay, R. N. Lester and S. D. Tanksley, 2002. Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161**:** 1713–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8**:** 175–185. [DOI] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8**:** 186–194. [PubMed] [Google Scholar]
- Feuillet, C., and B. Keller, 2002. Comparative genomics in the grass family: molecular characterization of grass genome structure and evolution. Ann. Bot. 89**:** 3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch, W. M., 1970. Distinguishing homologous from analogous proteins. Syst. Zool. 19**:** 99–113. [PubMed] [Google Scholar]
- Fitch, W. M., 2000. Homology: a personal view on some of the problems. Trends Genet. 16**:** 227–231. [DOI] [PubMed] [Google Scholar]
- Frankis, M. P., 1989. Generic inter-relationships in Pinaceae. Notes R. Bot. Gard. Edinb. 45**:** 527–548. [Google Scholar]
- Freeling, M., 2001. Grasses as a single genetic system. Reassessment 2001. Plant Physiol. 125**:** 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton, T. M., R. Van der Hoeven, N. T. Eannetta and S. D. Tanksley, 2002. Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14**:** 1457–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M. D., and K. M. Devos, 1998. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. USA 95**:** 1971–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, C., B. Walkemeier, H. Henselewski, A. Barakat, M. Delseny et al., 2003. Comparative mapping between potato (Solanum tuberosum) and Arabidopsis thaliana reveals structurally conserved domains and ancient duplications in the potato genome. Plant J. 34**:** 529–541. [DOI] [PubMed] [Google Scholar]
- Grant, D., P. Cregan and R. C. Shoemaker, 2000. Genome organization in dicots: genome duplication in Arabidopsis and synteny between soybean and Arabidopsis. Proc. Natl. Acad. Sci. USA 97**:** 4168–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8**:** 195–202. [DOI] [PubMed] [Google Scholar]
- Gordon, D., C. Desmarais and P. Green, 2001. Automated finishing with autofinish. Genome Res. 11**:** 614–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A. E., A. Fiebig and D. Preuss, 2002. Beyond the Arabidopsis genome: opportunities for comparative genomics. Plant Physiol. 129**:** 1439–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermstad, K. D., A. M. Reem, N. C. Wheeler and D. B. Neale, 1994. Inheritance of restriction fragment length polymorphisms, random amplified polymorphic DNAs and isozymes in coastal Douglas-fir. Theor. Appl. Genet. 89**:** 758–766. [DOI] [PubMed] [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, N. C. Wheeler and D. B. Neale, 1998. A sex-averaged linkage map in coastal Douglas-fir (Pseudotsuga menziesii [Mirb.] Franco) based on RFLP and RAPD markers. Theor. Appl. Genet. 97**:** 762–770. [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, K. S. Jech, N. C. Wheeler and D. B. Neale, 2001. a Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir: I. Timing of vegetative bud flush. Theor. Appl. Genet. 102**:** 1142–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, N. C. Wheeler, T. S. Anekonda, S. N. Aitken et al., 2001. b Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir: II. Spring and fall cold-hardiness. Theor. Appl. Genet. 102**:** 1152–1158. [Google Scholar]
- Jermstad, K. D., D. L. Bassoni, K. S. Jech, G. A. Ritchie, N. C. Wheeler et al., 2003. Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir: III. QTL by environment interactions. Genetics 165**:** 1489–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinlaw, C. S., and D. B. Neale, 1997. Complex gene families in pine genomes. Trends Plant Sci. 2**:** 356–359. [Google Scholar]
- Kirst, M., A. F. Johnson, C. Baucom, E. Ulrich, K. Hubbard et al., 2003. Apparent homology of expressed genes from wood-forming tissues of loblolly pine (Pinus taeda L.) with Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 100**:** 7383–7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, P. E., R. R. Klein, J. Vrebalov and J. E. Mullet, 2003. Sequence-based alignment of sorghum chromosome 3 and rice chromosome 1 reveals extensive conservation of gene order and one major chromosomal rearrangement. Plant J. 34**:** 605–621. [DOI] [PubMed] [Google Scholar]
- Kliebenstein, D. J., J. Gershenzon and T. Mitchell-Olds, 2001. Comparative quantitative trait loci mapping of aliphatic, indolic and benzylic glucosinolate production in Arabidopsis thaliana leaves and seeds. Genetics 159**:** 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komulainen, P., G. R. Brown, M. Mikkonen, A. Karhu, M. R. Garcia-Gil et al., 2003. Comparing EST-based genetic maps between Pinus sylvestris and Pinus taeda. Theor. Appl. Genet. 107**:** 667–678. [DOI] [PubMed] [Google Scholar]
- Krutovskii, K. V., S. S. Vollmer, F. C. Sorensen, W. Th. Adams, S. J. Knapp et al., 1998. RAPD genome maps of Douglas-fir. J. Hered. 89**:** 197–205. [Google Scholar]
- Lan, T. H., T. A. DelMonte, K. P. Reischmann, J. Hyman, S. P. Kowalski et al., 2000. An EST-enriched comparative map of Brassica oleracea and Arabidopsis thaliana. Genome Res. 10**:** 776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, D. A., and K. M. Devos, 2002. Trends in comparative genetics and their potential impacts on wheat and barley research. Plant Mol. Biol. 48**:** 729–740. [DOI] [PubMed] [Google Scholar]
- Leitch, I. J., L. Hanson, M. Winfield, J. Parker and M. D. Bennett, 2001. Nuclear DNA C-values complete familial representation in gymnosperms. Ann. Bot. 88**:** 843–849. [Google Scholar]
- Lukens, L., F. Zou, D. Lydiate, I. Parkin and T. Osborn, 2003. Comparison of a Brassica oleracea genetic map with the genome of Arabidopsis thaliana. Genetics 164**:** 359–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques, C. M., R. P. V. Brondani, D. Grattapaglia and R. Sederoff, 2002. Conservation and synteny of SSR loci and QTLs for vegetative propagation in four Eucalyptus species. Theor. Appl. Genet. 105**:** 474–478. [DOI] [PubMed] [Google Scholar]
- Mirny, L. A., and M. S. Gelfand, 2002. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J. Mol. Biol. 321**:** 7–20. [DOI] [PubMed] [Google Scholar]
- Morton, N. E., 1991. Parameters of the human genome. Proc. Natl. Acad. Sci. USA 88**:** 7474–7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, W. J., R. Stanyon and S. J. O'Brien, 2001. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2**:** 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, B., 1998. Nuclear DNA amounts in gymnosperms. Ann. Bot. 82**(Suppl. A):** 3–15. [Google Scholar]
- Ott, J., 1991 Analysis of Human Genetic Linkage. Johns Hopkins University Press, Baltimore/London.
- Paterson, A. H., J. E. Bowers, M. D. Burow, X. Draye, C. G. Elsik et al., 2000. Comparative genomics of plant chromosomes. Plant Cell 12**:** 1523–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, D. J., and J. Bousquet, 1998. a Sequence-tagged-site (STS) markers of arbitrary genes: development, characterization and analysis of linkage in Black spruce. Genetics 149**:** 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry, D. J., and J. Bousquet, 1998. b Sequence-Tagged-Site (STS) markers of arbitrary genes: the utility of black spruce-derived primers in other conifers. Theor. Appl. Genet. 97**:** 735–743. [Google Scholar]
- Prager, E. M., D. P. Fowler and A. C. Wilson, 1976. Rates of evolution of conifers. Evolution 30**:** 637–649. [DOI] [PubMed] [Google Scholar]
- Rydin, C., M. Källersjö and E. M. Friis, 2002. Seed plant relationships and the systematic position of Gnetales based on nuclear and chloroplast DNA: conflicting data, rooting problems, and the monophyly of conifers. Int. J. Plant Sci. 163**:** 197–214. [Google Scholar]
- Sankoff, D., and J. H. Nadeau (Editors), 2000 Comparative Genomics: Empirical and Analytical Approaches to Gene Order Dynamics, Map Alignment and the Evolution of Gene Families (Computational Biology Series, Vol. 1). Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Sasaki, T., and R. R. Sederoff, 2003. Genome studies and molecular genetics. The rice genome and comparative genomics of higher plants. Curr. Opin. Plant Biol. 6**:** 97–100. [Google Scholar]
- Schmidt, R., 2002. Plant genome evolution: lessons from comparative genomics at the DNA level. Plant Mol. Biol. 48**:** 21–37. [PubMed] [Google Scholar]
- Schubert, R., G. Mueller-Starck and R. Riegel, 2001. Development of EST-PCR markers and monitoring their intrapopulational genetic variation in Picea abies (L.) Karst. Theor. Appl. Genet. 103**:** 1223–1231. [Google Scholar]
- Sewell, M. M., B. K. Sherman and D. B. Neale, 1999. A consensus map for loblolly pine (Pinus taeda L.). I. Construction and integration of individual linkage maps from two outbred three-generation pedigrees. Genetics 151**:** 321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewell, M. M., D. L. Bassoni, R. A. Megraw, N. C. Wheeler and D. B. Neale, 2000. Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). I. Physical wood properties. Theor. Appl. Genet. 101**:** 1273–1281. [DOI] [PubMed] [Google Scholar]
- Sewell, M. M., M. F. Davis, G. A. Tuskan, N. C. Wheeler, C. C. Elam et al., 2002. Identification of QTLs influencing wood property traits in loblolly pine (Pinus taeda L.). II. Chemical wood properties. Theor. Appl. Genet. 104**:** 214–222. [DOI] [PubMed] [Google Scholar]
- Sherman, J. D., A. L. Fenwick, D. M. Namuth and N. L. V. Lapitan, 1995. A barley RFLP map: alignment of three barley maps and comparisons to Gramineae species. Theor. Appl. Genet. 91**:** 681–690. [DOI] [PubMed] [Google Scholar]
- Smilde, W. D., J. Haluškova, T. Sasaki and A. Graner, 2001. New evidence for the synteny of rice chromosome 1 and barley chromosome 3 H from rice expressed sequence tags. Genome 44**:** 361–367. [PubMed] [Google Scholar]
- Stam, P., 1993. Construction of integrated genetic linkage maps by means of a new computer package: JoinMap. Plant J. 3**:** 739–744. [Google Scholar]
- Stam, P., and J. W. Van Ooijen, 1995 JOINMAP Version 2.0: Software for the Calculation of Genetic Linkage Maps. CPRO-DLO, Wageningen, The Netherlands.
- Temesgen, B., D. B. Neale and D. E. Harry, 2000. Use of haploid mixtures and heteroduplex analysis enhance polymorphisms revealed by denaturing gradient gel electrophoresis. BioTechniques 28**:** 114–122. [DOI] [PubMed] [Google Scholar]
- Temesgen, B., G. R. Brown, D. E. Harry, C. S. Kinlaw, M. M. Sewell et al., 2001. Genetic mapping of expressed sequence tag polymorphism (ESTP) markers in loblolly pine (Pinus taeda L.). Theor. Appl. Genet. 102**:** 664–675. [Google Scholar]
- Yan, H. H., J. Mudge, D. J. Kim, D. Larsen, R. C. Shoemaker et al., 2003. Estimates of conserved microsynteny among the genomes of Glycine max, Medicago truncatula and Arabidopsis thaliana. Theor. Appl. Genet. 106**:** 1256–1265. [DOI] [PubMed] [Google Scholar]
- Wang, X.-Q., D. C. Tank and T. Sang, 2000. Phylogeny and divergence times in Pinaceae: evidence from three genomes. Mol. Biol. Evol. 17**:** 773–781. [DOI] [PubMed] [Google Scholar]
- Ware, D., and L. Stein, 2003. Comparison of genes among cereals. Curr. Opin. Plant Biol. 6**:** 121–127. [DOI] [PubMed] [Google Scholar]
- Ware, D. H., P. Jaiswal, J. Ni, I. V. Yap, X. Pan et al., 2002. Gramene, a tool for grass genomics. Plant Physiol. 130**:** 1606–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetten, R., Y. H. Sun, Y. Zhang and R. Sederoff, 2001. Functional genomics and cell wall biosynthesis in loblolly pine. Plant Mol. Biol. 47**:** 275–291. [PubMed] [Google Scholar]
- Zhang, J., H. G. Zheng, A. Aarti, G. Pantuwan, T. T. Nguyen et al., 2001. Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor. Appl. Genet. 103**:** 19–29. [Google Scholar]
- Zhu, H., D.-J. Kim, J.-M. Baek, H.-K. Choi, L. C. Ellis et al., 2003. Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol. 131**:** 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]