Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells (original) (raw)
Abstract
RNA interference (RNAi) was first recognized in Caenorhabditis elegans as a biological response to exogenous double-stranded RNA (dsRNA), which induces sequence-specific gene silencing. RNAi represents a conserved regulatory motif, which is present in a wide range of eukaryotic organisms. Recently, we and others have shown that endogenously encoded triggers of gene silencing act through elements of the RNAi machinery to regulate the expression of protein-coding genes. These small temporal RNAs (stRNAs) are transcribed as short hairpin precursors (∼70 nt), processed into active, 21-nt RNAs by Dicer, and recognize target mRNAs via base-pairing interactions. Here, we show that short hairpin RNAs (shRNAs) can be engineered to suppress the expression of desired genes in cultured Drosophila and mammalian cells. shRNAs can be synthesized exogenously or can be transcribed from RNA polymerase III promoters in vivo, thus permitting the construction of continuous cell lines or transgenic animals in which RNAi enforces stable and heritable gene silencing.
Keywords: RNAi, gene silencing, miRNA, shRNA, siRNA
An understanding of the biological role of any gene comes only after observing the phenotypic consequences of altering the function of that gene in a living cell or organism. In many cases, those organisms for which convenient methodologies for genetic manipulation exist blaze the trail toward an understanding of similar genes in less tractable organisms, such as mammals. The advent of RNA interference (RNAi) as an investigational tool has shown the potential to democratize at least one aspect of genetic manipulation, the creation of hypomorphic alleles, in organisms ranging from unicellular parasites (e.g., Shi et al. 2000) to mammals (Svoboda et al. 2000; Wianny and Zernicka-Goetz 2000).
Although Caenorhabditis elegans has, for some time, been well developed as a forward genetic system, the lack of methodologies for gene replacement by homologous recombination presented a barrier to assessing rapidly the consequences of loss of function in known genes. In an effort to overcome this limitation, Mello and Fire (Fire et al. 1998), building on earlier studies (Guo and Kemphues 1995), probed the utility of antisense RNA as a method for suppressing gene expression in worms. Through these efforts, they found that double-stranded RNA (dsRNA) was much more effective than antisense RNA as an inducer of gene silencing. Subsequent studies have shown that RNAi is a conserved biological response that is present in many, if not most, eukaryotic organisms (for review, see Bernstein et al. 2001b; Hammond et al. 2001b).
As a result of biochemical and genetic approaches in several experimental systems, the mechanisms underlying RNAi have begun to unfold (for review, see Bernstein et al. 2001b; Hammond et al. 2001b). These suggest the existence of a conserved machinery for dsRNA-induced gene silencing, which proceeds via a two-step mechanism. In the first step, the dsRNA silencing trigger is recognized by an RNase III family nuclease called Dicer, which cleaves the dsRNA into ∼21–23-nt siRNAs (small interfering RNAs). These siRNAs are incorporated into a multicomponent nuclease complex, RISC, which identifies substrates through their homology to siRNAs and targets these cognate mRNAs for destruction.
Although it was clear from the outset that RNAi would prove a powerful tool for manipulating gene expression in invertebrates, there were several potential impediments to the use of this approach in mammalian cells. Most mammalian cells harbor a potent antiviral response that is triggered by the presence of dsRNA viral replication intermediates. A key component of this response is a dsRNA-activated protein kinase, PKR, which phosphorylates EIF-2α, inducing, in turn, a generalized inhibition of translation (for review, see Williams 1997; Gil and Esteban 2000). In addition, dsRNA activates the 2‘5′ oligoadenylate polymerase/RNase L system and represses IκB. The ultimate outcome of this set of responses is cell death via apoptosis.
Therefore, it came as a welcome surprise that dsRNA could induce sequence-specific silencing in mammalian embryos, which apparently lack generalized responses to dsRNA (Svoboda et al. 2000; Wianny and Zernicka-Goetz 2000). Indeed, microinjection of dsRNA into mouse zygotes could specifically silence both exogenous reporters and endogenous genes to create anticipated phenotypes. Subsequently, these observations were extended to embryonic cell lines, such as embryonic stem cells and embryonal carcinoma cells, which do not show generic translational repression in response to dsRNA (Billy et al. 2001; Yang et al. 2001; Paddison et al. 2002). However, restriction of conventional RNAi to these few embryonic and cell culture systems would place a significant limitation on the utility of this approach in mammals.
Tuschl and colleagues first showed that short RNA duplexes, designed to mimic the products of the Dicer enzyme, could trigger RNA interference in vitro in Drosophila embryo extracts (Tuschl et al. 1999; Elbashir et al. 2001b,c). This observation was extended to mammalian somatic cells by Tuschl and coworkers (Elbashir et al. 2001a) and by Fire and colleagues (Caplen et al. 2001), who showed that chemically synthesized siRNAs could induce gene silencing in a wide range of human and mouse cell lines. The use of synthetic siRNAs to transiently suppress the expression of target genes is quickly becoming a method of choice for probing gene function in mammalian cells.
Dicer, the enzyme that normally produces siRNAs in vivo, has been linked to RNA interference both through biochemistry and through genetics (Bernstein et al. 2001a; Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001). Indeed, C. elegans animals that lack Dicer are RNAi-deficient, at least in some tissues. However, these animals also have additional phenotypic abnormalities. Specifically, they are sterile and show a number of developmental abnormalities that typify alterations in developmental timing. Indeed, the phenotypes of the Dicer mutant animals were similar to those previously observed for animals carrying mutations in the let-7 gene (Reinhart et al. 2000).
The let-7 gene encodes a small, highly conserved RNA species that regulates the expression of endogenous protein-coding genes during worm development. The active RNA species is transcribed initially as an ∼70-nt precursor, which is posttranscriptionally processed into a mature ∼21-nt form (Reinhart et al. 2000). Both in vitro and in vivo data from C. elegans (Grishok et al. 2001; Ketting et al. 2001; Knight and Bass 2001) and human cells (Hutvagner et al. 2001) have pointed to Dicer as the enzyme responsible for let-7 maturation and for the maturation of a similar small RNA, lin-4 (Grishok et al. 2001). Thus, at least some components of the RNAi machinery respond to endogenously encoded triggers to regulate the expression of target genes.
Recent studies have placed let-7 and lin-4 as the founding members of a potentially very large group of small RNAs known generically as micro-RNAs (miRNAs). Nearly 100 potential miRNAs have now been identified in Drosophila, C. elegans, and mammals (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Although the functions of these diverse RNAs remain mysterious, it seems likely that they, like let-7 and lin-4, are transcribed as hairpin RNA precursors, which are processed to their mature forms by Dicer (Lee and Ambros 2001; E. Bernstein, unpubl.).
Since the realization that small, endogenously encoded hairpin RNAs could regulate gene expression via elements of the RNAi machinery, we have sought to exploit this biological mechanism for the regulation of desired target genes. Here we show that short hairpin RNAs (shRNAs) can induce sequence-specific gene silencing in mammalian cells. As is normally done with siRNAs, silencing can be provoked by transfecting exogenously synthesized hairpins into cells. However, silencing can also be triggered by endogenous expression of shRNAs. This observation opens the door to the production of continuous cells lines in which RNAi is used to stably suppress gene expression in mammalian cells. Furthermore, similar approaches should prove efficacious in the creation of transgenic animals and potentially in therapeutic strategies in which long-term suppression of gene function is essential to produce a desired effect.
Results
Short hairpin RNAs trigger gene silencing in Drosophila cells
Several groups (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001) have shown that endogenous triggers of gene silencing, specifically small temporal RNAs (stRNAs) let-7 and lin-4, function at least in part through RNAi pathways. Specifically, these small RNAs are encoded by hairpin precursors that are processed by Dicer into mature, ∼21-nt forms. Moreover, genetic studies in C. elegans have shown a requirement for Argonaute-family proteins in stRNA function. Specifically, alg-1 and alg-2, members of the EIF2c subfamily, are implicated both in stRNA processing and in their downstream effector functions (Grishok et al. 2001). We have recently shown that a component of RISC, the effector nuclease of RNAi, is a member of the Argonaute family, prompting a model in which stRNAs may function through RISC-like complexes, which regulate mRNA translation rather than mRNA stability (Hammond et al. 2001a).
We wished to test the possibility that we might retarget these small, endogenously encoded hairpin RNAs to regulate genes of choice with the ultimate goal of subverting this regulatory system for manipulating gene expression stably in mammalian cell lines and in transgenic animals. Whether triggered by long dsRNAs or by siRNAs, RNAi is generally more potent in the suppression of gene expression in Drosophila S2 cells than in mammalian cells. We therefore chose this model system in which to test the efficacy of short hairpin RNAs (shRNAs) as inducers of gene silencing.
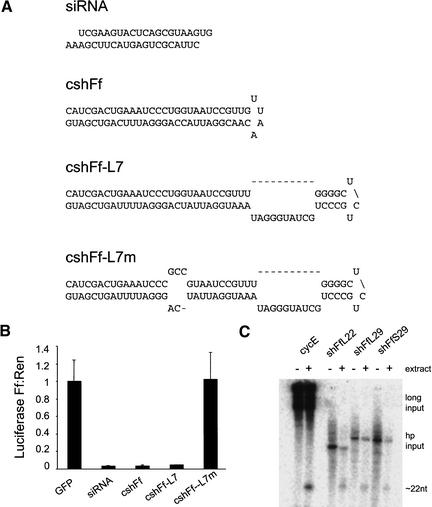
Neither stRNAs nor the broader group of miRNAs that has recently been discovered form perfect hairpin structures. Indeed, each of these RNAs is predicted to contain several bulged nucleotides within their rather short (∼30-nt) stem structures. Because the position and character of these bulged nucleotides have been conserved throughout evolution and among at least a subset of miRNAs, we sought to design retargeted miRNA mimics to conserve these predicted structural features. Only the let-7 and lin-4 miRNAs have known mRNA targets (Wightman et al. 1993; Slack et al. 2000). In both cases, pairing to binding sites within the regulated transcripts is imperfect, and in the case of lin-4, the presence of a bulged nucleotide is critical to suppression (Ha et al. 1996). We therefore also designed shRNAs that paired imperfectly with their target substrates. A subset of these shRNAs is depicted in Figure 1A.
Figure 1.
Short hairpins suppress gene expression in Drosophila S2 cells. (A) Sequences and predicted secondary structure of representative chemically synthesized RNAs. Sequences correspond to positions 112–134 (siRNA) and 463–491 (shRNAs) of Firefly luciferase carried on pGL3-Control. An siRNA targeted to position 463–485 of the luciferase sequence was virtually identical to the 112–134 siRNA in suppressing expression, but is not shown. (B) Exogenously supplied short hairpins suppress expression of the targeted Firefly luciferase gene in vivo. Six-well plates of S2 cells were transfected with 250 ng/well of plasmids that direct the expression of firefly and Renilla luciferase and 500 ng/well of the indicated RNA. Luciferase activities were assayed 48 h after transfection. Ratios of firefly to Renilla luciferase activity were normalized to a control transfected with an siRNA directed at the green fluorescent protein (GFP). The average of three independent experiments is shown; error bars indicate standard deviation. (C) Short hairpins are processed by the Drosophila Dicer enzyme. T7 transcribed hairpins shFfL22, shFfL29, and shFfS29 were incubated with (+) and without (−) 0–2-h Drosophila embryo extracts. Those incubated with extract produced ∼22-nt siRNAs, consistent with the ability of these hairpins to induce RNA interference. A long dsRNA input (cyclin E 500-mer) was used as a control. Cleavage reactions were performed as described in Bernstein et al. (2001a).
To permit rapid testing of large numbers of shRNA variants and quantitative comparison of the efficacy of suppression, we chose to use a dual-luciferase reporter system, as previously described for assays of RNAi in both Drosophila extracts (Tuschl et al. 1999) and mammalian cells (Caplen et al. 2001; Elbashir et al. 2001a). Cotransfection of firefly and Renilla luciferase reporter plasmids with either long dsRNAs or with siRNAs homologous to the firefly luciferase gene yielded an ∼95% suppression of firefly luciferase without effect on Renilla luciferase (Fig. 1B; data not shown). Firefly luciferase could also be specifically silenced by cotransfection with homologous shRNAs. Surprisingly, those shRNAs modeled most closely on the let-7 paradigm were the least effective inducers of silencing (data not shown). The inclusion of bulged nucleotides within the shRNA stem caused only a modest reduction in potency; however, the presence of mismatches with respect to the target mRNA essentially abolished silencing potential. The most potent inhibitors were those composed of simple hairpin structures with complete homology to the substrate. Introduction of G-U basepairs either within the stem or within the substrate recognition sequence had little or no effect (Fig. 1A,B; data not shown). Similarly, varying either the loop size from ∼4 to 23 bases or the loop sequence (e.g., to mimic let-7) also proved neutral (data not shown).
These results show that short hairpin RNAs can induce gene silencing in Drosophila S2 cells with potency similar to that of siRNAs (Fig. 1B). However, in our initial observation of RNA interference in Drosophila S2 cells, we noted a profound dependence of the efficiency of silencing on the length of the dsRNA trigger (Hammond et al. 2000). Indeed, dsRNAs of fewer than ∼200 nt triggered silencing very inefficiently. Silencing is initiated by an RNase III family nuclease, Dicer, that processes long dsRNAs into ∼22-nt siRNAs. In accord with their varying potency as initiators of silencing, long dsRNAs are processed much more readily than short RNAs by the Dicer enzyme (Bernstein et al. 2001a). We therefore tested whether shRNAs were substrates for the Dicer enzyme.
We had noted previously that let-7 (Ketting et al. 2001) and other miRNAs (E. Bernstein, unpubl.) are processed by Dicer with an unexpectedly high efficiency as compared with short, nonhairpin dsRNAs. Similarly, Dicer efficiently processed shRNAs that targeted firefly luciferase, irrespective of whether they were designed to mimic a natural Dicer substrate (let-7) or whether they were simple hairpin structures (Fig. 1C). These data suggest that recombinant shRNAs can be processed by Dicer into siRNAs and are consistent with the idea that these short hairpins trigger gene silencing via an RNAi pathway.
Short hairpin activated gene silencing in mammalian cells
RNAi is developing into an increasingly powerful methodology for manipulating gene expression in diverse experimental systems. However, mammalian cells contain several endogenous systems that were predicted to hamper the application of RNAi. Chief among these is a dsRNA-activated protein kinase, PKR, which effects a general suppression of translation via phosphorylation of EIF-2α (Williams 1997; Gil and Esteban 2000). Activation of these, and other dsRNA-responsive pathways, generally requires duplexes exceeding 30 bp in length, possibly to permit dimerization of the enzyme on its allosteric activator (e.g., Clarke and Mathews 1995).
Small RNAs that mimic Dicer products, siRNAs, presumably escape this limit and trigger specific silencing, in part because of their size. However, short duplex RNAs that lack signature features of siRNAs can efficiently induce silencing in Drosophila S2 cells but not in mammalian cells (A.A. Caudy, unpubl.). Endogenously encoded miRNAs may also escape PKR surveillance because of their size but perhaps also because of the discontinuity of their duplex structure. Given that shRNAs of <30 bp were effective inducers of RNAi in Drosophila S2 cells, we tested whether these RNAs could also induce sequence-specific silencing in mammalian cells.
Human embryonic kidney (HEK293T) cells were cotransfected with chemically synthesized shRNAs and with a mixture of firefly and Renilla luciferase reporter plasmids. As had been observed in S2 cells, shRNAs were effective inducers of gene silencing. Once again, hairpins designed to mimic let-7 were consistently less effective than were simple hairpin RNAs, and the introduction of mismatches between the antisense strand of the shRNA and the mRNA target abolished silencing (Fig. 2A; data not shown). Overall, shRNAs were somewhat less potent silencing triggers than were siRNAs. Whereas siRNAs homologous to firefly luciferase routinely yielded ∼90%–95% suppression of gene expression, suppression levels achieved with shRNAs ranged from 80%–90% on average. As we also observe with siRNAs, the most important determinant of the potency of the silencing trigger is its sequence. We find that roughly 50% of both siRNAs and shRNAs are competent for suppressing gene expression. However, neither analysis of the predicted structures of the target mRNA nor analysis of alternative structures in siRNA duplexes or shRNA hairpins has proved of predictive value for choosing effective inhibitors of gene expression.
Figure 2.
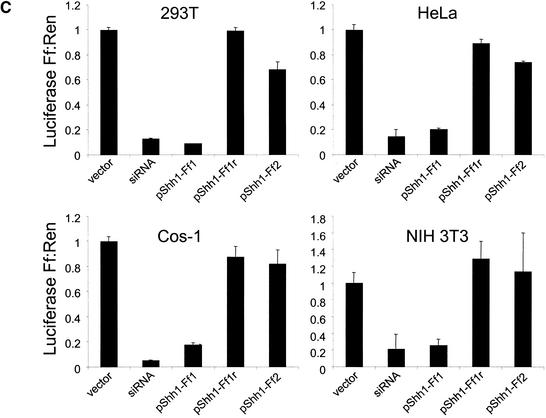
Short hairpins function in mammalian cells. HEK 293T, HeLa, COS-1, and NIH 3T3 cells were transfected with plasmids and RNAs as in Figure 1 and subjected to dual luciferase assays 48 h posttransfection. The ratios of firefly to Renilla luciferase activity are normalized to a control transfected with an siRNA directed at the green fluorescent protein (GFP). The average of three independent experiments is shown; error bars indicate standard deviation.
We have adopted as a standard, shRNA duplexes containing 29 bp. However, the size of the helix can be reduced to ∼25 nt without significant loss of potency. Duplexes as short as 22 bp can still provoke detectable silencing, but do so less efficiently than do longer duplexes. In no case do we observe a reduction in the internal control reporter (Renilla luciferase) that would be consistent with an induction of nonspecific dsRNA responses.
The ability of shRNAs to induce gene silencing was not confined to 293T cells. Similar results were also obtained in a variety of other mammalian cell lines, including human cancer cells (HeLa), transformed monkey epithelial cells (COS-1), murine fibroblasts (NIH 3T3), and diploid human fibroblasts (IMR90; Fig. 2; data not shown).
Synthesis of effective inhibitors of gene expression using T7 RNA polymerase
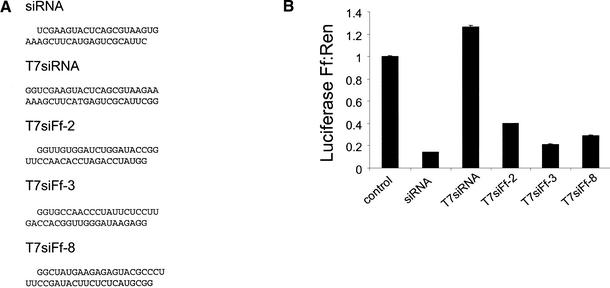
The use of siRNAs to provoke gene silencing is developing into a standard methodology for investigating gene function in mammalian cells. To date, siRNAs have been produced exclusively by chemical synthesis (e.g., Caplen et al. 2001; Elbashir et al. 2001a). However, the costs associated with this approach are significant, limiting its potential utility as a tool for investigating in parallel the functions of large numbers of genes. Short hairpin RNAs are presumably processed into active siRNAs in vivo by Dicer (see Fig. 1C). Thus, these may be more tolerant of terminal structures, both with respect to nucleotide overhangs and with respect to phosphate termini. We therefore tested whether shRNAs could be prepared by in vitro transcription with T7 RNA polymerase.
Transcription templates that were predicted to generate siRNAs and shRNAs similar to those prepared by chemical RNA synthesis were prepared by DNA synthesis (Fig. 3A,C). These were tested for efficacy both in S2 cells (data not shown) and in human 293 cells (Fig. 3B,D). Overall, the performance of the T7-synthesized hairpin or siRNAs closely matched the performance of either produced by chemical synthesis, both with respect to the magnitude of inhibition and with respect to the relative efficiency of differing sequences. Because T7 polymerase prefers to initiate at twin guanosine residues, however, it was critical to consider initiation context when designing in vitro transcribed siRNAs (Fig. 3B). In contrast, shRNAs, which are processed by Dicer (see Fig. 1C), tolerate the addition of these bases at the 5′ end of the transcript.
Figure 3.
siRNAs and short hairpins transcribed in vitro suppress gene expression in mammalian cells. (A) Sequences and predicted secondary structure of representative in vitro transcribed siRNAs. Sequences correspond to positions 112–134 (siRNA) and 463–491 (shRNAs) of firefly luciferase carried on pGL3-Control. (B) In vitro transcribed siRNAs suppress expression of the targeted firefly luciferase gene in vivo. HEK 293T cells were transfected with plasmids as in Figure 2. The presence of non-base-paired guanosine residues at the 5′ end of siRNAs significantly alters the predicted end structure and abolishes siRNA activity. (C) Sequences and predicted secondary structure of representative in vitro transcribed shRNAs. Sequences correspond to positions 112–141 of firefly luciferase carried on pGL3-Control. (D) Short hairpins transcribed in vitro suppress expression of the targeted firefly luciferase gene in vivo. HEK 293T cells were transfected with plasmids as in Figure 2.
Studies in Drosophila embryo extracts indicate that siRNAs possess 5′ phosphorylated termini, consistent with their production by an RNase III family nuclease (Bernstein et al. 2001a; Elbashir et al. 2001b). In vitro, this terminus is critical to the induction of RNAi by synthetic RNA oligonucleotides (Elbashir et al. 2001c; Nykanen et al. 2001). Chemically synthesized siRNAs are nonphosphorylated, and enzymatic addition of a 5′ phosphate group in vitro prior to transfection does not increase the potency of the silencing effect (A.A. Caudy, unpubl.). This suggests either that the requirement for phosphorylated termini is less stringent in mammalian cells or that a kinase efficiently phosphorylates siRNAs in vivo. RNAs synthesized with T7 RNA polymerase, however, possess 5′ triphosphate termini. We therefore explored the possibility of synthesizing siRNAs with T7 polymerase followed by treatment in vitro with pyrophosphatase to modify the termini to resemble those of siRNAs. Surprisingly, monophosphorylated siRNAs (data not shown) were as potent in inducing gene silencing as transcription products bearing triphosphate termini (Fig. 3B). This may suggest either that the requirement for monophosphorylated termini is less stringent in mammalian cells or that siRNAs are modified in vivo to achieve an appropriate terminal structure.
Considered together, our data suggest that both shRNAs and siRNA duplexes can be prepared by synthesis with T7 RNA polymerase in vitro. This significantly reduces the cost of RNAi in mammalian cells and paves the way for application of RNAi on a whole-genome scale.
Transcription of shRNAs in vivo by RNA polymerase III
Although siRNAs are an undeniably effective tool for probing gene function in mammalian cells, their suppressive effects are by definition of limited duration. Delivery of siRNAs can be accomplished by any of a number of transient transfection methodologies, and both the timing of peak suppression and the recovery of protein levels as silencing decays can vary with both the cell type and the target gene (Y. Seger and E. Bernstein, unpubl.). Therefore, one limitation on siRNAs is the development of continuous cell lines in which the expression of a desired target is stably silenced.
Hairpin RNAs, consisting of long duplex structures, have been proved as effective triggers of stable gene silencing in plants, in C. elegans, and in Drosophila (Kennerdell and Carthew 2000; Smith et al. 2000; Tavernarakis et al. 2000). We have recently shown stable suppression of gene expression in cultured mammalian cells by continuous expression of a long hairpin RNA (Paddison et al. 2002). However, the scope of this approach was limited by the necessity of expressing such hairpins only in cells that lack a detectable PKR response. In principle, shRNAs could bypass such limitations and provide a tool for evoking stable suppression by RNA in mammalian somatic cells.
To test this possibility, we initially cloned sequences encoding a firefly luciferase shRNA into a CMV-based expression plasmid. This was predicted to generate a capped, polyadenylated RNA polymerase II transcript in which the hairpin was extended on both the 5′ and 3′ ends by vector sequences and poly(A). This construct was completely inert in silencing assays in 293T cells (data not shown).
During our studies on chemically and T7-synthesized shRNAs, we noted that the presence of significant single-stranded extensions (either 5′ or 3′ of the duplex) reduced the efficacy of shRNAs (data not shown). We therefore explored the use of alternative promoter strategies in an effort to produce more defined hairpin RNAs. In particular, RNA polymerase III promoters have well-defined initiation and termination sites and naturally produce a variety of small, stable RNA species. Although many Pol III promoters contain essential elements within the transcribed region, limiting their utility for our purposes; class III promoters use exclusively nontranscribed promoter sequences. Of these, the U6 snRNA promoter and the H1 RNA promoter have been well studied (Lobo et al. 1990; Hannon et al. 1991; Chong et al. 2001).
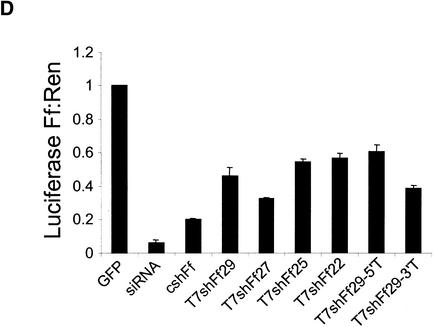
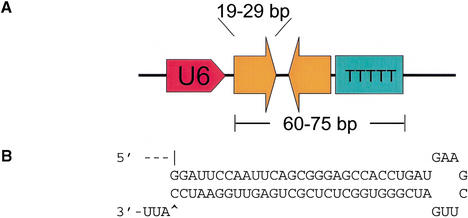
By placing a convenient cloning site immediately behind the U6 snRNA promoter, we have constructed pShh-1, an expression vector in which short hairpins are harnessed for gene silencing. Into this vector either of two shRNA sequences derived from firefly luciferase were cloned from synthetic oligonucleotides. These were cotransfected with firefly and Renilla luciferase expression plasmids into 293T cells. One of the two encoded shRNAs provoked effective silencing of firefly luciferase without altering the expression of the internal control (Fig. 4C). The second encoded shRNA also produced detectable, albeit weak, repression. In both cases, silencing was dependent on insertion of the shRNA in the correct orientation with respect to the promoter (Fig. 4C; data not shown). Although the shRNA itself is bilaterally symmetric, insertion in the incorrect orientation would affect Pol III termination and is predicted to produce a hairpin with both 5′ and 3′ single-stranded extensions. Similar results were also obtained in a number of other mammalian cell lines including HeLa, COS-1, NIH 3T3, and IMR90 (Fig. 4; data not shown). pShh1-Ff1 was, however, incapable of effecting suppression of the luciferase reporter in Drosophila cells, in which the human U6 promoter is inactive (data not shown).
Figure 4.
Transcription of functional shRNAs in vivo. (A) Schematic of the pShh1 vector. Sequences encoding shRNAs with between 19 and 29 bases of homology to the targeted gene are synthesized as 60–75-bp double-stranded DNA oligonucleotides and ligated into an _Eco_RV site immediately downstream of the U6 promoter. (B) Sequence and predicted secondary structure of the Ff1 hairpin. (C) An shRNA expressed from the pShh1 vector suppresses luciferase expression in mammalian cells. HEK 293T, HeLa, COS-1, and NIH 3T3 cells were transfected with reporter plasmids as in Figure 1, and pShh1 vector, firefly siRNA, or pShh1 firefly shRNA constructs as indicated. The ratios of firefly to Renilla luciferase activity were determined 48 h after transfection and represent the average of three independent experiments; error bars indicate standard deviation.
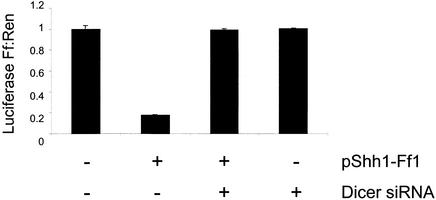
As a definitive test of whether the plasmid-encoded shRNAs brought about gene silencing via the mammalian RNAi pathway, we assessed the dependence of suppression on an essential component of the RNAi pathway. We transfected pShh1-Ff1 along with an siRNA homologous to human Dicer. Figure 5 shows that treatment of cells with Dicer siRNAs is able to completely depress the silencing induced by pShh1-Ff1. Addition of an unrelated siRNA had no effect on the magnitude of suppression by pShh1-Ff1 (data not shown). Importantly, Dicer siRNAs had no effect on siRNA-induced silencing of firefly luciferase (data not shown). These results are consistent with shRNAs operating via an RNAi pathway similar to those provoked by stRNAs and long dsRNAs. Furthermore, it suggests that siRNA-mediated silencing is less sensitive to depletion of the Dicer enzyme.
Figure 5.
Dicer is required for shRNA-mediated gene silencing. HEK 293T cells were transfected with luciferase reporter plasmids as well as pShh1-Ff1 and an siRNA targeting human Dicer either alone or in combination, as indicated. The Dicer siRNA sequence (TCA ACC AGC CAC TGC TGG A) corresponds to coordinates 3137–3155 of the human Dicer sequence. The ratios of firefly to Renilla luciferase activity were determined 26 h after transfection and represent the average of three independent experiments; error bars indicate standard deviation.
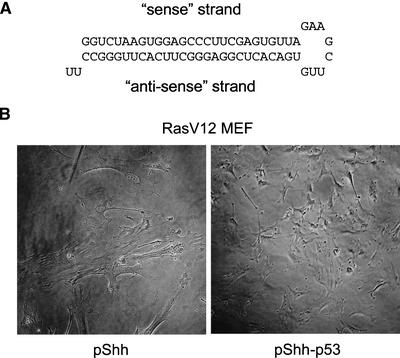
The ultimate utility of encoded short hairpins will be in the creation of stable mutants that permit the study of the resulting phenotypes. We therefore tested whether we could create a cellular phenotype through stable suppression. Expression of activated alleles of the ras oncogene in primary mouse embryo fibroblasts (MEFs) induces a stable growth arrest that resembles, as a terminal phenotype, replicative senescence (Serrano et al. 1997). Cells cease dividing and assume a typical large, flattened morphology. Senescence can be countered by mutations that inactivate the p53 tumor suppressor pathway (Serrano et al. 1997). As a test of the ability of vector-encoded shRNAs to stably suppress an endogenous cellular gene, we generated a hairpin that was targeted to the mouse p53 gene. As shown in Figure 6, MEFs transfected with pBabe-RasV12 fail to proliferate and show a senescent morphology when cotransfected with an empty control vector. As noted previously (Serrano et al. 1997), the terminally arrested state is achieved in 100% of drug-selected cells in culture by 8 d posttransfection. However, upon cotransfection of an activated ras expression construct with the pShh-p53, cells emerged from drug selection that not only fail to adopt a senescent morphology but also maintain the ability to proliferate for a minimum of several weeks in culture (Fig. 6). These data strongly suggest that shRNA expression constructs can be used for the creation of continuous mammalian cell lines in which selected target genes are stably suppressed.
Figure 6.
Stable shRNA-mediated gene silencing of an endogenous gene. (A) Sequence and predicted secondary structure of the p53 hairpin. The 5′ shRNA stem contains a 27-nt sequence derived from mouse p53 (nucleotides 166–192), whereas the 3′ stem harbors the complimentary antisense sequence. (B) Senescence bypass in primary mouse embryo fibroblasts (MEFs) expressing an shRNA targeted at p53. Wild-type MEFs, passage 5, were transfected with pBabe-RasV12 with control plasmid or with p53hp (5 μg each with FuGENE; Roche). Two days after transfection, cells were trypsinized, counted, and plated at a density of 1 × 105/10-cm plate in media containing 2.0 μg/mL of puromycin. Control cells cease proliferation and show a senescent morphology (left panel). Cells expressing the p53 hairpin continue to grow (right panel). Photos were taken 14 d posttransfection.
Discussion
The demonstration that short dsRNA duplexes can induce sequence-specific silencing in mammalian cells has begun to foment a revolution in the manner in which gene function is examined in cultured mammalian cells. These siRNAs (Elbashir et al. 2001a) mimic the products generated by Dicer (Bernstein et al. 2001a) in the initiation step of RNAi and presumably enter the silencing pathway without triggering nonspecific translational suppression via PKR. siRNAs can be used to examine the consequences of reducing the function of virtually any protein-coding gene and have proved effective in provoking relevant phenotypes in numerous somatic cell types from both humans and mice. However, a significant disadvantage of siRNAs is that their effects are transient, with phenotypes generated by transfection with such RNAs persisting for ∼1 wk. In C. elegans, RNAi has proved to be such a powerful tool, in part, because silencing is both systemic and heritable, permitting the consequences of altering gene expression to be examined throughout the development and life of an animal. We have therefore sought to expand the utility of RNAi in mammalian systems by devising methods to induce stable and heritable gene silencing. Previously, we have shown that expression of long (∼500-nt) dsRNAs could produce stable silencing in embryonic mammalian cells (Paddison et al. 2002); however, the utility of this approach was limited by its restriction to cells that lack endogenous, nonspecific responses to dsRNA, such as PKR.
Recently, a number of laboratories (Grishok et al. 2001; Hutvagner et al. 2001; Ketting et al. 2001; Knight and Bass 2001) have shown that there exist endogenously encoded triggers of RNAi-related pathways, which are transcribed as short hairpin RNAs (stRNAs, or generically miRNAs). Here, we have shown that short hairpin RNAs, modeled conceptually on miRNAs, are potent experimental tools for inducing gene silencing in mammalian somatic cells. These shRNAs can be provided exogenously or can be synthesized in vivo from RNA polymerase III promoters. Not only does this enable the creation of continuous cell lines in which suppression of a target gene is stably maintained by RNAi, but similar strategies may also be useful for the construction of transgenic animals. Thus, short-hairpin-activated gene silencing (SHAGging) provides a complement to the use of siRNAs in the study of gene function in mammalian cells. Finally, the ability to encode a constitutive silencing signal may permit the marriage of shRNA-induced silencing with in vivo and ex vivo gene delivery methods for therapeutic approaches based on stable RNAi in humans.
Materials and methods
Cell culture
HEK 293T, HeLa, COS-1, MEF, and IMR90 cells were cultured in DMEM (GIBCO BRL) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 1% antibiotic/antimycotic solution (GIBCO BRL). NIH 3T3 cells were cultured in DMEM supplemented with 10% heat-inactivated calf serum and 1% antibiotic/antimycotic solution.
RNA preparation
Both shRNAs and siRNAs were produced in vitro using chemically synthesized DNA oligonucleotide templates (Sigma) and the T7 Megashortscript kit (Ambion). Transcription templates were designed such that they contained T7 promoter sequences at the 5′ end. shRNA transcripts subjected to in vitro Dicer processing were synthesized using a Riboprobe kit (Promega). Chemically synthesized RNAs were obtained from Dharmacon, Inc.
Transfection and gene silencing assays
Cells were transfected with indicated amounts of siRNA, shRNA, and plasmid DNA using standard calcium phosphate procedures at 50%–70% confluence in 6-well plates. Dual luciferase assays (Promega) were carried out by cotransfecting cells with plasmids containing firefly luciferase under the control of the SV40 promoter (pGL3-Control, Promega) and Renilla luciferase under the control of the SV40 early enhancer/promoter region (pSV40, Promega). Plasmids were cotransfected using a 1:1 ratio of pGL3-Control (250 ng/well) to pRL-SV40. RNAi in S2 cells was performed as previously described (Hammond et al. 2000). For stable silencing, primary MEFs (a gift from S. Lowe, Cold Spring Harbor Laboratory, NY) were cotransfected using Fugene 6 with pBabe-Ha-rasV12 and pShh-p53 (no resistance marker), according to the manufacturer’s recommendations. Selection was for the presence of the activated Ha-rasV12 plasmid, which carries a puromycin-resistance marker. The pShh-p53 plasmid was present in excess, as is standard in a cotransfection experiment. We have now generated a version of the U6 promoter vector (pSHAG-1) that is compatible with the GATEWAY system (Invitrogen), and this can be used to transport the shRNA expression cassette into a variety of recipient vectors that carry _cis_-linked selectable markers. Furthermore, we have validated delivery of shRNAs using retroviral vectors. Updated plasmid information can be obtained at http://www.cshl.org/public/science/hannon.html.
Plasmids expressing hairpin RNAs
The U6 promoter region from −265 to +1 was amplified by PCR, adding 5′ _Kpn_I and 3′ _Eco_RV sites for cloning into pBSSK+. A linker/terminator oligonucleotide set bearing the U6 terminator sequence and linker ends of 5′ _Eco_RV and 3′ _Not_I was cloned into the promoter construct, resulting in a U6 cassette with an _Eco_RV site for insertion of new sequences. This vector has been named pShh1. Blunt-ended, double-stranded DNA oligonucleotides encoding shRNAs with between 19 and 29 bases of homology to the targeted gene were ligated into the _Eco_RV site to produce expression constructs. The oligonucleotide sequence used to construct Ff1 was: TCCAATTCAGCGGGAGCCACC TGATGAAGCTTGATCGGGTGGCTCTCGCTGAGTTGGA ATCCATTTTTTTT. This sequence is preceded by the sequence GGAT, which is supplied by the vector, and contains a tract of more than five Ts as a Pol III terminator.
In vitro Dicer assays
In vitro assays for Dicer activity were performed as described (Bernstein et al. 2001a).
Acknowledgments
We thank members of the Hannon laboratory for critical reading of the manuscript. P.J.P. is an Arnold and Mabel Beckman Anderson Fellow of the Watson School of Biological Sciences, and thanks Richard M. Paddison for academic support. A.A.C. is a George A. and Marjorie H. Anderson Fellow of the Watson School of Biological Sciences and a predoctoral fellow of the Howard Hughes Medical Institute. G.J.H. is a Rita Allen Foundation scholar. This work was supported in part by a grant from the NIH (RO1-GM62534, GJH) and by a grant from Genetica, Inc. (Cambridge, MA).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hannon@cshl.org; FAX (516) 367-8874.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.981002.
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001a;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Denli AM, Hannon GJ. The rest is silence. RNA. 2001b;7:1509–1521. [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci. 2001;98:14428–14433. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA. Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong SS, Hu P, Hernandez N. Reconstitution of transcription from the human U6 small nuclear RNA promoter with eight recombinant polypeptides and a partially purified RNA polymerase III complex. J Biol Chem. 2001;276:20727–20734. doi: 10.1074/jbc.M100088200. [DOI] [PubMed] [Google Scholar]
- Clarke PA, Mathews MB. Interactions between the double-stranded RNA binding motif and RNA: Definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA. 1995;1:7–20. [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001c;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Gil J, Esteban M. Induction of apoptosis by the dsRNA-dependent protein kinase (PKR): Mechanism of action. Apoptosis. 2000;5:107–114. doi: 10.1023/a:1009664109241. [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Ha I, Wightman B, Ruvkun G. A bulged lin-4/lin-14 RNA duplex is sufficient for Caenorhabditis elegans lin-14 temporal gradient formation. Genes & Dev. 1996;10:3041–3050. doi: 10.1101/gad.10.23.3041. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001a;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Hammond SM, Caudy AA, Hannon GJ. Post-transcriptional gene silencing by double-stranded RNA. Nat Rev Genet. 2001b;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Chubb A, Maroney PA, Hannon G, Altman S, Nilsen TW. Multiple cis-acting elements are required for RNA polymerase III transcription of the gene encoding H1 RNA, the RNA component of human RNase P. J Biol Chem. 1991;266:22796–22799. [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Kennerdell JR, Carthew RW. Heritable gene silencing in Drosophila using double-stranded RNA. Nat Biotechnol. 2000;18:896–898. doi: 10.1038/78531. [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL. A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science. 2001;293:2269–2271. doi: 10.1126/science.1062039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lobo SM, Ifill S, Hernandez N. cis-Acting elements required for RNA polymerase II and III transcription in the human U2 and U6 snRNA promoters. Nucleic Acids Res. 1990;18:2891–2899. doi: 10.1093/nar/18.10.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Paddison P, Caudy AA, Hannon GJ. Stable suppression of gene expression in mammalian cells by RNAi. Proc Natl Acad Sci. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Shi H, Djikeng A, Mark T, Wirtz E, Tschudi C, Ullu E. Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA. 2000;6:1069–1076. doi: 10.1017/s1355838200000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Smith NA, Singh SP, Wang MB, Stoutjesdijk PA, Green AG, Waterhouse PM. Total silencing by intron-spliced hairpin RNAs. Nature. 2000;407:319–320. doi: 10.1038/35030305. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Hayashi H, Schultz RM. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development. 2000;127:4147–4156. doi: 10.1242/dev.127.19.4147. [DOI] [PubMed] [Google Scholar]
- Tavernarakis N, Wang SL, Dorovkov M, Ryazanov A, Driscoll M. Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat Genet. 2000;24:180–183. doi: 10.1038/72850. [DOI] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA. Targeted mRNA degradation by double-stranded RNA in vitro. Genes & Dev. 1999;13:3191–3197. doi: 10.1101/gad.13.24.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wianny F, Zernicka-Goetz M. Specific interference with gene function by double-stranded RNA in early mouse development. Nat Cell Biol. 2000;2:70–75. doi: 10.1038/35000016. [DOI] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Williams BR. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21:7807–7816. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]