Protein O-fucosyltransferase 1 is an essential component of Notch signaling pathways (original) (raw)
Abstract
Notch receptor signaling regulates cell growth and differentiation, and core components of Notch signaling pathways are conserved from Drosophila to humans. Fringe glycosyltransferases are crucial modulators of Notch signaling that act on epidermal growth factor (EGF)-like repeats in the Notch receptor extracellular domain. The substrate of Fringe is EGF-_O_-fucose and the transfer of fucose to Notch by protein _O_-fucosyltransferase 1 is necessary for Fringe to function. _O_-fucose also occurs on Cripto and on Notch ligands. Here we show that mouse embryos lacking protein _O_-fucosyltransferase 1 die at midgestation with severe defects in somitogenesis, vasculogenesis, cardiogenesis, and neurogenesis. The phenotype is similar to that of embryos lacking downstream effectors of all Notch signaling pathways such as presenilins or RBP-Jκ, and is different from Cripto, Notch receptor, Notch ligand, or Fringe null phenotypes. Protein _O_-fucosyltransferase 1 is therefore an essential core member of Notch signaling pathways in mammals.
Notch receptor signaling determines cell fate and axial boundary formations during development in metazoa (1). Dysregulated Notch signaling leads to developmental defects or cancer (2). Thus it is important to identify the essential components of Notch signaling pathways. Notch receptors are transmembrane glycoproteins with 29–36 epidermal growth factor (EGF)-like repeats in their extracellular domain. Mammalian Notch ligands (Jagged and Delta) bind to Notch receptors and induce proteolytic release of Notch intracellular domain, which, in a complex with the transcriptional repressor RBP-Jκ (CBF1), activates target genes such as Hes5 (3). _O-_Fucose on Notch EGF repeats forms the substrate of Fringe β1,3-_N_-acetylglucosaminyltransferases that act in the Notch-expressing cell to focus Notch signaling at developmental boundaries (4, 5). Fucose is attached to Ser or Thr in EGF repeats containing the consensus sequence Cys-(Xaa)3–5-Ser(Thr)-Cys between the second and third Cys residues (6, 7). It is found on urokinase-type plasminogen activator (uPA) (8), on Notch receptors and their ligands (6, 9), on Cripto (10, 11), and on other proteins, including blood clotting factors. _O-_Fucose is required for signaling by uPA through the uPA receptor (12), for Fringe modulation of Notch receptor signaling in Drosophila (4, 5) and in coculture assays (4, 13–16), and for Cripto to mediate Nodal signaling (10, 11).
The first indication that fucose on Notch may function directly in Notch signaling was that Jagged1-induced Notch signaling is reduced in Lec13 mutant cells that have low fucose in glycoproteins but normal levels of Notch at the cell surface (4, 16). If decreased Notch signaling is due to reduced _O-_fucose on Notch, inactivation of protein _O-_fucosyltransferase 1 (_O_-FucT-1) should give a Notch receptor mutant phenotype. Consistent with this prediction, RNA interference (RNAi) of Drosophila OFUT1 was recently shown to cause both Fringe-dependent and Fringe-independent developmental defects typical of Notch receptor inactivation (17). Mammals have four Notch receptors, with the Notch1 receptor being most similar to Drosophila Notch. However, unlike Drosophila, mammals require Cripto to function earlier in development than any Notch receptor (18). Thus inactivation of mouse _O_-FucT-1 encoded by the Pofut1 gene (19) might be expected to elicit severe defects in posterior trunk development and lethality at about embryonic day 7.5 (E7.5) similar to _Cripto_−/− embryos (18), or defective somitogenesis, vasculogenesis, and cardiac development with death at ≈E10.5–E11.5 typical of _Notch1_−/− mutants (20–22), or skeletal defects and perinatal death similar to Lunatic fringe (Lfng) mutants or a severe Notch signaling phenotype like that of embryos lacking presenilins 1 and 2 (23, 24) or RBP-Jκ (25–27) required for signaling through all four mammalian Notch receptors. Here we report that _Pofut1_−/− mice have a phenotype most similar to mice in which all Notch signaling pathways are blocked due to global inactivation of Notch receptor signaling.
Materials and Methods
Gene Targeting.
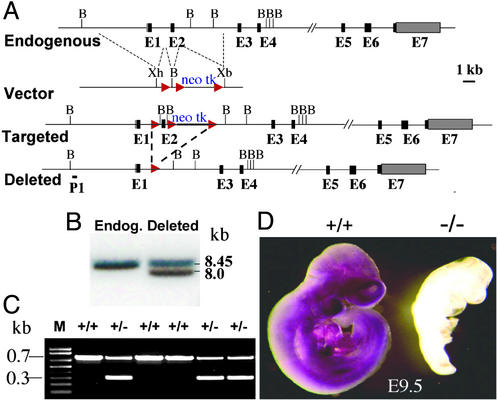
Bacterial artificial chromosome (BAC) clones containing the mouse Pofut1 gene were obtained by screening a mouse BAC library containing genomic DNA from 129svj mice (from Raju Kucherlapati, Brigham and Women's Hospital, Boston). PCR was used to generate a genomic DNA fragment containing exon 2 of the Pofut1 gene, and two flanking sequences of ≈6 kb (left) and ≈3.3 kb (right), respectively. They were inserted into the pFlox vector (from Jamey Marth, University of California at San Diego, La Jolla). After electroporation, WW6 embryonic stem (ES) cells (28) were selected with 250 μg/ml G418 (Gemini Bio-Products, Woodland, CA), and genomic DNA of survivors was subjected to PCR using primer 595 (5′-TGCAAAACCACACTGCTCGATCCG-3′) in the thymidine kinase gene and primer ssp9 (5′-CTTGGACAAATTGGATGTATCTGGAGGATATC-3′) located outside the right flanking sequence of the targeting vector. PCR-positive clones were confirmed by Southern analysis using a genomic DNA probe P1 upstream of the left flanking sequence in the targeting vector (see Fig. 2). A Cre recombinase-expressing plasmid, MC1 (from Jamey Marth), was introduced into targeted clones, and selection of deletion mutations was performed with 2 μM ganciclovir (Roche, Boulder, CO). Southern analysis was performed by using probe P1 (see Fig. 2) to identify deletions. Heterozygous WW6 ES cells were injected into C57BL/6 blastocysts, and male chimeras were mated with C57BL/6 females. Germ-line transmission occurred with three ES lines and heterozygous Pofut1+/− progeny were mated. PCR genotyping was performed with primers 644 (5′-GGGTCACCTTCATGTACAAGTGAGTG-3′) and 645 (5′-ACCCACAGGCTGTGCAGTCTTTG-3′) that span exon 2 of the Pofut1 gene. Embryo staging was calculated by using the morning a female was found to have a vaginal plug as E0.5.
Figure 2.
Targeting of the Pofut1 gene. (A) The endogenous Pofut1 gene contains 7 coding exons (black). A 560-bp region of genomic DNA containing exon 2 with a neomycin and thymidine kinase cassette between loxP sites (red) flanked by genomic DNA in the pFlox vector was used to obtain the targeted allele. Transfection with the Cre recombinase expression plasmid pMC1 gave ES cells with exon 2 deleted. B, _Bam_HI; Xh, _Xho_I; Xb; _Xba_I. (B) Southern analysis after digestion with _Bam_HI and hybridization with the genomic DNA probe P1 gave an 8.45-kb endogenous Pofut1 gene fragment with DNA from WW6 ES cells, and an 8.0-kb mutant band with DNA from a heterozygous clone with exon 2 deleted. (C) PCR genotyping with primers 644 and 645 of a litter from a cross between Pofut1+/− heterozygotes; bands are endogenous allele (700 bp) and deleted allele (300 bp). (D) Whole-mount in situ hybridization with a Pofut1 gene antisense probe of E9.5 control (+/+) and Pofut1 null (−/−) embryos.
Southern and Northern Analysis.
Genomic DNA from WW6 ES cell colonies was prepared by using DNAzol (Invitrogen), digested with _Bam_HI, electrophoresed, transferred to nylon membrane, hybridized for 2 h in Rapid-hyb buffer (Amersham Biosciences) at 65°C with the P1 probe labeled by using the Prime-It kit (Stratagene), and washed under stringent conditions at 65°C. Blots carrying mouse adult and embryonic total RNA from Seegene (Seoul, Korea) were probed with Pofut1 cDNA full-length coding sequence labeled by using the Prime-It kit. Hybridization was performed at 42°C overnight in Ultrahyb hybridization buffer (Ambion), and the blot was washed under stringent conditions at 65°C.
Whole-Mount Embryo in Situ Hybridization and Antibody Binding.
All RNA probes were from mouse gene coding sequences: Lunatic fringe [_Lfng_; 700 bp between _Bam_HI and _Pst_I sites of the coding sequence, from Thomas Vogt, Merck Research Laboratories, West Point, PA (29)]; Notch1 extracellular region, 4.7 kb, from Jeffrey Nye, Northwestern University, Chicago (30); Delta-like 1 (Dll1) ≈2.1 kb from Achim Gossler, Institut für Molekularbiologie, Hannover, Germany (31); Jagged1 ≈1.8 kb from Tim Mitsiadis, École Normale Superieur de Lyon, Lyon, France (32); Dll3 729 bp (33), Uncx4.1 ≈1.7 kb (33), and Hes5 ≈1.3 kb (34) from Ryoichiro Kageyama, Kyoto University, Kyoto; and myogenin ≈1.5 kb from Hanh Nguyen, Albert Einstein College Medicine, New York (35). Probes were transcribed and labeled by using the DIG (digoxigenin) RNA labeling kit (Roche). Purified probe was hydrolyzed to a size of ≈500 bp and hybridized as described (36) and per D. Henrique and David Ish-Horowicz, Cancer Research UK, London (personal communication). Platelet endothelial cell adhesion molecule 1 (PECAM-1) rat antibody MEC13.3 (PharMingen) was used to detect PECAM-1 in E9.5 embryos fixed in 4% paraformaldehyde according to the methods at http://cbi.swmed.edu/ryburn/sato/htmprotocols/immunowholemount.htm.
Sagittal Sections.
Embryos collected at E9.5 were fixed in Bouins' fixative (Polysciences). Paraffin embedding was performed and sections of 7 μm were stained with hematoxylin and eosin.
Results
Embryos Lacking _O_-FucT-1 Die at Midgestation.
The Pofut1 gene is increasingly expressed during embryonic development and is present in all adult mouse tissues (Fig. 1 A and B). In situ hybridization of embryos at E11.5 (Fig. 1C) and E9.5 (Fig. 2D) revealed essentially ubiquitous expression of the Pofut1 gene with some variation in expression level. At E9.5 expression was very low by Northern analysis and in situ hybridization. A Cre/_loxP_-mediated exon 2 deletion was introduced into the Pofut1 locus of WW6 ES cells (Fig. 2A). Southern analysis identified cells that had lost exon 2 and the selection cassette after transient expression of Cre recombinase (Fig. 2B). Three independent ES clones gave Pofut1+/− heterozygotes that developed normally, but no homozygotes were obtained among nine litters from Pofut1+/− heterozygous matings (Fig. 2C). At E9.5 homozygous _Pofut1_−/− embryos were severely growth retarded, and Pofut1 gene transcripts were not detected (Fig. 2D). Deletion of exon 2 generates a stop codon in exon 3 of the Pofut1 gene that presumably leads to nonsense-mediated decay of transcripts. _Pofut1_−/− embryos were present in the expected Mendelian ratio (112 of 466) and died at ≈E10.
Figure 1.
Expression of the mouse Pofut1 gene. (A and B) Northern analysis of adult mouse tissues (A) and embryos at different stages (B). (C) Whole-mount in situ hybridization of E11.5 wild-type embryos hybridized to antisense or sense Pofut1 coding region probe.
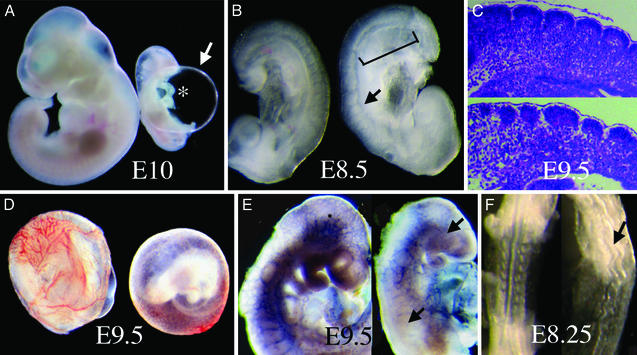
If _O-_FucT-1 is responsible for transferring the _O-_fucose required for Cripto to mediate Nodal signaling (10, 11), death of _Pofut1_−/− embryos should occur at ≈E7.5 and embryos should be devoid of posterior tissue (18). Therefore, _O_-fucose may not be required for Cripto to function before E10, or Cripto may be modified by an unrelated _O_-FucT. RT-PCR of E6.5 cDNA from _Pofut1_−/− embryos revealed no wild-type Pofut1 transcripts that might rescue a _Cripto_−/− phenotype. If _O-_FucT-1 were required merely to generate the substrate of Lunatic fringe (Lfng) (37, 38), or to modify the Notch ligands Delta-like1 (Dll1) (39), Delta-like3 (Dll3) (40, 41), Jagged1 (42) or Jagged2 (43), or Notch2 (44), death of null embryos would have occurred later in embryogenesis, or after birth, or not at all in the case of Notch4 (22) and Radical fringe (Rfng) mutants (45, 46). The time of death of _Pofut1_−/− embryos was slightly earlier than _Notch1-_null embryos (20, 21) and similar to mice lacking RBP-Jκ (25) or both presenilins 1 and 2 (23, 24), which are required for signaling by all Notch receptors. At ≈E10.5, resorption of _Pofut1_−/− embryos began, and it was complete by ≈E11.5.
Embryos Lacking _O_-FucT-1 Are Defective in Somitogenesis.
At E8.5 _Pofut1_−/− embryos (n > 20) were of normal size and appearance but somites adjacent to presomitic mesoderm (PSM) were fused (Fig. 3B). More anteriorly, five or six condensed somites of various sizes and shapes were present in _Pofut1_−/− embryos (Fig. 3B). This phenotype was similar to that of embryos lacking RBP-Jκ, in which four or five misshapen condensed somites develop above a region of fused somites (25). Sagittal sections revealed misshapen somites and disorganized fused epithelium in _Pofut1_−/− embryos (Fig. 3C Lower). The absence of presenilins 1 and 2 leads to a more severe defect with no condensed somites in E9 embryos (23). In embryos mutant for Notch1 16–20 somites develop (20, 21), but in Notch1/Notch4 double mutants few somites are formed (22). Somitogenesis is also disorganized, with fused somites and delayed development in embryos mutant for the genes encoding Notch ligands Dll1 (39) or Dll3 (40, 41), or the β1,3-_N_-acetylglucosaminyltranferase Lfng (37, 38).
Figure 3.
Morphological defects in _Pofut1_−/− embryos. (A) Control wild-type or heterozygote embryo (left) and _Pofut1_−/− null (right) embryo at E10 with distended pericardial sac (arrow) and unlooped heart (asterisk). (B) E8.5 control (left) and _Pofut1_−/− (right) embryos. Bracket indicates the posterior region with fused somites; arrow points to a defective somite. (C) Sagittal sections (7 μm) of somites in E9.5 control (Upper) and mutant (Lower) embryos. (D) Vascular defect in yolk sac of E9.5 _Pofut1-_null embryo (right). (D) PECAM-1 antibody staining of E9.5 control (left) and mutant (right) embryos. PECAM-1 is missing from large brain and intersomitic vessels in the mutant (arrows). (E) Kinked neural tube of E8.25 _Pofut1_−/− embryo (arrow; dorsal view).
Notch1 and Notch2 genes have overlapping expression patterns in PSM (27) and appear to function with Notch4 during somitogenesis (22) because inactivation of presenilins 1 and 2 or RBP-Jκ leads to more severe defects than inactivation of a single Notch receptor (27). To investigate gene expression during somitogenesis, whole-mount in situ hybridization was performed (Fig. 4). The Notch ligand gene Dll1 was expressed in PSM and in somites of control E8.75 embryos (n > 10; Fig. 4 A and B). Expression was decreased in PSM and absent in fused somites of _Pofut1_−/− embryos at E8.75 (n > 10; Fig. 4 A and B). This is similar to mutant embryos lacking RBP-Jκ (26, 27). Dll1 expression is only slightly decreased in PSM of Notch1 (26) and presenilins 1 and 2 null embryos (23). Dll3 was strongly expressed in PSM but not in somites of control E8.75 embryos (n = 8; Fig. 4C). In _Pofut1_−/− embryos (n = 4), Dll3 expression was significantly reduced in PSM (Fig. 4C), as observed in _Dll1_-null embryos (41). In control E8.75 embryos (n = 4) Jagged1 was expressed in PSM and in the forming somite (Fig. 4D). In E8.75 _Pofut1_−/− embryos (n = 10) Jagged1 expression was severely reduced and disorganized (Fig. 4D). A similar down-regulation of Jagged1 expression occurs in embryos lacking Notch1, Delta-like1, or RBP-Jκ (26).
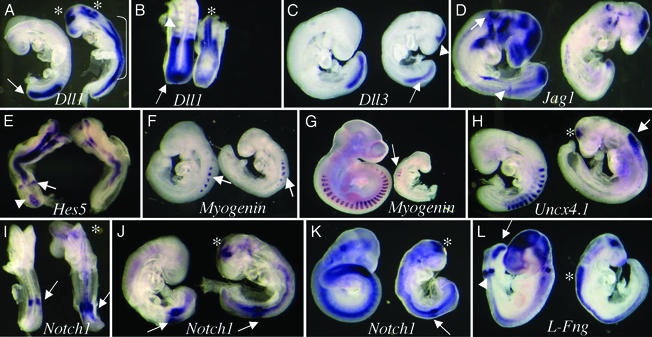
Figure 4.
Expression of Notch signaling pathway genes in _Pofut1_−/− embryos. Control wild-type or heterozygote embryo (left) and _Pofut1_−/− embryo (right) in all panels were probed together. (A and B) E8.75 lateral view (A) and E8.75 dorsal view (B) with a Dll1 probe. Dll1 was expressed in somites (arrowhead in B) and PSM (arrows) of control embryos and down-regulated in _Pofut1_−/− embryos. Dll1 was up-regulated in neural tube (bracket in A), fore-, and hind-brain (asterisks) of _Pofut1_−/− embryos. (C) Dll3 expression was down-regulated in PSM (arrow) but activated in the brain of E8.75 _Pofut1_−/− embryos (arrowhead). (D) Expression of Jagged1 (Jag1) in the posterior region of the forming somite (arrowhead) and in otic vesicle (arrow) and brain of E8.75 control embryos was down-regulated in _Pofut1_−/− embryos. (E) The Notch target Hes5 is expressed in the primitive streak (arrowhead) and forming somite (arrow) of E8.5 control (dorsal view). Hes5 expression was reduced and altered in E8.5 _Pofut1_−/− embryo. (F) Myogenin in anterior somites of control embryos at E8.75 (arrow) was also present in the condensed somites of _Pofut1_−/− embryos (arrow). (G) Myogenin was poorly and diffusely expressed in E9.5 _Pofut1_−/− embryos (arrow). (H) Uncx4.1 was expressed in the caudal compartment of formed somites in control but was missing from the somitic region of E8.75 _Pofut1_−/− embryos. Uncx4.1 was up-regulated in neural tube (arrow) and midbrain (asterisk) of E8.75 _Pofut1_−/− embryos. (I) At E8.25 Notch1 expression in the prospective somite of _Pofut1_−/− embryos (arrow) was higher and more diffuse than control (arrow). (J) At E8.75 Notch1 in the prospective somite (arrow) was similar but more diffuse in _Pofut1_−/− embryos (arrow). (K) At E9.0, Notch1 expression was greater in control compared with _Pofut1_−/− PSM (arrow). Notch1 was up-regulated in midbrain at all stages (asterisk in I, J, and K). (L) Lfng was expressed in PSM (arrow), the prospective somite (arrowhead), otic vesicle, and brain of E8.75 control embryos. In _Pofut1_−/− embryos Lfng expression was missing from somitic areas but markedly up-regulated in neural tube (asterisk).
A target gene of Notch signaling during somitogenesis, Hes5, is expressed in the primitive streak and the forming somite of E8.5 control embryos (n = 9; Fig. 4E). In _Pofut1_−/− embryos, Hes5 expression in somitic regions was reduced and more diffuse (n = 3; Fig. 4E). A similar down-regulation of Hes5 expression during somitogenesis occurs in Notch1 (26), presenilins 1 and 2 (23), and RBP-Jκ (26) mutants. Myogenin, a marker for myotome, was expressed in anterior somites in control embryos at E8.75 (n = 7; Fig. 4F). Expression was similar although slightly more diffuse in E8.75 _Pofut1_−/− embryos. By E9.5, when myogenin was expressed in all somites of control embryos, _Pofut1_−/− embryos expressed low levels of myogenin in ≈6 somites (Fig. 4G). At E9.5, mutants lacking RBP-Jκ have no myogenin expression (25), whereas _Notch1-_null embryos express myogenin at normal levels in their 16–20 somites (20). Uncx4.1, a transcriptional regulator of sclerotome whose expression is confined to the caudal half of formed somites, was missing in the fused somite region of E8.75 _Pofut1_−/− mutants (n = 7; Fig. 4H) as in RBP-Jκ (27) and presenilins 1 and 2 (23) null embryos. Uncx4.1 expression is reduced but not disorganized in _Notch1_−/− mutants (46).
If Notch receptors are inactivated in _Pofut1_−/− embryos because of the absence of _O-_fucose on EGF repeats, early Notch1 gene expression patterns should be normal. This was indeed the case. In control embryos, Notch1 was expressed in the prospective somite at E8.25 (n = 4; Fig. 4I) and E8.75 (n = 11; Fig. 4J). In _Pofut1_−/− embryos Notch1 expression was similar or higher than control in the prospective somite at E8.25 (n = 6; Fig. 4I) and E8.75 (n = 7; Fig. 4J). Therefore Notch1 was expressed well at stages when the Notch target gene Hes5 was poorly expressed (Fig. 4E), providing strong evidence that Notch1 receptor was present but not functional. Notch1 expression decreased by E9.0 (n = 16; Fig. 4K) indicating that Notch1 is a target of Notch signaling during somitogenesis, as observed previously (27). Embryos lacking presenilin 1 also have reduced Notch1 expression in PSM at E8.5 (23). The final panel in Fig. 4 shows that Lfng is severely down-regulated in Pofut1 mutants. Whereas control E8.75 embryos expressed Lfng in PSM and the forming somite (n = 12; Fig. 4L), in _Pofut1_−/− embryos (n = 6; Fig. 4L) Lfng expression was absent in PSM and the forming somite, similar to RBP-Jκ and Delta-like1 null embryos (27, 46). Lfng is well expressed in _Notch1_−/− and _Dll3_−/− mutants, although in a more diffuse pattern (27, 46).
Embryos Lacking _O_-FucT-1 Are Defective in Vasculogenesis and Cardiogenesis.
At E9.5, vascularization of the yolk sac had developed in controls but was essentially absent in _Pofut1_−/− embryos (n > 10; Fig. 3D), similar to mutants in Notch1 or Notch1/Notch4 (22) or presenilins 1 and 2 (23, 24). E9.5 _Pofut1_−/− embryos (n > 30) were very growth retarded, and a large pericardial sac ballooned around the heart (Fig. 3A), as in Notch1 (22), RBP-Jκ (25), and presenilins 1 and 2 (23) null embryos. This striking enlargement of the pericardial sac is not present in embryos lacking Notch2 (44) or Notch4 (22), Dll1 (39), Dll3 (40, 41), Jagged1 (42), Jagged2 (43), Lfng (37, 38), or Rfng (45, 46). Heart development was also retarded in _Pofut1_−/− embryos. At E8.25 the heart was not looped and was smaller and thinner than controls. From E8.75 to E10, heart development was arrested in _Pofut1_−/− embryos and looping never occurred (Fig. 3A). This arrested development was similar to that in embryos lacking RBP-Jκ (25) or presenilins 1 and 2 (23, 24) but was a more severe defect than observed in Notch1 mutants (22). Defective vascularization was evident in _Pofut1_−/− embryos after staining with antibodies to the endothelial cell marker PECAM-1 (Fig. 3E). Intersomitic blood vessels and large branching vessels in the head of control embryos were absent and remaining vessels were disorganized in _Pofut1_−/− embryos (n = 4), similar to _Notch1_−/−/_Notch4_−/− double mutants (22). Fusion of the chorion to the allantois was normal in _Pofut1_−/− embryos as in Notch receptor mutants (22), whereas embryos lacking RBP-Jκ (25) or presenilins 1 and 2 (23) may have defective chorioallantoic fusion and placentation.
Embryos Lacking _O_-FucT-1 Are Defective in Neurogenesis.
At E8.5 _Pofut1_−/− embryos had a markedly kinked neural tube (Fig. 3F), a characteristic of embryos lacking RBP-Jκ (25), presenilins 1 and 2 (23), or Notch1 (26). However, closure of the neuropore occurred normally in _Pofut1_−/− embryos as with _Notch1_−/− embryos (26), whereas embryos lacking RBP-Jκ (26) or presenilins 1 and 2 (23) often fail to close the neuropore. Notch1 and Notch3 are expressed in neural tube and developing brain and are proposed to have independent functions in inhibiting neuronal precursor cell formation (47). Therefore the expression of several genes inhibited by Notch signaling in neural tissues was investigated.
A striking effect of the Pofut1 mutation was the marked up-regulation of several Notch pathway genes in neural tube and brain. Compared with the lack of expression in control embryos, _Pofut1_−/− embryos had strongly up-regulated Dll1 in neural tube, fore- and hind-brain (Fig. 4 A and B), as observed in embryos null for RBP-Jκ (26) or presenilins 1 and 2 (23). Slight up-regulation of Dll1 in neural tube and brain also occurs in _Notch1_−/− mutants (26). Dll3 gene expression, like that of Dll1, was activated in the midbrain of _Pofut1_−/− embryos (Fig. 4C) similar to _Dll1_−/− mutants (27). Up-regulated Dll3 and Dll1 expression is also observed in embryos with Notch1 conditionally deleted solely in neuroepithelial cells that express Engrailed2 (47). Hes5 was strongly expressed in neural tube of E8.5 control embryos (n = 9; Fig. 4E). In _Pofut1_−/− embryos, Hes5 expression was reduced (n = 3; Fig. 4E). By E9.5, Hes5 was expressed throughout neural tube and brain in control embryos (n = 6) but was barely detectable in _Pofut1_−/− embryos (n = 4; data not shown). A similar down-regulation of Hes5 expression occurs in Notch1 (26), presenilins 1 and 2 (23), and RBP-Jκ (26) mutants. Uncx4.1 expression was confined to somites of control embryos but was up-regulated in neural tube and brain of _Pofut1_−/− embryos (Fig. 4H) similar to embryos lacking presenilins 1 and 2 (23). Strikingly, Notch1 expression was also up-regulated in _Pofut1_−/− brain at E8.5, E8.75, and E9 (Fig. 4 I_–_K), indicative of the Notch1 receptor being present but nonfunctional.
E8.75 control embryos expressed Lfng in neural tube, brain, and otic vesicle (n = 12; Fig. 4L). In _Pofut1_−/− embryos, Lfng expression was significantly up-regulated in neural tube and midbrain (n = 6; Fig. 4L), a phenotype not previously reported (27, 46). Lfng expression must therefore normally be inhibited by Notch receptor signaling in embryonic neural tissues. Our data suggest that in neuroepithelium Notch1 expression is normally repressed by an alternative Notch receptor such as Notch3 (47), which is not active in _Pofut1_−/− embryos. Thus, in the absence of _O-_FucT-1, several Notch pathway genes are up-regulated in brain. Notch receptors that inhibit neuronal precursor formation must therefore be inactive.
Discussion
The Notch signaling defects in _Pofut1_−/− mouse embryos and the results of down-regulation of _O_-FucT-1 by RNA interference in Drosophila (17) show that _O-_fucose on Notch clearly does not function solely as the substrate of Fringe. The somitogenic phenotype of _Pofut1_−/− mouse embryos is consistent with inactivation of Notch1, Notch2, and Notch4 in PSM (22, 27); the vasculogenic and cardiogenic phenotypes are consistent with inactivation of Notch1 and Notch4 during development (22); and the neurogenic phenotype is consistent with inactivation of Notch1 and Notch3 in neuroepithelium (47). The phenotype of _Pofut1_−/− embryos most closely resembles that of embryos lacking core downstream Notch signaling components such as presenilins 1 and 2 (23, 24) or RBP-Jκ (25, 26), and is stronger than a _Notch1_-null phenotype (20, 21, 26, 27, 46). _O-_FucT-1 is therefore a previously unrecognized core component of Notch signaling pathways. Furthermore, it seems likely that _O-_fucose is required on the extracellular domain of all mammalian Notch receptors for Notch signaling to occur. Whereas _O_-FucT-1 is predicted to act on Notch ligands (6), the phenotype of _Pofut1_−/− embryos suggests a cell-autonomous function for _O_-FucT-1 in the Notch-expressing cell, as was also found in Drosophila (17). However, it is also possible that ligand phenotypes are masked or precluded by the early death of _Pofut1_−/− embryos.
It is of interest that _Pofut1_−/− embryos do not have a _Cripto_−/− phenotype. Cripto encodes a membrane protein that is an essential component of Nodal signaling pathways (11). Inactivation of Nodal or Cripto gives rise to similar embryonic lethal phenotypes (18, 48). Conversion of the Thr that accepts _O-_fucose in the Cripto EGF repeat to Ala inactivates transforming growth factor-β/Nodal signaling in Xenopus (10) and in coculture assays (11). The lack of a _Cripto_−/− phenotype in _Pofut1_−/− embryos may reflect the existence of an _O_-FucT unrelated in sequence to _O-_FucT-1, or a less stringent requirement for _O_-fucose on Cripto in vivo than in reporter assays, or that Ala in place of Thr in the EGF repeat of Cripto inactivates Cripto by a mechanism unrelated to the absence of _O-_fucose.
A key challenge for the future is to determine how the lack of _O-_fucose on Notch extracellular domain results in inactivation of Notch receptors. Notch1 has >20 EGF repeats that contain a consensus sequence for O_-_fucosylation and most may be modified by Fringe (7). It is known that the action of Fringe in transferring GlcNAc to EGF-_O_-fucose causes changes in soluble ligand binding to Notch receptors (5, 13–15). We have shown that Lec13 mutant cells with low fucose in glycoproteins exhibit reduced Notch signaling for both Jagged1 and Delta1 ligands (refs. 4 and 16; J. Chen and P.S., unpublished results). Notch signaling is rescued by providing a cDNA encoding GDP-mannose-4,6-dehydratase to correct the Lec13 defect. Because both Notch1 and Notch2 receptors in Lec13 cells are expressed at normal levels on the cell surface, reduced signaling suggests reduced ligand binding. Consistent with this suggestion, soluble Jagged1 and Delta1 have reduced binding to Lec13 cells (D. Moloney, K. Uemura, and P.S., unpublished results). These data suggest a model in which both Jagged1 and Delta1 require EGF-_O-_fucose to bind to Notch. In Drosophila, Notch ligands bind at EGF repeats 11 and 12 (49). A comparison of Notch EGF11 and -12 sequences across species revealed a conserved _O_-fucose site at Thr/Ser between the second and third Cys residues [in the sequence Cys-(Xaa)4-Thr(Ser)-Cys] in Notch EGF repeat 12 of all mammalian, Drosophila, Xenopus, zebra fish, sea urchin, and sea squirt Notch receptors, and in the EGF12 equivalent (EGF6 or EGF9) of Lin12 and Glp1 in Caenorhabditis elegans, Caenorhabditis remanei, and Caenorhabditis briggsae, and this site was shown to be the only Thr to incorporate fucose in a mouse Notch1 EGF11,12 fragment (D. Moloney and P.S., unpublished results). Similar conclusions were reached from studies of a Notch1 fragment containing EGF repeats 11–15 that showed Fringe can act on EGF12-_O_-fucose (7). Notch ligands are also thought to bind to other regions of Notch, in particular to EGF repeats 24–29, which are the sites of Abruptex mutations in Drosophila (50). Particular EGF-_O-_fucose combinations or clusters may generate ligand-binding sites of various affinities and functions. We predict that the severe Notch signaling defects in _Pofut1_−/− embryos arise at least in part because Notch ligands require _O-_fucose to recognize Notch receptors at EGF-_O_-fucose sites on Notch.
Acknowledgments
We thank all those who provided plasmids and probes (see Materials and Methods), Harry Hou and Subha Sundaram for excellent technical assistance, and Ken Irvine, Robert Haltiwanger, Thomas Vogt, Jihua Chen, and Daniel Moloney for helpful discussions. This work was supported by National Cancer Institute Grants 30645 and 95022 to P.S. and by Albert Einstein Cancer Center Grant PO1 1333048.
Abbreviations
EGF
epidermal growth factor
_O_-FucT-1
protein _O-_fucosyltransferase 1
E_n_
embryonic day n
ES cells
embryonic stem cells
PECAM-1
platelet endothelial cell adhesion molecule 1
PSM
presomitic mesoderm
References
- 1.Baron M, Aslam H, Flasza M, Fostier M, Higgs J E, Mazaleyrat S L, Wilkin M B. Mol Membr Biol. 2002;19:27–38. doi: 10.1080/09687680110112929. [DOI] [PubMed] [Google Scholar]
- 2.Joutel A, Tournier-Lasserve E. Semin Cell Dev Biol. 1998;9:619–625. doi: 10.1006/scdb.1998.0261. [DOI] [PubMed] [Google Scholar]
- 3.Mumm J S, Kopan R. Dev Biol. 2000;228:151–165. doi: 10.1006/dbio.2000.9960. [DOI] [PubMed] [Google Scholar]
- 4.Moloney D J, Panin V M, Johnston S H, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine K D, Haltiwanger R S, Vogt T F. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 5.Bruckner K, Perez L, Clausen H, Cohen S. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 6.Panin V M, Shao L, Lei L, Moloney D J, Irvine K D, Haltiwanger R S. J Biol Chem. 2002;277:29945–29952. doi: 10.1074/jbc.M204445200. [DOI] [PubMed] [Google Scholar]
- 7.Shao L, Moloney D J, Haltiwanger R S. J Biol Chem. 2003;278:7775–7782. doi: 10.1074/jbc.M212221200. [DOI] [PubMed] [Google Scholar]
- 8.Harris R J, van Halbeek H, Glushka J, Basa L J, Ling V T, Smith K J, Spellman M W. Biochemistry. 1993;32:6539–6547. doi: 10.1021/bi00077a007. [DOI] [PubMed] [Google Scholar]
- 9.Moloney D J, Shair L H, Lu F M, Xia J, Locke R, Matta K L, Haltiwanger R S. J Biol Chem. 2000;275:9604–9611. doi: 10.1074/jbc.275.13.9604. [DOI] [PubMed] [Google Scholar]
- 10.Schiffer S G, Foley S, Kaffashan A, Hronowski X, Zichittella A E, Yeo C Y, Miatkowski K, Adkins H B, Damon B, Whitman M, et al. J Biol Chem. 2001;276:37769–37778. doi: 10.1074/jbc.M104774200. [DOI] [PubMed] [Google Scholar]
- 11.Yan Y T, Liu J J, Luo Y E C, Haltiwanger R S, Abate-Shen C, Shen M M. Mol Cell Biol. 2002;22:4439–4449. doi: 10.1128/MCB.22.13.4439-4449.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rabbani S A, Mazar A P, Bernier S M, Haq M, Bolivar I, Henkin J, Goltzman D. J Biol Chem. 1992;267:14151–14156. [PubMed] [Google Scholar]
- 13.Hicks C, Johnston S H, diSibio G, Collazo A, Vogt T F, Weinmaster G. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu K, Chiba S, Hosoya N, Kumano K, Saito T, Kurokawa M, Kanda Y, Hamada Y, Hirai H. Mol Cell Biol. 2000;20:6913–6922. doi: 10.1128/mcb.20.18.6913-6922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu K, Chiba S, Saito T, Kumano K, Takahashi T, Hirai H. J Biol Chem. 2001;276:25753–25758. doi: 10.1074/jbc.M103473200. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Moloney D J, Stanley P. Proc Natl Acad Sci USA. 2001;98:13716–13721. doi: 10.1073/pnas.241398098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okajima T, Irvine K D. Cell. 2002;111:893–904. doi: 10.1016/s0092-8674(02)01114-5. [DOI] [PubMed] [Google Scholar]
- 18.Ding J, Yang L, Yan Y T, Chen A, Desai N, Wynshaw-Boris A, Shen M M. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- 19.Wang Y, Shao L, Shi S, Harris R J, Spellman M W, Stanley P, Haltiwanger R S. J Biol Chem. 2001;276:40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 20.Conlon R A, Reaume A G, Rossant J. Development (Cambridge, UK) 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 21.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 22.Krebs L T, Xue Y, Norton C R, Shutter J R, Maguire M, Sundberg J P, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 23.Donoviel D B, Hadjantonakis A K, Ikeda M, Zheng H, Hyslop P S, Bernstein A. Genes Dev. 1999;13:2801–2810. doi: 10.1101/gad.13.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herreman A, Hartmann D, Annaert W, Saftig P, Craessaerts K, Serneels L, Umans L, Schrijvers V, Checler F, Vanderstichele H, et al. Proc Natl Acad Sci USA. 1999;96:11872–11877. doi: 10.1073/pnas.96.21.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka C, Nakano T, Wakeham A, de la Pompa J L, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak T W, Honjo T. Development (Cambridge, UK) 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 26.de la Pompa J L, Wakeham A, Correia K M, Samper E, Brown S, Aguilera R J, Nakano T, Honjo T, Mak T W, Rossant J, Conlon R A. Development (Cambridge, UK) 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- 27.Barrantes I B, Elia A J, Wunsch K, De Angelis M H, Mak T W, Rossant J, Conlon R A, Gossler A, de la Pompa J L. Curr Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- 28.Ioffe E, Liu Y, Bhaumik M, Poirier F, Factor S M, Stanley P. Proc Natl Acad Sci USA. 1995;92:7357–7361. doi: 10.1073/pnas.92.16.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnston S H, Rauskolb C, Wilson R, Prabhakaran B, Irvine K D, Vogt T F. Development (Cambridge, UK) 1997;124:2245–2254. doi: 10.1242/dev.124.11.2245. [DOI] [PubMed] [Google Scholar]
- 30.Nye J S, Kopan R, Axel R. Development (Cambridge, UK) 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 31.Bettenhausen B, Hrabe de Angelis M, Simon D, Guenet J L, Gossler A. Development (Cambridge, UK) 1995;121:2407–2418. doi: 10.1242/dev.121.8.2407. [DOI] [PubMed] [Google Scholar]
- 32.Mitsiadis T A, Henrique D, Thesleff I, Lendahl U. Development (Cambridge, UK) 1997;124:1473–1483. doi: 10.1242/dev.124.8.1473. [DOI] [PubMed] [Google Scholar]
- 33.Bessho Y, Sakata R, Komatsu S, Shiota K, Yamada S, Kageyama R. Genes Dev. 2001;15:2642–2647. doi: 10.1101/gad.930601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akazawa C, Sasai Y, Nakanishi S, Kageyama R. J Biol Chem. 1992;267:21879–21885. [PubMed] [Google Scholar]
- 35.Wright W E, Sassoon D A, Lin V K. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 36.Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- 37.Evrard Y A, Lun Y, Aulehla A, Gan L, Johnson R L. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- 38.Zhang N, Gridley T. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]
- 39.Hrabe de Angelis M, McIntyre J, II, Gossler A. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- 40.Kusumi K, Sun E S, Kerrebrock A W, Bronson R T, Chi D C, Bulotsky M S, Spencer J B, Birren B W, Frankel W N, Lander E S. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- 41.Dunwoodie S L, Clements M, Sparrow D B, Sa X, Conlon R A, Beddington R S. Development (Cambridge, UK) 2002;129:1795–1806. doi: 10.1242/dev.129.7.1795. [DOI] [PubMed] [Google Scholar]
- 42.Xue Y, Gao X, Lindsell C E, Norton C R, Chang B, Hicks C, Gendron-Maguire M, Rand E B, Weinmaster G, Gridley T. Hum Mol Genet. 1999;8:723–730. doi: 10.1093/hmg/8.5.723. [DOI] [PubMed] [Google Scholar]
- 43.Jiang R, Lan Y, Chapman H D, Shawber C, Norton C R, Serreze D V, Weinmaster G, Gridley T. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman J R, Tsujimoto Y. Development (Cambridge, UK) 1999;126:3415–3424. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- 45.Moran J L, Levorse J M, Vogt T F. Nature. 1999;399:742–743. doi: 10.1038/21560. [DOI] [PubMed] [Google Scholar]
- 46.Zhang N, Norton C R, Gridley T. Genesis. 2002;33:21–28. doi: 10.1002/gene.10081. [DOI] [PubMed] [Google Scholar]
- 47.Lutolf S, Radtke F, Aguet M, Suter U, Taylor V. Development (Cambridge, UK) 2002;129:373–385. doi: 10.1242/dev.129.2.373. [DOI] [PubMed] [Google Scholar]
- 48.Brennan J, Lu C C, Norris D P, Rodriguez T A, Beddington R S, Robertson E J. Nature. 2001;411:965–969. doi: 10.1038/35082103. [DOI] [PubMed] [Google Scholar]
- 49.Rebay I, Fleming R J, Fehon R G, Cherbas L, Cherbas P, Artavanis-Tsakonas S. Cell. 1991;67:687–699. doi: 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 50.de Celis J F, Bray S J. Development (Cambridge, UK) 2000;127:1291–1302. doi: 10.1242/dev.127.6.1291. [DOI] [PubMed] [Google Scholar]