Transcripts Targeted by the MicroRNA-16 Family Cooperatively Regulate Cell Cycle Progression (original) (raw)
Abstract
microRNAs (miRNAs) are abundant, ∼21-nucleotide, noncoding regulatory RNAs. Each miRNA may regulate hundreds of mRNA targets, but the identities of these targets and the processes they regulate are poorly understood. Here we have explored the use of microarray profiling and functional screening to identify targets and biological processes triggered by the transfection of human cells with miRNAs. We demonstrate that a family of miRNAs sharing sequence identity with miRNA-16 (miR-16) negatively regulates cellular growth and cell cycle progression. miR-16-down-regulated transcripts were enriched with genes whose silencing by small interfering RNAs causes an accumulation of cells in G0/G1. Simultaneous silencing of these genes was more effective at blocking cell cycle progression than disruption of the individual genes. Thus, miR-16 coordinately regulates targets that may act in concert to control cell cycle progression.
microRNAs (miRNAs) regulate mRNA translation and mRNA decay in plants and animals (49). Hundreds of human miRNAs are now known (4-6, 15, 19). In animals, miRNAs regulate thousands of genes with spatial and temporal specificity, helping to ensure the accuracy of gene expression programs (17, 38, 47). Understanding the precise biological functions of animal miRNAs will require the identification of their multiple targets and the pathways that they control.
Animal miRNAs generally share limited sequence complementarity with their targets. miRNA target recognition involves complementary base pairing of the target with the 5′ end (positions 1 to 8) of the miRNA guide strand seed region. However, the extent of seed region complementarity is not precisely determined and can be modified by 3′ pairing (8). Computational methods have been used to predict human miRNA targets (31, 34, 37, 45, 52). Most predicted miRNA target recognition sites lie in 3′ untranslated regions (3′UTRs), although coding region sites (CDS) may also be used (8, 36). Current estimates are that 30% or more of human mRNAs are regulated by miRNAs (36).
While thousands of miRNA targets have been predicted, relatively few have been experimentally validated. Available methods for validation are laborious and not easily amenable to high-throughput methodologies (4). Since a single miRNA can regulate hundreds of targets, the biological pathways regulated by miRNAs are not always obvious from an examination of their targets. There is a clear need for high-throughput, low-cost methods to experimentally determine miRNA targets, validate computational predictions, and decipher miRNA function.
One method to experimentally identify miRNA targets and their functions is microarray analysis (50). Although miRNAs may silence their targets via translational blocking (16), they also regulate target transcript levels. miRNAs in transfected cells down-regulate hundreds of mRNAs detectable by microarray profiling (38). These down-regulated transcripts have expression patterns that are complementary to that of the introduced miRNA and are also highly enriched within their 3′UTRs with hexamer, heptamer, and octamer motifs complementary to miRNA seed regions. This regulation resembles the “off-target” silencing of imperfectly matched targets by small interfering RNAs (siRNAs) (28, 29). Thus, both miRNAs and siRNAs can target partially complementary transcripts for degradation, resulting in transcript changes that can be monitored using microarrays. In fact, changes in transcript levels due to miRNA activity have been observed directly in vivo. The let-7 and lin-4 miRNAs trigger the degradation of their target mRNAs (2). Also, the depletion of miRNAs in mice and zebrafish led to the up-regulation of target mRNAs that were measured on microarrays (18, 35).
A potential advantage of using microarrays to analyze miRNA targets is the utility of expression profiles for predicting gene function (25). In this study, we have explored the use of miRNA expression profiles to analyze miRNA targets and functions. Included in our analysis are several miRNAs reported to have a role in cancer (9-12, 14, 21, 22, 27, 32, 40, 41). We use this approach to show that a family of miRNAs sharing seed region identity with miRNA-16 (miR-16) negatively regulates cell cycle progression from G0/G1 to S.
MATERIALS AND METHODS
Cell lines and chemicals.
HCT116 Dicerex5 and DLD-1 Dicerex5 cells were previously described (15). Wild-type HCT116 and DLD-1, HeLa, TOV21G, and SW1417 cells were from the American Type Culture Collection, Rockville, MD.
RNA duplexes.
RNA duplexes corresponding to mature miRNAs were designed as previously described (38). siRNA sequences were designed with an algorithm developed to increase the efficiency of the siRNAs in silencing while minimizing their off-target effects (28). siRNA and miRNA duplexes were ordered from Sigma-Proligo (Boulder, CO) and Dharmacon (Lafayette, CO), respectively. miRCURY locked-nucleic-acid (LNA) knockdown probes for miR-16 and miR-106b were obtained from Exiqon, Copenhagen, Denmark. Sequences of miRNA duplexes used for profiling and siRNA sequences corresponding to miR-16-down-regulated transcripts that trigger the accumulation of cells in G0/G1 are listed in Table S1 in the supplemental material.
Transfection with miRNA and siRNA.
Cells were plated 24 h prior to transfection. HCT116 cells were transfected in 6-well plates using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). DLD-1, HeLa, and TOV21G cells were transfected using SilentFect (Bio-Rad, Hercules, CA). SW1417 and HeLa cells were transfected using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA). Duplexes were used at final concentrations of 25 nM for DLD-1 cells and 100 nM for HeLa, TOV21G, HCT116, and SW1417 cells. We estimate that our siRNA conditions resulted in the transfection of >90% of the cells. When siRNA pools were used, three siRNAs targeting the same gene were pooled at equal molarities (final concentration of each siRNA, 33 nM; total siRNA concentration, 100 nM). Anti-miRNAs (anti-miRs) were used at 200 nM. Plasmids were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA).
Expression of miR-16.
A 945-base-pair HindIII/SacI genomic fragment spanning the miR-15a-miR-16 locus from human chromosome 13 was cloned into the Gateway entry vector pENTR1A (Invitrogen, Carlsbad, CA) modified to express exogenous sequences under an H1 promoter (pENTR1A H1). miR-16 was also expressed as a synthetic primary miRNA-like hairpin (46). In brief, a hairpin cassette encoding the miR-16 mature strand (http://microrna.sanger.ac.uk/cgi-bin/sequences/mirna_entry.pl?acc=MI0000070) plus a perfectly complementary passenger strand linked by a 19-base-pair human miR-16 loop and flanked by 25 base pairs of the human miR-30a primary transcript was cloned into pENTR1A H1 and transferred to pLenti6-BLOCK-iT-DEST (Invitrogen, Carlsbad, CA). Plasmid insert sequences were verified by DNA sequencing.
Cell cycle analysis.
Cells were transfected with miRNAs or siRNAs. Twenty-four hours after transfection, floating and adherent cells were harvested, combined, and processed. Alternatively, nocodazole (100 ng/ml; Sigma-Aldrich) was added and cells were further incubated for 16 to 20 h before harvesting. The supernatant from each well was combined with cells harvested from each well by trypsinization. Cells were collected by centrifugation, fixed with ice-cold 70% ethanol, washed with phosphate-buffered saline, and resuspended in 0.5 ml of phosphate-buffered saline containing propidium iodide (10 μg/ml) and RNase A (1 mg/ml). After a final incubation at 37°C for 30 min, cells were analyzed using a FACSCalibur flow cytometer (Becton Dickinson). A total of 10,000 events were counted for each sample. Data were analyzed using FlowJo software (Tree Star, Ashland, OR).
Microarray analysis.
Cells were transfected in 6-well plates, and RNA was isolated 6 to 24 h following transfection. Microarray analysis was performed as described previously (28).
Quantitative PCR and immunoblotting.
mRNA silencing was quantified by real-time PCR using an ABI PRISM 7900HT sequence detection system and Assays-on-Demand gene expression products (Applied Biosystems, Foster City, CA). The mRNA value for each gene was normalized relative to the β-glucuronidase (catalog no. 431088E) mRNA levels in each RNA sample. miRNA levels were determined using a quantitative primer extension PCR assay (43). Values were converted to copy numbers by comparison to standard curves generated using single-stranded mature miRNAs and are expressed as copies/20 pg of input RNA (approximately equivalent to the number of copies/cell). Immunoblotting was performed as described previously (29). Anti-CDK6 monoclonal antibody (DCS-90) was purchased from Abcam (Cambridge, MA).
Analysis methods. (i) Gene set annotation.
miRNA-down-regulated transcripts were identified in microarray gene expression signatures using a P value cutoff (P < 0.01). miRNA-down-regulated transcripts were defined by the intersection of down-regulated transcripts from two different Dicerex5 cell lines. Down-regulated transcripts were tested for enrichment relative to a background set using the hypergeometric distribution. miRNA target regulation was measured through enrichment with transcripts having annotated 3′UTRs containing miRNA hexamer seed strings (stretches of 6 contiguous bases complementary to miRNA seed region nucleotides [nt] 1 to 6, 2 to 7, or 3 to 8). Biological functions were detected through enrichment with transcripts corresponding to Gene Ontology (GO) biological process functional categories (http://www.geneontology.org/). The set of genes on the microarray was used as a background set.
(ii) Consensus miR-16-down-regulated transcripts.
HCT116 Dicerex5 cells and DLD-1 Dicerex5 cells were transfected with miR-16 duplexes, and gene expression signatures were determined at 24 h. The intersection signature (P < 0.01) between the two cell lines was identified. Transcripts in the intersection signature that were also regulated (P < 0.05) at 6 h in HCT116 Dicerex5 cells were defined as consensus miR-16-down-regulated transcripts (n = 116). To analyze the properties of miR-16-down-regulated transcripts, we focused on transcripts having sequence records denoting spans of ≥20 nucleotides (nt) for both CDS and 3′UTRs (n = 84). The background set for miR-16 transcriptional target comparisons comprised transcripts not down-regulated by miR-16 but with similar distributions of expression levels and having sequence records denoting spans of ≥20 nt for both CDS and 3′UTRs (n = 1,546).
RESULTS
Functional annotation of miRNA targets.
To test the utility of microarray profiling for the identification and functional annotation of miRNA targets, we devised and tested the experimental model described in the legend to Fig. 1A. We collected a compendium of microarray profiles of cells transfected with miRNAs (38). We selected 24 miRNAs (see Table S1 in the supplemental material) and then systematically examined expression profiles for the regulation of predicted miRNA targets and known biological pathways (Fig. 1A).
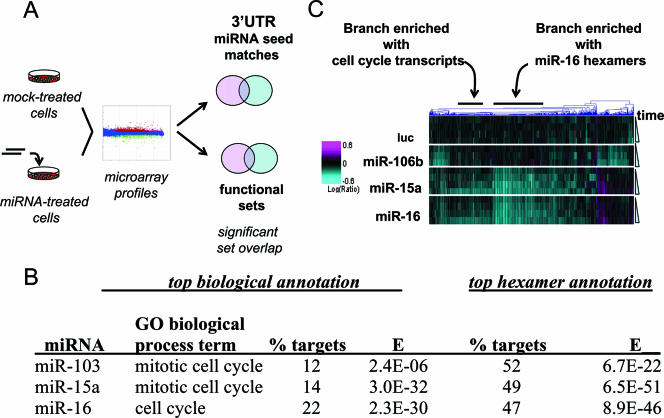
FIG. 1.
Microarray profiling identifies a family of miRNAs that regulates cell cycle transcripts. (A) Strategy for identifying pathways regulated by miRNAs. HCT116 or DLD-1 Dicerex5 cells were transfected with miRNAs, and RNA was isolated 24 h posttransfection. Gene expression profiles were determined by competitive hybridization of amplified mRNAs from miRNA-treated versus mock-treated cells. These profiles were then examined for enrichment with transcripts containing 3′UTR seed hexamer matches or those corresponding to biological annotation categories. (B) miRNA family whose expression signature scores highly for biological annotation. Expression signatures generated by 24 miRNAs were tested for enrichment with sequences annotated with GO biological process terms and 3′UTR seed hexamer matches. Shown are the three miRNAs whose expression signatures showed significant enrichment with GO biological process sequences [E < 1E(−2)]. The highest-scoring GO biological process term, the percentage of transcripts annotated with that term (percentage of targets), and the _E_ value for enrichment are shown. For 3′UTR hexamer annotation, the percentage of transcripts containing the top-ranked hexamer match and the _E_ value for enrichment are shown. There were 18,124 total annotated sequences on the microarray, of which ∼20% contained the top-ranked 3′UTR hexamer for miR-103, miR-15a, and miR-16. The miR-103, miR-15a, and miR-16 signatures comprised 215, 549, and 557 transcripts, respectively (Table S2 in the supplemental material). (C) Kinetic separation of miR-15, −16 target regulation, and the cell cycle gene regulation phenotype. HCT116 Dicerex5 cells were transfected with luciferase siRNA (luc) or miR-106b, miR-15a, or miR-16 duplexes. RNA samples were isolated 6, 10, 14, and 24 h after transfection and were compared to RNA from mock-transfected cells. Shown is a heat-map representation of regulated genes (columns) in different experiments (rows). Samples are arranged by increasing time after transfection, from top to bottom. The color bar represents log10 expression ratios (samples from treated cells/samples from mock-treated cells) of −0.6 (teal) to +0.6 (magenta). Shown are results for 1,394 transcripts regulated with _P_ of <0.05 and a log10 expression ratio of <0 in any one experiment. The 6-h signature transcripts were required additionally to be present at a majority of later time points. Individual branches of the dendrogram were tested for enrichment with transcripts annotated with the GO biological process term mitotic cell cycle and the miR-15a-miR-16 seed region hexamer beginning at position 2. The _E_ values for enrichment with cell cycle-related transcripts were 5.5E(−26) for the branch showing enrichment with mitotic cell cycle sequences and >1E(−2) for the branch showing enrichment with miR-16 hexamers. The E values for enrichment with miR-16 hexamers were 1.1E(−80) for the branch showing enrichment with miR-16 hexamers and >1E(−2) for the branch showing enrichment with mitotic cell cycle sequences.
We experimented with several cell lines, including Dicer hypomorphs of HCT116 and DLD-1 colon tumor cells (HCT116 Dicerex5 and DLD-1 Dicerex5, respectively) (15). These lines have homozygous disruption of the Dicer helicase domain and show reduced levels of many endogenous miRNAs (15). Perhaps because of the loss of endogenous miRNAs, the hypomorphs show ∼2-fold more intense expression changes following transfection with exogenous siRNAs or miRNAs than matched Dicer wild-type cells (see Fig. S1 in the supplemental material). We therefore used these Dicerex5 cells for subsequent experiments.
We transfected HCT116 Dicerex5 and DLD-1 Dicerex5 cells with 24 different miRNAs and determined expression profiles relative to those of mock-transfected cells (Fig. 1A). We measured expression profiles at 24 h, when mRNA silencing is maximal but secondary transcriptional effects due to protein depletion are minimal (28). Examples of expression profiles for cells transfected with miRNAs are shown in Fig. S2 in the supplemental material. These miRNAs represent several families of miRNAs sharing identity in the miRNA seed region (see Fig. S3 in the supplemental material). Patterns of down-regulated transcripts were largely specific to each family. The family-specific transcripts were highly enriched with hexamer sequence motifs complementary to the seed region for that family (not shown). Nearly identical expression profiles were obtained with miR-141 and miR-200a; miR-17-5p, miR-20a, and miR-106b; and miR-192 and miR-215. Thus, in most cases, miRNAs sharing seed region identity regulated the same transcripts. miR-15a, miR-16, and miR-195 also gave nearly identical expression profiles, but miR-103 gave an overlapping but distinct profile compared to other members of this family, consistent with the 1-nt offset of the miR-103 seed sequence (see Fig. S3 in the supplemental material). Since expression profiles for each miRNA were largely consistent between the two cell lines tested, we utilized an intersection of profiles from the two cell lines for further studies.
We analyzed down-regulated transcripts for enrichment with hexamer motifs matching the miRNA seed region (seed region hexamers) in their 3′UTRs. Virtually all (23/24) miRNA intersection signatures showed enrichment with seed region hexamers highly unlikely to happen by chance [expectation (E) < 1E(−20)]. We also measured highly significant enrichments with computationally predicted miRNA targets (34, 36, 52) in miRNA-down-regulated signatures.
We next asked whether transcripts down-regulated by miRNAs were enriched for association with known biological pathways (Fig. 1A). Only 3 of the 24 intersection signatures showed significant [E < 1E(−2)] enrichment with transcripts associated with a GO biological process term (Fig. 1B); the three corresponding miRNAs (miR-15a, miR-16, and miR-103) are members of a single miRNA family (see Fig. S3 in the supplemental material). All three miRNAs triggered expression signatures showing significant enrichment with transcripts annotated with the terms “mitotic cell cycle” or “cell cycle.” Functional enrichment was less significant in miR-103 signatures than in those for miR-15a and miR-16.
The transcript annotation results suggest that targets regulated by miR-15a and miR-16, and to a lesser extent miR-103, are involved in the regulation of the cell cycle and cell growth. Both mitotic cell cycle transcripts and putative targets of miR-15a and miR-16 were enriched in 24-h down-regulated signatures. It was unclear, however, whether mitotic cell cycle transcripts were direct or indirect targets of miR-15a and miR-16. To help distinguish between these possibilities, we performed a kinetic analysis of transcript regulation. We hypothesized that if mitotic cell cycle transcripts were direct targets, they would be regulated sooner and would be more enriched with miR-16 seed region hexamers than if they were indirect targets. As shown in Fig. 1C, transcripts enriched with miR-15a-miR-16 seed region hexamers in their 3′UTRs were regulated as early as 6 h following transfection. Mitotic cell cycle transcripts were not enriched with seed region hexamer matches in their 3′UTRs and were regulated later. Thus, it is likely that most mitotic cell cycle genes in miR-15a and miR-16 signatures are not direct targets of miR-15 and miR-16.
Cellular phenotypes triggered by miR-15a and miR-16.
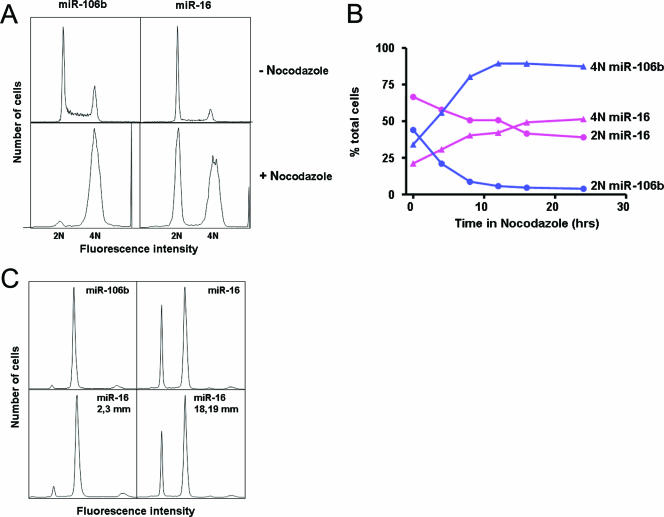
We next investigated whether the gene expression phenotypes described above were manifested in changes in cell cycle distribution. Mock and luciferase siRNA-treated (not shown) or miR-106b-transfected HCT116 Dicerex5 cells showed a normal cell cycle distribution (Fig. 2A). In contrast, cell cultures transfected with miR-16 had increased numbers of cells in G0/G1 (diploid DNA content) and corresponding decreases in numbers of cells in S and G2/M. This finding suggests that miR-16 induced the accumulation of cells in G0/G1 in this cell line. Similar effects were seen with DLD-1 Dicerex5 cells (not shown). The G0/G1 accumulation phenotype became clearer when the microtubule-depolymerizing drug nocodazole was added 24 to 28 h posttransfection to block cells from reentering the cell cycle after mitosis. This treatment caused nearly all miR-106b-transfected cells to accumulate in G2/M (tetraploid DNA content), whereas a large fraction (∼40%) of miR-16-transfected cells remained in G0/G1 (Fig. 2A).
FIG. 2.
miR-15 and miR-16 cause the accumulation of cells at the Golgi stage of the cell cycle. (A) miR-16 triggers the accumulation of cells at a stage of the cell cycle. HCT116 Dicerex5 cells were transfected with miR-106b or miR-16. −Nocodazole, cells were analyzed for cell cycle distribution 24 h posttransfection; +Nocodazole, cells were treated with nocodazole beginning at 20 h posttransfection and analyzed for cell cycle distribution 18 h later (46 h posttransfection). 2N, cells having diploid DNA content; 4N, cells having tetraploid DNA content. miR-106b gave a cell cycle profile indistinguishable from that of mock- or luciferase-transfected cells. (B) miR-16 causes the accumulation of cells at a stage of the cell cycle rather than cell cycle arrest. HCT116 Dicerex5 cells were transfected with miR-106b (blue) or miR-16 (magenta). Cells were treated with nocodazole 24 h after transfection and analyzed at the indicated times after nocodazole addition for cell cycle distribution. Shown are the fractions of total cells in G0/G1 (2N DNA; circles) and G2/M (4N DNA; triangles) with increasing time in nocodazole. (C) The miR-16 cell cycle phenotype is seed region dependent. HCT116 Dicerex5 cells were transfected with miR-106b, miR-16, or miR-16 containing mismatches at positions 2 and 3 (2,3 mm) or 18 and 19 (18,19 mm). Cells were treated with nocodazole and analyzed as described in the legend to panel A.
The fraction of miR-106b-transfected cells in G0/G1 decreased to a minimum and the fraction in G2/M reached a maximum ∼12 h after nocodazole addition (Fig. 2B). The fraction of miR-16-transfected cells accumulating in G1 decreased and the fraction in G2/M increased more slowly with time in nocodazole (Fig. 2B). This result suggests that the G0/G1 accumulation phenotype is reversible or that cells are not actually blocked but rather progress more slowly through G0/G1. It is also possible that the effects of miR-16 transfection are transient.
We next asked whether miR-16 caused the G0/G1 accumulation phenotype in cells having wild-type Dicer function. We found that miR-16 induced a similar cell cycle phenotype in wild-type HCT116 and DLD-1 cells (see Fig. S4 in the supplemental material). In other experiments, we found that miR-16 induced a similar cell cycle phenotype in A549 (lung cancer), MCF7 (breast cancer), and TOV21G (ovarian cancer) cells (not shown). In contrast, miR-16 did not induce a measurable cell cycle phenotype in HeLa cells (not shown). Thus, miR-16 triggered the G0/G1 accumulation phenotype in most but not all cells with wild-type Dicer function.
Cellular phenotypes and gene expression profiles triggered by miR-16 that result from miRNA target regulation should be dependent upon the seed region of the mature strand (38). To test this possibility, we tested the effects of miR-16 base pair mismatches on the cell cycle phenotype (Fig. 2C). The G0/G1 accumulation phenotype was reversed by paired seed region mismatches at positions 2 and 3 (and 4 and 5; not shown) but not by mismatches outside the seed region at positions 18 and 19 (and 19 and 20; not shown). We also tested the effects of seed region mismatches on the gene expression profiles triggered by miR-16 and other family members (Fig. S5 in the supplemental material). We identified a set (n = 116) of consensus transcripts down-regulated by miR-16 (see Table S4 in the supplemental material). Consensus miR-16-down-regulated transcripts were also down-regulated by miR-15a, miR-15b, and miR-195 but not miR-106b (see Fig. S5 in the supplemental material). Only a subset of these transcripts was regulated by miR-103 and miR-107. These transcripts were not regulated in cells transfected with miR-16 duplexes having mismatches at positions 2 and 3 or 4 and 5, but their regulation was unaltered in cells transfected with duplexes with mismatches at positions 18 and 19 and 19 and 20. Therefore, cell cycle and gene expression phenotypes induced by miR-16 family miRNAs are seed region dependent.
Cellular phenotypes are triggered by natural forms of miR-16 and reversed in loss-of-function experiments.
It is conceivable that the cellular phenotype induced by miR-16 is an artifact of the overexpression of the miR-16 mature form. To increase our confidence that miR-16 phenotypes were not artifactual, we determined whether the G0/G1 cell cycle accumulation phenotype could be induced by miR-16 expressed under more natural conditions (i.e., requiring processing from a precursor). We approached this problem in two ways. First, we expressed an miR-16-encoding short hairpin RNA (shRNA) (46). We also expressed miR-16 from a genomic fragment encoding the endogenous miR-15a-miR-16-1 locus. In both cases, HCT116 Dicerex5 cells were transfected with plasmids expressing precursor forms of miR-16 under the control of an H1 promoter, and cell cycle distributions were measured as described in the legend to Fig. 2. In these and other experiments, we found large variations in the absolute numbers of G0/G1 cells in identically treated samples in different experiments. When it was necessary to compare results obtained from different experiments, we normalized percentages of cells in G0/G1 so that the calculations for background cells and cells transfected with miR-16 (100 nM) gave 0% and 100% of cells in G0/G1, respectively (see the legend to Fig. 3A for details). These normalized G0/G1 values varied less than the raw values for identically treated samples between experiments. Both raw and normalized G0/G1 values are given in Table S3 in the supplemental material. Levels of miR-16 were measured in the same samples by using a quantitative primer extension PCR assay (43). This assay preferentially detects mature miR-16 over duplex and hairpin forms. We also determined cell cycle phenotypes and miR-16 levels induced by increasing concentrations of the miR-16 duplex.
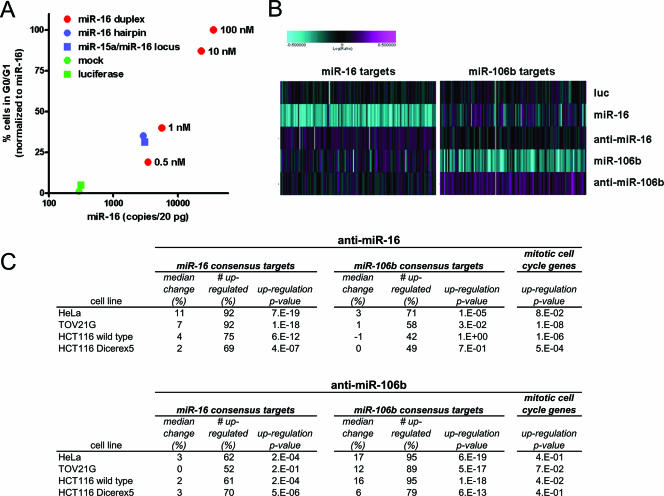
FIG. 3.
Natural forms and levels of miR-16 regulate cell cycle progression. (A) miR-16 expressed from hairpin precursors can trigger the phenotype of accumulation of cells at a stage of the cell cycle. HCT116 Dicerex5 cells were transfected with increasing concentrations of the miR-16 duplex (0.5, 1, 10, and 100 nM) or plasmids carrying miR-16 expressed as an shRNA (mR-16 hairpin) or from its endogenous locus on chromosome 13 (miR-16 locus). For transfections with plasmids, the DNA concentration was chosen to maximize transfection efficiency while minimizing toxicity (1.5 μg of DNA/2 × 105 to 3 × 105 cells in a 6-well dish). Cells were treated with nocodazole and analyzed as described in the legend to Fig. 2A. The percentages of cells in G0/G1 in different experiments were normalized so that background control and miR-16-transfected cells gave 0% and 100% of cells in G0/G1, respectively [(the percentage of G1 cells in the sample − the percentage of background G1 cells)/(the percentage of G1 miR-16-transfected cells − the percentage of background G1 cells) × 100]. For duplex transfections, we used mock-transfected cells to determine the percentage of background G1 cells. For plasmid transfections, we used cells transfected with an empty vector to determine the percentage of background G1 cells. miR-16 copy numbers (copies/20 pg of RNA) were determined by a quantitative primer extension PCR assay. miR-16 copy numbers shown are the means of quadruplicate determinations that typically differed from the mean by <15%. The results shown are representative of results from at least two experiments for each form of miR-16. (B) miR-16 and miR-106b targets are up-regulated by specific anti-miRs. HeLa cells were transfected with luciferase siRNA, miR-16, miR-106b, anti-miR-16, or anti-miR-106b. Microarray analysis was performed as described in the legend to Fig. 1C, and gene expression changes corresponding to consensus miR-16 (left panel)- and miR-106b (right panel)-down-regulated transcripts were examined. Shown is a heat-map depiction of gene expression changes in cells transfected with different miRNAs or anti-miRs. (C) Summary of miR-16 and miR-106b target regulation by specific anti-miRs in different cell lines. HeLa, TOV21G, HCT116 wild-type, and HCT116 Dicerex5 cells were transfected with anti-miR-16 (top panel) or anti-miR-106b (bottom panel). Microarray analysis was performed as described in the legend to Fig. 1C, and gene expression changes corresponding to miR-16 consensus and miR-106b consensus targets (see Table S4 in the supplemental material) were examined. For controls, we compared patterns of regulation of randomized sets of genes in luciferase siRNA-treated cells. Median change, median percentage of increase in regulation. Control sets gave 0.01% ± 0.6% change. # up-regulated, percentage of miR-16 or miR-106b consensus targets having a level of regulation of >0. Control sets gave 50% ± 5% of targets with a level of regulation of >0. Up-regulation P value, Wilcoxon signed-rank P values for the up-regulation of the indicated target sets. Mitotic cell cycle genes, transcripts down-regulated by the miR-16 duplex at 24 h and annotated with the GO biological process term mitotic cell cycle (Fig. 1C).
As shown in Fig. 3A, the percentage of cells in G0/G1 increased with rising concentrations of miR-16. The distinct accumulation of cells in G0/G1 was achieved with miR-16 duplex concentrations of 0.5 to 1 nM, which resulted in miR-16 levels of ∼3,500 to 5,600 copies/20 pg, respectively. This compared with ∼300 copies/20 pg measured with mock- or luciferase-transfected cells. Both the miR-16 shRNA and miR-16 locus consistently triggered a ∼2-fold increase in the number of cells in G0/G1 and an increase in miR-16 levels to ∼3,000 copies/20 pg. The miR-16 shRNA triggered identical results in HCT116 wild-type cells (not shown), suggesting that the processing of the hairpin does not require full Dicer activity. Transfection with empty vector and an shRNA for miR-106b did not result in the accumulation of cells in G0/G1 or miR-16 levels greater than those in mock-transfected cells (not shown). At equivalent miR-16 levels, the duplex was slightly less efficient at inducing the accumulation of cells in G0/G1, consistent with the idea that not all the duplex with which cells were transfected was accessible for incorporation into the RNA-induced silencing complex. The overexpression of miR-16 from hairpin precursors therefore triggered the accumulation of cells in G0/G1 with a level of efficiency similar to that of the miR-16 duplex.
It was also important to demonstrate that the cellular phenotype induced by miR-16 in gain-of-function experiments was reversed in loss-of-function experiments. We therefore performed loss-of-function analysis using LNA- and 2′-_O_-methyl-modified oligonucleotides (anti-miRs) to mediate the specific inhibition of miRNA function (26, 42). To demonstrate the effectiveness of transfection with anti-miRs, we first sought to demonstrate that endogenous miR-16 and miR-106b levels were reduced by this treatment. We found, however, that anti-miRs interfered with miRNA quantitation by a PCR assay (43). We therefore tested the ability of anti-miR-16 to block the accumulation of cells in G0/G1 induced by transfection with miR-16. Wild-type HCT116 cells were first transfected with miR-16 and then transfected with anti-miR-16 or anti-miR-106b 24 h later. We found that anti-miR-16, but not anti-miR-106b, abolished the accumulation of cells in G0/G1 induced by miR-16, indicating the effectiveness of our anti-miR-16 transfection protocols (not shown).
We then transfected wild-type HCT116 cells with LNA-modified anti-miR-16 and anti-miR-106b and measured levels of several miR-16 target transcripts using real-time PCR. These preliminary experiments showed increased expression of several miR-16 targets in anti-miR-16-transfected cells compared to anti-miR-106b-transfected cells, but the effects were small (<30% increase for each target).
To better quantify the effects of loss-of-function experiments, we used microarray profiling to measure the effects of anti-miRs on consensus miR-16 and miR-106b transcripts (see Table S4 in the supplemental material). We reasoned that small gene expression changes would assume greater statistical significance if measurements of many transcripts were combined. We transfected HeLa, TOV21G, HCT116 wild-type, and Dicerex5 cells with 2′-_O_-methyl oligonucleotide inhibitors of miR-16 and miR-106b (anti-miR-16 and anti-miR-106b, respectively) and examined changes in gene expression by using microarrays.
Consistent with the real-time PCR results, consensus miR-16-regulated transcripts were weakly up-regulated by anti-miR-16 but not anti-miR-106b. A heat map of results obtained with HeLa cells is shown in Fig. 3B. miR-16 and miR-106b consensus transcripts were weakly but detectably up-regulated by the specific anti-miR but not the nonspecific anti-miR. For other cells with wild-type Dicer function, the median regulation of consensus miR-16 and miR-106b targets was also slightly increased in cells transfected with the specific anti-miR (Fig. 3C).
It was of interest to determine the overlap of genes up-regulated in loss-of-function experiments and those down-regulated in gain-of-function experiments. The numbers of up-regulated miR-16 or miR-106b targets were higher in Dicer wild-type cells treated with the specific anti-miR (Fig. 3B and C). However, it was difficult to quantify the overlap in gene sets precisely because the magnitude of the regulations induced by anti-miRs was small. In HeLa cells, it appeared that nearly 100% of down-regulated targets were up-regulated by specific anti-miRs (Fig. 3B).
When considered as a group, miR-16 targets were more significantly up-regulated than miR-106b targets in cells treated with anti-miR-16 (Fig. 3C). Target regulation was greatly reduced in HCT116 Dicerex5 cells, as expected given the reduced miRNA levels in these cells. Likewise, anti-miR-106b-treated cells showed significant regulation of miR-106b, but not miR-16, consensus targets. Taken together, these results indicate that in cells with wild-type Dicer function, many, if not most, miR-16 and miR-106b targets down-regulated by miRNA duplexes in gain-of-function experiments were up-regulated by specific anti-miRs in loss-of-function experiments.
It was also important to determine the effect of anti-miRNAs on cell cycle progression. miR-16-regulated transcripts annotated with the GO biological process term mitotic cell cycle (Fig. 1B) were significantly up-regulated by anti-miR-16 in TOV21G and HCT116 wild-type cells (Fig. 3C). These transcripts were less regulated in HeLa cells, which do not show an miR-16 cell cycle phenotype, and HCT116 Dicerex5 cells, which have reduced endogenous levels of miR-16. Mitotic cell cycle transcripts were not significantly regulated in any cell line by anti-miR-106b (Fig. 3C). These results support the regulation of cell cycle progression by physiological levels of miR-16. However, analysis of the cell cycles of anti-miR-16-treated TOV21G and HCT116 wild-type cells did not reveal any obvious differences from that of control-treated cells (not shown). Thus, gene expression changes measured by microarray were not sufficient to drive a detectable cell cycle phenotype in these cells.
We hypothesized that cells having higher endogenous levels of miR-16 would be more susceptible to anti-miR-16-induced phenotypic changes detectable by flow cytometry. We therefore screened a number of transfectable cell lines for endogenous miR-16 levels by using a quantitative primer extension PCR assay (43). These experiments showed that SW1417 cells had elevated levels of endogenous miR-16 (∼1,500 copies/cell; not shown). The transfection of SW1417 cells with anti-miR-16 resulted in a significant decrease in numbers of cells in G0/G1 compared to mock-transfected cells [10% ± 1% decrease (three independent experiments); P < 1E(−3)]. In contrast, anti-miR-106b-transfected cells did not show significant differences from mock-transfected cells [5%_+_3% decrease (three independent experiments); _P_ > 5E(−2)]. Therefore, disruption of physiological miR-16 levels in certain cell types can alter cell cycle distribution.
Characteristics of miR-16-down-regulated transcripts.
Consensus miR-16-down-regulated transcripts (see Table S4 in the supplemental material) overlapped with but were not identical to computationally predicted miR-16 targets (see Fig. S6 in the supplemental material). Nearly 60% (65/110) of miR-16-down-regulated transcripts were not predicted by either of two different computational methods (34, 36). Conversely, >90% of the computational targets were not significantly down-regulated on microarrays. Moreover, most computational targets were unique to the particular method used (55% and 68% unique for TargetScan and PicTar, respectively). Similar results were found (not shown) with miR-16 targets predicted with other computational methods (31, 52). The poor overlap was unlikely to be due to a lack of target expression in HCT116 Dicerex5 cells, since the calculations were restricted to transcripts expressed at ∼≥1 copy per cell.
We next compared the properties of miR-16-down-regulated transcripts with those of a set of unregulated (background) transcripts selected to have similar distributions of expression levels. Hexamer motifs matching the miR-16 seed region (target sites) were found in both CDS and 3′UTRs. The numbers of transcripts with CDS target sites did not differ significantly between the miR-16-down-regulated transcripts and the background set (not shown). In contrast, nearly all miR-16-down-regulated transcripts had hexamer matches in their 3′UTRs, compared to less than half of the background set [P < 5E(−13)] (Fig. 4A).
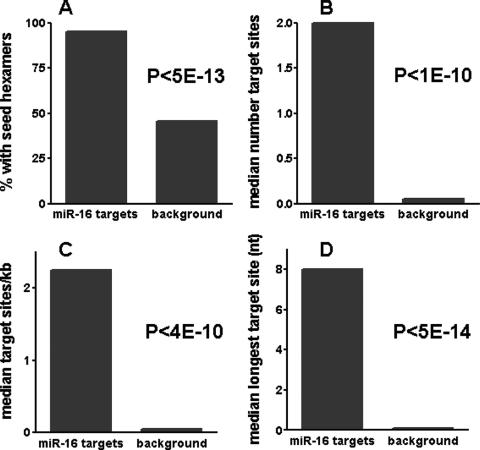
FIG. 4.
miR-16-down-regulated transcripts contain multiple miR-16 target sites. Properties of consensus miR-16-down-regulated transcripts (see Table S4 in the supplemental material) were compared with those of an expression level-matched background set. The significance of the differences between the groups is indicated (Wilcoxon rank-sum P values). (A) Nearly all miR-16-down-regulated transcripts contain miR-16 target sites in their 3′UTRs. Shown are the percentages of miR-16-down-regulated transcripts and background transcripts that contain sites matching miR-16 seed region hexamers in their 3′UTRs (miR-16 target sites). (B) miR-16-down-regulated transcripts contain multiple copies of miR-16 target sites in their 3′UTRs. Shown are the median numbers of miR-16 target sites in 3′UTRs of miR-16-down-regulated transcripts and background transcripts. (C) miR-16-down-regulated transcripts contain a higher density of miR-16 target sites in their 3′UTRs. Shown are the median densities of miR-16 target sites in the 3′UTRs of miR-16-down-regulated transcripts and background transcripts. (D) miR-16-down-regulated transcripts contain longer miR-16 target sites in their 3′UTRs. The longest target site per transcript was identified, and the median lengths of the longest target sites in the 3′UTRs of miR-16-down-regulated transcripts and background transcripts are shown.
The median number of target sites per transcript (Fig. 4B) was also higher for miR-16 transcriptional targets than for the background set [P < 1E(−10)]. The increased number of target sites per transcript for miR-16-down-regulated transcripts was partially attributable to longer 3′UTRs in these transcripts [median of ∼1,150 nt for miR-16-down-regulated transcripts versus ∼660 nt for the background set; P < 1E(−2)] but more significantly to a greater number of target sites per kilobase of the 3′UTRs [P < 4E(−10)] (Fig. 4C).
Furthermore, the longest target site per transcript was significantly longer for miR-16-down-regulated transcripts than for background transcripts [P < 5E(−14)] (Fig. 4D). The median length of the longest sites in individual miR-16 targets was 8 bases, versus 0 bases for background transcripts. Even when transcripts without hexamer matches were excluded from the background, the median length of the longest sites in individual background transcripts with hexamer matches was only 6 bases [not shown; P < 3E(−10)]. Thus, many miR-16-down-regulated transcripts have multiple target sites matching the miR-16 seed region, generally with at least one site showing extended complementarity to the miR-16 seed.
Many miR-16-down-regulated transcripts regulate G0/G1-to-S cell cycle progression.
We wished to determine which miR-16-down-regulated transcript(s) was essential for the G0/G1 cell cycle accumulation phenotype. We hypothesized that siRNA-mediated silencing of such transcripts would yield a phenocopy of the miR-16-induced phenotype (i.e., trigger the accumulation of cells in G0/G1). To test this idea, we transfected HCT116 Dicerex5 cells with siRNAs (pools of three siRNAs/target) targeting each miR-16-down-regulated transcript (n, 102 individual pools targeting the well-characterized genes from Table S4 in the supplemental material). We then analyzed transfected cells for their cell cycle distributions. Sequences of siRNAs triggering the accumulation of cells at a stage of the cell cycle are shown in Table S1 in the supplemental material. Raw and normalized G0/G1-cell accumulation values and target silencing data are presented in Table S3 in the supplemental material.
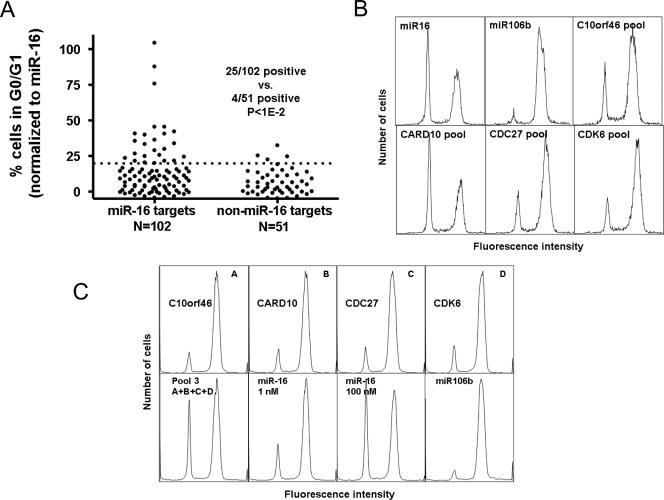
As shown in Fig. 5A, 25/102 siRNA pools (∼25%) targeting miR-16-down-regulated transcripts triggered the accumulation of ≥20% of the cells in G0/G1. In contrast, only 4/51 (∼8%) of siRNA pools targeting transcripts that did not match the miR-16 seed triggered G0/G1-cell accumulation (P < 0.01; Fisher's exact test). Examples of cell cycle phenotypes triggered by siRNA pools are depicted in Fig. 5B. These experiments provided further evidence that many miR-16 family targets regulate G0/G1 transition.
FIG. 5.
miR-16-down-regulated transcripts cooperatively regulate cell cycle progression. (A) miR-16-down regulated transcripts are enriched with targets whose disruption causes the accumulation of cells in G0/G1. HCT116 Dicerex5 cells were individually transfected with siRNA pools targeting 102 transcripts containing matches to the miR-16 seed region (miR-16 targets) and 51 transcripts that did not contain miR-16 seed region matches (non-miR-16 targets). Cell cycle phenotypes were determined as described in the legend to Fig. 2. The percentages of cells in G0/G1 in different experiments were normalized as described in the legend to Fig. 3A. Before normalization, the background control had an average of 6.4% + 1.1% (mean + standard deviation) of cells in G0/G1, and among miR-16-transfected cells, 41% + 5% were in G0/G1 (results are from seven independent experiments). Plotted are normalized percentages of cells in G0/G1 for each siRNA pool (dots). The dotted line indicates a cutoff of 20%, chosen statistically to maximize the recovery of miR-16 targets while maintaining the significance of the difference between miR-16 targets and non-miR-16 targets. Non-miR-16 targets tested are listed in Table S5 in the supplemental material. (B) Cells transfected with selected siRNA pools are phenocopies of miR-16-transfected cells. HCT116 Dicerex5 cells were transfected with individual miR-16, miR-106b (control), and siRNA pools corresponding to selected miR-16-down-regulated targets (three siRNAs, each at 33 nM; 100 nM total concentration). (C) Cooperative cell cycle regulation by miR-16-down-regulated targets. Cells were transfected with individual siRNAs at 0.25 nM corresponding to selected miR-16-down-regulated targets that induce the accumulation of cells in G0/G1. Cells were also transfected with these siRNAs together as a pool (pool 3, 0.25 nM [each] siRNAs; total concentration, 1 nM). Cells were transfected with miR-16 (1 nM and 100 nM) and miR-106b (100 nM) as controls. Cell cycle phenotypes were determined as described in the legend to Fig. 2.
The siRNA pool targeting CARD10 was the only one that induced the accumulation of cells in G0/G1 to the same extent as miR-16 (see Table S1 in the supplemental material; Fig. 5B). However, siRNA titration experiments showed that the phenotype induced by an individual CARD10 siRNA required greater CARD10 gene silencing than was achieved with miR-16 (not shown). Individual siRNAs targeting CDK6, CDC27, and C10orf46 genes also triggered less G0/G1-cell accumulation than miR-16, despite silencing their target mRNA more strongly than miR-16 (not shown and Table S1 in the supplemental material). HCT116 Dicerex5 cells transfected with miR-16 (100 nM) had CDK6, CARD10, CDC27, and C10orf46 gene transcript levels ranging from ∼45% to 75% of the maximum (i.e., levels in control-transfected cells; not shown). In contrast, cells transfected with the best siRNAs corresponding to CDK6, CARD10, CDC27, or C10orf46 genes (33 nM) had transcript levels of <40% of the maximum (see Table S1 in the supplemental material). Therefore, the silencing of individual miR-16-down-regulated targets can produce phenocopies of the miR-16-induced phenotype, but the effects are weaker.
One explanation for these findings is that the stronger cell cycle phenotype elicited by miR-16 resulted from the coordinate silencing of multiple targets. To test this possibility, we devised a strategy for comparing phenotypes caused by siRNAs corresponding to different targets alone and in combination (i.e., pools of siRNAs corresponding to different targets). If the miR-16 phenotype results from the coordinate silencing of different targets, then siRNAs corresponding to miR-16 targets should be more effective when added together than when tested individually. A potential limitation of this approach is that pooled siRNAs can compete for RNA-induced silencing complex binding, which may reduce silencing by the individual siRNAs (23).
We first determined whether the siRNA pools we tested were representative of the individual siRNAs comprising them. Individual siRNAs from the 25 siRNA pools that gave an accumulation of ≥20% of cells in G0/G1 were tested for their abilities to produce a phenocopy of the miR-16 phenotype. In some cases, additional siRNAs corresponding to the same targets were tested. For 24/25 pools, at least one member of the siRNA pool gave a phenotype as strong as that given by the pool itself (see Table S3 in the supplemental material).
We then performed experiments to determine whether pooled siRNAs corresponding to different miR-16 targets were more effective than the individual siRNAs at triggering arrest in G0/G1. For these experiments, we transfected cells with individual siRNAs at a concentration at which most gave phenotypes only slightly above the background (0.25 nM) (Fig. 5B). We tested whether a pool of siRNAs representing each target whose disruption caused ≥20% of cells to accumulate in G0/G1 (pool 1) (see Table S3 in the supplemental material) triggered a stronger phenotype than the individual siRNAs. To quantify these comparisons, we normalized percentages as described in the legend to Fig. 5A. None of the individual siRNAs triggered a normalized value of ≥20% of cells to accumulate in G0/G1 (at 0.25 nM), whereas a pool of all 24 siRNAs (pool 1; 0.25 nM [each]; total concentration, 6 nM) triggered a normalized value of 73% of cells to accumulate.
In another experiment, we compared individual siRNAs (0.25 nM) targeting the three targets that gave the strongest phenotypes as indicated in Fig. 5A (CARD10, KIAA0317, and C9orf91 genes) (Table S3 in the supplemental material) to a pool of all three siRNAs (pool 2; 0.25 nM [each]; total concentration, 0.75 nM). Each individual siRNA triggered a normalized value of <30% of cells to accumulate in G0/G1, but the pool of three siRNAs triggered a normalized value of ∼50% of cells to accumulate (see Table S3 in the supplemental material). These experiments supported our hypothesis that the robust cell cycle phenotype elicited by miR-16 results from coordinate silencing of multiple targets.
To address coordinate silencing and the G0/G1-cell accumulation phenotype more quantitatively, we focused on a more limited subset of targets (Fig. 5B). We first needed to demonstrate that the cell cycle phenotypes triggered by siRNAs corresponding to these targets resulted from the silencing of the intended targets rather than unintended targets (28). For CDK6, CARD10, CDC27, and C10orf46 genes, we identified at least two siRNAs that triggered the accumulation of cells in G0/G1. We then tested these siRNAs for their abilities to silence their target genes (see Table S3 in the supplemental material). Generally, siRNAs triggering phenotypes also silenced their targets well.
We next explored whether representative individual siRNAs corresponding to each target were more effective when pooled together than they were alone. Individual siRNAs targeting CDK6, CDC27, CARD10, and C10orf46 genes triggered minimal levels (normalized value of <12%) of cells to accumulate in G0/G1 when tested at 0.25 nM (Fig. 5C). However, when these siRNAs were pooled (pool 3; total concentration, 1 nM), a much stronger phenotype was observed (a normalized value of ∼61% of cells in G0/G1) (Fig. 5C). (Equivalent results were obtained when luciferase siRNA was added to individual siRNAs to maintain a total concentration of 1 nM.) Measurements of target silencing by quantitative PCR or immunoblotting showed that the silencing of target transcripts was maintained or slightly reduced when siRNAs were pooled (see Fig. S7 in the supplemental material).
The effects of the four siRNAs tested as described in the legend to Fig. 5C were greater than additive. If the effects were additive, we would have expected to find ∼34% more pool-transfected cells (normalized value) in G0/G1 than control-transfected cells (19% + 11% + 8.3% − 2.7% = ∼36%) (see Table S3 in the supplemental material). This compares with the normalized value of ∼61% of cells in G0/G1 that we actually observed (Fig. 5C; see Table S1 in the supplemental material). In four independent experiments, 67% ± 15% of pool-transfected cells were in G0/G1 versus the expected value of 37% ± 13% [P < 8.8E(−7); chi-square distribution]. Taken together, these findings demonstrate that miR-16 coordinately regulates targets that collaborate to regulate cell cycle progression from G0/G1 to S.
Levels of miR-16 family-down-regulated transcripts negatively correlate with miR-195 levels in human tumors.
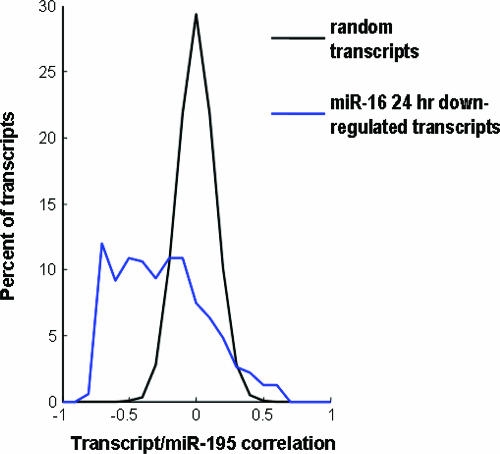
We wished to determine whether the phenotypes we showed for miR-16 family members in cell culture are relevant to an in vivo setting. We reasoned that steady-state levels of transcripts down-regulated by miR-16 family members would inversely correlate with miRNA levels. To examine this possibility, we compared levels of an miR-16 family member with levels of miR-16-down-regulated transcripts in a panel of human tumors and matched adjacent normal tissue samples (see Table S6 in the supplemental material). We chose miR-195 for comparison because it is the only member of the family for which we have reliable tumor atlas expression data. mRNA and miRNA expression levels in tumors and adjacent normal tissues were expressed as ratios of these levels to expression levels in a pool of normal samples of each tissue type. As shown in Fig. 6, we observed significant negative correlations between miR-195 levels and the levels of transcripts down-regulated by miR-16 at 24 h posttransfection. miR-16-down-regulated transcripts were significantly more likely to be negatively correlated with miR-195 levels than would be expected by chance [P < 1.5E(−12); Wilcoxon rank-sum P value for a difference in median correlation coefficient versus random permutations of expression ratios]. Thus, tumors with high levels of miR-195 tended to have low levels of transcripts that were down-regulated by transfection with miR-16, and vice versa. These results, therefore, show that gene expression changes triggered by miRNA transfection in our in vitro model reflect the relationship between levels of the transcripts and the miRNA in human tumors.
FIG. 6.
Levels of miR-16-down-regulated transcripts negatively correlate with miR-195 levels in human tumors. RNA was isolated from a series of 29 tumors and 28 adjacent uninvolved normal tissues. mRNA expression was measured using microarrays, and miR-195 levels were determined using a quantitative primer extension PCR assay. mRNA and miRNA expression levels in tumors and adjacent normal tissues were expressed as ratios of these levels to expression levels in a pool of normal samples from each tissue type. Correlations between expression level ratios for miR-195 and transcripts down-regulated 24 h after the transfection of tissue culture cells with miR-16 were calculated. As a control, correlations were also calculated for ∼200 random permutations of expression ratios (random transcripts).
DISCUSSION
Determining miRNA targets and function.
There are several reasons to believe that our current state of knowledge of miRNA targets is not mature. Current target predictions rely heavily on detecting seed region complementary motifs conserved in 3′UTR sequences across divergent species (31, 34, 37, 45, 52). Here, we used an experimental approach to screen for miRNA target sites, irrespective of their locations, levels of conservation across species, or degrees of complementarity. Functional annotation of these targets and follow-up experiments showed that the miR-16 family of microRNAs negatively regulates cell cycle progression by inducing G0/G1-cell accumulation. Most experimental miR-16 targets contain several seed matches in their 3′UTRs, which may extend to eight or more perfectly matched nucleotides. The presence of multiple seed matches in miR-16-down-regulated transcripts argues that these targets are regulated not because of secondary effects or artifacts of overexpression but because they are direct targets. Further evidence that they are direct targets comes from kinetic analysis of their expression and the reduction in their regulation when mismatches are introduced into the seed region of miR-16 duplexes. However, we would not have detected genes whose protein products are silenced without transcriptional regulation. It remains to be determined what fraction of miRNA targets are regulated solely at the protein level.
Microarrays detect the regulation of many transcripts not computationally predicted. Some of the miR-16 targets whose disruption triggered G0/G1-cell accumulation (Fig. 5A) were predicted by TargetScan (CDK6, CDC27, and C10orf46 genes) and/or PicTar (C10orf46, C9orf42, CARD10, PPP1R11, and RAB11FIP2 genes). However, most (18/25) targets whose disruption triggered the accumulation of cells in G0/G1 were not predicted by either computational method. Nonetheless, the four targets with which we demonstrated cooperative interactions (CDK6, CDC27, CARD10, and C10orf46 genes) were all predicted by at least one of the two methods.
Gain-of-function experiments are widely used for functional studies of miRNAs. It is important, however, to complement gain-of-function experiments with loss-of-function analyses. Loss-of-function experiments have revealed phenotypes associated with certain miRNAs (35). These previous studies revealed the down-regulation of miR-16 in many tissues by an anti-miR, though the functional consequences of this disruption were not noted. In our studies, we demonstrated the up-regulation of miR-16 and miR-106b targets upon the transfection of cells with anti-miRs targeting these miRNAs. However, the changes in the regulation of each individual target were small, perhaps in part because of the functional redundancy of the miR-16 family. Measurements of levels (copies/cell) of miR-16, miR-15a, miR-15b, and miR-195 in several cell lines showed that miR-16 usually made up ∼50 to 60% of this miRNA family (not shown). Thus, complete inhibition of miR-16 by using anti-miR-16 would have resulted in only a partial reduction in total levels of the family. The modest increase in target levels triggered by anti-miR-16 is therefore consistent with what would be expected given the functional redundancy of this miRNA family. Our experiments show, moreover, that microarray measurements offer an advantage in measuring small changes in gene expression resulting from the disruption of redundant miRNAs. When miRNA targets were considered as a group, we detected significant changes in regulation, despite the low magnitude of each individual measurement.
miR-16 family and cancer.
The locus corresponding to miR-15a and miR-16-1 at 13q14 is deleted in more than half of B-cell chronic lymphocytic leukemias (9). The undeleted miR-15a-miR-16-1 allele in chronic lymphocytic leukemia patients may also contain inactivating point mutations (10). These observations have led to the proposal that miR-15a and miR-16-1 may behave as tumor suppressors (9, 10).
However, several considerations suggest that the miR-15a-miR-16 locus does not fit the classical Knudson two-hit model for inactivation of a tumor suppressor gene (33). While deletion of one miR-15a-miR-16 allele is quite common, inactivating mutations in the remaining allele are rare (7). In addition, even if both miR-15a-miR-16 alleles were lost, other miR-16 family members would still be widely expressed (the miR-15b-miR-16-2 locus on chromosome 3, miR-195, miR-103, and miR-107). Tumors having deletions of the miR-15a-miR-16-1 locus do not commonly have simultaneous deletions at loci corresponding to these other family members (53). Thus, deletion and/or inactivation of the miR-15a-miR-16-1 locus could be compensated for to some extent by the expression of other miRNAs from this family.
In this study, we have shown that there is functional redundancy among members of the miR-16 family. Our data further suggest that relatively small alterations in miR-16 copy numbers can have phenotypic consequences. Taken together, these findings suggest that the miR-15a-miR-16-1 locus does not behave as a classical tumor suppressor gene, in which both alleles are lost or inactivated. Rather, the deletion of an miR-15a-miR-16-1 allele(s) likely regulates the dosage of this miRNA family. A current model suggests that miRNAs function as micromanagers that fine tune gene expression (3). In an extension of this view, copy number alterations of the miR-15a-miR-16-1 locus in tumors would further tune levels of gene expression by regulating the micromanagers.
It will be important to evaluate carefully the effects of miR-15a-miR-16-1 deletion in model systems more relevant to tumor biology than the one we used in this study. Clearly, further studies of miR-15a-miR-16 will greatly benefit from the construction of cell lines, tumors, and animals matched except for the expression of this locus. Nonetheless, our findings suggest that the deletion of the miR-15a-miR-16 locus may enable tumor cells to avoid cell cycle control mechanisms. Other functions have been attributed to miR-15 and miR-16. Cimmino et al. showed that miR-15 and miR-16 negatively regulate the antiapoptotic protein BCL2 (14). In our studies, BCL2 does not seem to play a major role in miR-16 function, as neither the BCL2 mRNA nor its protein product changes significantly following transfection with miR-16 (not shown). Furthermore, we do not see evidence for apoptosis following transfection with miR-15-miR-16 (14). This discrepancy between our results and those of previous studies may suggest cell type differences in miR-16 functions. It is well known that hematopoietic cells like those in studies by Cimmino et al. are especially susceptible to apoptosis. miR-16 has also been implicated in AU-rich-element-mediated mRNA instability (30). However, mismatches outside the seed region that disrupted miR-16 effects on transcript stability (30) did not reduce the accumulation of cells in G0/G1 (not shown). Thus, the function we demonstrate for miR-15-miR-16 does not depend on AU-rich elements.
miR-16 targets cooperatively regulate cell cycle progression.
As functions are assigned to miRNAs, it is important to determine which targets are responsible for their phenotypes. Although early genetic studies with miRNAs focused on relatively few targets for miRNAs (1), we now know that individual animal miRNAs are capable of regulating hundreds of targets (3). Here we have demonstrated that miR-16 regulates many targets that function to regulate cell cycle transition from G0/G1 to S. Since miR-16 family members are widely expressed, it is important to understand how they can regulate the highly orchestrated patterns of gene expression that occur during the cell cycle. Many regulators of cell cycle function are periodically expressed during the cell cycle (51), and it will be important to see whether the miR-16 family behaves likewise. For example, it is possible that miRNA loci could be differentially subject to posttranscriptional regulation at the Drosha processing step, which appears deficient in tumors (48).
The siRNA-mediated silencing of ∼25% of miR-16-down-regulated targets triggered the miR-16 phenotype. Among miR-16-down-regulated targets are many known cell cycle regulators that did not induce G0/G1-cell accumulation but may modify the effects of other targets (i.e., E2F7, CDC25A, CHEK1, WEE1, and CCNE1 genes). There also may be miRNA targets that induce the accumulation of cells in G0/G1 but escape detection because they are not regulated at the mRNA level. Thus, our experiments likely underestimate the number of miR-16 targets that regulate cell cycle progression. Taken together, our findings argue that miR-16 targets act in concert, rather than individually, to regulate G0/G1 progression.
Our data further argue that miR-16 targets may function synergistically. Simultaneous siRNA-mediated silencing of CDK6, CDC27, CARD10, and C10orf46 genes resulted in greater accumulation of cells in G0/G1 than silencing of any of the genes individually. The product of each of these genes individually has a known or predicted function in regulating cell cycle progression. CDK6 is a kinase that partners with the d-type cyclins, phosphorylates RB1, and regulates the G1 phase of the cell cycle (20). CDC27 is a component of the anaphase-promoting complex, which regulates mitosis and G1 phases of the cell cycle (13). CARD10 is an activator of NF-κB signaling that controls cell cycle progression and proliferation (24, 44). C10orf46 is a poorly characterized transcript encoding a predicted member of the cullin family of E3 ubiquitin ligases, which regulate cell cycle progression. Coordinate regulation of these and other cell cycle regulators suggests highly orchestrated cell cycle control by the miR-16 family of miRNAs.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank the Rosetta Inpharmatics Gene Expression Lab for microarray analysis; Rosetta Biosoftware for data analysis software; Jeff Bradshaw, Terry Ward, Matt Biery, Jane Guo, Kristine Niemeyer, Jill Magnus, and Sumi Kobayashi for technical assistance; and members of the Cancer Biology Department for invaluable discussions and support.
Rosetta Inpharmatics is a wholly owned subsidiary of Merck & Co.
Footnotes
▿
Published ahead of print on 22 January 2007.
REFERENCES
- 1.Ambros, V. 2004. The functions of animal microRNAs. Nature 431**:**350-355. [DOI] [PubMed] [Google Scholar]
- 2.Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122**:**553-563. [DOI] [PubMed] [Google Scholar]
- 3.Bartel, D. P., and C. Z. Chen. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5**:**396-400. [DOI] [PubMed] [Google Scholar]
- 4.Bentwich, I. 2005. Prediction and validation of microRNAs and their targets. FEBS Lett. 579**:**5904-5910. [DOI] [PubMed] [Google Scholar]
- 5.Bentwich, I., A. Avniel, Y. Karov, R. Aharonov, S. Gilad, O. Barad, A. Barzilai, P. Einat, U. Einav, E. Meiri, E. Sharon, Y. Spector, and Z. Bentwich. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37**:**766-770. [DOI] [PubMed] [Google Scholar]
- 6.Berezikov, E., V. Guryev, J. van de Belt, E. Wienholds, R. H. Plasterk, and E. Cuppen. 2005. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 120**:**21-24. [DOI] [PubMed] [Google Scholar]
- 7.Borkhardt, A., U. Fuchs, and T. Tuschl. 2006. MicroRNA in chronic lymphocytic leukemia. N. Engl. J. Med. 354**:**524-525. (Letter.) [DOI] [PubMed] [Google Scholar]
- 8.Brennecke, J., A. Stark, R. B. Russell, and S. M. Cohen. 2005. Principles of microRNA-target recognition. PLoS Biol. 3**:**e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calin, G. A., C. D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, L. Rassenti, T. Kipps, M. Negrini, F. Bullrich, and C. M. Croce. 2002. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 99**:**15524-15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calin, G. A., M. Ferracin, A. Cimmino, G. Di Leva, M. Shimizu, S. E. Wojcik, M. V. Iorio, R. Visone, N. I. Sever, M. Fabbri, R. Iuliano, T. Palumbo, F. Pichiorri, C. Roldo, R. Garzon, C. Sevignani, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353**:**1793-1801. [DOI] [PubMed] [Google Scholar]
- 11.Calin, G. A., C. G. Liu, C. Sevignani, M. Ferracin, N. Felli, C. D. Dumitru, M. Shimizu, A. Cimmino, S. Zupo, M. Dono, M. L. Dell'Aquila, H. Alder, L. Rassenti, T. J. Kipps, F. Bullrich, M. Negrini, and C. M. Croce. 2004. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. USA 101**:**11755-11760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calin, G. A., C. Sevignani, C. D. Dumitru, T. Hyslop, E. Noch, S. Yendamuri, M. Shimizu, S. Rattan, F. Bullrich, M. Negrini, and C. M. Croce. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 101**:**2999-3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Castro, A., C. Bernis, S. Vigneron, J. C. Labbe, and T. Lorca. 2005. The anaphase-promoting complex: a key factor in the regulation of cell cycle. Oncogene 24**:**314-325. [DOI] [PubMed] [Google Scholar]
- 14.Cimmino, A., G. A. Calin, M. Fabbri, M. V. Iorio, M. Ferracin, M. Shimizu, S. E. Wojcik, R. I. Aqeilan, S. Zupo, M. Dono, L. Rassenti, H. Alder, S. Volinia, C. G. Liu, T. J. Kipps, M. Negrini, and C. M. Croce. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 102**:**13944-13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cummins, J. M., Y. He, R. J. Leary, R. Pagliarini, L. A. Diaz, Jr., T. Sjoblom, O. Barad, Z. Bentwich, A. E. Szafranska, E. Labourier, C. K. Raymond, B. S. Roberts, H. Juhl, K. W. Kinzler, B. Vogelstein, and V. E. Velculescu. 2006. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA 103**:**3687-3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench, J. G., and P. A. Sharp. 2004. Specificity of microRNA target selection in translational repression. Genes Dev. 18**:**504-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farh, K. K., A. Grimson, C. Jan, B. P. Lewis, W. K. Johnston, L. P. Lim, C. B. Burge, and D. P. Bartel. 2005. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science 310**:**1817-1821. [DOI] [PubMed] [Google Scholar]
- 18.Giraldez, A. J., Y. Mishima, J. Rihel, R. J. Grocock, S. Van Dongen, K. Inoue, A. J. Enright, and A. F. Schier. 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312**:**75-79. [DOI] [PubMed] [Google Scholar]
- 19.Griffiths-Jones, S., R. J. Grocock, S. van Dongen, A. Bateman, and A. J. Enright. 2006. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34**:**D140-D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossel, M. J., and P. W. Hinds. 2006. From cell cycle to differentiation: an expanding role for cdk6. Cell Cycle 5**:**266-270. [DOI] [PubMed] [Google Scholar]
- 21.He, L., J. M. Thomson, M. T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S. W. Lowe, G. J. Hannon, and S. M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature 435**:**828-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hede, K. 2005. Studies define role of microRNA in cancer. J. Natl. Cancer Inst. 97**:**1114-1115. [DOI] [PubMed] [Google Scholar]
- 23.Holen, T., M. Amarzguioui, M. T. Wiiger, E. Babaie, and H. Prydz. 2002. Positional effects of short interfering RNAs targeting the human coagulation trigger tissue factor. Nucleic Acids Res. 30**:**1757-1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong, G. S., and Y. K. Jung. 2002. Caspase recruitment domain (CARD) as a bi-functional switch of caspase regulation and NF-kappaB signals. J. Biochem. Mol. Biol. 35**:**19-23. [DOI] [PubMed] [Google Scholar]
- 25.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102**:**109-126. [DOI] [PubMed] [Google Scholar]
- 26.Hutvagner, G., M. J. Simard, C. C. Mello, and P. D. Zamore. 2004. Sequence-specific inhibition of small RNA function. PLoS Biol. 2**:**E98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iorio, M. V., M. Ferracin, C. G. Liu, A. Veronese, R. Spizzo, S. Sabbioni, E. Magri, M. Pedriali, M. Fabbri, M. Campiglio, S. Menard, J. P. Palazzo, A. Rosenberg, P. Musiani, S. Volinia, I. Nenci, G. A. Calin, P. Querzoli, M. Negrini, and C. M. Croce. 2005. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 65**:**7065-7070. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, A. L., S. R. Bartz, J. Schelter, S. V. Kobayashi, J. Burchard, M. Mao, B. Li, G. Cavet, and P. S. Linsley. 2003. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 21**:**635-637. [DOI] [PubMed] [Google Scholar]
- 29.Jackson, A. L., J. Burchard, J. Schelter, B. N. Chau, M. Cleary, L. P. Lim, and P. S. Linsley. 2006. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA 12**:**1179-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jing, Q., S. Huang, S. Guth, T. Zarubin, A. Motoyama, J. Chen, F. Di Padova, S. C. Lin, H. Gram, and J. Han. 2005. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 120**:**623-634. [DOI] [PubMed] [Google Scholar]
- 31.John, B., A. J. Enright, A. Aravin, T. Tuschl, C. Sander, and D. S. Marks. 2004. Human microRNA targets. PLoS Biol. 2**:**e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson, S. M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K. L. Reinert, D. Brown, and F. J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell 120**:**635-647. [DOI] [PubMed] [Google Scholar]
- 33.Knudson, A. G. 1996. Hereditary cancer: two hits revisited. J. Cancer Res. Clin. Oncol. 122**:**135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krek, A., D. Grun, M. N. Poy, R. Wolf, L. Rosenberg, E. J. Epstein, P. MacMenamin, I. da Piedade, K. C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37**:**495-500. [DOI] [PubMed] [Google Scholar]
- 35.Krutzfeldt, J., N. Rajewsky, R. Braich, K. G. Rajeev, T. Tuschl, M. Manoharan, and M. Stoffel. 2005. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438**:**685-689. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, B. P., C. B. Burge, and D. P. Bartel. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120**:**15-20. [DOI] [PubMed] [Google Scholar]
- 37.Lewis, B. P., I. H. Shih, M. W. Jones-Rhoades, D. P. Bartel, and C. B. Burge. 2003. Prediction of mammalian microRNA targets. Cell 115**:**787-798. [DOI] [PubMed] [Google Scholar]
- 38.Lim, L. P., N. C. Lau, P. Garrett-Engele, A. Grimson, J. M. Schelter, J. Castle, D. P. Bartel, P. S. Linsley, and J. M. Johnson. 2005. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433**:**769-773. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Lu, J., G. Getz, E. A. Miska, E. Alvarez-Saavedra, J. Lamb, D. Peck, A. Sweet-Cordero, B. L. Ebert, R. H. Mak, A. A. Ferrando, J. R. Downing, T. Jacks, H. R. Horvitz, and T. R. Golub. 2005. MicroRNA expression profiles classify human cancers. Nature 435**:**834-838. [DOI] [PubMed] [Google Scholar]
- 41.O'Donnell, K. A., E. A. Wentzel, K. I. Zeller, C. V. Dang, and J. T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435**:**839-843. [DOI] [PubMed] [Google Scholar]
- 42.Orom, U. A., S. Kauppinen, and A. H. Lund. 2006. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 372**:**137-141. [DOI] [PubMed] [Google Scholar]
- 43.Raymond, C. K., B. S. Roberts, P. Garrett-Engele, L. P. Lim, and J. M. Johnson. 2005. Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11**:**1737-1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schottelius, A. J., and H. Dinter. 2006. Cytokines, NF-kappaB, microenvironment, intestinal inflammation and cancer. Cancer Treat. Res. 130**:**67-87. [DOI] [PubMed] [Google Scholar]
- 45.Sethupathy, P., B. Corda, and A. G. Hatzigeorgiou. 2006. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA 12**:**192-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Silva, J. M., M. Z. Li, K. Chang, W. Ge, M. C. Golding, R. J. Rickles, D. Siolas, G. Hu, P. J. Paddison, M. R. Schlabach, N. Sheth, J. Bradshaw, J. Burchard, A. Kulkarni, G. Cavet, R. Sachidanandam, W. R. McCombie, M. A. Cleary, S. J. Elledge, and G. J. Hannon. 2005. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 37**:**1281-1288. [DOI] [PubMed] [Google Scholar]
- 47.Stark, A., J. Brennecke, N. Bushati, R. B. Russell, and S. M. Cohen. 2005. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123**:**1133-1146. [DOI] [PubMed] [Google Scholar]
- 48.Thomson, J. M., M. Newman, J. S. Parker, E. M. Morin-Kensicki, T. Wright, and S. M. Hammond. 2006. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 20**:**2202-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valencia-Sanchez, M. A., J. Liu, G. J. Hannon, and R. Parker. 2006. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20**:**515-524. [DOI] [PubMed] [Google Scholar]
- 50.Wang, X., and X. Wang. 2006. Systematic identification of microRNA functions by combining target prediction and expression profiling. Nucleic Acids Res. 34**:**1646-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitfield, M. L., G. Sherlock, A. J. Saldanha, J. I. Murray, C. A. Ball, K. E. Alexander, J. C. Matese, C. M. Perou, M. M. Hurt, P. O. Brown, and D. Botstein. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13**:**1977-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie, X., J. Lu, E. J. Kulbokas, T. R. Golub, V. Mootha, K. Lindblad-Toh, E. S. Lander, and M. Kellis. 2005. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434**:**338-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, L., J. Huang, N. Yang, S. Liang, A. Barchetti, A. Giannakakis, M. G. Cadungog, A. O'Brien-Jenkins, M. Massobrio, K. F. Roby, D. Katsaros, P. Gimotty, R. Butzow, B. L. Weber, and G. Coukos. 2006. Integrative genomic analysis of protein kinase C (PKC) family identifies PKCiota as a biomarker and potential oncogene in ovarian carcinoma. Cancer Res. 66**:**4627-4635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]