Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation (original) (raw)
Abstract
Although genomic instability is a hallmark of human cancer cells, the mechanisms by which genomic instability is generated and selected for during oncogenesis remain obscure. In most human cancers, the pathway leading to the activation of the G1 cyclins is deregulated. Using budding yeast as a model, we show that overexpression of the G1 cyclin Cln2 inhibits the assembly of prereplicative complexes (pre-RCs) and induces gross chromosome rearrangements (GCR). Our results suggest that deregulation of G1 cyclins, selected for in oncogenesis because it confers clonal growth advantage, may also provide an important mechanism for generating genomic instability by inhibiting replication licensing.
Keywords: G1 cyclin, genomic instability, prereplicative complex, DNA replication, cell cycle control, oncogenesis
Cyclin-dependent kinases (Cdks) control cell cycle transitions in eukaryotic cells (Malumbres and Barbacid 2001; Ho and Dowdy 2002). In yeast, different cyclins act with a single catalytic subunit (Cdc28 in budding yeast, Cdc2 in fission yeast; Toone et al. 1997), whereas in multicellular eukaryotes, different cyclins act with different catalytic subunits at different times in the cell cycle (Ekholm and Reed 2000; Sherr 2000).
During the G1 phase, cells decide whether or not to proceed through the cell cycle. This decision (‘Start’ in yeast, ‘Restriction Point’ in mammalian cells) is executed by environmental cues acting through Cdks. In budding yeast, nutritional status and mating pheromone, a negative growth factor, both regulate the levels and activities of the G1 cyclins (Clns) to drive progression through Start. In mammalian cells, mitogens and negative growth factors such as TGF-β regulate the activity of Cdks to pass the Restriction Point.
Deregulation of the pathway leading to Cdk activation during the G1 phase occurs in most cancers by either inactivating inhibitors of the pathway like p16 and Rb or hyperactivating cyclins or kinase components of Cdks (Ekholm and Reed 2000; Sherr 2000; Malumbres and Barbacid 2001; Zhang et al. 2001; Ho and Dowdy 2002; Rane et al. 2002). This deregulation presumably contributes to oncogenesis by rendering cells less dependent on positive-acting growth factors and/or refractory to inhibition by negative growth factors.
Evidence suggests that deregulation of this pathway can also cause genomic instability, another cancer hallmark. For example, overexpression of cyclin E can lead to elevated levels of aneuploidy, whereas overexpression of cyclin D can lead to increased levels of gene amplification (Zhou et al. 1996; Spruck et al. 1999). The mechanisms by which deregulation of this pathway induces genomic instability are not known.
Cdks play a central role in limiting the initiation of DNA replication from multiple chromosomal replication origins to once per cell cycle in yeast and humans (Kelly and Brown 2000; Blow 2001; Diffley 2001; Bell and Dutta 2002). Two mutually exclusive steps in the process of initiation are separated temporally in the cell cycle because they are regulated in opposite ways by Cdks. First, the origin recognition complex (ORC), Cdc6, and Cdt1 act together to load the Mcm2-7 proteins into prereplicative complexes (pre-RCs) in a reaction known as “licensing.” Cdks inhibit this reaction and, therefore, pre-RCs can only assemble during the G1 phase, when Cdk levels are low. Cdks act through multiple targets to inhibit licensing. In budding yeast, for example, Cdc6, Cdt1, Mcm2-7, and ORC are all negatively regulated by Cdks. Cdc6 is targeted for SCFCDC4-dependent, ubiquitin-mediated proteolysis by Cdk phosphorylation; Cdt1 and Mcm2-7 are displaced from the nucleus to the cytoplasm by Cdk activation, and phosphorylation of the ORC plays a role in preventing pre-RC assembly (Drury et al. 1997, 2000; Elsasser et al. 1999; Labib et al. 1999; Calzada et al. 2000; Nguyen et al. 2000, 2001; Perkins et al. 2001; Tanaka and Diffley 2002). Cln–Cdc28 is primarily responsible for Cdc6 proteolysis (Drury et al. 2000) and contributes to Mcm2-7 relocalization (Labib et al. 1999). The S- and M-phase cyclins (Clb1–Clb6) also contribute to Mcm2-7 relocalization (Labib et al. 1999; Nguyen et al. 2000) and are responsible for additional negative regulation such as ORC phosphorylation (Nguyen et al. 2001). In metazoans, Cdks also prevent licensing by acting through multiple targets, although the detailed mechanisms are different (for review, see Diffley 2001).
Initiation is then triggered by Cdks in the S phase. The initiation protein Sld2/Drc1 has recently been identified as one crucial substrate in budding and fission yeasts (Masumoto et al. 2002; Noguchi et al. 2002). Critically, it is the presence of Cdks in S, G2, and M phases that prevents new pre-RC assembly until the next cell cycle. Oscillations between low and high Cdk activity, therefore, are crucial for normal DNA replication, suggesting a possible mechanism by which G1-cyclin deregulation could cause genomic instability. The lack of a proper “low Cdk” period in G1 may reduce the numbers of functional pre-RCs. Subsequent replication from an inadequate number of origins might then be responsible for chromosome loss and rearrangements. Previous work suggested that this is feasible. cdc14 mutants exhibit elevated levels of simple plasmid loss that can be suppressed by including multiple origins on the plasmid (Hogan and Koshland 1992). Suppression of plasmid loss by multiple origins is a phenotype shared with mutants in pre-RC components such as cdc6, suggesting that cdc14 mutants have a defect in pre-RC assembly (Hogan and Koshland 1992). However, unlike Cdc6, Cdc14 does not appear to play a direct, essential role in pre-RC assembly. Inactivation of Cdks in G2/M by overexpression of Sic1 is sufficient to bypass any requirement for Cdc14 in pre-RC assembly (Noton and Diffley 2000). Cdc14 is a protein phosphatase required for exit from mitosis (for review, see Bardin and Amon 2001; Jensen et al. 2002; Saunders 2002). The liberation of Cdc14 from its sequestration in the nucleolus at the end of mitosis is required to stabilize the Cdk inhibitor Sic1 and to activate the APC/C factor Cdh1. This promotes Cdk inactivation and allows mitotic exit. Moderate elevation of Sic1 levels suppresses the plasmid-loss phenotype of cdc14 mutants (Noton and Diffley 2000), strongly suggesting that a defect in Cdk inactivation, probably at the end of mitosis, in cdc14 mutants inhibits licensing without preventing exit from mitosis. Similarly, recent work has shown that deletion of Sic1 causes reduced origin activity, probably by inhibiting pre-RC assembly (Lengronne and Schwob 2002). In this paper we show that deregulation of G1 cyclins causes genomic instability by inhibiting pre-RC assembly in budding yeast.
Results
Cln2 deregulation causes increased plasmid loss
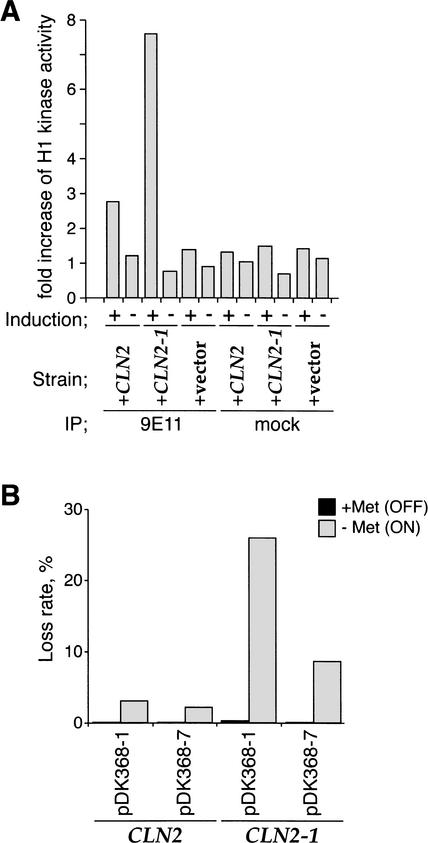
We have used the GAL1,10 and MET3 promoters to induce synthesis of the full-length Cln2 and a truncation (Cln2-1) that is functional as a cyclin but has had a C-terminal PEST domain removed (Hadwiger et al. 1989). Cln2-1 is more stable than full-length Cln2 and, consequently, accumulates to higher levels after induction. Figure 1A shows that expression of Cln2 and Cln2-1 from the GAL1,10 promoter leads to elevated levels of histone H1 kinase activity. This increase in activity is two- to threefold higher with Cln2-1 than with full-length Cln2, consistent with the fact that it is a more stable protein. To begin to investigate the effects of Cln2 deregulation on DNA replication, we have examined rates of plasmid loss. We used plasmids containing a centromere, ARS1 and either one (pDK368-1) or seven (pDK368-7) copies of H4 ARS (Hogan and Koshland 1992). Figure 1B shows that the plasmid loss rates for all three strains were low in the presence of methionine; however, promoter activation (−Met) caused a significant increase in the rate of pDK368-1 loss. This was especially true for Cln2-1, which caused a 200-fold increase in plasmid loss. For both Cln2 and Cln2-1, the rate of pDK368-7 loss was significantly lower. Suppression of plasmid loss rate by multiple replication origins strongly suggests that Cln2 overexpression induces plasmid loss by inhibiting the initiation of DNA replication.
Figure 1.
Cln2 overexpression induces plasmid loss that is suppressed by multiple origins. (A) Histone H1 kinase activity after Cln2 or Cln2-1 induction. YST66 (+CLN2), YST67 (+CLN2-1), and YST69 (+vector) were grown in YP-raffinose. Each culture was split into two, and the medium was changed to YP-galactose (Induction; +) or YP-glucose (Induction; −). Cells were collected after a 60-min incubation, and Myc-tagged Cln2 and Cln2-1 were immunoprecipitated with 9E11 anti-Myc antibody (9E11) or without the antibody (mock). The H1 kinase activity of the immunoprecipitates was measured and compared between the samples before and after the medium change. (B) Elevated plasmid loss rates after Cn2 or Cln2-1 expression. YST177 (CLN2, pDK368-1), YST178 (CLN2, pDK368-7), YST181 (CLN2-1, pDK368-1), and YST182 (CLN2-1, pDK368-7) were grown as described in Materials and Methods, and the rate of plasmid loss from each strain was measured. Plasmid loss rates after growing the cells in Cln-inductive and Cln-repressive conditions are shown by gray and black boxes, respectively.
Cln2 overexpression inhibits DNA rereplication induced by transient Sic1C70td overexpression
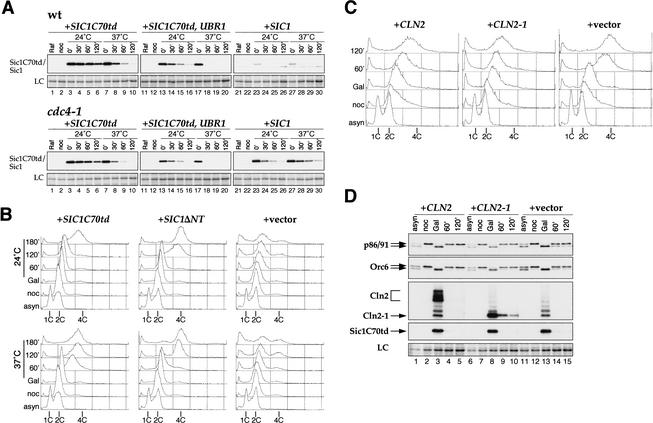
Overexpression of the B cyclin (Clb)-specific Cdk inhibitor Sic1 promotes the reassembly of pre-RCs at origins in nocodazole-arrested cells. Subsequent repression of Sic1 promotes an additional round of DNA replication in these cells. We wanted to determine whether overexpression of Cln2 could prevent pre-RC assembly and rereplication in this system. This approach has two advantages. Firstly, it allows us to distinguish direct effects of Cln expression on pre-RC formation from indirect effects caused by Clb activation. And secondly, it allows us to examine pre-RC formation in the absence of mitosis, which is inhibited by Cln expression (data not shown). However, this experiment is complicated by the fact that phosphorylation of Sic1 by Cln–Cdc28 targets it for ubiquitin-mediated degradation via the SCFCDC4 pathway (Verma et al. 1997; Nash et al. 2001). To circumvent this problem, we constructed a derivative of Sic1 lacking all Cdk consensus sites. The 70 amino acids at the C terminus of Sic1 (called Sic1C70), which does not contain any Cdk phosphorylation sites, contains the Cdk inhibitory domain and is sufficient to arrest the cell cycle when expressed from the GAL1,10 promoter (Hodge and Mendenhall 1999). Sic1C70 cannot be used in the rereplication assay described above, however, because it is an extremely stable protein and, thus, cannot be eliminated from cells to allow Clb reactivation. Therefore, we made an additional refinement and fused Sic1C70 to a temperature-sensitive degron cassette (Dohmen et al. 1994), producing Sic1C70td. To examine protein stability, cells were grown in raffinose-containing medium and arrested in G2/M phase with nocodazole. Sic1C70td synthesis was induced with galactose and repressed with glucose. Although Sic1C70td could be detected at 120 min after promoter shutoff at 24°C, most of the protein had disappeared at the same time point at 37°C (Fig. 2A, wt, lanes 6,10). We have previously shown that overexpression of Ubr1, the E3 ubiquitin ligase in the N-end-rule pathway of Saccharomyces cerevisiae, accelerates the degradation of proteins fused to the degron cassette (Labib et al. 2000). To increase the rate of Sic1C70td degradation, Ubr1 and Sic1C70td were simultaneously expressed from the GAL1,10 promoter. Ubr1 expression caused the degradation of Sic1C70td to be accelerated even at 24°C (Fig. 2A, cf. lanes 3–6 and 13–16). More importantly, Sic1C70td protein promptly disappeared at 30 min after repression at 37°C (Fig. 2A, cf. lanes 7–10 and 17–20). Either with or without Ubr1 overexpression, the degradation of Sic1C70td is entirely independent of SCFCDC4 (Fig. 2A).
Figure 2.
Cln2 can inhibit DNA rereplication induced by Sic1C70td. (A) Temperature-dependent proteolysis of Sic1C70td. YST78 [wild type (wt), +_SIC1C70td_], YST48 (wt, +SIC1C70td, UBR1), YLD11 (wt, +SIC1), YST71 (cdc4-1, +SIC1C70td), YST76 (cdc4-1, +SIC1C70td, UBR1), and YST27 (cdc4-1, +SIC1) were grown in YP-raffinose to early log phase (asyn) and arrested in G2/M with nocodazole (noc) at 24°C for 3 h. Then the medium was changed to YP-galactose plus nocodazole, and the cells were incubated for 2 h. The culture was then split into two, the medium was changed to YP-glucose plus nocodazole, and the cultures were incubated at 24°C or 37°C. Cells were collected every 30 min (0–120 min) and processed for immunoblotting. Sic1C70td–Myc, Sic1–Myc, and Myc–Ubr1 (data not shown) were detected with 9E10 anti-Myc antibody. (LC) Loading control. (B) Transient Sic1C70td expression induces rereplication in nocodazole-arrested cells. YST65 (+SIC1C70td), YST64 (+SIC1ΔNT), and YST57 (+vector) were grown as in A. Cells were collected at each time point and processed for flow cytometry. (C,D) Cln2 and Cln2-1 expression inhibits Sic1C70td-induced rereplication. YST66 (+CLN2), YST67 (+CLN2-1), and YST69 (+vector) were grown, and cells were collected as in A. Each sample was analyzed by flow cytometry (C) or immunoblotting (D). The second largest subunits of DNA polymerase I (p86/91), Orc6, Cln2–Myc, Cln2-1–Myc, and Sic1C70td–Myc were detected by probing with the appropriate antibodies. The ladder bands commonly observed in lanes 3, 8, and 13 of the Cln2/Cln2-1 panel are ubiquitinated Sic1C70td. LC, loading control from the PonceauS-stained membrane.
To determine whether Sic1C70td could induce DNA rereplication, cells harboring GAL–SIC1C70td and GAL–UBR1 were grown in raffinose-containing medium and arrested in G2/M with nocodazole at 24°C. Sic1C70td and Ubr1 were induced with galactose. The culture was split in two, Sic1C70td expression was turned off with glucose, and incubation was continued at either 24°C or 37°C. All medium used after the first arrest contained nocodazole to maintain the G2/M arrest. DNA rereplication in the Sic1C70td strain was observed by 60 min after glucose was added to these cells at 37°C (Fig. 2B), which is earlier than in the strain expressing Sic1ΔNT (rereplication occurs at between 60 and 120 min). At 24°C, DNA rereplication in the Sic1C70td strain was delayed until 180 min (Fig. 2B). The time of rereplication reflects the time at which the Sic1C70td protein disappears, that is, ∼180 min at 24°C and ∼60 min at 37°C (Fig. 2A; data not shown). Sic1ΔNT, a partially stabilized version of Sic1 whose degradation is dependent on SCFCDC4 (Noton and Diffley 2000), did not show a temperature dependence for the timing of DNA rereplication and disappearance of the protein (Fig. 2B, +SIC1ΔNT; data not shown).
Having established that Sic1C70td could be used to induce rereplication, we next asked whether CLN2 or CLN2-1 expression could prevent Sic1C70td-induced DNA rereplication. CLN2 or CLN2-1 was expressed from the GAL promoter, together with SIC1C70td and UBR1 in nocodazole-arrested cells at 24°C. Cells were then transferred to 37°C glucose-containing medium. Figure 2C shows that the vector-containing control strain (+vector) rereplicated DNA with the same kinetics as the experiment shown in Figure 2B, whereas CLN2 or CLN2-1 expression significantly inhibited rereplication. Some Cln2-expressing cells partially rereplicated their DNA, but even after 2 h the peak did not reach 4C. Expression of Cln2-1 was more effective at preventing rereplication. Figure 2D (lanes 3,8,13) shows that Sic1C70td protein levels were unaffected by Cln expression. Orc6 and the B subunit of DNA polymerase α (p86/91) are targets of Clb–Cdc28 but not Cln–Cdc28 (Foiani et al. 1995; Weinreich et al. 1999; data not shown). Figure 2D (lanes 3,8,13) shows that Orc6 and p86/91 were efficiently dephosphorylated after Sic1C70td expression even in the presence of Cln2 or Cln2-1. These data support the idea that inhibition of DNA rereplication was a direct effect of Cln2 expression rather than an indirect effect of activation of Clbs.
Cln2 overexpression inhibits pre-RC formation by preventing nuclear localization of Mcm2-7
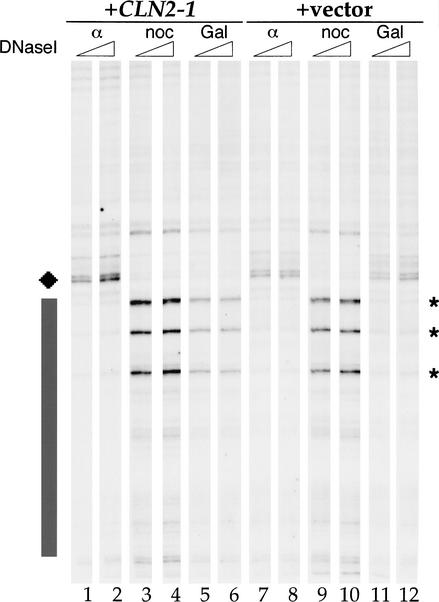
To determine whether the inhibition of rereplication correlates with inhibition of pre-RC assembly, we analyzed ARS305 by genomic footprinting, which can be used to quantify the efficiency of pre-RC assembly (Santocanale and Diffley 1996). The cultures of control (+vector) and GAL–CLN2-1 (+CLN2-1) strains used in Figure 2 were grown in raffinose-containing medium until early log phase and split into three (Fig. 3). One-third was arrested in G1 with α-factor and the remaining two-thirds were arrested in G2/M phase with nocodazole. One nocodazole-arrested culture was transferred to galactose- and nocodazole-containing medium, whereas the other was held in raffinose- and nocodazole-containing medium. In nocodazole-arrested cells, the origins are in the postreplicative state, as evidenced by three ORC-induced DNase I hypersensitive sites (asterisks). The pre-RC in α-factor-arrested cells is seen as a region of protection (gray box) and an additional hypersensitive site (diamond). In the control strain, the pre-RC-specific DNase I hypersensitive site and region of protection were efficiently reformed after galactose incubation in an essentially identical manner to that seen in G1-arrested cells (α; Fig. 3, cf. lanes 7,8 and 11,12). In contrast, pre-RCs were not efficiently reformed in the _GAL–CLN2-1_-expressing strain as indicated by the three ORC-induced hypersensitive sites that remain and the diminished protection (Fig. 3, lanes 3–6). These experiments show that Cln2-1 expression can inhibit pre-RC formation.
Figure 3.
Cln2 can inhibit pre-RC formation. DNase I genomic footprinting. YST67 (+CLN2-1) and YST69 (+vector) were grown in YP-raffinose to early log phase at 24°C. One-third of the culture was arrested at G1 in α-factor for 3.5 h (α). The remaining culture was arrested in G2/M with nocodazole for 3 h (noc), after which the medium of half of the culture was changed to YP-galactose and incubated at 24°C for a further 4 h (Gal). Genomic DNA was prepared, and primer–extension reactions were performed with an ARS305-specific primer. G1-specific DNase I hypersensitive sites (diamond), protection (black box), and G2-specific hypersensitive sites (*) are shown.
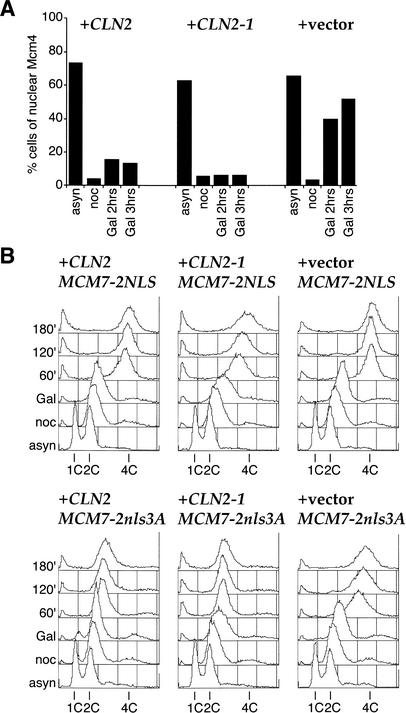
In budding yeast, the Mcm2-7 complex undergoes cell cycle-regulated changes in its subcellular localization (Hennessy et al. 1990; Chen et al. 1992; Dalton and Whitbread 1995; Labib et al. 1999; Nguyen et al. 2000). The complex enters the nucleus at the end of mitosis and exits the nucleus as it is displaced from chromatin during the S phase. Previous work has shown that this regulated nuclear localization does not require Cdc6 and, therefore, is independent of licensing. Relocalization of Mcm2-7 during the S phase is caused by Cdc28. Clb–Cdc28 is required for the export of chromatin-bound Mcm2-7, whereas Cln or Clb–Cdc28 can trigger the export of unbound Mcm2-7 (Labib et al. 1999). The specific requirement for Clb–Cdc28 in relocalizing the chromatin-bound fraction presumably reflects the fact that only Clb–Cdc28 can trigger replication, which is required to displace the Mcm2-7 complex from chromatin, a prerequisite for export. We were, therefore, interested in determining whether coexpression of Cln2 prevented the accumulation of Mcm2-7 in an experiment similar to the one described above. We used cells harboring Mcm4–GFP and expressed Sic1C70td together with Cln2 or Cln2-1. Cells were grown in raffinose-containing medium, arrested in G2/M phase with nocodazole, and then transferred to galactose-containing medium with nocodazole. Figure 4A shows that Mcm4–GFP accumulated in the nuclei in the control strain after Sic1C70td expression (Fig. 4A, “Gal 2hrs” and “Gal 3hrs” in +vector). This nuclear accumulation of Mcm4–GFP was inhibited by expression of Cln2 and, especially, Cln2-1 (Fig. 4A, +CLN2 and +CLN2-1).
Figure 4.
Prevention of nuclear localization of the Mcm2-7 complex by Cln2 is important for inhibition of DNA rereplication. (A) Cln2 or Cln2-1 expression blocks nuclear accumulation of Mcm2-7. YST66 (+CLN2), YST67 (+CLN2-1), and YST69 (+vector) were grown and treated as in Figure 3 except that samples were taken after either 2 or 3 h in YP-galactose. Samples were prepared for microscopy to examine which cells showed nuclear accumulation of Mcm4–GFP. (B) Cln2 or Cln2-1 expression cannot prevent rereplication when Cdk inhibition of Cdc6 and Mcm2-7 is bypassed. YST282 (+CLN2 MCM7-2NLS), YST283 (+CLN2 MCM7-2nls3A), YST284 (+CLN2-1 MCM7-2NLS), YST285 (+CLN2-1 MCM7-2nls3A), YST286 (+vector MCM7-2NLS), and YST287 (+vector MCM7-2nls3A) were grown as in Figure 2. Cells were collected and analyzed by flow cytometry.
Recent work has shown that cyclin-dependent kinases inhibit pre-RC formation primarily through three redundant targets: Orc, Cdc6, and Mcm2-7 (Nguyen et al. 2001). Cdk-dependent phosphorylation of Cdc6 targets it for ubiquitin-mediated proteolysis via SCFCDC4 (Drury et al. 1997, 2000; Elsasser et al. 1999; Calzada et al. 2000; Perkins et al. 2001). Cdks also prevent the accumulation of the Mcm2-7 complex and its associated Cdt1 subunit in the nucleus (Labib et al. 1999, 2001; Nguyen et al. 2000; Tanaka and Diffley 2002). Rereplication can be induced in nocodazole-arrested cells only when all three targets are bypassed. This can be done by mutating all of the potential Cdk sites in Orc2 and Orc6, by expressing a version of Cdc6 in which the SCFCDC4-targeting sequences in the N terminus have been removed (Cdc6ΔNT), and by adding a constitutive nuclear localization sequence (NLS) to one of the Mcm subunits (Mcm7). ORC appears to be primarily phosphorylated by Clb–Cdc28; however, previous work from our laboratory has indicated that relocalization of the Mcm2-7 complex to the cytoplasm and proteolysis of Cdc6 can both be triggered by Cln-associated forms of Cdc28. If this is true, then the ability of Cln2 to block Sic1C70td-induced rereplication should act only through Mcm2-7 and Cdc6, not through ORC. To address this, we asked whether bypassing CDK regulation of Mcm2-7 and Cdc6 is sufficient to promote rereplication in the presence of overexpressed Cln2. We have used strains containing Mcm7 fused to two copies of either wild-type or mutant SV40 T antigen nuclear localization sequences in which truncated Cdc6 is expressed from the GAL1,10 promoter. In the absence of Cln2 or Cln2-1 expression, transient Sic1C70td and Cdc6ΔNT expression induces efficient rereplication with both wild-type and mutant Mcm7-2NLS (+vector MCM7-2NLS and +vector MCM7-2nls3A). As expected based on the experiment in Figure 2C, overexpression of Cln2 and Cln2-1 prevents rereplication with Mcm7 fused to the mutant NLS (MCM7-2nls3A). However, the presence of the wild-type NLS on Mcm7 (MCM7-2NLS) allowed efficient rereplication even in the presence of Cln2 and Cln2-1. Therefore, Cln2 expression inhibits rereplication and, presumably, pre-RC formation at least in part by inhibiting the accumulation of the Mcm2-7 complex in the nucleus. Because these strains contain wild-type, phosphorylatable Orc2 and Orc6, Cln2 expression is unable to prevent rereplication through the ORC, consistent with the experiment in Figure 2D showing that Orc6 is efficiently dephosphorylated after Sic1C70td expression, even in the presence of Cln2 or Cln2-1.
Cln2-1 expression induces GCR that is suppressed by multiple origins
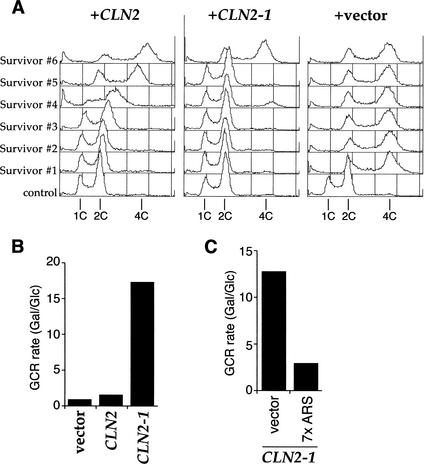
Survivors from Sic1-induced rereplication in nocodazole-arrested cells are converted from haploid to diploid (Noton 2000). To examine DNA rereplication, we recovered and analyzed survivors from the experiment in Figure 2C. In the case of the control strain, which did not express high levels of Cln2, 9 out of 10 survivors tested had diploidized, and only 1 remained haploid (Fig. 5A, +vector). In contrast, only 1 out of 10 diploidized in the GAL–CLN2-1 strain; the other 9 remained haploid (Fig. 5A, +CLN2-1). This is consistent with our observation that Cln2-1 expression efficiently blocks DNA rereplication. Cln2-1 expression did not significantly reduce viability relative to the vector control; however, Cln2 expression reduced viability by ∼50% (data not shown). Many of the survivors after Cln2 expression grew quite slowly. Survivors from the GAL–CLN2 strain showed an intermediate number of diploid survivors. Moreover, some of the survivors had DNA contents with nonintegral ploidy (Fig. 5A, +CLN2). For example, Survivor #4 had a main peak of DNA content around 2.5C. Analysis of the chromosomes in this survivor by pulse-field gel electrophoresis did not reveal any chromosome rearrangements; however, a subset of chromosomes appears to have been duplicated (data not shown).
Figure 5.
Cln2 expression can induce genomic instability. (A) Genome instability caused by Cln2 expression. An aliquot of the culture from the last time point of the rereplication assay in Figure 2C was spread onto fresh YP-glucose plates, and survivors were recovered. Each colony was grown in YP-glucose medium, fixed, and analyzed by flow cytometry to examine the ploidy. (B) Cln2 or Cln2-1 induces GCR. The GCR rates of YST196 (vector), YST192 (CLN2), and YST193 (CLN2-1) were measured as described in Materials and Methods. The ratios of the GCR rate between growth in YP-galactose and YP-glucose for each strain are shown. (C) Cln2-1-induced GCR is suppressed by multiple origins. The GCR rates of YST248 (vector) and YST252 (7× ARS) were measured as described in Materials and Methods. The ratios of the GCR rate between growth in YP-galactose and YP-glucose for each strain are shown.
We were interested in determining whether inhibition of pre-RC assembly caused by Cln expression could cause genome rearrangements. We used an assay developed by Kolodner and coworkers to measure the rate of gross chromosomal rearrangements (GCRs; Chen and Kolodner 1999). In this assay, the rate of simultaneous loss of two markers from the end of Chromosome V (CAN1 and URA3) is measured. Because the assay is performed in haploid cells and Chromosome V contains many essential genes, the loss of both markers cannot occur by simple chromosome loss. Because the probability of inactivating both genes by separate point mutations is extremely low (<10−12), loss of both markers invariably occurs by chromosome rearrangement, including chromosome truncation, internal deletion, or translocation, known collectively as gross chromosome rearrangements (GCR). We have tested whether overexpression of Cln2 affected the rate of GCR in this assay (Fig. 5B; Table 1). In the wild-type strain, the ratio of the GCR rate in cells grown in galactose (Gal) versus glucose (Glc) was 0.8. This increased approximately twofold in the GAL–CLN2 strain (Gal/Glc = 1.4). There was a much more dramatic increase in GCR rate in the GAL–CLN2-1 strain (Gal/Glc = 18.7). To test whether this GCR increase was related to origin activity, we introduced either an integrating plasmid or the same plasmid containing seven copies of ARS H4 between the CAN1 and URA3 markers. Figure 5C and Table 1 show that the increased GCR rate caused by Cln2-1 was significantly suppressed by seven copies of ARS H4 (Gal/Glc = 2.8) relative to the vector-alone control strain (Gal/Glc = 12.2). These experiments strongly suggest that GCR induced by Cln2-1 expression is caused by inhibition of origin firing, probably in the vicinity of the two markers.
Table 1.
GCR rates
| Strain | Relevant genotype | GCR rate | Fold increase in galactose | |
|---|---|---|---|---|
| In glucose | In galactose | |||
| YST196 | wta + GAL vector | 0.73 × 10−10 | 0.59 × 10−10 | 0.80 |
| YST192 | GAL–CLN2 | 1.49 × 10−10 | 2.14 × 10−10 | 1.43 |
| YST193 | GAL–CLN2-1 | 1.06 × 10−10 | 1.99 × 10−9 | 18.7 |
| YST248 | GAL–CLN2-1, sit1Δ::control fragment | 1.39 × 10−10 | 1.71 × 10−9 | 12.23 |
| YST252 | GAL–CLN2-1, sit1Δ::ARSH4(×7) | 3.54 × 10−10 | 1.00 × 10−9 | 2.83 |
Discussion
Genomic instability is a hallmark of cancer (Tlsty et al. 1995; Schar 2001). Not only do tumor cells often have highly rearranged genomes, but they also exhibit high rates of chromosome loss, ploidy changes, and recombination rates up to 10,000 times higher than primary cells (Mekeel et al. 1997; Schar 2001). However, the forces that drive genomic instability in tumor cells are still largely unknown. Our results show that deregulation of G1 cyclins in budding yeast can contribute to genomic instability by inhibiting licensing. We have shown that constitutive overexpression of Cln2 and Cln2-1 leads to elevated levels of both simple plasmid loss and gross chromosome rearrangements. Both phenotypes can be at least partially suppressed by incorporation of multiple origins into the test substrate. This supports the argument that the instability seen is caused by inefficient replication-origin firing. We have shown that Cln2-1 expression can prevent pre-RC assembly and Mcm2-7 nuclear accumulation even under conditions in which Clbs are inactive and known Clb–Cdk targets like ORC and DNA polymerase α-primase remain dephosphorylated. This indicates that at least one way in which Cln deregulation promotes genomic instability is by direct inhibition of licensing. We do not rule out the possibility that indirect inhibition of licensing by promoting inappropriate Clb activation either by Sic1 degradation or APC/C inactivation may also make important contributions to genomic instability. In this regard, we note that Sic1 deletion increases genomic instability by a similar mechanism (Nugroho and Mendenhall 1994; Lengronne and Schwob 2002). Finally, our experiments provide the first functional evidence indicating that Cln-dependent Cdc6 proteolysis and relocalization of Mcm2-7 can contribute to the block to rereplication.
Our experiments do not address how reducing origin licensing might contribute to genomic instability; however, we note that cdc6 mutants, which are defective in licensing, also exhibit high levels of chromosome rearrangements (Bruschi et al. 1995). We suggest two mechanisms by which reduced numbers of active origins might cause genomic instability. First, incompletely replicated chromosomes with duplicated centromeres that enter mitosis will generate broken chromosomes that would then be substrates for homologous recombination and nonhomologous end-joining. Second, forks that have stalled either randomly (e.g., at sites of endogenous DNA damage) or at specific pause sites may be recombinogenic. Such stalled forks may be “rescued” by forks from nearby origins arriving from the opposite direction. By reducing origin frequency and, therefore, increasing the interorigin distance, such stalled forks may persist for longer periods of time, thus increasing the likelihood of recombination. We note that these two models are not mutually exclusive and may both contribute to genomic instability. It is interesting to consider that differences in the density and distribution of potential replication origins may differentially affect GCR rates along chromosomes leading to potential hot spots for rearrangements.
Although virtually all tumor cells exhibit some form of genomic instability, and many inherited human diseases that increase genomic instability also predispose individuals to cancer, it has been argued that genome instability is unlikely to be selected for during oncogenesis because (1) genome instability, by itself, does not confer any growth advantage to cells; and (2) mutation rates in primary cells are sufficiently high to account for the number of mutations needed to transform cells (Tomlinson and Bodmer 1999). Our results suggest at least one way to resolve this apparent paradox. We propose that cyclin deregulation is selected for during cancer development because it confers growth advantage by rendering cells less mitogen-dependent and more resistant to negative growth factors, just as Cln2 overexpression renders cells insensitive to α-factor (data not shown). However, by also inhibiting licensing, genome instability is the inevitable by-product of this event.
Although our work has been done in budding yeast, there is reason to suggest that it will be relevant to human cells as well. Firstly, the premise that Cdks inhibit licensing appears to be true in human cells; treatment of human cells in G2 with specific Cdk inhibitors is sufficient to trigger reloading of the Mcm complex (Coverley et al. 1996, 1998), just as Cdk inhibition with Sic1 is sufficient to drive pre-RC assembly in yeast. Secondly, overexpression of cyclins D and E can induce genomic instability in human tissue culture cells (Zhou et al. 1996; Spruck et al. 1999). Thirdly, primary fibroblasts from Rb-deficient embryos exhibit an extended S phase, consistent with a defect in origin use (Classon et al. 2000).
Finally, it is interesting to consider the role of p53 in this process. Recent work has indicated that loss of p53, by itself, does not significantly increase genome instability (Paulson et al. 1998; Bunz et al. 2002). Wahl and colleagues have proposed that p53-deficient cells must undergo DNA replication under conditions that induce DNA strand breaks for instability to occur (Almasan et al. 1995; Paulson et al. 1998; Wahl and Carr 2001). Agents that interfere with replication and/or damage DNA can provide these breaks. We propose that deregulation of cyclins can also generate DNA damage. In such cells, loss of p53-mediated apoptosis might provide the growth advantage that selects for clonal expansion of the p53 mutant cells. In this regard, we note that cells in the central nervous system and lenses of Rb−/− mouse embryos have been shown to exhibit not only inappropriate entry into S phase but also elevated rates of apoptosis. This apoptosis can be suppressed by deletion of p53 (Morgenbesser et al. 1994; Macleod et al. 1996).
Materials and methods
Strains and media
The strains used in this study are listed in Table 2. All strains are derived from W303. For efficient DNA rereplication, three copies of the Sic1C70td plasmid were introduced into the strains used in rereplication assays (YST65, YST66, YST67, and YST69). To introduce the MCM7-2NLS or MCM-2nls3A mutation into these strains, the _Pvu_II fragments from pJL1208 or pKI1273 (Nguyen et al. 2000), containing mcm7Δ-2NLS or mcm7Δ-2nls3A, respectively, were subcloned into the _Sma_I site of pAUR101 (Takara Shuzo Co., Kyoto, Japan), and the plasmids obtained were integrated at the CDC47/MCM7 locus. To construct the 7× ARSH4 strain and its control, an _Sph_I–_Psh_AI fragment of pDK368-7 or pDK243 (Hogan and Koshland 1992) was cloned between _Sph_I and the filled-in _Nde_I sites of YIplac204, and then the _Sph_I–_Hin_dIII fragment derived from the SIT1 ORF was introduced into these plasmids. The plasmids obtained were introduced into YST193 after linearization with _Xho_I digestion, thus generating YST248 and YST252, respectively. Cells were grown in the rich medium YP (1% yeast extract, 2% Bacto peptone) supplemented with 2% sugar (glucose, galactose, or raffinose) or synthetic medium supplemented with the required amino acids and nucleotides. Cell cycle block and release experiments with α-factor, nocodazole, and temperature shift and induction and repression of the GAL or the MET promoter were performed as described previously (Diffley et al. 1994; Labib et al. 1999).
Table 2.
Yeast strains
| Strain | Genotype |
|---|---|
| W303-1a | MATa ade2-1 ura3-1 his3-11,15 trp1-1 leu2-3,112 can1-100 |
| YLD11 | W303-1a ura3::GAL1,10p–SIC1–Myc–His9 |
| YST27 | W303-1a cdc4-1 ura3::GAL1,10p–SIC1–Myc–His9 |
| YST48 | W303-1a ubr1Δ::GAL1,10p-Myc-UBR1::HIS3 ura3::GAL1,10p-SIC1C70td–Myc–His9 |
| YST57 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–Myc–His9(×1) |
| YST64 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p-SIC1ΔNT–Myc–His9(×1) |
| YST65 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) |
| YST66 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–CLN2–Myc–His9 |
| YST67 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–CLN2-1–Myc–His9 |
| YST69 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–Myc–His9 |
| YST71 | W303-1a cdc4-1 ura3::GAL1,10p–SIC1C70td–Myc–His9 |
| YST76 | W303-1a cdc4-1 ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9 |
| YST78 | W303-1a ura3::GAL1,10p–SIC1C70td–Myc–His9 |
| YST177 | W303-1a _ade3::hisG ura3::MET3p–CLN2–Myc–His9_pDK368-1 |
| YST178 | W303-1a _ade3::hisG ura3::MET3p–CLN2–Myc–His9_pDK368-7 |
| YST181 | W303-1a _ade3::hisG ura3::MET3p–CLN2-1–Myc–His9_pDK368-1 |
| YST182 | W303-1a _ade3::hisG ura3::MET3p–CLN2-1–Myc–His9_pDK368-7 |
| DRY1728 | W303-1a designer deletion (MATa arg4Δ::hisG his3-11,15 leu2-3,112 trp1Δ0::hisG ura3Δ0 CAN1) |
| YST192 | DRY1728 arg4Δ::ARG4 hxt13Δ::URA3 leu2::GAL1,10p–CLN2–Myc–His9 |
| YST193 | DRY1728 arg4Δ::ARG4 hxt13Δ::URA3 leu2::GAL1,10p–CLN2-1–Myc–His9 |
| YST196 | DRY1728 arg4Δ::ARG4 hxt13Δ::URA3 leu2::GAL1,10p–Myc–His9 |
| YST248 | DRY1728 arg4Δ::ARG4 hxt13Δ::URA3 leu2::GAL1,10p–CLN2-1–Myc–His9 sit1Δ::control fragment-TRP1 |
| YST252 | DRY1728 arg4Δ::ARG4 hxt13Δ::URA3 leu2::GAL1,10p–CLN2-1–Myc–His9 sit1Δ::ARSH4(×7)-TRP1 |
| YST282 | W303-1a CDC54/MCM4-GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–CLN2–Myc–His9 cdc47/mcm7Δ::MCM7-2NLS his3::GAL1,10p–CDC6ΔNT |
| YST283 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–CLN2–Myc–His9 cdc47/mcm7Δ::MCM7-2nls3A his3::GAL1,10p–CDC6ΔNT |
| YST284 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–CLN2-1–Myc–His9 cdc47/mcm7Δ::MCM7-2NLS his3::GAL1,10p–CDC6ΔNT |
| YST285 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–CLN2-1–Myc–His9 cdc47/mcm7Δ::MCM7-2nls3A his3::GAL1,10p–CDC6ΔNT |
| YST286 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–Myc–His9 cdc47/mcm7Δ::MCM7-2NLS his3::GAL1,10p–CDC6ΔNT |
| YST287 | W303-1a CDC54/MCM4–GFP ubr1Δ::GAL1,10p–Myc–UBR1::HIS3 ura3::GAL1,10p–SIC1C70td–Myc–His9(×3) leu2::GAL1,10p–Myc–His9 cdc47/mcm7Δ::MCM7-2nls3A his3::GAL1,10p–CDC6ΔNT |
H1 kinase assay
Soluble cell extracts were prepared by centrifugation after cell lysis with glass beads in lysis buffer (0.4 M sorbitol, 150 mM potassium acetate, 2 mM magnesium acetate, 20 mM PIPES at pH 6.8, 1 mM AEBSF, 1× Complete protease inhibitors from Boehringer Mannheim, 0.5 mM sodium orthovanadate, 10 mM sodium fluoride, and 20 mM β-glycerophosphate). Then 10 μg of 9E11 anti-myc antibody and protein G agarose beads (Sigma-Aldrich Chemie) or protein G beads alone was added to one-quarter of the extract and incubated at 4°C overnight. Half of the beads were washed with H1 kinase buffer (50 mM β-glycerophosphate, 20 mM EGTA, 15 mM magnesium acetate, and 1 mM DTT; pH was adjusted to 7.4). Then 6.5 μL of H1 kinase buffer and 2.5 μL of reaction mix (1 mg/mL Histone H1, 1 mM DTT, 15 mM magnesium acetate, 300 μM ATP, and 0.5 μCi/μL [γ-32P]ATP) were added to the washed beads and incubated at 25°C for 10 min; the reaction was stopped by adding 20 μL of sample buffer. The samples were analyzed by SDS-PAGE, and a phosphorimager (Molecular Dynamics) was used for further processing of the data.
Plasmid loss assay
YST177, 178, 181, and 182 were grown to mid-log phase in YP-glucose. Cells were diluted in synthetic medium lacking leucine and either with or without 2 mM methionine for the following 24 h to allow cells to grow for 5–10 generations. Cells before and after this incubation were spread onto synthetic medium supplemented with leucine, and the numbers of red and white colonies were counted. The plasmid loss rate per generation was calculated as described previously (Noton and Diffley 2000).
GCR assay
At least five independent colonies were picked from YP-raffinose plates and suspended in a small volume of water. Half of the suspension was inoculated into YP-glucose, and the rest was inoculated into YP-galactose. Cells were grown to stationary phase and then spread onto synthetic medium containing 60 μg/mL canavanine (CAN) and 1 mg/mL 5-fluoroorotic acid (5-FOA). The number of colonies formed on CAN + FOA plates was counted, and the median was used to calculate the GCR rate. The GCR rate was calculated as described previously (Reenan and Kolodner 1992; Chen and Kolodner 1999; Myung et al. 2001).
Immunoblotting
Western blot analysis was performed as described in Perkins et al. (2001). Myc-tagged Sic1, Sic1ΔNT, Sic1C70, Sic1C70td, Cln2, Cln2-1, and Ubr1 proteins were detected with the 9E10 monoclonal antibody; p86/91 was detected with the 6D2 antibody (Desdouets et al. 1998); and Orc6 was detected with the SB49 antibody (Weinreich et al. 1999; Seki and Diffley 2000). The secondary antibody in each case was anti-mouse HRP from Vector Labs. Membranes were stained with Ponceau-S and scanned as a loading control and to confirm even transfer during electroblotting.
Other methods
Flow cytometry, genomic footprinting, and microscopic observation of Mcm4–GFP were performed as described elsewhere (Perkins and Diffley 1998; Labib et al. 1999; Noton and Diffley 2000; Tanaka and Diffley 2002).
Acknowledgments
We thank Lucy Drury, Anne Early, Karim Labib, and Lil Noton for helpful discussions and advice, plasmids, and yeast strains. We also thank Kyungjae Myung and Richard D. Kolodner for help with the GCR assay, Yoshimi Tanaka for help with the H1 kinase assay, and Eileen Hogan for the sequence of pDK243. This work is supported by Cancer Research U.K. S.T. is the recipient of a JSPS Postdoctoral Fellowships for Research Abroad.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL John.Diffley@cancer.org.uk; FAX 44 0 20 7269-3801.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1011002.
References
- Almasan A, Linke SP, Paulson TG, Huang LC, Wahl GM. Genetic instability as a consequence of inappropriate entry into and progression through S-phase. Cancer Metast Rev. 1995;14:59–73. doi: 10.1007/BF00690212. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Amon A. Men and sin: What's the difference? Nat Rev Mol Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Blow JJ. New EMBO Member's Review: Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J. 2001;20:3293–3297. doi: 10.1093/emboj/20.13.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruschi CV, McMillan JN, Coglievina M, Esposito MS. The genomic instability of yeast cdc6-1/cdc6-1 mutants involves chromosome structure and recombination. Mol Gen Genet. 1995;249:8–18. doi: 10.1007/BF00290230. [DOI] [PubMed] [Google Scholar]
- Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, Kinzler KW, Vogelstein B, Lengauer C. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62:1129–1133. [PubMed] [Google Scholar]
- Calzada A, Sanchez M, Sanchez E, Bueno A. The stability of the Cdc6 protein is regulated by cyclin-dependent kinase/cyclin B complexes in Saccharomyces cerevisiae. J Biol Chem. 2000;275:9734–9741. doi: 10.1074/jbc.275.13.9734. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Chen Y, Hennessy KM, Botstein D, Tye B-K. CDC46/MCM5, a yeast protein whose subcellular localization is cell cycle-regulated, is involved in DNA replication at autonomously replicating sequences. Proc Natl Acad Sci. 1992;89:10459–10463. doi: 10.1073/pnas.89.21.10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Classon M, Salama S, Gorka C, Mulloy R, Braun P, Harlow E. Combinatorial roles for pRB, p107, and p130 in E2F-mediated cell cycle control. Proc Natl Acad Sci. 2000;97:10820–10825. doi: 10.1073/pnas.190343497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Wilkinson HR, Downes CS. A protein kinase-dependent block to reinitiation of DNA replication in G2 phase in mammalian cells. Exp Cell Res. 1996;225:294–300. doi: 10.1006/excr.1996.0179. [DOI] [PubMed] [Google Scholar]
- Coverley D, Wilkinson HR, Madine MA, Mills AD, Laskey RA. Protein kinase inhibition in G2 causes mammalian Mcm proteins to reassociate with chromatin and restores ability to replicate. Exp Cell Res. 1998;238:63–69. doi: 10.1006/excr.1997.3829. [DOI] [PubMed] [Google Scholar]
- Dalton S, Whitbread L. Cell-cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA-replication in budding yeast. Proc Natl Acad Sci. 1995;92:2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouets C, Santocanale C, Drury LS, Perkins G, Foiani M, Plevani P, Diffley JFX. Evidence for a Cdc6p-independent mitotic resetting event involving DNA polymerase α. EMBO J. 1998;17:4139–4146. doi: 10.1093/emboj/17.14.4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX. DNA Replication: Building the perfect switch. Curr Biol. 2001;11:R367–R370. doi: 10.1016/s0960-9822(01)00196-8. [DOI] [PubMed] [Google Scholar]
- Diffley JFX, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dohmen RJ, Wu P, Varshavsky A. Heat-inducible degron: A method for constructing temperature-sensitive mutants. Science. 1994;263:1273–1276. doi: 10.1126/science.8122109. [DOI] [PubMed] [Google Scholar]
- Drury LS, Perkins G, Diffley JFX. The Cdc4/34/53 pathway targets Cdc6p for proteolysis in budding yeast. EMBO J. 1997;16:5966–5976. doi: 10.1093/emboj/16.19.5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The cyclin dependent kinase Cdc28p regulates distinct modes of Cdc6p proteolysis during the budding yeast cell cycle. Curr Biol. 2000;10:231–240. doi: 10.1016/s0960-9822(00)00355-9. [DOI] [PubMed] [Google Scholar]
- Ekholm SV, Reed SI. Regulation of G1 cyclin-dependent kinases in the mammalian cell cycle. Curr Opin Cell Biol. 2000;12:676–684. doi: 10.1016/s0955-0674(00)00151-4. [DOI] [PubMed] [Google Scholar]
- Elsasser S, Chi Y, Yang P, Campbell JL. Phosphorylation controls timing of Cdc6p destruction: A biochemical analysis. Mol Biol Cell. 1999;10:3263–3277. doi: 10.1091/mbc.10.10.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M, Liberi G, Lucchini G, Plevani P. Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B subunit. Mol Cell Biol. 1995;15:883–891. doi: 10.1128/mcb.15.2.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadwiger J, Wittenberg C, Richardson H, Lopes M, Reed S. A family of cyclin homologs that control the G1 phase in yeast. Proc Natl Acad Sci. 1989;86:6255–6259. doi: 10.1073/pnas.86.16.6255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessy KM, Clark CD, Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes & Dev. 1990;4:2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Ho A, Dowdy SF. Regulation of G1 cell-cycle progression by oncogenes and tumor suppressor genes. Curr Opin Genet Dev. 2002;12:47–52. doi: 10.1016/s0959-437x(01)00263-5. [DOI] [PubMed] [Google Scholar]
- Hodge A, Mendenhall M. The cyclin-dependent kinase inhibitory domain of the yeast Sic1 protein is contained within the C-terminal 70 amino acids. Mol Gen Genet. 1999;262:55–64. doi: 10.1007/s004380051059. [DOI] [PubMed] [Google Scholar]
- Hogan E, Koshland D. Addition of extra origins of replication to a minichromosome suppresses its mitotic loss in cdc6 and cdc14 mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci. 1992;89:3098–3102. doi: 10.1073/pnas.89.7.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S, Geymonat M, Johnston LH. Mitotic exit: Delaying the end without FEAR. Curr Biol. 2002;12:R221–R223. doi: 10.1016/s0960-9822(02)00756-x. [DOI] [PubMed] [Google Scholar]
- Kelly TJ, Brown GW. Regulation of chromosome replication. Annu Rev Biochem. 2000;69:829–880. doi: 10.1146/annurev.biochem.69.1.829. [DOI] [PubMed] [Google Scholar]
- Labib K, Diffley JFX, Kearsey SE. G1-phase and B-type cyclins exclude the DNA-replication factor Mcm4 from the nucleus. Nat Cell Biol. 1999;1:415–422. doi: 10.1038/15649. [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JFX. Uninterrupted MCM2-7 function required for DNA replication fork progression. Science. 2000;288:1643–1647. doi: 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- Labib K, Kearsey SE, Diffley JFX. MCM2-7 proteins are essential components of prereplicative complexes, that accumulate co-operatively in the nucleus during G1-phase, and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell. 2001;12:3658–3667. doi: 10.1091/mbc.12.11.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Schwob E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G1. Mol Cell. 2002;9:1067–1078. doi: 10.1016/s1097-2765(02)00513-0. [DOI] [PubMed] [Google Scholar]
- Macleod KF, Hu Y, Jacks T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996;15:6178–6188. [PMC free article] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M. To cycle or not to cycle: A critical decision in cancer. Nat Rev Cancer. 2001;1:222–231. doi: 10.1038/35106065. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Mekeel KL, Tang W, Kachnic LA, Luo CM, DeFrank JS, Powell SN. Inactivation of p53 results in high rates of homologous recombination. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–74. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Irie K, Li JJ. Clb/Cdc28 kinases promote nuclear export of the replication initiator proteins Mcm2-7. Curr Biol. 2000;10:195–205. doi: 10.1016/s0960-9822(00)00337-7. [DOI] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Noguchi E, Shanahan P, Noguchi C, Russell P. CDK phosphorylation of Drc1 regulates DNA replication in fission yeast. Curr Biol. 2002;12:599–605. doi: 10.1016/s0960-9822(02)00739-x. [DOI] [PubMed] [Google Scholar]
- Noton EA. “The regulation of pre-replicative complex formation in the budding yeast cell cycle.” Ph.D. thesis. U.K: University College London; 2000. [Google Scholar]
- Noton EA, Diffley JFX. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol Cell. 2000;5:85–95. doi: 10.1016/s1097-2765(00)80405-0. [DOI] [PubMed] [Google Scholar]
- Nugroho TT, Mendenhall MD. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol Cell Biol. 1994;14:3320–3328. doi: 10.1128/mcb.14.5.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson TG, Almasan A, Brody LL, Wahl GM. Gene amplification in a p53-deficient cell line requires cell cycle progression under conditions that generate DNA breakage. Mol Cell Biol. 1998;18:3089–3100. doi: 10.1128/mcb.18.5.3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins G, Diffley JFX. Nucleotide dependent prereplicative complex assembly by Cdc6p, a homologue of eukaryotic and prokaryotic clamp-loaders. Mol Cell. 1998;2:23–32. doi: 10.1016/s1097-2765(00)80110-0. [DOI] [PubMed] [Google Scholar]
- Perkins G, Drury LS, Diffley JFX. Separate SCFCDC4 recognition elements target Cdc6 for proteolysis in S phase and mitosis. EMBO J. 2001;20:4836–4845. doi: 10.1093/emboj/20.17.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane SG, Cosenza SC, Mettus RV, Reddy EP. Germ line transmission of the Cdk4(R24C) mutation facilitates tumorigenesis and escape from cellular senescence. Mol Cell Biol. 2002;22:644–656. doi: 10.1128/MCB.22.2.644-656.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reenan RA, Kolodner RD. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: Evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santocanale C, Diffley JFX. ORC- and Cdc6-dependent complexes at active and inactive chromosomal replication origins in Saccharomyces cerevisiae. EMBO J. 1996;15:6671–6679. [PMC free article] [PubMed] [Google Scholar]
- Saunders WS. The FEAR factor. Mol Cell. 2002;9:207–209. doi: 10.1016/s1097-2765(02)00460-4. [DOI] [PubMed] [Google Scholar]
- Schar P. Spontaneous DNA damage, genome instability, and cancer—When DNA replication escapes control. Cell. 2001;104:329–332. doi: 10.1016/s0092-8674(01)00220-3. [DOI] [PubMed] [Google Scholar]
- Seki T, Diffley JFX. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ. The Pezcoller lecture: Cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Diffley JFX. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat Cell Biol. 2002;4:198–207. doi: 10.1038/ncb757. [DOI] [PubMed] [Google Scholar]
- Tlsty TD, Briot A, Gualberto A, Hall I, Hess S, Hixon M, Kuppuswamy D, Romanov S, Sage M, White A. Genomic instability and cancer. Mutat Res. 1995;337:1–7. doi: 10.1016/0921-8777(95)00016-d. [DOI] [PubMed] [Google Scholar]
- Tomlinson I, Bodmer W. Selection, the mutation rate and cancer: Ensuring that the tail does not wag the dog. Nat Med. 1999;5:11–12. doi: 10.1038/4687. [DOI] [PubMed] [Google Scholar]
- Toone WM, Aerne BL, Morgan BA, Johnston LH. Getting started: Regulating the initiation of DNA replication in yeast. Annu Rev Microbiol. 1997;51:125–149. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- Verma R, Annan RS, Huddleston MJ, Carr SA, Reynard G, Deshaies RJ. Phosphorylation of Sic1p by G1 Cdk required for its degradation and entry into S phase. Science. 1997;278:455–460. doi: 10.1126/science.278.5337.455. [DOI] [PubMed] [Google Scholar]
- Wahl GM, Carr AM. The evolution of diverse biological responses to DNA damage: Insights from yeast and p53. Nat Cell Biol. 2001;3:E277–E286. doi: 10.1038/ncb1201-e277. [DOI] [PubMed] [Google Scholar]
- Weinreich M, Liang C, Stillman B. The Cdc6p nucleotide-binding motif is required for loading Mcm proteins onto chromatin. Proc Natl Acad Sci. 1999;96:441–446. doi: 10.1073/pnas.96.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Taipale M, Heiskanen A, Laiho M. Ectopic expression of Cdk6 circumvents transforming growth factor-β mediated growth inhibition. Oncogene. 2001;20:5888–5896. doi: 10.1038/sj.onc.1204745. [DOI] [PubMed] [Google Scholar]
- Zhou P, Jiang W, Weghorst CM, Weinstein IB. Overexpression of cyclin D1 enhances gene amplification. Cancer Res. 1996;56:36–39. [PubMed] [Google Scholar]