A Novel Golgi Membrane Protein Is a Partner of the ARF Exchange Factors Gea1p and Gea2p (original) (raw)
Abstract
The Sec7 domain guanine nucleotide exchange factors (GEFs) for the GTPase ARF are highly conserved regulators of membrane dynamics and protein trafficking. The interactions of large ARF GEFs with cellular membranes for localization and/or activation are likely to participate in regulated recruitment of ARF and effectors. However, these interactions remain largely unknown. Here we characterize Gmh1p, the first Golgi transmembrane-domain partner of any of the high-molecular-weight ARF-GEFs. Gmh1p is an evolutionarily conserved protein. We demonstrate molecular interaction between the yeast Gmh1p and the large ARF-GEFs Gea1p and Gea2p. This interaction involves a domain of Gea1p and Gea2p that is conserved in the eukaryotic orthologues of the Gea proteins. A single mutation in a conserved amino acid residue of this domain is sufficient to abrogate the interaction, whereas the overexpression of Gmh1p can compensate in vivo defects caused by mutations in this domain. We show that Gmh1p is an integral membrane protein that localizes to the early Golgi in yeast and in human HeLa cells and cycles through the ER. Hence, we propose that Gmh1p acts as a positive Golgi-membrane partner for Gea function. These results are of general interest given the evolutionary conservation of both ARF-GEFs and the Gmh proteins.
INTRODUCTION
In eukaryotic cells, protein and membrane traffic between the organelles of the biosynthetic and endocytic pathways involves complex regulatory mechanisms. These mechanisms ensure the specificity and directionality of protein transport and balance the dynamic exchanges of membrane material within the cell. The small GTP-binding proteins of the ADP-ribosylation factor (ARF) family are central regulators of protein traffic and membrane dynamics in eukaryotic cells (Chavrier and Goud, 1999). ARF cycles between inactive GDP-bound and active, membrane-associated GTP-bound forms. ARF activation and binding to specific membranes are essential for the subsequent interactions with a number of effectors including coat proteins such as coatomer, AP-1 (adaptor protein 1), AP-3, AP-4, and GGAs (Donaldson and Jackson, 2000; Boehm and Bonifacino, 2001). ARF activation results in modulation of membrane structure and function through changes in both the protein and lipid contents of the membrane on which it is localized. In cells, guanine nucleotide exchange factors (GEFs) are required to activate ARF proteins. The activity of some of the ARF-GEFs is sensitive to brefeldin A (BFA), a fungal metabolite that perturbs Golgi structure and trafficking functions (Donaldson et al., 1992; Helms and Rothman, 1992). Gea1p, Gea2p, Sec7p, and Syt1p have been identified as ARF GEFs in the yeast Saccharomyces cerevisiae (reviewed inDonaldson and Jackson, 2000;Jackson and Casanova, 2000). Like all ARF GEFs identified to date, they possess a Sec7 domain that is sufficient to catalyze exchange of GDP for GTP in vitro (Chardin et al., 1996). Proteins containing Sec7 domains have been identified in a wide variety of organisms including intracellular pathogenic bacteria, yeast, plants, protozoa, worms, flies, and mammals (Nagai et al., 2002) and reviewed in Donaldson and Jackson, (2000) and Jackson and Casanova, (2000). The ARF GEFs Gea1p, Gea2p, and Sec7p are the major targets of BFA in the secretory pathway of yeast (Peyroche et al., 1999). The mechanism of BFA action has been determined and involves the Sec7 domain of these proteins (Peyroche et al., 1999).
Members of the Sec7 family can be subdivided in two major classes: the large (>100 kDa) ARF GEFs and the smaller (<100 kDa) ARF GEFs. The large yeast ARF GEFs Gea1p, Gea2p, and Sec7p have orthologues in higher eukaryotes. The yeast Gea1p and Gea2p proteins are 50% identical and at least partially functionally redundant: strains carrying a deletion of either GEA1 or_GEA2_ have no growth defect, whereas the double deletion strain is inviable (Peyroche et al., 1996). On the basis of sequence analysis, yeast Gea1p and Gea2p belong to a subfamily of ARF GEFs that also includes the mammalian GBF1 protein (Claude et al., 1999) and Arabidopsis GNOM/Emb30p (Shevell et al., 1994; Busch et al., 1996). GNOM has been implicated in the regulation of a BFA-sensitive specific endosomal trafficking pathway (Geldner et al., 2003). In contrast to Gea and Gnom proteins, the GEF activity of GBF1 protein is BFA resistant and exhibits in vitro specificity toward a different class of ARFs (Claude et al., 1999). Mammalian p200GEP/BIG1 and BIG2 are related to Sec7p. Smaller ARF GEFs such as ARNO, the cytohesins, and GRP1 exist solely in higher eukaryotes.
Sec7p is localized to the Golgi complex in yeast. It plays an important role in transport at multiple steps from the ER through the Golgi apparatus and is an important regulator of membrane dynamics in the ER-Golgi system of yeast (Franzusoff et al., 1991; Deitz et al., 2000). Yeast gea mutants have severe defects in protein transport through the ER-Golgi system and in Golgi structure (Peyroche et al., 1996,2001;Spang et al., 2001). Neither Gea1p nor Gea2p can replace Sec7p functionally in vivo, and vice versa, Sec7p cannot compensate for the loss of Gea1/2p, indicating that each must have a distinct role within the cell (Peyroche et al., 1996). The existence of these different ARF GEFs in the cell may determine specificity of coat recruitment.
The large ARF GEFs are peripherally associated membrane proteins found in both membrane-bound and soluble forms (Franzusoff et al., 1991; Peyroche et al., 1996; Claude et al., 1999; Yamaji et al., 2000; Spang et al., 2001; Kawamoto_et al.,_ 2002). The mechanisms by which the ARF GEFs are localized to membranes are of crucial importance because their location is thought to be a major determinant of the appropriate membrane sites of ARF activation. It is not known whether membrane recruitment of large ARF GEFs occurs by direct interaction with specific types of lipids or by a membrane-localized protein partner or both. No canonical lipid-interacting domain such as PH domains has been identified in any of the large ARF GEFs. The Sec7 domain has been exhaustively characterized. It has been shown that Gnom protein can homodimerize and interact with cyclophilin 5 via the DCB (Dimerization and cyclophilin-binding) domain conserved in all the members of the Gea/GBF/GNOM family (Grebe et al., 2000). However, the role of the rest of these large proteins is poorly understood and may be involved in membrane localization and regulatory functions. Hence, the identification of interacting partners of the large ARF GEFs should help to elucidate the mechanism of ARF GEF recruitment to the Golgi complex and the regulation of transport.
Here, we report the identification and characterization of a novel conserved Golgi-membrane protein named Gmh1p that interacts with Gea1/2p large ARF GEFs in yeast. Because Gmh1p has homologues in several eukaryotic organisms, we propose that Gmh1p participates in membrane-related functions of Gea/GNOM/GBF1 large ARF GEF proteins.
MATERIALS AND METHODS
Plasmids, Yeast Strains, and Media
Plasmids and yeast strains used in this study are listed in Tables1 and2, respectively. Yeast genetic techniques and media have been described by Sherman (1991).
Table 1.
Plasmids used in this study
| Plasmid | Description | Reference |
|---|---|---|
| YEp-ARF2 | 2μ, URA3, ARF2 | Peyroche et al. (1996) |
| pCLJ92 | 2μ, URA3, GEA1 | Peyroche et al. (1999) |
| pAP52 | 2μ, URA3, GMH1 | This study |
| pAP100 | 2μ, URA3, HA-GMH1 | This study |
| pAP101 | 2μ, URA3, GMH1-HA | This study |
| pRC7 | 2μ, LEU2, GMH1-HA6 | This study |
| pRC8 | 2μ, URA3, HA3-GMH1 | This study |
| pAP68 | 2μ, LEU2, GMH1 | This study |
| pRS-Emp47m | CEN, URA3, Emp47-myc | Lewis and Pelham (1996) |
| pRC11 | 2μ, LEU2, HA3-GMH1 | This study |
| pEM2 | pcDNA3-HA3-hGMH1 | This study |
| pAPDH15 | pACTII-GMH1 | This study |
| pSC6b | pASΔ-GEA1(aa749-1408) | This study |
| pSC4b | pASΔ-GEA1(aa749-1263) | This study |
| pSC37b | pACTII-h_GMH1_ | This study |
| pSC58 | pASΔ-GEA1(aa1-1263) | This study |
| pSC59 | pASΔ-GEA1 | This study |
| pSC64 | 2μ, URA3, tetO→GEA1(aa749-1408) | This study |
Table 2.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| APY022 | Matα ura3-52 leu2-3,112 his3-Δ200 lys2-801 ade2-101 gea2::HIS3 gea1-6 | Peyroche et al. (2001) |
| CJY045-5-3 | Mat a ura3-52 leu2 his3Δ200 lys2-801 trp1-Δ63 arf1::HIS3 | Peyroche et al. (1996) |
| SEY6210 | Mat α ura3-52 leu2-3,112 his3-Δ200 lys2-801 trp1-Δ901 suc2-Δ9 | From S. Emr |
| ALY013-1 | Mat a ste2::LEU2 STE2-WBP1::URA3 ura3-52 leu2-3,112 his3-Δ200 ade2-101 gea2::HIS3 gea1-6 | This study |
| ALY014-2 | Mat a ste2::LEU2 STE2-WBP1::URA3 ura3-52 leu2-3,112 his3-Δ200 ade2-101 | This study |
| SCY002 | Mat α ura3-52 leu2-3,112 his3-Δ200 lys2-801 ade2-101 GEA2-13MYC::KANMX. | This study |
| APY130-2 | Mat α ura3-52 leu2-3,112 his3-Δ200 lys2-801 ade2-101 GEA2-13MYC::KANMX. gmh1::HIS3 | This study |
| YPH499 | Mat a trp1Δ63 his3Δ200 ura3-52 leu2Δ1 ade2-101 lys2-801 | From P. Hieter |
| RCY006 | Mat a trp1Δ63 his3Δ200 ura3-52 leu2Δ1 ade2-101 lys2-801 GMH1-3HA::KANMX | This study |
| CHSY004-2 | Mat α trp1Δ63 his3Δ200 ura3-52 leu2Δ1 ade2-101 lys2-801 CHS5-Myc13::KANMX | This study |
| RSY253 | Matα his4-619 ura3-52 sec12-4 | From R. Schekman |
| C13-ABYS86 | Matα pra1-1 prb1-1 prc1-1 cps1-3 ura3Δ5 leu2-3,112 his-canR | From D.H. Wolf |
| Y190 | Mat a gal4Δ gal80Δ his3-Δ200 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GAL->lacZ LYS2::GAL-> HIS3 cyhR | From S. Elledge |
Antibodies and Immunoblotting
Western blot analysis was performed as previously described (Peyroche et al., 1996). Dilutions used for the primary antibodies were as follows: mouse monoclonal anti-HA.11 (Babco, Richmond, CA) 1:2000; anti-Myc mouse monoclonal (Santa Cruz Biotechnology, Santa Cruz, CA) 1:3000; anti-Myc rabbit polyclonal A14 (Santa Cruz) 1:4000; monoclonal anti-Dpm1p (Molecular Probes, Eugene, OR) 1:500; and monoclonal anti-ATPase 60 kDa (Molecular Probes) 1:500. Polyclonal antibodies were raised in rabbits against purified yeast ARF2 protein (Peyroche and Jackson, 2001). A Gmh1p-specific peptide NDLPAGPQGQRRRNN (amino acids 9–23) was selected for antibody generation. This peptide was synthesized with an additional C-terminal tyrosine, purified, coupled to OVA and injected into rabbits by Neosystem (Strasbourg, France). The sera anti-Arf2p and anti-Gmh1p was used to 1:1000 for Western blot analysis. HRP-conjugated anti-rabbit or anti-mouse (Promega, Madison, WI) were used as secondary antibodies (dilution 1:10000). Detection was performed with ECL chemiluminescent reagents (Amersham-Biosciences) (UK).
gea1-6 Suppressor Screen
A gea1-6 strain was transformed with a yeast genomic library carried in the 2-μm vector pFL44 (gift from F. Lacroute). Transformants (100,000) were grown at 24°C for 12 h and then shifted to 34°C. For growing colonies at 34°C, plasmids were isolated and sequenced.
Mating Assay
The plate mating assay is essentially as described by Letourneur et al. (1994). For quantitation, haploid strains were grown in YPD and resuspended to 5 μDO/ml, and 0.5 ml of each suspension was then preincubated at the appropriate temperature for 2 h. Mat a strains and Mat α SEY6210 testing strain were then mixed and incubated for 6 h at the same temperature to allow mating. Different dilutions of this mating mix were then spread on SC-Ade-Trp to select diploids. After 3 d at 30°C, diploid colonies were counted.
Immunofluorescence Analysis of Yeast Cells
Immunofluorescence analysis were carried out as previously described (Peyroche et al., 2001). Dilutions used for the primary antibodies were as follows: purified mouse monoclonal anti-HA.11 (Babco) 1:100; anti-Myc mouse monoclonal 9E10 (Santa Cruz) 1:100; affinity-purified anti-Anp1 (Jungmann and Munro, 1998) 1:250; and anti-Myc rabbit polyclonal A14 (Santa Cruz) 1:100.
Cell Culture and Transfection of HeLa Cells
HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing 4.5 g/l glucose (GIBCO, Scotland) supplemented with 10% fetal calf serum (GIBCO), 5 mM glutamine, and 5 mM sodium pyruvate (GIBCO) in a 5% humidified CO2 incubator. HeLa cells, plated on 12-mm round glass coverslips the day before transfection, were transfected using a calcium phosphate method (Jordan et al., 1996). For BFA-treated cells, BFA (5 mg/ml) purchased from the Alexis Corporation (San Diego, CA) was added 2 h before fixation.
Immunofluorescence and Confocal Microscopy of Human Cells
Cells were fixed with para-formaldehyde 3% in PBS and quenched in 50 mM glycine. After permeabilization in 0.2% saponin containing PBS and 1% BSA cells were double-labeled with antibodies with the affinity-purified mouse monoclonal HA-11 antibody (Babco) and affinity-purified antibodies raised against Rab6 (Goud et al., 1990) or GMAP-210 (Rios et al., 1994) or Rab1 (Saraste et al., 1995). Coverslips were mounted in Mowiol (Hoechst AG). Secondary antibodies were purchased from Jackson Corporation. Confocal laser scanning microscopy and immunofluorescence were performed using a Leica or a TCS4D confocal microscope (Leica Microsystems, Heidelberg, Germany).
Subcellular Fractionation, Membrane Association, and Protease Protection Assays
Yeast cells were harvested at midlogarithmic phase, washed twice, and resuspended in Spheroplast buffer (1.2 M sorbitol, 50 mM Tris-HCl, pH 7.5, 40 mM β-mercaptoethanol, 10 mM NaN3) to 20 μDO600 nm/ml. Cells were spheroplasted with zymolyase 100T (ICN) and disrupted in lysis buffer (20 mM HEPES-KOH, pH 7.4, 100 mM KCl, 2 mM DTT, 1 mM PMSF, Complete protease inhibitor cocktail; Boehringer-Mannheim) with a Dounce homogenizer. The cell lysate was centrifuged twice at 500 × g to remove cell debris. To verify that Gmh1p is an integral membrane protein, we proceeded as described in Matern et al. (2000). For differential centrifugation, gently lysed cells were prepared as abovementioned and submitted to a 13,000 × g centrifugation for 1 h. Supernatant fractions were then recentrifuged at 100,000 × g for 1 h. Aliquots of pellet (P13 and P100) and supernatant fractions were analyzed by Western blot after SDS-PAGE. Protease protection assay was performed as described in Yang et al. (1998).
Two-hybrid Analysis
pASΔ and pACTII were used to create fusions with Gal4-DNA binding domain and Gal4 transcription activation domain, respectively (seeTable 1). Y190 strain was transformed with the different plasmids and transformed cells were selected on SD minimal medium plates supplemented with adenine and histidine. The cells were then streaked on SD + adenine plates containing 10, 25, or 50 mM 3-aminotriazole to test expression of the first reporter gene HIS3. The cells were also streaked on SD +adenine +histidine plates to test for β-galactosidase production in an overlay plate assay. At least three independent experiments were performed for each two-hybrid analysis.
Immunoprecipitation Experiments
Yeast cells (40 μDO) were harvested at midlogarithmic phase and washed once with 10 mM NaN3 and once with lysis buffer (20 mM HEPES, pH 7.2, 100 mM KCl, 5 mM MgCl2, and 1% Triton X-100). The cells were disrupted with acid-washed glass beads (Sigma-Aldrich, St Louis, MO) in 0.1 ml of lysis buffer supplemented with Complete protease inhibitor cocktail (Roche-Boehringer, Mannheim, Germany) and Pefablock 1 mM (Roche). The cell lysate was centrifuged twice at 500 × g, and supernatant then was subjected to a 100,000 × g centrifugation at 4°C for 20 min. Cleared lysates were diluted 10-fold with lysis buffer lacking Triton X-100 and incubated with 25 μl of protein G-Sepharose (Amersham Pharmacia) coupled to anti-HA antibodies (12CA5) for at least 2 h at 4°C. The resin was then washed twice with buffer containing 200 mM KCl and 0.1% Triton X-100 and resuspended in 2× SDS-Laemmli buffer before SDS-PAGE analysis.
RESULTS
Overexpression of GMH1 Suppresses gea1-6_and_ arf1Δ Defects
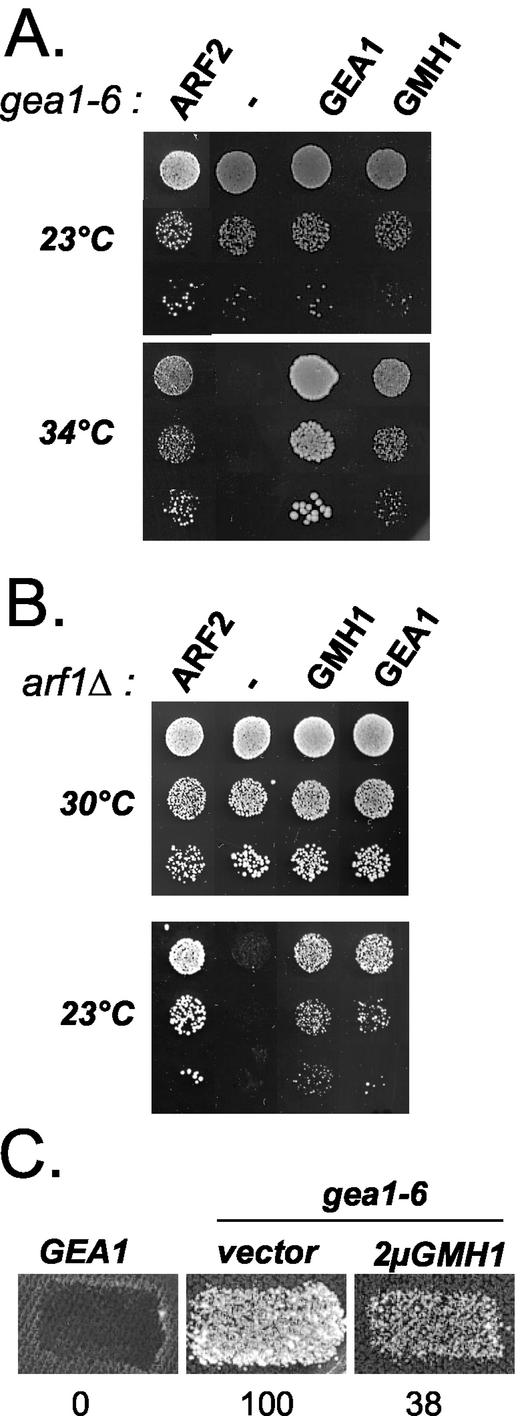
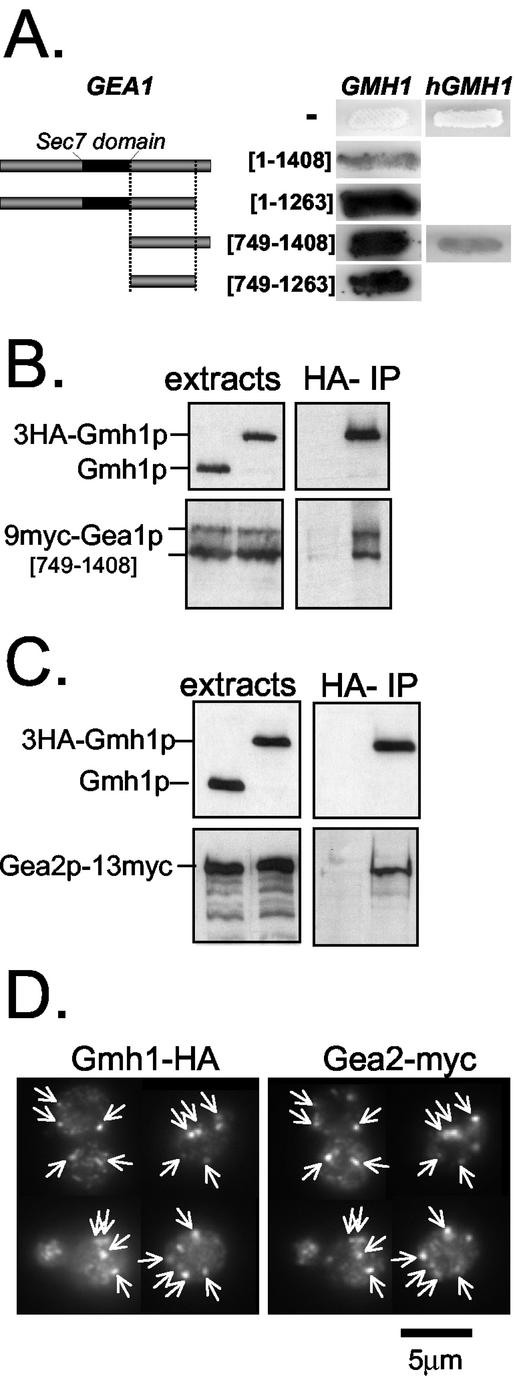
To gain insight into the function of Gea1/2p, we undertook a genetic approach. Because Gea1p and Gea2p have redundant functions for growth, temperature-conditional mutants of GEA1 were previously generated in a strain carrying a deletion of GEA2. We used the gea1-6 gea2_Δ mutant, herewith designated gea1-6 for simplicity (Peyroche et al., 2001). We expected that the overexpression of activators or stimulators of Gea1p-dependent functions would mitigate the growth defect and lead to an increased temperature tolerance. A multicopy suppressor screen was therefore performed to identify genes whose overexpression renders_gea1-6 mutant cells viable at nonpermissive temperature. For this purpose, gea1-6 mutant cells were transformed with a 2-μm–based yeast genomic library (Peyroche et al., 1996). Besides the GEA1 and GEA2 genes, several other genes, including ARF1 and ARF2 (Figure 1A and our unpublished results), were isolated. Another candidate was isolated and identified as the putative open reading frame YKR030w (Figure 1A). Suppression of_gea1-6_ growth defects by YKR030w is dose dependent because its expression from a centromere-based vector allows intermediate growth of_gea1-6_ mutant cells at 34°C (our unpublished results). However, unlike ARF1/2, YKR030w is not a general suppressor of GEA functions because its overexpression does not improve growth of another mutant strain (gea1-4) at nonpermissive temperature (our unpublished results). In the light of subsequent experiments, we named this gene GMH1 forgea1-6 membrane-associated high-copy suppressor.
Figure 1.
Overexpression of GMH1 rescues gea1-6 and_arf1_Δ mutant cells. APY022 gea1-6 (A) or CJY045-5-3_arf1_Δ (B) mutant cells were transformed either with multicopy vector alone or with the same vector containing ARF2, GEA1 or_GMH1(YKR030w_). Serial dilutions of the transformants were then spotted on selective medium and grown at indicated temperatures for 3 d. (C) Mat a wild-type (ALY014-2) or gea1-6 (ALY013-1) transformed either with multicopy vector alone or with the same vector containing_GMH1(YKR030w_) were grown on nonselective medium and replica plated to a lawn of wild-type cells Mat α SEY6210 and incubated at 33°C for 6 h. Mating was then analyzed by growth on medium selective for diploids. Numbers indicate the efficiency of mating in arbitrary units (average of three independent experiments).
To reinforce the potential functional link between the protein encoded by_GMH1_ and Gea1p, we investigated the effects of GMH1 overexpression in _arf1_Δ mutant cells. These mutant cells exhibit defects at multiple transport steps (Gaynor et al., 1998). In our strain background, these defects are accompanied by a marked delay of growth especially at low temperatures (Figure 1B). Strikingly, growth of arf1_Δ at either 16 or 23°C is almost completely restored when cells contain a multicopy vector containing either GEA1 or_GMH1 (Figure 1B and our unpublished results).
Conditional mutants gea2_Δ gea1 show different transport defects including in vitro retrograde transport defects (Peyroche_et al., 1996,2001;Spang et al., 2001). We used an in vivo assay to monitor defects in Golgi-to-ER traffic occurring in gea1-6 cells (Letourneur_et al.,_ 1994). In this assay, the cell surface α-factor receptor Ste2p is fused to the ER-resident protein Wbp1p. By virtue of this fusion, the receptor is retained in the ER and cannot mediate mating. Only mutant cells in which the fusion protein is mislocalized to the cell surface are competent for mating (Letourneur _et al.,_1994). We tested appropriate gea1-6 strain, along with negative control strains, for its ability to mate at different temperatures. The _gea2_Δ strain does not mate at any temperature (Figure 1C and our unpublished results). In contrast, the gea1-6 strain display temperature-dependent mating (Figure 1C and our unpublished results). This observation establishes for the first time that gea1-6 cells display in vivo retrograde transport defects. However, mating is efficiently reduced when GMH1 is overexpressed in gea1-6 cells, indicating that moderate overexpression of GMH1 cannot only restore growth defects but also at least partially restore retrograde transport defects of gea1-6 cells. Altogether, these genetic data suggest that Gmh1p could play a positive role in an essential Gea1/2p-controlled step of ARF activation.
To study the phenotype of the haploid disrupted strain, we deleted the entire GMH1 ORF in diploid cells that were then sporulated and dissected into tetrads. The GMH1_Δ haploids were viable, indicating that GMH1 is dispensable for vegetative growth in standard conditions (our unpublished results). However, strong overexpression of_GMH1 results in slightly slower growth of transformed cells that are hypersensitive to BFA and partially defective for retrograde transport as judged by the in vivo assay described above (our unpublished results).
Gmh1p Is a Member of a Phylogenetically Conserved Family of Proteins
The predicted protein sequence encoded by GMH1 is shown inFigure 2A. Yeast Gmh1p is a 273-residue protein with a calculated mass of 31.9 kDa and a p_K_ i equal to 9.2. Homologues were identified in fission yeast Schizosaccharomyces pombe, Caenorhabditis elegans, Arabidopsis thaliana, Drosophila melanogaster, and in mammals (Figure 2A). Gmh1p and its relatives from other species have similar length (244–275 amino acid residues) and contain several regions sharing identical sequence motifs (Figure 2A). No other Gmh1p homologues could be identified in S. cerevisiae on the basis of sequence analysis.
Figure 2.
Structure of Gmh1p and its putative orthologues. (A) Sequence comparison of Gmh1p and its putative orthologues. The amino acid sequence of Gmh1p from_S. cerevisiae_ (Sc; accession number Z28255) is aligned with those of related proteins present in H. sapiens (H.s; accession numberAF077038 for hGMH1a and AL080115 for hGMH1b), in R. norgevicus (R.n; accession number U96638), in C. elegans (C.e; accession numberZ48055), in A. thaliana (A.th; accession number AF378872), and in_D. melanogaster_ (D.m; accession number AE003680). Identical amino acid residues in the analogous positions of at least four proteins are in gray boxes. Lines indicate the predicted hydrophobic transmembrane domains (TDM1 to TDM5). Boxes indicate highly conserved motifs I, II, and III. (B) Hydropathy profiles of Gmh1p and hGMH1. Predicted transmembrane domains are numbered 1–5.
Hydropathy plot analysis of the yeast protein reveals five putative membrane-spanning helices flanked by an N-terminal hydrophilic domain (∼90 residues) and a short C-terminal domain (∼10 residues;Figure 2B). This overall topology is conserved in the putative orthologues (Figure 2B). Interestingly, regions of strong homology are not restricted to the hydrophobic domains, suggesting that nontransmembrane domains could play a key role in the functions of these proteins.
Gmh1p Is an Integral Membrane Protein
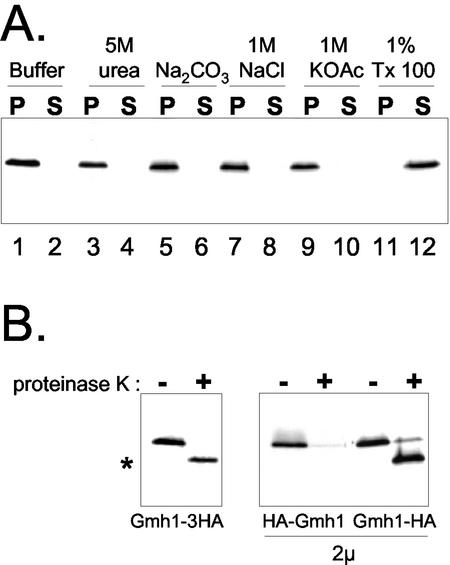
To confirm the membrane location of Gmh1p predicted by sequence analysis, we first performed biochemical fractionation experiments. For this purpose, the endogenous GMH1 locus was modified to lead to the expression of a 3HA tag at the C terminus of the protein. This HA-tagged version of Gmh1p is functional, based on the fact it suppresses the thermosensitive growth defect of gea1-6 when overexpressed (our unpublished results). Yeast cells expressing Gmh1p-3HA were spheroplasted and then lysed with a Dounce homogenizer. Cell homogenates were then subjected to centrifugation to separate membrane (pellet) and soluble (supernatant) fractions. The distribution of Gmh1p in the fractions was then analyzed by Western blotting. First, Gmh1p is efficiently pelleted from cell homogenates by centrifugation at 100,000 × g, indicating that Gmh1p is associated with membranes (Figure 3A, lanes 1 and 2). The pellet association was obtained even when Gmh1p was expressed from a multicopy vector (our unpublished results). To determine the nature of this association with the cellular membranes, cell homogenates were incubated with different buffers before centrifugation (Figure 3A). As shown inFigure 3A, only detergent treatment of the cell lysates led to a solubilization of Gmh1p. Altogether these results indicate that Gmh1p is embedded in cellular membranes.
Figure 3.
Gmh1p is an integral membrane protein. (A) Membrane association of HA-tagged Gmh1p expressed in yeast. Gently lysed RCY006 cells were treated as indicated and then submitted to 100,000 × g centrifugation. Pellet (P) and supernatant (S) fractions were then analyzed by immunoblotting with anti-HA antibodies. (B) Topology of Gmh1p. Gently lysed extracts were prepared from RCY006, YPH499/pAP100 or YPH499/pAP101 strains. Membrane pellets containing the proteins tagged at either Nor C terminus, as indicated, were digested (+) or not (-) with proteinase K and then analyzed by immunoblotting with anti-HA antibodies. Asterisk indicates the shorter polypeptide corresponding to a N-terminus truncated form of Gmh1p.
To determine the membrane topology of the protein, we performed proteinase K digestion of the membranes containing N- or C-terminally HA-tagged Gmh1p. The proteins were then analyzed by Western blotting with anti-HA antibodies (Figure 3B). When the tag is located at the C terminus of Gmh1p, the epitope was still detectable indicating that the C terminus of the protein was protected against protease digestion in native conditions. The C terminus was accessible to protease digestion after solubilization of Gmh1p by Triton X-100 (our unpublished results). The appearance of a shorter polypeptide after protease treatment in native conditions (Figure 3B) suggests that a part of the protein is accessible to proteinase action. Indeed, when the HA tag is located at the N-terminus of Gmh1p, HA-Gmh1p is no longer detectable after proteinase K digestion, indicating that the epitope is accessible to the protease action even in absence of detergent. These results indicate that the N-terminus of Gmh1p faces the cytosol and the C terminus faces the lumen, consistent with the presence of an uneven number of transmembrane domains in Gmh1p. In the light of these experiments, it appears that the most conserved regions in the GMH1 family are oriented toward the cytosol.
Gmh1p Is an Early Golgi Protein
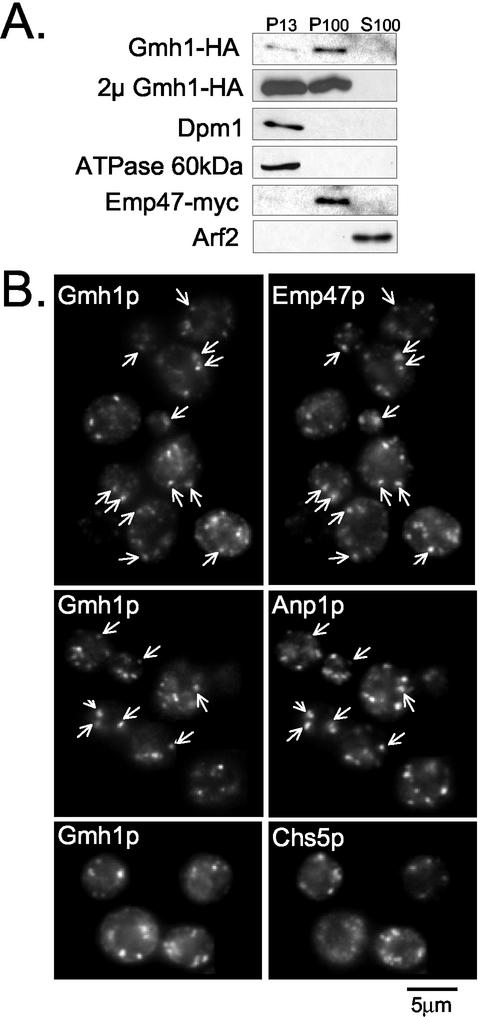
To investigate the intracellular localization of Gmh1p, both subcellular fractionation and indirect immunofluorescence were performed. On differential fractionation of cell lysates, Gmh1p was completely absent from the supernatant fraction and was exclusively detected in fractions pelleted at either 13,000 or 100,000 × g (Figures3A and4A). A proportion of Gmh1p–3HA sedimented at 13,000 × g but the majority of the protein sedimented at 100,000 × g in a fraction enriched for the early-medial Golgi protein Emp47p-myc (Schroder et al., 1995; Figure 4). Interestingly, high levels of expression of Gmh1p from a multicopy plasmid led to a higher proportion of the protein in the subcellular fraction sedimenting at 13,000 × g, which is enriched for ER harboring Dpm1p and for vacuolar membranes containing the 60-kDa subunit of the vacuolar ATPase (Figure 4A). It has already been reported that high levels of expression of Golgi proteins can lead to their accumulation in the ER (Munro, 1991; Yang et al., 1998).
Figure 4.
Intracellular localization of Gmh1p. (A) Analysis by sequential fractionation. RCY006 cells containing pRS-Emp47m were spheroplasted and gently lysed as indicated in MATERIALS AND METHODS. Lysates were submitted to sequential centrifugation at 13,000 and 100,000 × g. Aliquots of pellet (P13 and P100) and supernatant (S100) fractions were analyzed by western-blot with antibodies against the different proteins as indicated. The same experiment was performed with YPH499 cells containing HA-tagged Gmh1p on a 2-μm multicopy plasmid (from pAP100). (B) Analysis by immunofluorescence. HA-tagged Gmh1p (expressed from pRC7) was compared with a coexpressed Emp47-myc protein (from pRS-Emp47m) by double-immunofluorescence (top panel) or with endogenous Anp1p (middle panel). On the lower panel, double-immunofluorescence was performed from CHSY004-2 cells transformed with pRC7 to reveal Gmh1p and Chs5p with HA and Myc antibodies, respectively. Arrows indicate identical position in each pair of image.
To confirm and potentially extend the HA-Gmh1p fractionation data, we localized the protein in whole cells by indirect immunofluorescence. As shown in Figure 4B, Gmh1p exhibited primarily a punctate staining pattern typical for Golgi-localized proteins in yeast cells. On high expression of Gmh1p, a perinuclear ring staining characteristic of the ER compartment was observed and colocalized with the ER resident proteins Dpm1p and Kar2p (our unpublished results). The level of ER-like staining is higher when the cells reach stationary phase.
To determine which compartments the Gmh1p punctate staining pattern corresponded to, we performed double immunofluorescence experiments using several Golgi-localized markers. As shown inFigure 4B (top and middle panel), a significant colocalization was observed with the early-medial protein Emp47p and the _cis_-Golgi enzyme Anp1p. Some but not all of the structures containing Gmh1-HA also contain the C-terminally Myc-tagged Emp47p (Figure 4B, top panel). As shown in Figure 4B (middle panel), some spots decorated with Gmh1p correspond to Anp1p containing structures. Partial colocalization has often been seen for proteins that reside in the same region of the Golgi (Rayner and Munro, 1998;Wooding and Pelham, 1998). As localization at steady state in the Golgi results from very dynamic processes, such differences could reflect variations in the kinetics of delivery to and removal from compartments. There was clearly less obvious overlap of Gmh1p staining with CHS5-myc or Sec7p that have been shown to localized to late Golgi compartments (Figure 4B, bottom panel and our unpublished results;Franzusoff et al., 1991; Santos and Snyder, 2000).
Because endosomal/prevacuolar proteins also exhibit highly punctate staining patterns by indirect immunofluorescence microscopy, we next determined whether some of the punctate Gmh1p-decorated structures could also be present in endosomal structures. For this purpose, we examined Gmh1p immunolocalization in the class E _vp_s mutant vps27 Δ. Mutations in any of class E VPS genes lead to an exaggerated form of prevacuolar compartment that contains endocytosed proteins as well as Golgi proteins en route to the vacuole (Bryant_et al.,_ 1998). The Gmh1p staining pattern was unchanged in the vps27 mutant (our unpublished results), suggesting that a minority (if any) of Gmh1p resides in prevacuolar/endosomal compartments. Taken together our results indicate that Gmh1p preferentially locates to the early Golgi complex at steady state. Moreover, the same localization was observed for the human putative orthologue hGMH1b expressed in yeast (our unpublished results).
Gmh1p Cycles through the ER
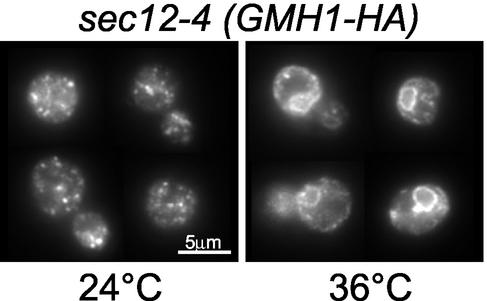
Golgi localization at steady state can be the result of a very dynamic process. For example, some Golgi membrane proteins such as Emp47p or Rer1p undergo retrograde transport from Golgi to ER (Schroder et al., 1995; Sato et al., 2001). To test whether Gmh1p could also follow a recycling pathway to the ER, we made use of the thermosensitive sec12 mutant. In_sec12_ cells, secretion is blocked before the budding of transport vesicles from the ER at nonpermissive temperature but retrograde transport from Golgi to ER still occurs under these conditions (Schroder et al., 1995; Lewis and Pelham, 1996). If Gmh1p localization results from a continuous shuttle between ER and Golgi, a specific block of forward transport should result in Gmh1p being trapped in the ER. During the incubation at nonpermissive temperature further protein synthesis was blocked by addition of cycloheximide. Cells were then fixed and stained for Gmh1p. A normal punctate staining is observed for Gmh1p at the permissive temperature (Figure 5). In contrast, at 36°C perinuclear staining typical of the yeast ER is seen for Gmh1p and Emp47p (Figure 5 and our unpublished results). This result demonstrates that Gmh1p recycles between Golgi and ER.
Figure 5.
Gmh1p cycles from the Golgi to the ER. The temperature-sensitive_sec12_-4 strain (RSY253) expressing_HA_ 3 -GMH1 from pRC8 was incubated at 25°C (permissive) or 36°C (nonpermissive) for 1 h after addition of cycloheximide (20 mg/ml). Cells were then processed for immunofluorescence using anti-HA antibodies.
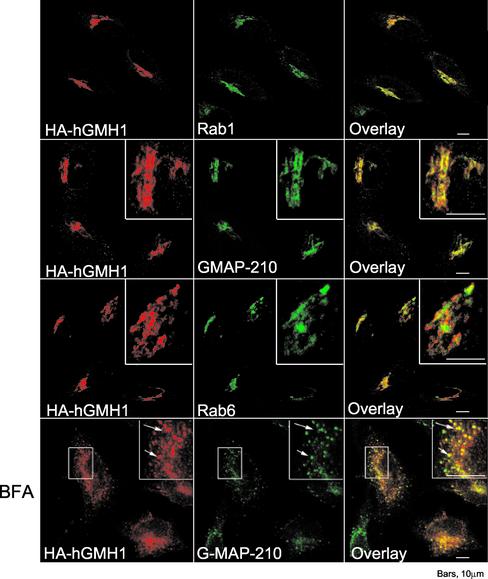
Human hGMH1 Localizes to the cis-Golgi in HeLa Cells
To determine whether the Golgi localization of Gmh1p is a common feature of the GMH1 family, we performed expression and immunolocalization of hGMH1 in HeLa cells. Figure 6 shows costaining of cells for hGMH1-HA and other well-characterized Golgi markers in human transfected cells. Fluorescence microscopy revealed for hGMH1 a strong compact juxtanuclear staining, a structure characteristic of the Golgi apparatus. In addition, we observed a number of punctate structures present in the Golgi region as well as more peripheral parts of the cytoplasm (Figure 6). To confirm that the compact region containing hGMH1-HA corresponded to a part of the Golgi apparatus, we compared the staining of HeLa cells with antibodies directed against the HA epitope and with a polyclonal serum directed against the_cis_-Golgi localized human GMAP-210 (Rios et al., 1994). There was almost complete overlap between GMAP-210 and hGMH1 (Figure 6). hGMH1 staining was also very similar to that of Rab1 (Figure 6), a small GTPase that localizes to early compartments of the secretory pathway (Saraste et al., 1995). In contrast, even if the overall localization of hGMH1-HA is similar to the one observed for Rab6 protein, after a close examination, Rab6, which is described to locate in medial/trans compartments in HeLa cells (Goud et al., 1990; Antony et al., 1992) could be readily distinguished from hGMH1 (Figure 6). It suggests that Rab6 and hGMH1 reside in adjacent compartments. It is not clear whether the small region of overlap represents partial colocalization of the two proteins or represents the failure to resolve two close but distinct fluorescence signals. Altogether, these observations led us to conclude that a very large fraction of hGMH1 is localized to the _cis_-Golgi in HeLa cells. BFA treatment of cells results in rapid loss of the Golgi as a distinct organelle due to the redistribution of the majority of cis/medial Golgi content and membrane into the ER (Lippincott-Schwartz et al., 1989,1990). By contrast, proteins of intermediate compartment (IC) are retained in what appears to be IC-derived elements upon treatment (Lippincott-Schwartz et al., 1990). After 2 h of BFA treatment, hGMH1 was not redistributed in a fine reticular pattern characteristic of ER labeling but was rather present in numerous small punctate structures scattered throughout the cytoplasm (Figure 6). These structures are also decorated by GMAP-210, which has been shown to be redistributed in IC-positive structures after BFA treatment (Rios et al., 1994). We conclude that hGMH1 is an early-Golgi protein that is induced to redistribute to the intermediate compartment by BFA. Some Golgi proteins that cycle through the ER/IC such as gp74 show the same type of redistribution upon BFA treatment (Alcalde et al., 1994).
Figure 6.
immunofluorescence localization of human hGMH1b in HeLa cells. HeLa cells transfected with HA-hGMH1b plasmid (pEM2) were fixed 20 h after transfection and processed for immunofluorescence. BFA (5 mg/ml) was added 2 h before fixation when indicated. Cells were double-labeled with a mAb against HA epitope and a polyclonal antibody against either Rab1, Rab6 or GMAP-210 as indicated. All pictures shown here represent stacks of four medial optical slices obtained by confocal microscopy. Arrows indicate identical position in each pair of image.
Gmh1p Physically Interacts with Gea1/2p
Allele-specific interactions usually indicate that the proteins encoded by these genes interact, although not necessary directly. To determine whether Gmh1p could be a physical partner of Gea1p, we undertook to analyze putative Gmh1p/Gea1p interactions using the two-hybrid system (Fields and Song, 1989). Several integral membrane proteins such as Yip1p and Yif1p have been successfully identified as prey or used as bait in such a system (Yang et al., 1998;Matern et al., 2000). We found that a Gal4 activating-domain fused to full-length Gmh1p could specifically activate transcription of reporter genes in the presence of full-length Gea1p fused to the Gal4 DNA-binding domain (Figure 7A and our unpublished results). Moreover, the study of different truncated versions of Gea1p fusions indicates that a C-terminal domain located downstream of the Sec7 domain of Gea1p (amino acid residues 749-1263) is sufficient for its binding to Gmh1p in the two-hybrid system (Figure 7A). We next wanted to determine whether this interaction was restricted to the yeast protein or was a common feature of Gmh1p family. For this purpose, we constructed a two-hybrid fusion with hGMH1. We established that hGMH1 could interact with the C-terminal portion of Gea1p[749–1408] in the two-hybrid system (Figure 7A), suggesting that structural requirements for the interaction with Gea1p are conserved in the human homolog hGMH1.
Figure 7.
Physical interaction between Gmh1p and Gea1/2p proteins. (A) Two-hybrid interactions of Gmh1 proteins with full-length and truncated forms of Gea1p. Y190 strain containing pAPDH15 (pACTII-GMH1) was transformed with plasmids containing different fragments of GEA1 fused to Gal4 DNA binding domain (corresponding respectively to pSC59, pSC58, pSC6b, and pSC4b). Alternatively, human Gmh1p (hGMH1) fused to Gal4 activation domain (pSC37b) was used as a bait. Numbers indicate first and last amino acid residues of Gea1p encompassed within the fusion protein. Coloration in presence of X-Gal assays for β-galactosidase activity. (B) Coimmunoprecipitation of Gmh1p and a C-terminal part of Gea1p. Coimmunoprecipitation experiments were performed from strain c13-ABYS86 expressing a myc-tagged version of Gea1p[749–1408] (from pSC64) and a wild-type or HA-tagged version of Gmh1p (from pAP68 or pRC11 respectively). One percent of total extracts and 10% or 35% of eluates were analyzed by immunoblotting either with anti-Gmh1p antibodies (top panel) or with A14 antimyc antibodies (lower panel). (C) Coimmunoprecipitation of Gmh1p and full-length Gea2p. Coimmunoprecipitation experiments were carried out with detergent-lysed cells extracts prepared from GEA2-myc SCY002 strain expressing Gmh1p or HA-Gmh1p (from pAP52 or pRC8, respectively). Eluates were analyzed as in B. (D) Gea2p and Gmh1p colocalize in yeast cells. Yeast cells expressing a chromosomal modified myc-tagged Gea2 protein (SCY002) were transformed with_HA_ 3 -GMH1 expressing plasmid (pRC11). Double immunofluorescence experiments were then performed with anti-Myc (A14) and anti-HA (HA-11) antibodies. Arrows indicate identical positions in each pair of image.
To corroborate the results of the two-hybrid analyses, which suggested specific physical interaction between Gmh1p and Gea1p, we prepared detergent-lysed cells and performed immunoprecipitation experiments. In these experiments, a myc-tagged version of Gea1p [749–1408] and HA-tagged version of Gmh1p were coexpressed in yeast cells. Cells expressing 9myc-Gea1p[749–1408] produced two major products detectable by immunoblot (Figure 7B). Two major bands are also observed for full-length Gea1p (unpublished data). As can be seen in Figure 7B, Gea1p[749–1408] was specifically eluted from Gmh1p-HA–loaded columns. To extend our observations, we performed immunoprecipitation experiments with a myc-tagged form of full-length Gea2p expressed at endogenous level. As shown in Figure 7C, the entire Gea2p-Myc is specifically immunoprecipitated with Gmh1p-HA. Altogether, these results strongly suggest that both Gea1p and Gea2p proteins can interact with Gmh1p and that this interaction involves the C-terminal part of the Gea proteins. Moreover, we show here that there is a substantial overlap of the Gea2p and Gmh1p signals in immunofluorescence experiments (Figure 7D), which is consistent with a physical interaction between the two proteins occurring in vivo.
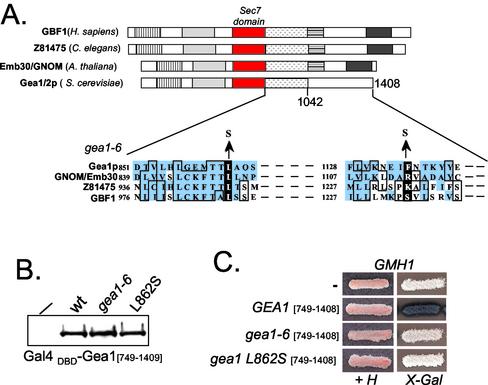
Gea1p–Gmh1p Interaction Is Abrogated in gea1-6_Mutant_
Strikingly, the two amino-acid substitutions present in the gea1-6 allele are located in the C-terminal region sufficient for interaction with Gmh1p (Figure 8A;Peyroche et al., 2001). This region includes a 300-amino-acid region located just downstream of the Sec7 domain and specifically shared by members of the Gea/GBF/GNOM family of ARF GEFs (Jackson and Casanova, 2000). One of the substitutions (that changes leucine 862 to a serine residue) is located in a highly conserved motif and affects one leucine residue shared by all the members of the Gea/GBF/GNOM family (Figure 8A). To address whether the interaction between Gmh1p and Gea1p is impaired in mutant forms of Gea1p, we performed two-hybrid analysis using different mutant forms of the C-terminal domain of Gea1p. We have verified by Western blot analysis that the level of expression of wild-type and mutant forms of Gea1p fusion proteins was equivalent under these experimental conditions (Figure 8B). Although the different hybrid proteins were expressed to similar levels, no binding activity was observed with the gea1-6 form of full-length Gea1p or its C-terminal domain (Figure 8C and our unpublished results), suggesting that one of the characteristic of the mutant form of GEA1 is to interact less efficiently with Gmh1p. Moreover, the single L862S substitution is sufficient to abrogate the interaction (Figure 8C). This observation is consistent with our previous results that indicate this interaction may be conserved in higher eukaryotes and therefore is likely to concern shared motifs in Gea/GBF/GNOM proteins. Because overexpression of Gmh1p can restore growth of gea1-6 cells at nonpermissive temperature, it is tempting to propose that the impairment of Gmh1p-Gea1p interaction is at least one of the growth-limiting factors in_gea1-6_ mutant cells.
Figure 8.
Physical interaction between Gmh1p and Gea1p is compromised in_gea1-6._ (A) The Gea/GBF/GNOM family. Representative members of this subfamily of Sec7-domain proteins are shown. The Sec7 domains are represented by red boxes. Regions showing significant level of sequence similarity are represented by drawn boxes. The position of the amino acid substitutions in the gea1-6 allele is indicated on the sequence alignments with three other ARF GEFs of the Gea/GBF/GNOM family. GenBank accession numbers for the sequences shown are Gea1p (Z49531); Gnom/Emb30 (U36433); C. elegans ARF GEF (Z81475); and GBF1 (AF068755). (B) Western-blot analysis of Gea1[749–1408] wild-type, gea1-6 or gea1 L862S expressed from pASΔ. Crude extracts of Y190 cells expressing the different fusions were loaded on a polyacrylamide gel and Gal4 (DBD) fusion proteins were revealed by immunoblot with anti-Gal4 (DBD) antibodies. (C) Gea1p-Gmh1p interaction is impaired with gea1 mutant forms. Y190 strain containing pAPDH15 (pACTII-GMH1) was transformed with Gea1[749–1408] wild-type, gea1-6 or gea1 L862S fused to Gal4 (DBD) in pASΔ. Transformants were streaked on selective medium (+H) and tested for expression of β-galactosidase on X-Gal containing plates.
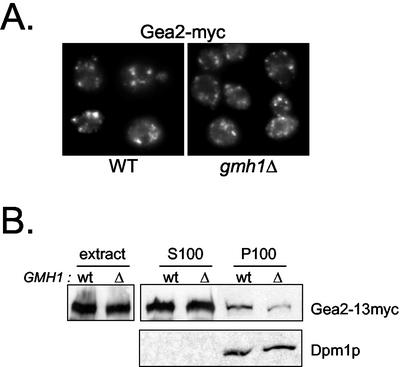
Effect of Loss of Gmh1p on the Distribution of Gea Proteins
If Gmh1p is a membrane receptor for Gea1/2p, we might expect that deletion of GMH1 would lead to mislocalization of Gea1p and/or Gea2p. We have tested for immuno-localization of Gea2p-myc in the absence of GMH1. We failed to detect massive changes in the membrane localization of Gea2p by immunofluorescence as shown in Figure 9A. We next tested for mislocalization of Gea2p in fractionation experiments. When we performed these experiments with gently lysed cells, most of Gea2p (>75%) was in the supernatant fractions. However, under these conditions, we observed a slight but reproducible reduction in the amount of Gea2p associated with the pellet fractions prepared from cells lacking Gmh1p (Figure 9B). It could reflect a more labile association with membranes of the pool of membrane-associated Gea2 protein. In summary, the majority of Gea protein remains associated with the membranes in the absence of Gmh1p, but at least a fraction of Gea proteins seems to be associated more weakly under these conditions.
Figure 9.
Membrane association of Gea2p in _gmh1_Δ cells. (A) Immunofluorescence of Gea2p in _gmh1_Δ yeast cells. A chromosomal modified myc-tagged Gea2 protein was introduced in wild-type cells (SCY002) or in _gmh1_Δ cells (APY130-2). Immunofluorescence experiments were then performed with anti-Myc (9E10). (B) Effect of GMH1 expression on membrane association of myc-tagged Gea2p by fractionation. Wild-type cells (SCY002) or _gmh1_Δ cells (APY130-2) were gently lysed (extracts) and then submitted to 100,000 × g centrifugation. Pellet (P100) and supernatant (S100) fractions were then analyzed by immunoblotting with antimyc antibodies (9E10) or anti-Dpm1p for control of loading.
DISCUSSION
Using a genetic screen to gain insight into the function of Gea ARF exchange factors, we isolated and characterized a new membrane protein Gmh1p, embedded in Golgi membranes in yeast. We cloned the human homologue, which we named hGMH1, and found that like its yeast counterpart, hGMH1 is localized to the Golgi in mammalian cells. We demonstrate that Gmh1p is a physical partner of the large ARF GEFs Gea1p and Gea2p. We propose that Gmh1p and its homologues act as Golgi-membrane partners of this family of ARF-GEFs.
Gmh1p Is a Novel Golgi Protein Conserved from Yeast to Human
We observe that Gmh1p and its human homolog hGMH1 are both localized to the Golgi apparatus in their respective organisms. Moreover, hGMH1 expressed in yeast also localizes to the yeast Golgi, indicating that the signals and/or the mechanisms that control the localization of Gmh1 proteins are conserved from yeast to human. Most of the conserved motifs in the Gmh1 family reside outside of the putative hydrophobic transmembrane domains, suggesting that the conservation of the overall topology is not the sole key of Gmh1p function and that the nonhydrophobic domains are important for the conserved function(s) of Gmh1p. We established that the N-terminal portion of Gmh1p faces the cytosol and the C-terminal segment faces the lumen. This orientation places the conserved motifs on the cytosolic face of the membrane, suggesting that conserved functions are related to cytosolic partners.
GMH1 Family and Protein Trafficking
The C. elegans GMH1 homolog is the product of the UNC-50 gene, which was identified in a screen for mutants resistant to an agonist of acetylcholine receptors, levamisole (Brenner, 1974;Lewis et al., 1980).UNC-50 appears to be required for cell-surface expression of assembled acetylcholine receptors (Lewis_et al.,_ 1987) but not for their synthesis, suggesting that UNC-50 promotes transport of these receptors to the plasma membrane (A. Gottschalk and W.R. Schafer, personal communication). A rat homolog of UNC-50 named UNCL has been cloned (Fitzgerald_et al.,_ 2000). In this study, the authors suggest that UNCL is localized to the ER and the inner nuclear membrane in SaOS-2 osteosarcoma cells. They showed that moderate expression of UNCL in_Xenopus_ oocyte or in COS cells can increase the number of active surface acetylcholine receptors. The data from UNC-50 and UNCL studies concerning the surface expression of nicotinic receptors are perfectly consistent with a role in protein trafficking for these proteins because membrane surface expression of oligomeric receptors requires functional endocytic and exocytic pathways.
GMH1 gene is dispensable for vegetative growth in yeast. However, in C. elegans, mutations in UNC-50 gene result in a phenotype of movement defects, indicating that the UNC-50 product is important for nervous system function. The ubiquitous expression observed for the rat homolog (Fitzgerald et al., 2000) and the existence of a yeast homolog favor a broader function not restricted to the nervous system. Moreover, genetic data in_C. elegans_ suggest that UNC-50 may have an essential function (Lewis et al., 1980). How can we explain the discrepancy between yeast and higher eukaryotes? One possibility is that another protein has related functions in yeast. Although, there is no obvious homolog of Gmh1p in the yeast genome, it is possible that a second protein shares a redundant function. Indeed, although the proteins Got1p and Sft2p display little direct sequence similarity, these two proteins share common global topology and are functional homologues (Conchon et al., 1999). Another possibility is that the function(s) assumed by Gmh1p is not essential in yeast, but is essential in higher eukaryotes. Indeed, whereas deletion of all members of p24 family of proteins in yeast has no detectable effect on growth (Springer_et al.,_ 2000), disruption of one member of the family resulted in early embryonic lethality in mice (Denzel et al., 2000). To seek clues to the function of GMH1, it would be interesting to isolate mutant yeast strains in which it is essential. Our preliminary results have led to the isolation of such a synthetic lethal mutation. These mutant cells display defects in protein trafficking (our unpublished results).
Gmh1p and Small GTPases
The open reading frame encoding Gmh1p has been isolated in two independent large scale two-hybrid screens as a putative interacting protein for the yeast small G proteins Arl3p and Ypt31p (Uetz_et al.,_ 2000; Ito_et al.,_ 2001). Arl3p is an ARF-like protein that has been implicated in ER-to-Golgi transport in yeast (Huang et al., 1999). Ypt31p belongs to the Ypt/Rab family and acts at multiple stages of transport (Benli et al., 1996;Jedd et al., 1997). Strikingly, genetic interactions have been observed between Ypt31/32p and ARF regulating factors (GEFs and GTPase-activating proteins) in yeast (Jones et al., 1999;Zhang et al., 2002). Gmh1p could thus participate in the cross-talk between ARF and Ypt activities. In the light of our present data, it was tempting to imagine that Gmh1p might interact with ARF proteins. However, we failed to display any direct interaction between Gmh1p and ARF proteins. We could not confirmed the published interactions between Gmh1p and Ypt31p or Arl3p in our two-hybrid system, but we observe a weak interaction between the Rho4 GTPase and Gmh1p (our unpublished data). Given the number of independent findings of Gmh1p interacting with different small G proteins, it remains an interesting possibility that it might interact with a GTPase.
Characterization of a Functional Domain of Gea1/2p
Unlike ARF1/2 or others gea1-6 suppressors we have identified, overexpression of GMH1 has no effect on gea1-4 cells growth. Importantly, the gea1-4 allele exhibits mutations in the Sec7 domain of Gea1p, whereas in gea1-6 the mutations are in the carboxy-terminal domain (Peyroche et al., 2001). All these genetic observations are in agreement with our biochemical data. Indeed, we show here that Gmh1p specifically interacts with the carboxy-terminal domain of Gea1/2p and fails to interact with the Sec7 domain (our unpublished observations). Gea1p and Gea2p are more than 1400 amino acid proteins, but, to date, only the 200 amino-acid Sec7 domain has been well characterized. One can imagine that one or more specific domains of the large ARF GEFs contain direct targeting information or is responsible for interaction with a partner in a hetero-oligomeric complex that contains such targeting information. Our data suggest that the C-terminal segment of Gea may help to localize the GEF by promoting contact with Gmh1p, which is anchored in the Golgi-membrane (but see below).
Gmh1p Is a Membrane Partner of Gea/GBF/GNOM Family Proteins
In yeast, there are three large ARF GEFs: Sec7p, Gea1p, and Gea2p. Can Gmh1p be a general partner for all these ARF GEFs? It is established that Sec7p and the pair Gea1/2p have nonredundant essential functions in yeast (Peyroche et al., 1996). They also show distinct localizations at steady state: whereas Sec7p is mainly associated with a late-Golgi compartment and colocalizes with Kex2p, a marker for the yeast TGN (Franzusoff et al., 1991), Gea1/2p are primarily early-Golgi proteins (Spang et al., 2001 and Figure 8). This difference is also observed in mammalian cells because BIG1 and BIG2 (Sec7p homologues) have been localized to the TGN, whereas GBF1 (a human Gea1/2p counterpart) is mainly associated with the _cis_-Golgi compartment (Mansour et al., 1999; Zhao et al., 2002). We observe that Gmh1p is an early-Golgi protein that colocalizes with Gea2p but not with Sec7p. This is also true for mammalian counterparts of Gmh1p and Gea1/2p. Indeed, we have established that hGMH1 is a_cis_-Golgi protein in human cells, and recent studies that GBF1 mostly colocalizes with _cis_-Golgi markers (Kawamoto et al., 2002; Zhao et al., 2002). These observations favor a preferential, if not specific, interaction of Gmh1p for Gea1/2p family proteins.
We have identified GMH1 because its overexpression rescues the thermosensitive growth defect of the gea1-6 mutant and have shown that Gmh1p interacts physically with Gea1/2p. Because the Gmh1p-Gea1p interaction is impaired in a gea1-6 mutant, we propose that overexpression of Gmh1p in gea1-6 cells overcomes the growth defect by helping this leaky interaction. These observations point to an involvement of Gmh1p in Gea/GNOM/GBF1 ARF-related functions. With regard to our results, different hypotheses can be proposed. Gmh1p could participate in the recruitment of Gea1/2p to Golgi membranes. In other words, Gmh1p could be a Golgi-specific receptor that facilitates specific docking of Gea proteins to the right cellular membrane. The existence of such a “receptor” has been proposed to achieve the correct localization of large ARF GEFs in the cell (Roth, 1999). The existence of a 90-kDa membrane protein that may be the receptor for Sec7p has been reported (Wolf et al., 1996) but has not been identified at the molecular level. There is no massive redistribution of Gea2p from membranes to cytosol in the absence of Gmh1p but at least a fraction of Gea proteins seems to be associated more weakly with membranes under these conditions. Hence, these results suggest that Gmh1p participates in the association of Gea proteins with membranes but show that Gmh1p is not absolutely required for this process. As mentioned above, these mitigated results could be explained by the existence of an alternative or complementary mode of localization for Gea proteins (i.e., interaction with a functionally redundant protein an/or with specific lipids).
We cannot exclude that the interaction with Gmh1p modulates Gea ARF GEF activity. Gmh1p might lead to a conformational change in the membrane-associated pool of Gea, making it more active, which in turn would lead to more efficient activation of ARF. This hypothesis is consistent with our observation that overexpression of GMH1 can rescue growth of cells with limited amount of Arfp. Even though we failed to show any direct interactions between ARF proteins and Gmh1p, we cannot rule out the possibility that Gmh1p interacts at the same time with ARF and Gea1/2p in facilitating ARF exchange on membranes. Another possibility is that Gmh1p is an effector of ARF that also interacts with one of its GEFs. There are at least two different large ARF GEFs families in the cells that localize to different compartments. An attractive hypothesis is that effector-GEF interactions may link activation of ARF at a particular location within the cell to activation of specific effectors, i.e., that GEFs may participate directly in effector selection as suggested by Pasqualato et al. (2001). Effector complexes containing a GEF have already been identified for Rab GTPases and are supposed to couple nucleotide exchange to effector recruitment (Horiuchi et al., 1997; Wurmser et al., 2000).
In summary, Gmh1p acts as a Golgi-membrane partner of some large ARF GEFs, and further studies will determine its precise role in ARF/ARF GEF function and more generally in membrane trafficking in eukaryotic cells.
Acknowledgments
We thank Sean Munro for antibodies against Anp1p, Michel Bornens for anti-GMAP-210 antiserum, Hugh Pelham for Emp47-myc plasmid, Randy Schekman for RSY253, and François Lacroute for yeast genomic library. We thank Armelle Lengronne for preliminary observations concerning retrograde defects of gea1-6 cells and Alexander Gottschalk for sharing results prior publication. S.C. was supported by a doctoral fellowship from the CEA. Part of the work was supported by a grant from the French Ministère de la Recherche (ACI Jeunes Chercheurs Grant 5168).
References
- Alcalde, J., Egea, G., and Sandoval, I.V. (1994). gp74 a membrane glycoprotein of the cis-Golgi network that cycles through the endoplasmic reticulum and intermediate compartment. J. Cell Biol. 124, 649-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony, C., Cibert, C., Geraud, G., Santa Maria, A., Maro, B., Mayau, V., and Goud, B. (1992). The small GTP-binding protein rab6p is distributed from medial Golgi to the trans-Golgi network as determined by a confocal microscopic approach. J. Cell Sci. 103, 785-796. [DOI] [PubMed] [Google Scholar]
- Benli, M., Doring, F., Robinson, D.G., Yang, X., and Gallwitz, D. (1996). Two GTPase isoforms, Ypt31p and Ypt32p, are essential for Golgi function in yeast. EMBO J. 15, 6460-6475. [PMC free article] [PubMed] [Google Scholar]
- Boehm, M., and Bonifacino, J.S. (2001). Adaptins: the final recount. Mol. Biol. Cell. 12, 2907-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner, S. (1974). The genetics of Caenorhabditis elegans. Genetics 77, 71-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, N.J., Piper, R.C., Gerrard, S.R., and Stevens, T.H. (1998). Traffic into the prevacuolar/endosomal compartment of Saccharomyces cerevisiae: a VPS45-dependent intracellular route and a VPS45-independent, endocytic route. Eur J. Cell Biol. 76, 43-52. [DOI] [PubMed] [Google Scholar]
- Busch, M., Mayer, U., and Jürgens, G. (1996). Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol. Gen. Genet. 250, 681-691. [DOI] [PubMed] [Google Scholar]
- Chardin, P., Paris, S., Antonny, B., Robineau, S., Bëraud-Dufour, S., Jackson, C.L., and Chabre, M. (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 384, 481-484. [DOI] [PubMed] [Google Scholar]
- Chavrier, P., and Goud, B. (1999). The role of ARF and Rab GTPases in membrane transport. Curr. Opin. Cell Biol. 11, 466-475. [DOI] [PubMed] [Google Scholar]
- Claude, A., Zhao, B.P., Kuziemsky, C.E., Dahan, S., Berger, S.J., Yan, J.P., Armold, A.D., Sullivan, E.M., and Melançon, P. (1999). GBF1. A novel Golgi-associated BFA-resistant guanine nucleotide exchange factor that displays specificity for ADP-ribosylation factor 5. J. Cell Biol. 146, 71-84. [PMC free article] [PubMed] [Google Scholar]
- Conchon, S., Cao, X., Barlowe, C., and Pelham, H.R. (1999). Got1p and Sft2p: membrane proteins involved in traffic to the Golgi complex. EMBO J. 18, 3934-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitz, S.B., Rambourg, A., Kepes, F., and Franzusoff, A. (2000). Sec7p directs the transitions required for yeast Golgi biogenesis. Traffic 1, 172-183. [DOI] [PubMed] [Google Scholar]
- Denzel, A., Otto, F., Girod, A., Pepperkok, R., Watson, R., Rosewell, I., Bergeron, J.J., Solari, R.C., and Owen, M.J. (2000). The p24 family member p23 is required for early embryonic development. Curr. Biol. 10, 55-58. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., Finazzi, D., and Klausner, R.D. (1992). Brefeldin A inhibits Golgi membrane-catalysed exchange of guanine nucleotide onto ARF protein. Nature 360, 350-352. [DOI] [PubMed] [Google Scholar]
- Donaldson, J.G., and Jackson, C.L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475-482. [DOI] [PubMed] [Google Scholar]
- Fields, S., and Song, O. (1989). A novel genetic system to detect protein-protein interactions. Nature 340, 245-246. [DOI] [PubMed] [Google Scholar]
- Fitzgerald, J., Kennedy, D., Viseshakul, N., Cohen, B.N., Mattick, J., Bateman, J.F., and Forsayeth, J.R. (2000). UNCL, the mammalian homologue of UNC-50, is an inner nuclear membrane RNA-binding protein. Brain Res. 877, 110-123. [DOI] [PubMed] [Google Scholar]
- Franzusoff, A., Redding, K., Crosby, J., Fuller, R.S., and Schekman, R. (1991). Localization of components involved in protein transport and processing through the yeast Golgi apparatus. J. Cell Biol. 112, 27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor, E.C., Chen, C.Y., Emr, S.D., and Graham, T.R. (1998). ARF is required for maintenance of yeast Golgi and endosome structure and function. Mol. Biol. Cell. 9, 653-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner, N. et al. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112, 219-230. [DOI] [PubMed] [Google Scholar]
- Goud, B., Zahraoui, A., Tavitian, A., and Saraste, J. (1990). Small GTP-binding protein associated with Golgi cisternae. Nature 345, 553-556. [DOI] [PubMed] [Google Scholar]
- Grebe, M., Gadea, J., Steinmann, T., Kientz, M., Rahfeld, J.U., Salchert, K., Koncz, C., and Jurgens, G. (2000). A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell 12, 343-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, J.B., and Rothman, J.E. (1992). Inhibition by brefeldin A of a Golgi membrane enzyme that catalyses exchange of guanine nucleotide bound to ARF. Nature 360, 352-354. [DOI] [PubMed] [Google Scholar]
- Horiuchi, H. et al. (1997). A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149-1159. [DOI] [PubMed] [Google Scholar]
- Huang, C.F., Buu, L.M., Yu, W.L., and Lee, F.J. (1999). Characterization of a novel ADP-ribosylation factor-like protein (yARL3) in Saccharomyces cerevisiae. J. Biol. Chem. 274, 3819-3827. [DOI] [PubMed] [Google Scholar]
- Ito, T., Chiba, T., Ozawa, R., Yoshida, M., Hattori, M., and Sakaki, Y. (2001). A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA 98, 4569-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, C.L., and Casanova, J.E. (2000). Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 10, 60-67. [DOI] [PubMed] [Google Scholar]
- Jedd, G., Mulholland, J., and Segev, N. (1997). Two new Ypt GTPases are required for exit from the yeast trans-Golgi compartment. J. Cell Biol. 137, 563-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, S., Jedd, G., Kahn, R.A., Franzusoff, A., Bartolini, F., and Segev, N. (1999). Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics 152, 1543-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan, M., Schallhorn, A., and Wurm, F.M. (1996). Transfecting mammalian cells: optimization of critical parameters affecting calcium-phosphate precipitate formation. Nucleic Acids Res. 24, 596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann, J., and Munro, S. (1998). Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with alpha-1,6-mannosyltransferase activity. EMBO J. 17, 423-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto, K., Yoshida, Y., Tamaki, H., Torii, S., Shinotsuka, C., Yamashina, S., and Nakayama, K. (2002). GBF1, a guanine nucleotide exchange factor for ADP-ribosylation factors, is localized to the cis-Golgi and involved in membrane association of the COPI coat. Traffic 3, 483-495. [DOI] [PubMed] [Google Scholar]
- Letourneur, F., Gaynor, E.C., Hennecke, S., Demolliere, C., Duden, R., Emr, S.D., Riezman, H., and Cosson, P. (1994). Coatomer is essential for retrieval of dilysine-tagged proteins to the endoplasmic reticulum. Cell 79, 1199-1207. [DOI] [PubMed] [Google Scholar]
- Lewis, J.A., Elmer, J.S., Skimming, J., McLafferty, S., Fleming, J., and McGee, T. (1987). Cholinergic receptor mutants of the nematode Caenorhabditis elegans. J. Neurosci. 7, 3059-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, J.A., Wu, C.H., Berg, H., and Levine, J.H. (1980). The genetics of levamisole resistance in the nematode_Caenorhabditis elegans._ Genetics 95, 905-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, M.J., and Pelham, H.R. (1996). SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell 85, 205-215. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Donaldson, J.G., Schweizer, A., Berger, E.G., Hauri, H.P., Yuan, L.C., and Klausner, R.D. (1990). Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell 60, 821-836. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Yuan, L.C., Bonifacino, J.S., and Klausner, R.D. (1989). Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell 56, 801-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour, S.J., Skaug, J., Zhao, X.H., Giordano, J., Scherer, S.W., and Melancon, P. (1999). p200 ARF-GEP1: a Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc. Natl. Acad. Sci. USA 96, 7968-7973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern, H., Yang, X., Andrulis, E., Sternglanz, R., Trepte, H.H., and Gallwitz, D. (2000). A novel Golgi membrane protein is part of a GTPase-binding protein complex involved in vesicle targeting. EMBO J. 19, 4485-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro, S. (1991). Sequences within and adjacent to the transmembrane segment of alpha-2,6-sialyltransferase specify Golgi retention. EMBO J. 10, 3577-3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, H., Kagan, J.C., Zhu, X., Kahn, R.A., and Roy, C.R. (2002). A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295, 679-682. [DOI] [PubMed] [Google Scholar]
- Pasqualato, S., Menetrey, J., Franco, M., and Cherfils, J. (2001). The structural GDP/GTP cycle of human Arf6. EMBO Rep. 2, 234-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche, A., Antonny, B., Robineau, S., Acker, J., Cherfils, J., and Jackson, C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3, 275-285. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Courbeyrette, R., Rambourg, A., and Jackson, C.L. (2001). The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J. Cell Sci. 114, 2241-2253. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., and Jackson, C.L. (2001). Functional analysis of ADP-ribosylation factor (ARF) guanine nucleotide exchange factors Gea1p and Gea2p in yeast. Methods Enzymol. 329, 290-300. [DOI] [PubMed] [Google Scholar]
- Peyroche, A., Paris, S., and Jackson, C.L. (1996). Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature 384, 479-481. [DOI] [PubMed] [Google Scholar]
- Rayner, J.C., and Munro, S. (1998). Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 273, 26836-26843. [DOI] [PubMed] [Google Scholar]
- Rios, R.M., Tassin, A.M., Celati, C., Antony, C., Boissier, M.C., Homberg, J.C., and Bornens, M. (1994). A peripheral protein associated with the cis-Golgi network redistributes in the intermediate compartment upon brefeldin A treatment. J. Cell Biol. 125, 997-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth, M.G. (1999). Snapshots of ARF1: implications for mechanisms of activation and inactivation. Cell 97, 149-152. [DOI] [PubMed] [Google Scholar]
- Santos, B., and Snyder, M. (2000). Sbe2p and sbe22p, two homologous Golgi proteins involved in yeast cell wall formation. Mol. Biol. Cell 11, 435-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraste, J., Lahtinen, U., and Goud, B. (1995). Localization of the small GTP-binding protein rab1p to early compartments of the secretory pathway. J. Cell Sci. 108, 1541-1552. [DOI] [PubMed] [Google Scholar]
- Sato, K., Sato, M., and Nakano, A. (2001). Rer1p, a retrieval receptor for endoplasmic reticulum membrane proteins, is dynamically localized to the Golgi apparatus by coatomer. J. Cell Biol. 152, 935-944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder, S., Schimmoller, F., Singer-Kruger, B., and Riezman, H. (1995). The Golgi-localization of yeast Emp47p depends on its dilysine motif but is not affected by the ret1–1 mutation in alpha-COP. J. Cell Biol. 131, 895-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Shevell, D.E., Leu, W.M., Gillmor, C.S., Xia, G., Feldmann, K.A., and Chua, N.H. (1994). EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell 77, 1051-1062. [DOI] [PubMed] [Google Scholar]
- Spang, A., Herrmann, J.M., Hamamoto, S., and Schekman, R. (2001). The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol. Biol. Cell 12, 1035-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer, S., Chen, E., Duden, R., Marzioch, M., Rowley, A., Hamamoto, S., Merchant, S., and Schekman, R. (2000). The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97, 4034-4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P. et al. (2000). A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623-627. [DOI] [PubMed] [Google Scholar]
- Wolf, J.R., Lasher, R.S., and Franzusoff, A. (1996). The putative membrane anchor protein for yeast Sec7p recruitment. Biochem. Biophys. Res. Commun. 224, 126-133. [DOI] [PubMed] [Google Scholar]
- Wooding, S., and Pelham, H.R. (1998). The dynamics of golgi protein traffic visualized in living yeast cells. Mol. Biol. Cell 9, 2667-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser, A.E., Sato, T.K., and Emr, S.D. (2000). New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151, 551-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji, R., Adamik, R., Takeda, K., Togawa, A., Pacheco-Rodriguez, G., Ferrans, V.J., Moss, J., and Vaughan, M. (2000). Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc. Natl. Acad. Sci. USA 97, 2567-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Matern, H.T., and Gallwitz, D. (1998). Specific binding to a novel and essential Golgi membrane protein (Yip1p) functionally links the transport GTPases Ypt1p and Ypt31p. EMBO J. 17, 4954-4963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C.J., Bowzard, J.B., Greene, M., Anido, A., Stearns, K., and Kahn, R.A. (2002). Genetic interactions link ARF1, YPT31/32 and TRS130. Yeast 19, 1075-1086. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Lasell, T.K., and Melanon, P. (2002). Localization of Large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi Compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell 13, 119-133. [DOI] [PMC free article] [PubMed] [Google Scholar]