Gene copy number variation spanning 60 million years of human and primate evolution (original) (raw)
Abstract
Given the evolutionary importance of gene duplication to the emergence of species-specific traits, we have extended the application of cDNA array-based comparative genomic hybridization (aCGH) to survey gene duplications and losses genome-wide across 10 primate species, including human. Using human cDNA arrays that contained 41,126 cDNAs, corresponding to 24,473 unique human genes, we identified 4159 genes that likely represent most of the major lineage-specific gene copy number gains and losses that have occurred in these species over the past 60 million years. We analyzed 1,233,780 gene-to-gene data points and found that gene gains typically outnumbered losses (ratio of gains/losses = 2.34) and these frequently cluster in complex and dynamic genomic regions that are likely to serve as gene nurseries. Almost one-third of all human genes (6696) exhibit an aCGH- predicted change in copy number in one or more of these species, and within-species gene amplification is also evident. Many of the genes identified here are likely to be important to lineage-specific traits including, for example, human-specific duplications of the AQP7 gene, which represent intriguing candidates to underlie the key physiological adaptations in thermoregulation and energy utilization that permitted human endurance running.
The primate order is thought to have first appeared ∼90 million years ago (Mya) and since that time has undergone dramatic evolutionary expansion, with perhaps as many as 300 different primate species estimated now to exist (Groves 2001). The primary genomic mechanisms thought to underlie this proliferation, as with other species, are gene duplication, single nucleotide substitution, and genome rearrangement. In addition to increasing gene expression via a dosage effect, gene duplication may also produce altered regulation of expression or altered function by mutation in the duplicate copy. From the classic work of Ohno (1970) to the present day (Hurles 2004), gene duplication is thought to be the central mechanism driving evolutionary change, a view that is being assessed more comprehensively by the wealth of comparative genomic data that are becoming available.
Foremost among these comparative genomic efforts is the sequencing of primate genomes, a valuable new resource that is allowing primate genomic evolution to be viewed in unprecedented breadth and detail (Sikela 2006). Human (finished), chimpanzee, and macaque (drafts) sequences have been reported (Human Genome Sequencing Consortium 2004; Chimpanzee Sequencing and Analysis Consortium 2005; Macaque Genome Sequencing and Analysis Consortium 2007), and draft genome sequences for several other primates are imminent. While these sequences are of great scientific benefit, draft sequences are known to have difficulty in correctly assembling highly similar sequences such as those that have arisen by recent duplication events (Cheung et al. 2003; She et al. 2004). This limitation is magnified by non-assembly-based reports that recent (<40 Mya) segmental duplications are abundant in primate genomes (Cheng et al. 2005), accounting for ∼5% of the human genome (Bailey et al. 2002), and raises the probability that conventional draft sequence assemblies can be expected to consistently underestimate the actual duplication repertoire of genomes.
A non-sequence-based method for studying copy number variation that avoids this limitation of draft sequencing is array-based comparative genomic hybridization (aCGH), which was initially used to detect DNA copy number changes between normal and disease (e.g., cancer) states (Pinkel et al. 1998; Pollack et al. 1999). More recently it has been used to look at normal variations both within (Iafrate et al. 2004; Sebat et al. 2004) and between species (Fortna et al. 2004; Goidts et al. 2006; Wilson et al. 2006) and has become one of the most widely used strategies for genome-wide studies of copy number variation. We previously reported the first genome-wide (and gene-based) cross-species aCGH study of human and great ape lineages, using full-insert cDNA arrays to detect lineage-specific (LS) gene copy number variations (Fortna et al. 2004). Subsequently, several other interspecies copy number variation analyses have been employed using various methods, including BAC aCGH (Goidts et al. 2006; Perry et al. 2006; Wilson et al. 2006), FISH (She et al. 2006), and computational analyses (Cheng et al. 2005; She et al. 2006; Macaque Genome Sequencing and Analysis Consortium 2007). By employing full cDNA inserts as targets, cDNA aCGH provides exclusively gene-based data and, because it uses sequences that are among the most highly conserved in the genome (i.e., gene-coding regions), is likely to more effectively minimize problems related to interspecies sequence divergence compared to other aCGH platforms. Results of our previous cross-species cDNA aCGH study (Fortna et al. 2004) supported this view, demonstrating that sequence divergence did not significantly interfere with aCGH accuracy even when the species being compared had diverged as much as 12–16 Mya (i.e., human and orangutan).
Based on the success of this earlier study, we have now applied cDNA aCGH to additional primate lineages that are even more evolutionarily distant from human. The 10 species compared (with estimated times at which they shared a last common ancestor, LCA, with human) are human, bonobo (5 Mya), chimpanzee (5 Mya), gorilla (7 Mya), orangutan (13 Mya), gibbon (18 Mya), macaque (24 Mya), baboon (24 Mya), marmoset (39 Mya), and lemur (60 Mya) (Jobling et al. 2004). cDNA aCGH was carried out as previously described (Fortna et al. 2004) using microarrays containing 41,126 human cDNAs, corresponding to 24,473 genes. Each cDNA aCGH experiment involved a pairwise comparison of two genomic DNAs, a reference sample (always human), and a test sample (one of the indicated primate species). Because the reference DNA is the same for all comparisons, cDNA aCGH data sets from every species surveyed could be interrelated to one another and an evolutionary portrait of gene copy number gain and loss for >24,000 human genes could be generated spanning much of human and primate evolutionary history.
Results and Discussion
Cross-species comparisons used at least three individuals from each of 10 primate species, and overall 1,233,780 gene-to-gene data points were analyzed. Use of the Treeview program (http://rana.lbl.gov/EisenSoftware.htm) permitted each gene (cDNA) to be visualized in the order in which it occurs in the genome, allowing copy number changes involving either single genes or blocks of multiple contiguous genes to be readily identified (Fig. 1). Using aCGH selection criteria described previously (Fortna et al. 2004), 4159 genes were predicted by cDNA aCGH to exhibit LS changes in copy number for these species (Fig. 2; Supplemental Table S1): 84 in human, 79 shared between Pan lineages (bonobo and chimp), 102 in gorilla, 117 in orangutan, 549 in gibbon, 369 in Old World monkeys (macaque and baboon), 543 in marmoset, and 1209 (increases) in lemur. Increases and decreases for the individual lineages of bonobo, chimp, macaque, and baboon were 21/2, 10/1, 48/37, and 78/91, respectively. We also identified a striking number of copy number increases (23) found among the African great apes (bonobo, chimp, and gorilla together) that were absent in human, orangutan, and other primates, suggesting that either the same genes increased independently among these three ape species (Fortna et al. 2004), or that additional gene copies were produced in the ancestor to humans and African great apes, and these were subsequently deleted in humans (Cheng et al. 2005). Overall, gene copy number increases markedly outnumbered decreases (1180/503), except for lemur (1209 increases vs. 3530 decreases) and, to a much lesser extent, baboon (79/91). This result suggests that sequence divergence, as reflected by a species exhibiting a disproportionate increase in the number of aCGH-predicted genes showing LS copy number losses, may not appreciably influence cDNA aCGH signals until divergence times increase from 39 million years (Myr) to 60 Myr. Because it is likely that sequence divergence may be responsible for a substantial fraction of the aCGH-predicted lemur decreases, lemur-specific decreases were omitted from any copy number calculations.
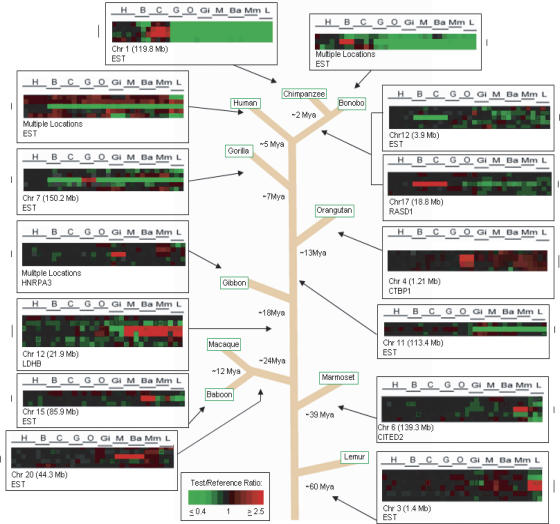
Figure 1.
Lineage-specific gene copy number changes among primate species. Examples of lineage-specific (LS) gene copy number changes across 10 primate species studied, as well as the divergence times in millions of years (Myr), from a last common ancestor with the human lineage, for each species: 5 Myr for chimpanzee, 5 Myr for bonobo, 2 Myr for chimp/bonobo split, 7 Myr for gorilla, 13 Myr for orangutan, 18 Myr for gibbon, 24 Myr for OWM, 39 Myr for NWM, and 60 Myr for lemurs (Jobling et al. 2004). Each vertical column represents data from one cDNA aCGH microarray experiment; the horizontal line represents data from one cDNA clone on the microarray, each of which is ordered according to human genome position. Arrows indicate to which primate lineage the LS change belongs. The estimated time frame of occurrence of LS changes is predicted using parsimony.
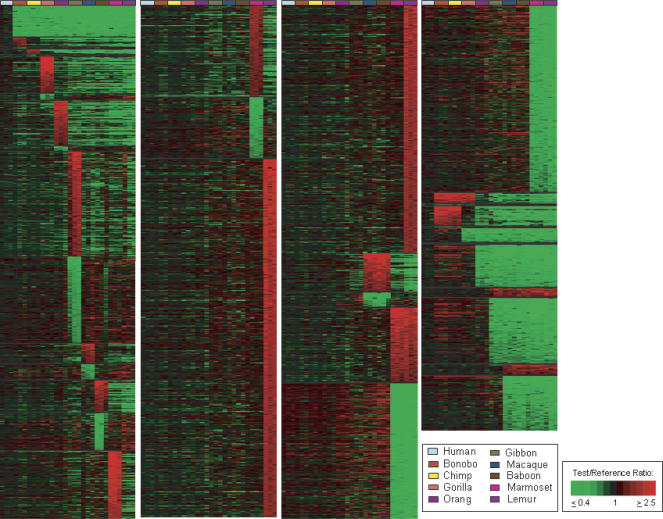
Figure 2.
Treeview image of genes showing lineage-specific copy number changes among 10 primate species. Treeview image of 7318 genes giving LS aCGH signatures are shown for each of 10 lineages, including human (blue-gray), bonobo (rust), chimp (yellow), gorilla (orange), orangutan (purple), gibbon (green), macaque (blue), baboon (brown), marmoset (magenta), and lemur (light purple), as well as Old World Monkeys (OWM), marmoset and lemur, African Great Apes, Pan lineage (bonobo and chimp together), and combined aCGH-predicted changes relative to the remaining extended primates for the following groups: human and Pan lineage, human and African great apes, human and great apes, and human and all apes (great and lesser). The LS signals are grouped according to lineage and within each lineage are ordered, highest to lowest, according to the log2 fluorescence ratio of the signal intensity of test sample to reference sample. Colors are displayed using a pseudocolor scale as shown. The green signals indicate LS decreases with respect to human, and the red signals indicate LS increases with respect to human.
A comparison of the list of LS genes for humans and great apes by Fortna et al. (2004) and the LS gene list presented here that includes five more distant primate lineages found that fewer human LS genes (84 vs. 134) were found in the larger primate study. Compared to the Fortna et al. HLS list, 31.3% of cDNAs are not found on the current HLS list because of technical factors (e.g., absent signals or reduced signal intensity). Perhaps most interestingly, a small but not insignificant subset of the “inconsistent” genes (17%) may reflect real biological effects, where the copy number changes were LS when human and great ape lineages were compared, but not LS once additional primate lineages were included.
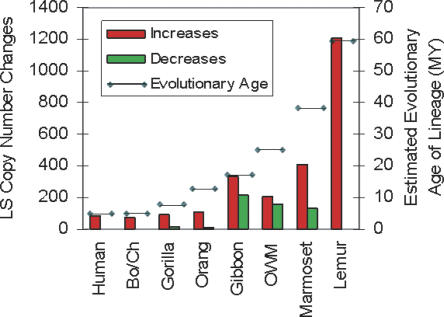
In order to gain insight into the relative rate of gene duplication and loss in each lineage, the age of each lineage was compared with the number of genes showing LS copy number changes. A generally consistent correlation (_r_2 = 0.93803 excluding gibbon) was found between the number of genes showing LS duplications for each species and their evolutionary age except for gibbon (Fig. 3), where a higher rate of gene copy number increase was found. This deviation may be, at least in part, related to the higher prevalence of chromosomal rearrangements in gibbons compared to the other primates studied (Jauch et al. 1992).
Figure 3.
LS increases and decreases and corresponding evolutionary age of each lineage. The total number of LS increases and decreases is shown as red and green bars, representing copy number gains and losses, respectively. The number of increases/decreases for each lineage is as follows: human 84/0, Pan lineage 75/4, gorilla 88/14, orangutan 107/10, gibbon 336/213, OWM (baboon and macaque combined) 211/158, marmoset 408/135, and lemur 1209/3530.The evolutionary age of each lineage in millions of years (depicted by the horizontal line) refers to the approximate divergence times from a last common ancestor with the human lineage and is the same as described in Figure 1.
In addition, gene copy number changes were also detected that were shared among related species, and likely due to gene duplications that occurred in the last common ancestor for each group. Among these were 27 genes predicted by aCGH to have elevated copy number in the Homo and Pan lineages relative to all others (Fig. 2; Supplemental Table S1). These expansions would be predicted by parsimony to occur 5–7 Mya, after divergence of the gorilla lineage but before the Homo and Pan lineages split. Also, 80 genes were predicted to have increased in copy number in human and African great apes and likely represent expansions that, by the same rationale, can be estimated to have occurred 7–13 Mya, during a time when humans and African great apes shared a common ancestor. Similarly, 124 genes were identified that were predicted to have increased in humans and the great apes relative to the other primates tested, events that would be expected to have occurred 13–18 Mya. Finally, 105 genes showed elevations in copy number in human and ape species relative to monkeys and prosimians, events that, by parsimony, can be estimated to have occurred 18–24 Mya.
The overall frequency of gene copy number variation between these lineages was also assessed by identifying all genes that showed a copy number change between humans and one or more of the other tested lineages. Using this strategy, 27.4% of cDNAs on the array, corresponding to 6696 different genes, were identified. Because aCGH has difficulty detecting small copy number changes in large gene families with closely related members, and because the criteria used for scoring positive copy number changes is conservative (i.e., threshold of >0.5), it is likely that this is a considerable underestimate of the actual gene copy number variation frequency between these lineages (Supplemental Table S2).
Accuracy of predicted copy number changes in primate lineages
As mentioned above, the contribution of sequence divergence to cross-species aCGH signals can be expected to increase with increasing evolutionary distance of the species being compared and, as a result, can pose potential challenges in identifying bona fide copy number changes. Because human cDNA arrays are used for all experiments, sequence divergence will tend to produce lower hybridization signals for the non-human primate (test) DNA compared to the human (reference) DNA. This feature can be expected to result in an artifactual inflation of the number of genes that show lower copy numbers in non-human primates (relative to human) and an underestimate of the number of genes that show elevated copy number in the non-human primates. However, the observation that copy number gains generally outnumbered losses for all species except the most evolutionarily distant, that is, lemur (Fig. 3; Supplemental Table S1), indicates that sequence divergence did not appear to significantly contribute to copy number decrease estimates (except for lemur), and suggests that gene duplication has had a much more dramatic impact on primate genome evolution than has gene copy number reduction. One of the most significant components of the strategy used here is its ability to reliably identify copy number gains in non-human species, and particularly in those that are evolutionarily distant from humans. The underlying rationale is that aCGH-predicted copy number increases in non-human primates must reflect gene copy number gains in those species that are of sufficient magnitude to overcome any reduction in hybridization signal due to interspecies sequence divergence. This feature makes it likely that the gene copy number increases reported here are accurate and robust and that gene copy number expansions in even more distant species may be identifiable by cDNA aCGH.
It should be noted, however, that the functional status of aCGH-predicted gene copies cannot be established by aCGH and will require additional studies to determine which gene copies are transcriptionally and functionally active and which are not. It is also important to recognize that, in principal, cDNA arrays have the potential to detect gene duplications that are part of segmental duplications and also those that occur via retroposed RNA copies, often producing processed pseudogenes. There are two reasons that suggest that the majority of copy number expansions reported here are not due to retroposed copies: (1) In our previous cDNA aCGH study of humans and great apes (Fortna et al. 2004), we found that the great majority (80%) of human sequences we predicted to be duplicated specifically in humans map to human segmental duplications predicted by Bailey et al. (2002). (2) Subsequently, a computational comparison of segmental duplication differences between human and chimpanzee genomes (Cheng et al. 2005) reported that 78% of the cDNAs we found that had cDNA aCGH-predicted changes in copy number between human and chimp were also detected by their analysis of segmental duplications in these species. Retroposed gene copies appear to reinsert in relatively random genomic locations, and the fact that the majority of our predicted gene copies do not appear to be randomly inserted but, rather, are preferentially associated with segmental duplications suggests that the majority of the duplication events we are detecting are bona fide gene duplications rather than processed pseudogenes.
To independently verify aCGH predictions, copy number gains and losses for a subset of genes—CA1 (Fig. 4); TERF1, DHFR, SEC13, AQP7 (Fig. 5); ALDH1B1, BMI1, SV2B, CBFB, KRT8, CDC42, and _GALNT1_—were calculated by quantitative-Real Time PCR (Q-PCR). Using a correlation coefficient value (_r_2) of >0.75 as a measure of confirmation between aCGH and Q-PCR results, nine of 12 genes (75%) showed Q-PCR values consistent with aCGH-predictions (Supplemental Table S3). Typically, values for all three genes that showed a correlation coefficient <0.75, (ALDH1B1, SV2B, and CBFB) could be raised above the 0.75 threshold by removal of one or two outlying data points, consistent with the possibility that small sequence variations in these three genes among different primate species may be influencing the Q-PCR results.
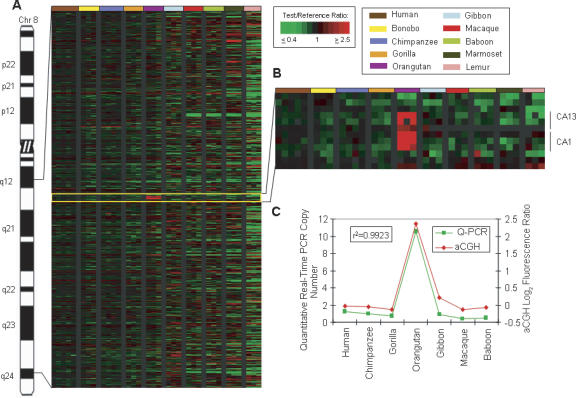
Figure 4.
Orangutan-specific amplification of carbonic anhydrase (CA) genes. (A) Human chromosome 8 is shown with a Treeview image corresponding to 8q12.1–q24.1. The Treeview image depicts the aCGH log2 fluorescence ratio in pseudocolor as shown, with green and red signals indicative of a copy number decrease and increase, respectively, relative to human. Each cDNA is ordered according to human genome position. All individuals from one species are color-coded as shown. (B) Enlarged Treeview image of an orangutan LS increase involving a block of six contiguous amplified cDNA signals, including three cDNAs corresponding to the carbonic anhydrase 13 (CA13) gene and three cDNAs correlating to the carbonic anhydrase 1 (CA1) gene. (C) The graph shows the aCGH log2 fluorescence ratio (red) plotted against the Q-PCR values (green) for the CA genes for 10 primate species. The correlation coefficient for the two data sets is _r_2 = 0.9923.
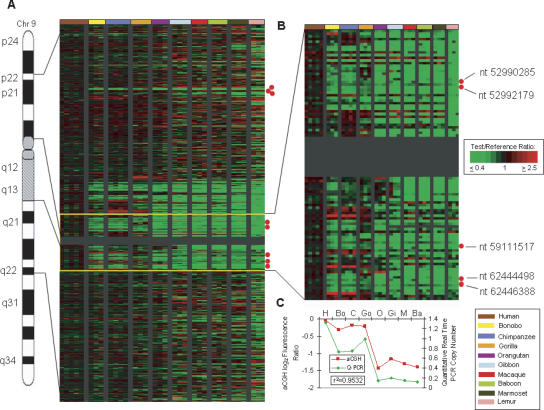
Figure 5.
Human LS amplification of the aquaporin 7 (AQP7) gene relative to other primates. (A) Human chromosome 9 is shown with the Treeview image corresponding to 9p22–9q22. The Treeview image depicts the aCGH log2 fluorescence ratio in pseudocolor as shown, with green and red signals indicative of a copy number decrease and increase, respectively, relative to human. Each cDNA is ordered according to human genome position. All individuals from one species are color-coded as shown. The red dots signify locations of _AQP7_-related cDNAs. (B) Enlarged Treeview image for the region adjacent to the C-band that includes five copies of AQP7. (C) The graph shows the aCGH log2 fluorescence ratio (red) plotted against the Q-PCR values (green) for the AQP7 gene for 10 primate species. The correlation coefficient for the two data sets is _r_2 = 0.9532.
Genome sequence assembly comparisons
Available genome assemblies provide independently generated data sets that can be used for comparison with aCGH predictions. Using sequences from LS data sets as BLAT queries (Kent 2002) against the respective genome assemblies, 84.5% of aCGH-based human LS predictions, 81.2% of chimp LS predictions, and 33.5% of rhesus LS predictions were consistent with the most recent available human (hg18), chimp (PanTro2), and rhesus (rheMac2) genome assemblies, respectively (Supplemental Table S4). The lower value obtained here for macaque may be due to assembly differences, polymorphism, and/or the possibility that a higher proportion of genes showing LS amplification in macaque fall into remaining sequence gaps. These results indicate that aCGH may provide a valuable complement to both current and future genome sequencing efforts that must rely on the computational assembly of shorter sequence reads and, as a result, are prone to misassembly of recently duplicated sequences.
Gene nurseries
Certain regions of the human genome are known to be extremely dynamic, a property that can facilitate rapid evolutionary innovation but can also predispose to genetic disease (Stankiewicz and Lupski 2002). It has been reported that such dynamic regions occur preferentially in or near pericentromeric regions in humans and subtelomeric regions in great apes (Fortna et al. 2004; Cheng et al. 2005). In agreement with this, we found an enrichment of genes predicted to exhibit human LS copy number increases, relative to all other non-human primate species tested, in pericentromeric regions (Supplemental Table S5). This finding supports the view that there has been a genome-wide expansion of pericentromeric gene duplications specifically in the human lineage, making these regions prime candidates to be human gene nurseries (Nahon 2003).
Among the most prominent human-specific cytogenetic features known are bands of constitutive heterochromatin, also called C-bands, located on four human chromosomes: 1q12, 9q12, 16q11.2, and Yq11.23. C-bands at these positions are absent in other primates, and it is noteworthy that regions adjacent to each of these exhibit high concentrations of human LS gene copy number increases in the genome (Supplemental Table S5). Indeed, regions at 1q21 and 9q13 that are adjacent to C-bands contain the two largest blocks of human LS gene increases in the genome. This correlation suggests that C-bands may be important facilitators of gene duplication in humans and that the adjacent regions are likely to be among the most active gene nurseries in the human genome. In contrast, genes showing chimp LS, Pan LS, gorilla LS, and African great ape LS duplications often map to subtelomeric regions, and in macaque LS duplications are often found near gaps, centromeres, and telomeres (Macaque Genome Sequencing and Analysis Consortium 2007).
Additional regions that also appear to be hotspots for gene duplication are the sites of lineage-specific chromosomal rearrangements. Among the most evolutionarily dynamic of these is the chromosome 2 (chr 2) fusion region, the site at which two ancestral ape chromosomes fused to produce human chr 2 (IJdo et al. 1991). Only the human genome exhibits this fusion on chr 2, and some of the most extreme lineage-specific copy number expansions we detect appear to have occurred in this region. For example, the largest gene copy number expansion in the Pan lineage (chimp and bonobo) compared to human was found at the chr 2 fusion region and includes copies of the PGM5 gene (Cheng et al. 2005), which also maps to the pericentric region of chr 9 and is involved in synthesis and breakdown of glucose (Edwards et al. 1995). Based on the similar average aCGH values for the PGM5 increase in chimp (avg. log2 ratio = 2.17) and bonobo (avg. log2 ratio = 1.86), it is plausible that all or most of this dramatic copy number expansion (BLAT analyses of human, chimp, and macaque genome assemblies produce four, 109, and one PGM5 hits, respectively) occurred prior to the chimp/bonobo split, which, if true, would imply that between 2 and 5 Mya, the PGM5 gene underwent a dramatic expansion from roughly four to >100 copies, an average rate of >25 copies/Myr.
Also within the fusion region is SLC35F5, a gene that shows the most extreme gorilla-specific gene amplification predicted by aCGH (avg. log2 ratio = 3.92). This gene is thought to encode an ion transporter and is very similar (BLAST value E = 2 × 10−33) to 1F218, a Caenorhabditis elegans gene that is required for axonal guidance (Schmitz et al. 2007). FISH analysis indicates that numerous (>30) gorilla-specific copies exist and are localized at the telomeric region of virtually all gorilla chromosomes (Fortna et al. 2004). The CXYorf1 gene shows a human LS amplification with copies located both in the chr 2 fusion region as well as at the telomeric regions of several other human chromosomes (1, 9, 15 16, X, and Y). In support of the aCGH data, BLAT analysis of sequence assemblies predicts there is a striking increase in copy number in human (seven copies) compared to chimp (one) and macaque (one). While the function of this gene in humans is unknown, a best reciprocal BLAST analysis between human and C. elegans identified the ddl-2 gene (daf 16-dependent longevity protein 2) that controls life span as part of the insulin FOXO pathway (Hansen et al. 2005), suggesting a possible link of CXYorf1 with human longevity. In this regard, it is noteworthy that the average log2 aCGH ratios for CXYorf1 (IMAGE cDNAs 811138 and 136933) in human (0.23), chimp (−1.27), and macaque (−2.11) roughly parallel the average life span for these species, with more copies being found in the more long-lived species.
Human disease and human evolutionary adaptation
It can be argued that an increase in genome instability will result in an increase in variation and, as a result, will fuel the emergence of both evolutionarily adaptive change as well as human disease. In support of this view, data presented here indicate that a disproportionate number of genes showing primate LS copy number change are also associated with “genomic disorders,” recurrent diseases that are due to regions of genome instability. Specifically, of 20 genomic regions thought to be related to genome instability (Stankiewicz and Lupski 2002), 14 (70%) were associated with clusters (five or more cDNAs in a row, with ≥80% classified as LS) of genes identified here that showed LS copy number changes. Interestingly, for some diseases, the evolutionary changes may involve genes that show LS copy number changes in one or more non-human primate lineages instead of, or in addition to, genes that show a human LS change. For example, the Williams-Beuren syndrome chromosome region genes (WBSCR19, 20C, 21, and 22), associated with Williams-Beuren Syndrome, a developmental disorder caused by gene deletions on 7q11.23, show variation in copy number among several primate species. Relative to human, WBSCR19 is elevated in copy number in chimp and bonobo and decreased in orangutan, gibbon, macaque, baboon, marmoset, and lemur; and WBSCR21 is elevated in gibbon, macaque, baboon, and marmoset.
DiGeorge syndrome is also a disease of genome instability, and the gamma-glutamyltransferase 2 and L4 (GGT2 and GGTL4) genes, found in the low copy repeats associated with DiGeorge, show a dramatic expansion (avg. log2 ratio = 2.23) in gorilla relative to all other primates tested. Given that the human genome is predicted to encode nine related genes, the strong gorilla aCGH signal we report implies that there may be >40 _GGT2/L4_-related genes encoded in the gorilla genome. GGT is thought to be important to toxin removal, and it is possible that the GGT2/L4 expansion may be related to the gorilla’s ability to eat a wide variety (>100 plant species) of toxin-laden plants.
Another example is the ceroid-lipofuscinosis, neuronal 3, juvenile (CLN3) gene, which when deleted causes Batten, Spielmeyer-Vogt disease, a fatal lysosomal storage disease and one of the most common recessively inherited neurodegenerative disorders of childhood (Mole et al. 1999). Data presented here indicate that the CLN3 gene has undergone copy number expansions specifically in both Pan lineages (avg log2 ratio = 1.18 for bonobo and chimp). These correlations suggest that evolutionarily dynamic regions (e.g., regions where clusters of primate genes occur that show LS copy number variations) could be disproportionately associated with genomic loci (or genes) that are commonly involved in human genetic diseases.
Biologically interesting genes that show LS copy number changes
Given that gene duplication followed by divergence and selection have been among the most important contributors to genome evolution and the emergence of species-specific traits, it is likely that many of the genes exhibiting LS copy number changes identified here are involved in the phenotypic differences that distinguish each of these primate lineages. A partial list of potentially important genes showing LS copy number changes can be found in Supplemental Table S6, and some of the more intriguing candidates are discussed here.
DUF1220 domains, centrosomes, and brain function
The NBPF15 gene is a member of the NBPF family (Vandepoele 2005) and encodes several DUF1220 protein domains, sequences that are highly amplified in the human lineage and may be involved in higher cognitive function in humans (Popesco et al. 2006). We noticed that the copy number of several DUF1220-encoding genes in the region at 1q21.1 is altered in certain individuals with mental retardation (de Vries et al. 2005; Redon et al. 2006; Sharp et al. 2006) and with autism spectrum disorder (Autism Genome Project Consortium 2007), providing additional support linking DUF1220 domains to cognition. Data reported here (IMAGE:843276 avg. log2 ratio = −2.114 for all non-human primates) is consistent with the view that DUF1220 (and NBPF15) sequences show human lineage-specific copy number increases even when more distant primate lineages are used for comparison. Interestingly, one DUF1220-encoding centrosomal gene, PDE4DIP (Verde et al. 2001), shows a dramatic (ninefold) increase in brain cortex expression in humans compared to chimp (Preuss et al. 2004) and is a homolog of CDK5RAP2, a non-DUF1220-encoding gene implicated in the evolutionary expansion of the brain that, when defective, is known to produce microencephaly (Bond et al. 2005). While it is plausible that DUF1220 domains are not directly involved in brain expansion (Popesco et al. 2006), these observations provide the first direct link between DUF1220 domains and the several centrosomal microencephaly disease genes that have been identified (Bond and Woods 2006). Interestingly, two more genes that encode centrosome-related proteins, NEK2 (avg. log2 ratio = −1.67 in non-human primates) and ANAPC1 (avg. log2 ratio = −0.92), also show a human lineage-specific increase in copy number. NEK2 is a cell cycle–regulated kinase thought to control centrosome structure, while ANAPC1, a ubiquitin ligase that is part of the anaphase promoting complex, is abundant in post-mitotic neurons of the adult brain (Gieffers et al. 1999) and directly involved in the degradation of NEK2 (Hayes et al. 2006). It has also been reported that changes in the timing of asymmetric cell division for neuronal precursors, a process that likely involves the centrosome, may be critical to the size of the neocortex (Kornack and Rakic 1998), a brain region that has undergone striking progressive expansion from monkey to ape to human. These findings, taken together, indicate that a surprisingly high number of centrosomal proteins are linked to human brain function and exhibit human lineage-specific copy number expansions, suggesting that the centrosome may play a key role in the evolutionary adaptations important to the neuronal development and function of the human brain.
AQP7 and human endurance running
It has been suggested that in order for humans to have successfully adapted to the open hot savanna, a series of anatomical and physiological changes occurred that resulted in humans becoming unusually adept at endurance running (Bramble and Lieberman 2004), a property that likely contributed to their eventual emergence as efficient diurnal endurance predators and/or scavengers (Carrier 1984). Among the most important of these changes were (1) the development of an exceptional sweating response that allowed the high levels of metabolic heat that would be generated by endurance running to be dissipated efficiently (sweating would also cool the expanding brain and may have contributed to the selection for human hairlessness, a trait that would facilitate evaporative cooling in a sweating animal), and (2) the development of a mechanism for maintaining large body stores of glycogen and fatty acids and an effective means for mobilizing these stores during prolonged periods of high energy demand (Carrier 1984).
These important phenotypic changes would be expected to leave traces in the human genome, and several factors have led us to speculate that the human lineage-specific copy number expansion of the aquaporin 7 (AQP7) gene may be central to one or both of these human adaptations. Aquaporins are thought to play a key role in water transport across membranes (Preston et al. 1992), and of the eight aquaporin family members that were tested here, the only one that showed a human LS copy number increase was AQP7 (avg. log2 ratio for non-human primates = −1.20). Interestingly, all human copies (five) are part of segmental duplications, each of which encompasses an entire ∼17-kb _AQP7_-like gene copy. The great majority of these map to the pericentromeric region of chr 9, one of the most evolutionarily dynamic regions of the human genome and the location of the greatest concentration of human LS gene copy number increases (Fortna et al. 2004) (Fig. 5; Supplemental Table S5).
It has been shown that AQP7 is abundantly expressed in fat cells and is an aquaglyceroporin, capable of transporting glycerol as well as water (Kondo et al. 2002). Glycerol transport is a major mechanism for using energy stored in fat, and, interestingly, AQP7 expression in fat cells is elevated during intense exercise, resulting in an increase in glycerol transport (Kondo et al. 2002). Consistent with this observation, _AQP7_-null mice show an inability to transport glycerol and a pronounced weight gain due to the accumulation of glycerol and triglycerides (Hara-Chikuma et al. 2005). These findings, taken together, suggest that the human lineage-specific duplications of the AQP7 gene may underlie the increased glycerol transport capability found in humans and, as a result, facilitated the development of the exceptional capacity for endurance running in humans.
In a similar manner, the genes underlying human’s enhanced capacity to sweat, which efficiently reduces heat load during endurance running, might also be expected to increase in expression as a function of exercise. Several additional human-specific copies of AQP7 retain long open reading frames and upstream regulatory regions (Kondo et al. 2002), making it plausible that they may also retain the AQP7 gene’s ability to be up-regulated as a function of exercise. These factors, and the observation that AQP7 can transport both glycerol and water, have led us to suggest the possibility that one or more of the additional human AQP7 copies may also be involved in exercise-induced sweating in humans.
CGB/LHB and gorilla reproduction
In gorilla, several additional genes were identified that showed gorilla-specific amplifications and are associated with biologically interesting processes. These include three adjacent cDNAs (IMAGE clones 1671903, 259973, and 2365721) corresponding to the chorionic gonadotropin/leutenizing hormone, beta-subunit (CGB/LHB) genes on human 9q13.11 that are essential to several key reproductive processes including maintenance of pregnancy, male gonad development, and sex determination. While the human genome is predicted to contain seven CGB/LHB genes, the strong lineage-specific aCGH signals obtained in gorilla (avg. log2 ratio = 2.93 for gorilla) indicate that the number of genes in gorilla will be unusually high (e.g., ∼50), with the great majority of these copies found exclusively in gorilla. While there does not appear to be an obvious gorilla-specific reproductive characteristic to which this expansion might be linked, it is noteworthy that a frameshift in two of the seven CGB/LHB genes has been identified in humans and great apes that creates a completely different protein (Hallast et al. 2006), raising the possibility that the gorilla-specific copy number expansion identified here may have functional effects quite distinct from those typically associated with the CGB/LHB genes.
Distant primate genes that show LS copy number changes
Numerous biologically intriguing genes were identified that showed LS copy number expansions in distant primate lineages (Supplemental Tables S7, S8), and a few are highlighted here. Genes on chr 6 that encode the histocompatibility antigen Class I (HLA-I) proteins are known to be increased in copy number in macaque relative to humans and great apes (Daza-Vamenta et al. 2004; Macaque Genome Sequencing and Analysis Consortium 2007), a finding that may partly explain the higher HLA complexity of the macaque and may have implications for the use of the macaque as a model of human immune function. cDNA aCGH data presented here indicate that the HLA copy number (for HLA-A, HLA-C, and HLA-F) is elevated in macaque, baboon, and marmoset relative to all other species tested including lemur (Supplemental Fig. S1). This suggests that either these sequences were expanded independently in each of these three species or that, by parsimony, an expansion occurred in the last common ancestor of macaque, baboon, and marmoset, followed by a reduction in copy number in the last common ancestor of humans and apes.
Regions of high genomic variability may produce genes that exhibit both intra- and interspecies copy number variation, and an example of this occurs with the pregnancy-specific glycoprotein (1–11) gene family that clusters at 19q12. These genes encode immunoglobulin-related proteins that are the most abundant fetal protein in the maternal circulation at term and have been postulated to be linked with maternal-fetal conflict (Haig 1993). aCGH data show dramatic fluctuation in PSG copy number among different primate lineages, most notably an overall increase in OWM lineages (avg. log2 ratio = 0.744) and a pronounced decrease in marmoset and lemur (avg. log2 ratio = −2.092). In addition, variation between human samples is also evident with two (log2 ratios of 0.495 and 0.607) of four samples showing increases relative to the reference human.
As mentioned previously (Fig. 4), a striking orangutan-specific amplification (avg. log2 ratio = 1.37) was found for the carbonic anhydrase (CA) genes that cluster at 8q21.2 in humans. CA proteins catalyze the reversible hydration of carbon dioxide, and, because they are involved in numerous biological processes (e.g., respiration, acid–base balance, bone resorption, and the formation of aqueous humor, cerebrospinal fluid, saliva, and gastric acid), the additional gene copies in orangutan, if functional, may have important effects on the physiology of this primate. Another gene increase was observed for the lactate dehydrogenase beta gene (LDHB) specifically in the monkey lineages (avg. log2 ratio = 1.89), a change that may be related to phenotypic effects on anaerobic glycolysis in these species. The highest predicted copy number increase specific for the marmoset was found for the SRP9 gene (avg. log2 ratio = 4.74), which is thought to be involved in signal recognition and transport of secretory proteins to the rough endoplasmic reticulum. Another gene demonstrating copy number variation among the primates is GALNT1 (UDP-N-acetyl-α-D-galactosamine: polypeptide-N-acetylgalactosaminyltransferase 1) located on 18q12.1, the product of which catalyzes the initial reaction in O-linked oligosaccharide biosynthesis. Both aCGH and Q-PCR results indicate that this gene is increased in copy number in marmoset and lemur (avg. log2 value = 1.70). Other lemur-specific increases were found for multiple cDNAs corresponding to the zinc finger 91 gene (ZNF91) (lemur avg. log2 ratio = 1.39) located at 4p14 in human and for a poliovirus receptor-related gene (PVRL3; lemur avg. log2 ratio = 1.44), also termed nectin, that is related to immunoglobulin-like adhesion molecules and maps to 3q13.12.
An example of the utility of this extended cross-species aCGH survey can be found with respect to the RANBP2 gene, which encodes a nuclear export protein. Previous genomic sequencing of chr 2 identified eight new _RANBP2_-like genes in humans, while only one copy is found in mouse (Ciccarelli et al. 2005; Hillier et al. 2005). Here we demonstrate that the human and great ape lineages show a significant copy number increase of RANBP2 sequences relative to all other primates tested (great ape avg. log2 = 0.11; other primates avg. log2 ratio = −2.18). These data suggest that a dramatic copy number expansion occurred for RANBP2 in a common ancestor of humans and great apes after the ancestral human/great ape lineage diverged from the gibbon/monkey lineages sometime between 13 and 18 Mya.
The work presented here provides a genome-wide, sequence-independent assessment of gene duplication and loss that covers much of primate evolutionary history. As such it should both aid in our understanding of gene and genome evolution as well as in the identification of genes underlying lineage-specific traits. Finally because genome-wide aCGH does not rely on sequence data to predict copy number differences, it should provide a valuable complement to genomic sequencing efforts that seek to generate the most accurate genome assemblies.
Methods
DNA
The DNA used for this study was derived from human (two females, two males), bonobo (three males), chimpanzee (two males, two females), gorilla (one male, two females), orangutan (three females), gibbon (three males), macaque (one male, two females), baboon (two males, one female), marmoset (three females), and lemur (two males, one female). Human and chimpanzee genomic DNA samples were isolated from blood cells using Super Quick-Gene kits from AGTC. One gorilla and two bonobo samples were isolated from cell lines using DNeasy Tissue kits from QIAGEN. The macaque genomic DNA samples, an orangutan sample, and a gorilla sample were isolated by other laboratories. The remaining DNA (gibbon, baboon, marmoset, and lemur) was obtained from the Coriell Institute and originally derived from primary fibroblast cell lines or whole blood samples.
Array CGH
Labeling of genomic DNA and hybridization to cDNA microarrays were performed according to the method previously described (Pollack et al. 1999, 2002). In brief, 4 μg of genomic DNA from test (hominoid DNA) and 4 μg of sex-matched normal human genomic DNA reference samples (with the exception of samples human 4, macaque 487, and baboon 976, which were not sex-matched) were DpnII-digested and random-primer-labeled, incorporating Cy5 (red) and Cy3 (green) fluorescent dyes, respectively. Test and reference samples were cohybridized to a cDNA microarray containing 41,126 nonredundant clones, representing 24,473 human genes (i.e., UniGene clusters); 34,244 cDNAs had single map positions, and 4857 had multiple map positions, with the remainder (2025) not yet assigned. Following hybridization, microarrays were imaged using a GenePix 4000B scanner (Axon Instruments). Fluorescence intensities for array elements were extracted using GenePix Pro 4.0 software, and uploaded into the Stanford Microarray Database (SMD) (Gollub et al. 2003) for subsequent analysis.
Array CGH data analysis
Fluorescence ratios were normalized for each experiment by setting the average log2 fluorescence ratio (test/reference) for all array elements equal to 0. We included for analysis only those genes that were reliably measured, having a fluorescence intensity/background >1.4 in the reference channel. Map positions for cDNA clones on the array were assigned using the UCSC GoldenPath assembly (http://genome.ucsc.edu), November 2002 freeze (hg13). This freeze was used because, of the map files available for the human cDNA arrays we used, this one retained a significantly greater number of genes that had multiple map locations (e.g., likely represented recently duplicated genes). Gene copy number ratios were visualized according to chromosome position using Treeview (http://rana.lbl.gov/EisenSoftware.htm). cDNAs with multiple genome map positions >1Mb apart were displayed in Treeview at each assigned map location.
Selection criteria for array data
To select lineage-specific cDNAs, the values used were the log2 of the red (test genomic DNA signal) to green (reference genomic DNA signal) normalized ratio (mean). The criteria for selection of LS cDNAs were similar to that described previously (Fortna et al. 2004) and were based on the following. First, a threshold of 0.5 was used, in which case at least two out of three (one could be missing or not meet the threshold) of the absolute values of the signal intensity ratio for the individuals in one species needed to be equal to or larger than 0.5 in the same direction (both positive or both negative), while at least two out of three (one could be missing or not meet the threshold) of the absolute values for all of the other individuals of the other species had to be less than the threshold and in the opposite direction. Second, the absolute value of the average of the intensity ratios for the nonhuman primate species compared to human was required to be at least 2.5 times greater than the absolute value of each of the remaining species average, including human versus human comparisons. For genes showing human LS changes, the absolute value of each species average of the non-human primate versus human comparisons had to be at least 2.5-fold greater than the average of the absolute value of the human versus human comparisons.
Gene copy number changes specific to multiple species
In cases in which the copy number was either increased or decreased in more than one primate species, the same criteria as above were used. However, the cDNA had to meet the 0.5 selection criterion for the species in question. Additionally, the species in question was required to be at least 2.5-fold greater than the average of the absolute values of the fluorescence ratios compared to the pairwise comparisons for the other species.
“Or-case”: Genes that show copy number variation between human and at least one other primate lineage
An analysis was conducted to detect cDNAs for which the log2 fluorescence signal was different in one or more species of primates relative to human. In order for the human aCGH signal to be considered different from one or more primate species, the human log2 aCGH ratio had to fall within the range of +0.5 to −0.5 and the non-human primate species had to exceed the threshold of 0.5 and had to have all (three out of three) individuals within the species meet or exceed a ratio of at least 2.5 times greater than that found in human.
Quantitative Real-Time PCR
Q-PCR, using an ABI 7300, was carried out on genes for each individual across species using optimal primer and fluorogenic probe sets that are unique to the exonic DNA sequence of the gene of interest. Optimal primers and probes were designed using PrimerExpress (ABI software). The amplicon sequence was used as a BLAT query (http://genome.ucsc.edu/cgi-bin/hgBlat) against the human March 2006 (hg18), chimpanzee March 2006 (PanTro2), and rhesus macaque January 2006 (rheMac2) assemblies to ensure that the primer/probe sets had no or minimal mismatches. The functionality of each primer pair was then verified using the UCSC database for in silico PCR (http://genome.ucsc.edu/cgi-bin/hgPcr?command=start). Gene copy number was determined by the number of cycles and amount of amplification product determined by Q-PCR. These assays were done in duplicate, and the copy numbers were normalized to CFTR, cystic fibrosis transmembrane conductance regulator, an ATP-binding cassette that was used as a control gene thought to represent one gene per haploid genome across humans and the great apes (Hallast et al. 2006). To further confirm CFTR copy number across primates, BLAT searches using the CFTR primer/probe sequences as a query were done for available human, chimp, and macaque genome assemblies. The CFTR sequence was also used as a BLAT query against the mouse and rat genomes, which both showed one copy per haploid genome.
The Q-PCR primer and probe sequences are as follows: ALDH1B1 F, 5′-CGGGACCGTGTGGGTAAAC-3′; ALDH1B1 R, 5′-AGATTCCTTAAACCCTCCAAATGG; and the probe 5′-6FAM CCTACAACATCGTCACCTGCCACA-TAMRA-3′; AQP7 F, 5′-CCATCACGGACCAGGAGAA-3′; AQP7 R, 5′-GACCACGAGGAT GCCTATCAC-3′; and the probe 5′-6FAM-CCAGCACTGCC AGGAACAGAGGC-TAMRA-3′; BMI1 F, 5′-GCCCAGCAGGAGG TATTCC-3′; BMI1 R, 5′-GATGAGGAGACTGCACTGGAGTAC-3′; and the probe 5′-6FAM-TCCACCTCTTCTTGTTTGCCTAGC CCC-TAMRA-3′; CAI F, 5′-AACGAGCCCCATTCACAAATT-3′; CA1 R, 5′-CCAGGGTAGGTCCAGAAATCC-3′; and the probe 5′-6FAM-TGACCCCTCTACTCTCCTTCCTTCATCCC-TAMRA-3′; CBFB F, 5′-CATTAGCACAACAGGCCTTTGA-3′; CBFB R, 5′-TCCATTTCCTCCCGATGAGA-3′; and the probe 5′-6FAM-AGGCTCGGAGAAGGACACGCGA-TAMRA-3′; CDC42 F, 5′-CCT ATCACTCCAGAGACTGCTGAA-3′; CDC42 R, 5′-GTGCAGA ACACTCCACATACTTGAC-3′; and the probe 5′-6FAM-AG CTGGCCCGTGACCTGAAGGC-TAMRA-3′; CFTR F, 5′-CGCGATTTATCTAGGCATAGGC-3′; CFTR R, 5′- TGTGATGAA GGCCAAAAATGG-3′; and the probe 5′-6FAM-TGCCTTCTCTT TATTGTGAGGACACTGCTCC-TAMRA-3′; DHFR F, 5′-CCC GCTGCTGTCATGGTT-3′; DHFR R, 5′-GCCGATGCCCATG TTCTG-3′; and the probe 5′-6FAM-TTCGCTAAACTGCATCGTC GCTGTGT-TAMRA-3′; GALNT1 F, 5′-TGGTGATTGTTTTCCACA ATGAG-3′; GALNT1 R, 5′-GGTGAGCGATTAATGACACTATGG-3′; and the probe 5′-6FAM-TTGGAGCACACTTCTGCGAACTG-TAMRA-3′; KRT8 F, 5′-CATCGAGATCGCCACCTACA-3′; KRT8 R, 5′-ACTCATGTTCTGCATCCCAGACT-3′; and the probe 5′-6FAM-TGGAGGGCGAGGAGAGCCGG-TAMRA-3′; SEC13L1 F, 5′-AGG GTCCTGTGTGGCAAGTG-3′; SEC13L1 R, 5′-TAGGAGCACGAT GCCAGGAT-3′; and the probe 5′-6FAM-CCTGGGCTCACCC CATGTACGG-TAMRA-3′; SV2B F, 5′-GGTATCCCTCACCCAGA TGATG-3′; SV2B R, 5′-CCCGAAGGCTGTCCATTCT-3′; and the probe 5′-6FAM-CAAGGCCAAGCAGGCCAAGATGG-TAMRA-3′; TERF1 F, 5′-GGGAAGAAGACAAGAATTTGAGATCT-3′; TERF1 R, 5′-GGGAAGAAGACAAGAATTTGAGATCT-3′; and the probe 5′-6FAM-TGAGGAAATATGGAGAGGGAAACTGGTCTAAA ATACTG-TAMRA-3′; and Znf91 F 5′- TCTGTAGCTTCCCTGTGA GGTCTT-3′; Znf91 R, 5′-AGGAGCACCCTGTGACATTCTATAA-3′; and the probe 5′-6FAM-TCTGGCTTTGGTGTTAAGGTAAT GTCCGC-TAMRA3′.
Computational analyses: BLAT comparisons
The sequences for GenBank accession numbers that corresponded to IMAGE clones whose signals were determined to be either HLS, chimp LS, or macaque LS by the abovementioned selection criteria were used as BLAT queries against the most recent human (hg18), chimp (PanTro2), and macaque (rheMac2) genome assemblies to determine if available primate genome assemblies were in agreement with the aCGH data set. BLAT hits were screened with a Perl (http://www.perl.org) script for scores of >200, to eliminate spurious matches. Those BLAT hits that met the score cutoff were then tallied for each genome, and the numbers were compared to determine whether or not the BLAT searches were in agreement with the proportions of human, chimp, and macaque LS increases and decreases discovered using aCGH.
Acknowledgments
We thank B. Soriano and J. Gaydos for offering their expertise regarding microarray analysis; M. Popesco, E. MacLaren, and A. Fortna for helpful discussions; and J. Chang, S. Williams, A. Komura, S. Glidewell, and S. Friedrichs for technical help. We also thank L. Lyons at UC, Davis, for providing genomic DNAs from Macaca mulatta; D.G. Smith at UC, Davis for a gorilla DNA sample; Yerkes National Primate Research Center (Atlanta, Georgia); the Coriell Institute (Camden, New Jersey) for bonobo, orangutan, and gorilla DNA samples; M. Goodman and D. Wildman at Wayne State University School of Medicine for an orangutan DNA sample; and the Human Genome Sequencing Consortium, the Chimpanzee Sequencing and Analysis Consortium, and the Macaque Genome Sequence and Analysis Consortium for generation and pre-publication release of genome sequence assemblies for these species. This work was supported by a Butcher foundation grant and NIH grant AA11853 (J.M.S.) and NIH grant CA97139 (J.R.P.).
Footnotes
[Supplemental material is available online at www.genome.org. Full array-based comparative genomic hybridization data for all primates surveyed has been deposited in the Stanford Microarray Database (SMD) at http://www.stanford.edu/microarray.]
References
- Autism Genome Project Consortium Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat. Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.A., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Gu Z., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Clark R.A., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Reinert K., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Samonte R.V., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Schwartz S., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Adams M.D., Myers E.W., Li P.W., Eichler E.E., Myers E.W., Li P.W., Eichler E.E., Li P.W., Eichler E.E., Eichler E.E. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. [DOI] [PubMed] [Google Scholar]
- Bond J., Woods C.G., Woods C.G. Cytoskeletal genes regulating brain size. Curr. Opin. Cell Biol. 2006;18:95–101. doi: 10.1016/j.ceb.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Bond J., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Roberts E., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Springell K., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Lizarraga S.B., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Scott S., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Higgins J., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Hampshire D.J., Morrison E.E., Leal G.F., Silva E.O., Morrison E.E., Leal G.F., Silva E.O., Leal G.F., Silva E.O., Silva E.O., et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat. Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- Bramble D.M., Lieberman D.E., Lieberman D.E. Endurance running and the evolution of Homo. Nature. 2004;432:345–352. doi: 10.1038/nature03052. [DOI] [PubMed] [Google Scholar]
- Carrier D.R. The energetic paradox of human running and hominid evolution. Curr. Anthropol. 1984;25:483–495. [Google Scholar]
- Cheng Z., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Ventura M., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., She X., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Khaitovich P., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Graves T., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Osoegawa K., Church D., DeJong P., Wilson R.K., Paabo S., Church D., DeJong P., Wilson R.K., Paabo S., DeJong P., Wilson R.K., Paabo S., Wilson R.K., Paabo S., Paabo S., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature. 2005;437:88–93. doi: 10.1038/nature04000. [DOI] [PubMed] [Google Scholar]
- Cheung J., Estivill X., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Estivill X., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Khaja R., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., MacDonald J.R., Lau K., Tsui L.C., Scherer S.W., Lau K., Tsui L.C., Scherer S.W., Tsui L.C., Scherer S.W., Scherer S.W. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 2003;4:R25. doi: 10.1186/gb-2003-4-4-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Ciccarelli F.D., von Mering C., Suyama M., Harrington E.D., Izaurralde E., Bork P., von Mering C., Suyama M., Harrington E.D., Izaurralde E., Bork P., Suyama M., Harrington E.D., Izaurralde E., Bork P., Harrington E.D., Izaurralde E., Bork P., Izaurralde E., Bork P., Bork P. Complex genomic rearrangements lead to novel primate gene function. Genome Res. 2005;15:343–351. doi: 10.1101/gr.3266405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Vamenta R., Glusman G., Rowen L., Guthrie B., Geraghty D.E., Glusman G., Rowen L., Guthrie B., Geraghty D.E., Rowen L., Guthrie B., Geraghty D.E., Guthrie B., Geraghty D.E., Geraghty D.E. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries B.B.A., Pfundt R., Leisink M., Koolen D.A., Vissers L.E.L.M., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Pfundt R., Leisink M., Koolen D.A., Vissers L.E.L.M., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Leisink M., Koolen D.A., Vissers L.E.L.M., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Koolen D.A., Vissers L.E.L.M., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Vissers L.E.L.M., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Janssen I.M., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Reijmersdal S., Nillesen W.M., Huys E.H.L.P., Leeuw N., Nillesen W.M., Huys E.H.L.P., Leeuw N., Huys E.H.L.P., Leeuw N., Leeuw N., et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards Y.H., Putt W., Fox M., Ives J.H., Putt W., Fox M., Ives J.H., Fox M., Ives J.H., Ives J.H. A novel human phosphoglucomutase (PGM5) maps to the centromeric region of chromosome 9. Genomics. 1995;30:350–353. doi: 10.1006/geno.1995.9866. [DOI] [PubMed] [Google Scholar]
- Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., Hink R., Burgers S., Hernandez-Boussard T., Burgers S., Hernandez-Boussard T., Hernandez-Boussard T., et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:937–954. doi: 10.1371/journal.pbio.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieffers C., Peters B.H., Kramer E.R., Dotti C.G., Peters J.M., Peters B.H., Kramer E.R., Dotti C.G., Peters J.M., Kramer E.R., Dotti C.G., Peters J.M., Dotti C.G., Peters J.M., Peters J.M. Expression of the CDH1-associated form of the anaphase-promoting complex in postmitotic neurons. Proc. Natl. Acad. Sci. 1999;96:11317–11322. doi: 10.1073/pnas.96.20.11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goidts V., Armengol L., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Armengol L., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Schempp W., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Conroy J., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Nowak N., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Muller S., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Cooper D.N., Estivill X., Enard W., Szamalek J.M., Estivill X., Enard W., Szamalek J.M., Enard W., Szamalek J.M., Szamalek J.M., et al. Identification of large-scale human-specific copy number differences by inter-species array comparative genomic hybridization. Hum. Genet. 2006;119:185–198. doi: 10.1007/s00439-005-0130-9. [DOI] [PubMed] [Google Scholar]
- Gollub J., Ball C.A., Binkley G., Demeter J., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Ball C.A., Binkley G., Demeter J., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Binkley G., Demeter J., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Demeter J., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Finkelstein D.B., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Hebert J.M., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Hernandez-Boussard T., Jin H., Kaloper M., Matese J.C., Jin H., Kaloper M., Matese J.C., Kaloper M., Matese J.C., Matese J.C., et al. The Stanford Microarray Database: Data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groves C. Primate taxonomy. Smithsonian Institution Press; Washington, DC: 2001. [Google Scholar]
- Haig D. Genetic conflicts in human pregnancy. Oxf. Rev. Reprod. Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- Hallast P., Rull K., Laan M., Rull K., Laan M., Laan M. The evolution and genomic landscape of CGB1 and CGB2 genes. Mol. Cell Endocrinol. 2006;260–262:2–11. doi: 10.1016/j.mce.2005.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M., Hsu A.L., Dillin A., Kenyon C., Hsu A.L., Dillin A., Kenyon C., Dillin A., Kenyon C., Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1:119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara-Chikuma M., Sohara E., Rai T., Ikawa M., Okabe M., Sasaki S., Uchida S., Verkman A.S., Sohara E., Rai T., Ikawa M., Okabe M., Sasaki S., Uchida S., Verkman A.S., Rai T., Ikawa M., Okabe M., Sasaki S., Uchida S., Verkman A.S., Ikawa M., Okabe M., Sasaki S., Uchida S., Verkman A.S., Okabe M., Sasaki S., Uchida S., Verkman A.S., Sasaki S., Uchida S., Verkman A.S., Uchida S., Verkman A.S., Verkman A.S. Progressive adipocyte hypertrophy in aquaporin-7-deficient mice: Adipocyte glycerol permeability as a novel regulator of fat accumulation. J. Biol. Chem. 2005;280:15493–15496. doi: 10.1074/jbc.C500028200. [DOI] [PubMed] [Google Scholar]
- Hayes M.J., Kimata Y., Wattam S.L., Lindon C., Mao G., Yamano H., Fry A.M., Kimata Y., Wattam S.L., Lindon C., Mao G., Yamano H., Fry A.M., Wattam S.L., Lindon C., Mao G., Yamano H., Fry A.M., Lindon C., Mao G., Yamano H., Fry A.M., Mao G., Yamano H., Fry A.M., Yamano H., Fry A.M., Fry A.M. Early mitotic degradation of Nek2A depends on Cdc20-independent interaction with the APC/C. Nat. Cell Biol. 2006;8:607–614. doi: 10.1038/ncb1410. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Graves T.A., Fulton R.S., Fulton L.A., Pepin K.H., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Graves T.A., Fulton R.S., Fulton L.A., Pepin K.H., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Fulton R.S., Fulton L.A., Pepin K.H., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Fulton L.A., Pepin K.H., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Pepin K.H., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Minx P., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Wagner-McPherson C., Layman D., Wylie K., Sekhon M., Layman D., Wylie K., Sekhon M., Wylie K., Sekhon M., Sekhon M., et al. Generation and annotation of the DNA sequences of human chromosomes 2 and 4. Nature. 2005;434:724–731. doi: 10.1038/nature03466. [DOI] [PubMed] [Google Scholar]
- Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Hurles M. Gene duplication: The genomic trade in spare parts. PLoS Biol. 2004;2:e206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Donahoe P.K., Qi Y., Scherer S.W., Lee C., Qi Y., Scherer S.W., Lee C., Scherer S.W., Lee C., Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- IJdo J.W., Baldini A., Ward D.C., Reeders S.T., Wells R.A., Baldini A., Ward D.C., Reeders S.T., Wells R.A., Ward D.C., Reeders S.T., Wells R.A., Reeders S.T., Wells R.A., Wells R.A. Origin of human chromosome 2: An ancestral telomere–telomere fusion. Proc. Natl. Acad. Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauch A., Wienberg J., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T., Wienberg J., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T., Stanyon R., Arnold N., Tofanelli S., Ishida T., Cremer T., Arnold N., Tofanelli S., Ishida T., Cremer T., Tofanelli S., Ishida T., Cremer T., Ishida T., Cremer T., Cremer T. Reconstruction of genomic rearrangements in great apes and gibbons by chromosome painting. Proc. Natl. Acad. Sci. 1992;89:8611–8615. doi: 10.1073/pnas.89.18.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling M.A., Hurles M.E., Tyler-Smith C., Hurles M.E., Tyler-Smith C., Tyler-Smith C. Human evolutionary genetics. Garland Science; New York: 2004. [Google Scholar]
- Kent W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo H., Shimomura I., Kishida K., Kuriyama H., Makino Y., Nishizawa H., Matsuda M., Maeda N., Nagaretani H., Kihara S., Shimomura I., Kishida K., Kuriyama H., Makino Y., Nishizawa H., Matsuda M., Maeda N., Nagaretani H., Kihara S., Kishida K., Kuriyama H., Makino Y., Nishizawa H., Matsuda M., Maeda N., Nagaretani H., Kihara S., Kuriyama H., Makino Y., Nishizawa H., Matsuda M., Maeda N., Nagaretani H., Kihara S., Makino Y., Nishizawa H., Matsuda M., Maeda N., Nagaretani H., Kihara S., Nishizawa H., Matsuda M., Maeda N., Nagaretani H., Kihara S., Matsuda M., Maeda N., Nagaretani H., Kihara S., Maeda N., Nagaretani H., Kihara S., Nagaretani H., Kihara S., Kihara S., et al. Human aquaporin adipose (AQPap) gene: Genomic structure, promoter analysis and functional mutation. Eur. J. Biochem. 2002;269:1814–1826. doi: 10.1046/j.1432-1033.2002.02821.x. [DOI] [PubMed] [Google Scholar]
- Kornack D.R., Rakic P., Rakic P. Changes in cell-cycle kinetics during the development and evolution of primate neocortex. Proc. Natl. Acad. Sci. 1998;95:1242–1246. doi: 10.1073/pnas.95.3.1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaque Genome Sequencing and Analysis Consortium Analyses of the rhesus macaque genome sequence. Science. 2007 (in press) [Google Scholar]
- Mole S.E., Mitchison H.M., Munroe P.B., Mitchison H.M., Munroe P.B., Munroe P.B. Molecular basis of the neuronal ceroid lipofuscinoses: Mutations in CLN1, CLN2, CLN3, and CLN5. Hum. Mutat. 1999;14:199–215. doi: 10.1002/(SICI)1098-1004(1999)14:3<199::AID-HUMU3>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Nahon J.-L. Birth of ‘human-specific’ genes during primate evolution. Genetica. 2003;118:193–208. doi: 10.1023/a:1024157714736. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene and genome duplication. Springer; Berlin: 1970. [Google Scholar]
- Perry G.H., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., Tchinda J., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., McGrath S.D., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., Zhang J., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., Picker S.R., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., Caceres A.M., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., Iafrate A.J., Tyler-Smith C., Scherer S., Eichler E., Tyler-Smith C., Scherer S., Eichler E., Scherer S., Eichler E., Eichler E., et al. Hotspots for copy number variation in chimpanzees and humans. Proc. Natl. Acad. Sci. 2006;103:8006–8011. doi: 10.1073/pnas.0602318103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Segraves R., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Sudar D., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Clark S., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Poole I., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Kowbel D., Collins C., Kuo W.L., Chen C., Zhai Y., Collins C., Kuo W.L., Chen C., Zhai Y., Kuo W.L., Chen C., Zhai Y., Chen C., Zhai Y., Zhai Y., et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Perou C.M., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Alizadeh A.A., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Eisen M.B., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Pergamenschikov A., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Williams C.F., Jeffrey S.S., Botstein D., Brown P.O., Jeffrey S.S., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat. Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Pollack J.R., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Sorlie T., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Perou C.M., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Rees C.A., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Jeffrey S.S., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Lonning P.E., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Tibshirani R., Botstein D., Borresen-Dale A.L., Brown P.O., Botstein D., Borresen-Dale A.L., Brown P.O., Borresen-Dale A.L., Brown P.O., Brown P.O. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc. Natl. Acad. Sci. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popesco M.C., Maclaren E.J., Hopkins J., Dumas L., Cox M., Meltesen L., McGavran L., Wyckoff G.J., Sikela J.M., Maclaren E.J., Hopkins J., Dumas L., Cox M., Meltesen L., McGavran L., Wyckoff G.J., Sikela J.M., Hopkins J., Dumas L., Cox M., Meltesen L., McGavran L., Wyckoff G.J., Sikela J.M., Dumas L., Cox M., Meltesen L., McGavran L., Wyckoff G.J., Sikela J.M., Cox M., Meltesen L., McGavran L., Wyckoff G.J., Sikela J.M., Meltesen L., McGavran L., Wyckoff G.J., Sikela J.M., McGavran L., Wyckoff G.J., Sikela J.M., Wyckoff G.J., Sikela J.M., Sikela J.M. Human lineage-specific amplification, selection, and neuronal expression of DUF1220 domains. Science. 2006;313:1304–1307. doi: 10.1126/science.1127980. [DOI] [PubMed] [Google Scholar]
- Preston G.M., Carroll T.P., Guggino W.B., Agre P., Carroll T.P., Guggino W.B., Agre P., Guggino W.B., Agre P., Agre P. Appearance of water channels in Xenopus oocytes expressing red cell CHIP28 protein. Science. 1992;256:385–387. doi: 10.1126/science.256.5055.385. [DOI] [PubMed] [Google Scholar]
- Preuss T.M., Caceres M., Oldham M.C., Geschwind D.H., Caceres M., Oldham M.C., Geschwind D.H., Oldham M.C., Geschwind D.H., Geschwind D.H. Human brain evolution: Insights from microarrays. Nat. Rev. Genet. 2004;5:850–860. doi: 10.1038/nrg1469. [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., Fiegler H., Shapero M.H., Carson A.R., Chen W., Shapero M.H., Carson A.R., Chen W., Carson A.R., Chen W., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz C., Kinge P., Hutter H., Kinge P., Hutter H., Hutter H. Axon guidance genes identified in a large-scale RNAi screen using the RNAi-hypersensitive Caenorhabditis elegans strain nre-1(hd20) lin-15b(hd126) Proc. Natl. Acad. Sci. 2007;104:834–839. doi: 10.1073/pnas.0510527104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M., Lundin P., Maner S., Massa H., Walker M., Chi M., Maner S., Massa H., Walker M., Chi M., Massa H., Walker M., Chi M., Walker M., Chi M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Stewart H., Price S.M., Blair E., Hennekam R.C., Price S.M., Blair E., Hennekam R.C., Blair E., Hennekam R.C., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- She X., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Jiang Z., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Clark R.A., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Liu G., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Cheng Z., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Tuzun E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Church D.M., Sutton G., Halpern A.L., Eichler E.E., Sutton G., Halpern A.L., Eichler E.E., Halpern A.L., Eichler E.E., Eichler E.E. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- She X., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rochhi M., Liu G., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rochhi M., Ventura M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rochhi M., Zhao S., Misceo D., Roberto R., Cardone M.F., Rochhi M., Misceo D., Roberto R., Cardone M.F., Rochhi M., Roberto R., Cardone M.F., Rochhi M., Cardone M.F., Rochhi M., Rochhi M., et al. A preliminary comparative analysis of primate segmental duplications shows elevated substitution rates and a great-ape expansion of intrachromosomal duplications. Genome Res. 2006;16:576–583. doi: 10.1101/gr.4949406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikela J.M. The jewels of our genome: The search for the genomic changes underlying the evolutionarily unique capacities of the human brain. PLoS Genet. 2006;2:646–655. doi: 10.1371/journal.pgen.0020080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P., Lupski J.R., Lupski J.R. Genome architecture, rearrangements and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- Vandepoele K., Van Roy N., Staes K., Speleman F., van Roy F., Van Roy N., Staes K., Speleman F., van Roy F., Staes K., Speleman F., van Roy F., Speleman F., van Roy F., van Roy F. A novel gene family NBPF: Intricate structure generated by gene duplications during primate evolution. Mol. Biol. Evol. 2005;22:2265–2274. doi: 10.1093/molbev/msi222. [DOI] [PubMed] [Google Scholar]
- Verde I., Pahlke G., Salanova M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S.L., Conti M., Pahlke G., Salanova M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S.L., Conti M., Salanova M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S.L., Conti M., Zhang G., Wang S., Coletti D., Onuffer J., Jin S.L., Conti M., Wang S., Coletti D., Onuffer J., Jin S.L., Conti M., Coletti D., Onuffer J., Jin S.L., Conti M., Onuffer J., Jin S.L., Conti M., Jin S.L., Conti M., Conti M. Myomegalin is a novel protein of the golgi/centrosome that interacts with a cyclic nucleotide phosphodiesterase. J. Biol. Chem. 2001;276:11189–11198. doi: 10.1074/jbc.M006546200. [DOI] [PubMed] [Google Scholar]
- Wilson G.M., Flibotte S., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Flibotte S., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Jones S., Thornton K., Clark A.G., Holt R.A., Thornton K., Clark A.G., Holt R.A., Clark A.G., Holt R.A., Holt R.A. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]