Targeted recruitment of a histone H4-specific methyltransferase by the transcription factor YY1 (original) (raw)
Abstract
Methylation of specific residues within the N-terminal histone tails plays a critical role in regulating eukaryotic gene expression. Although great advances have been made toward identifying histone methyltransferases (HMTs) and elucidating the consequences of histone methylation, little is known about the recruitment of HMTs to regulatory regions of chromatin. Here we report that the sequence-specific DNA-binding transcription factor Yin Yang 1 (YY1) binds to and recruits the histone H4 (Arg 3)-specific methyltransferase, PRMT1, to a YY1-activated promoter. Our data confirm that histone methylation does not occur randomly but rather is a targeted event and provides one mechanism by which HMTs can be recruited to chromatin to activate gene expression.
Keywords: Histone methyltransferase, PRMT1, transcription factor YY1
In eukaryotes, DNA is tightly bound to histones, forming repeating units of DNA–protein particles called nucleosomes. Each nucleosome contains a nucleosomal core particle, consisting of 146 base pairs of supercoiled DNA wrapped twice around a complex of eight histone molecules. The histone core complex consists of two molecules each of histones H2A, H2B, H3, and H4. All core histones contain a C-terminal globular domain and an N-terminal lysine- and arginine-rich domain, where many posttranslational modifications occur. Modifications of core histones by acetylation, phosphorylation, methylation, ubiquitination, and poly-ADP-ribosylation have been described. Many thousands of different combinations of histone modification are possible, providing an abundance of regulatory potential (Berger 2001, 2002). In fact, it was proposed that the combinatorial nature of histone modification forms a “histone code” that is read by other proteins to bring about distinct downstream cellular responses (Strahl and Allis 2000; Jenuwein and Allis 2001). Understanding the nature and mechanisms of histone modification is, therefore, clearly a definite prerequisite to elucidating the complex mechanisms of gene regulation in eukaryotic cells.
Many studies suggest that hyperacetylation of histones generally correlates with transcriptionally active chromatin, perhaps by increasing the accessibility of nucleosomal DNA to transcription factors, whereas hypoacetylation of histones correlates with transcriptional silencing (Kuo and Allis 1998). In comparison, phosphorylation of histones, at least in Ser 10 of histone H3, may lead to either gene activation or chromatin condensation during mitosis (Cheung et al. 2000). Similar to modifications by acetylation and phosphorylation, a series of recent studies provide strong evidence that histone methylation has a profound effect on gene regulation in all eukaryotic cells (Zhang and Reinberg 2001; Kouzarides 2002).
The most abundant, and most studied, histone methylation events take place on lysine residues located in the N terminus of histone H3. Additionally, at least one lysine methylation occurs in the N-terminal tail of histone H4. Most, but not all, enzymes responsible for histone lysine methylation belong to the SET domain family of histone methyltransferases (HMTs). Although in some cases histone lysine methylation can be a marker for transcriptional activation, in other situations, histone lysine methylation can have a repressive function in vivo (Zhang and Reinberg 2001; Kouzarides 2002; Santos-Rosa et al. 2002).
Unlike lysine methylation, the function and significance of histone arginine methylation are less well understood. The first arginine-HMT identified was the coactivator-associated arginine methyltransferase 1 (CARM1; Chen et al. 1999). CARM1, also known as protein arginine methyltransferase 4 (PRMT4), methylates histone H3 at Arg 2, Arg 17, Arg 26, and several C-terminal residues in vitro. In a separate attempt to isolate enzymes that methylate core histones, Zhang and colleagues purified PRMT1, which accounts for most of the type I protein arginine methyltransferase activity in cells, as an H4-specific Arg 3 HMT (Wang et al. 2001). Interestingly, the HMT activity of PRMT1 is required for PRMT1 to function as a coactivator of nuclear hormone receptors, suggesting that the HMT activity of PRMT1 is involved in transcriptional activation. Similarly, a study by Allis and colleagues showed that PRMT1 is the major, if not exclusive, H4 Arg 3-methyltransferase in human 293 cells (Strahl et al. 2001). It is now clear from these studies that PRMT1 is an H4-specific arginine HMT and that methylation of histone H4 Arg 3 could result in transcriptional activation. However, it is unclear how PRMT1 is targeted to specific promoters or transcriptional regulatory regions.
YY1 (Yin Yang 1) is a 414-amino-acid Krüppel-related zinc finger transcription factor that binds to the CGC CATNTT consensus DNA element located in promoters and enhancers of many cellular and viral genes (Shi et al. 1997; Thomas and Seto 1999). Like many transcription factors, YY1 requires coactivators and corepressors to function properly. Interestingly, YY1 appears to be equally effective as an activator and as a repressor depending on its relative concentration, its binding partners, and on promoter context. Although reports of the number of genes that might be regulated by YY1 are ever-increasing, the exact mechanisms by which this factor regulates transcription are still unclear.
Over the years, a number of models have been proposed to explain the mechanisms of YY1 action. Of these different models, the recruitment of histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzymes by YY1 is especially appealing and has recently gained the most attention. In the current study, we uncovered an additional histone modification enzyme that could be recruited by YY1 to regulate transcription. Specifically, we found that YY1 binds to PRMT1 and recruits PRMT1 to DNA. The interaction between YY1 and PRMT1 most likely occurs via a bridging protein, the double-stranded RNA-binding protein 76 (DRBP76). A purified YY1 complex possesses histone H4-specific methyltransferase activity, and results from chromatin immunoprecipitation (ChIP) assays suggest that YY1 directs histone H4 methylation at a YY1-activated, but not a YY1-repressed, promoter. Together, these results provide an example for targeted recruitment of HMT by a sequence-specific-binding transcription factor and provide a plausible explanation of how one single transcription factor can regulate a diverse number of genes.
Results
Purification of a YY1 complex
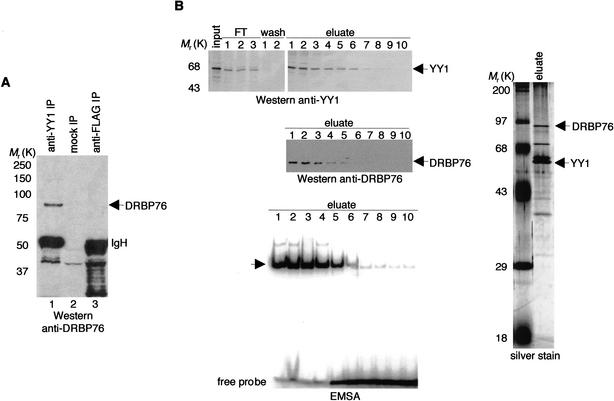
To explore how YY1 regulates transcription, we purified a complex containing YY1 by immunoaffinity chromatography using an extract prepared from HeLa cells infected with recombinant adenovirus-expressing Flag epitope-tagged YY1. We found that at least four polypeptides associated specifically with Flag–YY1 (Fig. 1A, lane 2). As a control, a mock purification was performed on cells infected with an adenovirus expressing green fluorescent protein (GFP; Fig. 1A, lane 1). Mass spectrometric analysis revealed that the 90-kD YY1-associated protein was either the human interleukin enhancer binding factor 3 (ILF3) or DRBP76, an alternatively spliced product of ILF3 (Fig. 1B; Patel et al. 1999; Duchange et al. 2000). Based on the molecular mass of the protein and Western blot results with an anti-DRBP76 specific antibody (Fig. 1C), we concluded that the 90-kD YY1-associated protein was DRBP76.
Figure 1.
YY1 physically associates with DRBP76. (A) Silver-stained SDS-PAGE of the Flag–YY1 complex. “Control” indicates an anti-Flag immunopurified sample prepared from HeLa cells transduced with adenovirus expressing GFP. In addition to Flag–YY1, arrows indicate proteins copurified, in approximately stoichiometrically equivalent amounts, with Flag–YY1. (B) Amino acid sequence of DRBP76 and ILF3. Each dash (–) indicates an amino acid in ILF3 that is identical to the corresponding one in DRBP76. Peptide sequences obtained by microsequencing are underlined. (C) An immunoblot of the purified Flag–YY1 complex using an anti-DRBP76 antibody.
Endogenous YY1 and DRBP76 associate in vivo
To determine whether YY1 interacts with DRBP76 under normal physiological conditions, we tested whether the proteins could be coimmunoprecipitated from a nuclear extract without overexpression of either protein. Indeed, a significant fraction of DRBP76 was coprecipitated by an anti-YY1 antibody, as detected via Western blot analysis with the anti-DRBP76 antibody (Fig. 2A, lane 1). In contrast, no DRBP76 was detected in anti-Flag precipitates or in a precipitate where no primary antibody was used (Fig. 2A, lanes 2,3).
Figure 2.
Interaction of endogenous YY1 and DRBP76. (A) Anti-YY1 and anti-Flag immunoprecipitates from HeLa whole-cell extracts were separated by SDS-PAGE, transferred onto a membrane, and probed with an anti-DRBP76 antibody. “Mock IP” indicates a reaction carried out identically but without the primary antibody. (B) HeLa nuclear extracts were fractionated first on a P11 phosphocellulose column, and the active fraction, as monitored by Western blots and EMSAs, was subsequently fractionated on a Q-Sepharose column. Active fractions eluted from the second column were pooled and loaded onto a nickel affinity column. Final active fractions (1–5) were pooled, separated by SDS-PAGE, and analyzed by silver staining and separately by Coomassie blue staining followed by mass spectrometry. For simplicity, the Western blots and EMSA results obtained from the initial two purification steps are not shown here.
Taking advantage of the fact that YY1, with its unusually long stretch of consecutive histidines (Shi et al. 1991), binds strongly to a nickel affinity column, we purified an endogenous native YY1 complex from HeLa cells using a simple conventional chromatography procedure (Fig. 2B). In-gel tryptic digestion of the 90-kD YY1-associated protein followed by sequencing by microcapillary HPLC ion trap mass spectrometry again revealed that the 90-kD protein was DRBP76. This result unequivocally confirms that DRBP76 partners with YY1 in vivo.
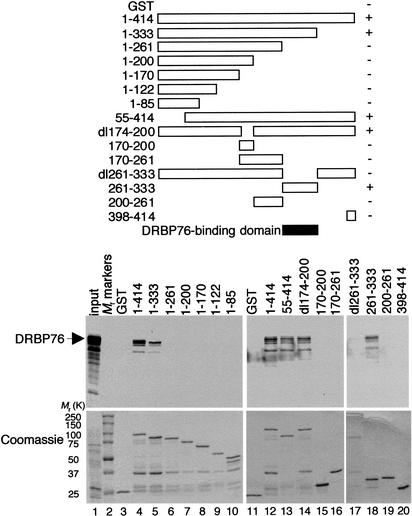
DRBP76 binds YY1 through residues 261–333 of YY1
To identify the YY1 domain that interacts with DRBP76, we tested the ability of different GST–YY1 fusion proteins to bind DRBP76. DRBP76 was captured by full-length YY1 (1–414) fused to GST, but not by the GST polypeptide alone (Fig. 3, lanes 3,4,11,12). Extensive analysis of YY1 segments indicated that DRBP76 interacted with residues 261–333 of YY1. Interestingly, this same conserved region of YY1 binds HDACs (Yao et al. 2001).
Figure 3.
Mapping of the DRBP76-binding domain in YY1. Schematic diagram of full-length or various truncated GST–YY1 fusion proteins (top panel). For simplicity, the GST portions of the fusion proteins are not shown. The ability of each GST–YY1 fusion protein to bind DRBP76 is indicated (+ or −). The shaded bar represents the deduced DRBP76-binding domain. Autoradiographs of in vitro translated DRBP76 protein captured by GST–YY1 fusion proteins are shown in the bottom panel. The input lane was loaded with one-tenth the amount of 35S-labeled proteins used in the binding reactions. The gel was stained with Coomassie blue prior to autoradiography to show approximately equal amounts of GST fusion proteins in each lane.
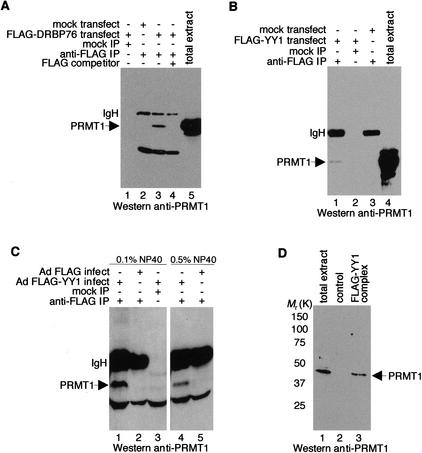
Interaction between YY1 and PRMT1
ILF3 is known to bind specifically to PRMT1 (Tang et al. 2000). Because DRBP76 is nearly identical to ILF3, we predicted that DRBP76 would also interact with PRMT1, and we sought to determine whether YY1 interacts with PRMT1, possibly through DRBP76. As expected, anti-Flag antibody coprecipitated PRMT1 from extracts of cells that expressed Flag–DRBP76 (Fig. 4A, lane 3). Intriguingly, anti-Flag antibody also specifically coprecipitated PRMT1 from extracts of cells transfected with a Flag–YY1 expression vector or infected with recombinant adenovirus that expressed Flag–YY1 (Fig. 4B, lane 1, C, lane 1). The interaction between PRMT1 and YY1 remains intact under stringent immunoprecipitation conditions, suggesting that PRMT1 binds avidly to YY1 in vivo (Fig. 4C, lane 4).
Figure 4.
YY1 binds to PRMT1. HeLa cells were either transfected with plasmids encoding the indicated proteins (A,B) or transduced with recombinant adenovirus expressing Flag–YY1, Flag, or GFP (C,D). Cell extracts were immunoprecipitated with an anti-Flag antibody (A_–_C) or purified through an anti-Flag affinity column (D) and immunoblotted with an anti-PRMT1 antibody. “Mock IP” indicates reactions carried out identically but without the anti-Flag antibody. “Flag competitor” corresponds to the addition of excess Flag peptide immunogen.
To address the likelihood that YY1–DRBP76–PRMT1 exists as a tricomplex, we performed Western blot analysis on the Flag–YY1 complex where we identified DRBP76. Although at this time we cannot rule out the possibility that the YY1–PRMT1 interaction takes place without DRBP76, the presence of PRMT1 in the purified Flag–YY1 complex (Fig. 4D, lane 3) clearly favors the idea that DRBP76 acts as a bridging protein between YY1 and PRMT1.
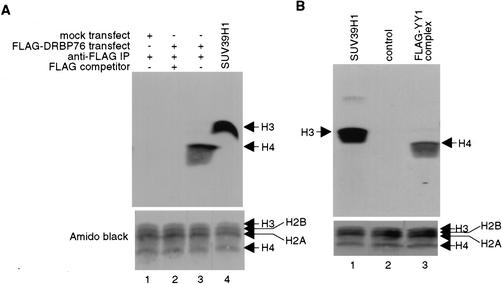
Recruitment of histone H4-specific methyltransferase activity by YY1
Because PRMT1 selectively methylates Arg 3 of histone H4 (Strahl et al. 2001; Wang et al. 2001), we determined whether the YY1–PRMT1 interaction could result in the recruitment of H4-specific HMT enzymatic activity by YY1. Immunoprecipitates from Flag–DRBP76 transfected cells were prepared using an anti-Flag antibody, and assayed for HMT activity. As shown in Figure 5A, the Flag–DRBP76 immunocomplex contains unique H4-specific HMT activity (Fig. 5A, lane 3). Similarly, a Flag–YY1 immunocomplex efficiently methylated H4, but not H2A, H2B, or H3 (Fig. 5B, lane 3), thus reinforcing the idea that YY1 specifically binds PRMT1 and recruits H4-specific HMT activity.
Figure 5.
A purified YY1 complex methylates histone H4. Anti-Flag immunoprecipitates obtained either from HeLa cells transfected with plasmids expressing Flag–DRBP76 (A) or from immunopurification of Flag–YY1 (B) were assayed for methylase activity in the presence of core histones. “Flag competitor” corresponds to the addition of excess Flag peptide immunogen. Negative controls include immunoprecipitates from mock transfected cells and anti-Flag immunopurified materials from cells transduced with adenovirus expressing the GFP. Purified recombinant SUV39H1 was used as a positive control for histone H3 methylation. Each blot was stained with Amido black to ensure proper protein transfer.
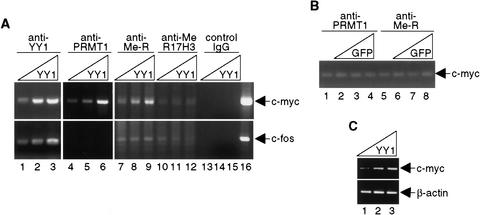
Recruitment of PRMT1 to a YY1-inducible promoter
Earlier reports suggested that methylation of Arg 3 of histone H4 by PRMT1 resulted in transcriptional activation (Strahl et al. 2001; Wang et al. 2001). We therefore hypothesized that YY1 could recruit PRMT1 to YY1-activated, but not YY1-repressed, promoters. To test this, ChIP assays were performed with the mouse c-myc promoter, which is known to be positively regulated by YY1 (Riggs et al. 1993). For comparative purposes, ChIP assays also were done using the YY1-repressed c-fos promoter (Gualberto et al. 1992; Natesan and Gilman 1993). Chromatin prepared from untreated NIH3T3 cells, or NIH3T3 cells expressing various amounts of YY1, was immunoprecipitated with anti-YY1, anti-PRMT1, or anti-methyl arginine antibodies. PCR analyses showed that PRMT1 associated with the c-myc promoter (Fig. 6A, lanes 4–6), but not the c-fos promoter. Further, the amount of PRMT1 present on the c-myc promoter was directly proportional to the amount of YY1 expressed in cells. Importantly, increased arginine methylation occurred in response to increased expression of YY1 at the c-myc, but not at the c-fos promoter (Fig. 6A, lanes 7–9). Consistent with our observations that YY1 binds PRMT1, and that the YY1-containing complex possesses H4-specific HMT activity, methylation of histone H3 at Arg 17 was not affected by YY1 abundance (Fig. 6A, lanes 10–12). Neither PRMT1 nor arginine methylation level changed at the c-myc promoter by overexpression of the GFP protein (Fig. 6B). Semiquantitative reverse transcriptase PCR (RT–PCR) analysis (Wang et al. 2002) was performed to confirm activation of c-myc by YY1 (Fig. 6C). Together, our results provide firm evidence that YY1 specifically targets histone H4 for methylation.
Figure 6.
YY1-dependent recruitment of PRMT1. (A,B) Cross-linked chromatin from NIH3T3 cells nontransduced or transduced with YY1- or GFP-expressing adenovirus was immunoprecipitated with the indicated antibodies and analyzed by PCR using primers specific for DNA surrounding YY1 sites in the c-myc and c-fos promoters. Identical results were obtained from multiple experiments. (C) Semiquantitative RT–PCR was performed to analyze expression of the c-myc gene in YY1-expressing cells. PCR products of cDNA samples are shown.
Further experimental evidence showing that YY1 recruits PRMT1, through DRBP76, to activate transcription
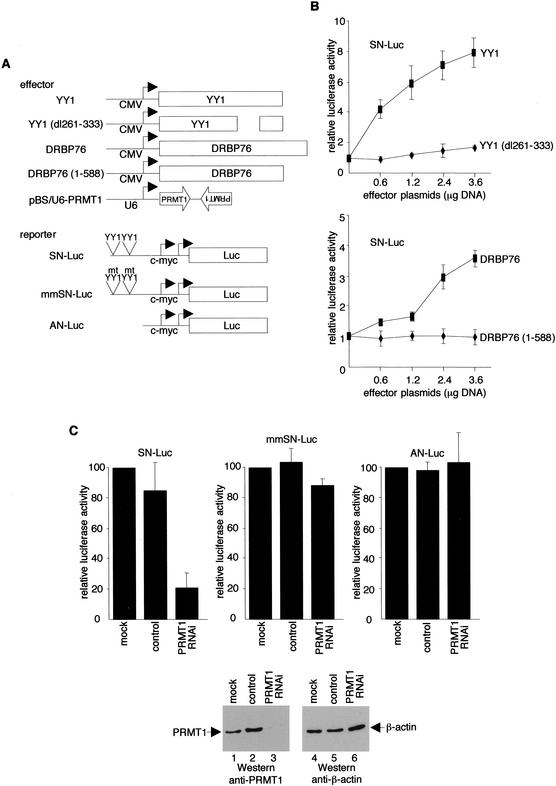
To obtain additional evidence that YY1 recruits PRMT1 via DRBP76, we performed transfection-reporter assays with a c-myc promoter luciferase construct (Fig. 7A). Consistent with earlier studies (Riggs et al. 1993), a plasmid that expresses wild-type YY1 activated the c-myc promoter in a dose-dependent fashion (Fig. 7B, top graph). Importantly, in agreement with the observation that YY1 interacts with DRBP76 through residues 261–333, the ability to activate the c-myc promoter was completely abolished by a YY1 mutant containing a deletion of residues 261–333. In a similar experiment, wild-type, but not a C-terminally truncated, DRBP76 activated the c-myc promoter (Fig. 7B, bottom graph).
Figure 7.
DRBP76- and PRMT1-dependent activation by YY1. (A) Schematic diagram of effector and reporter plasmids. (B,C) Expression plasmids and reporter plasmids were transfected together into HeLa cells as indicated. Luciferase activities are the averages ± S.D. from three separate experiments. Western blots were performed to monitor protein expression.
Finally, to prove that the ability of YY1 to bind PRMT1 correlates with activation, we employed a DNA vector-based RNAi method to suppress PRMT1 expression in HeLa cells, and determined the consequences of YY1-dependent transcription on the c-myc promoter. siRNA synthesized from the BS/U6 template (Sui et al. 2002) was targeted to PRMT1 and efficiently inhibited endogenous PRMT1, but not the control protein β-actin, with expression monitored by immunoblotting analysis (Fig. 7C, bottom, lanes 3,6). Mock transfected cells or cells transfected with the BS/U6 vector had no effect on the PRMT1 gene product (Fig. 7C, bottom, lanes 1,2). In total agreement with the observation that YY1 targets PRMT1 to activate the c-myc promoter, luciferase activity was significantly reduced from the c-myc promoter reporter (SN-Luc) in the presence of the pBS/U6–PRMT1 construct (Fig. 7C, top left panel). Expression of siRNA from the pBS/U6–PRMT1 plasmid had no effect on c-myc promoter reporters with mutated (mmSN-Luc) or deleted (AN-Luc) YY1-binding sites (Fig. 7C, top middle and right panels). Taken together, these results provide strong supporting evidence that YY1 recruits PRMT1 to activate transcription.
Discussion
YY1 is a protein with dual potential and may either activate or repress the transcription of a long list of genes (Shi et al. 1997; Thomas and Seto 1999). YY1 was the first sequence-specific DNA-binding transcription factor shown to recruit HDACs to repress transcription (Yang et al. 1996). In the study presented here, we found that YY1 recruits an additional histone-modifying enzyme, PRMT1. In this respect, YY1 is similar to the retinoblastoma (Rb) protein that not only binds HDACs but also forms a complex with HMT to target histone methylation (Brehm et al. 1998; Magnaghi-Jaulin et al. 1998; Nielsen et al. 2001; Vandel et al. 2001). Unlike YY1, Rb targets Lys 9 of histone H3 for methylation by associating with SUV39H1 and consequently repressing transcription. A recent study also showed that the transcription factor E2F-6 contributes to gene silencing by recruiting HMTase1 and G9a to methylate Lys 9 of H3 independent of Rb (Ogawa et al. 2002). In addition, HP1 recruits SUV39H1 to heterochromatin and simultaneously associates with MITR, HDAC4, and HDAC5 to repress transcription (Zhang et al. 2002). To date, YY1 is the only human sequence-specific DNA-binding transcription factor that associates with HDACs for transcriptional repression and with PRMT1 for transcriptional activation. An important question that needs to be addressed in the near future is whether recruitment of multiple histone modification enzymes by a single factor is a general feature for many sequence-specific DNA-binding transcription factors.
ILF3, with an apparent molecular mass of 110 kD, is one of several proteins that can bind to the antigen receptor response element 2 (ARRE-2) sequence (Li et al. 1992). Several double-stranded RNA-binding proteins, including NF90, MMP4, TCP80, and DRBP76, are either polymorphic variants or products of alternative splicing of the ILF3 transcript. In addition to double-stranded RNA binding, NF90 is involved in specific DNA binding (Corthesy and Kao 1994; Kao et al. 1994; Sakamoto et al. 1999) and TCP80 might regulate protein translation (Xu and Grabowski 1999). The product of the MMP4 gene, which was initially isolated as an incomplete cDNA of DRBP76, was identified as an M-phase phosphoprotein, suggesting that it may have a special function in cell division (Matsumoto-Taniura et al. 1996). In addition, DRBP76 has been reported to serve as a substrate for the interferon-induced protein kinase, PKR, and contribute to a role for PKR in cell-cycle regulation. Our finding that DRBP76, like ILF3, interacts with PRMT1 provides an additional function for this protein not previously described.
We have precisely mapped the DRBP76-interacting domain to residues 261–333 of YY1. However, we do not yet know the exact PRMT1-interacting domain within DRBP76. Earlier studies demonstrated that the C-terminal region of ILF3 (residues 622–910) is responsible for the interaction between PRMT1 and ILF3 (Tang et al. 2000). The absence of a DRBP76 C terminus corresponding to residues 701–910 of ILF3 suggests that the minimal PRMT1-interacting domain may reside within residues 622–700 of DRBP76. Our finding that a DRBP76 C-terminal deletion mutant, DRBP76 (1–588) failed to activate the c-myc promoter supports this arrangement.
It is well known that many proteins besides histone H4 can be methylated by PRMT1 (McBride and Silver 2001). Consistent with a recent report (Gabellini et al. 2002), we found that nucleolin, a protein that induces chromatin decondensation by binding to histone H1, copurified with YY1 (data not shown). Additionally, a human homolog of the yeast protein RRP5 was identified in our Flag–YY1 complex (data not shown). Like DRBP76, both nucleolin and RRP5 contain conserved RNA-binding domains, a hallmark of many PRMT1 substrates (Wada et al. 2002). In future studies, it will be important to determine whether nucleolin and RRP5 can also serve as substrates for YY1-bound PRMT1 and to determine the biological role of YY1 in these potential PRMT1 substrates.
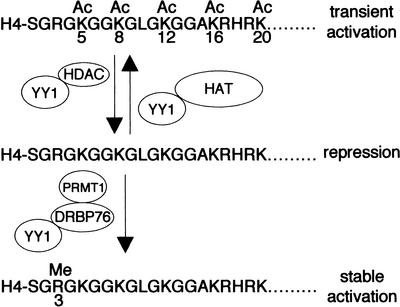
It is interesting to note that the HMT-interacting domain of YY1 is identical to one of the HDAC-interacting domains. At this time, we do not know if the interaction of YY1 with HMT and HDACs is mutually exclusive or synergistic. In vitro, the acetylated histone H4 tail is a poor substrate for PRMT1 (Wang et al. 2001). It is conceivable then, that YY1 could facilitate H4 Arg 3 methylation by targeting PRMT1 to chromatin regions undergoing active histone deacetylation (Fig. 8). Perhaps deacetylation of lysines in histone tails by YY1-bound HDACs conveniently accelerates the rate or efficiency with which YY1-targeted PRMT1 can methylate Arg 3 of H4. The challenge ahead is to determine the signal that dictates the recruitment of these two different activities by YY1.
Figure 8.
A model of gene activation and repression by YY1 through recruitment of histone-modifying enzymes.
Materials and methods
Plasmids
pBS/U6–PRMT1 was constructed by inserting an oligodeoxynucleotide corresponding to nucleotides 977–998 of PRMT1 cDNA [5′-GGCGAGGAGATCTTCGGCACCAA-3′ (forward) and 5′-AGCTTTGGTGCCGAAGATCTCCTCGCC-3′ (reverse)] into pBS/U6 (Sui et al. 2002) between the _Apa_I and _Hin_dIII sites. The second inverted sequence [5′-AGCTTTGGTGCCGAA GATCTCCTCGCCCTTTTTG-3′ (forward) and 5′-AATTCAAA AAGGGCGAGGAGATCTTCGGCACCAA-3′ (reverse)] then was inserted into the intermediate plasmid between the _Hin_dIII and _Eco_RI sites to generate the final product. Details of all other plasmid constructions are available upon request.
Immunochemical reagents and techniques
Anti-Flag and anti-β-actin antibodies were obtained from Sigma Biochemical. Anti-DRBP76 and anti-YY1 antibodies were obtained from BD Biosciences and Santa Cruz Biotechnology, respectively. Anti-PRMT1, anti-methyl arginine, and anti-R17H3 antibodies were obtained from Abcam. Immunoprecipitations were performed in a solution of PBS containing NP-40 and protease inhibitors, as described (Laherty et al. 1997). Immunocomplexes were washed six times with the same buffer, and immunoprecipitated proteins were removed from protein A beads by boiling in gel loading buffer. For Western blot analyses, proteins were resolved on SDS–polyacrylamide gels and transferred to nitrocellulose membranes. After blocking with nonfat dried milk, the membranes were probed with antibodies and developed with the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Purification of YY1 complexes
A nuclear extract was prepared from HeLa cells transduced with recombinant adenovirus expressing Flag–YY1. Affinity purification of the Flag–YY1 complex with anti-Flag antibody was performed according to the protocol used for purification of a Flag–PCAF complex (Ogryzko et al. 1998). Purified samples were concentrated, resolved by SDS-PAGE, and analyzed by silver staining. A Coomassie blue-stained sample was prepared in parallel and the band corresponding to p90 was excised; samples were sequenced by peptide mass spectrometry. For purification of an endogenous YY1 complex, a HeLa cell nuclear extract was prepared and fractionated on 20 mL of P11 phosphocellulose (Whatman) using a 0.1–1 M KCl step gradient in a buffer containing 20 mM Tris-HCl (pH 8.0), 4 mM EDTA, 10% glycerol, 1 mM PMSF, and 0.5 mM DTT. Fractions containing active YY1 were loaded onto a Q Sepharose column (Pharmacia), washed, and eluted using a 0.1–1 M linear gradient. Active fractions from this second column were combined and loaded onto a Ni-NTA resin column (Novagen). The final column was washed with 50 mM imidazole, and the YY1 complex was eluted with 300 mM imidazole.
Electrophoretic mobility shift assays (EMSAs)
Single-stranded oligodeoxynucleotides corresponding to a consensus YY1-binding site (5′-AGGGTCTCCATTTTGAAGC-3′ and its complement) were labeled individually with γ32P-ATP and T4 polynucleotide kinase, heated together to 65°C, and allowed to anneal by slow cooling to room temperature. Each 12 μL reaction mixture contained 12 mM Hepes (pH 7.9), 10% glycerol, 5 mM MgCl2, 60 mM KCl, 1 mM DTT, 0.5 mM EDTA, 50 μg/mL bovine serum albumin, 0.05% NP-40, 0.1 μg poly(dIdC), purified proteins, and 5 fmole radiolabeled DNA. Reactions were incubated for 10 min at room temperature, separated on 4% nondenaturing polyacrylamide gels, dried, and autoradiographed.
GST pull-down assays
GST, GST–YY1, and GST–YY1 deletion mutants were expressed and purified as described (Yang et al. 1997). 35S-DRBP76 was prepared using the coupled transcription-translation rabbit reticulocyte lysate system (Promega). Equimolar quantities of GST or GST–YY1 conjugated to glutathione-Sepharose beads were incubated with radiolabeled DRBP76. Binding reactions, washing conditions, electrophoretic analysis, and subsequent autoradiography were performed as previously described (Yang et al. 1997).
Methyltransferase assay
For each reaction, 4 μg of chicken core histones were incubated with 0.55 μCi of _S_-adenosyl-L-[methyl-3H]methionine (Perkin Elmer) in MTase buffer (50 mM Tris at pH 8.0) for 1 h at 30°C in a final volume of 30 μL. The reactions were terminated by the addition of SDS gel loading buffer followed by heating for 10 min at 95°C. Samples were separated by SDS-PAGE and transferred onto PVDF membranes. Membranes were exposed to X-ray film in the presence of a Kodak transcreen LE.
ChIP assay
ChIP assays were performed essentially as described (Weinmann et al. 2001). Briefly, mouse NIH3T3 cells were treated with formaldehyde, and chromatin was purified on CsCl-equilibrium gradients. After ultracentrifugation, DNA containing fractions were dialyzed and kept frozen at −80°C in aliquots. Equal amounts of purified chromatin were incubated overnight with different antibodies and collected on protein A beads. Cross-linked products were reversed by heating overnight at 65°C, and the immunoprecipitated DNA was purified by proteinase K treatment. Primers for PCR amplification of the c-myc promoter DNA were CTCATTCGTTCGTCCTTCCCC CTTTC and CTTCTTCTCGTTTCCCCGCCCTCTGC. For the c-fos promoter, primers were CCGTCAATCCCTCCCTC CTTTAC and CGCCTCAGCTGGCCGTTTATAG. Identities of the amplified DNA products were verified by subcloning and DNA sequencing.
Transfection and luciferase assay
HeLa cells were transfected with different combinations of plasmids directing the synthesis of various effector proteins or siRNA plus a luciferase reporter using the Fugene 6 transfection reagent (Roche). All transfections were normalized to equal amounts of DNA with parental expression vectors. Forty-eight hours later, cells were collected and luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega).
Acknowledgments
We thank Kathryn Calame, Nathalie Duchange, Harvey Herschman, Thomas Jenuwein, Ganes Sen, and Yang Shi for plasmids; Bill Lane and his colleagues at the Harvard Microsequencing Facility for their assistance with protein microsequencing; Nancy Olashaw and Xiang-Jiao Yang for discussion and critical reading of the manuscript; and the Moffitt Cancer Center Core Facility for technical support. This work was supported by grants from the NIH (GM64850, GM58486) to E.S.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL setoe@moffitt.usf.edu; FAX (813) 979-7264.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1068003.
References
- Berger SL. An embarrassment of niches: The many covalent modifications of histones in transcriptional regulation. Oncogene. 2001;20:3007–3013. doi: 10.1038/sj.onc.1204324. [DOI] [PubMed] [Google Scholar]
- ————— Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature. 1998;391:597–601. doi: 10.1038/35404. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- Corthesy B, Kao PN. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J Biol Chem. 1994;269:20682–20690. [PubMed] [Google Scholar]
- Duchange N, Pidoux J, Camus E, Sauvaget D. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene. 2000;261:345–353. doi: 10.1016/s0378-1119(00)00495-9. [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–348. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Gualberto A, LePage D, Pons G, Mader SL, Park K, Atchison ML, Walsh K. Functional antagonism between YY1 and the serum response factor. Mol Cell Biol. 1992;12:4209–4214. doi: 10.1128/mcb.12.9.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kao PN, Chen L, Brock G, Ng J, Kenny J, Smith AJ, Corthesy B. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269:20691–20699. [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Laherty CD, Yang WM, Sun JM, Davie JR, Seto E, Eisenman RN. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell. 1997;89:349–356. doi: 10.1016/s0092-8674(00)80215-9. [DOI] [PubMed] [Google Scholar]
- Li C, Lusis AJ, Sparkes R, Tran SM, Gaynor R. Characterization and chromosomal mapping of the gene encoding the cellular DNA binding protein HTLF. Genomics. 1992;13:658–664. doi: 10.1016/0888-7543(92)90138-i. [DOI] [PubMed] [Google Scholar]
- Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, HarelBellan A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature. 1998;391:601–605. doi: 10.1038/35410. [DOI] [PubMed] [Google Scholar]
- Matsumoto-Taniura N, Pirollet F, Monroe R, Gerace L, Westendorf JM. Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell. 1996;7:1455–1469. doi: 10.1091/mbc.7.9.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride AE, Silver PA. State of the Arg: Protein methylation at arginine comes of age. Cell. 2001;106:5–8. doi: 10.1016/s0092-8674(01)00423-8. [DOI] [PubMed] [Google Scholar]
- Natesan S, Gilman MZ. DNA bending and orientation-dependent function of YY1 in the c-fos promoter. Genes & Dev. 1993;7:2497–2509. doi: 10.1101/gad.7.12b.2497. [DOI] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Kotani T, Zhang X, Schiltz RL, Howard T, Yang XJ, Howard BH, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- Patel RC, Vestal DJ, Xu Z, Bandyopadhyay S, Guo W, Erme SM, Williams BRG, Sen GC. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J Biol Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- Riggs KJ, Saleque S, Wong KK, Merrell KT, Lee JS, Shi Y, Calame K. Yin-yang 1 activates the c-myc promoter. Mol Cell Biol. 1993;13:7487–7495. doi: 10.1128/mcb.13.12.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Morisawa K, Ota K, Nie J, Taniguchi T. A binding protein to the DNase I hypersensitive site II in HLA-DR α gene was identified as NF90. Biochemistry. 1999;38:3355–3361. doi: 10.1021/bi982099g. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are trimethylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, et al. Methylation of histone H4 at arginine 3 occurs in vivo and is mediated by the nuclear receptor coactivator PRMT1. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- Sui G, Soohoo C, Affar EB, Gay F, Shi Y, Forrester WC, Shi Y. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Kao PN, Herschman HR. Protein-arginine methyltransferase I, the predominant protein-arginine methyltransferase in cells, interacts with and is regulated by interleukin enhancer-binding factor 3. J Biol Chem. 2000;275:19866–19876. doi: 10.1074/jbc.M000023200. [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: Are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Inoue K, Hagiwara M. Identification of methylated proteins by protein arginine N-methyltransferase 1, PRMT1, with a new expression cloning strategy. Biochim Biophys Acta. 2002;1591:1–10. doi: 10.1016/s0167-4889(02)00202-1. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang ZQ, Xia L, Feng Q, Erdjument-Bromage H, Strahl BD, Briggs SD, Allis CD, Wong J, Tempst P, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293:853–857. doi: 10.1126/science.1060781. [DOI] [PubMed] [Google Scholar]
- Wang HL, Wang J, Xiao SY, Haydon R, Stoiber D, He TC, Bissonnette M, Hart J. Elevated protein expression of cyclin D1 and Fra-1 but decreased expression of c-myc in human colorectal adenocarcinomas overexpression β-catenin. Int J Cancer. 2002;101:301–310. doi: 10.1002/ijc.10630. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Bartley SM, Zhang T, Zhang MQ, Farnham PJ. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol Cell Biol. 2001;21:6820–6832. doi: 10.1128/MCB.21.20.6820-6832.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YH, Grabowski GA. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid β-glucosidase and other mRNAs. Mol Genet Metab. 1999;68:441–454. doi: 10.1006/mgme.1999.2934. [DOI] [PubMed] [Google Scholar]
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CL, McKinsey TA, Olson EN. Association of class II histone deacetylases with heterochromatin protein 1: Potential role for histone methylation in control of muscle differentiation. Mol Cell Biol. 2002;22:7302–7312. doi: 10.1128/MCB.22.20.7302-7312.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Reinberg D. Transcription regulation by histone methylation: Interplay between different covalent modifications of the core histone tails. Genes & Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]