A Functional Relationship between NuMA and Kid Is Involved in Both Spindle Organization and Chromosome Alignment in Vertebrate Cells (original) (raw)
Abstract
We examined spindle morphology and chromosome alignment in vertebrate cells after simultaneous perturbation of the chromokinesin Kid and either NuMA, CENP-E, or HSET. Spindle morphology and chromosome alignment after simultaneous perturbation of Kid and either HSET or CENP-E were no different from when either HSET or CENP-E was perturbed alone. However, short bipolar spindles with organized poles formed after perturbation of both Kid and NuMA in stark contrast to splayed spindle poles observed after perturbation of NuMA alone. Spindles were disorganized if Kid, NuMA, and HSET were perturbed, indicating that HSET is sufficient for spindle organization in the absence of Kid and NuMA function. In addition, chromosomes failed to align efficiently at the spindle equator after simultaneous perturbation of Kid and NuMA despite appropriate kinetochore-microtubule interactions that generated chromosome movement at normal velocities. These data indicate that a functional relationship between the chromokinesin Kid and the spindle pole organizing protein NuMA influences spindle morphology, and we propose that this occurs because NuMA forms functional linkages between kinetochore and nonkinetochore microtubules at spindle poles. In addition, these data show that both Kid and NuMA contribute to chromosome alignment in mammalian cells.
INTRODUCTION
The mitotic spindle is a microtubule-based structure responsible for accurate chromosome segregation during cell division (Hyman and Karsenti, 1996; Compton, 2000; Sharp et al., 2000a). Chromosome attachment to and movement on the spindle have been extensively documented and involve both poleward and away from the pole forces (Mitchison, 1989a; Gorbsky, 1992; Rieder and Salmon 1994, 1998; Khodjakov et al., 1999). Before sister chromatid separation, these antagonistically acting forces cause alignment of chromosomes at the spindle equator, a conspicuous event that defines the metaphase stage of mitosis. Unfortunately, our understanding of the mechanisms driving chromosome alignment is limited because it is currently unknown how forces driving chromosome alignment are generated or how chromosomes sense their position on the spindle (Mitchison, 1989a; Kapoor and Compton, 2002).
One model for chromosome alignment is based on the oscillatory movements displayed by both mono- and bioriented chromosomes in many cell types (Bajer, 1982; Skibbens et al., 1993; Khodjakov et al. 1999). This model relies on the polar ejection force, a force acting on chromosome arms to push chromosomes away from spindle poles (Rieder et al., 1986), to limit poleward chromosome movement driven by kinetochore activity (Skibbens et al., 1993; Rieder and Salmon, 1994). The polar ejection force antagonizes poleward chromosome movement, creating tension that causes attached kinetochores on mono-oriented chromosomes to shift into neutral permitting the polar ejection force to push the chromosome away from the pole. Likewise, leading kinetochores on bioriented chromosomes experience tension created by antagonism from both the sister kinetochore and polar ejection force which causes the leading kinetochore to shift into neutral allowing the sister kinetochore and polar ejection force to move the chromosome away from the pole. The consequence of this interaction of antagonistically acting forces is that the duration of chromosome movement away from the pole exceeds movement toward the pole, which favors steady-state positioning of bioriented chromosomes at the spindle equator (Skibbens et al., 1993).
Recent experiments demonstrate that the kinesin-related protein Kid generates a significant fraction of polar ejection force (Tokai et al., 1996; Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001). Perturbation of Kid activity caused chromosome arms to project toward spindle poles instead of perpendicular to the spindle axis (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001) and permitted chromosomes to be dragged into the polar region of cells with monopolar spindles (Levesque and Compton, 2001). Perturbation of Kid function in cultured somatic cells also suppressed oscillatory chromosome movement, however, all chromosomes aligned efficiently at the spindle equator in >80% of cells that progressed to anaphase and all but one or a few chromosomes aligned efficiently in the <20% of cells that did not enter anaphase. These data reveal the existence of mechanisms for directing chromosome alignment to the spindle equator that act independently of Kid activity and chromosome oscillation. Herein, we examine the contribution of other spindle-associated proteins to chromosome alignment in mitosis. We find that simultaneous perturbation of Kid and NuMA significantly impaired chromosome alignment and affected both spindle morphology and size. These results indicate that both Kid and NuMA contribute to chromosome alignment and suggest that the functional linkage of kinetochore and nonkinetochore microtubules at spindle poles is important for several aspects of spindle morphology.
MATERIALS AND METHODS
Cell Culture
The human CFPAC-1 cell line was maintained in Iscove's modified DME containing 10% fetal calf serum, 50 IU/ml penicillin, and 50 μg/ml streptomycin. Cells were grown at 37°C in a humidified incubator with a 5% CO2 atmosphere.
Antibodies
Mad2-specific antibodies were prepared by immunizing rats with the C-terminal 203 amino acids of the Mad2 protein. A segment of the human Mad2 cDNA (GenBank accession no. BE311763) encoding the COOH-terminal 203 amino acids of Mad2 was inserted into the pRSET-C vector in the appropriate reading frame to generate a 6His-Mad2 fusion protein. The 6His-Mad2 fusion protein was purified on nickel-agarose and injected into rats (Covance Research, Richmond, CA). The resulting serum was specific for Mad2 as judged by immunoblot and immunofluorescence microscopy.
Tubulin was detected with the DM1α mouse monoclonal antibody (Sigma-Aldrich, St. Louis, MO). Kinetochores were detected with a human anticentromere antibody (ACA-m) provided by Kevin Sullivan (Scripps Research Institute, San Diego, CA). Centrosomes were detected with a human anti-centrosome antibody provided by J.B. Rattner (University of Calgary, Calgary, Alberta, Canada). CENP-E was detected with a rabbit polyclonal antibody provided by Tim Yen (Fox Chase Cancer Center, Philadelphia, PA). NuMA-, HSET-, and Kid-specific antibodies used for injections were as described previously (Gaglio et al., 1995; Mountain et al., 1999; Levesque and Compton, 2001). For immunofluorescence staining, NuMA was detected using a human NuMA-specific antibody provided by D. Pettijohn (University of Colorado, Boulder, CO). Kid was detected using a rabbit polyclonal antibody raised against 91-amino acid segment of the Kid protein that spans the neck region (Vale and Fletterick, 1997). The region of the Kid cDNA (GenBank accession no. R56446) encoding the neck domain was polymerase chain reaction amplified using the forward primer (CGGGATCCCGGCCCCTGAGAGACGCTTCTACC) containing a _Bam_HI site and the reverse primer (GGAATTCCGTCCATGCTGCTTAGCTTCTGTAGG) containing an _Eco_RI site. The polymerase chain reaction product was inserted into the _Bam_HI and _Eco_RI sites of the pGEX-5X-1 vector, which results in the in-frame fusion of glutathione _S_-transferase and amino acids 346–436 of Kid. The recombinant protein was expressed in BL21 cells by induction with 1 mM isopropyl β-d-thiogalactoside and purified using glutathione-Sepharose 4B (Amersham Biosciences, Piscataway, NJ). The column eluate was dialyzed against phosphate-buffered saline and used to immunize two rabbits. The resulting serum was specific for Kid as judged by immunofluorescence and immunoblot.
Antibody Microinjection and Indirect Immunofluorescence Microscopy
Kid-, NuMA-, CENP-E-, and HSET-specific IgGs were purified from whole rabbit serum by affinity chromatography using protein A-conjugated agarose (Roche Diagnostics, Indianapolis, IN). IgG fractions were exchanged into antibody microinjection buffer (100 mM KCl, 10 mM KPO4, pH 7.0) by gel filtration using PD-10 Sepharose (Amersham Biosciences) and concentrated to 3 (anti-CENP-E), 10 (anti-HSET), or 20 (anti-NuMA, anti-Kid) mg/ml. Human CFPAC-1 cells were grown on photo-etched alphanumeric glass coverslips (Bellco Glass, Vineland, NJ). Antibodies were injected into the cell nucleus during G1 and analyzed in the subsequent mitosis as described previously (Levesque and Compton, 2001).
Indirect immunofluorescence microscopy was performed as described previously (Gordon et al., 2001; Levesque and Compton, 2001). Fluorescent images were captured with a Hamamatsu Orca II cooled charge-coupled device camera mounted on an Axioplan 2 microscope (Carl Zeiss, Thornwood, NY) equipped for epifluorescence. A series of 0.5-μm optical sections were collected in the z-plane for each channel (4,6-diamidino-2-phenylindole [DAPI], fluorescein, and/or Texas Red) and deconvolved using the Openlab software (Improvision, Boston, MA) to eliminate extraneous fluorescence background. Selected planes from the deconvolved z-series were overlayed to generate the final image.
Pixel intensities of Kid staining were measured from selected focal planes from z-series through control and injected cells taken from the same coverslip at the same exposures. Region of interest boxes of equal size were drawn at the spindle pole, metaphase plate, and in a blank region outside the cell. Average pixel intensities were determined using the area measurements tool in the Openlab software package.
Calcium-stable Kinetochore Fibers
Methods for calcium treatment have been described previously (Mitchison et al., 1986; Kapoor et al., 2000). Cells were permeabilized in a calcium-containing buffer (100 mM PIPES, pH 6.8, 1 mM MgCl2, 1 mM CaCl2, 0.5% Triton X-100) for 5 min at room temperature followed by fixation in the same buffer containing 1% glutaraldehyde for 10 min at room temperature. The cells were processed for immunofluorescence and fluorescent images were captured as described above, except a series of 0.2-μm optical sections were collected in the z-plane for the DAPI and fluorescein isothiocyanate channels. The deconvolved tubulin images were used to measure kinetochore fiber lengths. Kinetochore fibers were measured as pairs (both kinetochore fibers attached to an individual chromosome) that were distinguished based on proximity of plus-ends. Only kinetochore fiber pairs for which microtubule plus- and minus-ends of both kinetochore fibers were clearly delineated were measured. Lengths were determined using the straight line calibration tool in the Openlab software package.
Time-Lapse Microscopy
Methods for time-lapse videomicroscopy and chromosome velocity measurements were described previously (Gordon et al., 2001; Levesque and Compton, 2001). Chromosome velocities were obtained from the digital time-lapse record of each cell. The microscopy system used for time-lapse recordings was calibrated using a stage micrometer under the same conditions used for image acquisition. Individual chromosome movement was tracked by frame-by-frame analysis of digital images using Openlab software (Improvision). The straight line calibration tool in the Openlab software package was used to determine the distance traveled by an individual chromosome at the point of its centromere between different time points. Velocities were then calculated by dividing the total distance traveled (in micrometers) by the time interval in which the measurements were made (in minutes). In most cells, the spindle equator was used as a frame of reference and was assigned as the position where a bulk of the chromosomes were positioned. In other cells, the cell midline was used as a frame of reference. Chromosomes were judged to be making directed movements when the chromosome was displaced by ≥2 μm in a linear manner. Displacement of this magnitude is easily distinguishable from Brownian motion (Alexander and Rieder, 1991).
Measurements of metaphase plate width and spindle length were made using the straight line calibration tool in the Openlab software package as described above. Spindle length was measured from centrosome to centrosome. The width of the metaphase plate was measured across the chromosome mass at three different locations in each cell in multiple focal planes.
Transmission Electron Microscopy
CFPAC-1 cells were grown on alphanumeric glass coverslips and were injected as described above. The position of mitotic cells was determined by phase-contrast microscopy and noted for subsequent examination by electron microscopy. Cells were incubated in MTSB (4 mM glycerol, 100 mM PIPES, pH 6.9, 1 mM EGTA, and 5 mM MgCl2) for 1 min, extracted in MTSB + 2% Triton X-100 for 5 min, and washed in MTSB for 2 min at room temperature. Cells were fixed in 1% glutaraldehyde in 0.1 M Na-cacodylate buffer overnight. After fixation, cells were rinsed twice with cacodylate buffer for 30 min at room temperature, and en bloc stained in 2% uranyl acetate for 30 min at room temperature. Cells were dehydrated through a graded series of ethanols and propylene oxide, and flat embedded in epon (LX112)/araldite(502). The glass coverslip was removed by etching in cold concentrated hydrofluoric acid, as described by Moore (1975) and Rieder and Bowser (1987). The areas containing the injected cells or uninjected control cells were identified under a dissecting microscope, cut out of the flat embedded rectangle, and remounted onto epoxy blanks. Sections (90–200 nm) were stained with 2% aqueous uranyl acetate for 45 min at 50°C and with Reynold's lead citrate for 20 min at room temperature. Electron micrographs were taken at 80 for 100 kV on a JEOL (Tokyo, Japan) 100CX.
Online Supplemental Material
Supplemental video material is available at www.molbiolcell.org. Time-lapse videomicroscopy reveals chromosome movement at normal velocities in a human CFPAC-1 cell injected with antibodies to both Kid and NuMA. These and other data show that chromosomes interact with spindle microtubules appropriately after injection with antibodies to both Kid and NuMA; however, they do not align at the spindle equator.
RESULTS
Chromosome Alignment Requires NuMA and Kid Activities
During metaphase in human CFPAC-1 cells, chromosomes aligned at the spindle equator with the metaphase plate spanning ∼8 μm on spindles averaging 35 μm in length from pole to pole (Figure 1A and Table 1). Injection of antibodies against Kid, the chromokinesin responsible for generating a majority of the polar ejection force, did not alter spindle size or chromosome alignment at the spindle equator, but it caused an increase in the width of the metaphase plate of ∼10% (Figure 1B and Table 1). Inhibition of Kid activity caused chromosome arms to project toward spindle poles in both human cells and frog egg extracts (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001), which accounts for the 10% increase in the width of the metaphase plate.
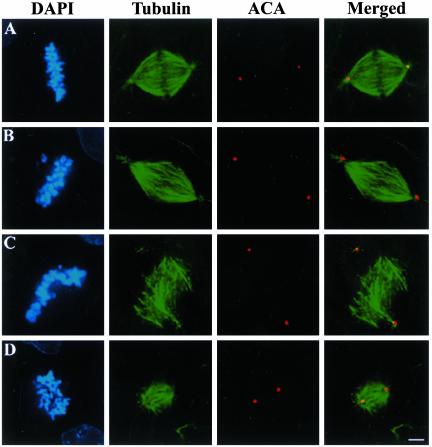
Figure 1.
Chromosome alignment and spindle organization in cells injected with antibodies to NuMA and Kid. Uninjected CFPAC-1 cells (A) and cells microinjected with either Kid-specific antibodies alone (B), NuMA-specific antibodies alone (C), or both NuMA- and Kid-specific antibodies (D) were stained with tubulin-specific antibodies, human anti-centrosome antibodies (ACA), and the DNA-specific dye DAPI as indicated. Bar, 10 μm.
Table 1.
Spindle characteristics
| Treatment | Pole to pole distance, μm (n)a | Metaphase plate width, μm (n)b | Chromosome velocities, μm/min (n)c |
|---|---|---|---|
| Uninjected | 34.0 ± 4.9 (18) | 8.1 ± 1.7 (18) | 3.8 ± 1.2 (131) |
| α-Kid injected | 35.5 ± 5.7 (11) | 9.0 ± 1.7 (11) | 3.6 ± 1.2 (80) |
| α-NuMA injected | N/A | 8.2 ± 2.4 (17) | 3.6 ± 1.0 (101) |
| α-Kid, α-HSET injected | 32.9 ± 3.7 (13) | 8.8 ± 1.5 (13) | 3.7 ± 1.2 (41) |
| α-Kid, α-NuMA injected | 19.7 ± 3.7 (17) | 14.3 ± 2.7 (17) | 3.5 ± 1.0 (56) |
| α-Kid, α-NuMA, α-HSET injected | N/A | 20.9 ± 6.5 (23) | 2.7 ± 0.9 (53) |
To identify other spindle-associated proteins that participate in chromosome alignment, we simultaneously injected cells with antibodies to Kid and various other spindle-associated proteins. We reasoned that defects in chromosome alignment may only become evident if Kid and any compensating protein were perturbed simultaneously. We initiated these experiments by microinjection of antibodies against the spindle pole organizing protein NuMA. We previously showed that injection of antibodies to NuMA caused the dislocation of centrosomes from the spindle and splaying of microtubule minus ends (Gaglio et al., 1995; Gordon et al., 2001). Despite such gross spindle abnormalities, chromosomes displayed typical oscillatory movement at normal velocities (Gordon et al., 2001) and aligned in the middle of the microtubule array (Figure 1C and Table 1). In contrast, chromosomes displayed a significantly broader distribution on the spindle after simultaneous injection of antibodies to Kid and NuMA (Figure 1D and Table 1). Chromosomes failed to align despite the organization of bipolar spindles and fixation of cells (for immunofluorescence microscopy) >2 h after nuclear envelope breakdown. Chromosomes were distributed >70% of the spindle in cells injected with antibodies to both Kid and NuMA in contrast to control cells or cells injected with Kid antibodies alone, which had chromosomes distributed over ∼23–25% of the spindle. We used three-dimensional image reconstruction to examine these spindles in all possible orientations, which eliminated the possibility that chromosomes looked misaligned as a consequence of viewing the spindle at a skewed angle.
To verify that both Kid and NuMA were perturbed by the simultaneous injection of both antibodies we examined the localization of each protein after single and double antibody injection (Figure 2). In uninjected control cells, NuMA associates with spindle microtubules at the polar ends of the spindle in a characteristic crescent-shaped pattern (Figure 2A), and Kid associates with chromosomes and spindle microtubules (Figure 2D). After injection of either NuMA antibodies alone (Figure 2B) or both Kid and NuMA antibodies (Figure 2C) NuMA was observed in aggregates and not associated with spindle microtubules in a crescent-like pattern. As we showed previously (Gaglio et al., 1995; Gordon et al., 2001), the perturbation of NuMA alone by antibody injection resulted in spindles that lack tightly focused poles with the minus ends of numerous microtubule bundles lacking any detectable NuMA (Figure 2B, arrowheads). Moreover, injection of the antibody against the DNA binding domain of Kid prevented it from localizing to chromosomes but not spindle microtubules when injected alone (Figure 2E) or in combination with NuMA antibodies (Figure 2F). Consistent with our previous data (Levesque and Compton, 2001), the injection of Kid antibodies reduced the average pixel intensity for Kid staining on chromosomes 3- to 10-fold relative to control cells, but had no detectable effect on the average pixel intensity for Kid staining at spindle poles relative to control cells. Thus, Kid and NuMA were perturbed by the simultaneous injection of antibodies to both proteins in a manner indistinguishable from the injection of each antibody alone.
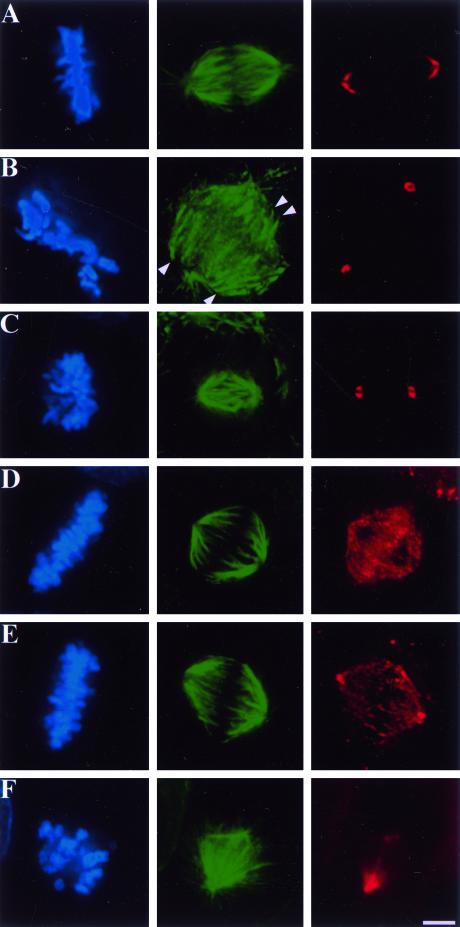
Figure 2.
Localization of both Kid and NuMA is disrupted in cells injected with both Kid- and NuMA-specific antibodies. Uninjected CFPAC-1 cells (A and D) and cells injected with NuMA- (B), Kid- (E), or both NuMA- and Kid-specific antibodies (C and F) were stained with the DNA-specific dye DAPI (blue), tubulin-specific antibodies (green), NuMA-specific antibodies (red, A–C), and Kid-specific antibodies (red, D–F). Arrowheads in B indicate minus ends of spindle microtubules. Bar, 10 μm.
To test whether the lack of efficient chromosome alignment was specific to the perturbation of both Kid and NuMA, we simultaneously injected antibodies to Kid and either HSET or CENP-E. HSET is a kinesin-related protein that cooperates with NuMA and cytoplasmic dynein in organizing microtubule minus ends at spindle poles (Walczak et al., 1998; Mountain et al., 1999; Gordon et al., 2001). We previously demonstrated that injection of HSET-specific antibodies perturbed HSET function, which disrupted spindle pole organization in the absence of either centrosomes or NuMA activity (Mountain et al., 1999; Gordon et al., 2001). However, perturbation of HSET alone did not significantly alter spindle organization, chromosome movement and alignment, or anaphase onset (Mountain et al., 1999; Gordon et al., 2001). Similarly, simultaneous injection of antibodies to Kid and HSET had no detectable effect on chromosome movement and alignment or spindle organization although the metaphase plate was ∼10% broader than control cells similar to when cells were injected with Kid antibodies alone (Figure 3F and Table 1). We also injected cells with antibodies to both Kid and CENP-E, a kinetochore-associated kinesin-related protein (Yen et al., 1991). Perturbation of CENP-E function by antibody injection does not alter spindle organization, but delays anaphase onset and blocks the alignment of one or a few chromosomes without altering the alignment or extent and velocity of movement of all other chromosomes (Schaar et al., 1997; McEwen et al., 2001). Cells injected with antibodies to both Kid and CENP-E had normal bipolar spindles (our unpublished data), but were delayed in anaphase onset and displayed a metaphase-like alignment of most chromosomes although one or a few chromosomes were located very close to the spindle pole (Figure 4B). This arrangement of chromosomes was not significantly different from cells injected with CENP-E antibodies alone (Figure 4A). Thus, simultaneous perturbation of Kid and either HSET or CENP-E did not significantly alter chromosome alignment, which demonstrates that the lack of chromosome alignment induced by perturbation of both Kid and NuMA is a specific effect related to the activities of those two proteins.
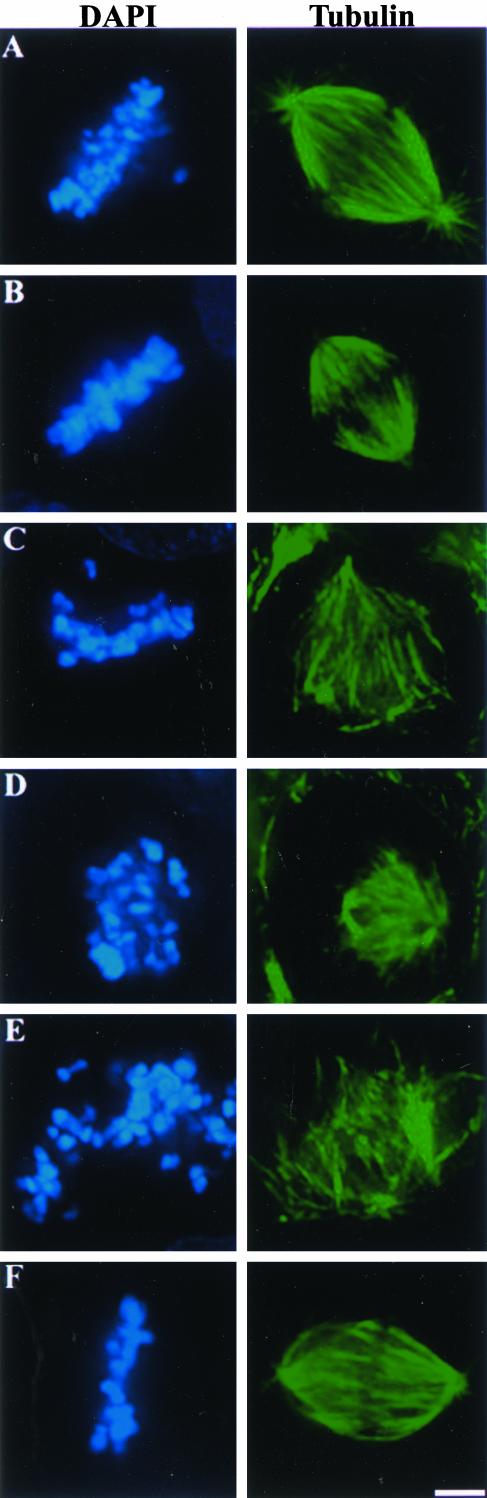
Figure 3.
HSET stabilizes spindle poles after perturbation of NuMA and Kid. Uninjected CFPAC-1 cells (A) or cells injected with either Kid- (B), NuMA- (C), NuMA- and Kid- (D), NuMA-, Kid-, and HSET- (E), or Kid- and HSET-specific antibodies (F) were stained with tubulin-specific antibodies and the DNA-specific dye DAPI as indicated. Bar, 10 μm.
Figure 4.
CENP-E and Kid do not cooperate to direct chromosome alignment. CFPAC-1 cells injected with either CENP-E- (A) or Kid- and CENP-E-specific antibodies (B) were stained with the DNA-specific dye DAPI to visualize chromosome arrangements in mitosis. Bar, 10 μm.
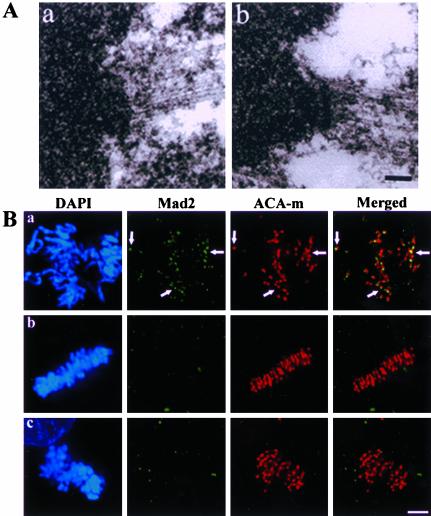
One explanation for the lack of chromosome alignment on spindles in cells injected with antibodies to both Kid and NuMA is that chromosomes failed to interact appropriately with spindle microtubules. We evaluated that possibility using four assays. First, we examined injected cells during mitosis by electron microscopy and observed microtubules interacting with kinetochores in a manner similar to control cells (Figure 5A). Second, we stained cells for the checkpoint protein Mad2 as a marker for microtubule attachment to kinetochores (Hoffman et al., 2001). Whereas control prometaphase cells had significant numbers of Mad2-positive kinetochores, both control metaphase cells and cells injected with antibodies to both Kid and NuMA displayed few, if any, detectable Mad2-positive kinetochores (Figure 5B). Third, we measured interkinetochore distances as a readout of bioriented poleward force production and observed no significant difference between the average interkinetochore distances in cells injected with antibodies to both Kid and NuMA (2.7 ± 0.5 μm, n = 183) and uninjected control metaphase cells (2.8 ± 0.5 μm, n = 100)
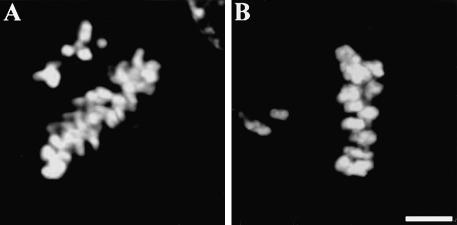
Figure 5.
Kinetochore-microtubule interactions in cells injected with NuMA- and Kid-specific antibodies. (A) Kinetochores of uninjected CFPAC-1 cells (a) and cells injected with both NuMA- and Kid-specific antibodies (b) were visualized by transmission electron microscopy. Bar, 200 nm. (B) Uninjected prometaphase (a) and metaphase (b) cells along with cells injected with both NuMA- and Kid-specific antibodies (c) were stained with rat Mad2-specific antibodies (Mad2), human anti-centromere antibodies (ACA-m), and DAPI as indicated. Arrows highlight Mad2-positive kinetochores. Bar, 10 μm.
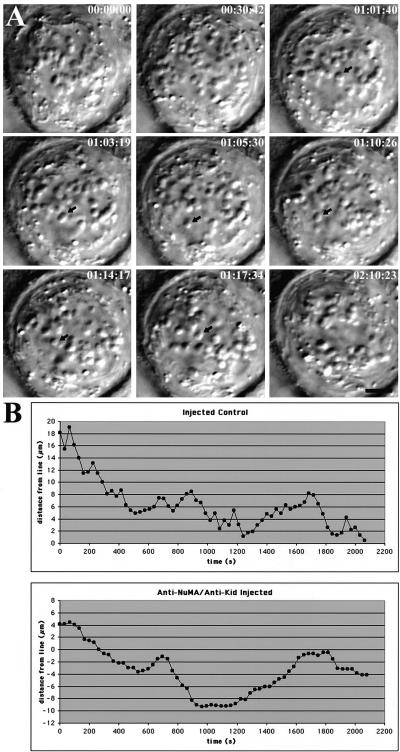
Finally, we monitored chromosome movement by time-lapse differential interference contrast microscopy in cells injected with antibodies to both Kid and NuMA (Figure 6). We previously showed that individual chromosomes in cells injected with antibodies to NuMA alone displayed directed movement at normal velocities with typical oscillatory movement (Gordon et al., 2001). We also showed that individual chromosomes in cells injected with Kid antibodies alone displayed directed movements to the spindle equator at velocities equivalent to control mitotic cells, although chromosomes remained at the metaphase plate and did not undergo detectable oscillatory movements (Levesque and Compton, 2001). Individual chromosomes in cells injected with antibodies to both Kid and NuMA also displayed directed movement traversing distances up to ∼12.5 μm with average velocities of 3.5 ± 1.0 μm/min (n = 56), which was not significantly different from control cells (Figure 6 and Table 1). However, unlike chromosomes in cells injected with either NuMA or Kid antibodies alone, chromosomes in cells injected with both Kid and NuMA antibodies failed to form a tight metaphase plate despite observation for more than 2 h after nuclear envelope breakdown. Individual chromosomes underwent bidirectional movement (Figure 6, A, arrow; and B), but directional switching was separated by pauses of variable duration unlike chromosome oscillation in control cells that display a “saw-toothed” displacement tracing due to directional instability (Skibbens et al., 1993; Figure 6B). Indirect immunofluorescence microscopy performed after time-lapse videomicroscopy of the cell shown in Figure 6 verified that it had a short, bipolar spindle with unaligned chromosomes (Figure 3D). Together, these data demonstrate that kinetochore-microtubule interactions occur appropriately on spindles in cells injected with antibodies to both Kid and NuMA. Those interactions generate forces sufficient to maximally stretch sister kinetochores and power chromosome movement at velocities indistinguishable from control cells. Thus, the lack of chromosome alignment generated by injection of antibodies to both Kid and NuMA is not the consequence of improper kinetochore-microtubule attachment or impaired kinetochore function.
Figure 6.
Chromosome movement in a CFPAC-1 cell microinjected with NuMA- and Kid-specific antibodies. (A) Selected differential interference contrast images from a video record of a mitotic cell that has been microinjected with NuMA- and Kid-specific antibodies. Times are indicated in hours:minutes:seconds. Arrow indicates a chromosome undergoing bidirectional movement. Bar, 10 μm. Supplemental video is available online at http://www.molbiolcell.org. (B) Traces of individual chromosome movements in a cell injected with either preimmune serum or NuMA- and Kid-specific antibodies as indicated. Distance (micrometers) of the centromeric region of the chromosome from the cell midline is plotted versus time (seconds).
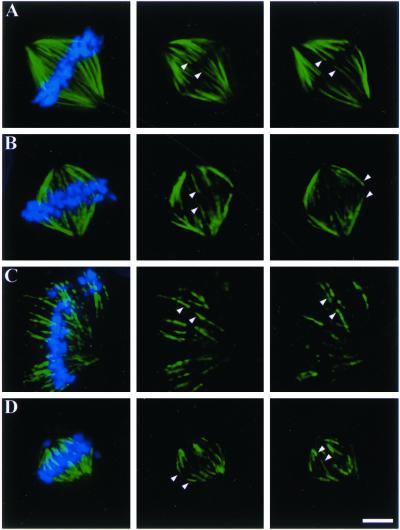
Collectively, these data indicate that Kid and NuMA participate in chromosome alignment during mitosis in cultured mammalian cells. However, there are two possible explanations for the lack of chromosome alignment after perturbation of these two proteins. First, chromosomes may be positioned appropriately at the middle of sister kinetochore fibers of equal length, but kinetochore fiber minus ends may not be efficiently organized at poles causing misalignment of entire chromosome-kinetochore fiber ensembles. Second, chromosomes may fail to be positioned appropriately at the middle of sister kinetochore fibers. To distinguish between these two possibilities we measured the lengths of sister kinetochore fibers associated with individual chromosomes (Figure 7 and Table 2). Kinetochore microtubules were revealed by depolymerization of nonkinetochore microtubules by extraction of cells in calcium-containing buffer before fixation. In uninjected control cells at metaphase, sister kinetochore fibers on individual chromosomes were, on average, 10.5 and 9.0 μm in length (Figure 7A and Table 2). The ratio of the lengths of sister kinetochore fibers in control cells averaged 1.2, very near the expected value of 1.0 for chromosomes aligned at the spindle equator associated with sister kinetochore fibers of equal length. The fact that the sum of the lengths of sister kinetochore fibers and interkinetochore distances was shorter than the pole-to-pole distance measured for control cells in Table 1 was expected as the measurements in this analysis were derived from calcium-treated cells and were based on positions of kinetochore fiber minus ends and not centrosomes as in Figure 1. In cells injected with antibodies to either Kid or NuMA the ratio of sister kinetochore lengths averaged 1.2 in both cases (Figure 7, B and C, and Table 2), indicating that perturbation of either Kid or NuMA activity alone did not significantly alter the positioning of individual chromosomes on their associated kinetochore fibers. Importantly, individual chromosomes were associated with sister kinetochore fibers of approximately equal length in cells injected with NuMA antibodies despite the disruption of spindle pole organization (Figure 7C; see also Figure 3B in Gordon et al., 2001). Cells injected with antibodies to both Kid and NuMA had well defined kinetochore fibers, although these kinetochore fibers were less tightly focused at spindle poles compared with control cells. Sister kinetochore fibers on individual chromosomes in cells injected with both Kid and NuMA antibodies were, on average, 7.5 and 3.8 μm in length (Figure 7D and Table 2). The ratio of the lengths of kinetochore fibers in these cells averaged 2.2, indicating that many chromosomes in cells injected with antibodies to both Kid and NuMA were associated with sister kinetochore fibers of unequal length. Thus, defects in chromosome alignment occurred because chromosomes failed to localize appropriately at the middle of their associated kinetochore fibers.
Figure 7.
Kinetochore fiber lengths in cells injected with NuMA- and Kid-specific antibodies. Uninjected CFPAC-1 cells (A), or cells injected with Kid-specific antibodies alone (B), NuMA-specific antibodies alone (C), or both NuMA- and Kid-specific antibodies (D) were permeabilized in the presence of 0.1 mM calcium and then were fixed and stained using tubulin-specific antibodies and the DNA-specific dye DAPI. Left panels show overlay of DNA and tubulin signals from the merged z-series to represent the entire spindle. Right two panels show individual focal planes in the tubulin channel to demonstrate kinetochore fiber pairs (arrowheads). Bar, 10 μm.
Table 2.
Kinetochore fiber lengths
| Treatment | L1, μm (n)a | L2, μm (n)a | L1/L2 (n)b |
|---|---|---|---|
| Uninjected (25 cells) | 10.5 ± 1.9 (74) | 9.0 ± 1.5 (74) | 1.2 ± 0.1 (74) |
| α-Kid (10 cells) | 9.9 ± 3.0 (39) | 8.5 ± 2.6 (39) | 1.2 ± 0.2 (39) |
| α-NuMA (10 cells) | 11.2 ± 2.9 (39) | 9.3 ± 2.3 (39) | 1.2 ± 0.2 (39) |
| α-Kid, α-NuMA injected (10 cells) | 7.5 ± 2.0 (42) | 3.8 ± 1.3 (42) | 2.2 ± 0.9 (42) |
Spindle Pole Organization and Spindle Size Are Influenced by NuMA and Kid
We also observed two other consequences of injection of antibodies to both Kid and NuMA. First, spindles were, on average, 42% shorter (pole-to-pole) than control cells or cells injected with Kid antibodies alone (Figures 1 and 3, and Table 1). Time-lapse differential interference contrast microscopy sequences indicate that spindles in cells injected with antibodies to both Kid and NuMA assembled with short pole-to-pole distances and did not arise because spindles formed with normal length but subsequently shortened (our unpublished data). Second, centrosomes remained associated with the spindle, microtubule minus ends were focused at the poles, and spindles were bipolar indicating that inhibition of Kid function restored spindle pole organization to cells lacking NuMA function (compare Figures 1C to D and 3C to D). We hypothesized that spindle poles were organized in the absence of Kid and NuMA function by the activity of the kinesin-related protein HSET, which has been shown to cooperate with NuMA and cytoplasmic dynein in spindle organization (Walczak et al., 1998; Mountain et al., 1999; Gordon et al., 2001). To test this, we simultaneously injected cells with antibodies to NuMA, Kid, and HSET, and found that 90% of cells injected with all three antibodies displayed disorganized poles and unaligned chromosomes (Figure 3E and Table 1). The efficiency of pole disruption by all three antibodies closely approached the 100% efficiency of pole disruption after NuMA antibody injection (Figure 3C), but starkly contrasted bipolar spindle organization in 86% of cells injected with antibodies to both NuMA and Kid (Figure 3D). Chromosome velocity was significantly reduced in cells injected with all three antibodies consistent with our previous results showing that perturbation of both HSET and NuMA suppressed chromosome velocities (Gordon et al., 2001). Thus, HSET is sufficient to organize spindle poles after perturbation of the activities of both Kid and NuMA.
DISCUSSION
We used antibody injection to simultaneously perturb the functions of the chromokinesin Kid and the spindle pole organizing protein NuMA in cultured mammalian cells. Although we previously demonstrated that antibody injection efficiently perturbed the activities of each of these proteins (Gaglio et al., 1995; Gordon et al., 2001; Levesque and Compton, 2001), our conclusions herein are subject to the caveats inherent to the technique of antibody microinjection, including the potential of partial inhibition of the target protein. In particular, there is potential for partial inhibition of Kid because our antibodies specifically block Kid from targeting to chromosomes, but not spindles. However, several observations support the view that antibody injection completely and specifically inhibited the activities of both Kid and NuMA. First, defects in chromosome alignment and spindle morphology generated by injection of antibodies to either Kid or NuMA are equivalent to the effects observed upon the complete depletion of either of these proteins from extracts prepared from frog eggs (Merdes et al., 1996; Antonio et al., 2000; Funabiki and Murray, 2000). Second, the relationship between Kid and NuMA is specific because simultaneous injection of antibodies to Kid and either HSET or CENP-E showed no significant differences from injection of each antibody alone. Third, both Kid and NuMA were displaced from their normal cellular locations to a similar degree after injection of either one or both antibodies. Fourth, addition of Kid antibodies to a mammalian mitotic extract lacking chromosomes had no effect on the organization of microtubule asters, suggesting that Kid's primary role in spindle organization relies on its association with chromosomes (our unpublished data). Finally, it is unlikely that some nonspecific effect associated with antibody injection would restore spindle bipolarity to cells where NuMA function has been perturbed.
Spindle Organization
NuMA is required to tether centrosomes to spindle poles and to organize microtubule minus ends at spindle poles in vertebrate cells (Gaglio et al., 1995; Merdes et al., 1996; Gordon et al., 2001). Surprisingly, we show that perturbation of Kid function permits centrosomes to remain tethered to the spindle and microtubule minus ends to remain focused at spindle poles despite the perturbation of NuMA function. Moreover, we show that the kinesin-related protein HSET is responsible for organizing microtubules into short, bipolar spindles after perturbation of both Kid and NuMA.
Kid generates polar ejection force by associating with chromosome arms and translocating toward the plus ends of nonkinetochore spindle microtubules (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001; Yajima et al., 2003). Therefore, it stands to reason that as Kid pushes chromosome arms toward the spindle equator it generates an equal and opposite force that pushes nonkinetochore microtubules outward toward spindle poles. NuMA most likely stabilizes spindle organization in the face of this outward force on nonkinetochore microtubules by physically cross-linking nonkinetochore microtubules to kinetochore microtubules at spindle poles. Several previous observations support this idea. First, electron microscopy, used alone to track individual microtubules or coupled to chromosome micromanipulation, has shown that minus ends of nonkinetochore microtubules are physically linked to kinetochore microtubules near spindle poles (Nicklas et al., 1982; Mastronarde et al., 1993). Next, immunogold electron microscopy indicates that NuMA cross-links spindle microtubules at spindle poles (Dionne et al., 1999) and in vitro experiments show that NuMA can bind directly to microtubules and cross-link adjacent microtubules into bundles (Merdes et al., 1996; Haren and Merdes, 2002). Finally, it was recently shown that kinetochore fiber minus ends can be recruited to spindle poles through a mechanism that relies on the activity of NuMA to cross-link kinetochore microtubule minus ends to nonkinetochore astral microtubules (Khodjakov et al., 2003).
In this context, spindle organization is disrupted upon perturbation of NuMA alone because forces acting on spindle microtubules are imbalanced and the remaining proteins that cross-link spindle microtubules lack sufficient rigidity to maintain spindle architecture under the stress of spindle forces. However, perturbation of Kid function reduces the force acting on nonkinetochore microtubules rendering the cross-linking activity of HSET sufficient to maintain spindle architecture in the absence of NuMA function. Thus, NuMA may act as part of a hypothetical spindle matrix that cross-links spindle microtubules and bears the load of spindle forces (Gordon et al., 2001). Alternatively, NuMA may participate in the generation of an inwardly directed force on nonkinetochore microtubules that antagonizes the outwardly directed force of Kid. NuMA could generate an inward force on nonkinetochore microtubules as an equal and opposite reaction to the force it exerts as it pulls kinetochore microtubules poleward along nonkinetochore microtubules or as a consequence of the pull that kinetochore motors place on kinetochore microtubules. Such an inwardly directed force is consistent with the observation that HSET has been shown to generate an inwardly directed force that antagonizes the activity of Eg5 (Mountain et al., 1999), and HSET and NuMA have been shown to act redundantly in some circumstances (Gordon et al., 2001). Moreover, the short spindles that we observed after perturbation of Kid and NuMA activities could arise because the inwardly directed force generated by HSET is overemphasized. Regardless of the precise nature of NuMA action, an implication of the functional cross-linking of kinetochore and nonkinetochore microtubules is that forces acting on different subpopulations of spindle microtubules are integrated at spindle poles due to the mechanical connections between microtubules at that site.
Chromosome Alignment
We also show that Kid and NuMA contribute to chromosome alignment during mitosis in mammalian cells. The lack of chromosome alignment was a specific effect of perturbation of Kid and NuMA because chromosome alignment defects were not detected after the simultaneous perturbation of Kid and either HSET or CENP-E. Because perturbation of Kid and NuMA had no detectable effect on kinetochore microtubule interactions, kinetochore function, or chromosome velocity, these results indicate that Kid and NuMA do not power chromosome alignment directly. Rather, Kid and NuMA must generate positional cues that instruct the motors driving chromosome movement to align chromosomes at the spindle equator. Currently, there is no evidence to suggest that NuMA cooperates with Kid in generating polar ejection force. Thus, we speculate that NuMA generates a positional cue for chromosome alignment through a different mechanism than Kid and its associated polar ejection force. Because chromosomes aligned properly after perturbation of either Kid or NuMA, but not after perturbation of both Kid and NuMA, these results indicate that the positional cues generated by Kid and NuMA must act independently, and redundantly, to direct chromosome alignment.
Kid generates a significant fraction of polar ejection force (Antonio et al., 2000; Funabiki and Murray, 2000; Levesque and Compton, 2001), and the mechanism by which polar ejection force acts as a positional cue to direct chromosome alignment has been well documented (Figure 8; see INTRODUCTION; Rieder and Salmon, 1994, 1998; Khodjakov et al., 1999). The nature of the positional cue provided by NuMA is unknown. One possibility is that perturbation of NuMA alters the activity of other spindle proteins such as cytoplasmic dynein. Cytoplasmic dynein has been shown to have a role in chromosome movement and positioning in some systems (Sharp et al., 2000b; Howell et al., 2001), and NuMA has been shown to associate with cytoplasmic dynein and its associated activating complex dynactin (Merdes et al., 1996). Mislocalization of cytoplasmic dynein as a result of NuMA perturbation might perturb chromosome positioning when coupled to the lack of polar ejection force. However, it is not obvious how force from a motor such as cytoplasmic dynein would vary depending on position within the spindle (i.e., act as a positional cue), and how perturbation of that motor would alter chromosome positioning without altering bidirectional chromosome movement or velocity as we observed here.
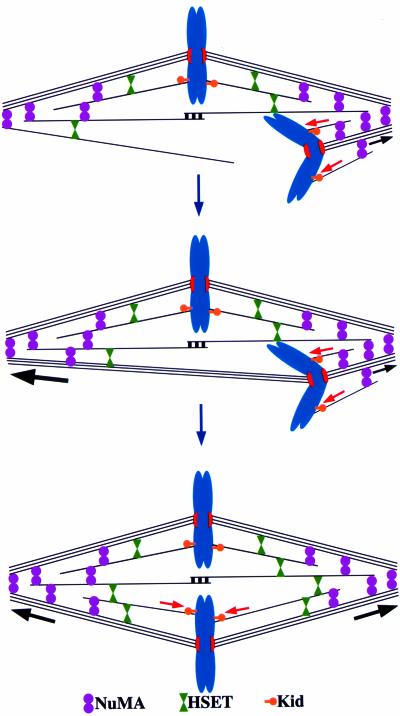
Figure 8.
Kid and NuMA participate in chromosome alignment in mammalian cells. Both Kid and NuMA contribute to chromosome alignment by providing positional cues. Kid provides a significant fraction of polar ejection force (red arrows) that directs chromosomes to the spindle equator because that is the site where polar ejection force is equal between the two poles. NuMA may contribute to chromosome alignment through a traction fiber-based mechanism (black arrows). The magnitude of poleward traction force is proposed to be high on long kinetochore fibers (large black arrow) and small on short kinetochore fibers (small black arrow). Bioriented chromosomes tend to align at the spindle equator in response to poleward traction forces because that is the site where poleward force is balanced between the poles.
Another possibility is that NuMA provides a positional cue for chromosome alignment as a component of a traction fiber-type mechanism (Figure 8; Östergren, 1951). The traction fiber model posits that kinetochore microtubules are actively translocated poleward and depolymerized at their minus ends, a behavior manifested as poleward microtubule flux (Mitchison, 1989b). If the magnitude of poleward force generated by the traction fiber varied as a function of kinetochore microtubule length, then that force could act as a positional cue to direct chromosome alignment because the spindle equator would be the position where poleward traction forces would be equal between the poles. We favor this model because poleward microtubule flux is a conserved feature of spindles in most cell types (Mitchison, 1989a; Mitchison and Salmon, 1992; Desai et al., 1998; LaFountain et al., 2001; Brust-Mascher and Scholey, 2002; Maddox et al., 2002) and NuMA is localized at spindle poles, the likely position of factors needed to generate poleward microtubule flux (Waters et al., 1996). Furthermore, by examining multivalent chromosome position and kinetochore microtubule lengths in grasshopper spermatocytes, Hays et al. (1982) concluded that “the magnitude of traction force on a kinetochore fiber is a linear function of fiber length.” This conclusion is in line with a role for traction fiber forces directing chromosome alignment to the spindle equator, although it is important to note that alternative interpretations of this data have been proposed (Rieder and Salmon, 1994). Currently, evidence implicating NuMA's involvement in a traction fiber mechanism is only circumstantial. NuMA may act directly to promote either poleward microtubule translocation or microtubule minus end disassembly or it may act indirectly as a scaffold to maintain spindle organization as microtubules translocate poleward and depolymerize at their minus ends.
Supplementary Material
[MBC Video]
Acknowledgments
We thank J.B. Rattner, Kevin Sullivan, D. Pettijohn, and Tim Yen for providing antibodies used in this work. We also thank Tarun Kapoor for helpful discussions and comments on the manuscript. This study was supported by a grant from the National Institutes of Health (GM-51542). Time-lapse microscopy equipment was purchased using a grant from the Fannie E. Rippel Foundation.
V⃞
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
References
- Alexander, S.P., and Rieder, C.L. (1991). Chromosome motion during attachment to the vertebrate spindle: initial saltatory-like behavior of chromosomes and quantitative analysis of force production by nascent kinetochore fibers. J. Cell Biol. 113, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonio, C., Ferby, I., Wilhelm, H., Jones, M., Karsenti, E., Nebreda, A.R., and Vernos, I. (2000). Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102, 425–435. [DOI] [PubMed] [Google Scholar]
- Bajer, A. (1982). Functional autonomy of monopolar spindle and evidence for oscillatory movement in mitosis. J. Cell Biol. 93, 33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brust-Mascher, I., and Scholey, J.M. (2002). Microtubule flux and sliding in mitotic spindles or Drosophila embryos. Mol. Biol. Cell 13, 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, D.A. (2000). Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114. [DOI] [PubMed] [Google Scholar]
- Desai, A., Maddux, P.S., Mitchison, T.J., and Salmon, E.D. (1998). Anaphase A chromosome movement and poleward spindle microtubule flux occur at similar rates in Xenopus extract spindles. J. Cell Biol. 141, 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionne, M.A., Howard, L., and Compton, D.A. (1999). NuMA is a component of an insoluble matrix at mitotic spindle poles. Cell Motil. Cytoskeleton 42, 189–203. [DOI] [PubMed] [Google Scholar]
- Funabiki, H., and Murray, A.W. (2000). The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 102, 411–424. [DOI] [PubMed] [Google Scholar]
- Gaglio, T., Saredi, A., and Compton, D.A. (1995). NuMA is required for the organization of microtubules into aster-like mitotic arrays. J. Cell Biol. 131, 693–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky, G.J. (1992). Chromosome motion in mitosis. Bioessays 14, 73–80. [DOI] [PubMed] [Google Scholar]
- Gordon, M.B., Howard, L., and Compton, D.A. (2001). Chromosome movement in mitosis requires microtubule anchorage at spindle poles. J. Cell Biol. 152, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haren, L., and Merdes, A. (2002). Direct binding of NuMA to tubulin is mediated by a novel sequence motif in the tail domain that bundles and stabilizes microtubules. J. Cell Sci. 115, 1815–1824. [DOI] [PubMed] [Google Scholar]
- Hays, T.S., Wise, D., and Salmon, E.D. (1982). Traction force on a kinetochore at metaphase acts as a linear function of kinetochore fiber length. J. Cell Biol. 93, 374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, D.B., Pearson, C.G., Yen, T.J., Howell, B.J., and Salmon, E.D. (2001). Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell 12, 1995–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell, B.J., McEwen, Canman, J.C., Hoffman, D.B., Farrar, E.M., Rieder, C.L., and Salmon, E.D. (2001). Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint activation. J. Cell Biol. 155, 1159–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A., and Karsenti, E. (1996). Morphogenetic properties of microtubules and mitotic spindle assembly. Cell 84, 401–410. [DOI] [PubMed] [Google Scholar]
- Kapoor, T.M., and Compton, D.A. (2002). Searching for the middle ground: mechanisms of chromosome alignment during mitosis. J. Cell Biol. 157, 551–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor, T.M., Mayer, T.U., Coughlin, M.L., and Mitchison, T.J. (2000). Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150, 975–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., Gabashvili, I.S., and Rieder, C.L. (1999). “Dumb” versus “smart” kinetochore models for chromosome congression during mitosis in vertebrate somatic cells. Cell Motil. Cytoskeleton 43, 179–185. [DOI] [PubMed] [Google Scholar]
- Khodjakov, A., Copenagle, L., Gordon, M.B., Compton, D.A., and Kapoor, T.M. (2003). Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 160, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaFountain, J.R., Oldenbourg, R., Cole, R.W., and Rieder, C.L. (2001). Microtubule flux mediates poleward motion of acentric chromosome fragments during meiosis in insect spermatocytes. Mol. Biol. Cell 12, 4054–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque, A.A., and Compton, D.A. (2001). The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J. Cell Biol. 154, 1135–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox, P., Desai, A., Oegema, K., Mitchison, T.J., and Salmon, E.D. (2002). Poleward microtubule flux is a major component of spindle dynamics and anaphase A in mitotic Drosophila embryos. Curr. Biol. 12, 1670–1674. [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N., McDonald, K.L., Ding, R., and McIntosh, J.R. (1993). Interpolar spindle microtubules in PTK cells. J. Cell Biol. 123, 1475–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen, B.F., Chan, G.K.T., Zubrowski, B., Savoian, M.S., Sauer, M.T., and Yen, T.J. (2001). CENP-E is essential for reliable bioriented spindle attachment, but chromosome alignment can be achieved via redundant mechanisms in mammalian cells. Mol. Biol. Cell 12, 2776–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merdes, A., Ramyar, K., Vechio, J.D., and Cleveland, D.W. (1996). A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell 87, 447–458. [DOI] [PubMed] [Google Scholar]
- Mitchison, T.J. (1989a). Chromosome alignment at mitotic metaphase: balanced forces or smart kinetochores? In: Cell Movement. Volume 2: Kinesin, Dynein, and Microtubule Dynamics, ed. F.D. Warner and J.R. McIntosh, New York: Alan R. Liss, Inc., 421–430. [Google Scholar]
- Mitchison, T.J. (1989b). Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 109, 637–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J., and Salmon, E.D. (1992). Poleward kinetochore fiber movement occurs during both metaphase and anaphase-A in newt lung cell mitosis. J. Cell Biol. 119, 569–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison, T.J., Evans, L., Schulze, E., and Kirschner, M. (1986). Sites of microtubule assembly and disassembly in the mitotic spindle. Cell 45, 515–527. [DOI] [PubMed] [Google Scholar]
- Moore, M.J. (1975). Removal of glass coverslips from cultures flat embedded in epoxy resins using HF. Microscopy 104, 205. [DOI] [PubMed] [Google Scholar]
- Mountain, V., Simerly, C., Howard, L., Ando, A., Schatten, G., and Compton, D.A. (1999). The kinesin-related protein, HSET, opposes the activity of Eg5 and cross-links microtubules in the mammalian mitotic spindle. J. Cell Biol. 147, 351–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas, R.B., Kubai, D.F., and Hays, T.S. (1982). Spindle microtubules and their mechanical associations after micromanipulation in anaphase. J. Cell Biol. 95, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östergren, G. (1951). The mechanism of co-orientation in bivalents and multivalents. Hereditas 37, 85–156. [Google Scholar]
- Rieder, C.L., and Bowser, S. (1987). Correlative LM and EM on the same epoxy section. In: Correlative Microscopy in Biology, ed. M.A. Hyat, New York: Academic Press, 249–277.
- Rieder, C.L., Davison, E.A., Jensen, L.C.W., Cassimeris, L., and Salmon, E.D. (1986). Oscillatory movements of monooriented chromosomes and their position relative to the spindle pole result from the ejection properties of the aster and half spindle. J. Cell Biol. 103, 581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and Salmon, E.D. (1994). Motile kinetochores and polar ejection forces dictate chromosome position on the vertebrate mitotic spindle. J. Cell Biol. 124, 223–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., and Salmon, E.D. (1998). The vertebrate cell kinetochore and its roles during mitosis. Trends Cell Biol. 8, 310–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaar, B.T., Chan, G.K.T., Maddox, P., Salmon, E.D., and Yen, T.J. (1997). CENP-E function at kinetochores is essential for chromosome alignment. J. Cell Biol. 139, 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000a). Microtubule motors in mitosis. Nature 407, 41–47. [DOI] [PubMed] [Google Scholar]
- Sharp, D.J., Rogers, G.C., and Scholey, J.M. (2000b). Cytoplasmic dynein is required for poleward chromosomes movement during mitosis in Drosophila embryos. Nat. Cell Biol. 2, 922–930. [DOI] [PubMed] [Google Scholar]
- Skibbens, R.V., Skeen, V.P., and Salmon, E.D. (1993). Directional instability of kinetochore motility during chromosome congression and segregation in mitotic newt lung cells: a push-pull mechanism. J. Cell Biol. 122, 859–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokai, N., Fujimoto-Nishiyama, A., Toyoshima, Y., Yonemura, S., Tsukita, S., Inoue, J., and Yamamoto, T. (1996). Kid, a novel kinesin-like DNA binding protein, is localized to chromosomes and the mitotic spindle. EMBO J. 15, 457–467. [PMC free article] [PubMed] [Google Scholar]
- Vale, R.D., and Fletterick, R.J. (1997). The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol. 13, 745–777. [DOI] [PubMed] [Google Scholar]
- Walczak, C.E., Vernos, I., Mitchison, T.J., Karsenti, E., and Heald, R. (1998). A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8, 903–913. [DOI] [PubMed] [Google Scholar]
- Waters, J.C., Mitchison, T.J., Rieder, C.L., and Salmon, E.D. (1996). The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 7, 1547–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima, J., Edamatsu, M., Watai-Nishii, J., Tokai-Nishizumi, N., Yamamoto, T., and Toyoshima, Y.Y. (2003). The human chromokinesin Kid is a plus end-directed microtubule-based motor. EMBO J. 22, 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen, T.J., Compton, D.A., Wise, D., Zinkowski, R.P., Brinkley, B.R., Earnshaw, W.C., and Cleveland, D.W. (1991). CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J. 10, 1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[MBC Video]