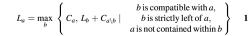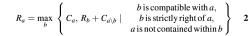Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies (original) (raw)
Abstract
The spliced alignment of expressed sequence data to genomic sequence has proven a key tool in the comprehensive annotation of genes in eukaryotic genomes. A novel algorithm was developed to assemble clusters of overlapping transcript alignments (ESTs and full-length cDNAs) into maximal alignment assemblies, thereby comprehensively incorporating all available transcript data and capturing subtle splicing variations. Complete and partial gene structures identified by this method were used to improve The Institute for Genomic Research Arabidopsis genome annotation (TIGR release v.4.0). The alignment assemblies permitted the automated modeling of several novel genes and >1000 alternative splicing variations as well as updates (including UTR annotations) to nearly half of the ∼27 000 annotated protein coding genes. The algorithm of the Program to Assemble Spliced Alignments (PASA) tool is described, as well as the results of automated updates to Arabidopsis gene annotations.
INTRODUCTION
Expressed sequence tags (ESTs) and complete complementary DNA (cDNA) sequences have been powerful tools for gene discovery and studies of gene expression since long before complete genome sequences were a reality (1). Combined with the availability of complete genomic sequences, the transcribed sequences delineate the structures of genes by resolving introns and exons via gapped alignments (2,3). Genome annotation projects rely heavily upon the transcribed sequences to locate expressed genes within the genome and accurately annotate gene structures using both manual and automated methods. Abundantly expressed genes provide a high level of redundancy in the EST populations and, in many cases, yield evidence for the existence of alternative splicing isoforms (4,5).
Various efforts currently exist to assemble the expressed sequence populations into a unique index of genes, including the Gene Indices at The Institute for Genomic Research (TIGR) (6), Unigene at the National Center for Biotechnology Information (NCBI) (7), and STACKdb at the South African National Bioinformatics Institute (SANBI) (8), reviewed in Bouck et al. (9). Depending on the stringency of the clustering and alignment methods employed in generating the unique data set, subtle alternative splice variants may be lost and chimeric assemblies can be generated.
Several popular cDNA alignment programs are available for aligning ESTs and cDNAs to genomic sequences, including sim4 (10), gap2 (2), spidey (11), BLAT (12) and GeneSeqer (13). In addition, tools have been developed to incorporate EST alignments into gene predictions (14–16). A recent effort to predict gene structures and alternative splicing variations has focused on the assembly of transcript–genome alignments (5). The genome sequence alignment-based method of consolidating the transcript sequences offers advantages over the conventional clustering and sequence assembly since the alignment-deduced spliced orientation can be exploited to prevent the assembly of overlapping transcripts aligning to opposite strands, minimizing the artificial construction of chimeric assemblies. In addition, when the genome sequence is of unambiguous high fidelity, the co-assembly of transcripts mapped to distinct closely related paralogs is prevented. Sequence errors prevalent in ESTs that may puzzle raw sequence assembly are less likely to confound alignment assembly, since clustering relies on more mismatch-tolerant cDNA–genome alignments and short unaligned regions at termini due to low sequence quality or vector contamination are disregarded.
Alignments of full-length cDNAs (FL-cDNAs) have proved very useful in resolving gene structures and improving annotations in Arabidopsis (17,18). Until recently, efforts to incorporate EST alignments into gene structure annotations have mostly involved manual inspection and refinements. A recent independent analysis of Arabidopsis EST alignments in comparison to the genome annotation (release v.3.0, July 2002) identified nearly 1000 genes with structures in conflict with EST alignments, in addition to hundreds of alternative splicing variations and non-consensus splice sites, all indicating the need for improvements in the genome annotation (19). Our latest efforts to refine the Arabidopsis gene structure annotations combine both EST and FL-cDNA alignments to update conflicting gene structures and annotate alternative splicing variations, as well as to identify and annotate substantially supported non-consensus splice sites.
We developed a novel dynamic programming algorithm to assemble cDNA alignments and identify alternative splice variants implemented in a tool named the Program to Assemble Spliced Alignments (PASA). PASA creates unique maximal assemblies based on EST and cDNA alignments by merging sets of compatible overlapping alignments. This algorithm was employed to consolidate and maximize the untranslated regions (UTRs) of Arabidopsis FL-cDNAs and integrate EST evidence in regions where no FL-cDNA alignments were available, providing maximal substrates for updating and improving the Arabidopsis genome annotation. These transcript alignment assembly-based gene structure annotation updates were performed as part of the TIGR complete Arabidopsis genome re-annotation effort (20).
MATERIALS AND METHODS
Alignment assembly algorithm
The goal of the assembly algorithm is to find, for each alignment a, the largest assembly that contains a, i.e. the assembly containing a together with the maximum number of other ESTs and cDNAs. The maximal alignment assembly is then used as the substrate for creating gene models or modifying existing gene models. Note that many different alignments may have the same largest assembly. Indeed, when the alignments to a specific genomic region are all consistent with one another, a single assembly will contain all of them.
In the remainder of this section, for simplicity, we consider just one strand of the genomic sequence and treat it as a line marked with integers, with low positions to the left. We also use the term cDNAs to refer collectively to FL-cDNAs and ESTs. An alignment can be considered as a series of intervals that correspond to the positions for which the cDNA aligns to the genome; for example, an alignment might consist of the intervals [(50,100),(150,170)] for a cDNA that is aligned in two places, spanning nucleotide positions 50–100 and 150–170. The span of an alignment will be defined as the range from its beginning to end, e.g. the span of our example is 50–170. We assume that each cDNA has been aligned unambiguously with the genomic sequence. Thus, the terms cDNA and alignment are interchangeable.
We compute the largest assemblies by dynamic programming. First, all alignments are sorted into ascending order by their beginning positions along the genomic sequence. Next, each pair of overlapping alignments is tested for compatibility, where alignments are compatible if they have the same orientation and are identical in their region of overlap. All overlapping cDNAs in an assembly must be compatible.
Let La denote the maximum number of cDNAs in a contiguous assembly that ends at alignment a, i.e. it includes a, compatible alignments contained in the span of a and alignments that end strictly before the end of a, but not alignments that strictly contain a. For compatible overlapping alignments a and b, let C a_\_b denote the number of _a_-compatible alignments contained in the span of a (including a itself) but not contained in b and let Ca denote the number of _a_-compatible alignments contained in the span of a. Then we have:
The Ca values for the alignments are easily identified during the phase when overlapping alignments are tested for compatibility. From these alignment lists, the values of C a_\_b can be calculated by a merge operation. Thereafter, the values of La can be computed in a simple left-to-right scan using equation 1. During this computation, each alignment a retains a pointer, pa, to the alignment b that achieves the maximum that defines La.
Containments between alignments a and b prohibit any direct comparisons between the two alignments during the La calculations. As specified in the equation, alignment a cannot be contained within alignment b and, since alignment b must be strictly before alignment a, alignment b cannot be contained within alignment a. The reason that contained alignments are treated specially is illustrated in the follow ing simple example (where | represents an inferred exon boundary).
a:------| |-----||-------
b: -| |-----||--
c: ---| |----| |-----
Alignment a is compatible with alignment b and alignment b is compatible with alignment c. Note, however, that alignment a is not compatible with alignment c. This non-transitivity of the compatibility relationship prevents the simple chaining of compatible alignments. Compatibility of non-contained alignments is transitive, however, so that we can chain those alignments together, keeping track of the containments within them.
The maximum of all the La values, La*, represents the largest number of assembled alignments; in other words, the assembly containing the greatest number of compatible cDNAs. Starting from alignment a*, a trace back of the pa pointers, together with the alignments for the C a_\_b values, yields the cDNAs comprising the maximum assembly.
If any alignments are not included in the maximum assembly, conflicting alignments exist, indicating alternative splicing isoforms or overlapping transcripts corresponding to different genes; pairwise alignment incompatibilities are derived from alternative acceptors or donors, unspliced introns, skipped exons or opposite spliced orientations. In order to include every cDNA in its maximal assembly, further efforts are made to obtain the maximal assembly for the cDNAs not in the a* assembly.
Let _a_′ be a cDNA not yet included in a maximal assembly. Tracing back the pa pointers gives the largest assembly to the left of _a_′, but unfortunately these pointers do not provide the largest assembly to the right of _a_′. In order to identify the alignments to the right of _a_′ that should be in its maximal assembly, we perform a reciprocal scan from right to left, computing:
where Ra represents the maximum number of cDNAs in an assembly containing a consisting only of cDNAs ending at a from its right. As before, we retain a pointer qa to the alignment b that achieves the maximum value.
The total number of cDNAs in the largest assembly containing any alignment a is then equal to max_b_{Lb + Rb – Cb | b contains a}. Note that it is necessary to subtract Cb since these cDNAs have been counted in both Lb and Rb. The cDNAs that comprise this assembly can be obtained by tracing back the px and qx pointers and including the corresponding C x_\_y cDNAs. Thus, the remainder of the PASA algorithm searches all alignments not yet included in a maximal assembly for the one with maximum Lb + Rb – Cb value. The corresponding assembly (with the new alignments it contains) is added to the collection of maximal assemblies and the process is repeated until all alignments have been included in at least one assembly.
Taking into account the spliced orientation of the transcript alignments prevents overlapping transcripts from opposite strands being merged into a single assembly. Since many EST sequences align as single segments lacking introns, their orientations cannot be inferred from splice sites and hence remain ambiguous. Alignments of ambiguous orientation have the potential to merge bridging transcripts from opposite orientations, creating a chimeric assembly. To prevent this, all computations described above are performed twice, setting ambiguous orientations first to the forward strand and secondly to the reverse strand. The maximal assembly is obtained from each computation and the larger maximal assembly is assumed correct. The single segment EST alignments are the only alignments of ambiguous orientation and these generally serve to extend the termini of alignment assemblies. Single segment alignments of full-length cDNAs also lack spliced orientations due to the lack of introns, but are assumed to be correctly oriented in the GenBank sequence record, so these alignments are not allowed ambiguous orientations.
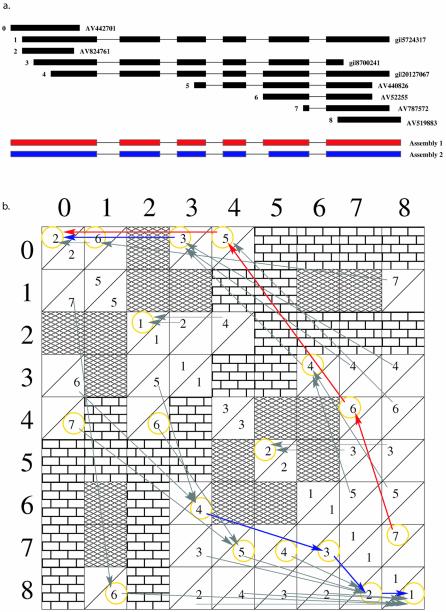
Assuming the comparison of two alignments takes O(1) time, the overall complexity of the algorithm as implemented is O(_n_3), where n represents the number of alignments being assembled. The computations can be thought of as occurring within a matrix in which alignments are the rows and columns, ordered by their beginning position in the genome. Figure 1 illustrates a real example of Arabidopsis transcript alignment assemblies within the context of a matrix.
Figure 1.
Pictorial representation of the PASA algorithm. Overlapping Arabidopsis transcript sequence alignments are ordered by their beginning position and assigned indices 0–8 as shown in (a). The matrix shown in (b) provides the ordered alignments along the rows and columns according to their index positions. The predetermined containments (chain links) and incompatibilities (bricks) present obstacles within the matrix, disallowing any direct comparisons between two alignments during La or Ra calculations. To compute La, each alignment a is compared with all compatible preceding alignments b, generating the value max[Ca, Lb + C _a_\_b_] stored in the upper left of the matrix cell at [row _b_][column _a_], with an arrow drawn to the L b value that yielded the max. L a is then the maximum upper left value in column a and is shown circled in yellow. After all L a values are computed, the maximum one is found at [row 7][column 8] and the arrows traced back from it (indicated in red) identify the alignments comprising the maximal assembly (alignments 8, 7, 4 and 0) with their contained alignments (5, 6 and 2). The result is the red assembly 1 shown in (a). An examination of assembly 1 indicates that it lacks alignments 1 and 3. The trace back from the forward scan at La where a = 3 provides the maximal assembly containing a originating from the left of a (trace back drawn in blue), but does not identify the alignments to the right of a that are in the a maximal assembly. To find the maximal alignment assembly containing alignment 3, the reverse scan computations were performed, calculating max[Ca, Lb + C _a_\_b_], where b > a, and storing the score in the lower right of cell [row _b_][column _a_]. Ra is the maximum lower right value in column a and is shown circled in yellow. The trace forward from Ra, where a = 3, is shown with blue arrows. Combined with the trace back from La where a = 3, this yields the maximal assembly containing alignment 3, shown as assembly 2, namely alignments 0–3, 6–8. This assembly is also the maximal assembly containing alignment 1. In general, maximal assemblies for missing alignments are found in order of decreasing La + Ra – Ca value until all missing alignments are accounted for within maximal alignment assemblies; this was not done here for brevity. Note that alignment 6 is regarded as contained in alignment 1, even though it extends a few bases into intron 4. PASA has a parameter, called ‘fuzz distance’, which specifies the length of mismatches to discount at transcript ends, where sequence alignment quality is often poor.
Alignment assembly and annotation refinement pipeline
Complete cDNAs, potentially incomplete cDNAs and EST sequences were downloaded from the GenBank nucleotide database. All sequences were screened and trimmed for low quality sequence regions and poly(A) tails using the TIGR Gene Indices sequence cleaning protocols (6) implemented in the SeqClean tool (available at http://www.tigr.org/tdb/tgi/software). After removing the irrelevant transcript sequence regions, the sequences were aligned against the complete Arabidopsis genome sequence (available at ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/SEQUENCES/ATH1_bacs.seq, with redundant overlapping BAC sequence regions masked) using the cDNA alignment programs BLAT (12), sim4 (10) and GeneSeqer (13) in order of program speed. After first aligning the cDNA sequences using BLAT, each of the alignments were validated requiring the [GT,GC]/AG consensus donor/acceptor splice sites at all introns and a near-complete, near-perfect alignment requiring at least 90% of the sequence aligned with at least 95% sequence identity. Given the average intron length of ∼170 bp for Arabidopsis genes, alignments inferring introns of length greater than a generous 2 kb were excluded, effectively excluding alignments with false terminal extensions which otherwise appear to be high quality alignments. If a BLAT alignment failed the validation tests, the BLAT-aligned region was extracted together with 5 kb regions of flanking genomic sequence and realigned using sim4. The sim4 alignments failing the validation tests were then realigned similarly using the GeneSeqer program. The non-validating alignments were disregarded for the purposes of alignment assembly and, more importantly, automated annotation updates (described below). These non-validating alignments are expected to be manually examined separately from the automated processes described here.
Prior to assembly, the validated alignments were grouped into clusters of overlapping cDNAs. Each cDNA cluster was subjected to the PASA algorithm to generate a set of unique, maximal cDNA alignment assemblies. The assemblies within each cluster were further divided into subclusters representing conflicting complete or partial transcripts likely corresponding to the same gene (i.e. alternative or anomalous splicing isoforms of the same gene). Assemblies subcategorized into subclusters required the same spliced orientation and at least 50% overlap of a neighboring alignment.
The cDNA alignment assemblies were compared to the current TIGR Arabidopsis gene structure annotations. The alignment assemblies were assigned to annotated genes by examining overlaps between coordinates of gene annotations and those of the alignment assemblies. Stringent overlap criteria were required to assign FL-cDNA-containing assemblies to gene products, forcing any required structural updates to occur on the most appropriate gene products. In this case, the alignment assembly and gene product must overlap by at least 40% of either length and include the same spliced orientation. The EST-containing assemblies are given greater latitude for gene annotation assignments, requiring the same spliced orientation, as determined from the genome alignments, and any overlap with the existing gene annotation. Alignment assemblies matching multiple adjacent gene annotations were excluded from automated annotation updates and were examined for the prospect of merging multiple annotations, in which case annotation updates were performed manually.
Assemblies containing FL-cDNAs are expected to contain all of the information required to fully and accurately reconstruct the structure of the corresponding gene de novo. Gene models were created based on FL-cDNA-containing assemblies as previously described for individual FL-cDNAs (17). Gene models constructed based on FL-cDNA assemblies replaced the existing annotated gene structures where inconsistencies were identified. The structures of alternative splicing isoforms were automatically annotated in cases where multiple FL-cDNA assemblies mapped to the same gene and represented novel transcripts.
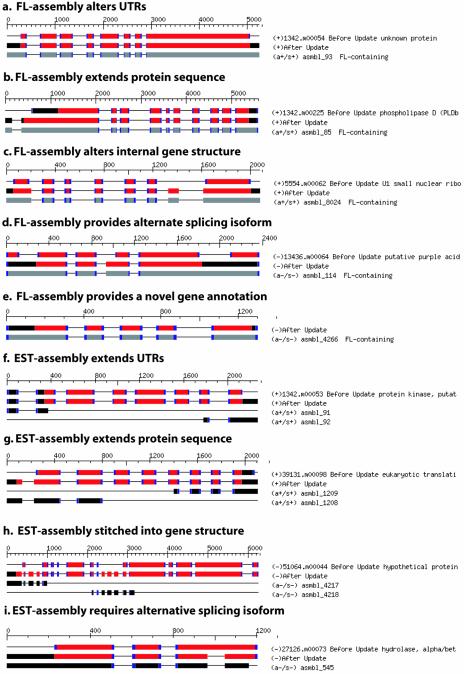
The cDNA alignment assemblies lacking a FL-cDNA component are assumed to be incomplete, lacking sufficient data to fully reconstruct a gene model, but containing partial structural information yielding components of one or more exons. These assemblies lacking FL-cDNAs were used to update conflicting annotated gene structures by ‘stitching’ the alignment exon components into the existing annotated structure. The stitching was performed by anchoring the alignment termini into overlapping exons of the annotated gene structure and replacing the overlapping structural components with those of the alignment assembly (example in Fig. 3h, asmbl_4218). Unanchored alignment termini, if they exist, serve to terminate the stitched gene structure, replacing the corresponding terminus of the annotated gene, often altering the protein coding sequence and yielding UTRs (example in Fig. 3h, asmbl_4217).
Figure 3.
Examples of annotation updates using alignment assemblies. The types of gene structure updates provided by alignment assemblies are classified into several distinct categories. FL-cDNA containing assemblies are presumed to encode the full-length gene product including UTRs. Existing annotated gene structures can therefore be replaced by gene structures inferred from the FL-cDNA-containing alignment assemblies. Those assemblies lacking FL-cDNAs are presumed to encode only partial gene structures and are stitched into existing annotated gene structures, providing significant alterations to gene structures or simply adding or extending UTR annotations. The protein coding segments of gene structures are shown in red and UTRs are shown in black. Alignment assemblies lacking a FL-cDNA are shown in black, whereas those containing FL-cDNAs are shown in gray. Boundaries consistent with the original gene structure annotation are highlighted in blue.
Automated annotation update validation criteria
In addition to the alignment validation requirements for inclusion in an assembly (described above), the tentatively updated gene structures were required to pass stringent validation tests prior to being committed to the annotation database. These additional validation criteria were implemented as a prophylactic measure to minimize the corruption of existing annotated gene structures via alignments of incompletely processed mRNA or artifact-containing transcripts. Because most of the Arabidopsis gene structures have been manually curated and already incorporate contemporary cDNA and protein spliced alignment evidence, the expectation is that the existing Arabidopsis annotated gene structures are mostly correct, requiring minimal updates to become fully consistent with the transcript alignments. Thus, the encoded proteins would change only moderately as a result of the automated updates.
FL-cDNA-containing assemblies were required to encode a protein along at least 40% of the tentative cDNA sequence. In cases where distinct FL-cDNA alignment assemblies mapped to the same location providing evidence for alternative splicing isoforms, the smaller isoforms were required to encode a protein with a length of at least 70% of the longest isoform. The smaller isoforms were aligned to the longest isoform using the Grasta alignment program [modified Fasta (21), available at http://www.tigr.org/software] and required to share at least 70% identity across at least 70% of the shorter protein length, parameters carefully decided upon after manually inspecting several hundred tentative annotation updates.
Prior to updating any existing annotated gene structure, the annotated protein sequence was compared to the tentatively updated protein and required to pass the same set of validation tests as described above: the tentative updated protein was required to have at least 70% of the length of the annotated protein and align with at least 70% identity across 70% of the annotated protein length. Finally, all tentative gene structure updates were permitted no more than two adjacent non-coding UTR exons in order to prevent assemblies containing centrally located unspliced introns or other splicing aberration, which severely truncate the protein sequence, from updating gene structures inaccurately.
Software implementation
PASA was implemented in Perl. The spliced alignments generated using BLAT, sim4 and GeneSeqer were parsed using Perl scripts and stored in a MySQL database. The alignment assemblies generated by PASA were also stored in the MySQL database along with results from comparisons to the Arabidopsis genome annotation. The alignment assemblies and tentative annotation updates were made navigable via a series of CGI scripts interfaced to the MySQL database to allow TIGR annotators to thoroughly inspect the results of the alignment assembly and proposed annotation updates prior to committing changes to the database. Hundreds of examples were manually inspected in this fashion, leading to parameter optimization and improvements to validation protocols. The proposed gene structure updates were finally committed to the TIGR Arabidopsis thaliana annotation database prior to generating release v.4.0 of the Arabidopsis genome annotation. The PASA tool, annotation pipeline, associated software, source code, sequences and data sets are available at: http://www.tigr.org/tdb/e2k1/ath1/pasa_annot_updates/pasa_annot_updates.shtml. In addition to the Perl implementation of PASA, a more flexible version written in C++ is now available.
The current version of PASA and the related annotation pipeline does not examine EST GenBank annotations, clone pair information, 5′ or 3′ EST clustering data or identified polyadenylation sites in order to unambiguously assign orientations to unspliced EST sequences. We expect to include these additional features in an enhanced version of the software. This is expected to improve the accuracy of transcript mapping and alignment assembly and may prove to be particularly beneficial for organisms such as mouse or human, where EST sequences are many times more plentiful.
RESULTS AND DISCUSSION
cDNA alignment assembly
PASA was developed to improve the quality of Arabidopsis genome annotation through the comprehensive incorporation of cDNA and EST data available in the public domain. At the time of this analysis, ∼180 000 Arabidopsis ESTs have been deposited in GenBank. The earliest sequences were generated by consortia in France (22,23) and the USA (24) as a program to facilitate gene discovery and chromosome mapping prior to the complete sequencing of the Arabidopsis genome (reviewed in 25,26). Even with the completed genome, Arabidopsis EST sequences are continuing to be generated as a tool to analyze tissue-specific gene expression (27,28) and, more pragmatically, as a strategy to identify full-length insert cDNA clones (18).
The current collection of 177 973 Arabidopsis ESTs, 27 414 complete cDNAs and 3217 potentially partial cDNAs, totaling 208 604 expressed transcript sequences, were examined and aligned to the complete Arabidopsis genome. The complete cDNAs consist of the newly available FL-cDNAs (18) coupled with mRNA records in GenBank presumed to provide complete CDS sequences. Due to their complete protein coding properties, both the complete mRNA records and the FL-cDNAs are herein collectively referred to as the FL-cDNAs. The transcripts were aligned to the genome using a hierarchical alignment protocol employing BLAT (12), sim4 (10) and GeneSeqer (13). Both BLAT and sim4 were used because of their speed and accuracy. GeneSeqer, although significantly slower than sim4 and BLAT, tends to excel where others fail, particularly in regard to micro-exon finding in Arabidopsis (29).
The ‘Blast-like Alignment Tool’, BLAT, was used as the primary alignment program for mapping the transcripts to the genome, and was found to map 98% of the transcript sequences to the Arabidopsis nuclear genome (Table 1). The highest scoring BLAT alignments for each transcript were presumed to be the correct map locations. Each of these BLAT alignments was examined extensively and any transcript alignment lacking the defined validation requirements (see Materials and Methods) was realigned using sim4, followed by GeneSeqer as needed. Approximately 90% of the mapped transcripts were found to pass the stringent validation requirements, providing substrates for further analyses and annotation updates.
Table 1. Reducing transcript sequence alignments to alignment assemblies.
| Stage of transcript alignment and assembly pipeline | No. of transcripts, alignments or assemblies |
|---|---|
| Transcript sequences obtained from GenBank (28/01/03) | 177 973 ESTs |
| 27 414 FL-cDNAs | |
| 3217 partial? cDNAs | |
| Total 208 604 transcript sequences | |
| Transcripts mapped to genome using BLAT | 204 053 transcript sequences |
| BLAT, sim4 or GeneSeqer validating alignments | 182 540 transcript alignments |
| Alignment assemblies using PASA | 18 643 multi-element assemblies |
| 6522 singleton assemblies | |
| Total 25 165 alignment assemblies |
The novel algorithm implemented in PASA was designed to assemble the validating cDNA alignments to provide evidence-based templates for improving gene structure annotations. The alignment assemblies serve to consolidate the cDNA alignments, maximize the UTRs of full-length transcripts, identify alternative splicing isoforms and provide complete and partial models of gene structures. Assembling the cDNA alignments affords several advantages over conventional raw sequence assembly (5,19); by taking into account the genomic location and spliced orientation of transcript alignments, chimeric sequence assembly is greatly reduced. In addition, subtle splicing variations identified by gapped sequence alignments, which are often lost during direct sequence assembly, are retained in alignment assemblies (19). In contrast to the TAP algorithm (5), which assembles adjacent splice pairs (introns) using a local scoring scheme, our algorithm assembles complete cDNA alignments so that all features in the same cDNA, no matter how distant, are inevitably retained in the assembled products. This prevents chimeric assembly between conflicting cDNA alignments where conflicting introns are separated by several intervening identical introns. PASA assembled the 182 540 validating transcript alignments into 25 165 alignment assemblies (Table 1).
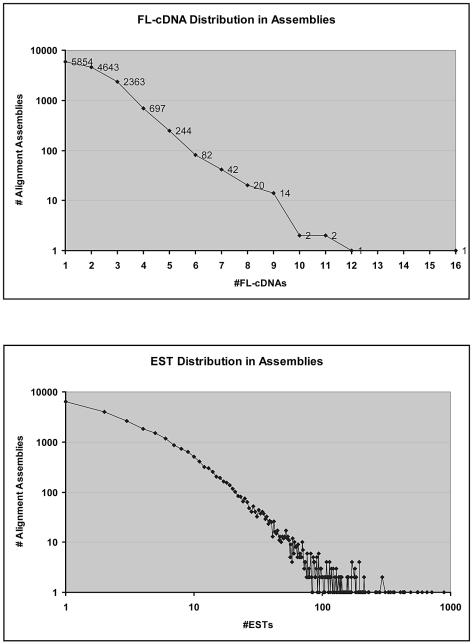
The distribution of ESTs and complete cDNAs within the alignment assemblies is illustrated in Figure 2. Of the 13 965 FL-cDNA-containing assemblies, 5854 contained only a single FL-cDNA, whereas the remainder contained multiple FL-cDNAs, often of varied lengths. Just over half of the assemblies containing FL-cDNAs (7890 of 13 965) were extended by ESTs, with 5208 of these assemblies extended by at least 20 bp on either terminus, demonstrating the value of combining EST alignments with FL-cDNAs to maximize gene structure annotations, especially those of UTRs. The ESTs and FL-cDNAs are non-randomly represented among the assemblies, with more highly expressed genes likely corresponding to the larger alignment assemblies; approximately half of all ESTs are built into fewer than 10% of the assemblies containing ESTs. The distribution of FL-cDNAs within alignment assemblies was more uniform, with ∼45% of the FL-cDNAs built into just fewer than 25% of the assemblies containing them. The gene most highly represented by ESTs and complete cDNAs corresponds to the Rubisco small subunit (At1g67090), matched by an alignment assembly containing 16 complete cDNAs and 889 ESTs.
Figure 2.
Distribution of ESTs and FL-cDNAs within alignment assemblies. The ESTs and FL-cDNAs are non-randomly distributed within alignment assemblies, with relatively small numbers of assemblies containing large numbers of transcript alignments.
Genome annotation comparison and annotation updates
The original annotation of the Arabidopsis genome was generated manually with the assistance of computational tools (30). Gene discovery and modeling often involved the painstaking process of manually editing gene models to become more consistent with the exons and splice junctions supported by transcript and protein alignments. Given the complete genome sequence, we now are aware of the extensive duplications within the Arabidopsis genome, responsible for a large number of the paralogous genes now known to exist. In the absence of a complete genome sequence during the early phase of the genome annotation, accurate mapping of transcript alignments was confounded by close paralogs that were undiscovered at that time. Also, much of the wealth of expressed sequence data currently available did not exist. Just prior to publication of the complete Arabidopsis genome (30), only ∼3300 mRNA records were available in GenBank, of which ∼450 were partial sequences. Slightly more than half of the currently available EST sequences were available in 2000, and these partial mRNA sequences could improve only components of gene structures due to their lack of complete protein coding sequence.
The influx of FL-cDNA sequences since genome completion has provided an invaluable resource for studying and annotating gene structure as well as providing evidence for novel genes. We recently improved the Arabidopsis genome annotation using ∼5000 full-length cDNAs obtained from Ceres Inc. (17). Also, since the time of genome sequence completion, the RIKEN group has released more than 20 000 Arabidopsis FL-cDNA sequences to the community (18), expanding a resource for improving gene annotations. Effective computational methods are essential to keep pace with the speed at which valuable annotation resources are being made available and ensure that genome annotations accurately reflect the most recent experimental evidence.
As described earlier, the transcript alignment assemblies were generated to both consolidate the available transcript sequences and to provide maximal structural templates for improving gene structure annotations. These high quality alignments provide the best evidence for gene identification and resolution of gene structure and provide a strong basis to create new gene models or update existing gene models that are found to be in conflict with the experimental evidence. The alignment assemblies were treated as two distinct classes of data. (i) FL-cDNA-containing alignment assemblies (FL assemblies) are expected to contain all information required for resolving a complete gene structure; all exons, introns and, possibly, UTRs. (ii) Alignment assemblies lacking a FL-cDNA alignment (non-FL assemblies), constructed from overlapping EST and partial cDNA alignments, are expected to encode components of the gene structure; complete or partial exons which may or may not code for the protein sequence, including UTR sequences.
The complete set of 25 165 alignment assemblies were compared to our TIGR Arabidopsis whole genome annotation (release 3.0, including subsequent annotation changes). Of the complete set of assemblies, 3856 were found to be previously incorporated into the genome annotation. Another 16 542 assemblies were found capable of providing automated updates to 14 247 gene structures. The types of updates provided by the alignment assemblies are illustrated in Figure 3, and the distribution of assemblies providing structural updates are enumerated in Table 2. The vast majority of all updates simply added to or extended the UTRs of already annotated genes, representing nearly 80% of all automated updates. This finding is not surprising given that most major gene structure updates committed to the annotation between the genome publication and annotation release v.3.0 (July 2002) were based on the newly available FL- cDNA sequences using methods already described (17). Consolidating the full-length cDNA sequences and including the EST alignments served to maximally extend the UTR lengths in most of these cases, thus automating the process of incorporating non-full-length transcript sequences into the gene structure annotation and annotating alternative splicing isoforms, both of which were previously performed manually.
Table 2. Distribution of structure updates corresponding to assembly type.
| Structure update class | No. of FL assemblies | No. of EST assemblies |
|---|---|---|
| Alters UTR annotations | 8844 (8800) | 4174 (3444) |
| Extends CDS structure, elongating protein | 309 (309) | 275 (274) |
| Alters internal gene structure | 732 (732) | 905 (853) |
| Provides alternate splicing isoform | 529 (502) | 701 (645) |
| Provides novel gene annotation | 73 (73) | NA |
In addition to the UTR annotation updates, incorporating the remaining complete cDNA sequence and EST alignment assemblies into the annotation yielded 583 genes with elongated proteins and 1585 genes with substantial structure updates, altering introns and often modifying the corresponding protein sequences. Alternatively spliced isoforms were modeled in cases where conflicting sources of alignment data corresponded to a single gene. Alternative splicing isoforms can be modeled relatively easily using FL assemblies due to their full protein coding attribute, requiring only multiple conflicting FL assemblies to correspond to the same gene. Annotation of alternative splicing isoforms based solely on alignments of ESTs or partial cDNAs is complicated by the lack of inherent full coding potential. In this case, the annotated gene is used as a template for creating an alternative splicing isoform by stitching the non-FL assembly into a copy of that gene model; then the stitched copy provides the alternative splicing isoform. Alternative splicing isoforms created in this fashion provide a best approximation of a gene model containing the splice variation(s). The FL assemblies provided for the new annotation of 529 isoforms, while assemblies lacking FL-cDNAs accounted for the annotation of 701 additional isoforms.
These numbers do not include the alternatively spliced isoforms that failed to meet our criterion that smaller isoforms are required to encode a protein with a length of at least 70% of the longer isoform and have no more than two UTR exons. It is possible that many such isoforms are genuine by-products of splicing regulation, as observed in the well-studied case of Drosophila sex determination in which the predominant mRNA products of the Sx1 gene in males are known to be non-functional (31–33).
The alternatively spliced isoforms are described in more detail below.
Splicing variations
Unlike the human genome, where millions of EST alignments indicate that approximately half of the genes are alternatively spliced (5,34,35), the study of alternative splicing in Arabidopsis has been mostly limited to the study of individual genes (36–39). Although ∼5% of the Arabidopsis EST and mRNA sequences have been estimated to represent splicing variants (40), few alternative splicing isoforms were previously annotated in the Arabidopsis genome annotation. The incorporation of 5000 Ceres full-length cDNA sequences into the Arabidopsis genome annotation uncovered the identification of ∼100 splicing variants (17). In addition, a very recent independent analysis identified several hundred splicing variants based on EST alignments using the GeneSeqer alignment program (19). Our analysis of the currently available transcript sequences allowed for the automated annotation of 1230 splicing isoform variants, now yielding a total of 1188 genes encoding 2501 isoforms. The isoform classifications and distributions are provided in Table 3, and can be navigated at the TIGR website (http://www.tigr.org/tdb/e2k1/ath1/altsplicing/splicing_variations.shtml). The more conventional alternative splicing isoforms containing alternative donor and/or acceptor splice sites for the same intron account for nearly half of the genes encoding splicing variations, whereas many others contain unspliced introns or skipped exons yielding alternative transcript isoforms.
Table 3. Distribution of genes according to splice variation.
| Splice variation classification | No. of genes containing isoform type |
|---|---|
| Alternate acceptor and/or donor | 549 |
| Unspliced introns | 386 |
| Alternate terminal exons | 61 |
| Exon skipping | 53 |
| Start or end within intron | 288 |
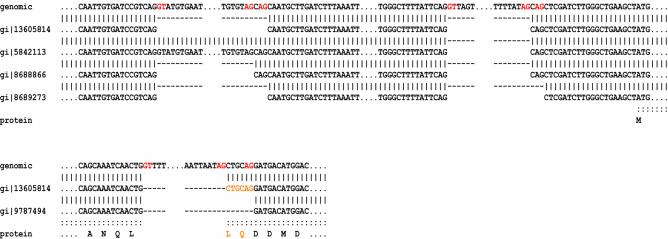
Of the 1188 genes containing alternative splicing variations, 1079 have only two isoforms, 95 have three isoforms and 13 have four annotated isoforms. The single gene At2g32700, WD-40 repeat protein, has the maximum of five annotated splicing isoforms. The ESTs and FL-cDNAs supporting these five splicing variations are illustrated in Figure 4.
Figure 4.
Five splicing isoforms supported by transcript sequence alignments. The cDNA alignments supporting the five splicing variations identified for the WD-40 repeat gene (At2g32700) are illustrated. For the purpose of comparison, FL-cDNA gi|13605814 is presumed to provide the representative gene structure. EST gi|5842113 contains an unspliced intron within the upstream UTR. EST gi|8688866 provides an alternative AG acceptor splice site within the upstream UTR which extends the spliced transcript length by 3 bp. EST gi|8689273 provides an alternative AG acceptor splice site corresponding to a different upstream UTR exon which removes 3 bp from the spliced transcript length. EST gi|9787494 provides an alternative AG acceptor splice site at a protein coding exon, deleting 6 bp corresponding to two codons of the translated sequence. Only one of the five isoforms encodes a variant protein sequence, while the remainder encode variations restricted to the upstream UTR region.
An assumption underlying the automated annotation methods exploiting expressed sequenced data is that the sequences represent fully processed mRNAs. It is unknown whether the unspliced introns represent unprocessed mRNAs or a variation in mRNA processing intended to provide transcript and protein variations. Special care was taken to exclude annotation updates from occurring if the original gene annotation appeared to be corrupted by the incorporation of potentially unprocessed or artifact-containing expressed sequence data. Of the 386 genes with isoforms containing unspliced introns, 190 genes contained the unspliced intron in their UTR regions, not affecting the encoded protein sequence. 218 genes contained transcript isoforms with unspliced introns which overlapped the protein coding region of a sibling isoform, yielding distinct protein products derived from a single gene. There were 22 genes belonging to both categories, containing examples of unspliced introns within the UTRs and other unspliced introns overlapping the protein coding region.
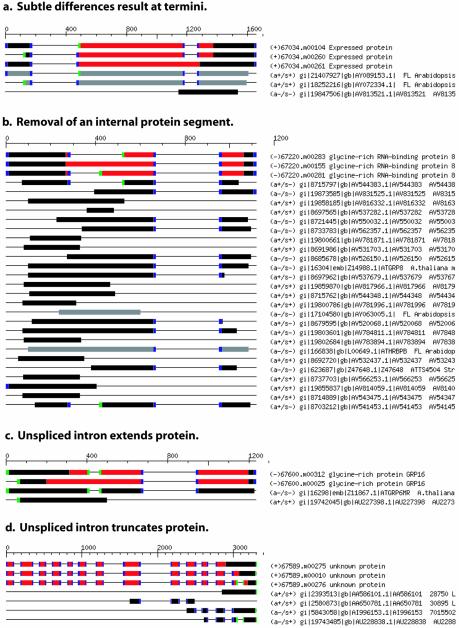
Unspliced introns found within transcript sequences are often thought to result from incomplete mRNA processing, which may be an artifact of experimental methods employed in EST or FL-cDNA generation. Consistent with this hypothesis, approximately half (99 of 218) of the genes with unspliced intron-containing isoforms encode truncated proteins; a stop codon encountered within the unspliced intron truncates the ORF. However, the remainder of these genes encode variant proteins supporting a role of unspliced introns in providing alternative proteins. There are 61 genes containing unspliced introns in which the intron encodes an integral number of codons and intron splicing removes an internal segment of the protein sequence. Of the remaining examples, 38 genes encode isoforms with subtly altered protein termini and 21 proteins are substantially extended due to the unspliced intron(s); gene At3g54890, a putative light harvesting chlorophyll a/b-binding protein, contains isoforms which fall into two categories: accession gi|23303771 provides a spliced intron which removes an integral number of codons and accession gi|23302896 infers a spliced intron which alters the reading frame and truncates the protein. Additional examples of genes encoding isoforms with unspliced introns classified into the categories described above are included in Figure 5.
Figure 5.
Unspliced introns impact on protein products. Unspliced introns have variable effects on translation products. (a) The lack of splicing of the second intron yields a protein of similar length, albeit a different C-terminus. (b) Two different overlapping introns, varying at the donor splice junction, encode an integer number of codons and splicing removes internal segments from the protein. (c) Intron splicing alters the reading frame, providing a different and shorter C-terminus. (d) The lack of splicing truncates the protein sequence due to a stop codon encountered within the unspliced intron sequence.
Merged and split gene structures
Gene prediction programs are known to sometimes merge distinct genes or to incorrectly split single genes into separate gene predictions. Since many of the original gene structure annotations were based on computational gene predictions, some of these gene structures were expected to be either merged or split based on FL-cDNA evidence. This is further supported by the finding that the number of annotated Arabidopsis genes differed from the number of FL-cDNA identified genes (18). Genes likely to require merging were identified by analyzing annotated genes which overlapped by single alignment assemblies. Since the automated methods employed were restricted to assemblies anchored to single genes, the merging of genes based on transcript alignments remained a manual process requiring inspection by TIGR annotators. 150 annotated genes were merged after analyzing FL assembly alignments and an additional six genes merged after analyzing non-FL assembly alignments.
FL-cDNAs matching an unexpectedly small region of a gene structure annotation may suggest a required gene splitting event, but, on the other hand, could otherwise be indicative of a cDNA sequence which is not full length. Large numbers of 3′ EST sequences terminating within a central portion of an annotated gene would also provide evidence for required gene splitting, but given the relatively small number of EST sequences available and their non-uniform distribution among Arabidopsis genes, this was not explored. To examine potential cases of required gene splitting, annotated genes only partially matching FL assemblies were examined. A total of 122 FL assemblies encoded proteins which were less than 70% of the length of the currently annotated protein, suggesting that the current gene annotation is inaccurate and gene splitting may be necessary. Upon manual examination, nearly all of these FL assemblies appeared not to be full length, initiating within the protein coding region of the corresponding annotated gene. A tell-tale sign that these FL-cDNAs were not full length was the absence of intervening stop codons within the presumed 5′ UTR after identification of the longest ORF within the FL assembly-based cDNA sequence. Although no gene annotations were split based on the work described here, the splitting of annotated Arabidopsis genes based on transcript and protein alignments continues to be an ongoing effort as part of the TIGR Arabidopsis genome re-annotation (20).
Novel genes
Given the high gene density within the Arabidopsis genome and given our recent efforts to re-annotate the genome, the discovery of novel genes (those previously undescribed in the genome annotation) is becoming increasingly rare. Nevertheless, 73 FL assemblies could not be anchored to existing TIGR gene annotations but were found to provide alignments consistent with complete gene structures. These FL assemblies were converted into models for novel genes and incorporated into the TIGR Arabidopsis genome annotation.
ESTs and FL-cDNAs failing automated incorporation into gene annotations
The statistics provided above are based on only those expressed transcript sequences built into the latest Arabidopsis gene structure annotation, resulting from automated processes described here coupled with our previous efforts to improve gene structure annotations. These numbers do not reflect the total number of splicing variations that can be inferred from transcript sequence alignment. There were 4767 alignment assemblies containing 15 061 transcript alignments which, in the context of the alignment assemblies, were not found suitable for automated annotation updates and will need to be examined in greater detail. These unincorporated alignment assemblies mostly include non-FL assemblies which could not be stitched properly into existing gene models or aligned to genomic regions currently classified as intergenic and, given the lack of inherent full-length protein coding capacity, were not automatically converted into complete gene model annotations. The remaining assemblies failing incorporation largely included FL assemblies found to drastically alter existing annotations or provide gene structures which vary considerably from the norm (as inferred from the majority of the FL-cDNA-based annotations) and were not employed in automated annotation updates due to the relatively stringent criteria employed. For example, two FL-cDNAs (gi|21539500 and gi|23198329) correspond to gene At2g43400 (putative electron transfer flavoprotein ubiquinone oxidoreductase) inferring 17 individual exons, only half of which can encode a protein due to a stop codon encountered within exon 10. Gene predictions along with numerous homologous protein alignments, plus both rice and wheat TIGR Gene Index assembly alignments, predict an intron within this region and, in all likelihood, both FL-cDNA sequences contain a single unspliced intron. While modeling a severely truncated protein in this region based on the FL-cDNA alignments may reflect the generation of a non-functional by-product of regulated splicing (41), we have chosen to avoid generating annotations to mRNAs that are not presumed to encode functional products.
Efforts to clone and sequence FL-cDNAs concentrate on long insert clones and may unfortunately select for unspliced transcript inserts even when fully spliced transcript inserts are available, as a result of the length-based selection protocol. Given the importance of FL-cDNAs for annotation efforts and functional studies, obtaining the most biologically relevant product may be at odds with obtaining the longest insert cDNA clone. Our alignments of FL-cDNAs with the Arabidopsis genomic sequence have identified several such suspect clones, and future studies will be required to better understand the structure of these genes.
While the stringent alignment validation measures employed did purposefully prevent automated annotation updates from occurring based on low quality alignments, there were several legitimate cDNAs which failed to conform to the validation criteria and were excluded from automated updates. For example, the gene for MADS affecting flowering 1 protein (MAF1) (At1g77080) (42) contains an intron of length 2470 bp, which is extraordinarily large for an Arabidopsis gene. In addition, three alternative splicing variations of MAF1 are supported by alignments of transcript sequences gi|11545546, gi|11545544 and gi|13649968. All isoforms were annotated correctly during previous efforts employing less stringent validation requirements (17). Another gene eluding these automated annotation updates is the Agamous gene (At4g18960), which has both an extraordinarily large intron of 2999 bp and lacks an ATG start codon, replaced by an ACG codon (43). Future software enhancements should allow for automated annotation updates under certain exceptions to validation criteria, although the importance of manual examination and curation of the unusual and unexpected cannot be overstated.
The Arabidopsis transcriptome
The gene structure annotation improvements described above, including the annotation of alternative splicing isoforms, non-consensus splice sites and UTRs, are included in the latest release of the TIGR Arabidopsis genome annotation, the fourth Arabidopsis annotation release (release v.4.0, April 2003) provided by TIGR since the beginning of the Arabidopsis genome re-annotation effort in January 2001, directly following the completion of the genome sequence (30). This latest Arabidopsis genome annotation contains 27 170 protein coding genes, 18 272 of which are matched by EST and/or cDNA sequences. Of the 27 395 complete cDNA sequences currently available, 24 964 are now incorporated into the annotation, supporting the complete gene structures of 12 053 genes. UTRs have been annotated on 17 069 genes, yielding 16 216 upstream UTRs of 129 bp average length and 17 754 downstream UTRs of 235 bp average length. Analysis of the remaining expressed sequence data which failed to be incorporated is continuing and may serve to provide more complex updates to gene structures or lead to the annotation of new genes. The latest TIGR genome annotation and associated data are available at TIGR (ftp://ftp.tigr.org/pub/data/a_thaliana/ath1).
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank the TIGR Information Technology group for their support, particularly Susan Lo, Michael Heaney and Billy Lee. Thanks are due to Mihai Pop for fruitful algorithmic input. Finally, we would like to give special thanks to Jim Kent, Liliana Florea, Webb Miller and, especially, Xiaoqiu Huang for providing the community with useful sets of tools for interrogating sequence data and providing a foundation for the work described here. Additional thanks are due to Volker Brendel for making the GeneSeqer program available and for providing a thorough review of the manuscript. A.L.D. and S.L.S. were supported in part by NIH grant R01-LM06845-04. The Arabidopsis genome re-annotation at TIGR is supported by the National Science Foundation (Cooperative Agreement DBI 9813586).
REFERENCES
- 1.Adams M.D., Kelley,J.M., Gocayne,J.D., Dubnick,M., Polymeropoulos,M.H., Xiao,H., Merril,C.R., Wu,A., Olde,B., Moreno,R.F. et al. (1991) Complementary DNA sequencing: expressed sequence tags and human genome project. Science, 252, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 2.Huang X., Adams,M.D., Zhou,H. and Kerlavage,A.R. (1997) A tool for analyzing and annotating genomic sequences. Genomics, 46, 37–45. [DOI] [PubMed] [Google Scholar]
- 3.Bailey L.C. Jr, Searls,D.B. and Overton,G.C. (1998) Analysis of EST-driven gene annotation in human genomic sequence. Genome Res., 8, 362–376. [DOI] [PubMed] [Google Scholar]
- 4.Wolfsberg T.G. and Landsman,D. (1997) A comparison of expressed sequence tags (ESTs) to human genomic sequences. Nucleic Acids Res., 25, 1626–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kan Z., Rouchka,E.C., Gish,W.R. and States,D.J. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res., 11, 889–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quackenbush J., Cho,J., Lee,D., Liang,F., Holt,I., Karamycheva,S., Parvizi,B., Pertea,G., Sultana,R. and White,J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res., 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wheeler D.L., Church,D.M., Federhen,S., Lash,A.E., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Sequeira,E., Tatusova,T.A. et al. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res., 31, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christoffels A., van Gelder,A., Greyling,G., Miller,R., Hide,T. and Hide,W. (2001) STACK: Sequence Tag Alignment and Consensus Knowledgebase. Nucleic Acids Res., 29, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouck J., Yu,W., Gibbs,R. and Worley,K. (1999) Comparison of gene indexing databases. Trends Genet., 15, 159–162. [DOI] [PubMed] [Google Scholar]
- 10.Florea L., Hartzell,G., Zhang,Z., Rubin,G.M. and Miller,W. (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res., 8, 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wheelan S.J., Church,D.M. and Ostell,J.M. (2001) Spidey: a tool for mRNA-to-genomic alignments. Genome Res., 11, 1952–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Usuka J., Zhu,W. and Brendel,V. (2000) Optimal spliced alignment of homologous cDNA to a genomic DNA template. Bioinformatics, 16, 203–211. [DOI] [PubMed] [Google Scholar]
- 14.Birney E. and Durbin,R. (2000) Using GeneWise in the Drosophila annotation experiment. Genome Res., 10, 547–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salamov A.A. and Solovyev,V.V. (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res., 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeh R.F., Lim,L.P. and Burge,C.B. (2001) Computational inference of homologous gene structures in the human genome. Genome Res., 11, 803–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haas B.J., Volfovsky,N., Town,C.D., Troukhan,M., Alexandrov,N., Feldmann,K.A., Flavell,R.B., White,O. and Salzberg,S.L. (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol., 3, RESEARCH0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seki M., Narusaka,M., Kamiya,A., Ishida,J., Satou,M., Sakurai,T., Nakajima,M., Enju,A., Akiyama,K., Oono,Y. et al. (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science, 296, 141–145. [DOI] [PubMed] [Google Scholar]
- 19.Zhu W., Schlueter,S.D. and Brendel,V. (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol., 132, 469–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wortman J.R., Haas,B.J., Hannick,L.I., Smith,R.K.,Jr, Maiti,R., Ronning,C.M., Chan,A.P., Yu,C., Ayele,M., Whitelaw,C.A. et al. (2003) Annotation of the Arabidopsis genome. Plant Physiol., 132, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pearson W.R. and Lipman,D.J. (1988) Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofte H., Desprez,T., Amselem,J., Chiapello,H., Rouze,P., Caboche,M., Moisan,A., Jourjon,M.F., Charpenteau,J.L., Berthomieu,P. et al. (1993) An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J., 4, 1051–1061. [DOI] [PubMed] [Google Scholar]
- 23.Cooke R., Raynal,M., Laudie,M., Grellet,F., Delseny,M., Morris,P.C., Guerrier,D., Giraudat,J., Quigley,F., Clabault,G. et al. (1996) Further progress towards a catalogue of all Arabidopsis genes: analysis of a set of 5000 non-redundant ESTs. Plant J., 9, 101–124. [DOI] [PubMed] [Google Scholar]
- 24.Newman T., de Bruijn,F.J., Green,P., Keegstra,K., Kende,H., McIntosh,L., Ohlrogge,J., Raikhel,N., Somerville,S., Thomashow,M. et al. (1994) Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol., 106, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delseny M., Cooke,R., Raynal,M. and Grellet,F. (1997) The Arabidopsis thaliana cDNA sequencing projects. FEBS Lett., 405, 129–132. [DOI] [PubMed] [Google Scholar]
- 26.Rounsley S., Lin,X. and Ketchum,K.A. (1998) Large-scale sequencing of plant genomes. Curr. Opin. Plant Biol., 1, 136–141. [DOI] [PubMed] [Google Scholar]
- 27.White J.A., Todd,J., Newman,T., Focks,N., Girke,T., de Ilarduya,O.M., Jaworski,J.G., Ohlrogge,J.B. and Benning,C. (2000) A new set of Arabidopsis expressed sequence tags from developing seeds. The metabolic pathway from carbohydrates to seed oil. Plant Physiol., 124, 1582–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Asamizu E., Nakamura,Y., Sato,S. and Tabata,S. (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res., 7, 175–180. [DOI] [PubMed] [Google Scholar]
- 29.Volfovsky N., Haas,B.J. and Salzberg,S.L. (2003) Computational discovery of internal micro-exons. Genome Res., 13, 1216–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature, 408, 796–815. [DOI] [PubMed] [Google Scholar]
- 31.Bell L.R., Maine,E.M., Schedl,P. and Cline,T.W. (1988) Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell, 55, 1037–1046. [DOI] [PubMed] [Google Scholar]
- 32.Bell L.R., Horabin,J.I., Schedl,P. and Cline,T.W. (1991) Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell, 65, 229–239. [DOI] [PubMed] [Google Scholar]
- 33.Lopez A.J. (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu. Rev. Genet., 32, 279–305. [DOI] [PubMed] [Google Scholar]
- 34.Brett D., Hanke,J., Lehmann,G., Haase,S., Delbruck,S., Krueger,S., Reich,J. and Bork,P. (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett., 474, 83–86. [DOI] [PubMed] [Google Scholar]
- 35.Modrek B., Resch,A., Grasso,C. and Lee,C. (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res., 29, 2850–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vonarx E.J., Howlett,N.G., Schiestl,R.H. and Kunz,B.A. (2002) Detection of Arabidopsis thaliana AtRAD1 cDNA variants and assessment of function by expression in a yeast rad1 mutant. Gene, 296, 1–9. [DOI] [PubMed] [Google Scholar]
- 37.Murphy T.M. and Gao,M.J. (2001) Multiple forms of formamidopyrimidine-DNA glycosylase produced by alternative splicing in Arabidopsis thaliana. J. Photochem. Photobiol. B, 61, 87–93. [DOI] [PubMed] [Google Scholar]
- 38.Lazar G. and Goodman,H.M. (2000) The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol. Biol., 42, 571–581. [DOI] [PubMed] [Google Scholar]
- 39.Xiao Y.L., Malik,M., Whitelaw,C.A. and Town,C.D. (2002) Cloning and sequencing of cDNAs for hypothetical genes from chromosome 2 of Arabidopsis. Plant Physiol., 130, 2118–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brett D., Pospisil,H., Valcarcel,J., Reich,J. and Bork,P. (2002) Alternative splicing and genome complexity. Nature Genet., 30, 29–30. [DOI] [PubMed] [Google Scholar]
- 41.Mount S.M. (2000) Genomic sequence, splicing and gene annotation. Am. J. Hum. Genet., 67, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ratcliffe O.J., Nadzan,G.C., Reuber,T.L. and Riechmann,J.L. (2001) Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol., 126, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Riechmann J.L., Ito,T. and Meyerowitz,E.M. (1999) Non-AUG initiation of AGAMOUS mRNA translation in Arabidopsis thaliana. Mol. Cell. Biol., 19, 8505–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]