The PX-BAR membrane-remodeling unit of sorting nexin 9 (original) (raw)
Abstract
Sorting nexins (SNXs) form a family of proteins known to interact with components in the endosomal system and to regulate various steps of vesicle transport. Sorting nexin 9 (SNX9) is involved in the late stages of clathrin-mediated endocytosis in non-neuronal cells, where together with the GTPase dynamin, it participates in the formation and scission of the vesicle neck. We report here crystal structures of the functional membrane-remodeling unit of SNX9 and show that it efficiently tubulates lipid membranes in vivo and in vitro. Elucidation of the protein superdomain structure, together with mutational analysis and biochemical and cell biological experiments, demonstrated how the SNX9 PX and BAR domains work in concert in targeting and tubulation of phosphoinositide-containing membranes. The study provides insights into the SNX9-induced membrane modulation mechanism.
Keywords: BAR domain, membrane transport, PX domain, sorting nexin, tubulation
Introduction
The generation of vesicular carriers for intracellular trafficking is characterized by an elaborate system of integral and peripheral proteins, which together with membrane lipids, package cargo for delivery to target destinations. The modulation of membrane shape into round or tubular extensions, and the subsequent sealing and scission of the leaving vesicle, are key events in such progressions. Proteins with BAR (Rvs/amphiphysin/Bin) domains, and related F-BAR domains, were recently acknowledged as important proteins with membrane-modulating activities (Peter et al, 2004; Gallop and McMahon, 2005; Itoh and De Camilli, 2006). When assembled on the lipid surface, these proteins can create or stabilize membrane curvature, and thereby facilitate the generation of vesicular or tubular structures. The BAR domain is a dimer of subdomains, consisting of three or four long helices forming a protein structure with a crescent shape that contributes to the bending of the membrane into a fixed curvature by scaffolding (Peter et al, 2004; Gallop et al, 2006; Masuda et al, 2006; Shimada et al, 2007). The generation of high curvature, as found in small vesicles or narrow tubes, requires in addition that lipid molecules are displaced by partial insertion of the protein into the bilayer (Zimmerberg and Kozlov, 2006). In some BAR proteins, like amphiphysin and endophilin, such function is accomplished by N-terminal extensions that fold as amphipatic helices that embed into the membrane to create a ‘wedge effect' (Gallop and McMahon, 2005; Itoh and De Camilli, 2006). Electron micrographs showed that spherical liposomes incubated in vitro with recombinant amphiphysin 1 BAR domain are converted to tubules with a continuous and tightly spaced arrangement of thin rings (Takei et al, 1999), indicating well-ordered oligomerization of the BAR domains on the membrane surface. Also the EFC/F-BAR domain induces tubular membranes coated with rings/spirals of protein oligomers (Shimada et al, 2007). This suggests that the tubulation mechanism might be accompanied by BAR domain oligomerization.
BAR domain proteins possess additional domains that either bind to other proteins required for the vesiculation process, or which assist in their localization to certain lipid structures. In a large number of proteins, BAR domains are flanked by PX (phox homology) domains or PH (pleckstrin homology) domains (Habermann, 2004). Both these domains show affinity for various phosphorylated phosphatidylinositols (phosphoinositides), but may also be involved in protein–protein interactions (Seet and Hong, 2006; Lemmon, 2007). It is thought that the combination of the BAR domain with PX or PH domains adds to the specificity in localization and action on defined cellular membranes. The PX-BAR proteins are found within a collection of proteins designated as sorting nexins (SNXs) (Worby and Dixon, 2002; Seet and Hong, 2006). The SNX protein family, which consists of over 30 members in mammals, is involved in diverse vesicular trafficking events. Twelve of the SNX proteins have the PX-BAR domain combination, which in all cases is located at the C-terminus of the protein. This is in contrast to the BAR-PH proteins, which have their lipid-binding domains at the N-terminus.
SNX1, SNX2, and SNX9 are the best characterized of the PX-BAR proteins. SNX1 and SNX2 have been shown to participate in retromer function for sorting and formation of tubular endosomal structures involved in the recycling of material to the trans-Golgi network (Carlton et al, 2004; Griffin et al, 2005; Rojas et al, 2007). SNX9, which in addition to PX-BAR also contains an SH3 domain and a long unstructured motif domain (LC domain), participates in the formation of clathrin-coated vesicles at the plasma membrane in non-neuronal cells (Lundmark and Carlsson, 2003, 2004; Soulet et al, 2005). Together with its two paralogs SNX18 and SNX30, SNX9 forms a sub-family of PX-BAR proteins (K Haberg, R Lundmark, and SR Carlsson, in preparation). These proteins are the only ones with an SH3 domain in combination with PX-BAR. SNX9 comes into play at a relatively late stage in clathrin vesicle formation and recruits the large GTPase dynamin to the neck of the vesicle. Dynamin is thought to be directly responsible for the scission reaction that releases the vesicle from the membrane (Hinshaw, 2000; Praefcke and McMahon, 2004). SNX9 binds dynamin via its SH3 domain, and adheres to the clathrin coat by binding motifs in the LC domain for both clathrin and its adaptor protein AP-2. Although dynamin itself has membrane-tubulating activity in vitro (Takei et al, 1999), it is uncertain if this activity is alone responsible for formation of the narrow tubular neck before scission takes place (Roux et al, 2006). It is possible that the PX-BAR unit of SNX9 cooperates with dynamin for the tubulation and scission reaction to release the coated vesicle. In addition to its proposed direct role in clathrin-mediated endocytosis, SNX9 has been suggested to be involved in a number of plasma membrane-related events such as receptor downregulation, clathrin-independent endocytosis, and actin dynamics (Howard et al, 1999; Lin et al, 2002; MaCaulay et al, 2003; Yeow-Fong et al, 2005; Childress et al, 2006; Badour et al, 2007; Yarar et al, 2007).
In order to understand how PX-BAR proteins perform their functions in the progression of membrane deformation, it is of importance to reveal the details of this structural unit. We show here that PX-BAR of SNX9, together with flanking subdomains, form a single superdomain with efficient membrane-tubulation activity, and mutational analysis revealed important features for the function of combined lipid-binding domains.
Results and discussion
PX-BAR is a functional membrane-binding unit of SNX9 active in membrane tubulation
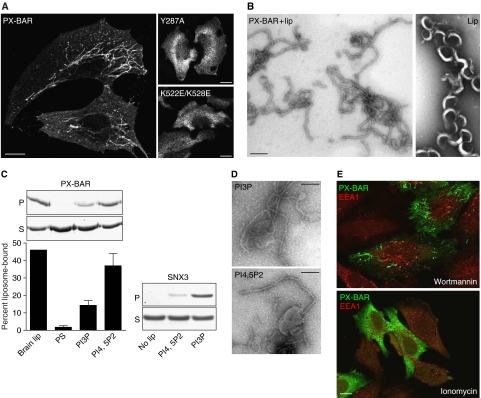
It was previously shown that recombinant PX-BAR (residues 185–595) from SNX9 had membrane-binding activity in vitro (Lundmark and Carlsson, 2003). When expressed in HeLa cells as a myc-tagged protein, PX-BAR produced large numbers of long tubules (Figure 1A). Earlier structural/functional studies on PX domain and BAR domain proteins have identified amino acids critical for lipid binding and/or tubulation activity (Cheever et al, 2001; Peter et al, 2004). Mutations of the corresponding residues in the PX domain (Y287A) or in the BAR domain (K522E/K528E) of SNX9 totally abolished the tubulation capacity, showing that both domains are involved in the membrane-modulating function. The tubulating activity could be reproduced in vitro by incubation of recombinant PX-BAR with liposomes made from brain-derived phospholipids (Figure 1B). The generated lipid tubules had a uniform diameter of approximately 20 nm, but no distinct protein pattern could be visualized by electron microscopy.
Figure 1.
Membrane binding and tubulation by SNX9 PX-BAR. (A) Epifluorescence micrographs of HeLa cells transfected with myc-tagged wild-type (PXBAR) and mutated (Y287A and K522E/K528E) PX-BAR, stained with anti-myc antibodies. While wild-type protein created distinct elongated tubular structures, the PX and BAR domain mutants gave diffuse cytoplasmic staining. Scale bars are 10 μm. (B) Electron micrographs of liposomes and tubules. Liposomes were made from brain lipid extract and passed through 800-nm filters by extrusion. Liposomes were mixed with 10 μM purified PX-BAR (PXBAR+lip) or buffer (lip), and processed for electron microscopy. Scale bar is 100 nm. (C) Liposomes were made from brain lipid extract (brain lip), a 10:10:40:40 (w/w) mixture of phosphatidylserine, cholesterol, phosphatidylethanolamine, and phosphatidylcholine (PS), or with 5% of PtdIns-3-phosphate (PI3P) or PtdIns-4,5-bisphosphate (PI4,5P2) in the PS mixture at the expense of phosphatidylserine. Liposomes were extruded through 800-nm filters and mixed with 0.7 μM protein as indicated, or protein was incubated without liposomes (no lip). Samples were centrifuged and supernatants (S) and pellets (P) were analyzed by SDS–PAGE and quantitated by densitometry. The bars show the means (±s.e.m.) from four experiments. (D) Electron micrographs of PI4,5P2- and PI3P-containing liposomes generated as in panel C and incubated with 10 μM purified PX-BAR. Scale bar is 100 nm. (E) Epifluorescence micrographs of HeLa cells transfected with myc-tagged PX-BAR construct and treated with wortmannin or ionomycin as indicated. Cells were stained with anti-myc antibodies and costained with antibodies for EEA1. Scale bar is 10 μm.
The quantitative dominating phosphoinositides involved in endosomal trafficking are PI(3)P and PI(4,5)P2 (Roth, 2003). When liposomes with defined lipid composition were used in binding experiments with PX-BAR, we found that the protein showed higher affinity to PI(4,5)P2- than to PI(3)P-containing liposomes (Figure 1C), consistent with the role of SNX9 in clathrin-mediated endocytosis. As a control, the PX domain of SNX3 (Xu et al, 2001) showed strongest binding to liposomes with PI(3)P as expected (Figure 1C). PX-BAR of SNX9 was able to induce tubules from both PI(4,5)P2- and PI(3)P-containing liposomes in vitro (Figure 1D). Consistently, however, tubulation was more efficient when the liposomes contained PI(4,5)P2, possibly due to the fact that the binding was stronger to these liposomes. The results indicate that SNX9 uses restricted phosphoinositides to achieve a robust initial binding required for membrane tubulation.
In order to confirm the preference of SNX9 PX-BAR for PI(4,5)P2 in cells, we treated transfected cells with agents known to affect the metabolism of specific phosphoinositides, and scored for PX-BAR tubulation efficiency. Ionomycin is a calcium ionophore that induces the activation of phospholipase C, resulting in hydrolysis of PI(4,5)P2 (Varnai and Balla, 1998). A brief treatment with ionomycin gave a complete redistribution of PX-BAR from a tubular to a cytoplasmic appearance (Figure 1E). Wortmannin, which is an inhibitor of PI3 kinases (Vanhaesebroeck et al, 2001), had no effect on the distribution of PX-BAR. As a control, wortmannin was found to markedly affect the localization of the PI(3)P-binding protein EEA1, which appeared on more elongated structures after treatment. As expected, EEA1 was found not to be affected by ionomycin. The results show that, although PX-BAR has the capacity to bind and tubulate PI(3)P-liposomes in vitro, the localization in cells is dependent on PI(4,5)P2-containing membranes, which suggests that this is the primary phosphoinositide for SNX9 function.
The crystal structure of SNX9 PX-BAR
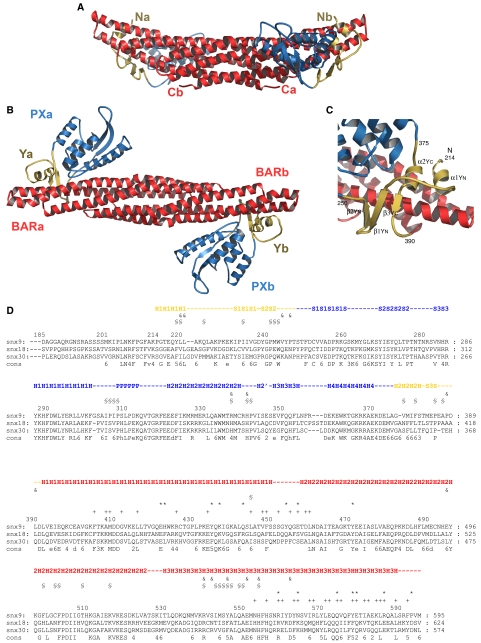
The crystal structure of recombinant SNX9 PX-BAR (residues 204–595) represents the protein homodimer (Figure 2A and B). SNX9 PX-BAR dimerizes via a canonical symmetric crescent-shaped BAR dimer formation similar to that observed in other BAR domains (Tarricone et al, 2001; Peter et al, 2004; Gallop et al, 2006; Masuda et al, 2006). The distal parts of the curved BAR dimer are symmetrically decorated with PX domains and small subdomains. The regions flanking the SNX9-PX domain are located in proximity and form a single subdomain of two parallel packed α-helices and an antiparallel β-sheet of three β-strands twining about the BAR domain arm (Figure 2C). Based on its shape and its role in interconnecting the PX and BAR domains, we call this subdomain the ‘yoke domain' (Y domain), which is split into YN (residues 214–250) and YC (residues 375–390) (Figure 2C). The recently described BAR-PH structure of APPL1 (Li et al, 2007) showed no similar interconnecting domain between the BAR and PH domains, which instead contacted via a small surface at the tip of the BAR domain. The yoke subdomain in SNX9 unites the membrane-binding domains, forming a single superdomain for membrane sculpting. The extensive interdomain contacts between BAR, PX and Y provide proper domains inter-orientation and define the curved shape of the entire protein construct. The structure-based sequence alignment (Figure 2D) of the SNX9, SNX18, and SNX30 PX-BARs demonstrate high degree of structural conservation in the proteins composing the subfamily.
Figure 2.
(A) Side view of the SNX9 PX-BAR dimer structure ribbon diagram. BAR domains are shown in red, PX in blue, and the novel Yoke (Y) subdomain in yellow. N- and C-termini are labeled. (B) Top view of the SNX9 PX-BAR dimer structure. The domains are labeled. (C) Ribbon representation of SNX9 Yoke (Y) subdomain structure. The Yoke domain consists of two parts: YN derived from amino-acid residues 214–250 and YC from 375–390. Secondary structure elements are labeled. (D) Sequence alignment of human SNX9 (Q9Y5X1), SNX18 (AAH67860), and SNX30 (ABN09670) PX-BAR domains. The proteins represent a subfamily of PX-BAR SNXs. Secondary structure elements corresponding to the SNX9 structure are monitored. Coloring corresponds to that from panels A–C. Helices are labeled with H, β-strands with S, and P denotes a proline-rich motif. The amino-acid residues involved in the dimerization are labeled (+, hydrophobic bar–bar contacts; *, H-bond bar–bar contacts), as well as residues involved in interdomain interaction (§, hydrophobic bar-px:y contacts; &, H-bonds bar-px:y contacts). Symbols of consensus sequence are: 2, E/Q; 3, T/S; 4, K/R; 5, Y/F; and 6, hydrophobic.
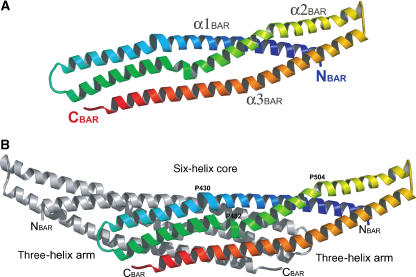
The SNX9 BAR domain. The SNX9-BAR monomer consists of three long α-helices connected via loops (Figure 3A). The three helices form a left-handed bent coiled-coil structure within an SNX9-BAR monomer. Upon dimerization, the three-helical coiled-coils from both the monomers weave into a six-helical coiled-coil forming the dimer core, whereas the extremity of the monomers form three-helix bundle arms extended from the dimer core (Figure 3B). The dimer contacts via extensive hydrophobic and polar interactions typical for coiled-coil helical bundles, and the interface contact area is 3191 Å2. The core BAR regions can be superimposed with the reported BAR domain structures with an r.m.s.d. ∼2 Å. In spite of similar domain architecture and conserved core structure, the shape and curvature of different BARs varies significantly from 150 Å for arfaptin 2 (Tarricone et al, 2001) to 600 Å for PHC-F-BAR (Shimada et al, 2007). SNX9-BAR exhibits an intermediate curved crescent-like shape that would fit a circle of diameter 400 Å. As shown for the known BAR domain structures, the curvature of the BAR dimers results from the manner in which the monomers interact and from the kinks in the helices forming the dimers (Peter et al, 2004; Casal et al, 2006; Masuda et al, 2006). In SNX9-BAR the α1 is kinked in the middle at the position of the Pro430–Leu431–-Pro432 sequence (Figure 3B). This insert allows bending of helix1 in the direction opposite to the main curvature pattern of the BAR dimer. The kink is located at a position corresponding to the helical insert in the α1 of endophilin, which is believed to be incorporated into the membrane, facilitating membrane curvature (Masuda et al, 2006). The α2 is kinked at two sites: the first is at the position of Pro482, which is aligned in the space with the Pro–Leu–Pro kink of α1, and the second is at position Pro504. The kinks allow bending of the SNX9-BAR arms in a direction perpendicular to the dimer symmetry axis. The described kinks structurally determine the SNX9-BAR arm orientations and consequently the curvature of the SNX9-BAR. The bent SNX9 BAR dimer conformation is constrained by the extensive (contact area is 1661 Å2) hydrophobic and polar contacts with the PX and Y domains.
Figure 3.
(A) SNX9 BAR domain monomer in rainbow colors from the N-terminus in blue to C-terminus in red. α-helices are labeled. (B) SNX9 BAR domain. The core and arm regions are monitored, and the proline amino-acid residues at the α-helices kinks are labeled.
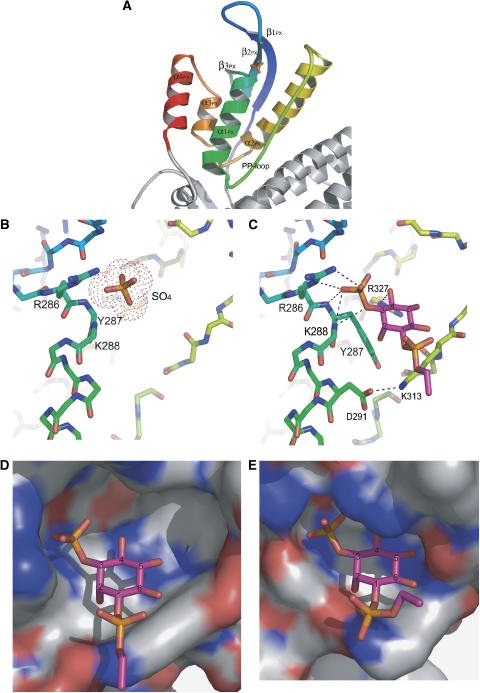
The SNX9 PX domain. The PX domain (residues 251–372) represents the conventional PX domain fold but with an extra C-terminal α4-helix. It consists of an N-terminal three-stranded β-sheet (β1–β3) and a C-terminal helical bundle comprised of four α-helices (α1–α4), where α1 and α2 are connected via a long loop containing a Pro-rich motif (PP-loop) (Figure 4A). The SNX9-PX PIP-binding pocket represents a positively charged cavity formed by the α1, β2, and α2 with the adjacent PP-loop.
Figure 4.
(A) Ribbon diagram of the SNX9 PX domain in rainbow colors, from PX N-terminus (251) in blue to PX C-terminus (372) in red. Secondary structure elements are labeled. (B) SO4 ion bound to the SNX9 PX domain PIP-binding pocket. Amino-acid residues contacting the ion are shown and labeled. (C) PI(3)P molecule bound to SNX9 PX domain PIP-binding pocket. Amino-acid residues contacting the PI(3)P are shown and labeled. H-bonds are depicted as dashed lines. (D) PI(3)P molecule in the SNX9 PX domain PIP-binding pocket shown as a molecular surface. The pocket is large enough to accommodate a PIP molecule phosphorylated at the fourth and fifth positions. (E) The PI(3)P molecule fills the SNX3 PI(3)P-binding pocket (PDB ID 1OCU).
The SO4-bound SNX9 PX-BAR structure revealed an anion-binding site in the SNX9-PX PIP-binding pocket occupied by the sulfate ion (Figure 4B). The ion-binding site is structurally conserved among PX domains that bind phosphoinositides with head groups phosphorylated at the third position (p40phox, Grd19p, and CISK-PX) (Bravo et al, 2001; Zhou et al, 2003; Xing et al, 2004) and corresponds to the PI3P third phosphate moiety-binding site in the Grd19p–PI(3)P (Zhou et al, 2003) and p40phox–PI(3)P (Bravo et al, 2001) complex structures. The site is formed by the side chain of Arg286 and the main-chain nitrogens of Tyr287 and Lys288. Arg286 is conserved among most of PX domains and has been characterized as the binding determinant of the 3-phosphate moiety of PIPs (Bravo et al, 2001; Zhou et al, 2003; Xing et al, 2004) The observed SO4 ion bound to SNX9-PX may mimic the phosphate moiety of a bound PIP.
The SNX9 PX-BAR crystals were soaked in solutions containing different PIPs (see Materials and methods), but only C4PtdIns3P soaking resulted in SNX9-PX-BAR complex formation. The PI(3)P-bound SNX9 PX-BAR structure proved the functionality of the described anion binding site. The observed PI(3)P molecule occupies the SNX9-PX PIP-binding pocket in a manner similar to that observed in the Grd19p–PI(3)P and p40phox–PI(3)P complexes (Figure 4C), where the third phosphate moiety is accommodated in the conserved anion-binding pocket. Lys313 is strictly conserved among PX domains and located close to the 1-phosphate group of the PIP ligand, and contacts via hydrophobic and Van der Waals interactions with the inositol ring. The ring also contacts the side chain of the conserved aromatic Tyr287 via hydrophobic stacking interactions. Mutations of these residues greatly decrease PX domain affinities to PIPs (Bravo et al, 2001). This structural conservation might indicate the potential role of the Lys313 and Tyr287 residues in the inositol ring positioning.
Thus, the SNX9 PX-BAR crystal structure shows that the SNX9-PX domain PIP-binding pocket is preformed for PI(3)P binding, but the pocket has extra room to accommodate a PIP, which is also phosphorylated at the 4 and 5 positions (Figure 4D; Supplementary Figure S1A). This is in contrast to the situation in Grd19p, where the pocket appears to be restricted to PI(3)P binding (Figure 4E). The crystal structure of PI3K-Ca-PX, which specifically binds PI(4,5)P2 (Stahelin et al, 2006), revealed a sulfate ion mimicking a plausible position of the 4-phosphate, consistent with the inositol ring orientation observed in PX-PI(3)P bound structures. The ion is hydrogen bonded to the Arg327 side chain located on α2, and this residue is conserved among PXs regardless of PIP specificity. In the PI(3)P-binding PXs, including SNX9, the Arg327 coordinates the 4-and 5-OH groups of PI(3)P, but the Arg327 side chain may adopt a different rotomer conformation in the presence of the 4- and 5-phosphate moieties. It was previously suggested that SNX9 is promiscuous in PIP lipid binding (Lundmark and Carlsson, 2004) and this was confirmed in the present study (Supplementary Figure S2). However, in the liposome assay condition used here, which was optimized for tubule formation, binding was consistently stronger to PI(4,5)P2- than to PI(3)P-containing liposomes. It is likely that SNX9 PX-BAR in the crystalin state was unable to bind PI(4,5)P2 due to the crystal packing, restricting the flexibility of the loops involved in PIP-binding pocket formation. It is also possible that protein assembly during tubulation affects the affinity and specificity in PIP binding.
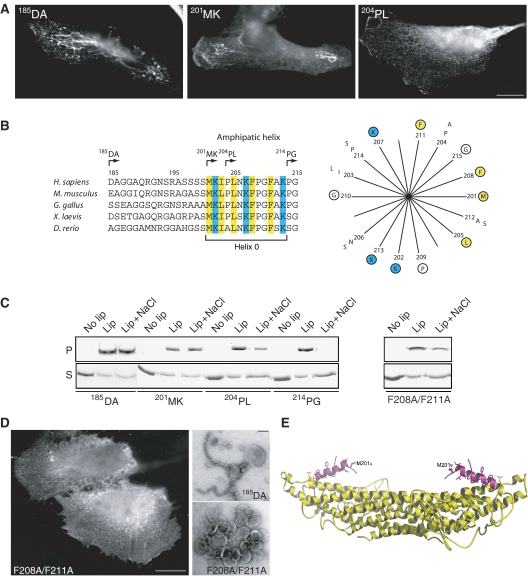
Amphipatic helix important for membrane tubulation
The structurally resolved N-terminal α-helix begins at P214, whereas the P204-K213 stretch at the N-terminus appears to be flexible or unordered. This finding, together with the fact that the crystallizable construct (204PL; for designations see Figure 5B) was shorter than the tubulating protein (185DA), led us to investigate a series of truncation variants to test for the importance of the N-terminus for tubulating activity. When the myc-tagged protein was expressed in HeLa cells, no difference in tubulating activity was found between 201MK and 185DA (Figure 5A). This indicates that the sequence from D185 to S200, which is not conserved between species (Figure 5B), is not directly involved in membrane modulation, but is probably functioning as a bridge to lead the rest of the full-length protein (SH3 and LC domains) to the convex side of the structure when SNX9 is bound to the membrane. Removal of three additional residues from 201MK to yield the 204PL construct resulted in a striking loss of tubulating activity (Figure 5A), although occasionally tubes could be seen in a few cells. Removal of the remaining residues to P214 (construct 214PG) rendered the protein completely inactive in tubulation. Control experiments with an inserted linker sequence ruled out that the effect was due to the close position of the myc tag in the shorter variants (results not shown). The results indicate that the sequence 201-MKIPLNKFPGFAK is critical for tubulation activity.
Figure 5.
Membrane tubulation is dependent on an amphipatic helix upstream of the YN part of yoke sub-domain. (A) Epifluorescence micrographs of HeLa cells transfected with myc-tagged 185DA, 201MK, and 204PL protein constructs stained with anti-myc antibodies. Scale bar is 10 μm. (B) Sequence comparison of the potential amphipatic helix region (helix 0) in SNX9 from selected species as indicated. Numbers refer to amino-acid position in human SNX9. Arrows depict the N-terminus of protein constructs 185DA, 201MK, 204PL, and 214PG. Amino acids M201-G215 are presented as a helical wheel with positively charged amino acids in blue and hydrophobic amino acids in yellow. Conserved amino acids are encircled. (C) Liposome binding assay where indicated proteins were incubated with liposomes generated from total brain lipids (lip), or incubated without liposomes (no lip). Where indicated (lip+NaCl), 0.5 M NaCl was added to the protein/liposome mixture. Samples were centrifuged and supernatants (S) and pellets (P) were analyzed by SDS–PAGE. The F208A/F211A protein was made by mutation of the 185DA construct. (D) Epifluorescence micrograph of myc-tagged amphipatic helix mutant F208A/F211A expressed in HeLa cells (left panel, scale bar is 10 μm). Electron microscopic analysis of liposomes incubated with purified proteins as indicated (right panel, scale bar is 100 nm). Note the inability of the F208A/F211A mutant to tubulate membranes both in vivo and in vitro. (E) A 3D model of the N-terminal SNX9 helix 0. Structurally resolved amino acids are shown in yellow and the modeled part is in magenta.
Theoretical analysis of the highly conserved M201-K213 sequence suggested that it may form an amphipatic helix upon membrane contact (Figure 5B and E). If so, this structure (designated helix 0) might functionally resemble the N-helix in N-BAR proteins, which has been shown to be inserted into the lipid bilayer to create curvature (Peter et al, 2004; Gallop et al, 2006; Itoh and De Camilli, 2006). Insertion of SNX9-helix 0 into the membrane should make the membrane-bound protein resistant to high salt concentrations due to a hydrophobic effect. Experimental analysis showed, as predicted, that membrane-bound 185DA and 201MK were resistant to 0.5 M NaCl (Figure 5C). 214PG, although still binding efficiently to liposomes, had lost the hydrophobic property and was almost completely dissolved by salt, whereas 204PL showed intermediate sensitivity. That hydrophobic amino acids in helix 0 are important for membrane insertion and tubulation was verified by mutating F208 and F211 to Ala in the 185DA context. The results from both salt sensitivity and tubulation experiments with recombinant protein, together with expression of myc-tagged protein in cells (Figure 5C and D), confirmed that helix 0 is acting by insertion of hydrophobic amino acids, most likely to create a wedge effect similar to what has been suggested for N-BAR proteins. However, since we presently have no experimental proof for a helix (which may be formed only when the protein is contacting the membrane), it cannot be ruled out that the M201-K213 sequence exerts its function through some alternative conformational structure to create membrane curvature. SNX9, SNX18, and SNX30 show similar properties in their corresponding sequences with most of their positive and hydrophobic amino acids in the same positions (see Figure 2D), and are therefore likely to function similarly. Although conserved regions upstream of the PX domain can be found in other PX-BAR proteins, such as in SNX1 and SNX2, these do not resemble the sequences predicted as amphipatic helices in the SNX9 subfamily members. It is possible that PX-BAR proteins have adopted different strategies to modulate or stabilize membrane curvature.
The membrane-binding surface
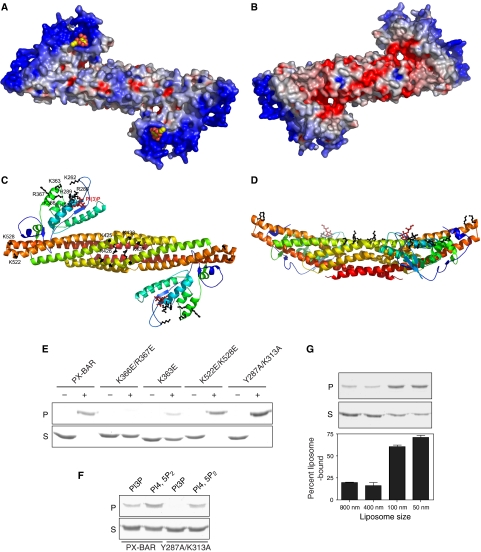
The crystal structures with the biochemical data showed that the SNX9 PX-BAR represents a single functional membrane binding module efficient in membrane tubulation. The SNX9 PX-BAR electrostatic surface potential distribution calculated using GRASP (Nicholls et al, 1991) showed that the concave surface of the dimer is mostly positively charged (Figure 6A), whereas the convex side of the protein is more negative (Figure 6B). This allows proper initial orientation of the protein relative to the negatively charged membrane surface. The positive charge is not equally distributed on the SNX9 PX-BAR concave surface. The highly cationic patches are located on the PX domain surface (Figure 6C). Interestingly, in addition to the described PIP-binding pocket, the positive patch extends continuously to a second basic pocket, which is formed by amino-acid residues R286, K288, H289 from α1; K363, K366, R367 from α4; and K262 from the β1–β2 loop. These amino acids are all conserved among SNX9 subfamily members. Mutations in this second positively charged pocket demonstrated its importance for liposome binding. K363E and K366E/R367E SNX9 PX-BAR mutants showed significantly reduced binding capacity to brain liposomes, whereas mutations in the primary PIP-binding pocket (Y287A/K313A) had little effect on binding to this type of liposome (Figure 6E). However, all the described mutants totally abolished tubulation in vivo (see Figure 1A, and results not shown). Control experiments showed that the Y287A/K313A mutant indeed displayed decreased binding when liposomes with defined lipid composition were used (Figure 6F), confirming that the mutated site has a role in phosphoinositide binding. Since protein binding to PI(4,5)P2 was not completely abolished, it is possible that also other sites have affinity to this phospholipid. The SNX9 BAR domain also possesses well-pronounced positively charged surface patches. Conserved basic residues (R426, K425, K433, and K437) from both monomers are clustered in the middle of the dimer. The tops of the BAR dimer arms expose conserved basic residues K522 and K528. Mutation of these residues leads to loss of the tubulation activity in vivo, but does not affect the binding to brain liposomes (Figures 1A and 6E). These data indicate that the second positively charged pocket plays a critical role in the initial steps of SNX9 membrane binding, whereas membrane deformation requires the full set of intact PX-BAR superdomain structure, including the primary PIP-binding pocket and the positively charged patches on the BAR domain. At present, it is not known if the second site has preference for PIPs or other negatively charged phospholipids, and further studies are required to solve this issue. As we can see from the membrane-interacting surface analysis, the interdomain orientation and dimerization of SNX9 aligns the membrane binding surface of the BAR domain with the PIP-binding pockets of PX domains, and the curved shape of the entire protein construct facilitates simultaneous membrane interaction of both domains (Figure 6D).
Figure 6.
(A, B) Electrostatic potential surface of SNX9 PX-BAR. (A) The concave face of the dimer is mostly positively charged. PI(3)P moieties are shown in yellow and red. (B) The convex face is negatively charged. (C) Subfamily conserved basic amino-acid residues (black) forming clusters on the concave face of SNX9 PX-BAR dimer. The residues from one monomer are labeled. (D) Side view of panel C. (E) Membrane binding of PX-BAR mutants. The indicated proteins (0.7 μM) were incubated together with liposomes from total brain lipids (+) or without liposomes (−), and samples were centrifuged and the supernatant (S) and pellet (P) fractions were analyzed by SDS–PAGE. (F) Indicated proteins were incubated with PI(3)P- or PI(4,5)P2-containing liposomes made as in Figure 1C, and the samples were processed as in panel E. (G) The PX-BAR unit of SNX9 is curvature sensitive. Liposomes were made from a 30:70 (w/w) mixture of phosphatidylserine and phosphatidylcholine and extruded through filters of pore size 800, 400, 100, and 50 nm. PX-BAR (3 μM) was added and the samples were centrifuged, and the supernatants (S) and pellets (P) were analyzed by SDS–PAGE and quantitated by densitometry. The bars show the means from two experiments, with the maximum value indicated for each set.
When phosphoinositide-liposomes of different sizes were used in binding assays, we were unable to detect a binding preference dependent on the intrinsic curvature of the membrane (results not shown). Most likely, this reflects the efficient tubulating capacity of PX-BAR, which is expected to result in high binding regardless of starting liposome size. However, when phosphoinositide-free liposomes were used, a clear curvature-sensing effect was found (Figure 6G). PX-BAR was unable to bind 800 and 400-nm liposomes consisting of only phosphatidylserine and phosphatidylcholine, but when the size was decreased to 100 and 50 nm, a significant interaction was achieved. This result conforms to the shape of the BAR domain, and indicates that SNX9 requires sufficient initial binding strength in order to modulate the membrane into tubules. With phosphoinositides in the membrane, the binding via the PX domain is strong enough to force the lipids into contact with the positively charged amino acids at the concave face of the BAR domain. Without phosphoinositides, the membrane has to be of correct geometry from the start to display a suitable binding surface. In certain BAR proteins such as amphiphysin and endophilin, which have no phosphoinositide requirement, the tubulation capacity is enhanced by highly charged areas at the distal ends of the BAR domain or by the extra segments at the amino termini (Farsad et al, 2001; Peter et al, 2004; Gallop et al, 2006; Masuda et al, 2006). SNX9 uses in addition a phosphoinositide interaction mediated by the PX domain to overcome the thermodynamic barrier for bending the membrane by the BAR domain. This strategy, which adds another level of specificity, may be a common theme used by PX-BAR and BAR-PH proteins.
Based on the results in the present report, we propose a model for the membrane-modulating activity of SNX9 PX-BAR to generate 20-nm tubes. PX-BAR dimers approach the negatively charged membrane by interactions via the second lipid-binding site on the PX domains on the concave surface, followed by binding of PIP into the phosphoinositide-binding pocket. The specific PIP binding leads to the tightening of the PX-BAR–membrane interactions, initiating membrane deformation. When contacting the membrane, helix 0 forms and inserts partially into the membrane, creating membrane curvature. In order to stabilize the tubes and to create directionality, SNX9 PX-BAR dimers might oligomerize on the lipid surface by lateral contacts (R Lundmark and SR Carlsson, unpublished data, 2007; Yarar et al, 2007). Since the diameter of the generated membrane tubes is smaller than the curvature of the concave side of the PX-BAR structure, it follows that the elongated protein must assemble on the surface at an angle that is not perpendicular to the direction of the tube. It is possible that the oligomerization property of SNX9 PX-BAR is the major driving force behind the generation of narrow lipid tubules. Further studies are required to investigate this possibility in more detail.
The lack of curvature sensitivity for phosphoinositide-containing membranes indicates that the initial binding of SNX9 is to membrane formations with lower curvature in vivo, such as clathrin-coated membrane buds containing PI(4,5)P2. Subsequently, an enhancement of electrostatic interactions and PIP binding, together with membrane insertion and assembly into higher-order structures, might generate the 20-nm neck previously observed for highly invaginated clathrin-coated structures (Kosaka and Ikeda, 1983). This action of SNX9 is likely coordinated with the function of dynamin, which is also specific for PI(4,5)P2 (Salim et al, 1996). Dynamin is recruited to the membrane by SNX9 (Lundmark and Carlsson, 2004) and has been shown to play an essential role in the fission reaction at the neck of the forming vesicle (Hinshaw, 2000). Indeed, SNX9 has been found to be an efficient stimulator of dynamin's GTPase activity. This function is dependent both on the SH3 domain and the C-terminal part of SNX9, and is enhanced by PI(4,5)P2-containing membranes (Soulet et al, 2005).
Materials and methods
Protein expression and purification
cDNA fragments corresponding to amino acids 185–595, 201–595, 204–595, or 214–595 of human SNX9 were inserted into the pGEX-6P-2 vector (GE Healthcare). Mutagenesis was performed on plasmids containing the cDNA fragment 185–595 using QuickChange (Stratagene). All constructs were verified by sequencing.
Different SNX9 protein constructs were expressed and purified using essentially the same protocol. Proteins were expressed in BL21 pLysS cells grown in LB medium, or in LeMaster medium (LeMaster and Richards, 1985) supplemented with 50 mg/l seleno-methionine, at 20°C for 20 h after induction with 0.05 mM IPTG at an OD600 nm of 0.5. The proteins were purified using glutathione–Sepharose (GE Healthcare) followed by on-column cleavage of the GST-tag with PreScission protease (GE Healthcare), and gel-filtrated on a Sephacryl S-300 HR column (GE Healthcare) in 200 mM Tris–HCl pH 7.4, 1 mM EDTA. All PX-BAR proteins, including the mutants, behaved as dimers in gel filtration. For purification of Se–Met-containing protein, 20 mM DTT was added to all the buffers. SNX3 (Xu et al, 2001) was purified in 150 mM NaCl, 20 mM Hepes, pH 7.4, after bacterial expression from full-length cDNA inserted into pGEX-6P-2. The GST tag was removed with PreScission protease.
Liposome binding and tubulation
Generation of liposomes from total brain lipids (Folch fraction I, Sigma) or synthetic lipids (Avanti Polar Lipids and Echelon), and liposomes of a specified diameter, was performed as previously described (Peter et al, 2004). Liposome binding assays for lipid specificity and curvature sensitivity were performed essentially as described (Peter et al, 2004). Briefly, proteins (0.7–3 μM) were incubated together with liposomes (1 mg/ml) in 200 mM NaCl, 20 mM Hepes, pH 7.4 at room temperature for 5 min, followed by centrifugation at 100 000 g for 20 min and analysis of the pellet and supernatant by SDS–PAGE. For the experiment shown in Figure 6E, the buffer contained 1.5 mM MgCl2. Salt sensitivity of liposome-bound protein was examined by the addition of 500 mM NaCl and incubation for another 5 min before centrifugation. Samples for electron microscopy were prepared by incubation of liposomes with 5–10 μM of protein, and after incubation at room temperature for 10 min the samples were spread on electron microscopy grids and stained with uranyl acetate.
Cell transfection and microscopy
SNX9 cDNA encoding different regions was inserted into a modified pCMV vector after an myc epitope sequence (_MEQKLISEEDL_NGIEFGS), using appropriate restriction enzymes. HeLa cells were grown in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum and transfected using Lipofectamine (Invitrogen) for transient protein expression, and analyzed 16–20 h after transfection. For manipulations of the cellular phosphoinositide content, cells were treated with 100 nM wortmannin for 30 min, or 10 μM ionomycin for 5 min (both drugs from Sigma), before processing for microscopy.
For immunofluorescence analysis, HeLa cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min, washed, and blocked in 5% goat serum, 0.05% saponin in PBS before staining with rabbit anti-myc antibodies (Cell Signaling), or mouse anti-EEA1 (BD Transduction laboratories), and secondary fluorescent antibodies (Alexa 488 and 568; Molecular Probes) in 1% goat serum, 0.1% saponin in PBS using standard protocols. Epifluorescent images were taken using a Zeiss Axioimager Z1 system with AxioVision software.
Crystallization, X-ray data collection, and structure determination
The protein was crystallized in hanging drops by the vapor diffusion method as described elsewhere (Pylypenko et al, in press). Briefly, in the initial crystallization experiments, the 185DA-SNX9 PX-BAR protein preparation produced a granular crystal like pellet. Analysis of the pellet by SDS–PAGE and mass spectroscopy showed that the crystallized protein was N-terminally truncated at amino acid 204. Subsequent protein construct optimization in combination with macro seeding technique resulted in 500 × 200 × 100-μm crystals. The optimized fragment 204PL-SNX9 PX-BAR crystallized in two crystal forms P212121 and C2221. The data sets were collected from cryoprotected (mother liquid supplemented with 20% of glycerol) frozen crystals at 100 K on MAR CCD detector at the PXI beamline of SLS (Villingen) synchrotron radiation source and processed with XDS (Kabsch, 1993; Table I). SNX9 PX-BAR crystal structure was determined by single anomalous dispersion method using P212121 form crystal grown from the Se–Met-substituted protein. According to the calculated Matthews coefficient, the asymmetric unit contained two protein molecules, consistent with the biological unit representing a dimer. The positions of 22 selenium atoms were found using SHELXD (Schneider et al, 2000). The heavy atom position refinement and phasing were performed in CNS (Brunger et al, 1998) using data to 3.2 Å resolution with the subsequent density modification procedure. The resulting electron density map clearly reflected the protein secondary structure elements arrangement allowing the protein model building. Several rounds of model building in O (Jones et al, 1991) were followed by simulated annealing refinement using the Hendrickson-Lattman coefficients maximum likelihood as the refinement target in CNS. The final refinement runs including simulated annealing, energy minimization and _B_-factor refinement were performed using the structure factor maximum likelihood target in CNS.
Table 1.
X-ray data collection, phasing, and refinement statistics
| Crystal | Se–Met | SO4 bound | PI(3)P bound |
|---|---|---|---|
| Space group | P212121 | C2221 | C2221 |
| Unit cell parameters a, b, c (Å) | 65.6, 117.5, 145.8 | 61.5, 147.4, 115.2 | 64.06, 144.18, 118.25 |
| Number of molecules in the AU | 2 | 1 | 1 |
| Matthews coefficient (Å3/Da) | 3.03 | 2.82 | 2.94 |
| Solvent content (%) | 59.4 | 56.3 | 58.2 |
| Wavelength (Å) | 0.97952 | 0.97949 | 0.97955 |
| Resolution (Å) | 19.9–3.2 | 18–2.45 | 18–3.0 |
| Number of unique reflections | 35702 | 20901 | 12284 |
| Redundancy | 7.7 | 9.1 | 4.6 |
| Completeness | 99.2 (99.8) | 99.7 (99.9) | 99.4 (99.8) |
| _R_mrgd (%) | 6.9 (21.9) | 3.8 (26.1) | 5.7 (37.2) |
| I/σ | 21.5 (6.6) | 22.9 (5.7) | 22.3 (4.7) |
| Wilson _B_-factor | 63.8 | 78.9 | 69.6 |
| Phasing power SAD phasing with CNS centric/acentric | 2.43/2.05 | ||
| FOM after SAD phasing | 0.36 | ||
| FOM after density modification | 0.86 | ||
| Modeled amino-acid residues | Mol A: 214–257, 268–311, 321–595; mol B: 214–257, 268–317, 322–595 | 214–595 | 214–595 |
| _R_cryst/_R_free (%) | 23/30 | 24/28 | 23/29 |
| R.m.s.d. from ideal geometry bonds (Å)/angles (deg) | 0.009/1.2 | 0.007/1.25 | 0.008/1.45 |
| Ramachandran plot region most favored/additional allowed/generously allowed/disallowed (%) | 87.3/12.1/0.3/0.3 | 89.5/9.4/0.9/0.3 | 86.8/11.7/1.5/0 |
| Average _B_-factors (Å2) | 64.3 | 74.5 | 61.1 |
| PDB ID | 2RAI | 2RAJ | 2RAK |
| mol, molecule; PDB, Protein Data Bank. |
The addition of 0.1 M ammonium sulfate to the cryoprotectant solution improved the C2221 form crystals' diffraction resolution to 2.4 Å. The structure of the C2221 crystal form was determined by molecular replacement method using a monomer of the Se–Met Snx9-PX-BAR as a search model. The asymmetric unit of the C2221 crystal form contained one protein molecule; the molecules in the protein biological dimer are related by crystallographic symmetry. The molecular replacement solution was refined in CNS using rigid-body refinement, simulated annealing, energy minimization, and _B_-factor refinement procedures. The resulting difference electron density maps revealed a strong peak of additional electron density in the PX domain phosphoinositide-binding pocket that was modeled as a SO4 ion. Several iterative cycles of the model rebuilding and refinement resulted in the final SO4-bound SNX9-PX-BAR model.
In order to obtain PIP–protein complexes, the crystals were soaked in the cryoprotectant solution containing 5 mM of C4PtdIns(3,4)P2, Di-C4PtdIns(3,5)P2, C4PtdIns(4,5)P2, or C4PtdIns3P (Echelon), but only C4PtdIns3P soaking resulted in complex formation. The C2221 crystal soaked in the C4-PI(3)P solution slightly changed the crystal unit-cell parameters probably due to the ligand binding. The structure was solved by molecular replacement method with the previously described SNX9 PX-BAR structure as a search model. The initial difference electron density maps revealed a clear electron density reflecting structural features of the PI(3)P molecule bound in the PX domain phosphoinositide-binding pocket. The PI(3)P-bound SNX9 PX-BAR model was iteratively rebuild and refined as described for the SO4-bound structure.
Data quality, refinement statistics, and models geometry are given in Table I. The atomic coordinates and diffraction data have been deposited at the Protein Data Bank under accession codes 2RAI, 2RAJ, and 2RAK. Figures were prepared using the program PyMol (http://www.pymol.org).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank W Hong for the SNX3 plasmid, The Kempe Foundation for microscope equipment, and N Bleimling and G Holtermann for invaluable technical assistance. We thank the staff at beamline PXII, SLS Villingen, and Dr W Blankenfeldt for help with X-ray data collection. We thank Professor Roger S Goody for reading the manuscript and providing very useful comments. This work, as part of an award under the European Young Investigator Awards scheme (EURYI) to AR, was supported by funds from the DFG (grant RA 1364/1–1) and the EC Sixth Framework Programme that is coordinated by the European Science Foundation as well as by grants from PE Lindahls (The Royal Swedish Academy of Sciences) and Magn. Bergvalls foundations (to RL), and The Swedish Research Council and Cancerfonden (to SRC).
References
- Badour K, McGavin MK, Zhang J, Freeman S, Vieira C, Filipp D, Julius M, Mills GB, Siminovitch KA (2007) Interaction of the Wiskott–Aldrich syndrome protein with sorting nexin 9 is required for CD28 endocytosis and cosignaling in T cells. Proc Natl Acad Sci USA 104: 1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo J, Karathanassis D, Pacold CM, Pacold ME, Ellson CD, Anderson KE, Butler PJ, Lavenir I, Perisic O, Hawkins PT, Stephens L, Williams RL (2001) The crystal structure of the PX domain from p40(phox) bound to phosphatidylinositol 3-phosphate. Mol Cell 8: 829–839 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Carlton J, Bujny M, Peter BJ, Oorschot VM, Rutherford A, Mellor H, Klumperman J, McMahon HT, Cullen PJ (2004) Sorting nexin-1 mediates tubular endosome-to-TGN transport through coincidence sensing of high-curvature membranes and 3-phosphoinositides. Curr Biol 14: 1791–1800 [DOI] [PubMed] [Google Scholar]
- Casal E, Federici L, Zhang W, Fernandez-Recio J, Priego EM, Miguel RN, DuHadaway JB, Prendergast GC, Luisi BF, Laue ED (2006) The crystal structure of the BAR domain from human Bin1/amphiphysin II and its implications for molecular recognition. Biochemistry 45: 12917–12928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M (2001) Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol 3: 613–618 [DOI] [PubMed] [Google Scholar]
- Childress C, Lin Q, Yang W (2006) Dimerization is required for SH3PX1 tyrosine phosphorylation in response to epidermal growth factor signalling and interaction with ACK2. Biochem J 394: 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P (2001) Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol 155: 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallop JL, McMahon HT (2005) BAR domains and membrane curvature: bringing your curves to the BAR. Biochem Soc Symp 438: 223–231 [DOI] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT (2006) Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J 25: 2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin CT, Trejo J, Magnuson T (2005) Genetic evidence for a mammalian retromer complex containing sorting nexins 1 and 2. Proc Natl Acad Sci USA 102: 15173–15177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann B (2004) The BAR-domain family of proteins: a case of bending and binding? EMBO Rep 5: 250–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw JE (2000) Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol 16: 483–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard L, Nelson KK, Maciewicz RA, Blobel CP (1999) Interaction of the metalloprotease disintegrins MDC9 and MDC15 with two SH3 domain-containing proteins, endophilin I and SH3PX1. J Biol Chem 274: 31693–31699 [DOI] [PubMed] [Google Scholar]
- Itoh T, De Camilli P (2006) BAR, F-BAR (EFC) and ENTH/ANTH domains in the regulation of membrane–cytosol interfaces and membrane curvature. Biochim Biophys Acta 1761: 897–912 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Pt 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystogr 26: 795–800 [Google Scholar]
- Kosaka T, Ikeda K (1983) Reversible blockage of membrane retrieval and endocytosis in the garland cell of the temperature-sensitive mutant of Drosophila melanogaster, shibirets1. J Cell Biol 97: 499–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeMaster DM, Richards FM (1985) 1H-15N heteronuclear NMR studies of Escherichia coli thioredoxin in samples isotopically labeled by residue type. Biochemistry 24: 7263–7268 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2007) Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp 74: 81–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Mao X, Dong LQ, Liu F, Tong L (2007) Crystal structures of the BAR-PH and PTB domains of human APPL1. Structure 15: 525–533 [DOI] [PubMed] [Google Scholar]
- Lin Q, Lo CG, Cerione RA, Yang W (2002) The Cdc42 target ACK2 interacts with sorting nexin 9 (SH3PX1) to regulate epidermal growth factor receptor degradation. J Biol Chem 277: 10134–10138 [DOI] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR (2003) Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J Biol Chem 278: 46772–46781 [DOI] [PubMed] [Google Scholar]
- Lundmark R, Carlsson SR (2004) Regulated membrane recruitment of dynamin-2 mediated by sorting nexin 9. J Biol Chem 279: 42694–42702 [DOI] [PubMed] [Google Scholar]
- MaCaulay SL, Stoichevska V, Grusovin J, Gough KH, Castelli LA, Ward CW (2003) Insulin stimulates movement of sorting nexin 9 between cellular compartments: a putative role mediating cell surface receptor expression and insulin action. Biochem J 376: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N (2006) Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J 25: 2889–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Pylypenko O, Ignatev A, Lundmark R, Rasmuson E, Carlsson SR, Rak A (2007) Combinatorial approach in crystallization of PX-BAR membrane remodelling unit of the human Sorting Nexin 9. J Struct Biol (in press) [DOI] [PubMed] [Google Scholar]
- Rojas R, Kametaka S, Haft CR, Bonifacino JS (2007) Interchangeable but essential functions of SNX1 and SNX2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol Cell Biol 27: 1112–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MG (2003) Phosphoinositides in constitutive membrane traffic. Physiol Rev 84: 699–730 [DOI] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P (2006) GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature 441: 528–531 [DOI] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, Waterfield MD, Panayotou G (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J 15: 6241–6250 [PMC free article] [PubMed] [Google Scholar]
- Schneider TR, Karcher J, Pohl E, Lubini P, Sheldrick GM (2000) Ab initio structure determination of the antibiotic mersacidin. Acta Crystallogr D Biol Crystallogr 56: 705–713 [DOI] [PubMed] [Google Scholar]
- Seet LF, Hong W (2006) The Phox (PX) domain proteins and membrane traffic. Biochim Biophys Acta 1761: 878–896 [DOI] [PubMed] [Google Scholar]
- Shimada A, Niwa H, Tsujita K, Suetsugu S, Nitta K, Hanawa-Suetsugu K, Akasaka R, Nishino Y, Toyama M, Chen L, Liu ZJ, Wang BC, Yamamoto M, Terada T, Miyazawa A, Tanaka A, Sugano S, Shirouzu M, Nagayama K, Takenawa T et al. (2007) Curved EFC/F-BAR-domain dimers are joined end to end into a filament for membrane invagination in endocytosis. Cell 129: 761–772 [DOI] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL (2005) SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell 16: 2058–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin RV, Karathanassis D, Bruzik KS, Waterfield MD, Bravo J, Williams RL, Cho W (2006) Structural and membrane binding analysis of the Phox homology domain of phosphoinositide 3-kinase-C2alpha. J Biol Chem 281: 39396–39406 [DOI] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, De Camilli P (1999) Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol 1: 33–39 [DOI] [PubMed] [Google Scholar]
- Tarricone C, Xiao B, Justin N, Walker PA, Rittinger K, Gamblin SJ, Smerdon SJ (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature 411: 215–219 [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Leevers SJ, Ahmadi K, Timms J, Katso R, Driscoll PC, Woscholski R, Parker PJ, Waterfield MD (2001) Synthesis and function of 3-phosphorylated inositol lipids. Annu Rev Biochem 70: 535–602 [DOI] [PubMed] [Google Scholar]
- Varnai P, Balla T (1998) Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Dixon JE (2002) Sorting out the cellular functions of sorting nexins. Nat Rev Mol Cell Biol 3: 919–931 [DOI] [PubMed] [Google Scholar]
- Xing Y, Liu D, Zhang R, Joachimiak A, Songyang Z, Xu W (2004) Structural basis of membrane targeting by the Phox homology domain of cytokine-independent survival kinase (CISK-PX). J Biol Chem 279: 30662–30669 [DOI] [PubMed] [Google Scholar]
- Xu Y, Hortsman H, Seet L, Wong SH, Hong W (2001) SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat Cell Biol 3: 658–666 [DOI] [PubMed] [Google Scholar]
- Yarar D, Waterman-Storer CM, Schmid SL (2007) SNX9 couples actin assembly to phosphoinositide signals and is required for membrane remodeling during endocytosis. Dev Cell 13: 43–56 [DOI] [PubMed] [Google Scholar]
- Yeow-Fong L, Lim L, Manser E (2005) SNX9 as an adaptor for linking synaptojanin-1 to the Cdc42 effector ACK1. FEBS Lett 579: 5040–5048 [DOI] [PubMed] [Google Scholar]
- Zhou CZ, de La Sierra-Gallay IL, Quevillon-Cheruel S, Collinet B, Minard P, Blondeau K, Henckes G, Aufrere R, Leulliot N, Graille M, Sorel I, Savarin P, de la TF, Poupon A, Janin J, van Tilbeurgh H (2003) Crystal structure of the yeast Phox homology (PX) domain protein Grd19p complexed to phosphatidylinositol-3-phosphate. J Biol Chem 278: 50371–50376 [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM (2006) How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7: 9–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures