Activation of the Flt3 signal transduction cascade rescues and enhances type I interferon–producing and dendritic cell development (original) (raw)
Abstract
Flt3 ligand (Flt3L) is a nonredundant cytokine in type I interferon–producing cell (IPC) and dendritic cell (DC) development, and IPC and DC differentiation potential is confined to Flt3+ hematopoietic progenitor cells. Here, we show that overexpression of human Flt3 in Flt3− (Flt3−Lin−IL-7Rα−Thy1.1−c-Kit+) and Flt3+ (Flt3+Lin−IL-7Rα−Thy1.1−c-Kit+) hematopoietic progenitors rescues and enhances their IPC and DC differentiation potential, respectively. In defined hematopoietic cell populations, such as Flt3− megakaryocyte/erythrocyte-restricted progenitors (MEPs), enforced Flt3 signaling induces transcription of IPC, DC, and granulocyte/macrophage (GM) development–affiliated genes, including STAT3, PU.1, and G-/M-/GM-CSFR, and activates differentiation capacities to these lineages. Moreover, ectopic expression of Flt3 downstream transcription factors STAT3 or PU.1 in Flt3− MEPs evokes Flt3 receptor expression and instructs differentiation into IPCs, DCs, and myelomonocytic cells, whereas GATA-1 expression and consecutive megakaryocyte/erythrocyte development is suppressed. Based on these data, we propose a demand-regulated, cytokine-driven DC and IPC regeneration model, in which high Flt3L levels initiate a self-sustaining, Flt3-STAT3– and Flt3-PU.1–mediated IPC and DC differentiation program in Flt3+ hematopoietic progenitor cells.
Differentiation of hematopoietic stem cells (HSCs) to mature hematopoietic cells is characterized by progressive loss of developmental options and restriction to one lineage (1). In both mice and men, HSCs as well as multiple developmental intermediates with limited cellular expansion and restriction to specific mature cell types have been identified. These in-clude myeloid progenitors, as clonal common myeloid progenitors (CMPs) that give rise to either granulocyte/macrophage progenitors (GMPs) or megakaryocyte/erythrocyte progenitors (MEPs; references 2 and 3), and clonal common lymphoid progenitors (CLPs; references 4 and 5), which robustly produce the respective mature cell types. Hematopoietic differentiation is regarded as a multi-linear, unidirectional process, and regeneration and expansion of specific lineages are largely regulated extrinsically by different hematopoietic cytokines. However, it is unclear whether under physiologic conditions cytokines are capable to instruct HSCs and subsequent progenitors to differentiate to lineage-restricted progenitors (extrinsic lineage determination). Alternatively, HSCs and subsequent progenitors commit to lineage-restricted progenitors by intrinsic differentiation programs (intrinsic lineage determination), and restricted progenitors are con-secutively stimulated by hematopoietic cytokines produced upon demand (6, 7).
On the molecular level, access to lineage developmental options and readiness to receive lineage-permissive and -instructive signals might be determined by graded, relative expression levels of diverse transcription factors and cytokine receptors (8, 9). Indeed, it has been demonstrated that genetic deletion or overexpression of different single transcription factors is sufficient to reprogram committed progenitors or mature cells to alternative hematopoietic lineages (10): _Pax5_-deficient pre–B cells lose B cell differentiation potential and mature into T and myelomonocytic cells, but reexpression of Pax5 restores B cell commitment (11, 12); ectopic expression of GATA-1 instructs HSCs and CMPs and converts CLPs and GMPs to the megakaryocyte/erythrocyte lineage, respectively (13); and enforced expression of C/EBPα and C/EBPβ in B cells leads to macrophage differentiation (14). Furthermore, it has been shown that artificial expression of GM-CSF receptor and stimulation with the cognate ligand redirect CLPs and early T cell progenitors to myeloid lineage outcomes (15–17). This proves that hematopoietic lineage instruction can be mediated extrinsically by cytokines, at least in these experimental settings.
DCs are important regulators of innate and adaptive immune responses and are involved in initiation of immunity as well as in maintenance of self-tolerance (18–20). DCs are cells of the hematopoietic system and are continuously replenished from hematopoietic stem and progenitor cells (1). In mice, multiple DC subsets that differ in maturation state, phenotype, location, and in some functions were identified (21). Here, for simplicity, these will grossly be divided into CD11c+B220+ natural type I interferon–producing cells (IPCs; also called plasmacytoid cells or plasmacytoid pre-DCs) and CD11c+B220− “conventional” DCs, consisting of CD11c+CD8α−CD4−CD11b+, CD11c+CD8α−CD4+CD11b+, and CD11c+CD8α+CD4−CD11b− subpopulations (21). Although it was suggested that IPCs as well as conventional CD11c+CD8α+ DCs are derived from lymphoid-committed progenitors (21), it was demonstrated recently that any of the IPCs and conventional DCs can be generated via lymphoid and myeloid progenitors (22–27). Specifically, all IPCs and conventional DCs are generated by mouse CMPs, GMPs, CLPs, and pro–T1 cells, whereas IPC and DC differentiation potential is lost once definitive MEP or B cell commitment occurs (22–27). Thus, in contrast to other hematopoietic lineages, IPC and DC potentials are conserved along lymphoid and myeloid developmental pathways.
Flt3 is a receptor tyrosine kinase with homology to c-Kit (the receptor for stem cell factor) and c-fms (the receptor for M-CSF; reference 28) that has a nonredundant role in steady-state differentiation of IPCs and DCs in vivo: Flt3 ligand (Flt3L)-deficient mice and mice with hematopoietic system–confined deletions of STAT3, a transcription factor activated in the Flt3 signaling cascade, as well as mice that are treated with flt3 tyrosine kinase inhibitors, show massively reduced IPCs and DCs (29–31). Conversely, injection or conditional expression of Flt3L in mice increases IPCs and DCs, with up to 30% of mouse spleen cells expressing CD11c (32–34). Furthermore, Flt3L as a single cytokine is capable of inducing differentiation of IPCs and DCs in mouse bone marrow cell cultures (35).
Flt3 is expressed in mouse short-term HSCs and multipotent progenitors (36, 37) in most CLPs and CMPs, at lower levels on fractions of GMPs and pro–T1 cells, as well as on mature steady-state IPCs and DCs. It is down-regulated on pro–B cells, further downstream T cell progenitors, and absent on MEPs (33, 38). To determine what might define IPC and DC developmental potential in lymphoid- and myeloid-committed cells, we and others showed that in vitro and in vivo IPC, DC, and Langerhans cell differentiation potential is confined to Flt3-expressing hematopoietic progenitors (33, 38, 39). Furthermore, we de-monstrated that injection of Flt3L expands Flt3+ but not downstream Flt3− progenitors and drives IPC and DC development along both lymphoid and myeloid differentiation pathways (33).
Based on these data, we postulate that high environmental Flt3L levels and consecutive Flt3 signaling might be both the earliest event and a continuous regulator that determines IPC and DC developmental outcomes in bone marrow hematopoietic progenitor cells. Thus, we were interested to test whether enforced Flt3 expression and signaling would be sufficient to instruct IPC and DC development from Flt3− progenitor cells and enhance IPC and DC development from Flt3+ progenitor cells, respectively.
RESULTS
Enforced expression of human Flt3 in Flt3− and Flt3− progenitors rescues and enhances IPC and DC developmental potential, respectively
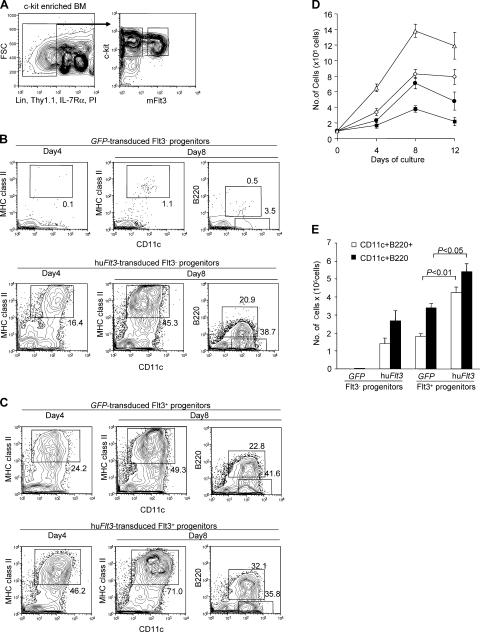
We used a bicistronic retroviral transduction system to transduce human Flt3 into progenitor cells. The constructs carrying either GFP or hu_Flt3-GFP_ are shown in Fig. S1 A, available at http://www.jem.org/cgi/content/full/jem.20051645/DC1. Lin−IL-7Rα−Thy1.1−c-Kit+ bone marrow cells that contain a heterogeneous fraction of progenitors, including multipotent progenitors and myeloid-committed cells, and are devoid of Thy1.1+ HSCs, IL-7Rα+ lymphoid–committed cells, and mature lineage cells, were sorted into Flt3− (Flt3− progenitors also containing Flt3− MEPs) and Flt3+ (Flt3+ progenitors containing multipotent progenitors and Flt3+ CMPs) cell fractions (Fig. 1 A; references 33, 36–38) and consecutively retrovirally transfected as described in Materials and methods. Fig. S1 B shows typical 18-h coculture transduction efficiencies (18–26%) in Flt3− and Flt3+ progenitors as determined by GFP expression.
Figure 1.
Enforced expression of human Flt3 in Flt3− and Flt3+ progenitors rescues and enhances IPC and DC developmental potential, respectively. (A) Contour plots indicate sorting gates for mouse bone marrow, c-Kit–enriched Flt3−Lin−IL-7Rα−Thy1.1−c-Kit+ (Flt3− progenitors), and Flt3+Lin−IL-7Rα−Thy1.1−c-Kit+ (Flt3+ progenitors) cells. (B) Flow cytometric analysis of GFP+ cells derived from control-_GFP_– and hu_Flt3-GFP_–transduced Flt3− progenitors cultured for 4 or 8 d in huFlt3L-Ig– and SCF-supplemented media. (C) Flow cytometric analysis of GFP+ cells derived from control_-GFP_– and hu_Flt3-GFP_–transduced Flt3+ progenitors cultured for 4 or 8 d in huFlt3L-Ig– and SCF-supplemented media. (B and C) Numbers at gates represent percentages of total plotted cells. Results are from one representative experiment out of six each. (D) Graph depicts numbers of cells derived from 105 retrovirus-transduced progenitor cells cultured with huFlt3L-Ig and SCF at days 4, 8, and 12 of culture. ▪, _GFP_-transduced Flt3− progenitors; •, hu_Flt3_-transduced Flt3− progenitors; ⋄, _GFP_-transduced Flt3+ progenitors; △, hu_Flt3_-transduced Flt3+ progenitors. Each point represents mean values ± SD from three independent experiments. (E) Bars show total IPC (CD11c+B220+) and DC (CD11c+B220−) yields per 105 retrovirus-transduced progenitor cells at day 8. The data represents mean values ± SD from three independent experiments. Statistical analysis was performed by Student's t test.
To study the effects of enforced hu_Flt3_ expression on IPC and DC development, Flt3− and Flt3+ progenitors were transduced with control-GFP or hu_Flt3-GFP_ and cultured in human Flt3L-Ig fusion protein (huFlt3L-Ig)– and stem cell factor (SCF)-supplemented media. Cultures were analyzed for cell numbers and the presence of IPCs and DCs at days 4, 8, and 12. Freshly isolated Flt3− progenitors did not express CD11c or MHC class II (not depicted). As expected, unmanipulated Flt3− progenitors (not depicted) as well as _GFP_-transduced Flt3− progenitors gave rise to no or very few CD11c+ MHC class II+ (up to 1.1%) and CD11c+B220+ (up to 0.5%) cells (Fig. 1 B, top). In contrast, hu_Flt3-GFP_–transduced Flt3− progenitors differentiated into CD11c+ MHC class II+ and CD11c+B220+ cells that increased in relative numbers from day 4 to 8 of culture (Fig. 1 B, bottom). Similarly, _GFP_-transduced as well as hu_Flt3_-transduced Flt3+ progenitor cells gave rise to CD11c+ MHC class II+ and CD11c+B220+ cells, with hu_Flt3_-transdced Flt3+ progenitor cells producing slightly higher relative numbers of both cell types (Fig. 1 C). To quantify absolute CD11c+ cell production, cell numbers were determined at days 4, 8, and 12 of culture. Numbers peaked at day 8 of culture, with hu_Flt3_-transduced Flt3+ progenitors producing the highest cell numbers (∼14-fold expansion of input cells), followed by intermediate expansion (6–9-fold) of both _GFP_-transduced Flt3+ and hu_Flt3_-transduced Flt3− progenitors, and low expansion (3–4-fold) of _GFP_-transduced Flt3− progenitors (Fig. 1 D). The total cellular expansion was paralleled by a peak expansion of CD11c+ MHC class II+ and CD11c+B220+ cells at day 8 of culture (Fig. 1, B–E). Interestingly, hu_Flt3_-transduced Flt3+ progenitors produced significantly higher total numbers of both CD11c+ MHC class II+ and CD11c+B220+ cells compared with _GFP_-transduced Flt3+ progenitors (Fig. 1 E), with somewhat higher relative CD11c+B220+ cell numbers (Fig. 1, C and E).
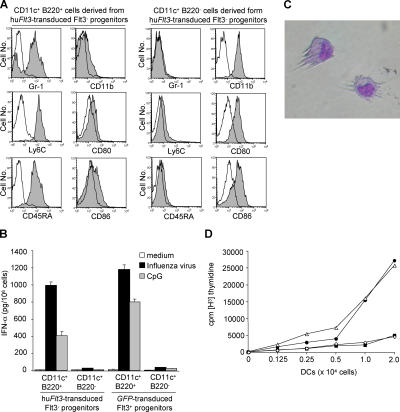
Next, we evaluated IPC- and DC-associated surface antigen expression and function of CD11c+B220+ and CD11c+B220− cells derived from different progenitor cell populations at day 8 of culture. Consistent with typical mouse IPC and DC phenotypes, CD11c+B220+ cells expressed Gr-1, Ly6C, and CD45RA, whereas CD11c+B220+ cells expressed CD11b and intermediate levels of CD80 and CD86, respectively (Fig. 2 A). Furthermore, CD11c+B220+ cells produced substantial amounts of IFN-α upon stimulation with either influenza virus or CpG, whereas CD11c+B220− cells did not (Fig. 2 B). CD11c+B220− cells, but not CD11c+B220+ cells, displayed typical DC morphology (Fig. 2 C) and were efficient stimulators in allogeneic MLR cultures, as evaluated by thymidine incorporation (Fig. 2 D). Therefore, CD11c+B220+ cells phenotypically and functionally were typical IPCs, whereas CD11c+B220− cells were typical conventional DCs. Collectively, these results indicate that huFlt3 signaling rescues and enhances the development of functional IPCs and DCs from Flt3− and Flt3+ hematopoietic progenitor cell populations, respectively. In addition, it suggests that strong Flt3 signaling slightly skews development toward an IPC phenotype.
Figure 2.
IPCs and DCs developed in culture from hu_Flt3_-transduced Flt3− progenitors are functional. (A) Contour plots indicate surface markers (closed histogram) and respective isotype controls (open histogram) on CD11c+B220+ and CD11c+B220− cells derived from huFlt3L-Ig– and SCF-cultured hu_Flt3_-transduced Flt3− progenitors. CD11c+B220+ and CD11c+B220− cells show typical phenotypes of IPCs and DCs, respectively. Results are from one representative experiment out of three. (B) Sorted CD11c+B220+ IPCs but not CD11c+B220− DCs, both derived from either hu_Flt3_-transduced Flt3− progenitors or _GFP_-transduced Flt3+ progenitors, produce IFN-α upon influenza virus or CpG stimulation. Culture supernatants were collected after 24 h and analyzed by ELISA. Results are from one representative experiment out of three. (C) About half of day 8 progeny from hu_Flt3_-transduced Flt3− progenitors display typical DC morphology. Giemsa-stained cytospin, photographed at a magnification of 40. (D) In vitro–generated CD11c+B220− cells from retrovirus-transduced progenitors are efficient stimulators of allogeneic T cells. Graph depicts thymidine incorporation of 2 × 105 allogeneic BALB/c spleen CD4+ T cells incubated with graded numbers (x axis) of sorted CD11c+B220− DCs derived from hu_Flt3_-transduced Flt3− progenitors (•), CD11c+B220+ IPCs derived from hu_Flt3_-transduced Flt3− progenitors (▪), or CD11c+B220− DCs derived from _GFP_-transduced Flt3+ progenitors (△), and CD11c+B220+ IPCs derived from _GFP_-transduced Flt3+ progenitors (⋄). Results are from one representative experiment out of three.
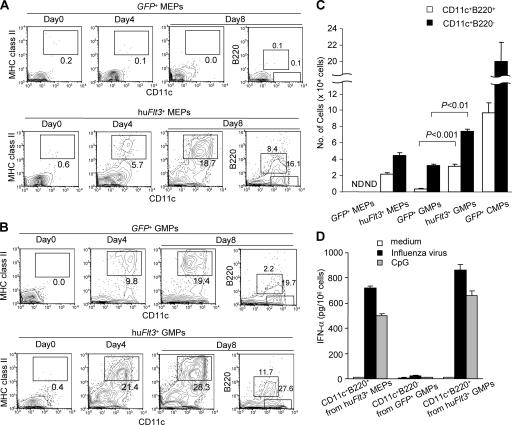
Enforced expression of hu_Flt3_ is sufficient to rescue IPC and DC development from MEPs and enhances IPC and DC development from GMPs
In normal mouse and human hematopoiesis, IPC and DC developmental potentials are maintained from Flt3+ CMPs to downstream Flt3+ GMPs, but are lost in Flt3− MEPs (23–27, 33, 38). Thus, we tested whether enforced hu_Flt3_ expression in MEPs would be sufficient to rescue IPC and DC development. As comparator cell population, we used hu_Flt3_-transduced GMPs and GFP_-transduced CMPs (GFP +-CMPs). As reported previously for unmanipulated MEPs, GFP_-transduced MEPs (GFP +-MEPs) gave rise to no or very few CD11c+ cells (Fig. 3 A, top). In contrast, hu_Flt3 +-MEPs gave rise to CD11c+B220+ cells and CD11c+B220− cells at day 8 in huFlt3L-Ig– and SCF-supplemented cultures, at least as efficient as GFP +-GMPs (Fig. 3, A–C). Similarly as from hu_Flt3_-transduced Flt3+ progenitor cells, CD11c+B220+ and CD11c+B220− cell development was significantly enhanced from hu_Flt3_-transduced GMPs (Fig. 3, B and C). Upon influenza virus or CpG stimulation, hu_Flt3 +-MEP– and hu_Flt3_ +-GMP–derived CD11c+B220+ cells produced IFN-α (Fig. 3 D), suggesting that these cells are functional.
Figure 3.
Enforced expression of hu_Flt3_ is sufficient to rescue IPC and DC development from MEPs and enhances IPC and DC development from GMPs. (A and B) Flow cytometric analysis of GFP+ cells from control-GFP_– and hu_Flt3_-transduced MEPs (A) and GMPs (B). Retrovirus-transduced cells were cultured for 4 or 8 d in huFlt3L-Ig– and SCF-supplemented media. Numbers at gates represent percentages of total plotted cells. Results are from one representative experiment out of three. (C) Bars show total IPC (CD11c+B220+) and DC (CD11c+B220−) yields per 5 × 104 retrovirus-transduced MEPs, GMPs, and CMPs at day 8. The data represents mean values ± SD from three independent experiments. Statistical analysis was performed by Student's t test. (D) IPCs developed from hu_Flt3 +-MEPs and hu_Flt3_ +-GMPs are functional. Bar graph shows IFN-α production by sorted CD11c+B220+ cells derived from day 8 culture of hu_Flt3_ +-MEPs or hu_Flt3_ +-GMPs stimulated with influenza virus or CpG. Culture supernatants were collected after 24 h and analyzed by ELISA. Results are from one representative experiment out of three.
As reported previously for untransduced CMPs (22–27, 38), GFP +-CMPs generated both IPCs and DCs. The efficacy of GFP +-CMPs to produce these offspring cells was about threefold higher than that observed from hu_Flt3_ +-GMPs (Fig. 3 C). Thus, enforced huFlt3 expression is sufficient to rescue IPC and DC developmental potential in MEPs to levels comparable to their developmental counterparts, GFP +-GMPs. Furthermore, huFlt3 expression enhances IPC and DC development from GMPs, but not to levels observed in the upstream CMP population.
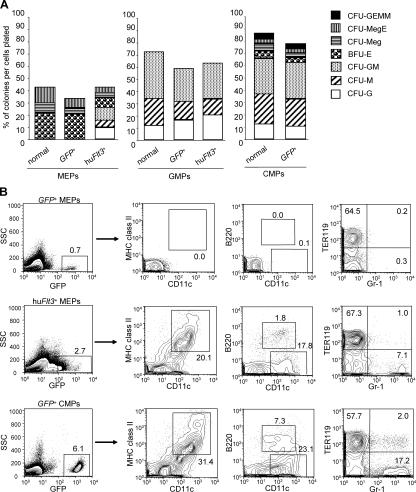
Enforced expression of hu_Flt3_ permits myelomonocytic development from hu_Flt3_ +-MEPs but not megakaryocyte/erythrocyte development from hu_Flt3_ +-GMPs
Because huFlt3 signaling in MEPs activated IPC and DC development, a differentiation option normally confined to Flt3+ progenitors as CMPs and GMPs, we were interested to test whether huFlt3 signaling in MEPs would also reestablish myeloid CFU activity. GFP +-transduced CMPs, GMPs, and MEPs gave rise to their respective colony types, but with somewhat lower plating efficacy as compared with freshly isolated CMPs, GMPs, and MEPs (Fig. 4 A; reference 2). hu_Flt3_ +-MEPs gave rise to not only erythroid-affiliated colonies but also granulocyte/macrophage (GM)-affiliated colonies, including CFU-G, CFU-M, and CFU-GM (Fig. 4 A). Compared with CMPs and GMPs, the myelomonocytic colony–forming efficiency of hu_Flt3_ +-MEPs was lower; however, the diversity of colony formation resembled that of CMPs with the exception that no CFU-Mix colonies developed. No substantial difference in CFU activity was observed in hu_Flt3_ +-GMPs compared with GFP +-GMPs (Fig. 4 A). Thus, huFlt3 signaling in MEPs reestablishes myelomonocytic CFU activity, whereas huFlt3 signaling in GMPs does not affect their overall CFU activity and particularly does not reestablish megakaryocyte/erythrocyte read out.
Figure 4.
Enforced expression of hu_Flt3_ permits myelomonocytic development from hu_Flt3_+-MEPs but not megakaryocyte/erythrocyte development from hu_Flt3_+-GMPs. (A) Bar graph shows myeloid colony–forming activity in normal and retrovirus-transduced myeloid progenitors (MEPs, GMPs, and CMPs). CFU-GEMM, CFU-granulocyte/erythroid/macrophage/megakaryocyte; CFU-MegE, CFU-megakaryocyte/erythroid; CFU-Meg, CFU-megakaryocyte; BFU-E, burst-forming units/erythroid; CFU-GM, CFU-granulocyte/macrophage; CFU-M, CFU-macrophage; CFU-G, CFU-granulocyte. Results are from one representative experiment out of three. 200 GFP+ cells were plated each. (B) Enforced expression of hu_Flt3_ in MEPs is sufficient to rescue IPC, DC, and myelomonocytic cell development in vivo. Contour plots show GFP expression in day 7 spleen progeny of GFP +-MEPs, hu_Flt3_ +-MEPs, and GFP +-CMPs. Numbers at gates represent percentages of total plotted cells. Results are from one representative experiment out of four.
Enforced expression of hu_Flt3_ in MEPs is sufficient to rescue IPC, DC, and myelomonocytic cell development in vivo
To test the robustness of Flt3 transduction–mediated effects observed in vitro, we compared in vivo reconstitution activity of GFP +-MEPs, hu_Flt3_ +-MEPs, and GFP +-CMPs. 2 × 104 cells of each progenitor population combined with 2 × 105 cells of host bone marrow cells were transplanted into lethally irradiated mice, and spleen progeny cells were analyzed on day 7. As reported previously for MEPs, GFP +-MEPs produced ∼0.7% of nucleated GFP+ spleen cells that consisted mostly of Ter119+ erythroid cells, but no DCs, IPCs, or Gr-1+ myeloid cells (Fig. 4 B, top; references 2, 23, and 25). In contrast, hu_Flt3_ +-MEPs gave rise to ∼2.7% of nucleated GFP+ spleen cells containing CD11c+ MHC class II+ conventional DCs, low numbers of CD11c+B220+ IPCs, as well as Ter119+ erythroid and Gr-1+ myeloid cells (Fig. 4 B, middle). As expected, GFP +-CMPs gave rise to ∼6.0% of nucleated GFP+ spleen cells that contained DCs, IPCs, as well as erythroid and myeloid cells (Fig. 4 B, bottom; references 2, 23, and 25). This formally demonstrates that enforced expression of hu_Flt3_ is sufficient to rescue in vivo IPC, DC, and myelomonocytic cell development from MEPs.
HuFlt3 signaling in MEPs induces activation of DC and myeloid development–associated genes
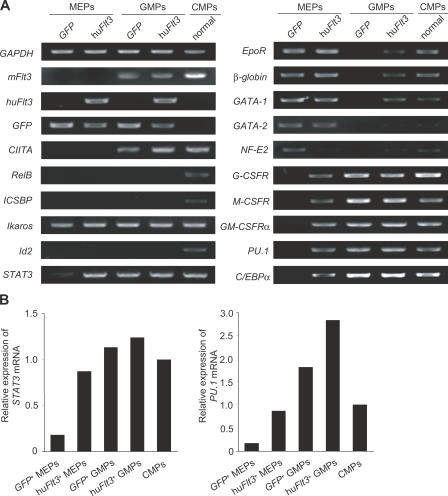
To evaluate the immediate consequences of huFlt3 signaling on gene transcription profiles that might be involved in the consecutive activation of IPC, DC, and myelomonocytic developmental options, we analyzed the transcription of a panel of IPC, DC, and myeloid development–associated genes by RT-PCR in hu_Flt3_ +-MEPs. STAT3, an indispensable transcription factor for Flt3L-mediated DC development (30), was hardly detectable in GFP +-MEPs but was clearly up-regulated in hu_Flt3_ +-MEPs. Furthermore, hu_Flt3_ +-MEPs but not GFP +-MEPs expressed myelomonocytic development–associated genes, such as the cytokine receptors for G-CSF, M-CSF, and GM-CSF, as well as the transcription factors C/EBPα and PU.1, similar to that found in GFP +-GMPs, hu_Flt3_ +-GMPs, and normal CMPs (Fig. 5 A; references 2 and 40). However, transcription factors RelB, ICSBP, and Id2 could not be detected in any of the retrovirus-transduced MEPs and GMPs, whereas they were detectable in unmanipulated CMPs (Fig. 5 A). Real-time RT-PCR revealed low-level STAT3 expression in GFP +-MEPs and similar high levels of STAT3 expression in hu_Flt3_ +-MEPs, GFP +-GMPs, hu_Flt3_ +-GMPs, and CMPs (Fig. 5 B). Furthermore, PU.1 expression levels in hu_Flt3_ +-MEPs increased to levels found in CMPs, whereas expression levels of this gene were somewhat higher in GM-committed GFP +-GMPs and hu_Flt3_ +-GMPs (Fig. 5 B).
Figure 5.
HuFlt3 signaling in MEPs induces activation of DC and myeloid development–associated genes. (A) RT-PCR analysis of myeloid lineage differentiation–affiliated genes, including cytokine receptors and transcription factors in retrovirus-transduced myeloid progenitors. The PCR products were electrophoresed on agarose gel and visualized with ethidium bromide. Results are from one representative experiment out of three. (B) Expression levels of STAT3 and PU.1 mRNA were assessed by real-time RT-PCR. Data were normalized by the level of 18s rRNA expression in each sample. (A and B) cDNA products equivalent to RNAs from 200 progenitors were analyzed. Results are from one representative experiment out of three.
Consistent with their maintained Meg/E-developmental potential, Meg/E-related genes, such as EpoR, β_-globin_, GATA-1, and GATA-2, were still transcribed in hu_Flt3_ +-MEPs, although the expression of NF-E2 decreased (Fig. 5 A). Interestingly, hu_Flt3_ +-GMP showed some transcription of Meg/E development–associated genes as β_-globin_ and GATA-1. However, GATA-2 and NF-E2 transcripts were not detectable, and hu_Flt3_ +-GMPs were not able to give rise to Meg/E lineage colonies (Figs. 4 A and 5 A). Collectively, these results demonstrate on a molecular level that MEPs have a latent IPC, DC, and myelomonocytic lineage potential that is inducible by enforced Flt3 signaling.
HuFlt3 signaling in MEPs leads to downstream STAT3 phosphorylation
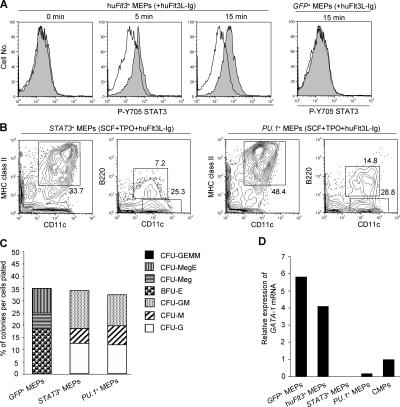
To test activation of STAT3 in hu_Flt3_ +-MEPs, we performed intracellular phospho-STAT3 staining in retrovirus-transduced MEPs that were Flt3L deprived and then stimulated with huFlt3L-Ig. Indeed, phosphorylation of STAT3 was detected in hu_Flt3_ +-MEPs but not in GFP +-MEPs, indicating that as expected (30) STAT3 is a downstream activated transcription factor of enforced huFlt3 signaling (Fig. 6 A).
Figure 6.
HuFlt3 signaling in MEPs leads to downstream STAT3 phosphorylation; enforced STAT3 or PU.1 expression instructs MEPs to differentiate into IPCs, DCs, and GM lineage cells and suppresses GATA-1 expression. (A) Contour plots show phosho-STAT3 expression (closed histogram) and respective isotype controls (open histogram) in cytokine-deprived and consecutively 5- and 15-min huFlt3L-Ig–stimulated hu_Flt3_ +-MEPs by intracellular staining. Results are from one representative experiment out of three. (B) Flow cytometric analysis of STAT3 +-MEPs and PU.1 +-MEPs cultured for 8 d in huFlt3L-Ig–, SCF-, and TPO-supplemented media. Results are from one representative experiment out of three. (C) Bar graph shows myeloid colony–forming activity in GFP +-MEPs, STAT3 +-MEPs, and PU.1 +-MEPs. Results are from one representative experiment out of three. (D) Expression of GATA-1 mRNA was assessed by real-time RT-PCR. Data were normalized by the level of 18s rRNA expression in each sample. cDNA products equivalent to RNAs from 200 progenitors were analyzed. Results are from one representative experiment out of three.
Enforced STAT3 or PU.1 expression instructs MEPs to differentiate into IPCs, DCs, and GM lineage cells
Given the importance of STAT3 for IPC and DC development, and the finding that STAT3 and PU.1 were up-regulated in hu_Flt3_ +-MEPs, we tested whether STAT3 and PU.1 could directly activate IPC, DC, and myelomonocytic development from MEPs. Mouse STAT3 and PU.1 were transduced into MEPs using retrovirus expression vectors, respectively (Fig. S2, available at http://www.jem.org/cgi/content/full/jem.20051645/DC1). However, survival of cells was low when cultured in SCF alone or SCF and huFlt3L-Ig (not depicted). To possibly support the survival of MEPs, we first added thrombopoietin (TPO), followed by TPO and huFlt3L-Ig to SCF in consecutive cultures. STAT3 +-MEPs and PU.1 +-MEPs differentiated into CD11c+B220+ and CD11c+B220− cells at day 8 in both SCF as well as TPO (not depicted) and, with even higher efficacy in SCF, TPO, and huFlt3L-Ig, supplemented cultures (Fig. 6 B). Interestingly, enforced expression of STAT3 or PU.1 in MEPs in turn led to the up-regulation of mouse Flt3 mRNA levels (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20051645/DC1).
To test myeloid CFU activity of these cells, STAT3 +-MEPs and PU.1 +-MEPs were plated in methylcellulose assays. Both STAT3 +-MEPs and PU.1 +-MEPs gave rise to CFU-G, CFU-M, and CFU-GM colonies, but not to Meg/E-affiliated colonies (Fig. 6 C).
Finally, we evaluated the expression of GATA-1, a nonredundant transcription factor, for megakaryocyte/erythrocyte development by real-time RT-PCR. GATA-1 expression was down-regulated in STAT3 +-MEPs and PU.1 +-MEPs compared with that in GFP +-MEPs, huFlt3 +-MEPs, and CMPs (Fig. 6 D). Thus, enforced expression of STAT3 and PU.1 in MEPs reprogrammed them to differentiate into IPCs, DCs, and myelomonocytic cell lineages and inhibited Meg/E lineage potentials, indicating that strong Flt3 downstream signals were capable of inducing complete lineage conversion.
DISCUSSION
A standing question in early hematopoiesis is whether cytokine signaling is sufficient to induce cell fate decisions. Here, we showed that enforced expression of hu_Flt3_ in Flt3− progenitors rescued their potential to differentiate into functional IPCs and DCs with comparable in vitro differentiation efficiency as Flt3+ progenitors (Figs. 1 and 2). We also demonstrated that enforced expression of hu_Flt3_ in MEPs, which under normal conditions cannot give rise to IPCs and DCs (23–27) and are contained in Flt3− progenitor cells, induced in vitro IPC and DC differentiation comparable to that observed from GFP +-GMPs (Fig. 3). Furthermore, hu_Flt3_ +-MEPs differentiated into IPCs and DCs upon in vivo transfer, the most informative assay available to prove the robustness of in vitro observations (Fig. 4 B). Thus, these data demonstrate that enforced expression and signaling of hu_Flt3_ in Flt3− progenitors deliver an instructive signal to activate latent IPC and DC differentiation programs.
Overexpression of hu_Flt3_ in total Flt3+ progenitors, in Flt3-expressing GMPs, and in CLPs (of which ∼70% are Flt3+) led to a gain of higher relative and absolute (two- to threefold) numbers of IPCs and DCs in vitro (Figs. 1, D and E, and 3 C, and not depicted). Thus, beyond activation, increased Flt3 signaling also enhanced IPC and DC development. The gain in offspring cells was consistently higher for IPCs than for DCs (Figs. 1 E and 3 C), in line with our previous findings that after 10 d of in vivo Flt3L injection, spleen IPCs and DCs were expanded on average 28- and 21-fold, respectively (33). This suggests that a continuous strong Flt3 signal might induce a shift toward relatively higher IPC levels.
We previously found that Flt3 is expressed in lymphoid- and myeloid-committed progenitor cells, and in vivo Flt3L application mediates the expansion of both cell types without changing their biology (33). The enforced expression of hu_Flt3_ in MEPs not only led to a gain of IPC and DC developmental capacity, but, with the exception of mixed colony formation, also to a gain of CFU activity of upstream myeloid progenitors as well as to differentiation of erythroid and myelomonocytic cells in vivo (Fig. 4 A). In contrast, huFlt3 signaling in GMPs did not activate megakaryocyte/erythrocyte potential (Fig. 4 A). This implies that beyond activation and enhancement of IPC and DC development, Flt3 signaling is not immediately deterministic but primarily opens access to an IPC, DC, and myelomonocytic differentiation program. Thus, we propose that final IPC and DC lineage outcome might be a gradual process, depending on continuous strong Flt3 signaling.
What are the Flt3 signaling–initiated downstream molecular events? It was shown that hematopoietic system–confined deletion of STAT3 transcription factor leads to the inhibition of Flt3-driven IPC and DC development (30). Furthermore, human Flt3 transfection and stimulation with Flt3L in mouse myeloid 32Dcl3 cells leads to the induction of PU.1 and C/EBPα expression (41). These transcription factors are indispensable for granulocyte and monocyte development (42), and it was shown that PU.1 cooperatively with C/EBPα activates myeloid development–associated cytokine receptor genes, including G-CSFR, M-CSFR, and GM-CSFR (42). Interestingly, PU.1_-deficient mice, in addition to other hematopoietic defects, lack either CD8α− or both CD8α− and CD8α+ DCs, depending on the type of PU.1 deletion (43, 44). Here, we showed that enforced huFlt3 signaling in MEPs results in enhanced expression of IPC, DC, and GM lineage development related transcription factors STAT3, PU.1, and C/EBPα, as well as expression of G-, M-, and GM-CSF cytokine receptors (Fig. 5). Thus, at least in terms of these RNA transcripts, hu_Flt3 +-MEPs but not GFP +-MEPs resembled the gene expression profiles of CMPs (2, 40).
Importantly, enforced expression of STAT3 or PU.1 in Flt3− MEPs was again sufficient to permit the development of both IPCs and DCs (Fig. 6 B). This, however, was only possible once TPO was added to SCF or SCF and Flt3L in cultures. Thus, TPO possibly substitutes for a survival signal otherwise delivered by Flt3. In addition, or alternatively, TPO might be involved in the phosphorylation of overexpressed STAT3 (45). Interestingly, enforced expression of STAT3 or PU.1 in MEPs led to the up-regulation of mouse Flt3 mRNA levels (Fig. S3). This in turn likely allowed culture-supplemented human Flt3L to cross-reactively stimulate _STAT3_- or _PU.1-_transduced cells via mouse Flt3. These results suggest a self-sustaining effect of Flt3 signaling–induced Flt3 transcription via downstream STAT3 and PU.1.
As enforced expression of hu_Flt3_ in MEPs did not terminate megakaryocyte/erythrocyte differentiation potential, whereas hu_Flt3_ expression in GMPs did not lead to a gain of these differentiation potentials (Fig. 4 A), how can Flt3 signaling be integrated in megakaryocyte/erythrocyte versus IPC, DC, and GM lineage commitment? By using PU.1gfp reporter mice, PU.1 expression was recently mapped in early hematopoietic progenitor cells (46). It was shown that PU.1+Flt3+ CMPs contain high myelomonocytic developmental potential, whereas PU.1−Flt3− CMPs and PU.1− MEPs have high megakaryocyte/erythrocyte potential (46). The data presented here suggests that Flt3 might be critical in PU.1 regulation, although this likely will not be an exclusive event. GATA-1 is a nonredundant transcription factor for megakaryocyte and erythrocyte development (47). DNA binding activity of GATA-1 can be suppressed by enforced PU.1 expression, resulting in a differentiation block and apoptotic cell death of an erythroid cell line (48). Conversely, GATA-1 inhibits the binding of PU.1 to c-Jun, a critical coactivator of myeloid gene transactivation by PU.1 (49). Furthermore, GATA-1 interferes with DNA binding activity of STAT3 and inhibits TPO-dependent growth of the Ba/F3 cell line (50). Thus, as suggested previously for PU.1 and GATA-1 (51, 52), relative dosage of gene transcription and protein levels will likely determine lineage outcomes. Indeed, STAT3 and PU.1 expression levels in hu_Flt3_ +-MEPs were increased to levels of normal CMPs and were somewhat lower than those observed in GFP +-GMPs or hu_Flt3_ +-GMPs (Fig. 5). Thus, it is possible that MEPs with relatively lower hu_Flt3_ and consecutive STAT3 and PU.1 expression do not fully inhibit GATA-1, whereas high Flt3 expressing and signaling cells develop to IPC, DC, or GM lineages. Of note, enforced expression of STAT3 and PU.1 in MEPs suppressed GATA-1 expression and inhibited megakaryocyte/erythrocyte development (Fig. 6, C and D). Hu_Flt3_ overexpression in GMPs in turn induced some EpoR, β_-globin_, and GATA-1 mRNA expression; however, this was not sufficient to reactivate megakaryocyte/erythrocyte development as demonstrated for enforced high-level GATA-1 expression in GMPs (Figs. 4 A and 5 A; reference 13).
Because overexpression of hu_Flt3_ in Flt3− progenitors does not occur under physiologic conditions, what do these findings imply for normal hematopoiesis? Flt3 is expressed on short-term HSCs, multipotent progenitors, CLPs, CMPs, and GMPs, and in vivo injection of Flt3L resulted in increased numbers of these cells as well as IPCs and DCs, whereas MEPs and their progeny remained unchanged (32, 33). The data presented here demonstrate that enforced Flt3 cytokine receptor signaling is sufficient to activate as well as enhance IPC and DC differentiation programs, suggesting that instructive cytokine signaling might indeed occur in hematopoiesis. Thus, we speculate that once Flt3+ short-term HSCs and their offspring Flt3+ cells are exposed to Flt3L-rich environments, these cells will be instructed to differentiate into IPCs and DCs. This might be enhanced by a self-sustaining process in which Flt3 downstream transcription factors STAT3 and PU.1 in turn maintain Flt3 receptor expression. However, Flt3 signaling does not immediately silence other developmental options. It is likely that most Flt3-expressing progenitors will not continuously be stimulated via Flt3L but will receive and activate alternative signals, and thus consecutively acquire myeloid or lymphoid, but not IPC or DC, cell fates. Beyond previous studies, our data further emphasize that IPC and DC development does not fit into a deterministic “lymphoid” nor “myeloid” lineage, but rather a “Flt3-permissive” developmental model, where Flt3-expressing progenitors maintain IPC and DC differentiation options in response to Flt3L as long as no competing signal shuts these down. It will be of interest to test whether downstream dividing Flt3+ common IPC and DC progenitors with silenced alternative developmental programs exist and which critical factors are involved in final IPC or DC lineage termination.
MATERIALS AND METHODS
Mice.
C57BL/6 (CD45.2), C57BL/Ka-Thy1.1 (CD45.1), and BALB/c mice (Charles River Laboratories) were maintained at the Institute for Research in Biomedicine animal facility in accordance with the Swiss Federal Veterinary Office guidelines.
Flow cytometry and cell sorting.
Hematopoietic progenitors were isolated as described previously with minor modifications (2, 37). Bone marrow cells were immunomagnetically preenriched for c-Kit+ cells using APC-conjugated c-Kit antibodies (2B8; eBioscience) and APC microbeads (Miltenyi Biotec). Cells were then stained with monoclonal antibodies as indicated below. Flt3+ and Flt3− hematopoietic progenitors were sorted as lineage− (CD3ε, 145-2C11; CD4, GK1.5; CD8, 53-6.7; B220, RA3-6B2; CD19, MB19-1; CD11b, M1/70; Gr-1, RB6-8C5; and TER119, TER119), IL-7Rα− (A7R34), Thy1.1− (19XE5), c-Kit+, and Flt3+or− (A2F10.1) cells. Thus, Flt3+ and Flt3− hematopoietic progenitors did not contain Thy1.1+ HSCs or IL-7Rα+ lymphoid progenitors. Myeloid progenitors were sorted as Lin−Sca-1− (E13-161-7) c-Kit+CD34+ (RAM34) FcγRlo (2.5G2; CMPs), Lin−Sca-1+c-Kit+CD34+FcγRhigh (GMPs), and Lin−Sca-1−c-Kit+CD34−FcγRlo (MEPs) cells. For IPC and DC analysis and sorting, additional monoclonal antibodies against the following antigens were used: CD11c (N418), MHC class II (I-A/I-E; M15/114.15.2), Ly6C (AL-21), CD45RA (A20.1.7), CD80 (16-10A1), and CD86 (GL-1). Cells were sorted and analyzed using a FACSCalibur and FACSAria (Becton Dickinson).
Retroviral transduction of hematopoietic progenitors.
The full length of human Flt3, mouse STAT3, and PU.1 cDNA was inserted into a retroviral expression vector, pMYs-IRES-GFP, respectively (53). These constructs were transiently transfected into Phoenix-Ampho cells by LipofectAMINE (Invitrogen). The amphotropic retrovirus supernatants were used to transduce GP+E-86 cells. After 2 d, the brightest GFP-expressing GP+E-86 cells were FACS sorted and expanded. For transduction of hematopoietic progenitor cells, GP+E-86 cells were 20-Gy irradiated and plated in 24-well plates at 1.5 × 105 cells per well for 24 h. Progenitor cells were transduced by coculture with GP+E-86 for 18 h in IMDM (Invitrogen) containing 2% FCS, 4 μg/ml polybrene (Sigma-Aldrich), 100 ng/ml human Flt3L-Ig fusion protein (huFlt3L-Ig), 10 ng/ml mSCF (R&D Systems), and 10 ng/ml mIL-11 (R&D Systems). Transduced cells were removed by gentle pipetting and then subjected to further assays.
In vitro IPC and DC differentiation assays.
Retrovirus-transduced Flt3+ and Flt3− progenitors as well as CMPs, GMPs, and MEPs were cultured in IMDM, supplemented with 10% FCS, 10−4 M 2-ME, sodium pyruvate, and antibiotics, 100 ng/ml huFlt3L-Ig, and 10 ng/ml mSCF as indicated. Half of the media was replaced every 3 d and new cytokines were added. Human Flt3L-Ig fusion protein was produced in Drosophila cells as described previously (54).
In vitro myeloid colony formation assays.
For myeloid colony–forming assays, GFP +-CMPs, GFP +-GMPs, and GFP +-MEPs were sorted after viral transduction and were cultured in IMDM-based methylcellulose media (Methocult H4100; StemCell Technologies Inc.), containing 30% FCS, 1% bovine serum albumin, 2 mM l-glutamine, and 50 μM 2-ME, 10 ng/ml mSCF, 10 ng/ml mIL-3 (R&D Systems), 10 ng/ml mIL-11, 10 ng/ml mGM-CSF (R&D Systems), 10 ng/ml mTpo (R&D Systems), 1 U/ml hEpo (Roche), and 100 ng/ml huFlt3L-Ig. Colonies were determined and enumerated under an inverted microscope consecutively from day 3 to 8. In some cases, to confirm colony types, colonies were picked using fine-drawn pasteur pipettes, spun on slides, Giemsa stained, and evaluated by light microscopy.
In vivo reconstitution assays.
2 × 104 CD45.2 GFP +-MEPs, huFlt3 +-MEPs, or GFP +-CMPs each were injected intravenously into lethally irradiated (2 × 6 Gy in a 4-h interval from a Cesium 137 source; Biobeam 8000; STS GmbH) congenic mice (CD45.1) with 2 × 105 recipient-type CD45.1 whole bone marrow cells. Mice were killed on day 7. The progeny of donor-derived cells were isolated as described previously (33) and evaluated by FACS analysis.
MLR.
Graded numbers of sorted, irradiated (25 Gy) IPCs or DCs were plated in U-bottom 96-well plates with 2 × 105 immunomagnetically selected (CD4 microbeads; Miltenyi Biotec) BALB/c spleen CD4+ T cells in a final volume of 200 μl RPMI 1640 supplemented with 10% FCS. Cells were cultured for 5 d and pulsed with 1 μCi [3H]thymidine (Amersham Biosciences) per well during the last 16 h of culture. [3H]thymidine incorporation was measured on a β-plate counter (MicroBeta TriLux; EG&G WALLAC).
IFN-α production.
To evaluate IFN-α production, sorted CD11c+B220+ IPCs or conventional CD11c+B220− DCs derived from retrovirus-transduced progenitors were cultured for 24 h at 105 cells/200 μl in U-bottom 96-well plates in RPMI 1640 supplemented with 10% FCS, 2-ME, penicillin G, and streptomycin. Either 40 HAU/ml influenza virus (strain A/Beijing/353/89; provided by I. Julkunen, National Public Health Institute, Helsinki, Finland) or 1 μM CpG-A-ODN (ggTGCATCGATGCAgggggG; lowercase letters indicate base with phosphorothioate-modified backbones) was added at start of culture and again at 12 h. Culture supernatants were assayed using an IFN-α ELISA kit (Performance Biomedical Laboratories).
RT-PCR analysis.
Total RNA was extracted from sorted progenitors as indicated using TRIzol reagent (Invitrogen) followed by DNase I (Invitrogen) treatment. The cDNA was synthesized using random hexamers as well as SuperScript II reverse transcriptase (Invitrogen) and amplified using specific primers as described previously (13). For real-time PCR, cDNA products equivalent to RNAs from 200 progenitors were amplified using an Applied Biosystems 7900HT Fast Real-Time PCR System. The data were normalized by the level of 18s rRNA expression in each sample. Taqman probes for mouse STAT3, PU.1, GATA-1, Flt3, and 18s rRNA were purchased from Applied Biosystems.
Intracellular phosoho-STAT3 staining.
Retroviral-transduced MEPs were cytokine starved for 24 h in 1% FCS-IMDM. Cells were then incubated with or without 100 ng/ml huFlt3L-Ig and analyzed at indicated times for phosho-STAT3 by FACS according to the manufacturer's instructions (Cell Signaling).
Statistical analysis.
Results of experiments are reported as mean ± SD. Differences were analyzed using Student's t test.
Online supplemental material.
Fig. S1 shows the diagrams of pMY-IRES-GFP and pMY-hu_Flt3_-IRES-GFP bicistronic retroviral expression vector constructs and virus transduction efficacy in progenitor cells. Fig. S2 shows the diagrams of pMY-m_STAT3_-IRES-GFP and pMY-m_PU.1_-IRES-GFP bicistronic retroviral expression vector constructs. Fig. S3 shows the analysis of mouse Flt3 mRNA expression in _GFP_-, hu_Flt3_-, _STAT3_-, and _PU.1_-transduced MEPs, as well as CMPs. Figs. S1–S3 are available at http://www.jem.org/cgi/content/full/jem.20051645/DC1.
Supplemental Material
[Supplemental Material Index]
Acknowledgments
We thank F. Sallusto for critical reading and discussion; T. Kitamura for providing retrovirus expression vector; D. Jarrossay for technical assistance with cell sorting; and G. Bosshard and K. Karjalainen for help in preparation of recombinant human Flt3L-Ig fusion protein. Congenic C57BL/Ka-Thy1.1 CD45.1 mouse breeder pairs were kindly provided by I.L. Weissman.
This work is supported in part by a Swissbridge grant (to M.G. Manz), the Swiss National Science Foundation (grants 3100-63885 to A. Lanzavecchia and 3100-102221 to M.G. Manz), and the European Commission FP6 “Network of Excellence” initiative under contract number LSHB-CT-2004-512074 DC-THERA.
The authors have no conflicting financial interests.
Abbreviations used: CLP, common lymphoid progenitor; CMP, common myeloid progenitor; Flt3 ligand, Flt3L; GM, granulocyte/macrophage; GMP, GM progenitor; HSC, hematopoietic stem cell; IPC, type I interferon-producing cell; MEP, megakaryocyte/erythrocyte progenitor; SCF, stem cell factor; TPO, thrombopoietin.
N. Onai and A. Obata-Onai contributed equally to this work.
References
- 1.Kondo, M., A.J. Wagers, M.G. Manz, S.S. Prohaska, D.C. Scherer, G.F. Beilhack, J.A. Shizuru, and I.L. Weissman. 2003. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu. Rev. Immunol. 21:759–806. [DOI] [PubMed] [Google Scholar]
- 2.Akashi, K., D. Traver, T. Miyamoto, and I.L. Weissman. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 404:193–197. [DOI] [PubMed] [Google Scholar]
- 3.Manz, M.G., T. Miyamoto, K. Akashi, and I.L. Weissman. 2002. Prospective isolation of human clonogenic common myeloid progenitors. Proc. Natl. Acad. Sci. USA. 99:11872–11877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galy, A., M. Travis, D. Cen, and B. Chen. 1995. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 3:459–473. [DOI] [PubMed] [Google Scholar]
- 5.Kondo, M., I.L. Weissman, and K. Akashi. 1997. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 91:661–672. [DOI] [PubMed] [Google Scholar]
- 6.Metcalf, D. 1998. Lineage commitment and maturation in hematopoietic cells: the case for extrinsic regulation. Blood. 92:345–347. [PubMed] [Google Scholar]
- 7.Cantor, A.B., and S.H. Orkin. 2001. Hematopoietic development: a balancing act. Curr. Opin. Genet. Dev. 11:513–519. [DOI] [PubMed] [Google Scholar]
- 8.Orkin, S.H. 2000. Diversification of haematopoietic stem cells to specific lineages. Nat. Rev. Genet. 1:57–64. [DOI] [PubMed] [Google Scholar]
- 9.Akashi, K., X. He, J. Chen, H. Iwasaki, C. Niu, B. Steenhard, J. Zhang, J. Haug, and L. Li. 2003. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 101:383–389. [DOI] [PubMed] [Google Scholar]
- 10.Graf, T. 2002. Differentiation plasticity of hematopoietic cells. Blood. 99:3089–3101. [DOI] [PubMed] [Google Scholar]
- 11.Nutt, S.L., B. Heavey, A.G. Rolink, and M. Busslinger. 1999. Com-mitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 401:556–562. [DOI] [PubMed] [Google Scholar]
- 12.Rolink, A.G., S.L. Nutt, F. Melchers, and M. Busslinger. 1999. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 401:603–606. [DOI] [PubMed] [Google Scholar]
- 13.Iwasaki, H., S. Mizuno, R.A. Wells, A.B. Cantor, S. Watanabe, and K. Akashi. 2003. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 19:451–462. [DOI] [PubMed] [Google Scholar]
- 14.Xie, H., M. Ye, R. Feng, and T. Graf. 2004. Stepwise reprogramming of B cells into macrophages. Cell. 117:663–676. [DOI] [PubMed] [Google Scholar]
- 15.Kondo, M., D.C. Scherer, T. Miyamoto, A.G. King, K. Akashi, K. Sugamura, and I.L. Weissman. 2000. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 407:383–386. [DOI] [PubMed] [Google Scholar]
- 16.King, A.G., M. Kondo, D.C. Scherer, and I.L. Weissman. 2002. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc. Natl. Acad. Sci. USA. 99:4508–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwasaki-Arai, J., H. Iwasaki, T. Miyamoto, S. Watanabe, and K. Akashi. 2003. Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J. Exp. Med. 197:1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banchereau, J., and R.M. Steinman. 1998. Dendritic cells and the control of immunity. Nature. 392:245–252. [DOI] [PubMed] [Google Scholar]
- 19.Lanzavecchia, A., and F. Sallusto. 2001. The instructive role of dendritic cells on T cell responses: lineages, plasticity and kinetics. Curr. Opin. Immunol. 13:291–298. [DOI] [PubMed] [Google Scholar]
- 20.Steinman, R.M., D. Hawiger, and M.C. Nussenzweig. 2003. Tolerogenic dendritic cells. Annu. Rev. Immunol. 21:685–711. [DOI] [PubMed] [Google Scholar]
- 21.Shortman, K., and Y.J. Liu. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151–161. [DOI] [PubMed] [Google Scholar]
- 22.Traver, D., K. Akashi, M. Manz, M. Merad, T. Miyamoto, E.G. Engleman, and I.L. Weissman. 2000. Development of CD8alpha-positive dendritic cells from a common myeloid progenitor. Science. 290:2152–2154. [DOI] [PubMed] [Google Scholar]
- 23.Manz, M.G., D. Traver, T. Miyamoto, I.L. Weissman, and K. Akashi. 2001. Dendritic cell potentials of early lymphoid and myeloid progenitors. Blood. 97:3333–3341. [DOI] [PubMed] [Google Scholar]
- 24.Wu, L., A. D'Amico, H. Hochrein, M. O'Keeffe, K. Shortman, and K. Lucas. 2001. Development of thymic and splenic dendritic cell populations from different hemopoietic precursors. Blood. 98:3376–3382. [DOI] [PubMed] [Google Scholar]
- 25.Shigematsu, H., B. Reizis, H. Iwasaki, S. Mizuno, D. Hu, D. Traver, P. Leder, N. Sakaguchi, and K. Akashi. 2004. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 21:43–53. [DOI] [PubMed] [Google Scholar]
- 26.Chicha, L., D. Jarrossay, and M.G. Manz. 2004. Clonal type I interferon–producing and dendritic cell precursors are contained in both human lymphoid and myeloid progenitor populations. J. Exp. Med. 200:1519–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karsunky, H., M. Merad, I. Mende, M.G. Manz, E.G. Engleman, and I.L. Weissman. 2005. Developmental origin of interferon-alpha-producing dendritic cells from hematopoietic precursors. Exp. Hematol. 33:173–181. [DOI] [PubMed] [Google Scholar]
- 28.Lyman, S.D., and S.E. Jacobsen. 1998. c-kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities. Blood. 91:1101–1134. [PubMed] [Google Scholar]
- 29.McKenna, H.J., K.L. Stocking, R.E. Miller, K. Brasel, T. De Smedt, E. Maraskovsky, C.R. Maliszewski, D.H. Lynch, J. Smith, B. Pulendran, et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497. [PubMed] [Google Scholar]
- 30.Laouar, Y., T. Welte, X.Y. Fu, and R.A. Flavell. 2003. STAT3 is required for Flt3L-dependent dendritic cell differentiation. Immunity. 19:903–912. [DOI] [PubMed] [Google Scholar]
- 31.Tussiwand, R., N. Onai, L. Mazzucchelli, and M.G. Manz. 2005. Inhibition of natural type I interferon-producing and dendritic cell development by a small molecule receptor tyrosine kinase inhibitor with Flt3 affinity. J. Immunol. 175:3674–3680. [DOI] [PubMed] [Google Scholar]
- 32.Maraskovsky, E., K. Brasel, M. Teepe, E.R. Roux, S.D. Lyman, K. Shortman, and H.J. McKenna. 1996. Dramatic increase in the numbers of functionally mature dendritic cells in Flt3 ligand-treated mice: multiple dendritic cell subpopulations identified. J. Exp. Med. 184:1953–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karsunky, H., M. Merad, A. Cozzio, I.L. Weissman, and M.G. Manz. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller, G., V.G. Pillarisetty, A.B. Shah, S. Lahrs, and R.P. DeMatteo. 2003. Murine Flt3 ligand expands distinct dendritic cells with both tolerogenic and immunogenic properties. J. Immunol. 170:3554–3564. [DOI] [PubMed] [Google Scholar]
- 35.Gilliet, M., A. Boonstra, C. Paturel, S. Antonenko, X.L. Xu, G. Trinchieri, A. O'Garra, and Y.J. Liu. 2002. The development of murine plasmacytoid dendritic cell precursors is differentially regulated by FLT3-ligand and granulocyte/macrophage colony-stimulating factor. J. Exp. Med. 195:953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adolfsson, J., O.J. Borge, D. Bryder, K. Theilgaard-Monch, I. Astrand-Grundstrom, E. Sitnicka, Y. Sasaki, and S.E. Jacobsen. 2001. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 15:659–669. [DOI] [PubMed] [Google Scholar]
- 37.Christensen, J.L., and I.L. Weissman. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Amico, A., and L. Wu. 2003. The early progenitors of mouse dendritic cells and plasmacytoid predendritic cells are within the bone marrow hemopoietic precursors expressing Flt 3. J. Exp. Med. 198:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mende, I., H. Karsunky, I.L. Weissman, E.G. Engleman, and M. Merad. 2005. Flk2+ myeloid progenitors are the main source of Langerhans cells. Blood. In press. [DOI] [PMC free article] [PubMed]
- 40.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I.L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell. 3:137–147. [DOI] [PubMed] [Google Scholar]
- 41.Mizuki, M., J. Schwable, C. Steur, C. Choudhary, S. Agrawal, B. Sargin, B. Steffen, I. Matsumura, Y. Kanakura, F.D. Bohmer, et al. 2003. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 101:3164–3173. [DOI] [PubMed] [Google Scholar]
- 42.Friedman, A.D. 2002. Transcriptional regulation of granulocyte and monocyte development. Oncogene. 21:3377–3390. [DOI] [PubMed] [Google Scholar]
- 43.Anderson, K.L., H. Perkin, C.D. Surh, S. Venturini, R.A. Maki, and B.E. Torbett. 2000. Transcription factor PU.1 is necessary for development of thymic and myeloid progenitor-derived dendritic cells. J. Immunol. 164:1855–1861. [DOI] [PubMed] [Google Scholar]
- 44.Guerriero, A., P.B. Langmuir, L.M. Spain, and E.W. Scott. 2000. PU.1 is required for myeloid-derived but not lymphoid-derived dendritic cells. Blood. 95:879–885. [PubMed] [Google Scholar]
- 45.Bacon, C.M., P.J. Tortolani, A. Shimosaka, R.C. Rees, D.L. Longo, and J.J. O'Shea. 1995. Thrombopoietin (TPO) induces tyrosine phosphorylation and activation of STAT5 and STAT3. FEBS Lett. 370:63–68. [DOI] [PubMed] [Google Scholar]
- 46.Nutt, S.L., D. Metcalf, A. D'Amico, M. Polli, and L. Wu. 2005. Dynamic regulation of PU.1 expression in multipotent hematopoietic progenitors. J. Exp. Med. 201:221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantor, A.B., and S.H. Orkin. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 21:3368–3376. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, P., X. Zhang, A. Iwama, C. Yu, K.A. Smith, B.U. Mueller, S. Narravula, B.E. Torbett, S.H. Orkin, and D.G. Tenen. 2000. PU.1 inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 96:2641–2648. [PubMed] [Google Scholar]
- 49.Nerlov, C., E. Querfurth, H. Kulessa, and T. Graf. 2000. GATA-1 interacts with the myeloid PU.1 transcription factor and represses PU.1-dependent transcription. Blood. 95:2543–2551. [PubMed] [Google Scholar]
- 50.Ezoe, S., I. Matsumura, K. Gale, Y. Satoh, J. Ishikawa, M. Mizuki, S. Takahashi, N. Minegishi, K. Nakajima, M. Yamamoto, et al. 2005. GATA transcription factors inhibit cytokine-dependent growth and survival of a hematopoietic cell line through the inhibition of STAT3 activity. J. Biol. Chem. 280:13163–13170. [DOI] [PubMed] [Google Scholar]
- 51.Kulessa, H., J. Frampton, and T. Graf. 1995. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9:1250–1262. [DOI] [PubMed] [Google Scholar]
- 52.DeKoter, R.P., and H. Singh. 2000. Regulation of B lymphocyte and macrophage development by graded expression of PU. 1. Science. 288:1439–1441. [DOI] [PubMed] [Google Scholar]
- 53.Kitamura, T., Y. Koshino, F. Shibata, T. Oki, H. Nakajima, T. Nosaka, and H. Kumagai. 2003. Retrovirus-mediated gene transfer and expression cloning: powerful tools in functional genomics. Exp. Hematol. 31:1007–1014. [PubMed] [Google Scholar]
- 54.Wallny, H.J., G. Sollami, and K. Karjalainen. 1995. Soluble mouse major histocompatibility complex class II molecules produced in Drosophila cells. Eur. J. Immunol. 25:1262–1266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]