3′ end mRNA processing: molecular mechanisms and implications for health and disease (original) (raw)
Abstract
Recent advances in the understanding of the molecular mechanism of mRNA 3′ end processing have uncovered a previously unanticipated integrated network of transcriptional and RNA-processing mechanisms. A variety of human diseases impressively reflect the importance of the precision of the complex 3′ end-processing machinery and gene specific deregulation of 3′ end processing can result from mutations of RNA sequence elements that bind key specific processing factors. Interestingly, more general deregulation of 3′ end processing can be caused either by mutations of these processing factors or by the disturbance of the well-coordinated equilibrium between these factors. From a medical perspective, both loss of function and gain of function can be functionally relevant, and an increasing number of different disease entities exemplifies that inappropriate 3′ end formation of human mRNAs can have a tremendous impact on health and disease. Here, we review the mechanistic hallmarks of mRNA 3′ end processing, highlight the medical relevance of deregulation of this important step of mRNA maturation and illustrate the implications for diagnostic and therapeutic strategies.
Keywords: cancer, immunity, influenza, thalassemia, thrombosis, polyadenylation
Introduction
Messenger RNA 3′ end processing is a well-orchestrated process that involves components of the transcription, the splicing and the translation machinery. The medical importance of 3′ end processing is illustrated by an increasing number of different disease entities, which are caused by inappropriate 3′ end processing. In this review, we discuss key mechanistic features of 3′ end formation, although the reader is referred to several excellent reviews that cover the basic aspects of 3′ end formation and its regulation in more detail (Zhao and Manley, 1996; Colgan and Manley, 1997; Edwalds-Gilbert et al, 1997; Keller and Minvielle-Sebastia, 1997; Wahle and Kuhn, 1997; Barabino and Keller, 1999; Zhao et al, 1999; Edmonds, 2002; Proudfoot et al, 2002; Gilmartin, 2005; Weiner, 2005; Rosonina et al, 2006). We then focus on the medical perspective of 3′ end processing and illustrate how disease can be caused either by mutations of important RNA sequence elements and/or by the pathological expression of the proteins interacting with such sequence elements and between each other.
The eukaryotic mRNA 3′ end-processing machinery
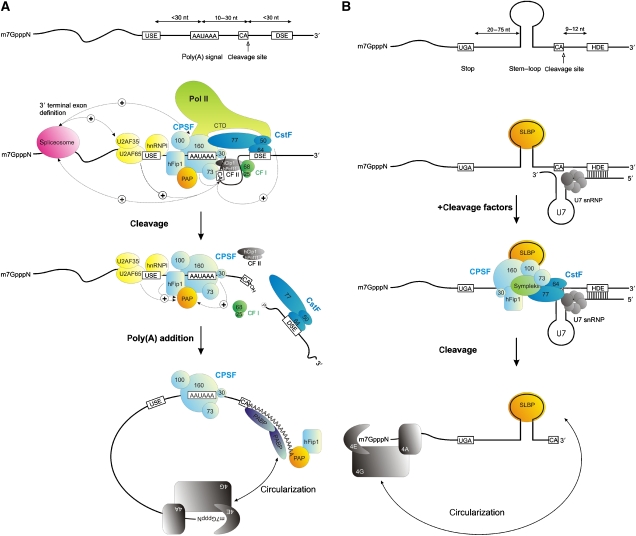
With the exception of some histone mRNAs, all eukaryotic mRNAs possess poly(A) tails at their 3 end, which are produced by a two-step reaction involving endonucleolytic cleavage and subsequent poly(A) tail addition. The specificity and efficiency of 3′ end processing is determined by the binding of multiprotein complexes to specific elements at the 3′ end of the pre-mRNA. Most cellular pre-mRNAs contain two core elements (Figure 1A). The canonical polyadenylation signal AAUAAA (or less frequently AUUAAA) upstream of the cleavage site is recognized by the multimeric cleavage and polyadenylation specificity factor (CPSF, light blue in Figure 1A) consisting of at least five subunits (CPSF 160, CPSF 100, CPSF 73, CPSF 30 and hFip1). This RNA–protein interaction determines the site of cleavage 10–30 nt downstream, preferentially immediately 3′ of a CA dinucleotide. The second canonical sequence element is characterized by a high density of G/U or U residues and is located up to 30 nt downstream of the cleavage site. This downstream sequence element (DSE) is bound by the 64-kDa subunit of the heterotrimeric cleavage-stimulating factor (CstF, dark blue in Figure 1A) that promotes the efficiency of 3′ end processing. Furthermore, accessory sequences that have initially been identified in (retro-) viral polyadenylation signals (Gilmartin et al, 1995; Graveley et al, 1996) can function as upstream sequence elements (USEs) (Moreira et al, 1995, 1998; Brackenridge and Proudfoot, 2000; Natalizio et al, 2002; Legendre and Gautheret, 2003; Danckwardt et al, 2004, 2007; Hall-Pogar et al, 2005, 2007; Hu et al, 2005; Xie et al, 2005) to facilitate 3′ end processing by serving as an additional anchor for the (canonical) 3′ end-processing machinery (Danckwardt et al, 2007; Hall-Pogar et al, 2007), or by recruiting canonical 3′ end factors directly (Moreira et al, 1995, 1998). Moreover, in vitro studies have identified a UGUA motif to be present in one or more copies at variable distances upstream of the cleavage site. These sequence motifs are thought to recruit the heterodimeric cleavage factor CFIm (Brown and Gilmartin, 2003; Venkataraman et al, 2005). These elements may also represent a primary determinant of poly(A) site recognition in the absence of the highly conserved A(A/U)UAAA motif (Venkataraman et al, 2005). Unlike the replication-dependent histone pre-mRNA processing elements (see below), conventional poly(A) sites thus exhibit a wide range of both sequence and spatial variability.
Figure 1.
_Cis_-acting sequence elements and _trans_-acting factors involved in mammalian 3′ end processing. (A) 3′ end processing of polyadenylated mRNAs. After assembly of multiprotein complexes at the respective RNA recognition motifs (upper panel), the primary transcript is endonucleolytically cleaved at the cleavage site by CPSF 73. This is followed by the addition of adenine residues to the 3′ end to form a poly(A) tail that is bound by PABPN1. The interaction of PAP with this protein and with CPSF is critical to establish the processive action of the polymerase for the synthesis of approximately 250 A residues. Following polyadenylation, the interaction of PABPN1 with the poly(A) tail is characterized by a rapid on–off rate, and PABN1 is exchanged by cytoplasmic PAPB (PABPC) at the time of nuclear export. PABPC interacts with the translation initiation factor elF4G as part of the initiation complex thus generating a translation competent pseudocircular ribonucleoprotein particle. (B) In contrast to mRNAs with a poly(A) site, 3′ end processing of replication-dependent histone mRNAs requires conserved sequence elements and structural elements. The signal for 3′ end processing of this small class of pre-mRNAs consists of a conserved stem–loop element that is positioned upstream of the cleavage site and a purine-rich HDE. The HDE is recognized by base pairing with the 5′ end of the U7 small nuclear RNA, which is incorporated into a U7 snRNP. The stem–loop is bound by the SLBP, which functions in 3′ end processing, translation and in the coupling of message stability to DNA replication and the cell cycle. After recruitment of the 3′ end-processing apparatus, the site of endonucleolytic cleavage occurs 3′ of a CA dinucleotide 9–12 nucleotides upstream of the intermolecular U7/HDE RNA duplex. The SLBP also serves to establish a pseudo-circularization together with initiation factor elF4G. Stimulatory interactions are highlighted (+). Cis-acting RNA elements and trans-acting factors are summarized inTables I and II, USE=upstream sequence element, AAUAAA=poly(A) signal, DSE=downstream sequence element, HDE=Histone downstream element, CstF=cleavage stimulating factor (blue complex), CPSF=cleavage/polyadenylation specificity factor (light blue complex), Pol II=RNA polymerase II (light green complex) with CTD (C-terminal domain), CF I=cleavage factor I (green complex), CF II=cleavage factor II (grey complex), PAP=poly(A)-polymerase (yellow complex), PABPN1=nuclear poly(A)-binding protein, PABPC=cytoplasmic poly(A)-binding protein (dark blue complex), 4A, 4E, 4G=translation initiation factors (grey complex), SLBP=stem loop binding protein (yellow complex).
After assembly of the basal 3′ end-processing machinery, the endonucleolytic cleavage reaction is thought to be catalyzed by CPSF 73 (Ryan et al, 2004; Dominski et al, 2005; Mandel et al, 2006). Subsequently, a nuclear poly(A) polymerase (PAP) adds ∼250 A-nucleotides to the 3′ end in a template-independent manner. The length of the poly(A) tail is similar in different mRNAs, and is thought to be determined by an interaction between the nuclear poly(A)-binding protein (PABPN1, PABP2), CPSF and PAP (Kuhn and Wahle, 2004). The binding of PABPN1 to the RNA is unstable and upon nuclear export PABPN1 is replaced by the cytosolic poly(A)-binding protein (PABPC; for review see Mangus et al, 2003; Kuhn and Wahle, 2004), which interacts with the translation initiation factor eIF4G, stimulating translation and regulating mRNA stability (Sachs et al, 1997; Kahvejian et al, 2001; Kuhn and Wahle, 2004). Furthermore, PABPC can interact with the translation termination factor eRF3 (Cosson et al, 2002), implicating a role of the poly(A) tail in translation termination and possibly ribosome recycling.
In contrast to bulk mRNAs, 3′ processing of the replication-dependent histone mRNAs requires both conserved sequence and structural elements (Figure 1B; for review see Marzluff and Duronio, 2002; Gilmartin, 2005; Marzluff, 2005). The signal for 3′ end processing of this class of pre-mRNAs consists of a conserved stem–loop that is positioned upstream of the cleavage site, and a purine-rich histone downstream element (HDE). The HDE is recognized by base pairing with the 5′ end of the U7 small nuclear RNA, which is incorporated into a ribonucleoprotein (RNP) of the Sm class (U7 snRNP). The stem–loop is bound by stem–loop-binding protein (SLBP) that functions in 3′ end processing, translation and in coupling of histone mRNA stability to DNA replication and the cell cycle. The site of endonucleolytic cleavage occurs after a CA dinucleotide of the histone pre-mRNA sequence, and is located 9–12 nucleotides upstream of the intermolecular U7/HDE RNA duplex. Thus, the replication-dependent histone mRNAs display a far more rigid composition of _cis_-acting sequence elements than mRNAs with poly(A) sites. Despite the distinct mechanisms that specify the site of 3′ end processing, it is now becoming increasingly clear that both classes of transcripts share a common catalytic core consisting of the CPSF and CstF subcomplexes (Kolev and Steitz, 2005) likely including CPSF 73 as the endonuclease (Ryan et al, 2004; Dominski et al, 2005; Mandel et al, 2006).
The shared components point to a common evolutionary origin of the different 3′ end formation machineries. However, whereas poly(A) site processing is a two-step process coupled to other steps of the gene expression pathway (transcription and splicing; Minvielle-Sebastia and Keller, 1999; Adamson et al, 2005; see next section), the synthesis of mature (inherently intronless) histone mRNAs requires only one RNA-processing reaction. In addition, histone mRNA 3′ end processing seems to be incompatible with splicing (Pandey et al, 1990) and does not appear to depend on transcription (Adamson and Price, 2003). Thus, by sharing the same catalytic core machinery, but using specific 3′ end-processing sites, the histone pre-mRNAs have developed an efficient way to meet their needs of replication-dependent gene expression.
An integrated network of co-transcriptional mRNA-processing events controls gene expression
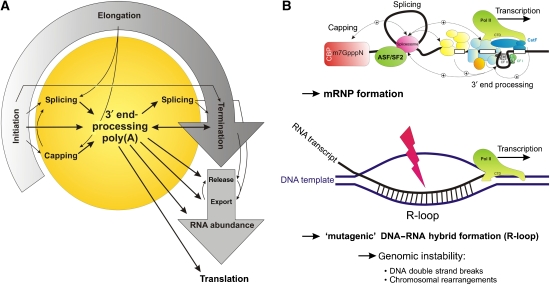
Producing a mature RNA from a mammalian gene requires a complex and tightly coupled series of molecular mechanisms (Hirose and Manley, 2000; Maniatis and Reed, 2002; Proudfoot et al, 2002; Proudfoot, 2004); as a nascent pre-mRNA transcript emerges from the elongating RNA polymerase II (Pol II) both extensive constitutive and alternative splicing events occur co-transcriptionally (Lopez, 1998; Hirose and Manley, 2000; Proudfoot et al, 2002), giving rise to a perplexingly high diversity within the transcriptome (Black, 2000, 2003; Graveley, 2001; Shin and Manley, 2004). Eventually, transcription termination stops the elongating polymerase triggered by recognition of poly(A) signals in the nascent transcript (Proudfoot et al, 2002; Rosonina et al, 2006), and both CPSF and CstF are transferred by the C-terminal domain (CTD) of Pol II to their specific pre-mRNA-binding sites (see above) to produce the mRNA 3′ end. The phosphorylation of serine residues within the CTD critically regulates gene expression (Ahn et al, 2004): it coordinates the recruitment of RNA-processing factors (including the 5′ capping complex, the spliceosome and 3′ processing machinery), and regulates chromatin organization by histone methylation (Hampsey and Reinberg, 2003).
As proposed for the coupling between transcription and pre-mRNA 3′ end processing at poly(A) sites, the extensive crosstalk between the different pre-mRNA-processing activities (capping, splicing and polyadenylation) has recently emerged to be critical for both gene expression and, interestingly, genome integrity (Figure 2); proteins that bind to the cap structure of pre-mRNAs interact with splicing factors and promote recognition of the cap-proximal splice site (Colot et al, 1996; Lewis et al, 1996). Conversely, splicing factors that associate with the 3′ terminal intron also interact with downstream polyadenylation factors to mutually promote both 3′ end cleavage/polyadenylation and terminal intron splicing (Niwa et al, 1990; Wassarman and Steitz, 1993; Lutz et al, 1996; Gunderson et al, 1997; Vagner et al, 2000; Li et al, 2001; McCracken et al, 2002, 2003; Millevoi et al, 2002, 2006; Awasthi and Alwine, 2003; Kyburz et al, 2006; Danckwardt et al, 2007). It should be noted, however, that the effect of these interactions might also be inhibitory when factors are bound at different positions such as within the 3′ terminal exon or within the 3′ UTR. These interactions ensure that the correct splice sites are recognized, and that 3′ end processing is timely, accurate and efficient. Moreover, the extensive integration between the different co-transcriptional mechanisms is believed to protect chromosomes from potentially deleterious effects that could arise from interaction between the nascent RNA and template DNA during transcription (Li and Manley, 2006; Figure 2B). Finally, functional polyadenylation signals and polyadenylation factors are required for efficient transcription termination (Proudfoot et al, 2002; Buratowski, 2005; Rosonina et al, 2006; Kaneko et al, 2007) and release of the polyadenylated mRNAs for export from the nucleus (Reed and Hurt, 2002). Therefore, the efficiency of polyadenylation can have significant quantitative effects on gene expression in general, and defects of mRNA 3′ end formation can profoundly affect cell viability, growth and development.
Figure 2.
Integrated networks of co-transcriptional mRNA processing to regulate gene expression and to maintain genomic stability. (A) Transcription initiation, elongation and termination (circular arrow) are tightly coupled to mRNA processing steps such as capping, splicing and 3′ end processing (inner circle). Appropriate 3′ end processing is functionally interconnected with transcription and mRNA capping and splicing, and impacts on post-transcriptional mechanisms (mRNA release, export, abundance and translation). Loss or gain of function of 3′ end processing thus critically interferes with other gene expression steps. (B) Co-transcriptional mRNA processing is believed to promote packaging of the nascent RNA transcript (formation of an ‘inert' RNP particle, upper panel) and thus to prevent the accumulation of co-transcriptional R-loops (lower panel), which can lead to DNA double strand breaks and chromosomal rearrangements. Disruption of co-transcriptional RNA processing is therefore thought to result in genomic instability (for review see Li and Manley, 2006).
The medical relevance of errors of 3′ end processing is exemplified by different inherited and acquired human disorders. In the following section, we focus on a group of disorders that highlight the most characteristic features of 3′ end processing and show how alterations of sequence elements and of protein components of the 3′ end processing machinery result in human pathology.
Mutations of sequence elements
Loss of function—no flexibility in cis?
Loss-of-function mutations of globin mRNA 3′ end processing are a well-recognized cause of thalassemias. The thalassemias are a heterogenous group of very common human genetic disorders that result from defects in hemoglobin production. The globin genes were the first human genes to be cloned (Maniatis et al, 1976), and thus represent the earliest medically important genes that illustrate how different steps of the gene expression pathway can be inactivated by naturally occurring mutations. Thalassemias are characterized by highly variable phenotypes that are determined by the extent of hemolysis and ineffective erythropoiesis—ranging from a complete lack of symptoms to severe, transfusion-dependent anemia.
This remarkable phenotypic diversity reflects the heterogeneity of mutations of the globin loci and modulation by a variety of modifiers (for review see Weatherall, 2001). In addition, general surveillance mechanisms of gene expression, such as nonsense-mediated mRNA decay (NMD), represent potent phenotypic modifiers as has been exemplified by the identification of β-thalassemia alleles with an unusual dominant mode of inheritance (for review see Hentze and Kulozik, 1999; Holbrook et al, 2004; Maquat, 2004; Neu-Yilik et al, 2004).
The understanding of 3′ end mRNA processing has been markedly advanced by studies of thalassemia patients with different mutations that result in an alteration of the AAUAAA hexanucleotide. Such mutations have been identified in both the α-globin (Higgs et al, 1983; Harteveld et al, 1994) and β-globin genes (Orkin et al, 1985; Jankovic et al, 1990; Rund et al, 1992; van Solinge et al, 1996), and invariably inactivate or severely inhibit gene expression. Similar mutations have been observed in, for example, the Foxp3 gene causing IPEX syndrome (Bennett et al, 2001), a rare fatal disorder characterized by immune dysregulation, polyendocrinopathy, enteropathy and X-linked inheritance.
However, it is important to note that poly(A) signal mutations do not always cause disease. This is exemplified by individuals who carry an AAUAAC to AGUAAC mutation of the arylsulfatase A gene poly(A) signal. Null mutations of this gene cause metachromatic leucodystrophy, a most serious neurodegenerative disorder. The AAUAAC to AGUAAC mutation reduces, but does not completely inactivate, mRNA and enzyme expression. Carriers of this hypomorphic mutation do not develop disease symptoms but a state referred to as ‘pseudodeficiency', exemplifying that biochemically severe mutations can remain clinically silent (Barth et al, 1993; Harvey et al, 1998).
The complexity of effects of poly(A) site mutations is further reflected by mutations of the lysosomal alpha-galactosidase A (alpha-GalA) gene. Human alpha-GalA is one of the rare mammalian genes that bears its polyadenylation signal within the coding sequence and lacks a 3′ untranslated region (Bishop et al, 1988). An AA dinucleotide deletion within this poly(A) site results in aberrant 3′ end formation, with generation of multiple non-functional transcripts, and in complete inactivation of the gene (functional null allele). Affected patients develop Fabry disease, a severe X-linked recessive inborn error of glycosphingolipid catabolism.
These examples underscore the functional importance of the highly conserved poly(A) signal and show that there is little sequence flexibility with regard to the hexanucleotide signal.
Gain of function—too much of a good thing
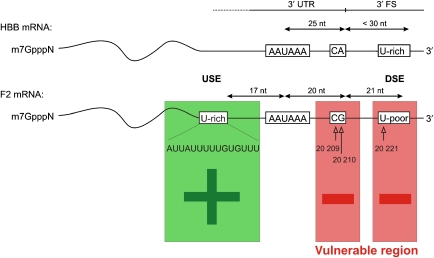
Clinically relevant gain-of-function mutations stimulating 3′ end processing were first identified in the prothrombin (coagulation factor II; F2) gene (Gehring et al, 2001; Danckwardt et al, 2004). Such mutations cause raised prothrombin plasma concentrations, which disturb the finely tuned balance between pro- and anticoagulatory activities, and result in an increased risk to develop thrombosis (referred to as thrombophilia). The F2 20210*A mutation affects approximately 1–2% of the general Caucasian population and hence represents a common cause of thrombophilia (Poort et al, 1996). This mutation affects the most 3′ nucleotide of the F2 mRNA, where it is endonucleolytically cleaved and polyadenylated (Figure 3; Gehring et al, 2001). Physiological 3′ cleavage of the F2 mRNA occurs 3′ of a CG dinucleotide, whereas most mRNAs are cleaved 3′ of a CA dinucleotide. The CG dinucleotide has been shown to be less efficient in promoting the cleavage reaction in vitro (Chen et al, 1995), and the CG → CA mutation increases F2 3′ end processing efficiency in cell lines and in transgenic mice (Gehring et al, 2001; Danckwardt et al, 2004; Kuwahara et al, 2004). Hence, the G → A mutation at position 20 210 reverts the physiologically inefficient F2 cleavage site into the mechanistically most efficient CA dinucleotide, which increases cleavage site recognition and results in an approximately twofold enhancement of prothrombin mRNA and protein expression. Increased mRNA 3′ end formation efficiency thus emerged as a novel molecular principle causing pathological gene expression and explains the role of F2 20210*A in the pathogenesis of thrombophilia. Furthermore, this mutation represents a paradigm of a quantitatively subtle change of gene expression that can cause functionally and clinically most significant consequences.
Figure 3.
The human prothrombin 3′ end-processing signal shows an unusual architecture of non-canonical sequence elements, which are susceptible to clinically relevant gain-of-function mutations. A sequence comparison of the 3′ end-processing signals of efficiently processed mRNAs such as SV40 late or β-globin (HBB) with F2 (lower lane) revealed an inefficient F2 cleavage dinucleotide context and a uridine-poor DSE. In contrast, the F2 3′ untranslated region contains a uridine-rich USE that promotes 3′ end processing and hence balances F2 mRNA expression. Sequences encompassing the cleavage site and the 3′-flanking region represent a vulnerable region for clinically relevant gain-of-function mutations.
Subsequently, two further, albeit rare, thrombosis-related mutations of F2 3′ end processing were identified (Balim et al, 2003; Schrijver et al, 2003; Danckwardt et al, 2004, 2006b; Soo et al, 2005) and shown to increase 3′ end formation efficiency (Figure 3; Danckwardt et al, 2004, 2006b). One of these mutations is a C → T exchange that occurs at the penultimate position 20 209 of the F2 3′ UTR (F2 20209*T). The other mutation introduces an additional U-residue 11 nucleotides 3′ of the cleavage site into the putative CstF-binding site in the F2 3′-flanking region (F2 20221*T). The detailed analysis of F2 3′ end formation determinants in patients with thrombophilia has thus identified an unusual architecture of non-canonical sequence elements, which explains the susceptibility of the F2 3′ end processing region to gain-of-function mutations (Danckwardt et al, 2004, 2006a); (1) the F2 3′ end formation signal contains the least efficient dinucleotide at the physiological cleavage site. Mutations such as F2 20210*A and F2 20209*T can revert this physiological inefficiency to pathologically increased efficiency. (2) The F2 putative CstF-binding site displays an unusually low density of uridine residues when compared with efficiently 3′ end-processed mRNAs such as β-globin and SV 40. Consequently, the introduction of (an) additional uridine residue(s) into the F2 3′-flanking sequence by either the natural F2 20221*T mutation or the experimental insertion at adjacent sites enhances 3′ end processing, presumably by facilitating interaction of CstF with the pre-mRNA (Danckwardt et al, 2004).
Based on these data, the question arose how efficient gene expression in general and 3′ end-processing efficiency in particular can be maintained for a gene with inefficient canonical 3′ end-processing elements, but which encodes a highly abundant protein that makes up approximately 5% (≈3 g/l) of the total blood plasma protein content. This question could be resolved by the identification of a stimulatory USE within the F2 3′ UTR, which compensates for the weak functional activities of the cleavage site and the downstream U-rich element (DSE) in the F2 3′-flanking sequence (Figure 3). The F2 3′ end-processing signal thus appears to be balanced by an unusual architecture of weak and strong stimulatory sequence elements (Danckwardt et al, 2004, 2006a).
This architecture in general and the USE in particular have elicited considerable interest, because USEs play key roles in important physiological functions such as blood coagulation (prothrombin; Danckwardt et al, 2004), innate immunity (complement C2; Moreira et al, 1998), inflammation (cyclooxygenase-2 (Cox-2); Hall-Pogar et al, 2005) and in the maintenance of cell structure (lamin B2; Brackenridge and Proudfoot, 2000) and the intercellular matrix (collagen; Natalizio et al, 2002). Biocomputational analyses predict that USE-dependent 3′ end processing may be common amongst cellular mRNAs (Legendre and Gautheret, 2003; Hu et al, 2005; Danckwardt et al, 2007), and characterize a novel class of transcripts that are regulated by alternative 3′ end processing (Hall-Pogar et al, 2005). In some RNAs, USEs functionally compensate for the lack of a DSE by recruiting CstF (Moreira et al, 1995, 1998). Furthermore, novel USE-interacting _trans_-acting factors have been identified (hnRNPI/PTB, U2AF35, U2AF65, PSF, U2A1, p54nrb), which are thought to stabilize protein–mRNA interactions with the canonical 3′ end-processing machinery (CPSF and CstF) and thus to promote 3′ end processing (Moreira et al, 1995, 1998; Danckwardt et al, 2007; Hall-Pogar et al, 2007). Moreover, the USE-dependent recruitment of splicing factors involved in 3′ end processing indicates that sequence elements in the 3′ UTR contribute to the integrated network of different mRNA-processing steps (Hirose and Manley, 2000; Maniatis and Reed, 2002; Proudfoot et al, 2002).
The physiological importance of USE-dependent 3′ end-processing signals has not yet been fully elucidated. However, because of different cofactor requirements (Danckwardt et al, 2007; Hall-Pogar et al, 2007), it is tempting to speculate that 3′ end formation of some of these mRNAs might be differentially regulated. USE-dependent 3′ end formation may thus play an important role in the regulated physiology of processes such as blood coagulation or inflammatory processes (see below).
Role of trans-acting factors
Regulated alternative 3′ end processing in immunity and inflammation
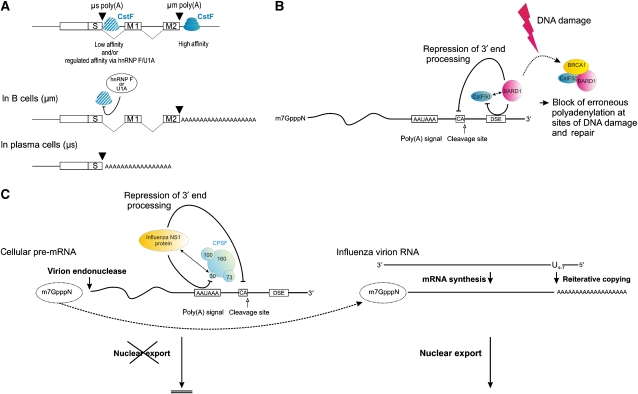
Alternative poly(A) site selection has been identified to represent an important and evolutionary conserved regulatory mechanism for spatial (tissue specificity, that is, Cox-2 or calcitonin/calcitonin gene-related peptide; Feng et al, 1993; Lukiw and Bazan, 1997; Lou and Gagel, 1998; Hall-Pogar et al, 2005) and temporal control of gene expression (i.e., immunoglobulin class switch; Alt et al, 1980; Early et al, 1980; Rogers et al, 1980; Cushley et al, 1982; Takagaki et al, 1996; Takagaki and Manley, 1998; for review see Zhao and Manley, 1996; Edwalds-Gilbert et al, 1997; Barabino and Keller, 1999; Zhao et al, 1999). This is underlined by the observation that about half of the human mRNAs contain more than one polyadenylation site (Tian et al, 2005; Yan and Marr, 2005). Alternative poly(A) site choices are usually regulated by various _cis_-and _trans_-acting determinants, and primarily influenced by (1) the intrinsic strength of sequence elements, (2) the concentration or activity of polyadenylation factors and/or (3) by tissue- or stage-specific regulatory factors (Barabino and Keller, 1999). In this respect, the regulation of IgM heavy-chain expression during B-cell differentiation represents an intriguing example (Figure 4A; Alt et al, 1980; Early et al, 1980; Rogers et al, 1980): In this mRNA, alternative poly(A) site selection has been proposed to be regulated by the concentration of the CstF 64-kDa subunit. According to this model, increased CstF-64 switches the IgM heavy-chain expression from a membrane-bound form (μm) to the secreted form (μs) by activation of an alternative upstream μs-specific poly(A) site in plasma cells (Takagaki et al, 1996). The low affinity upstream CstF-64-binding site would thus be favored under conditions of high CstF-64 concentration, whereas the high-affinity site of the membrane-bound form (μm) is used when the CstF-64 concentration is low.
Figure 4.
Regulated and alternative 3′ end processing in development and disease. (A) During B-cell differentiation, alternative poly(A) site selection effects a switch of the IgM heavy-chain expression from a membrane-bound form (μm) to the secreted form (μs). In this example, CstF-64 binding to the site of the RNA giving rise to secreted IgM (μs) is favored either by high CstF-64 concentrations (Takagaki et al, 1996) or under conditions of low hnRNP F and/or low U1A concentrations (Veraldi et al, 2001; Phillips et al, 2004) in plasma cells (lower lane). In contrast, the high-affinity site of the membrane bound form (μm) is used in B cells (upper lane), where the CstF-64 concentration is low or when high concentrations of U1A and/or hnRNP F inhibit CstF-64 binding to the secretory μs-specific poly(A) site (modified after Barabino and Keller, 1999; boxes indicate exons). (B) The BRCA1-associated protein BARD1 physically interacts with CstF-50, thereby repressing the polyadenylation machinery (Kleiman and Manley, 1999). Both BARD1 and CstF-50 also interact with Pol II (not shown), and BARD1 has been proposed to sense sites of DNA damage and repair. The BARD1-mediated inhibition of polyadenylation thus prevents inappropriate RNA processing during transcription at such compromised sites. Consequently, challenging cells with DNA-damaging agents results in a transient inhibition of 3′ end formation by enhanced formation of a CstF/BARD1/BRCA1 complex. Furthermore, a tumor-associated germline mutation in BARD1 (Gln564His) decreases its affinity to CstF-50 and renders the protein inactive in polyadenylation inhibition. These findings link 3′ end RNA processing with DNA repair, and loss of wild-type BARD1 could therefore lead to defective control of gene expression as a result of inappropriate polyadenylation (Kleiman and Manley, 2001). (C) In influenza A virus-infected cells, the highly abundant NS1 protein interacts with the cellular 30-kDa subunit of CPSF (Nemeroff et al, 1998) and PABPN1 (not shown) (Chen et al, 1999). This prevents binding of the CPSF complex to its RNA substrates and selectively inhibits 3′ end processing and nuclear export of host pre-mRNAs (adopted from; Nemeroff et al, 1998). In contrast, the 3′ terminal poly(A) sequence on viral mRNAs is produced by the viral transcriptase, which reiteratively copies a stretch of 4–7 uridines in the virion RNA templates. In addition, an endonuclease intrinsic to the viral polymerase cleaves cellular capped RNAs to generate capped fragments that serve as primers for the viral mRNA synthesis (so-called ‘cap-snatching mechanism'; Rao et al, 2003). Thus, by interfering with the activity of an essential 3′ end-processing factor, influenza has devised an efficient way to specifically shut off cellular gene expression and to facilitate viral gene expression (further detail see Nemeroff et al, 1998; Chen et al, 1999; Rao et al, 2003).
Subsequent publications, however, showed that CstF-64 levels do not change in primary human splenic B cells during differentiation (Martincic et al, 1998), and additional factors have been suggested to play an essential role in activating the secretory μs-specific poly(A) site (Figure 4A; Edmonds, 2002); in one case, hnRNP F has been proposed to compete with CstF-64 for binding sequences downstream of the μs-specific poly(A) site (Veraldi et al, 2001). Furthermore, the U1A protein was reported to inhibit the secretory poly(A) site by a dual mechanism: U1A binding to cognate binding sites upstream to the 3′ end processing signal was reported to inhibit polyadenylation step at the μs-specific poly(A) site (Phillips et al, 2001), whereas binding downstream inhibited the cleavage reaction by sterically inhibiting CstF-64 RNA binding at the μs-specific poly(A) site (Phillips et al, 2004). In any case, these findings illustrate that alternative 3′ end processing may be regulated by a number of different mechanisms to tightly control appropriate poly(A) site selection.
A similar mechanism seems to underlie the regulated expression of the transcription factor NF-ATc during T-cell differentiation (Chuvpilo et al, 1999). Two longer isoforms of NF-ATc mRNA are synthesized in naïve T cells, whereas a shorter isoform is found in effector cells. This switch is mediated by activation of a proximal poly(A) site by upregulation of CstF-64, which occurs upon T-cell stimulation. Interestingly, LPS stimulation has recently been shown to increase CstF-64 expression in macrophages, which promotes alternative polyadenylation of several mRNAs (Shell et al, 2005), establishing a possible link between the induction of an acute-phase response and alternative polyadenylation. Such a link is also suggested by differences in poly(A) site use in the pancreatitis-associated protein II/regenerating gene III (PAP II/Reg III), which is expressed during the acute-phase response following acute pancreatitis. An elongated isoform of the PAP II/Reg III mRNA, which is processed at a downstream poly(A) site, appears to be specifically expressed during the early hours of regeneration, whereas the shorter isoform is expressed for a much longer period (Honda et al, 2002).
Further links between acute-phase reactions and alternative polyadenylation have since been demonstrated for the inducible and tissue-specific expression Cox-2 mRNA in liver and colon cells (Ristimaki et al, 1996; Hall-Pogar et al, 2005), in human neocortex in Alzheimer disease (Lukiw and Bazan, 1997) and for tropoelastin expression in sun-damaged skin (Schwartz et al, 1998).
Cancer and 3′ end formation
Mutations that drive uncontrolled cell-cycle progression are critical events in tumorigenesis (Kastan and Bartek, 2004). Interestingly, PAP activity is regulated in a cyclin-dependent manner (Colgan et al, 1996, 1998) and is critical for early development (Ballantyne et al, 1995). Furthermore, depletion of CstF-64 can lead to cell-cycle arrest and result in apoptosis (Takagaki and Manley, 1998). Similarly, defective PAP causes cell-cycle arrest in the G0–G1 phases (Zhao and Manley, 1996). PAP mRNA is overexpressed in human carcinomas of the breast, colon, ovary and pancreas (Pendurthi et al, 1997), and polyadenylation activity is significantly enhanced in aggressive acute leukemias and Burkitt lymphoma compared with less aggressive chronic leukemias and normal lymphocytes (Trangas et al, 1984); a similar situation applies to aggressive forms compared to less progressive forms of breast cancer (Scorilas et al, 2000). The activity of PAP thus likely reflects the proliferative activity of cells and serves as an indicator for dedifferentiation. Therefore, it may serve as a prognostic biomarker in different forms of malignancy (Pangalis et al, 1985; Sasaki et al, 1990; for review see Jacob et al, 1989; Scorilas, 2002).
It is not known whether enhanced PAP activity per se contributes to tumorigenesis or whether it represents a coincidental (epi-) phenomenon to meet the requirements of rapidly proliferating cells. The latter mechanism has been suggested to account for the overexpression of a neoplasma-associated ‘Neo'-PAP (isotype), which is thought to be regulated by a mechanism distinct from that utilized by PAP in normal cells (Topalian et al, 2001). Notably, PAP isotype antibodies in the serum of cancer patients and rat models seem to have a predictive potential for cancer disease progression and outcome (Stetler et al, 1981). However, iso-PAP antibodies are not cancer-specific and have also been observed in patients with rheumatic disorders (Stetler et al, 1987).
A more specific causal link between mRNA 3′ end processing and DNA repair and tumor suppression is suggested by the observation that the BRCA1-associated protein BARD1 physically interacts with CstF-50 to inhibit the polyadenylation machinery (Kleiman and Manley, 1999; Figure 4B). Both BARD1 and CstF-50 also interact with Pol II, and BARD1 has been proposed to sense sites of DNA damage and repair. The BARD1-mediated inhibition of polyadenylation may thus prevent inappropriate RNA processing during transcription of damaged DNA loci. Challenging cells with DNA-damaging agents has been shown to transiently inhibit 3′ end formation by enhanced formation of CstF/BARD1/BRCA1 complexes. Furthermore, a tumor-associated germline mutation in BARD1 (Gln564His) decreases its affinity for CstF-50 and renders the protein inactive in polyadenylation inhibition. These findings highlight an intriguing link between 3′ end RNA processing and DNA repair (Kleiman and Manley, 2001).
Notably, mutations of _cis_-acting determinants of 3′ end processing can also elicit unregulated cell-cycle progression and can be observed during carcinogenesis; polymorphisms in the acetyltransferase 1 gene (NAT1) affecting the poly(A) signal (or located in immediate vicinity) are believed to modulate _N_- or _O_-acetylation of carcinogens. These polymorphisms have thus been implicated to represent an important genetic determinant of colorectal cancer risk (Bell et al, 1995).
Idiopathic hypereosinophilic syndrome and hFip1
The hypereosinophilic syndrome represents a severe hematologic disorder, with sustained overproduction of eosinophils in the bone marrow, eosinophilia, tissue infiltration, and organ damage. A DNA rearrangement involving a chromosomal deletion of 800 kb and fusion of the hFip1 (FIP1L1) and PDGFRα genes might be the cause of this syndrome (Cools et al, 2003). The corresponding chimeric protein, hFip1-PDGFRα, contains the N-terminus of hFip1, an integral subunit of the CPSF complex, and the C-terminal kinase domain of PDGFRα. The expression of the hFip1-PDGFRα fusion protein in hematopoietic cells constitutively activates the PDGFRα kinase and transforms cells. As constitutive activation of tyrosine kinases is a key feature of the pathogenesis of myeloproliferative disorders, it is tempting to speculate that the N-terminal domain of hFip1 could mediate growth factor-independent dimerization and thus result in the activation of the fusion protein. Alternatively, the N-terminal domain of hFip1 could directly stimulate the kinase domain that is the therapeutic target for the highly specific kinase inhibitor imatinib.
Furthermore, the deletion that creates the hFip1-PDGFRα fusion gene also causes a functional depletion of hFip1 that, under normal conditions, stimulates 3′ end processing (Kaufmann et al, 2004). Therefore, loss of hFip activity and an interference with 3′ end processing may contribute to or even alternatively explain the observed phenotype.
Ocular muscle dystrophy and the nuclear poly(A)-binding protein
The medical relevance of mutations affecting 3′ end processing factors is further highlighted by oculopharyngeal muscular dystrophy (OPMD). This disorder is an adult-onset disease with slowly progressive muscle weakness chiefly affecting the eyelids, resulting in ptosis, and the pharyngeal muscles resulting in dysphagia. OPMD is usually inherited as an autosomally dominant trait, but a more rare allelic autosomal recessive form also exists.
Both forms are typically caused by short trinucleotide repeat [(GCG)8–13] expansions in the coding region of the nuclear poly(A)-binding protein 1 (PABPN1, PABP2; Brais et al, 1998). Normally, the polyalanine stretch encoded by this trinucleotide comprises 10 alanines, which is expanded to 12–17 alanines in autosomal-dominant OPMD. This expansion results in an increase of self-association, misfolding and filamentous nuclear aggregation of the PABPN1 protein in skeletal muscle. It is unknown why only muscle is affected phenotypically despite the ubiquitous expression of PAPBN1. In vitro, the mutant protein is fully active and OPMD cells do not display a severe polyadenylation defect (Calado et al, 2000; Kuhn and Wahle, 2004). It is likely therefore that the subtle clinical phenotype is caused by a quantitatively minor disturbance of the protein's function in polyadenylation, which may be difficult to detect in vitro or in transfected cells. Alternatively, the polyalanine stretch may increase the binding affinity to other proteins, which could consequently be inactivated by co-sequestration. Finally, PABPN1 has also been suggested to play a role in the transcription of muscle-specific genes, which could help to explain why other tissues are unaffected (Kim et al, 2001).
First come, first served: influenza A and its connection to the cellular 3′ end apparatus
Influenza A virus infection provides another interesting link between the disturbance of protein interactions within the polyadenylation machinery and human disease. The influenza A NS1 protein interacts with the cellular 30-kDa subunit of CPSF, which prevents binding of the entire CPSF complex to the RNA substrate (Nemeroff et al, 1998; Figure 4C). Efficient polyadenylation depends on the interaction of CPSF 30 with PABPN1, which together stimulate PAP from a distributive to a processive mode of action. Importantly, the influenza a virus protein NS1 represents one of the most abundant proteins synthesized in infected cells (Lazarowitz et al, 1971), and regulates several post-transcriptional processing steps (Fortes et al, 1994; Lu et al, 1994). In influenza virus-infected cells, the highly abundant NS1 protein sequesters CPSF 30 and inhibits 3′ end cleavage and polyadenylation of the host pre-mRNAs. Interestingly, the NS1 protein also targets PABPN1, which inhibits the processive synthesis of long poly(A) tails catalyzed by PAP (Chen et al, 1999). As mRNA processing represents a prerequisite for cytoplasmic export (Stutz and Rosbash, 1998), the uncleaved host pre-mRNAs are retained in the nucleus. In contrast, viral RNAs are still exported. Thus, by interfering with the activity of an essential 3′ end processing factor, influenza has devised an efficient way to specifically shut off cellular gene expression and to facilitate viral gene expression that does not depend on the cellular 3′ end processing apparatus (Nemeroff et al, 1998; Chen et al, 1999).
Similar mechanisms have been identified to contribute to a preferential viral gene expression in HSV-1-infected cells through ICP27 (a multifunctional regulator of HSV gene expression, also known as IE63) that functions post-transcriptionally to activate weaker viral poly(A) sites and to inhibit splicing of host cell mRNAs (McLauchlan et al, 1989, 1992; Sandri-Goldin and Mendoza, 1992).
Perspective
The 3′ UTR has clearly emerged as a hot spot for post-transcriptional gene regulation, controlling important cellular functions such as, for example, morphogenesis, cell differentiation, metabolism, cell proliferation and apoptosis, by controlling mRNA translation, stability, localization as well as 3′ end processing (Ambros, 2004; Calin et al, 2005; Croce and Calin, 2005; He et al, 2005; Lu et al, 2005; O'Donnell et al, 2005). More recently, sequence determinants and 3′ end-processing factors have emerged as common targets of mutations resulting in human pathology (see Tables I and II) (Chen et al, 2006a, 2006b).
Table 1.
_Cis_-acting RNA elements involved in 3′ end processing and their medical relevance
| Signal | Physiological function | Medical relevance |
|---|---|---|
| Poly(A) signal (AAUAAA, and less frequently AUUAAA) | Highly conserved _cis_-acting hexanucleotide, essential for 3′ end formation, localized in the 3′ UTR and recognized by CPSF through the CPSF-160 subunit, determines the site of cleavage and polyadenylation 10–30 nt downstream | Loss-of-function • Alpha-thalassemias (Higgs et al, 1983; Harteveld et al, 1994) • Beta-thalassemias (Orkin et al, 1985; Jankovic et al, 1990; Rund et al, 1992; van Solinge et al, 1996) • IPEX syndrome (Bennett et al, 2001) • Metachromatic leukodystrophies (Gieselmann et al, 1989) • Fabry disease (Yasuda et al, 2003) • Acetyltransferase 1 polymorphism in colorectal cancer (Bell et al, 1995) • Insulin-like growth factor 1 deficiency (Bonapace et al, 2003) • X-linked severe combined immunodeficiency (Hsu et al, 2000) |
| Cleavage site | In most mRNAs represented by a CA-dinucleotide 10–30 nt downstream of the poly(A) signal | Gain-of-function Thrombophilia (Gehring et al, 2001; Ceelie et al, 2004; Danckwardt et al, 2004, 2006b, 2007; Kuwahara et al, 2004) |
| Downstream G/U-rich or U-rich sequence element (DSE) | Less stringently conserved sequence element, characterized by high density of G/U or U residues, essential for 3′ end formation, localized in the 3′ flanking region up to 30 nt downstream of the cleavage site, recognized by CstF through CstF-64 subunit, stimulates the cleavage reaction | Gain-of-function • Thrombophilia, prothrombin (Danckwardt et al, 2004) • Thrombophilia, fibrinogen gamma (Uitte de Willige et al, 2005, 2007) |
| Upstream sequence element (USE) | Auxiliary sequence motif within the 3′ UTR, characterized by (combinations) of highly conserved sequence elements (Legendre and Gautheret, 2003; Hu et al, 2005; Xie et al, 2005), promotes 3′ end processing by serving as an additional anchor for the (canonical) 3′ end-processing machinery or by stabilizing the RNA interaction of both CPSF and CstF components, recognized by hnRNP1/PTB (Moreira et al, 1998), U2AF35, U2AF65 (Danckwardt et al, 2007), PSF, U2A1, p54nrb (Hall-Pogar et al, 2007). Of note, p54nrb and PSF have also been shown to facilitate pre-mRNA 3′ processing and transcription termination most probably independently from USEs (Kaneko et al, 2007) | No disease-associated mutations have yet been identified; important for the physiological 3′ end processing of for example • Complement C2 (Moreira et al, 1995, 1998) • Lamin B (Brackenridge and Proudfoot, 2000) • Cyclooxygenase 2 (Hall-Pogar et al, 2005, 2007) • Collagen (Natalizio et al, 2002) • Prothrombin (Danckwardt et al, 2004, 2007) |
| Additional sequence elements (i.e., (UGUA)n) | Present in one ore more copies at a variable distance upstream of the cleavage site (Hu et al, 2005), recruits the heterodimeric cleavage factor CFIm (Brown and Gilmartin, 2003; Venkataraman et al, 2005) | Still unknown |
| Alternative poly(A) signal (recognition) | About half of the human RNAs contain (multiple) alternative poly(A) signals (Edwalds-Gilbert et al, 1997; Tian et al, 2005; Yan and Marr, 2005). Alternative recognition is influenced (1) by intrinsic strength of sequence elements, (2) by the concentration or activity of polyadenylation factors and (3) by tissue- or stage-specific regulatory factors (for review see Colgan and Manley, 1997; Barabino and Keller, 1999; Zhao et al, 1999) | • IgM heavy-chain pre-mRNA during B-cell differentiation (stage specificity) (Alt et al, 1980; Early et al, 1980; Rogers et al, 1980; Cushley et al, 1982; Takagaki et al, 1996; Phillips et al, 2001, 2004; Veraldi et al, 2001) • Regulated expression of the transcription factor NF-ATc during T-cell differentiation (stage specificity) (Chuvpilo et al, 1999) • LPS stimulation of macrophages results in alternative polyadenylation via upregulation of CstF-64 (Shell et al, 2005) • Calcitonin/calcitonin-related peptide (tissue specificity) (Lou and Gagel, 1998) • Cyclooxygenase 2 (Feng et al, 1993; Ristimaki et al, 1996; Lukiw and Bazan, 1997; Hall-Pogar et al, 2005) • Pancreatitis-associated protein II/regenerating gene III (Honda et al, 2002) • Fanconi anemia group c (FACC) (Strathdee et al, 1992) • Modulation of polyadenylation at weak viral polyadenylation sites in HSV-1-infected cells (McLauchlan et al, 1989, 1992; Sandri-Goldin and Mendoza, 1992; McGregor et al, 1996) • Synthesis of the protein elastin in sun-damaged skin (Schwartz et al, 1998) • NADPH quinone oxidoreductase (Pan et al, 1995) • Set, putative oncogene associated with myeloid leukogenesis (von Lindern et al, 1992) • For further detailed and comprehensive survey of mRNAs that undergo alternative poly(A) selection see (Edwalds-Gilbert et al, 1997) |
| CPSF, cleavage and polyadenylation specificity factor; CstF, cleavage-stimulating factor; DSE, downstream sequence element; HSV-1, herpes simplex virus-1; LPS, lipopolysaccharide; NADPH, nicotinamide adenine dinucleotide phosphate dehydrogenase; PAP, poly(A) polymerase; UTR, untranslated region. |
Table 2.
Essential and auxiliary _trans_-acting factors involved in poly(A) mRNA 3′ end processing, and their impact on health and disease
| Factor | Physiological function | Medical relevance |
|---|---|---|
| CPSF (cleavage and polyadenylation specificity factor) | Five subunits (CPSF 30, 73, 100, 160 and hFip1) plays a key role in pre-mRNA 3′ end formation, involved in the RNA recognition step, and determine the site of cleavage 10–30 nt downstream CPSF 160 subunit recognizes the AAUAAA signal sequence, interacts with CstF-77, PAP and RNA Pol II-CTD (Murthy and Manley, 1995) CPSF 73 subunit represents the 3′-processing endonuclease (Ryan et al, 2004; Dominski et al, 2005; Mandel et al, 2006) | • CPSF 30 represents the cellular target of the influenza A virus NS1 protein. In influenza virus-infected cells, this prevents CPSF binding to the RNA substrate and inhibits 3′ end cleavage and polyadenylation of host pre-mRNAs. Thus, the NS1 protein selectively inhibits the nuclear export of cellular (host) mRNAs (Nemeroff et al, 1998) • CPSF-73 specifically induced by HIV-1-Tat protein may represent a mechanism by which this virus increases mRNA processing, causing an increase in both cellular and viral gene expression (Calzado et al, 2004) |
| CstF (cleavage-stimulating factor) | Three subunits (CstF-50, -64, -77), stimulates 3′ end cleavage and promotes the efficiency of 3′ end processing 64-kDa subunit (CstF-64) recognizes the downstream U or G/U-rich element (DSE) of the RNA 3′ flanking sequence CstF-64 represents target for many regulatory mechanisms of 3′ end processing (McLauchlan et al, 1989, 1992; Sandri-Goldin and Mendoza, 1992; Takagaki et al, 1996; Takagaki and Manley, 1998; Kleiman and Manley, 1999, 2001; Veraldi et al, 2001; Phillips et al, 2004; Shell et al, 2005; Gawande et al, 2006) CstF-64 increases fivefold during the G0 to S phase transition and results in a functionally relevant increment of the CstF trimer (Martincic et al, 1998), which may account for the accumulation of poly(A)-plus mRNA when cells leave G0 and enter S (Getz et al, 1976) | • CstF-50–BARD1 interaction links mRNA 3 end formation to DNA damage and tumor suppression (Kleiman and Manley, 1999, 2001) • CstF-64 reduction causes cell-cycle arrest in G0/G1 phase, depletion results in apoptotic cell death (Takagaki and Manley, 1998) • CstF-64 regulates alternative processing of IgM heavy-chain pre-mRNA during B-cell differentiation. Upregulation is proposed to result in a switch of IgM heavy-chain mRNA from a membrane-bound form to the secreted form (Takagaki et al, 1996). Furthermore, the CstF-64–RNA interaction has been shown to be sterically inhibited by U1A (Phillips et al, 2004) and/or hnRNP F (Veraldi et al, 2001). Antagonistic effects with CstF-64 may thus represent an alternative explanation for the regulation of alternative processing of IgM heavy-chain pre-mRNA during B-cell differentiation without changing CstF-64 levels (Martincic et al, 1998) • CstF-64 confers increased polyadenylation at weak viral polyadenylation sites in HSV-1-infected cells (McLauchlan et al, 1989, 1992; Sandri-Goldin and Mendoza, 1992) • CstF-64 expression upregulated in LPS-stimulated macrophages results in alternative polyadenylation (Shell et al, 2005) CstF-64–RNA interaction modulated by competitive binding of sex-lethal protein to confer sex-specific poly(A) site switching (in Drosophila) (Gawande et al, 2006) |
| PAP (poly(A) polymerase) | Polymerase that creates the 3′ poly(A) tail of mRNAs, also required for the endoribonucleolytic cleavage reaction at some polyadenylation sites, exhibits both nonspecific and CPSF/AAUAAA-dependent polyadenylation activity PAP activity regulated in a cell cycle-dependent manner (Ballantyne et al, 1995; Colgan et al, 1996) U1A inhibits PAP activity on various substrates (i.e., of the U1A mRNA; Gunderson et al, 1994, and of the IgM secretory mRNA; Phillips et al, 2001) U1 snRNP inhibits poly(A) sites and PAP (Furth and Baker, 1991; Gunderson et al, 1998) | Loss-of-function • Cell arrest in G0–G1 phase (Zhao and Manley, 1996) • Overexpression of U1A in vivo results in the selective inhibition of the IgM secretory mRNA (Phillips et al, 2001) Gain-of-function • Confers high proliferative activity • Overexpression in human carcinomas (Pendurthi et al, 1997; Scorilas et al, 2000) • Hematological malignancies (Trangas et al, 1984; Pangalis et al, 1985; Sasaki et al, 1990) • Neo-PAP overexpression in tumors (Topalian et al, 2001) • Increased seizure susceptibility (Kioka et al, 1997) Autoantibodies against PAP • In patients with tumors (Stetler et al, 1981) and • Rheumatic autoimmune disorders (Stetler et al, 1987) |
| PABPC (PABP1) | Binds the poly(A)-tail in the cytoplasm and interacts with eIF4G to stimulate transcription, interacts with deadenylases and controls RNA degradation | Still unknown |
| PABPN1 (=PABP2) | Interacts with CPSF and confers processivity of PAP to control the size of the poly(A) tail to about 250 nt, binds to poly(A)-tail in the nucleus, is replaced by PABPC during nuclear export | Loss-of-function • Oculopharyngeal muscular dystrophy (Brais et al, 1998) • In influenza A virus-infected cells the viral NS1A protein targets PABN1, which inhibits the processive synthesis of long poly(A) tails on cellular RNAs catalyzed by the poly(A) polymerase (PAP) (Chen et al, 1999) |
| hFip1 | Integral subunit of CPSF, contributes to the CPSF-mediated stimulation of PAP (Kaufmann et al, 2004) | Loss-of-function • Observed in the Idiopathic hypereosinophilic syndrome (Cools et al, 2003) |
| CF Im (cleavage factor I) | Consists of two subunits (25, 68 kDa) interaction with the RNA is one of the earliest steps in the assembly of the 3′ end-processing complex and facilitates the recruitment of other processing factors, required for 3′ RNA cleavage and polyadenylation processing | Still unknown |
| CF IIm (cleavage factor II) | Still unknown | Still unknown |
| RNA Pol II (polymerase II) | Required for transcription, transfers CPSF and CstF to their specific signals, phosphorylation of serine residues within the CTD regulates gene expression (Hirose and Manley, 1998; Hampsey and Reinberg, 2003) | Still unknown |
| CTD, C-terminal domain; DSE, downstream sequence element; HIV-1, human immunodeficiency virus-1; HSV-1, herpes simplex virus-1; LPS, lipopolysaccharide; PAP, poly(A) polymerase. |
With the ongoing identification of novel sequence elements that impact on 3′ end processing (Moreira et al, 1995, 1998; Brackenridge and Proudfoot, 2000; Natalizio et al, 2002; Danckwardt et al, 2004, 2007; Hall-Pogar et al, 2005, 2007), additional mutations located in these regions or in close proximity to the poly(A) signal altering 3′ end processing are likely to be found. Possibly, some of these may account for a number of polymorphisms linked to disease, which have not yet been functionally characterized. Such polymorphisms represent risk factors for cancer (Zheng et al, 2004; Zhang et al, 2004a, 2004b) that strikingly influence their treatment modalities (Cox et al, 2004; Stoehlmacher et al, 2004). Other 3′ UTR polymorphisms have been associated with the risk to develop type II diabetes (Shin et al, 2003), insulin resistance (Pizzuti et al, 2002), coronary artery disease (Chen et al, 2003), Alzheimer disease (Lambert et al, 2003), end-stage renal disease (Bensen et al, 2003), the Fukuyama-type congenital muscular dystrophy (Kobayashi et al, 1998), and a variety of clinically relevant polymorphisms known to affect the immunoregulation (Hennig et al, 2002; Kuroki et al, 2002; Seegers et al, 2002), although their molecular mode of action is still unknown.
While some of the established mutations affect the efficiency of constitutive 3′ end mRNA processing, others could conceivably affect the regulation of this critical step. Regulated 3′ end mRNA processing efficiency and regulated alternative 3′ end mRNA processing are becoming increasingly topical both in biology and in medicine, because both aspects of regulation add to the functional complexity of the transcriptome and are already known to be altered in human disease.
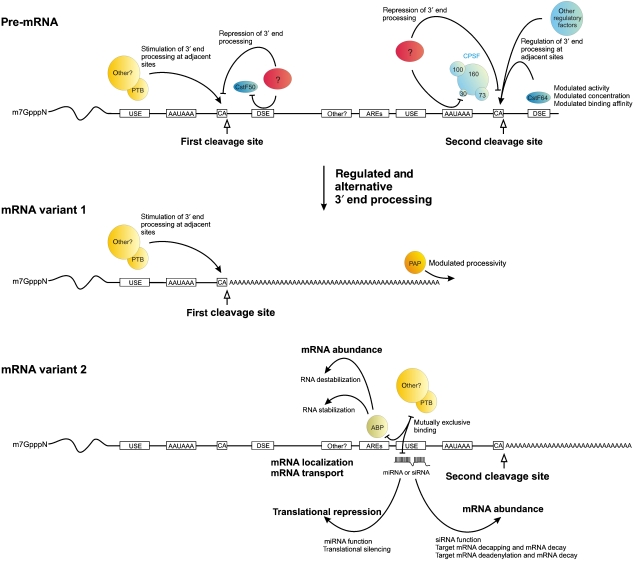
Such mechanisms of regulated 3′ end mRNA processing may include the recruitment of critical proteins to the 3′ end-processing machinery in response to specific stimuli, as has previously been proposed for the control of splicing (Shin and Manley, 2004). Furthermore, specific sequence determinants of 3′ end processing may serve as target sites for competitive binding of mutually exclusive protein complexes or miRNAs, thus enabling a complex layer of post-transcriptional regulation (Figure 5). Similar mechanisms have recently been shown to account for SXL-dependent poly(A) site choice in the Drosophila female germline (Gawande et al, 2006), or to regulate 3′ end-processing efficiencies in different tissues (Zhu et al, 2006, 2007). Finally, the activation of alternative, distal polyadenylation sites may result in the inclusion of additional mRNA sequence motifs in a transcript. Such sequence motifs may be recognized by proteins that regulate the stability of the mRNA (Wilson and Treisman, 1988; Shyu and Wilkinson, 2000; Barreau et al, 2005; Gilat and Shweiki, 2007). Alternatively, downstream activation of alternative polyadenylation sites may modulate the accessibility for miRNAs (Stark et al, 2005), thus resulting in an additional layer of regulation of mRNA degradation and translation (Cohen and Brennecke, 2006; Giraldez et al, 2006; Legendre et al, 2006).
Figure 5.
Regulated and alternative 3′ end processing modulates the temporal and spatial diversity of gene expression. About half of the human pre-mRNAs contain (multiple) alternative poly(A) signals. A large number of these pre-mRNAs has alternative, mostly tandem, arrays of poly(A) sites within the 3′ UTR. A smaller set of pre-mRNAs bears alternative poly(A) signals within intronic or exonic regions. In both the cases, endogenous and exogenous factors can modulate pre-mRNA poly(A) site selection by interfering with constitutive and/or auxiliary 3′ end-processing factors/subunits (upper panel; depicted are various scenarios on a single pre-mRNA). This results in various polyadenylated mRNAs that either code for identical (tandem terminal poly(A) sites) or C-terminally modified (internal poly(A) sites) proteins (see also Figure 4A). Furthermore, alternatively 3′ end-processed mRNAs can display different 3′ UTR properties (middle and lower panel; mRNA variants 1 and 2, respectively). This diversity can affect mRNA abundance, mRNA localization, mRNA transport and translation of the respective mRNA variant (lower lane). Importantly, interaction of AU-rich elements (AREs) with the respective binding proteins (ABP) has a dual function and can result in both mRNA stabilization and destabilization. Mutually exclusive binding of different _trans_-acting factors allows a complex modulation of different processing activities. Auxiliary 3′ end-processing sequences (USE) may represent target sites for _trans_-acting factors that modulate 3′ end formation efficiencies.
From a medical perspective, it will be interesting to explore polymorphisms of components of the 3′ end-processing machinery as biomarkers that influence therapeutic decisions. This is already exemplified by the introduction of the common prothrombin 20 210 mutation into clinical algorithms for secondary thrombosis prophylaxis (Hirsh and Lee, 2002). Even more interesting, a better understanding of the molecular pathology of specific disorders may lead to approaches to correct deregulated RNA processing events. This is exemplified in OPMD (see above). In animal models of this form of muscular dystrophy, the detrimental impact of aggregates of the nuclear poly(A)-binding protein in skeletal muscle could be reduced by chaperone-guided approaches or by doxycyclin treatment (Davies et al, 2006). Finally, the emerging knowledge about USEs stimulating mRNA 3′ end formation (Moreira et al, 1995, 1998; Brackenridge and Proudfoot, 2000; Natalizio et al, 2002; Danckwardt et al, 2004, 2007; Hall-Pogar et al, 2005, 2007) may have useful implications for the development of safe retroviral gene therapy strategies (Valsamakis et al, 1991; Zaiss et al, 2002). USEs improve the efficiency of the poly(A) signal (Gilmartin et al, 1995), increase transgene expression efficiency and decrease the risk of downstream activation of potentially harmful cellular genes by reducing readthrough transcription (Schambach et al, 2007).
In sum, the mechanisms of 3′ end mRNA processing and of its regulation are highly relevant both in biology and in medicine, and deserve to be met with an increasing level of awareness.
Acknowledgments
We thank the members of the Molecular Medicine Partnership Unit for discussions. We apologize for the limited citation of the primary literature outside of the main focus of this review. This work was supported by the Fritz Thyssen Stiftung, the Deutsche Forschungsgemeinschaft and by the ‘Young Investigator Award' fellowship from the University of Heidelberg (to SD).
References
- Adamson TE, Price DH (2003) Cotranscriptional processing of Drosophila histone mRNAs. Mol Cell Biol 23: 4046–4055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson TE, Shutt DC, Price DH (2005) Functional coupling of cleavage and polyadenylation with transcription of mRNA. J Biol Chem 280: 32262–32271 [DOI] [PubMed] [Google Scholar]
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Alt FW, Bothwell ALM, Knapp M, Siden E, Mather E, Koshland M, Baltimore D (1980) Synthesis of secreted and membrane-bound immunoglobulin mu heavy chains is directed by mRNAs that differ at their 3-prime ends. Cell 20: 293–301 [DOI] [PubMed] [Google Scholar]
- Ambros V (2004) The functions of animal microRNAs. Nature 431: 350–355 [DOI] [PubMed] [Google Scholar]
- Awasthi S, Alwine JC (2003) Association of polyadenylation cleavage factor I with U1 snRNP. RNA 9: 1400–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balim Z, Kosova B, Falzon K, Bezzina Wettinger S, Colak Y (2003) Budd–Chiari syndrome in a patient heterozygous for the point mutation C20221T of the prothrombin gene. J Thromb Haemost 1: 852–853 [DOI] [PubMed] [Google Scholar]
- Ballantyne S, Bilger A, Astrom J, Virtanen A, Wickens M (1995) Poly(A) polymerases in the nucleus and cytoplasm of frog oocytes: dynamic changes during oocyte maturation and early development. RNA 1: 64–78 [PMC free article] [PubMed] [Google Scholar]
- Barabino SM, Keller W (1999) Last but not least: regulated poly(A) tail formation. Cell 99: 9–11 [DOI] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB (2005) AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth ML, Fensom A, Harris A (1993) Prevalence of common mutations in the arylsulphatase A gene in metachromatic leukodystrophy patients diagnosed in Britain. Hum Genet 91: 73–77 [DOI] [PubMed] [Google Scholar]
- Bell DA, Stephens EA, Castranio T, Umbach DM, Watson M, Deakin M, Elder J, Hendrickse C, Duncan H, Strange RC (1995) Polyadenylation polymorphism in the acetyltransferase 1 gene (NAT1) increases risk of colorectal cancer. Cancer Res 55: 3537–3542 [PubMed] [Google Scholar]
- Bennett CL, Brunkow ME, Ramsdell F, O'Briant KC, Zhu Q, Fuleihan RL, Shigeoka AO, Ochs HD, Chance PF (2001) A rare polyadenylation signal mutation of the FOXP3 gene (AAUAAA → AAUGAA) leads to the IPEX syndrome. Immunogenetics 53: 435–439 [DOI] [PubMed] [Google Scholar]
- Bensen JT, Langefeld CD, Li L, Mccall CE, Cousart SL, Dryman BN, Freedman BI, Bowden DW (2003) Association of an IL-1A 3′UTR polymorphism with end-stage renal disease and IL-1 alpha expression. Kidney Int 63: 1211–1219 [DOI] [PubMed] [Google Scholar]
- Bishop DF, Kornreich R, Desnick RJ (1988) Structural organization of the human alpha -galactosidase A gene: further evidence for the absence of a 3′ untranslated region. Proc Natl Acad Sci USA 85: 3903–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL (2000) Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103: 367–370 [DOI] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Bonapace G, Concolino D, Formicola S, Strisciuglio P (2003) A novel mutation in a patient with insulin-like growth factor 1 (IGF1) deficiency. J Med Genet 40: 913–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brackenridge S, Proudfoot NJ (2000) Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol 20: 2660–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brais B, Bouchard JP, Xie YG, Rochefort DL, Chretien N, Tome FM, Lafreniere RG, Rommens JM, Uyama E, Nohira O, Blumen S, Korczyn AD, Heutink P, Mathieu J, Duranceau A, Codere F, Fardeau M, Rouleau GA, Korcyn AD (1998) Short GCG expansions in the PABP2 gene cause oculopharyngeal muscular dystrophy. Nat Genet 18: 164–167 [DOI] [PubMed] [Google Scholar]
- Brown KM, Gilmartin GM (2003) A mechanism for the regulation of pre-mRNA 3′ processing by human cleavage factor Im. Mol Cell 12: 1467–1476 [DOI] [PubMed] [Google Scholar]
- Buratowski S (2005) Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17: 257–261 [DOI] [PubMed] [Google Scholar]
- Calado A, Tome FM, Brais B, Rouleau GA, Kuhn U, Wahle E, Carmo-Fonseca M (2000) Nuclear inclusions in oculopharyngeal muscular dystrophy consist of poly(A) binding protein 2 aggregates which sequester poly(A) RNA. Hum Mol Genet 9: 2321–2328 [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG et al. (2005) A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353: 1793–1801 [DOI] [PubMed] [Google Scholar]
- Calzado MA, Sancho R, Munoz E (2004) Human immunodeficiency virus type 1 Tat increases the expression of cleavage and polyadenylation specificity factor 73-kilodalton subunit modulating cellular and viral expression. J Virol 78: 6846–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceelie H, Spaargaren-van Riel CC, Bertina RM, Vos HL (2004) G20210A is a functional mutation in the prothrombin gene; effect on protein levels and 3′-end formation. J Thromb Haemost 2: 119–127 [DOI] [PubMed] [Google Scholar]
- Chen F, MacDonald CC, Wilusz J (1995) Cleavage site determinants in the mammalian polyadenylation signal. Nucleic Acids Res 23: 2614–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN (2006a) A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes I: general principles and overview. Hum Genet 120: 1–21 [DOI] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN (2006b) A systematic analysis of disease-associated variants in the 3′ regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3′ UTR variants. Hum Genet 120: 301–333 [DOI] [PubMed] [Google Scholar]
- Chen Q, Reis SE, Kammerer C, Craig WY, LaPierre SE, Zimmer EL, McNamara DM, Pauly DF, Sharaf B, Holubkov R, Bairey Merz CN, Sopko G, Bontempo F, Kamboh MI (2003) Genetic variation in lectin-like oxidized low-density lipoprotein receptor 1 (LOX1) gene and the risk of coronary artery disease. Circulation 107: 3146–3151 [DOI] [PubMed] [Google Scholar]
- Chen Z, Li Y, Krug RM (1999) Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J 18: 2273–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuvpilo S, Zimmer M, Kerstan A, Glockner J, Avots A, Escher C, Fischer C, Inashkina I, Jankevics E, Berberich-Siebelt F, Schmitt E, Serfling E (1999) Alternative polyadenylation events contribute to the induction of NF-ATc in effector T cells. Immunity 10: 261–269 [DOI] [PubMed] [Google Scholar]
- Cohen SM, Brennecke J (2006) Developmental biology. Mixed messages in early development. Science 312: 65–66 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Prives C, Manley JL (1996) Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature 384: 282–285 [DOI] [PubMed] [Google Scholar]
- Colgan DF, Murthy KG, Zhao W, Prives C, Manley JL (1998) Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J 17: 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot HV, Stutz F, Rosbash M (1996) The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev 10: 1699–1708 [DOI] [PubMed] [Google Scholar]
- Cools J, DeAngelo DJ, Gotlib J, Stover EH, Legare RD, Cortes J, Kutok J, Clark J, Galinsky I, Griffin JD, Cross NCP, Tefferi A, Malone J, Alam R, Schrier SL, Schmid J, Rose M, Vandenberghe P, Verhoef G, Boogaerts M et al. (2003) A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 348: 1201–1214 [DOI] [PubMed] [Google Scholar]
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G (2002) Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 22: 3301–3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DG, Pontes C, Guino E, Navarro M, Osorio A, Canzian F, Moreno V (2004) Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer 91: 339–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM, Calin GA (2005) miRNAs, cancer, and stem cell division. Cell 122: 6–7 [DOI] [PubMed] [Google Scholar]
- Cushley W, Coupar BE, Mickelson CA, Williamson AR (1982) A common mechanism for the synthesis of membrane and secreted immunoglobulin alpha, gamma and mu chains. Nature 298: 77–79 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Gehring NH, Neu-Yilik G, Hundsdoerfer P, Pforsich M, Frede U, Hentze MW, Kulozik AE (2004) The prothrombin 3′end formation signal reveals a unique architecture that is sensitive to thrombophilic gain-of-function mutations. Blood 104: 428–435 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hartmann K, Gehring NH, Hentze MW, Kulozik AE (2006a) 3′ end processing of the prothrombin mRNA in thrombophilia. Acta Haematol 115: 192–197 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Hartmann K, Katz B, Hentze M, Levy Y, Eichele R, Deutsch V, Kulozik A, Ben-Tal O (2006b) The prothrombin 20209 C>T mutation in Jewish–Moroccan Caucasians: molecular analysis of gain-of-function of 3′end processing. J Thromb Haemost 4: 1078–1085 [DOI] [PubMed] [Google Scholar]
- Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE (2007) Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J 26: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JE, Sarkar S, Rubinsztein DC (2006) Trehalose reduces aggregate formation and delays pathology in a transgenic mouse model of oculopharyngeal muscular dystrophy. Hum Mol Genet 15: 23–31 [DOI] [PubMed] [Google Scholar]
- Dominski Z, Yang XC, Marzluff WF (2005) The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123: 37–48 [DOI] [PubMed] [Google Scholar]
- Early P, Rogers J, Davis M, Calame K, Bond M, Wall R, Hood L (1980) Two mRNAs can be produced from a single immunoglobulin mu gene by alternative RNA processing pathways. Cell 20: 313–319 [DOI] [PubMed] [Google Scholar]
- Edmonds M (2002) A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol 71: 285–389 [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res 25: 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Sun W, Xia Y, Tang WW, Chanmugam P, Soyoola E, Wilson CB, Hwang D (1993) Cloning two isoforms of rat cyclooxygenase: differential regulation of their expression. Arch Biochem Biophys 307: 361–368 [DOI] [PubMed] [Google Scholar]
- Fortes P, Beloso A, Ortin J (1994) Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J 13: 704–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furth PA, Baker CC (1991) An element in the bovine papillomavirus late 3′ untranslated region reduces polyadenylated cytoplasmic RNA levels. J Virol 65: 5806–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawande B, Robida MD, Rahn A, Singh R (2006) Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J 25: 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Frede U, Neu-Yilik G, Hundsdoerfer P, Vetter B, Hentze MW, Kulozik AE (2001) Increased efficiency of mRNA 3′ end formation: a new genetic mechanism contributing to hereditary thrombophilia. Nat Genet 28: 389–392 [DOI] [PubMed] [Google Scholar]
- Getz MJ, Elder PK, Benz EW Jr, Stephens RE, Moses HL (1976) Effect of cell proliferation on levels and diversity of poly(A)-containing mRNA. Cell 7: 255–265 [DOI] [PubMed] [Google Scholar]
- Gieselmann V, Polten A, Kreysing J, von Figura K (1989) Arylsulfatase A pseudodeficiency: loss of a polyadenylylation signal and _N_-glycosylation site. Proc Natl Acad Sci USA 86: 9436–9440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilat R, Shweiki D (2007) A novel function for alternative polyadenylation as a rescue pathway from NMD surveillance. Biochem Biophys Res Commun 353: 487–492 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM (2005) Eukaryotic mRNA 3′ processing: a common means to different ends. Genes Dev 19: 2517–2521 [DOI] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J, Graveley BR (1995) CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev 9: 72–83 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF (2006) Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Graveley BR (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107 [DOI] [PubMed] [Google Scholar]
- Graveley BR, Fleming ES, Gilmartin GM (1996) RNA structure is a critical determinant of poly(A) site recognition by cleavage and polyadenylation specificity factor. Mol Cell Biol 16: 4942–4951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunderson SI, Beyer K, Martin G, Keller W, Boelens WC, Mattaj LW (1994) The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell 76: 531–541 [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Polycarpou-Schwarz M, Mattaj IW (1998) U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell 1: 255–264 [DOI] [PubMed] [Google Scholar]
- Gunderson SI, Vagner S, Polycarpou-Schwarz M, Mattaj IW (1997) Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev 11: 761–773 [DOI] [PubMed] [Google Scholar]
- Hall-Pogar T, Liang S, Hague LK, Lutz CS (2007) Specific _trans_-acting proteins interact with auxiliary RNA polyadenylation elements in the COX-2 3′-UTR. RNA 13: 1103–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Pogar T, Zhang H, Tian B, Lutz CS (2005) Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res 33: 2565–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M, Reinberg D (2003) Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113: 429–432 [DOI] [PubMed] [Google Scholar]
- Harteveld CL, Losekoot M, Haak H, Heister GA, Giordano PC, Bernini LF (1994) A novel polyadenylation signal mutation in the alpha 2-globin gene causing alpha thalassaemia. Br J Haematol 87: 139–143 [DOI] [PubMed] [Google Scholar]
- Harvey J, Carey W, Morris C (1998) Importance of the glycosylation and polyadenylation variants in metachromatic leukodystrophy pseudodeficiency phenotype. Hum Mol Genet 7: 1215–1219 [DOI] [PubMed] [Google Scholar]
- He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM (2005) A microRNA polycistron as a potential human oncogene. Nature 435: 828–833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig BJ, Hellier S, Frodsham AJ, Zhang L, Klenerman P, Knapp S, Wright M, Thomas HC, Thursz M, Hill AV (2002) Association of low-density lipoprotein receptor polymorphisms and outcome of hepatitis C infection. Genes Immun 3: 359–367 [DOI] [PubMed] [Google Scholar]
- Hentze MW, Kulozik AE (1999) A perfect message: RNA surveillance and nonsense-mediated decay. Cell 96: 307–310 [DOI] [PubMed] [Google Scholar]
- Higgs DR, Goodbourn SE, Lamb J, Clegg JB, Weatherall DJ, Proudfoot NJ (1983) Alpha-thalassaemia caused by a polyadenylation signal mutation. Nature 306: 398–400 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hirsh J, Lee AY (2002) How we diagnose and treat deep vein thrombosis. Blood 99: 3102–3110 [DOI] [PubMed] [Google Scholar]
- Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE (2004) Nonsense-mediated decay approaches the clinic. Nat Genet 36: 801–808 [DOI] [PubMed] [Google Scholar]
- Honda H, Nakamura H, Otsuki M (2002) The elongated PAP II/Reg III mRNA is upregulated in rat pancreas during acute experimental pancreatitis. Pancreas 25: 192–197 [DOI] [PubMed] [Google Scholar]
- Hsu AP, Tsai EJ, Anderson SM, Fischer RE, Malech H, Buckley RH, Puck JM (2000) Unusual X-linked SCID phenotype due to mutation of the poly-A addition signal of IL2RG (abstract 206). Am J Hum Genet 67: A206 [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B (2005) Bioinformatic identification of candidate _cis_-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob ST, Terns MP, Maguire KA (1989) Polyadenylate polymerases from normal and cancer cells and their potential role in messenger RNA processing: a review. Cancer Res 49: 2827–2833 [PubMed] [Google Scholar]
- Jankovic L, Efremov GD, Petkov G, Kattamis C, George E, Yang KG, Stoming TA, Huisman TH (1990) Two novel polyadenylation mutations leading to beta(+)-thalassemia. Br J Haematol 75: 122–126 [DOI] [PubMed] [Google Scholar]
- Kahvejian A, Roy G, Sonenberg N (2001) The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 66: 293–300 [DOI] [PubMed] [Google Scholar]
- Kaneko S, Rozenblatt-Rosen O, Meyerson M, Manley JL (2007) The multifunctional protein p54nrb/PSF recruits the exonuclease XRN2 to facilitate pre-mRNA 3′ processing and transcription termination. Genes Dev 21: 1779–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kaufmann I, Martin G, Friedlein A, Langen H, Keller W (2004) Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J 23: 616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W, Minvielle-Sebastia L (1997) A comparison of mammalian and yeast pre-mRNA 3′-end processing. Curr Opin Cell Biol 9: 329–336 [DOI] [PubMed] [Google Scholar]
- Kim YJ, Noguchi S, Hayashi YK, Tsukahara T, Shimizu T, Arahata K (2001) The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum Mol Genet 10: 1129–1139 [DOI] [PubMed] [Google Scholar]
- Kioka T, Yamagami S, Mui K, Onishi H (1997) Nuclear polyadenylate polymerase activity in the brain of seizure-prone EL mice. Psychiatry Clin Neurosci 51: 151–155 [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL (1999) Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285: 1576–1579 [DOI] [PubMed] [Google Scholar]
- Kleiman FE, Manley JL (2001) The BARD1–CstF-50 interaction links mRNA 3′ end formation to DNA damage and tumor suppression. Cell 104: 743–753 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Nakahori Y, Miyake M, Matsumura K, Kondo-Iida E, Nomura Y, Segawa M, Yoshioka M, Saito K, Osawa M, Hamano K, Sakakihara Y, Nonaka I, Nakagome Y, Kanazawa I, Nakamura Y, Tokunaga K, Toda T (1998) An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature 394: 388–392 [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA (2005) Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs 10.1101/gad.1371105. Genes Dev 19: 2583–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn U, Wahle E (2004) Structure and function of poly(A) binding proteins. Biochim Biophys Acta 1678: 67–84 [DOI] [PubMed] [Google Scholar]
- Kuroki K, Tsuchiya N, Tsao BP, Grossman JM, Fukazawa T, Hagiwara K, Kano H, Takazoe M, Iwata T, Hashimoto H, Tokunaga K (2002) Polymorphisms of human CD19 gene: possible association with susceptibility to systemic lupus erythematosus in Japanese. Genes Immun 3 (Suppl 1): S21–S30 [DOI] [PubMed] [Google Scholar]
- Kuwahara M, Kurachi S, Kurachi K (2004) Molecular mechanism of prothrombin G20210A variant: critical new role of exon splicing enhancer in poly (A) tailing. Blood 104: 1944 (abstract) [Google Scholar]
- Kyburz A, Friedlein A, Langen H, Keller W (2006) Direct interactions between subunits of CPSF and the U2 snRNP contribute to the coupling of pre-mRNA 3′ end processing and splicing. Molecular Cell 23: 195–205 [DOI] [PubMed] [Google Scholar]
- Lambert J-C, Luedecking-Zimmer E, Merrot S, Hayes A, Thaker U, Desai P, Houzet A, Hermant X, Cottel D, Pritchard A, Iwatsubo T, Pasquier F, Frigard B, Conneally PM, Chartier-Harlin M-C, DeKosky ST, Lendon C, Mann D, Kamboh MI, Amouyel P (2003) Association of 3′-UTR polymorphisms of the oxidised LDL receptor 1 (OLR1) gene with Alzheimer's disease. J Med Genet 40: 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Compans RW, Choppin PW (1971) Influenza virus structural and nonstructural proteins in infected cells and their plasma membranes. Virology 46: 830–843 [DOI] [PubMed] [Google Scholar]
- Legendre M, Gautheret D (2003) Sequence determinants in human polyadenylation site selection. BMC Genomics 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre M, Ritchie W, Lopez F, Gautheret D (2006) Differential repression of alternative transcripts: a screen for miRNA targets. PLoS Comput Biol 2: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Izaurralde E, Jarmolowski A, McGuigan C, Mattaj IW (1996) A nuclear cap-binding complex facilitates association of U1 snRNP with the cap-proximal 5′ splice site. Genes Dev 10: 1683–1698 [DOI] [PubMed] [Google Scholar]
- Li X, Manley JL (2006) Cotranscriptional processes and their influence on genome stability. Genes Dev 20: 1838–1847 [DOI] [PubMed] [Google Scholar]
- Li Y, Chen ZY, Wang W, Baker CC, Krug RM (2001) The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7: 920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez AJ (1998) Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet 32: 279–305 [DOI] [PubMed] [Google Scholar]
- Lou H, Gagel RF (1998) Alternative RNA processing—its role in regulating expression of calcitonin/calcitonin gene-related peptide. J Endocrinol 156: 401–405 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR (2005) MicroRNA expression profiles classify human cancers. Nature 435: 834–838 [DOI] [PubMed] [Google Scholar]
- Lu Y, Qian XY, Krug RM (1994) The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev 8: 1817–1828 [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Bazan NG (1997) Cyclooxygenase 2 RNA message abundance, stability, and hypervariability in sporadic Alzheimer neocortex. J Neurosci Res 50: 937–945 [DOI] [PubMed] [Google Scholar]
- Lutz CS, Murthy KG, Schek N, O'Connor JP, Manley JL, Alwine JC (1996) Interaction between the U1 snRNP-A protein and the 160-kDa subunit of cleavage–polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev 10: 325–337 [DOI] [PubMed] [Google Scholar]
- Mandel CR, Kaneko S, Zhang H, Gebauer D, Vethantham V, Manley JL, Tong L (2006) Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A (2003) Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Kee SG, Efstratiadis A, Kafatos FC (1976) Amplification and characterization of a beta-globin gene synthesized in vitro. Cell 8: 163–182 [DOI] [PubMed] [Google Scholar]
- Maniatis T, Reed R (2002) An extensive network of coupling among gene expression machines. Nature 416: 499–506 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2004) Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99 [DOI] [PubMed] [Google Scholar]
- Martincic K, Campbell R, Edwalds-Gilbert G, Souan L, Lotze MT, Milcarek C (1998) Increase in the 64-kDa subunit of the polyadenylation/cleavage stimulatory factor during the G0 to S phase transition. Proc Natl Acad Sci USA 95: 11095–11100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF (2005) Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol 17: 274–280 [DOI] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol 14: 692–699 [DOI] [PubMed] [Google Scholar]
- McCracken S, Lambermon M, Blencowe BJ (2002) SRm160 splicing coactivator promotes transcript 3′-end cleavage. Mol Cell Biol 22: 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Longman D, Johnstone IL, Caceres JF, Blencowe BJ (2003) An evolutionarily conserved role for SRm160 in 3′-end processing that functions independently of exon junction complex formation. J Biol Chem 278: 44153–44160 [DOI] [PubMed] [Google Scholar]
- McGregor F, Phelan A, Dunlop J, Clements J (1996) Regulation of herpes simplex virus poly (A) site usage and the action of immediate-early protein IE63 in the early–late switch. J Virol 70: 1931–1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J, Phelan A, Loney C, Sandri-Goldin RM, Clements JB (1992) Herpes simplex virus IE63 acts at the posttranscriptional level to stimulate viral mRNA 3′ processing. J Virol 66: 6939–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLauchlan J, Simpson S, Clements JB (1989) Herpes simplex virus induces a processing factor that stimulates poly(A) site usage. Cell 59: 1093–1105 [DOI] [PubMed] [Google Scholar]
- Millevoi S, Geraghty F, Idowu B, Tam JL, Antoniou M, Vagner S (2002) A novel function for the U2AF 65 splicing factor in promoting pre-mRNA 3′-end processing. EMBO Rep 3: 869–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Loulergue C, Dettwiler S, Karaa SZ, Keller W, Antoniou M, Vagner S (2006) An interaction between U2AF 65 and CF I(m) links the splicing and 3′ end processing machineries. EMBO J 25: 4854–4864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Keller W (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol 11: 352–357 [DOI] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev 12: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira A, Wollerton M, Monks J, Proudfoot NJ (1995) Upstream sequence elements enhance poly(A) site efficiency of the C2 complement gene and are phylogenetically conserved. EMBO J 14: 3809–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy KG, Manley JL (1995) The 160-kDa subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev 9: 2672–2683 [DOI] [PubMed] [Google Scholar]
- Natalizio BJ, Muniz LC, Arhin GK, Wilusz J, Lutz CS (2002) Upstream elements present in the 3′-untranslated region of collagen genes influence the processing efficiency of overlapping polyadenylation signals. J Biol Chem 277: 42733–42740 [DOI] [PubMed] [Google Scholar]
- Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM (1998) Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell 1: 991–1000 [DOI] [PubMed] [Google Scholar]
- Neu-Yilik G, Gehring NH, Hentze MW, Kulozik AE (2004) Nonsense-mediated mRNA decay: from vacuum cleaner to Swiss army knife. Genome Biol 5: 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, Rose SD, Berget SM (1990) In vitro polyadenylation is stimulated by the presence of an upstream intron. Genes Dev 4: 1552–1559 [DOI] [PubMed] [Google Scholar]
- O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435: 839–843 [DOI] [PubMed] [Google Scholar]
- Orkin SH, Cheng TC, Antonarakis SE, Kazazian HH Jr (1985) Thalassemia due to a mutation in the cleavage-polyadenylation signal of the human beta-globin gene. EMBO J 4: 453–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan SS, Forrest GL, Akman SA, Hu LT (1995) NAD(P)H:quinone oxidoreductase expression and mitomycin C resistance developed by human colon cancer HCT 116 cells. Cancer Res 55: 330–335 [PubMed] [Google Scholar]
- Pandey NB, Chodchoy N, Liu TJ, Marzluff WF (1990) Introns in histone genes alter the distribution of 3′ ends. Nucleic Acids Res 18: 3161–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pangalis GA, Trangas T, Papanastasiou CJ, Roussou PA, Tsiapalis CM (1985) Poly(A)-polymerase activity in chronic lymphocytic leukemia of the B cell type. Acta Haematol 74: 31–34 [DOI] [PubMed] [Google Scholar]
- Pendurthi UR, Alok D, Rao LVM (1997) Binding of factor VIIa to tissue factor induces alterations in gene expression in human fibroblast cells: upregulation of poly(A) polymerase. Proc Natl Acad Sci USA 94: 12598–12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Jung S, Gunderson SI (2001) Regulation of nuclear poly(A) addition controls the expression of immunoglobulin M secretory mRNA. EMBO J 20: 6443–6452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C, Pachikara N, Gunderson SI (2004) U1A inhibits cleavage at the immunoglobulin M heavy-chain secretory poly(A) site by binding between the two downstream GU-rich regions. Mol Cell Biol 24: 6162–6171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzuti A, Argiolas A, Di Paola R, Baratta R, Rauseo A, Bozzali M, Vigneri R, Dallapiccola B, Trischitta V, Frittitta L (2002) An ATG repeat in the 3′-untranslated region of the human resistin gene is associated with a decreased risk of insulin resistance. J Clin Endocrinol Metab 87: 4403–4406 [DOI] [PubMed] [Google Scholar]
- Poort SR, Rosendaal FR, Reitsma PH, Bertina RM (1996) A common genetic variation in the 3′-untranslated region of the prothrombin gene is associated with elevated plasma prothrombin levels and an increase in venous thrombosis. Blood 88: 3698–3703 [PubMed] [Google Scholar]
- Proudfoot N (2004) New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Proudfoot NJ, Furger A, Dye MJ (2002) Integrating mRNA processing with transcription. Cell 108: 501–512 [DOI] [PubMed] [Google Scholar]
- Rao P, Yuan W, Krug RM (2003) Crucial role of CA cleavage sites in the cap-snatching mechanism for initiating viral mRNA synthesis. EMBO J 22: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Hurt E (2002) A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108: 523–531 [DOI] [PubMed] [Google Scholar]
- Ristimaki A, Narko K, Hla T (1996) Downregulation of cytokine-induced cyclooxygenase-2 transcript isoforms by dexamethasone: evidence for post-transcriptional regulation. Biochem J 318 (Pt 1): 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J, Early P, Carter C, Calame K, Bond M, Hood L, Wall R (1980) Two mRNAs with different 3′ ends encode membrane-bound and secreted forms of immunoglobulin mu chain. Cell 20: 303–312 [DOI] [PubMed] [Google Scholar]
- Rosonina E, Kaneko S, Manley JL (2006) Terminating the transcript: breaking up is hard to do. Genes Dev 20: 1050–1056 [DOI] [PubMed] [Google Scholar]
- Rund D, Dowling C, Najjar K, Rachmilewitz E, Kazazian H Jr, Oppenheim A (1992) Two Mutations in the {beta}-globin polyadenylylation signal reveal extended transcripts and new RNA polyadenylylation sites. Proc Natl Acad Sci USA 89: 4324–4328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K, Calvo O, Manley JL (2004) Evidence that polyadenylation factor CPSF-73 is the mRNA 3′ processing endonuclease. RNA 10: 565–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs AB, Sarnow P, Hentze MW (1997) Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 89: 831–838 [DOI] [PubMed] [Google Scholar]
- Sandri-Goldin RM, Mendoza GE (1992) A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev 6: 848–863 [DOI] [PubMed] [Google Scholar]
- Sasaki R, Minowada J, Bollum FJ, Miura Y (1990) Polyadenylic acid polymerase activity in chronic myelogenous leukemia. Leuk Res 14: 273–278 [DOI] [PubMed] [Google Scholar]
- Schambach A, Galla M, Maetzig T, Loew R, Baum C (2007) Improving transcriptional termination of self-inactivating gamma-retroviral and Lentiviral vectors. Mol Ther 15: 1167–1173 [DOI] [PubMed] [Google Scholar]
- Schrijver I, Lenzi TJ, Jones CD, Lay MJ, Druzin ML, Zehnder JL (2003) Prothrombin gene variants in non-Caucasians with fetal loss and intrauterine growth retardation. J Mol Diagn 5: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E, Gelfand JM, Mauch JC, Kligman LH (1998) Generation of a tropoelastin mRNA variant by alternative polyadenylation site selection in sun-damaged human skin and ultraviolet B-irradiated fibroblasts. Biochem Biophys Res Commun 246: 217–221 [DOI] [PubMed] [Google Scholar]
- Scorilas A (2002) Polyadenylate polymerase (PAP) and 3′ end pre-mRNA processing: function, assays, and association with disease. Crit Rev Clin Lab Sci 39: 193–224 [DOI] [PubMed] [Google Scholar]
- Scorilas A, Talieri M, Ardavanis A, Courtis N, Dimitriadis E, Yotis J, Tsiapalis CM, Trangas T (2000) Polyadenylate polymerase enzymatic activity in mammary tumor cytosols: a new independent prognostic marker in primary breast cancer. Cancer Res 60: 5427–5433 [PubMed] [Google Scholar]
- Seegers D, Zwiers A, Strober W, Pena AS, Bouma G (2002) A TaqI polymorphism in the 3′UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes Immun 3: 419–423 [DOI] [PubMed] [Google Scholar]
- Shell SA, Hesse C, Morris SM Jr, Milcarek C (2005) Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem 280: 39950–39961 [DOI] [PubMed] [Google Scholar]
- Shin C, Manley JL (2004) Cell signalling and the control of pre-mRNA splicing. Nat Rev Mol Cell Biol 5: 727–738 [DOI] [PubMed] [Google Scholar]
- Shin HD, Kim LH, Park BL, Jung HS, Cho YM, Moon MK, Lee HK, Park KS (2003) Polymorphisms in fatty acid-binding protein-3 (FABP3)—putative association with type 2 diabetes mellitus. Hum Mutat 22: 180. [DOI] [PubMed] [Google Scholar]
- Shyu A-B, Wilkinson MF (2000) The double lives of shuttling mRNA binding proteins. Cell 102: 135–138 [DOI] [PubMed] [Google Scholar]
- Soo PY, Patel RK, Best S, Arya R, Thein SL (2005) Detection of prothrombin gene polymorphism at position 20209 (PT20209C/T): Pilot study in a black population in the United Kingdom. Thromb Haemost 93: 179–180 [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM (2005) Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell 123: 1133–1146 [DOI] [PubMed] [Google Scholar]
- Stetler DA, Reichlin M, Berlin CM, Jacob ST (1987) Antibodies against nuclear poly(A) polymerases in rheumatic autoimmune diseases. J Clin Immunol 7: 24–28 [DOI] [PubMed] [Google Scholar]
- Stetler DA, Rose KM, Jacob ST (1981) Anti-poly(A) polymerase antibodies in sera of tumor-bearing rats and human cancer patients. Proc Natl Acad Sci USA 78: 7732–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoehlmacher J, Park DJ, Zhang W, Yang D, Groshen S, Zahedy S, Lenz HJ (2004) A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer 91: 344–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathdee CA, Duncan AM, Buchwald M (1992) Evidence for at least four Fanconi anaemia genes including FACC on chromosome 9. Nat Genet 1: 196–198 [DOI] [PubMed] [Google Scholar]
- Stutz F, Rosbash M (1998) Nuclear RNA export. Genes Dev 12: 3303–3319 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL (1998) Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell 2: 761–771 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL (1996) The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS (2005) A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian SL, Kaneko S, Gonzales MI, Bond GL, Ward Y, Manley JL (2001) Identification and functional characterization of Neo-Poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol Cell Biol 21: 5614–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trangas T, Courtis N, Pangalis GA, Cosmides HV, Ioannides C, Papamichail M, Tsiapalis CM (1984) Polyadenylic acid polymerase activity in normal and leukemic human leukocytes. Cancer Res 44: 3691–3697 [PubMed] [Google Scholar]
- Uitte de Willige S, de Visser MCH, Houwing-Duistermaat JJ, Rosendaal FR, Vos HL, Bertina RM (2005) Genetic variation in the fibrinogen gamma gene increases the risk for deep venous thrombosis by reducing plasma fibrinogen {gamma}′ levels. Blood 106: 4176–4183 [DOI] [PubMed] [Google Scholar]
- Uitte de Willige S, Rietveld IM, De Visser MC, Vos HL, Bertina RM (2007) Polymorphism 10034C>T is located in a region regulating polyadenylation of FGG transcripts and influences the fibrinogen gamma′/gammaA mRNA ratio. J Thromb Haemost 5: 1243–1249 [DOI] [PubMed] [Google Scholar]
- Vagner S, Vagner C, Mattaj IW (2000) The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev 14: 403–413 [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A, Zeichner S, Carswell S, Alwine J (1991) The human immunodeficiency virus type 1 polyadenylylation signal: a 3′ long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci USA 88: 2108–2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Solinge WW, Lind B, van Wijk R, Hart HC, Kraaijenhagen RJ (1996) Clinical expression of a rare beta-globin gene mutation co-inherited with haemoglobin E-disease. Eur J Clin Chem Clin Biochem 34: 949–954 [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veraldi KL, Arhin GK, Martincic K, Chung-Ganster LH, Wilusz J, Milcarek C (2001) hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol Cell Biol 21: 1228–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A, Grosveld G (1992) Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol Cell Biol 12: 3346–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Kuhn U (1997) The mechanism of 3′ cleavage and polyadenylation of eukaryotic pre-mRNA. Prog Nucleic Acid Res Mol Biol 57: 41–71 [DOI] [PubMed] [Google Scholar]
- Wassarman KM, Steitz JA (1993) Association with terminal exons in pre-mRNAs: a new role for the U1 snRNP? Genes Dev 7: 647–659 [DOI] [PubMed] [Google Scholar]
- Weatherall DJ (2001) Phenotype–genotype relationships in monogenic disease: lessons from the thalassaemias. Nat Rev Genet 2: 245–255 [DOI] [PubMed] [Google Scholar]
- Weiner AM (2005) E Pluribus Unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol Cell 20: 168–170 [DOI] [PubMed] [Google Scholar]
- Wilson T, Treisman R (1988) Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature 336: 396–399 [DOI] [PubMed] [Google Scholar]
- Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Marr TG (2005) Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res 15: 369–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Shabbeer J, Osawa M, Desnick RJ (2003) Fabry disease: novel alpha-galactosidase A 3′-terminal mutations result in multiple transcripts due to aberrant 3′-end formation. Am J Hum Genet 73: 162–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss A-K, Son S, Chang L-J (2002) RNA 3′ readthrough of oncoretrovirus and lentivirus: implications for vector safety and efficacy. J Virol 76: 7209–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Cui Y, Kuang G, Li Y, Wang N, Wang R, Guo W, Wen D, Wei L, Yu F, Wang S (2004a) Association of the thymidylate synthase polymorphisms with esophageal squamous cell carcinoma and gastric cardiac adenocarcinoma. Carcinogenesis 25: 2479–2485 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shi Q, Sturgis EM, Spitz MR, Hong WK, Wei Q (2004b) Thymidylate synthase 5′- and 3′-untranslated region polymorphisms associated with risk and progression of squamous cell carcinoma of the head and neck. Clin Cancer Res 10: 7903–7910 [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Manley JL (1996) Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol 16: 2378–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SL, Augustsson-Balter K, Chang B, Hedelin M, Li L, Adami H-O, Bensen J, Li G, Johnasson J-E, Turner AR, Adams TS, Meyers DA, Isaacs WB, Xu J, Gronberg H (2004) Sequence variants of Toll-like receptor 4 are associated with prostate cancer risk: results from the cancer prostate in Sweden study. Cancer Res 64: 2918–2922 [DOI] [PubMed] [Google Scholar]
- Zhu H, Hasman RA, Barron VA, Luo G, Lou H (2006) A nuclear function of Hu proteins as neuron-specific alternative RNA processing regulators. Mol Biol Cell 17: 5105–5114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Zhou HL, Hasman RA, Lou H (2007) Hu proteins regulate polyadenylation by blocking sites containing U-rich sequences. J Biol Chem 282: 2203–2210 [DOI] [PubMed] [Google Scholar]