Atypical ubiquitin chains: new molecular signals. ‘Protein Modifications: Beyond the Usual Suspects' Review Series (original) (raw)
Abstract
Ubiquitin (Ub) is a small protein modifier that regulates many biological processes, including gene transcription, cell-cycle progression, DNA repair, apoptosis, virus budding and receptor endocytosis. Ub can be conjugated to target proteins either as a monomer or as Ub chains that vary in length and linkage type. The various types of Ub modification are linked to distinct physiological functions in cells. MonoUb, for example, regulates DNA repair and receptor endocytosis, whereas lysine 48-linked Ub chains label proteins for proteasomal degradation. More recently, the importance of chains conjugated through the other six lysines in Ub, known as atypical Ub chains, has been revealed. Atypical chains can be homotypic, sequentially using the same lysine residue in Ub for conjugation; mixed-linkage, utilizing several distinct lysines to connect consecutive Ub moieties; or heterologous, connecting Ub with other Ub-like modifiers. Here, we describe recent progress in the understanding of atypical Ub chain assembly and their recognition by Ub-binding domains, and we discuss further their functional roles in vivo.
Keywords: atypical chain, signalling, ubiquitin, ubiquitin-like protein, UBD, Ubl
Introduction
Protein modification by ubiquitin (Ub)—a process known as ubiquitination or ubiquitylation—is involved in the regulation of numerous cellular functions, including protein stability, cell-cycle progression, gene transcription, receptor transport, immune responses and viral infection (Haglund & Dikic, 2005; Mukhopadhyay & Riezman, 2007). In general, these modifications are controlled by a three-step reaction initialized by E1-mediated activation of Ub, followed by conjugation to an E2 enzyme and finally substrate targeting by an E3 Ub ligase (Hershko & Ciechanover, 1998; Pickart & Eddins, 2003). Similar to the process of phosphorylation and dephosphorylation, ubiquitination is also reversible, and the cleavage of Ub from substrates is carried out by specific deubiquitinating enzymes (DUBs; Hochstrasser, 1995).
Ubiquitin is one of the most versatile molecular signals in the cell because of its ability to modify substrate proteins in its monomeric form (monoubiquitination) or to be conjugated to preceding Ub moieties and consequently form many types of Ub chain (polyubiquitination; Haglund & Dikic, 2005; Welchman et al, 2005). Monoubiquitination has been shown to control numerous cellular processes such as receptor transport, viral budding and DNA repair (Hicke, 2001; Di Fiore et al, 2003; Haglund et al, 2003), whereas Ub chains that form through their lysine (Lys) residues at position 48 (Lys 48) are known to regulate protein stability (Hershko & Ciechanover, 1998). Ub can also be conjugated through other lysine residues on its surface (Lys 6, Lys 11, Lys 27, Lys 29, Lys 33 and Lys 63), forming Ub chains of various lengths and shapes in vitro and in vivo (Johnson et al, 1995; Peng et al, 2003; Kim et al, 2007). Different Ub linkages result in various conformations of Ub chain and create a range of molecular signals in the cell.
In addition to Ub itself, many ubiquitin-like (Ubl) proteins have been identified, including small Ub-like modifier (SUMO), interferon-stimulated gene 15 (ISG15), autophagy 8 (ATG8) and neural precursor cell expressed, developmentally downregulated 8 (NEDD8; Kerscher et al, 2006). Ubls have a similar structural-fold to Ub, but use specific conjugation machineries and are recognized by distinct Ubl-binding domains. So far, SUMO2/3 and NEDD8 are the only Ubls known to participate in chain formation (Tatham et al, 2001; Knipscheer et al, 2007; Matic et al, 2008; Xirodimas et al, 2008). Ubls are implicated in the regulation of many cellular processes such as gene transcription, signal transduction, autophagy and cell-cycle control (Kerscher et al, 2006).
The conserved ubiquitin β-grasp fold has had numerous functions during evolution and is used, for example, as a protein domain integrated in the coding sequence of several cellular proteins. These ubiquitin-like domains (ULDs) adopt a Ub-like fold within the tertiary structure of the host protein (Tanaka et al, 1990; Hartmann-Petersen & Gordon, 2004; Ikeda et al, 2007). ULDs can intrinsically regulate the conformational status of the host protein, as well as participate in the reorganization of protein complexes (Tanaka et al, 1990; Hartmann-Petersen & Gordon, 2004; Ikeda et al, 2007).
Classification of atypical ubiquitin chains
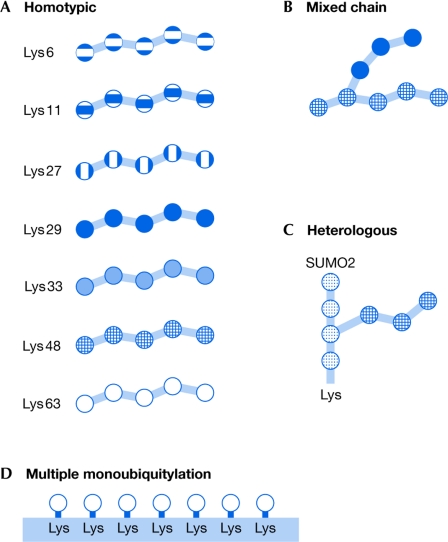
Atypical Ub chains include all variations of multimeric Ub structure with the exception of classical Lys 48 polyubiquitination, which is considered to be a typical Ub chain based on its original discovery as a destruction tag for proteasomal degradation. To distinguish various types of atypical Ub chain, we have divided them into several classes (Fig 1). Chains formed by the conjugation of a single type of lysine residue in sequential Ub molecules are homotypic, whereas those assembled through several distinct lysines in the Ub monomers are mixed-linkage chains. As a consequence of using divergent lysines for conjugation, mixed-linkage Ub chains form bifurcations, such as chains containing two different types of linkage—Lys 6/11, Lys 27/29, Lys 29/48 or Lys 29/33 (Kim et al, 2007). Moreover, the integration of other Ubl modifiers, such as SUMO and NEDD8, into Ub chains gives rise to heterologous Ub chains; so far, the heterologous chains are the least studied. Finally, many monoUbs attached to a substrate, when packed spatially in close proximity, can be considered as atypical Ub signals with acquired Ub multivalency.
Figure 1.
A schematic model of possible ubiquitin chain formations on a target protein. (A) Homotypic atypical and typical chains, such as lysine 6 (Lys 6)-, Lys 11-, Lys 27-, Lys 29-, Lys 33-, Lys 48- and Lys 63-linked ubiquitin (Ub) chains. (B) Mixed-linkage atypical chains are formed by the use of different lysines for sequential Ub conjugation, leading to the formation of bifurcated chains, for example, Lys 48/Lys 29 forks. (C) Heterologous chains are formed between Ub and ubiquitin-like proteins, for example the small ubiquitin-like modifier (SUMO) 2. (D) Multiple monoubiquitination moieties represent a subtype of multivalent chain-like Ub signals owing to the spatial organization of multiple monoUb molecules attached to the substrate.
Assembly of various types of ubiquitin chain
During ubiquitination, Ub is initially activated by an ATP-dependent E1 enzyme before it is passed to one of several distinct Ub-conjugating enzymes (E2s). The E2 subsequently acts to either transfer Ub to a HECT (homologous to the E6AP carboxyl terminus)-type Ub ligase (E3), or to catalyse substrate ubiquitination in conjunction with RING (really interesting new gene)-type or U-box E3 enzymes. An exception to this is the process of coupled monoubiquitination, through which E2 enzymes can catalyse the ubiquitination of Ub-binding domain (UBD)-containing proteins independently of E3 (Hoeller et al, 2006, 2007).
Ubiquitin ligases seem to be the crucial determinants of substrate selection and are also considered to be important components in controlling Ub-chain formation on a substrate. For example, different HECT-type E3s can specify both the linkage of a Ub chain and the process of its assembly. E6AP can form a Lys 48-linked chain on its active cysteine residue (Scheffner & Whitaker, 2003), whereas KIAA10 catalyses Lys 48- and Lys 29-linked chains as free entities (Wang et al, 2006). In budding yeast, the Ub ligase Rsp5 has been shown to modify specific substrates either with monoUb, Lys 48- or Lys 63-linked chains (Kee et al, 2005). Correspondingly, the mammalian RING-type ligase CBL mediates multiple monoubiquitination and Lys 11-, Lys 48- and Lys 63-oligoubiquitination of activated epidermal growth factor receptor (EGFR; Haglund et al, 2003; Mosesson et al, 2003; Huang et al, 2006), as well as Lys 48-linked polyubiquitination of SRC kinases (Thien & Langdon, 2005).
Besides the E3s, E2 enzymes can also be structured to control lysine specificity during Ub chain assembly. Although most E2s are designed to ubiquitinate substrates with no inherent selectivity for a specific acceptor lysine residue, some stimulate Ub chain formation linked through a defined lysine in Ub. Structural studies of the MMS2–UBC13 complex have given rise to a model describing the specific elongation of Lys 63-linked chains. In this model, the catalytically active E2 enzyme UBC13, when conjugated with Ub, binds to MMS2, a pseudo-E2 enzyme with the ability to bind an acceptor Ub. As a consequence of this interaction, the acceptor Ub on MMS2 is positioned in such a way that only Lys 63 is accessible for Ub transfer from UBC13, hence forming Lys 63-linked chains (VanDemark et al, 2001). Conversely, other E2s, such as UBCH5, can catalyse the formation of Ub chains that lack specificity for any lysine residue of Ub (Brzovic et al, 2006).
Individual E3 ligases commonly interact with several E2 enzymes to correctly modify target substrates. In the case of the E3 ligase breast cancer susceptibility gene 1 (BRCA1), interaction with the E2s UBCH6, UBE2E2, UBCM2 and UBE2W supports monoubiquitination, whereas MMS2–UBC13 and UBE2K catalyse Lys 63- or Lys 48-linked chain formation on BRCA1, respectively (Christensen et al, 2007). Moreover, certain pairs of E2s and E3s can synthesize Ub chains containing different isopeptide linkages (Kim et al, 2007; Windheim et al, 2008). The E2 UBCH5, together with the E3 ligases CHIP and MDM2, induce the assembly of homotypic chains by using all seven possible linkages, as well as mixed-linkage chains joined through adjacent lysines at Lys 6/11, Lys 27/29 or Lys 29/33 (Kim et al, 2007). By contrast, in the presence of UBCH13/UEV1A and UBCH1 the same E3s can induce Lys 63 and Lys 48 chains, respectively. Thus, various combinations of E2 and E3 enzymes can direct the synthesis of diverse types of atypical Ub chain during in vitro reactions (Sidebar A). It is likely that additional factors, including subcellular compartmentalization of selected E2 and E3 pairs, are detrimental to the formation of some Ub chains in vivo.
The original idea that Ub chains are assembled by the sequential addition of Ub monomers to a lysine residue in the target protein is now challenged by evidence indicating that Ub polymers can be formed on E2/E3 enzymes before target substrate conjugation (Wu-Baer et al, 2003; Ben-Saadon et al, 2006; Li et al, 2007). E3 enzymes often form different types of Ub chain at their active sites (Hochstrasser, 2006), an activity that was originally thought to be a mechanism for controlling the abundance of the ligases by marking them for degradation (Brown et al, 2002). However, self-ubiquitination through mixed-linkage Ub chains can have a regulatory role, as in the case of the Polycomb E3 RING-type ligases Ring1B and BMI1 (Ben-Saadon et al, 2006). Moreover, it was shown that Lys 63-linked self-ubiquitination of TNF receptor-associated factor 6 (TRAF6) is crucial for IκB kinase complex activation in vivo (Lamothe et al, 2007). E2 enzymes can also assemble divergent Ub chains at their active sites, which can then be transferred to a substrate. One such example is the UBE2G2 E2 enzyme that, together with the E3 ligase GP78, mediates substrate ubiquitination by using Ub chains pre-assembled at the catalytic cysteine of UBE2G2 (Li et al, 2007). In addition, Ub can be transferred from one E2 to another, thus forming diUb chains presumably owing to E2 dimerization. Interestingly, dimerization of CDC34 (cell division cycle 34), which is induced by thioester formation between Ub and CDC34, is essential for the synthesis of Lys 48-linked Ub chains by this enzyme (Varelas et al, 2003).
Quantitative analyses of atypical chains
The analysis of the formation and dynamic changes of atypical Ub chains represents the main bottleneck in understanding the structural complexity and functional diversity of these modifications in vivo. Most of the collected data are derived from biochemical experiments performed with purified proteins and synthetic Ub chains or from studies using transfection of tagged versions of lysine-to-arginine mutant forms of Ub in cells. The problem is that there are no controlled comparisons between different experimental conditions—for example, extent of protein overexpression or influence of endogenous Ub on the specific lysine-to-arginine mutants—and current conclusions often rely on several assumptions, which do not always represent physiological Ub modifications in vivo. In addition, antibodies with preferences for Ub chains are not sufficiently sensitive and specific for comparative analyses of distinct Ub modifications in cells.
The most powerful analytical method for measuring global changes in Ub linkages is the use of mass spectrometry (Kirkpatrick et al, 2005). The first large-scale analysis of all ubiquitinated proteins in yeast expressing epitope-tagged Ub identified 1,075 candidate substrates (Peng et al, 2003). All seven lysine residues have been found to participate in the formation of Ub–Ub linkages, with a relative abundance order of Lys 48 > Lys 11 and Lys 63 >> Lys 6, Lys 27, Lys 29 and Lys 33. Ub peptides with mixed linkages of Lys 29 and Lys 33 were also identified, although in a low proportion compared with conjugated monoUb or different types of homotypic chain (Peng et al, 2003). Such quantitative data are valuable, but not complete owing to intrinsic technical limitations. For example, mass spectrometry analyses are inherently biased towards more abundant species and might give an incorrect impression of the complexity of less abundant Ub chains. As pointed out by Kirkpatrick and colleagues, the absence of evidence should not necessarily be construed as evidence of absence (Kirkpatrick et al, 2005).
Recent development of the Ub-AQUA (absolute quantification of Ub) method has provided a new tool for more precise quantitative characterization of the various forms of Ub chain (Kirkpatrick et al, 2006). In the AQUA method, all seven ubiquitinated -GG peptides are labelled with a stable isotope and then quantified as internal standards, which can be distinguished in a mass spectrometer. Quantitative analysis of in vitro ubiquitinated cyclin B1 revealed complex chain topology, including Ub linked through lysine residues (Lys 63, Lys 11 and Lys 48). Interestingly, even in the absence of Lys 48 the remaining Ub chains were able to mediate degradation of cyclin B1 in the proteasome (Kirkpatrick et al, 2006). In accordance with this observation, homotypic Ub chains linked through Lys 63, but not mixed-linkage chains, can mediate proteasomal degradation in vitro as rapidly as Lys 48-linked Ub chains (Kim et al, 2007).
However, a significant obstacle in quantitative in vivo analysis of Ub chains and their recognition by Ub-binding proteins is the low abundance of substrates, the modifications of which vary markedly under certain conditions in cells. Such modified proteins often escape detection by current proteomics technologies, indicating the need for rapid development of more sensitive proteomics platforms.
Recognition of ubiquitin chains by ubiquitin-binding domains
More than 20 families of Ub-binding domain (UBD), which bind to monoUb and Ub chains, have been identified (Hicke et al, 2005; Hurley et al, 2006; Husnjak et al, 2008; Iha et al, 2008; Schreiner et al, 2008; Wagner et al, 2008). The in vitro binding affinity between a UBD and Ub is usually within _K_d = 10–500 μM, which is relatively low for a physiological interaction. Conversely, the level of free Ub in cells is fairly high—2 μM in muscle tissues and 10–20 μM in different cultured cells (Haas & Bright, 1985; Haas, 1988; Riley et al, 1988)—which raises the question how UBD-containing proteins can selectively bind to conjugated Ub and not to free Ub. It is likely that physiological Ub–UBD interactions are regulated by the avidity of binding, which might be mediated through multivalent interactions between ubiquitinated substrates and Ub-binding proteins. This can be achieved in several ways, including multiplication of UBDs in adaptor proteins, oligomerization of Ub-binding proteins, secondary Ub-independent interactions brought about in signalling complexes or by an accumulation of interacting partners in defined cellular compartments. For example, the Ub-interacting motif (UIM) domains in EPS15, epsin and HRS bind to monoUb with relatively low affinities (_K_d > 100 μM; Hurley et al, 2006). However, these interactions are physiologically relevant for the sorting of ubiquitinated cargoes in the endosome, probably as a result of the oligomerization of endosomal Ub receptors and the presence of many mono-Ubs on transport cargoes (Hicke, 2001; Di Fiore et al, 2003; Haglund et al, 2003). Interestingly, the UIM domain of HRS folds into a double-sided α-helix with the ability to simultaneously bind to two Ub molecules, one on each side (Hirano et al, 2006).
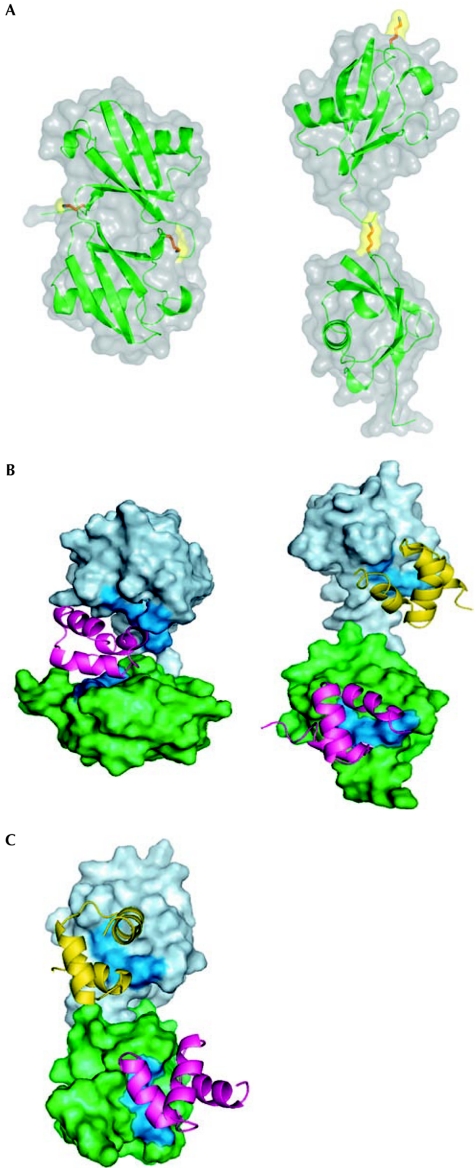
One of the important challenges in the field is to understand why and how some UBDs preferentially bind to Ub chains with linkage specificity, whereas others are more or less promiscuous with regard to linkage type. Initial clues have come from the realization that Ub chains have different conformations that provide specific surfaces for interactions with UBDs (Fig 2). For example, Lys 48-linked Ub polymers adopt a compact conformation, whereas Lys 63 diUb acquires an extended shape, similar to a linear chain (Varadan et al, 2002, 2004; Fig 2A). In a comprehensive study involving more than 30 Ub-associated (UBA) domains, it was shown that one subgroup preferentially binds to Lys 48-linked chains, a second subgroup primarily recognizes Lys 63-linked chains and a third subgroup binds to Ub chains without any preference for linkage specificity (Raasi et al, 2005). Interestingly, all of the UBAs that recognize Ub chains also bind to monoUb (Raasi & Pickart, 2002). Conformational studies of the UBA domains in HR23a and MUD1 in solution bound to Lys 48-linked diUb, indicating that UBA domains bind between the two monoUb molecules in a ‘sandwich-like' manner (Trempe et al, 2005; Varadan et al, 2005; Fig 2B, left panel). By contrast, the open conformation of the Lys 63-linked diUb allows interaction with the human HR23a UBA domain independent of additional interactions, similar to the binding to linear monoUb (Varadan et al, 2004; Fig 2B, right panel). The UBA domain of ubiquilin 1 (PLIC-1) binds to Lys 48-linked diUb by using independent surfaces of each of the Ub molecules (Fig 2C), which is distinct from the mode of binding of the UBA2 domain of HR23a (Fig 2B, left panel; Zhang et al, 2008). A similar situation to the UBA domain of PLIC-1 is observed with a UIM of S5a, which also recognizes both Lys 48- and Lys 63-linked chains (Haririnia et al, 2007).
Figure 2.
Structural models indicating differences between Lys 48 and Lys 63 signals. (A) Lysine 48 (Lys 48)-linked di-ubiquitin (diUb; left panel) and Lys 63-linked diUb (right panel) show two different conformations, which indicate distinct binding properties for interaction with ubiquitin-binding domains (UBDs). Structures of other atypical chains have not yet been determined. (B) Conformational differences between alternatively linked polyubiquitin chains allow them to interact with UBDs in distinct binding modes: a sandwich-like mode (left panel) and as multiple independent UBD-binding sites (right panel). Shown here are structural models of the complexes of Lys 48- and Lys 63-linked diUb (left and right panels, respectively) with the UBA2 domain of HR23a. The distal and proximal Ubs are shown in surface representation, coloured light blue and green, respectively; the UBA domains are shown as ribbon. The hydrophobic patch residues (Leu 8–Ile 44–Val70) on the Ub surface are painted blue. (C) Lys 48-linked diUb can bind to UBDs at several binding sites, as observed in the case of the UBA domain of ubiquilin-1 (PLIC-1). UBA, ubiquitin-associated.
More recently, RPN13/ARM1 was identified as a new Ub receptor at the proteasome and shown to stoichiometrically bind to Lys 48-linked diUb through its pleckstrin-like receptor for ubiquitin (Pru) domain (Husnjak et al, 2008; Schreiner et al, 2008). Fluorescence spectroscopy analysis revealed that the Pru domain binds to Lys 48-linked diUb with a _K_d of 90 nM and monoUb with a _K_d of 300 nM, which is the highest affinity observed among known UBDs (Husnjak et al, 2008). In this particular case, the loops of the Pru domain, rather than secondary structural elements, create a large interaction surface with Ub (1,256 Å2), thus providing the rationale for the observed high affinity.
Recent studies have reported the characterization of new UBDs that show specificity for Lys 63-linked chains in vivo. A UBD found in NEMO, optineurin and A20 binding inhibitor of NF-κB activation (ABIN) proteins—commonly named UBAN (UBD in ABIN proteins and NEMO)—is involved in the regulation of NF-κB signalling pathways (Ea et al, 2006; Wu et al, 2006; Zhu et al, 2007; Bloor et al, 2008; Wagner et al, 2008). In all these cases, the UBAN domain binds preferentially to Lys 63-linked chains or linear Ub polymers, with the exception of the UBAN domain in ABIN3, which was found to interact with monoUb as potently as with Ub chains (Wagner et al, 2008). Nuclear magnetic resonance analysis of the UBAN domain in ABIN1 clarified that UBAN recognizes an extended surface of Ub, suggesting that the UBAN domain has an elongated shape that prefers the linear conformation of Lys 63 chains (Wagner et al, 2008). In addition, the Ub-binding zinc-finger (UBZ) domain of the adaptor protein TAX1BP1 was recently shown to target Lys 63-ubiquitinated forms of TRAF6 and the receptor-interacting protein RIP in tumour necrosis factor-α (TNFα) and interleukin-1 (IL-1)-treated cells, respectively (Iha et al, 2008). However, in vitro TAX1BP1 can bind to monoUb, Lys 63- and Lys 48-linked chains. TAX1BP1 is constitutively associated with the deubiquitinating enzyme A20, which removes Ub chains from interacting proteins leading to the inhibition of NF-κB signalling (Iha et al, 2008).
Physiological roles of atypical ubiquitin chains
Although all lysine residues in Ub have been shown to participate in chain formation in vivo under different conditions, only a few studies have addressed their biological significance in cells. The most-studied of the atypical Ub chains is the Lys 63-linked chain, which has a crucial role in signal transduction through the NF-κB pathway, as well as in receptor endocytosis and DNA-repair processes (reviewed by Haglund & Dikic, 2005; Hayden & Gosh, 2008). Proteins modified with Lys 63-linked chains are recognized by numerous UBDs (see above), mediating interactions essential for NF-κB activation in response to IL-1 and TNFα stimulation. This topic has been already discussed in detailed in several excellent recent reviews (for example, Hayden & Gosh, 2008).
In DNA-damage response pathways, Lys 63-linked Ub chains seem to have important functions during the recruitment of repair machineries to the sites of DNA damage. The RING-type E3-ligase BRCA1 forms a multimeric protein complex required for the repair of double-stranded DNA breaks (Bennett & Harper, 2008). As BRCA1 is not a Ub-binding protein, the recruitment of this complex to the sites of damage depends on the receptor-associated protein 80 (RAP80), which contains two UIM domains that preferentially bind to Lys 63-linked Ub chains on histones H2A and H2AX (Bennett & Harper, 2008).
Lys 29/Lys 33-linked mixed chains have been recently implicated in the regulation of AMP-activated protein kinase (AMPK)-related kinases. AMPK family member 5 (ARK5/NUAK1) and MAP/microtubule-affinity-regulating kinase (MARK) 4 kinases are polyubiquitinated in vivo through Lys 29/Lys 33-coupled chains (Al-Hakim et al, 2008). This event blocks their kinase activation by interfering with phosphorylation of the activation-loop residues (Al-Hakim et al, 2008). In addition, Lys 29-linked Ub chains formed by the E3 ligase ITCH/AIP4 have been implicated in the lysosomal degradation of proteins (Chastagner et al, 2006).
The formation of mixed-linkage Ub chains is implicated in the activation of the Polycomb protein Ring1B Ub ligase complex (Ben-Saadon et al, 2006). Ring1B forms a complex with BMI1, a protein with no detectable ubiquitinating activity. However, BMI1 stimulates the monoubiquitinating activity of the Ring1B ligase complex towards histone H2A. Ring1B catalyses its auto-ubiquitination through atypical Ub chains that involve Lys 6-, Lys 27- and Lys 48-linkage types on the same Ub molecule; this modification does not lead to the degradation of Ring1B, but is instead essential for the efficient monoubiquitination of histone H2A in vitro (Ben-Saadon et al, 2006).
Heterologous chains—between Ub and Ubl modifiers—also exist and seem to have important physiological roles. A recent study has shown that RING finger protein 4 (RNF4), an E3 Ub ligase that contains several SUMO-interacting motifs, can specifically target SUMOylated promyelocytic leukaemia (PML) proteins for ubiquitination, consequently generating SUMO-Ub heterologous chains (Fig 1D; Tatham et al, 2008). This leads to the recognition and recruitment of SUMOylated PML proteins for degradation in the proteasome (Lallemand-Breitenbach et al, 2008; Tatham et al, 2008). The formation of such heterologous chains is of clinical relevance as treatment with arsenic trioxide triggers SUMO-dependent polyubiquitination and degradation of the PML-RARα fusion protein that causes acute promyelocytic leukaemia (APL; Lallemand-Breitenbach et al, 2008; Tatham et al, 2008).
Conclusion and future challenges
Atypical Ub chains represent new, albeit poorly understood, molecular signals. They can form a sizeable range of modifications through variations in their length and linkage type. Although experimental data have indicated the existence of a large variety of atypical chains in vivo, there is, however, only sparse physiological evidence about their functional role in the cell (Sidebar A). Ongoing work in this area seeks to develop a more complete understanding of the structure and function of polyubiquitin chains, and to define the role and specificity of Ub-binding proteins. An important issue to resolve will be to discriminate atypical Ub chains that encode a valid physiological signal from the ones that are formed during the ubiquitination reaction but that lack any specific biological role. Moreover, it will be important to reveal molecular and atomic details of how each of the various chains is synthesized and attached to substrates, and what determines the specificity of their interactions with Ub-binding proteins in vivo. The most challenging task will be to solve three-dimensional structures of a ubiquitinated substrate in complex with a Ub-binding protein. As more studies combine the analysis of biological mechanisms with biophysical and structural studies, future prospects in the field of atypical Ub chains are both exciting and promising.
Sidebar A | In need of answers.
- How are different types of ubiquitin polymer synthesized?
- What are the functions of atypical chains in vivo?
- How are the heterologous chains of Ub and Ubl formed in vivo?
- How are different atypical chains recognized by ubiquitin-binding domains?

Fumiyo Ikeda

Ivan Dikic

Acknowledgments
We thank D. Hoeller, D. McEwan, C. Grabbe, D. Komander, S. Raasi, D. Fushman, K. Walters, R. Hay, A. Goldberg, A. Haas and A. Ciechanover for discussions and critical reading of the manuscript. We are also grateful to D. Komander and D. Fushman for help with creating Fig 2. We apologize to investigators whose important contributions were not included in this review owing to space limitations. F.I. is supported by the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad. I.D. acknowledges support from the Deutsche Forschungsgemeinschaft, the German-Israeli Foundation and the Boehringer Ingelheim Foundation.
References
- Al-Hakim AK, Zagorska A, Chapman L, Deak M, Peggie M, Alessi DR (2008) Control of AMPK-related kinases by USP9X and atypical Lys 29/Lys 33-linked polyubiquitin chains. Biochem J 411: 249–260 [DOI] [PubMed] [Google Scholar]
- Bennett EJ, Harper JW (2008) DNA damage: ubiquitin marks the spot. Nat Struct Mol Biol 15: 20–22 [DOI] [PubMed] [Google Scholar]
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A (2006) The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell 24: 701–711 [DOI] [PubMed] [Google Scholar]
- Bloor S, Ryzhakov G, Wagner S, Butler PJ, Smith DL, Krumbach R, Dikic I, Randow F (2008) Signal processing by its coil zipper domain activates IKKγ. Proc Natl Acad Sci USA 105: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Hostager BS, Bishop GA (2002) Regulation of TRAF2 signaling by self-induced degradation. J Biol Chem 277: 19433–19438 [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 21: 873–880 [DOI] [PubMed] [Google Scholar]
- Chastagner P, Israël A, Brou C (2006) Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep 7: 1147–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE (2007) E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol 14: 941–948 [DOI] [PubMed] [Google Scholar]
- Di Fiore PP, Polo S, Hofmann K (2003) When ubiquitin meets ubiquitin receptors: a signalling connection. Nat Rev Mol Cell Biol 4: 491–497 [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ (2006) Activation of IKK by TNFα requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22: 245–257 [DOI] [PubMed] [Google Scholar]
- Haas A (1988) Immunochemical probes of ubiquitin pool dynamics. In Ubiquitin, M Rechsteiner (ed), pp 173–206. New York, NY, USA: Plenum Publishing Corporation [Google Scholar]
- Haas AL, Bright PM (1985) The immunochemical detection and quantitation of intracellular ubiquitin-protein conjugates. J Biol Chem 260: 12464–12473 [PubMed] [Google Scholar]
- Haglund K, Dikic I (2005) Ubiquitylation and cell signaling. EMBO J 24: 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I (2003) Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol 5: 461–466 [DOI] [PubMed] [Google Scholar]
- Haririnia A, D'Onofrio M, Fushman D (2007) Mapping the interactions between Lys 48 and Lys 63-linked di-ubiquitins and a ubiquitin-interacting motif of S5a. J Mol Biol 368: 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Gordon C (2004) Integral UBL domain proteins: a family of proteasome interacting proteins. Semin Cell Dev Biol 15: 247–259 [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S (2008) Shared principles in NF-κB signaling. Cell 132: 344–362 [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A (1998) The ubiquitin system. Annu Rev Biochem 67: 425–479 [DOI] [PubMed] [Google Scholar]
- Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP (2005) Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6: 610–621 [DOI] [PubMed] [Google Scholar]
- Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S (2006) Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol 13: 272–277 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (1995) Ubiquitin, proteasomes, and the regulation of intracellular protein degradation. Curr Opin Cell Biol 7: 215–223 [DOI] [PubMed] [Google Scholar]
- Hochstrasser M (2006) Lingering mysteries of ubiquitin-chain assembly. Cell 124: 27–34 [DOI] [PubMed] [Google Scholar]
- Hoeller D et al. (2006) Regulation of ubiquitin-binding proteins by monoubiquitylation. Nat Cell Biol 8: 163–169 [DOI] [PubMed] [Google Scholar]
- Hoeller D, Hecker CM, Wagner S, Rogov V, Dotsch V, Dikic I (2007) E3-independent monoubiquitination of ubiquitin-binding proteins. Mol Cell 6: 891–898 [DOI] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell 21: 737–748 [DOI] [PubMed] [Google Scholar]
- Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Hofmann K, Walters K, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iha H et al. (2008) Inflammatory cardiac valvulitis in TAX1BP1-deficient mice through selective NF-κB activation. EMBO J 27: 629–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F, Hecker CM, Rozenknop A, Nordmeier RD, Rogov V, Hofmann K, Akira S, Dotsch V, Dikic I (2007) Involvement of the ubiquitin-like domain of TBK1/IKK-i kinases in regulation of IFN-inducible genes. EMBO J 26: 3451–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Kee Y, Lyon N, Huibregtse JM (2005) The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J 24: 2414–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Ann Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL (2007) Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem 282: 17375–17386 [DOI] [PubMed] [Google Scholar]
- Kirkpatrick DS, Denison C, Gygi SP (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol 7: 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol 8: 700–710 [DOI] [PubMed] [Google Scholar]
- Knipscheer P, van Dijk WJ, Olsen JV, Mann M, Sixma TK (2007) Noncovalent interaction between Ubc9 and SUMO promotes SUMO chain formation. EMBO J 26: 2797–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H (2008) Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10: 547–555 [DOI] [PubMed] [Google Scholar]
- Lamothe B, Besse A, Campos AD, Webster WK, Wu H, Darnay BG (2007) Site-specific Lys 63-linked tumor necrosis factor receptor-associated factor 6 auto-ubiquitination is a critical determinant of IκB kinase activation. J Biol Chem 282: 4102–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Tu D, Brunger AT, Ye Y (2007) A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446: 333–337 [DOI] [PubMed] [Google Scholar]
- Matic I, van Hagen M, Schimmel J, Macek B, Ogg SC, Tatham MH, Hay RT, Lamond AI, Mann M, Vertegaal AC (2008) In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol Cell Proteomics 7: 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, Yarden Y (2003) Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem 278: 21323–21326 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D, Riezman H (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315: 201–205 [DOI] [PubMed] [Google Scholar]
- Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP (2003) A proteomics approach to understanding protein ubiquitination. Nat Biotechnol 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ (2003) Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta 23: 55–72 [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM (2002) Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem 277: 8951–8959 [DOI] [PubMed] [Google Scholar]
- Raasi S, Varadan R, Fushman D, Pickart CM (2005) Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol 12: 708–714 [DOI] [PubMed] [Google Scholar]
- Riley DA, Bain JL, Ellis S, Haas AL (1988) Quantitation and immunocytochemical localization of ubiquitin conjugates within rat red and white skeletal muscles. J Histochem Cytochem 36: 621–632 [DOI] [PubMed] [Google Scholar]
- Scheffner M, Whitaker NJ (2003) Human papillomavirus-induced carcinogenesis and the ubiquitin-proteasome system. Semin Cancer Biol 13: 59–67 [DOI] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters K, Groll M (2008) Ubiquitin docking at the proteasome via a novel PH domain interaction. Nature (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Suzuki T, Chiba T (1990) The ligation systems for ubiquitin and ubiquitin-like proteins. Mol Cells 265: 503–512 [PubMed] [Google Scholar]
- Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276: 35368–35374 [DOI] [PubMed] [Google Scholar]
- Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 [DOI] [PubMed] [Google Scholar]
- Thien CB, Langdon WY (2005) c-Cbl and Cbl-b ubiquitin ligases: substrate diversity and the negative regulation of signalling responses. Biochem J 391: 153–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA (2005) Mechanism of Lys 48-linked polyubiquitin chain recognition by the Mud1 UBA domain. EMBO J 24: 3178–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C (2001) Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105: 711–720 [DOI] [PubMed] [Google Scholar]
- Varadan R, Walker O, Pickart C, Fushman D (2002) Structural properties of polyubiquitin chains in solution. J Mol Biol 324: 637–647 [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D (2004) Solution conformation of Lys 63-linked diubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279: 7055–7063 [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D (2005) Structural determinants for selective recognition of a Lys 48-linked polyubiquitin chain by a UBA domain. Mol Cell 18: 687–698 [DOI] [PubMed] [Google Scholar]
- Varelas X, Ptak C, Ellison MJ (2003) Cdc34 self-association is facilitated by ubiquitin thiolester formation and is required for its catalytic activity. Mol Cell Biol 23: 5388–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S et al. (2008) Ubiquitin binding mediates the NF-κB inhibitory potential of ABIN proteins. Oncogene [doi:10.1038/sj.onc.1211042] [DOI] [PubMed] [Google Scholar]
- Wang M, Cheng D, Peng J, Pickart CM (2006) Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J 25: 1710–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchman RL, Gordon C, Mayer RJ (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol 6: 599–609 [DOI] [PubMed] [Google Scholar]
- Windheim M, Peggie M, Cohen P (2008) Two different classes of E2 ubiquitin-conjugating enzymes are required for the mono-ubiquitination of proteins and elongation by polyubiquitin chains with a specific topology. Biochem J 409: 723–729 [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD (2006) Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation. Nat Cell Biol 8: 398–406 [DOI] [PubMed] [Google Scholar]
- Wu-Baer F, Lagrazon K, Yuan W, Baer R (2003) The BRCA1/BARD1 heterodimer assembles polyubiquitin chains through an unconventional linkage involving lysine residue K6 of ubiquitin. J Biol Chem 278: 34743–34746 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Sundqvist A, Nakamura A, Shen L, Botting C, Hay RT (2008) Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep 9: 280–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Raasi S, Fushman D (2008) Affinity makes the difference: nonselective interaction of the UBA domain of ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol 377: 162–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Wu CJ, Zhao Y, Ashwell JD (2007) Optineurin negatively regulates TNFα-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Curr Biol 17: 1438–1443 [DOI] [PubMed] [Google Scholar]