Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications (original) (raw)
Abstract
The biosynthesis of RNA includes its post-transcriptional modifications, and the crucial functions of these modifications have supported their conservation within all three kingdoms. For example, the modifications located within or adjacent to the anticodon of the transfer RNA (tRNA) enhance the accuracy of codon binding, maintain the translational reading frame and enable translocation of the tRNA from the A-site to the P-site of the ribosome. Although composed of different chemistries, the more than 70 known modifications of tRNA have in common their ability to reduce conformational dynamics, and to bring order to the internal loops and hairpin structures of RNA. The modified nucleosides of the anticodon domain of tRNA restrict its dynamics and shape its architecture; therefore, the need of the ribosome to constrain or remodel each tRNA to fit the decoding site is diminished. This concept reduces an entropic penalty for translation and provides a physicochemical basis for the conservation of RNA modifications in general.
Keywords: entropy, order, pre-structured, rRNA, tRNA
Introduction
The efficient and exact translation of genomic information into proteins is of fundamental importance in biology. Some 40 different transfer RNAs (tRNAs) decode all of the messenger RNA (mRNA) codons into the 22 amino acids of proteins, with the exception of the translational stop codons (Fig 1; Ambrogelly et al, 2007; Szymański & Barciszewski, 2007). In his ‘wobble hypothesis', Francis Crick proposed that the many anticodons of different tRNAs underwent an induced fit to conform to the needs of the ribosome for a uniform structure in binding to the mRNA codons (Crick, 1966). However, the remodelling of the anticodon architecture produces an entropic penalty that would necessitate the repeated investment of energy for each tRNA every time it responds to its codon; thereby the accuracy and speed of translation would be compromised. In the Gibbs–Helmholtz equation, Δ_G_ = Δ_H_ – T_Δ_S, G represents the Gibbs free energy, H represents the enthalpy and T represents the absolute temperature. The entropy term Δ_S_ is a measure of the motional energy—which is often equated with disorder—within a system (for example, tRNA). A decrease in entropy is affected by a decrease in motional energy. For more than 30 years, modifications have been known to restrict the motional dynamics of tRNAs, and the anticodon-domain modifications are most effective (Fig 2; Schmidt et al, 1987). Modified nucleosides also alter the architecture of the anticodon stem and loop domain (ASL) towards that of the canonical structure with a ‘U-turn', as first seen in the X-ray crystallographic structure of yeast tRNAPhe (Fig 2; Kim et al, 1974). The crystallography structures were confirmed by solution nuclear magnetic resonance (NMR) and thermodynamic studies on the ASLs of Escherichia coli and mammalian tRNAs (Vendeix et al, 2008; Agris et al, 2007; Stuart et al, 2003; Cabello-Villegas et al, 2002; Sundaram et al, 2000). In restricting the conformational dynamics of the ASL of tRNA, modifications decrease the Δ_S_. Here, I propose that this lowering of entropic energy and shaping of the anticodon reduces the need for the ribosome continually to remodel each anticodon loop for codon binding.
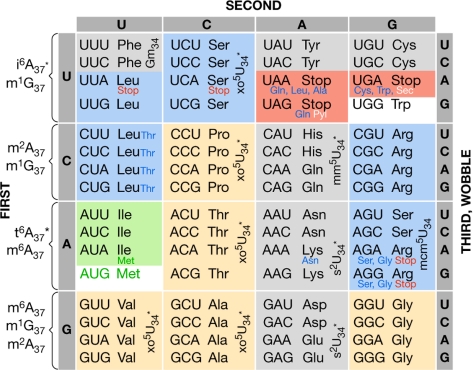
Figure 1.
Universal genetic code. The 64 codes are associated with the transfer RNA (tRNA) modifications that are important for decoding and/or translocation. Twofold degenerate amino-acid codes are highlighted in grey and fourfold degenerate codes are highlighted in tan. Amino acids with six codons are highlighted in blue. The threefold degenerate codons of Ile are highlighted in green, whereas the single codons of Met and Trp are highlighted in white. The three stop codons are highlighted in orange. Non-canonical codon use by some organisms and the mitochondrion is shown by using a small font for the amino acids (blue) or translational stop codons (red). The modified nucleoside abbreviations are defined in the text. Selenocysteine (Sec) and pyrrolysine (Pyl) codons are denoted in white. In the mitochondrion, tRNAMet responds to AUG and AUA, which is not used as an Ile codon (Agris et al, 2007; Szymański & Barciszewski, 2007; Björk et al, 1987).
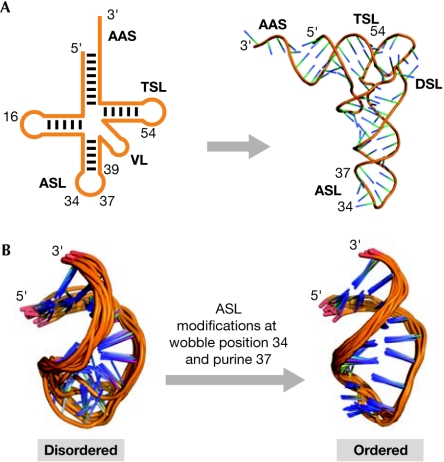
Figure 2.
Modifications order the anticodon stem and loop domain of transfer RNA. (A) The secondary structure and tertiary folding of cytoplasmic transfer RNAs (tRNAs). The physical and functional domains of the tRNA structure are the amino-acid-accepting stem (AAS), and the stem and loop domains designated dihydrouridine (DSL), anticodon (ASL), variable (VL) and thymidine (TSL). tRNA can fold into its tertiary structure before modification. pre-tRNA will fold with the help of Mg2+ and the aid of the most conserved of the modifications (Helm, 2006; Nobles et al, 2001). These modifications occur outside of the anticodon domain. (B) Modification of the anticodon stem and loop domain (ASL) of tRNA and its effect on dynamics. The ASL (left) is unmodified and disordered. Extensive modifications at wobble position 34 and purine 37 restrain the dynamics of the anticodon loop, and direct its conformation towards that of the canonical structure shown on the right.
Bifunctional tRNA architecture
In general, tRNA isoacceptors are aminoacylated with one amino acid with great accuracy; therefore, each aminoacyl-tRNA synthetase recognizes one or more tRNA species as being chemically and structurally distinct (Ibba & Söll, 2000). However, the accuracy is considerably improved by the ability of some synthetases to proofread and then to edit, if incorrect, the amino acid that is bound to the tRNA (Schimmel & Ribas de Pouplana, 2001). During aminoacylation, the distal portion of the amino-acid-accepting stem (AAS) of tRNA (Fig 2) is distorted with the 3′-terminal adenosine placed inside the active site of the enzyme (Fukunaga & Yokoyama, 2005). In recognizing their cognate tRNAs, some synthetases distort the anticodon loop (Nakanishi et al, 2005). This suggests a mechanism by which a transient change in structure communicates through the tRNA to the active site of the enzyme that the cognate anticodon has been recognized (Ghosh & Vishveshwara, 2007; Rogers et al, 1993). Once aminoacylated, the coaxial stems of the AAS and the ribothymidine stem and loop domain (TSL) of all tRNAs with the exception of the initiator methionyl-tRNAMet are bound by elongation factor-GTP, and the tRNA transported to the decoding site of the ribosome, the A-site (Fig 3). The isoaccepting tRNAs then decode their appropriate amino-acid codons on the ribosome. Here, I focus on the modifications of the anticodon domain and their physicochemical contributions to codon binding.
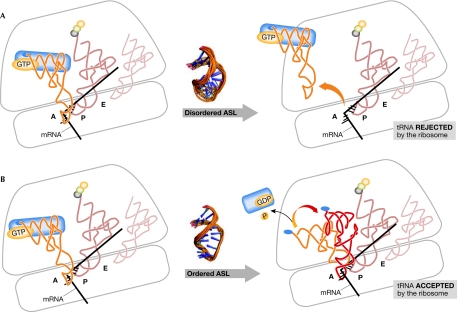
Figure 3.
Anticodon recognition and transfer RNA accommodation on the ribosome. (A) The complex of aminoacylated transfer RNA (tRNA) bound to elongation factor/GTP enters the A-site (left). The anticodon of the tRNA makes contact with the messenger RNA (mRNA) codon; however, the tRNA with a disordered anticodon stem and loop domain (ASL) cannot contact the codon correctly and is rejected by the ribosome (right). An aminoacyl-tRNA having successfully received the growing peptide now occupies the P-site, while the tRNA from which it received the nascent protein occupies the ribosome's exit or E-site. (B) Accommodation of aminoacyl-tRNA in the A-site. A complex of aminoacylated tRNA with elongation factor and GTP enters the A-site (left). Modifications have restricted the dynamics and shaped the architecture of the ASL of the tRNA. With recognition that the tRNA has the correct anticodon, the 16S ribosomal RNA (rRNA) nucleosides A1492, A1493 and G530 hydrogen bond to the backbones of the mRNA and the anticodon. The small ribosomal subunit and the tRNA undergo conformational changes. The conformation of the tRNA above the ASL changes, but the architecture of the ASL and its interaction with the codon remain unchanged (right). GTP is hydrolysed and the elongation factor-GDP leaves the ribosome.
ASLs vary in sequence and modification; yet, every anticodon domain entering the ribosomal A-site must have a similar global conformation. This architecture conforms to the structural restraints of the ribosome, and thereby maintains a rapid and consistent processivity for translation. The ribosome sustains a notable rate of synthesis (20–40 peptide bonds per second; reviewed in Lovmar & Ehrenberg, 2006). In addition, mRNA is translated with high fidelity (1 error per 1 × 103−1 × 104 amino acids incorporated; Kurland, 1992). The accuracy of protein synthesis coupled with its speed requires a constant evaluation of the precision with which a tRNA anticodon is selected (Cochella et al, 2007). The precision of the anticodon–codon interaction is evaluated by kinetic and induced-fit mechanisms of proofreading in the decoding site of the ribosome. On entering the A-site of the ribosome and binding to the codon, the canonical shape of the ASL of the tRNA remains relatively unchanged (Valle et al, 2003; Steitz, 2008; Vendeix et al, 2008). There is little difference in anticodon conformation between the solution structures of the ASLs with their natural modifications and the same ASLs on the ribosome in response to their synonymous codons (Vendeix et al, 2008; Weixlbaumer et al, 2007; Murphy et al, 2004). Therefore, it seems that the architecture of the seven-member anticodon loop of all tRNAs is pre-structured and ordered by modification to conform to the A-site requirements of the ribosome. Posttranscriptional modifications can structure the ASL and place a restraint on its dynamics that reduces its entropic energy. However, the anticodon retains a uniqueness of chemistry, including that contributed by modifications, which enables it to respond to its respective codons.
The cognate anticodon readily forms a minihelix with the codon. The minihelix is evaluated and stabilized by the eight or nine hydrogen bonds between A1492, A1493 and G530 of 16S ribosomal RNA (rRNA) and the backbones of the codon and anticodon (Ogle et al, 2001). As A1492, A1493 and G530 are unable to form many of these hydrogen bonds when the non-cognate anticodon interacts with the codon, the non-cognate tRNA is released (Fig 3). When these interactions are successful, the small subunit converts from an ‘open' to a ‘closed' conformation. However, near-cognate anticodons—for example, those with a single mismatch in formation of the second or third base pair with the codon—might still trigger a conformational change in the small subunit. After the anticodon–codon minihelix is proofread, the conformation of tRNA is altered above the ASL—that is, above nucleosides 27 to 43 (Fig 3; Valle et al, 2003; Frank et al, 2005). A second proofreading event and substantial barrier is surmounted in the conformational change of the ternary complex, GTP hydrolysis and the release of the elongation factor with GDP (Fig 3; Cochella et al, 2007). For this to occur, a correct interaction of the anticodon with the codon is communicated from the ASL to the structural neighbourhood of the ‘hinge' region of the tRNA where the conformational change takes place and GTP is hydrolysed (Cochella et al, 2007).
Modifications order the anticodon of tRNAs
Posttranscriptional modifications are one of the processing events that result in functional tRNA molecules. tRNAs have more than 70 chemically distinct modifications (Rozenski et al, 1999; Agris, 1996). tRNA modifications are found across the kingdoms and throughout evolution, although at the expense of maintaining a large amount of genomic information, and by means of the use of energy and materials (Agris, 1996, 2004). Modified nucleosides are found in all of the biological and structural domains of tRNA, the AAS, and the dihydrouridine stem and loop domain (DSL), ASL, variable loop domain (VL) and TSL (Fig 2). Most modified nucleosides are localized to internal and terminal loops (Sprinzl & Vassilenko, 2005). Modifications contribute to tRNA folding, Mg2+ binding, intron removal, protein recognition, codon recognition, fidelity of the translational reading frame and other functions in translation (Fig 1; Agris et al, 2007; Agris, 2004; Björk et al, 1987). In general, tRNA genes are transcribed, sized, spliced and modified into functional RNA. Some of the site-specific modifications of tRNA are highly conserved in type and location, and their syntheses precede the sizing of the tRNA; however, modification before sizing is not obligatory. By contrast, the synthesis of pseudouridine in the anticodon of tRNATyr, Ψ35, requires the presence of the intron (Johnson & Abelson, 1983). The precise order of the many nucleoside modifications in the processing of tRNAs is not known, but, with few exceptions, they contribute to folding, thermal stability and restricted dynamics (Helm, 2006).
Modifications at the anticodon wobble position 34 (such as inosine) and at the conserved purine 37, 3′-adjacent to the anticodon (such as N6-isopentenyladenosine) were the first to be associated with the recognition of specific codons by tRNA (Crick, 1966; reviewed in Nishimura & Watanabe, 2006). The anticodon of tRNA resides in a relatively large seven-residue loop. Modifications maintain an open-loop conformation for presentation of the anticodon to the codon by negating canonical and non-canonical intra-loop hydrogen bonding (Olejniczak & Uhlenbeck, 2006; Dao et al, 1994). Such an open seven-membered loop would be expected to be dynamic; however, several investigations (see Agris, 2004, and references therein) now support the hypothesis (Agris, 1991) that numerous modifications at wobble position 34 and conserved purine 37 of tRNA shape the architecture of the ASL and constrain its dynamics, decreasing the motional (entropic) energy (Table 1). The anticodon loops of many tRNAs are dynamic and disordered in solution, particularly those that are pyrimidine-rich (Agris, 1991). The dipole–dipole and hydrophobic interactions of pyrimidines are far less stable than those of purines (Saenger, 1984). When anticodon loop bases stack as a consequence of modifications at wobble position 34 and purine 37, the resulting structure is ordered and the conformational dynamics of the anticodon are restricted (Fig 2). Base stacking removes the hydrophobic nucleobase from the environment of the aqueous solvent and, in so doing, the surrounding water molecules are able to become more organized with hydrogen bonding to the ASL backbone. Therefore, modification results in an ordered ASL and a lowering of the entropy.
Table 1.
Modified nucleoside relative entropic contributions to transfer RNAs
| Modified nucleosidea | ΔΔ_SR_bR | References |
|---|---|---|
| D17,18 | −/− − | Sipa et al, 2007 |
| Cm32 | − | Ashraf et al, 2000 |
| f5C34 | − − | H. Lusic, P.F. Agris & A. Deiters, unpublished data |
| Gm34 | +/− | Schmidt et al, 1987; Ashraf et al, 2000 |
| Q34 | + | Morris et al, 1999 |
| cmo5U34 | ++ | Vendeix et al, 2008 |
| s2U8 | ++ | Testa et al, 1999; Sipa et al, 2007 |
| mcm5s2U34 | + | Bajji & Davis, 2000 |
| mcm5U34 | ++ | Bajji & Davis, 2000 |
| mnm5s2U34 | + | Durant et al, 2005; Vendeix et al, 2008c |
| mnm5U34 | ++ | Yarian et al, 2000 |
| i6A37 | + | Cabello-Villegas et al, 2002;Kierzek & Kierzek, 2001 |
| t6A37 | + | Yarian et al, 2000; Vendeix et al, 2008c |
| ms2t6A37 | +/− | Durant et al, 2005; Vendeix et al, 2008c |
| m1G37 | + | Ashraf et al, 2000 |
| yW37 | + | Schmidt et al, 1987 |
| Ψ39 | + | Yarian et al, 1999; Ashraf, 2000;Bajji & Davis, 2000;Tworowska & Nikonowicz, 2006 |
| m5C40 | − | Ashraf et al, 2000 |
| m5U54 (T54) | 0 | Schmidt et al, 1987; Sengupta et al, 2000 |
| Ψ55 | 0 | Sengupta et al, 2000 |
The modifications of uridine at wobble position 34 of tRNA are of particular interest because they contribute a degree of order (Table 1) and are crucial to the specificity of codon recognition (Fig 1). Few sequenced cytoplasmic tRNAs show an unmodified U34 (Sprinzl & Vassilenko, 2005). The 2-thiouridine (s2U34) derivatives reinforce base pairing to codons ending in A. They are strictly gauche+, C3′-endo and anti, and are found in tRNAs with weakly stacked pyrimidine-rich anticodons, such as those for Lys, Glu and Gln. The 5-methylaminomethyluridine (mnm5U34) and 5-methoxycarbonylmethyluridine (mcm5U34) contribute order in the absence of the thio-moiety (Table 1) and are found in pyrimidine-rich anticodon domains. They restrict tRNA to recognition of codons ending in A and G, and are important for translocation (Phelps et al, 2004; Agris, 2004). Uridine 5-oxyacetic acid (cmo5U34) restricts motional dynamics within the anticodon domain (Table 1); however, its function contrasts greatly with that of the other uridine modifications. The cmo5U34, and presumably other 5-oxy-derivatives (xo5U34, where x is H, methyl or acetic acid), enables the one isoaccepting tRNA species for each of Ala, Leu, Pro, Ser, Thr and Val that has the modification to read three and sometimes all four of the synonymous codons, NNA/G/U/C (Näsvall et al, 2004, 2007; Weixlbaumer et al, 2007; Vendeix et al, 2008). With the deletion of two of the three genes for the tRNAPro isoacceptors in Salmonella, the one remaining tRNAPro with a cmo5U34 was able to maintain cell viability (Näsvall et al, 2004). However, in the presence of all three isoacceptors, the removal of one of the modification enzymes in the synthetic pathway for cmo5U34 led to poor growth of the culture. In addition, this modification is crucial for the reading of the Ala, Pro and Val codons ending in G (Näsvall et al, 2007; Vendeix et al, 2008). Therefore, cmo5U34 expands the ability of these six tRNAs to read theoretically 24 of the 61 amino-acid codes. In contrast to cmo5U34, which occurs in six different sets of isoaccepting tRNAs, 5-formylcytidine (f5C34) contributes disorder to the ASL of the one mitochondrial tRNAMet (Table 1; Païs de Barros et al, 1996). Mitochondrial tRNAMet decodes both the AUA and AUG codons. The f5C34 might allow the mitochondrial tRNAMet to respond to the two codons in both the P-sites and the A-sites of the ribosome. The 2′-O-methylations of G and C (Gm34 and Cm32) seem to introduce some dynamics to the 5′-side of purine-rich anticodon loops that already have considerable order owing to the stacking properties of the purines (Table 1). A polymer composed of the parent compound (7-deazaguanylic acid) of the modified nucleoside queosine, Q, showed few thermodynamic differences when compared to polyG (Seela et al, 1982). However, molecular-dynamics simulations of the anticodon-loop structures of tRNAs containing Q (Asn, Asp, His and Tyr) indicated that the modification restricts anticodon dynamics (Morris et al, 1999).
Purine-37 modifications are 3′-adjacent to the anticodon and can be complex. They negate intra-loop base pairing and thereby ensure the correct width to the anticodon loop (Dao et al, 1994). In negating intra-loop base pairing, the purine-37 modification lowers the melting temperature of an ASL compared with that of the unmodified RNA (Stuart et al, 2000). However, the augmented stacking properties of these modifications order the 3′-side of the anticodon domain (Table 1). This is apparent not only in the X-ray crystallographic structures of tRNAs, but also in the solution-structure analyses of modified ASLs in comparison to their unmodified counterparts. The diverse chemistries of N6-methyladenosine, N6-isopentenyladenosine, N6-threonylcarbamoyladenosine, 2-methylthio-N6-threonylcarbamoyladenosine, N1-methylguanosine and wyeosine (m6A37, i6A37, t6A37, ms2t6A37, m1G37 and yW37) bring order to the ASL while negating intra-loop base pairing (Table 1). However, the purine 37 is the only nucleoside within the anticodon domain that shows any substantial movement from its geometry in the unbound ASL to that of the ASL bound to the codon in the A-site (Vendeix et al, 2008; Weixlbaumer et al, 2007; Murphy et al, 2004). The purine 37 nucleoside moves to a position above the third base of the anticodon and the first base of the codon (Weixlbaumer et al, 2007; Murphy et al, 2004). The position maintains the 3′-stack of the anticodon domain and, at the same time, becomes a hydrophobic platform that stabilizes the first base pair of the anticodon–codon interaction. Many of the modifications of purine 37 increase its hydrophobicity (Agris, 1996) and, therefore, enhance its ability to stabilize this crucial first base pair—particularly when the pair is an A•U or U•A. Almost without exception, tRNAs that have a A36 also have an i6A37 or a derivative at position 37; tRNAs that have a U36 frequently have a t6A37 or one of its derivatives at position 37. This observation holds for the tRNAs of Phe, Leu, Ser, Tyr, Cys and Trp, and of Ile, Met, Thr, Asn, Lys and Ser, respectively; this tendency is therefore related to decoding and not to the recognition of the tRNAs by cognate aminoacyl-tRNA synthetase. The modified purine 37 facilitates codon binding (Agris, 2004) and is important for maintaining the translational reading frame (Urbonavicius et al, 2001).
The contribution of modified nucleosides to the ordering of RNA structure is evident for modifications in loops, whereas their enthalpic (Δ_H_) contributions are readily observed in the base-paired regions of the RNA. Investigations of RNA duplexes show that certain modifications enhance the Δ_H_ contribution to the thermal stability of base pairing, such as the s2U in pairing with A (Testa et al, 1999). However, significant changes in the enthalpy term of the free-energy determination are not always evident from the study of unpaired and naturally occurring modifications in the anticodon loop (Ashraf et al, 2000) or the unpaired terminus of a duplex (Kierzek & Kierzek, 2001). For example, when s2U34 at the wobble position of ASLLys was studied, it did not contribute a significant change in enthalpy; however, it did reduce the conformational dynamics of the loop and enhanced the codon binding (Ashraf et al, 1999). Ψ occurs in the anticodon loop at position 38, in the anticodon stem at residue 39 and sometimes at 31, 35 or 40. The entropic contribution of Ψ seems to be dependent on location, stem or loop, and context. Ψ39, in the ASL stem and adjacent to the loop (Fig 2), has been most extensively studied. It contributes to the thermal stability of the anticodon stem of tRNA and to the order of the anticodon domain (Table 1); however, in the absence of other modifications, it does not contribute to codon binding (Yarian et al, 1999). Ψ55 is commonly found in the TSL with T54. Although extremely common, neither Ψ55 nor T54 results in a significant entropic contribution to the tRNA (Table 1). T54, which makes a reverse Hoogsten pair with A58—often N1-methyladenosine—across the TSL loop, modestly increased the thermal stability of the TSL (Sengupta et al, 2000). Consistent with our understanding of the contribution of the 2-thio group to the stability of U•A base pairs, tRNAs of thermophilic organisms are further modified from T54 to s2T54 and show an increased thermal stability (Shigi et al, 2002). The modification 5-methylcytosine is found at position 40 (m5C40) within the ASL stem, and at position 49 of the TSL stem. The modifications do not seem to contribute order to either of these domains (Table 1). In contrast to the many modifications that constrain the molecular dynamics of tRNA, dihydrouridine located in the DSL (Fig 2) increases molecular motion (Table 1). D, which is the only non-aromatic nucleoside found in nucleic acids, is unable to stack and is so strongly C2′-endo that it transfers the conformation to nucleosides that are 3′-adjacent (Stuart et al, 1996). The tRNAs of the psychrophilic organisms growing at temperatures of – 5 °C to 12 °C are supplemented with D in comparison to mesophiles (Dalluge et al, 1997), therefore taking advantage of the contribution of the nucleoside to dynamics.
Conclusion
The wobble position and purine-37 modifications reduce conformational dynamics and shape the anticodon-domain architecture. However, their distinctive chemistries either restrict codon recognition to one or two codons, or expand recognition to three or even four synonymous codons. With an ordered structure, the recognition and binding of the tRNA anticodon to cognate and wobble codons occurs accurately, rapidly and with a reduced entropic penalty to the ribosome. The saving of energy and an efficient use of time are important for the cell to respond as quickly as possible to signals and environmental changes. Modifications are not exclusive to tRNA, and their evolution and maintenance was an investment that reduced the need for a repetitive expenditure of energy in moulding the functional form of RNA.

Paul F. Agris
Acknowledgments
I acknowledge the advice of Dr F.A.P. Vendeix, and the contributions of Drs D. Davis, A. Deiters, B. Nawrot and E. Nikonowicz in helping to collate the thermodynamic data. Studies cited from P.F.A. and co-workers were supported by the National Science Foundation (MCB0548602) and the National Institutes of Health (2-RO1-GM23037).
References
- Agris PF (1991) Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 73: 1345–1349 [DOI] [PubMed] [Google Scholar]
- Agris PF (1996) The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Progr Nucl Acid Res Mol Biol 53: 79–129 [DOI] [PubMed] [Google Scholar]
- Agris PF (2004) Decoding the genome. A modified view. Nucleic Acids Res 32: 223–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Vendeix F, Graham WD (2007) tRNA's Wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 [DOI] [PubMed] [Google Scholar]
- Ambrogelly A, Palioura S, Söll D (2007) Natural expansion of the genetic code. Nat Chem Biol 3: 29–35 [DOI] [PubMed] [Google Scholar]
- Ashraf SS, Sochacka E, Cain R, Guenther R, Malkiewicz A, Agris PF (1999) Single atom modification (O®S) of tRNA confers ribosome binding. RNA 5: 188–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf SS, Guenther RH, Ansari G, Malkiewicz A, Sochacka E, Agris PF (2000) Role of modified nucleosides of yeast tRNAPhe in ribosomal binding. Cell Biochem Biophys 33: 241–252 [DOI] [PubMed] [Google Scholar]
- Bajji AC, Davis DR (2000) Synthesis and biophysical characterization of tRNA(Lys,3) anticodon stem–loop RNAs containing the mcm5s2U nucleoside. Org Lett 2: 3865–3868 [DOI] [PubMed] [Google Scholar]
- Björk GR, Ericson JU, Gustafsson CED, Hagervall TG, Jonsson YH, Wikstrom PM (1987) Transfer RNA modification. Ann Rev Biochem 56: 263–285 [DOI] [PubMed] [Google Scholar]
- Cabello-Villegas J, Winkler ME, Nikonowics EP (2002) Solution conformations of unmodified and A37 N6-dimethylallyl modified anticodon stem–loops of Escherichia coli tRNAPhe. J Mol Biol 319: 1015–1034 [DOI] [PubMed] [Google Scholar]
- Cochella L, Brunelle JL, Green R (2007) Mutational analysis reveals two independent molecular requirements during transfer RNA selection on the ribosome. Nat Struct Mol Biol 14: 30–36 [DOI] [PubMed] [Google Scholar]
- Crick FHC (1966) Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 19: 548–555 [DOI] [PubMed] [Google Scholar]
- Dalluge JJ, Hamamoto T, Horikoshi K, Morita RY, Stetter KO, McCloskey JA (1997) Posttranscriptional modification of tRNA in psychrophilic bacteria. J Bacteriol 179: 1918–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao V, Guenther R, Malkiewicz A, Nawrot B, Sochacka E, Kraszewski A, Jankowska J, Everett K, Agris PF (1994) Ribosome binding of DNA analogs of tRNA requires base modifications and supports the “extended anticodon”. Proc Natl Acad Sci USA 91: 2125–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durant PC, Bajji AC, Sundaram M, Kuman RK, Davis DR (2005) Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: the effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 44: 8078–8089 [DOI] [PubMed] [Google Scholar]
- Frank J, Sengupta J, Gao H, Li W, Valle M, Zavialov A, Ehrenberg M (2005) The role of tRNA as a molecular spring in decoding, accommodation, and peptidyl transfer. FEBS Lett 579: 959–962 [DOI] [PubMed] [Google Scholar]
- Fukunaga R, Yokoyama S (2005) Aminoacylation complex structures of leucyl-tRNA synthetase and tRNALeu reveal two modes of discriminator-base recognition. Nat Struct Mol Biol 12: 915–922 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Vishveshwara S (2007) A study of communication pathways in methionyl-tRNA synthetase by molecular dynamics simulations and structure network analysis. Proc Natl Acad Sci USA 104: 15711–15716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M (2006) Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res 34: 721–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Söll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69: 617–650 [DOI] [PubMed] [Google Scholar]
- Johnson PF, Abelson J (1983) The yeast tRNATyr gene intron is essential for correct modification of its tRNA product. Nature 302: 681–687 [DOI] [PubMed] [Google Scholar]
- Kierzek E, Kierzek R (2001) Influence of N6-isopentenyladenosine (i(6)A) on thermal stability of RNA duplexes. Biophys Chem 91: 135–140 [DOI] [PubMed] [Google Scholar]
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AHJ, Seeman NC, Rich A (1974) Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 185: 435–440 [DOI] [PubMed] [Google Scholar]
- Kurland CG (1992) Translational accuracy and the fitness of bacteria. Annu Rev Genet 26: 29–50 [DOI] [PubMed] [Google Scholar]
- Lovmar M, Ehrenberg M (2006) Rate, accuracy and cost of ribosomes in bacterial cells. Biochimie 88: 951–961 [DOI] [PubMed] [Google Scholar]
- Morris RC, Brown KG, Elliott MS (1999) The effect of queuosine on tRNA structure and function. J Biomol Struct Dyn 16: 757–774 [DOI] [PubMed] [Google Scholar]
- Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF (2004) The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 11: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Nakanishi K, Ogiso Y, Nakama T, Fukai S, Nureki O (2005) Structural basis for anticodon recognition by methionyl-tRNA synthetase. Nat Struct Mol Biol 12: 931–932 [DOI] [PubMed] [Google Scholar]
- Näsvall SJ, Chen P, Björk GR (2004) The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA 10: 1662–1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Näsvall SJ, Chen P, Björk R (2007) The wobble hypothesis revisited: uridine-5-oxyacetic acid is critical for reading of G-ending codons. RNA 13: 2151–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura S, Watanabe K (2006) The discovery of modified nucleosides from the early days to the present: a personal perspective. J Biosci 31: 465–475 [DOI] [PubMed] [Google Scholar]
- Nobles KN, Yarian CS, Guenther RH, Agris PF (2001) Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res 30: 4751–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle JM, Brodersen DE, Clemons WM Jr, Tarry MJ, Carter AP, Ramakrishnan V (2001) Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897–902 [DOI] [PubMed] [Google Scholar]
- Olejniczak M, Uhlenbeck OC (2006) tRNA residues that have coevolved with their anticodon to ensure uniform and accurate codon recognition. Biochimie 88: 943–950 [DOI] [PubMed] [Google Scholar]
- Païs de Barros JP, Keith G, El Adlouni C, Glasser AL, Mack G, Dirheimer G, Desgrés J (1996) 2′-O-methyl-5-formylcytidine (f5Cm), a new modified nucleotide at the ‘wobble' of two cytoplasmic tRNAs Leu (NAA) from bovine liver. Nucleic Acids Res 24: 1489–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps SS, Malkiewicz A, Agris PF, Joseph S (2004) Modified nucleotides in tRNA(Lys) and tRNA(Val) are important for translocation. J Mol Biol 338: 439–444 [DOI] [PubMed] [Google Scholar]
- Rogers MJ, Weygand-Durasević I, Schwob E, Sherman JM, Rogers KC, Adachi T, Inokuchi H, Söll D (1993) Selectivity and specificity in the recognition of tRNA by E. coli glutaminyl-tRNA synthetase. Biochimie 75: 1083–1090 [DOI] [PubMed] [Google Scholar]
- Rozenski J, Crain PF, McCloskey JA (1999) The RNA Modification Database: 1999 update. Nucleic Acids Res 27: 196–197. http://library.med.utah.edu/RNAmods [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger W (1984) Principles of Nucleic Acid Structure. New York, NY, USA: Springer [Google Scholar]
- Schimmel P, Ribas de Pouplana L (2001) Formation of two classes of tRNA synthetases in relation to editing functions and genetic code. Cold Spring Harb Symp Quant Biol 66: 161–166 [DOI] [PubMed] [Google Scholar]
- Schmidt PG, Sierzputowska-Gracz H, Agris PF (1987) Internal motions in yeast phenylalanine transfer RNA from 13C NMR relaxation rates of modified base methyl groups: a model-free approach. Biochemistry 26: 8529–8534 [DOI] [PubMed] [Google Scholar]
- Seela F, Tran-Thi QH, Franzen D (1982) Poly(7-deazaguanylic acid), the homopolynucleotide of the parent nucleoside of queuosine. Biochemistry 21: 4338–4343 [DOI] [PubMed] [Google Scholar]
- Sengupta R, Vainauskas S, Yarian C, Sochacka E, Malkiewicz A, Guenther RH, Koshlap KM, Agris PF (2000) Modified constructs of tRNA's TYC-domain to probe substrate conformational requirements of m1A58 and m5U54-tRNA methyltransferases. Nucleic Acids Res 28: 1374–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigi N, Suzuki T, Tamakoshi M, Oshima T, Watanabe K (2002) Conserved bases in the TPsi C loop of tRNA are determinants for thermophile-specific 2-thiouridylation at position 54. J Biol Chem 277: 39128–39135 [DOI] [PubMed] [Google Scholar]
- Sipa K, Sochacka E, Kazmierczak-Baranska J, Maszewska M, Janicka M, Nowak G, Nawrot B (2007) Effect of base modifications on structure, thermodynamic stability, and gene silencing activity of short interfering RNA. RNA 13: 1301–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS (2005) Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res 33: D139–D140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA (2008) A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9: 242–253 [DOI] [PubMed] [Google Scholar]
- Stuart JW, Basti MM, Smith WS, Forrest B, Guenther R, Sierzputowska-Gracz H, Nawrot B, Malkiewicz A, Agris PF (1996) Structure of the trinucleotide D-acp3U-A with coordinated Mg2+ demonstrates that modified nucleosides contribute to regional conformations of RNA. Nucleosides Nucleotides 15: 1009–1028 [Google Scholar]
- Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF (2000) Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry 39: 13396–13404 [DOI] [PubMed] [Google Scholar]
- Stuart JW, Koshlap KM, Guenther R, Agris PF (2003) Naturally-occurring modification restricts the anticodon domain conformational space of tRNAPhe. J Mol Biol 334: 901–918 [DOI] [PubMed] [Google Scholar]
- Sundaram M, Durant PC, Davis DR (2000) Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 39: 12575–12584 [DOI] [PubMed] [Google Scholar]
- Szymański M, Barciszewski J (2007) The genetic code 40-years on. Acta Biochim Pol 54: 51–54 [PubMed] [Google Scholar]
- Testa SM, Disney MD, Turner DH, Kierzek R (1999) Thermodynamics of RNA–RNA duplexes with 2- or 4-thiouridines: implications for antisense design and targeting a group I intron. Biochemistry 38: 16655–16662 [DOI] [PubMed] [Google Scholar]
- Tworowska I, Nikonowicz EP (2006) Base pairing within the psi32,psi39-modified anticodon arm of Escherichia coli tRNAPhe. J Am Chem Soc 128: 15570–15571 [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Björk GR (2001) Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J 20: 4863–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M, Zavialov A, Li W, Stagg SM, Sengupta J, Nielsen RC, Nissen P, Harvey SC, Ehrenberg M, Frank J (2003) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Vendeix FAB, Dziergowska A, Gustilo EM, Graham WD, Sproat B, Malkiewicz A, Agris PF (2008) Wobble-position modifications contribute order to tRNA's anticodon for ribosome-mediated codon binding. Biochemistry 47: 6117–6129 [DOI] [PubMed]
- Weixlbaumer A, Murphy FV, Dziergowska A, Malkiewicz A, Vendeix FAP, Agris PF, Ramakrishnan V (2007) Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines. Nat Struct Mol Biol 14: 498–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian CS, Cain R, Basti MM, Ansari G, Guenther RH, Sochacka E, Malkiewicz A, Agris PF (1999) Structural and functional roles of the N1- and N3-protons of Ψ at tRNA's position 39. Nucleic Acids Res 27: 3543–3549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian C, Marszalek M, Sochacka E, Malkiewicz M, Guenther R, Miskiewicz A, Agris PF (2000) tRNALysUUU species require anticodon domain modified nucleosides for Watson–Crick and wobble codon binding. Biochemistry 39: 13390–13395 [DOI] [PubMed] [Google Scholar]