A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation (original) (raw)
Abstract
Decreased expression of specific microRNAs (miRNAs) occurs in human tumors, which suggests a function for miRNAs in tumor suppression. Herein, levels of the miR-17-5p/miR-20a miRNA cluster were inversely correlated to cyclin D1 abundance in human breast tumors and cell lines. MiR-17/20 suppressed breast cancer cell proliferation and tumor colony formation by negatively regulating cyclin D1 translation via a conserved 3′ untranslated region miRNA-binding site, thereby inhibiting serum-induced S phase entry. The cell cycle effect of miR-17/20 was abrogated by cyclin D1 siRNA and in cyclin D1–deficient breast cancer cells. Mammary epithelial cell–targeted cyclin D1 expression induced miR-17-5p and miR-20a expression in vivo, and cyclin D1 bound the miR-17/20 cluster promoter regulatory region. In summary, these studies identify a novel cyclin D1/miR-17/20 regulatory feedback loop through which cyclin D1 induces miR-17-5p/miR-20a. In turn, miR-17/20 limits the proliferative function of cyclin D1, thus linking expression of a specific miRNA cluster to the regulation of oncogenesis.
Introduction
MicroRNAs (miRNAs) are 21–22 nucleotide molecules that regulate the stability or translational efficiency of targeted mRNAs. Derived from nuclear precursor RNAs, initial processing occurs by the internuclease Drosha to release pre-miRNAs of 60–70 nucleotides in length from pri-miRNA. Subsequent transport to the cytoplasm by exportin-5 results in processing by the internuclease Dicer to generate the ∼22-nucleotide mature miRNA (Yi et al., 2003; Lund et al., 2004; Zhang et al., 2004). The base pairing interactions between miRNAs and their target mRNAs, often within the 3′ untranslated region (UTR) of target genes, results in the degradation of target mRNAs (Ambros, 2004; Cullen, 2004) or inhibition of their translation (Lai, 2002). To date, 533 miRNAs have been identified in humans (miRBase Sequence Database version 10.0, released in August 2007; http://microrna.sanger.ac.uk/sequences/). It has been proposed that, as each vertebrate miRNA may bind to as many as 200 gene targets, miRNAs potentially control the expression of about one-third of human mRNAs (Krek et al., 2005).
Several independent lines of evidence support a role for miRNAs in human cancer (Calin et al., 2005; Croce and Calin, 2005, 2006; He et al., 2005, 2007). miRNA-encoding genes are frequently located at fragile sites, as well as in minimal regions of loss of heterozygosity, minimal regions of amplification, and in common breakpoint regions involved in cancers (Calin et al., 2004). Aberrant expression of miRNAs or mutations of miRNA genes have been described in many types of tumors. Let-7 abundance is reduced in several cancers, including lung cancer (Takamizawa et al., 2004), and let-7 was reported to regulate tumor growth by targeting the ras gene (Johnson et al., 2005). miR-15a and miR16-1 were deleted and/or down-regulated in ∼70% of patients with chronic lymphocytic leukemia (Calin et al., 2002). miR-15a/16-1 induced apoptosis by inhibiting BCL-2 (Cimmino et al., 2005). The miR-34 family is an important component of the p53 tumor suppressor network (He et al., 2007). The human miR-17/20 cluster's genomic location, chromosome 13q31, correlates with loss of heterozygosity in several different cancers, including breast cancer (Eiriksdottir et al., 1998; Lin et al., 1999). The expression and function of miRNA varies by cell type. The miR-17/20 cluster functions as a tumor suppressor in the human B cell line P493-6 by decreasing E2F1 expression, thereby inhibiting _myc_-induced cell proliferation (O'Donnell et al., 2005). In contrast, in both lung cancer and lymphomas, expression of this miRNA cluster was increased, enhancing cell growth (Hayashita et al., 2005; He et al., 2005). As the same miRNA performs different functions through distinct pathways dependent on the tissue or cell type, it is important to understand the mechanisms by which miRNA regulates the cell cycle and thereby tumorigenesis.
The onset and progression of tumorigenesis involves evasion of apoptotic signals, sustained cellular proliferation, and the ability to promote tumor neoangiogenesis. Cyclin D1 overexpression is found in ∼50% of human breast cancers (Fu et al., 2004). Antisense to cyclin D1 abrogated the growth of ErbB2-induced breast tumors, and _cyclin D1_−/− mice are resistant to the induction of tumors either in the mammary gland or the skin induced by oncogenic ras or ErbB2 (Lee et al., 2000; Fu et al., 2004). A subset of miRNAs are regulated by oncogenic myc and ras (Johnson et al., 2005; O'Donnell et al., 2005), but the requirement for cell cycle proteins in the regulation of miRNA is still unknown. Furthermore, the understanding of the mechanism by which specific oncogenes regulate miRNA expression and function is limited.
By a large-scale miRnome analysis using miRNA chips on mouse mammary tumor virus (MMTV)–cyclin D1 transgenic mice and cyclin D1 knockout mice, we determined a subset of miRNAs controlled by cyclin D1. These studies identified a novel regulatory mechanism in which cyclin D1 induces a miRNA signature that includes miR-17-5p and miR-20a through the binding of the miR-17/20 promoter region. miR-17/20 in turn attenuates cyclin D1 abundance via a negative feedback loop to regulate cyclin D1 through a conserved 3′ UTR miRNA binding site. Given the central role for cyclin D1 in cellular differentiation and tumorigenesis, the studies herein, which demonstrate that miRNAs determine cyclin D1 abundance, have broad implications for cancer biology. This work demonstrates additional regulatory mechanisms between oncogenes and tumor suppressor miRNAs that act in concert with other factors to limit oncogene function.
Results
miR-17/20 down-regulates cyclin D1 expression and suppresses breast cancer cell proliferation
The miR-17/20 cluster located on chromosome 13q31 encodes six miRNAs within an ∼1-kb region. Amplification and overexpression of miR-17/20 has been described in B cell lymphomas (He et al., 2005) and lung cancers (Hayashita et al., 2005). miR-17-5p and miR-20a are key important components of the miR-17/20 cluster. To characterize the miR-17/20 expression in breast cancer cell lines, Northern blot analysis was performed of nine breast cancer cell lines (MDA-MB-231, BT-474, MDA-MB-453, Hs578T, MDA-MB-486, SKBR-3, MDA-MB-361, MCF-7, and T-47D), one nontumorigenic breast epithelial cell line (MCF-10A), and two nonbreast tumor cell lines (HeLa and 293T; Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1). The expression of miR-17-5p and miR-20a was low to undetectable in at least six of the nine breast cancer cell lines compared with either nontumorigenic breast epithelial cell (MCF-10A) or nonbreast cancer cell lines (HeLa and 293T). The reduced expression of miR-17/20 in breast cancer cells suggested the miRNA cluster may have a unique function during breast tumorigenesis.
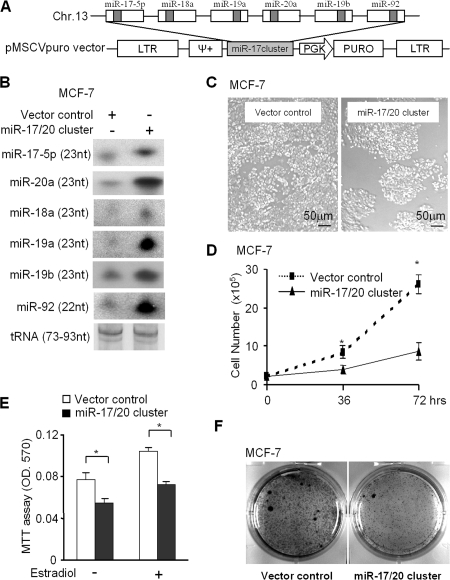
To further investigate the role of miR-17/20 in breast cancer, a retroviral vector encoding the miR-17/20 cluster was used to infect MCF-7 and the NAFA cell line, which is derived from MMTV-ErbB2 transgenic mice breast tumors (Fig. 1 A). Northern blotting confirmed the increased expression of the components in the miR-17/20 cluster (miR-17-5p, 18a, 19a, 19b, 20a, and 92) in the transduced cells (Fig. 1 B). Cellular proliferation assays were performed on the miR-17/20–transduced cells. miR-17/20 expression inhibited MCF-7 cell proliferation and cell growth by ∼60% after 72 h (Fig. 1, C and D). To determine if miR-17/20 regulated estrogen-dependent cellular proliferation, MCF-7 cells were treated with estrogen (17β-estradiol). Both estrogen-dependent and -independent cell proliferation was inhibited by miR-17/20 (Fig. 1 E). Furthermore, miR-17/20 inhibited MCF-7 contact-independent growth determined through colony formation assays (Fig. 1 F).
Figure 1.
miR-17/20 inhibits breast cancer cell proliferation. (A) The genomic region of miR-17/20 cluster on chromosome 13q31 and the vector structure for miRNA17/20 overexpression. (B) Northern blot analysis demonstrating the increased expression of the six members of miR-17/20 cluster in the miR-17/20–transduced MCF-7 cells. tRNA served as loading control. (C and D) Cellular proliferation assay; (E) an MTT assay; and (F) a colony formation assay consistently showed the inhibition of MCF-7 cell proliferation by miR-17/20 overexpression. Cellular proliferation assays and the MTT assay were performed in three independent experiments (data are equal to mean ± SEM). *, P < 0.01.
miR-17/20 arrests the cell cycle at the G1 phase in a _cyclin D1_–dependent manner
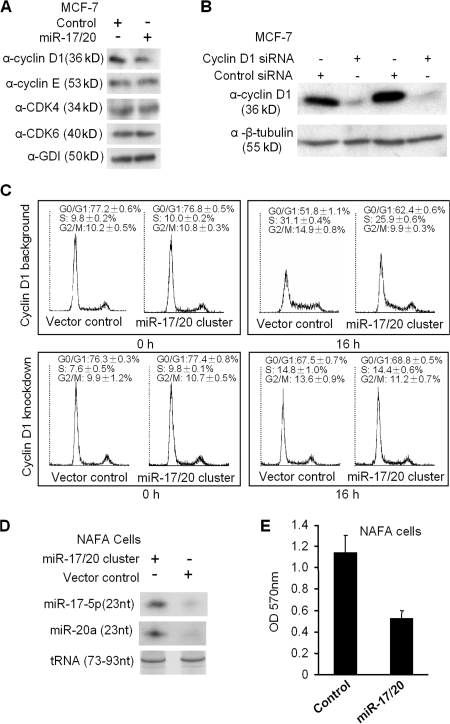
To determine the mechanism by which miR-17/20 suppressed breast cancer cell proliferation, the abundance of key cell cycle regulatory proteins was assessed. Cyclin D1 abundance is rate limiting for MCF-7 cell DNA synthesis. Cyclin D1 expression was inhibited ∼50% by miR-17/20 overexpression, whereas cyclin E, Cdk 4, and Cdk 6 expressions were unaltered in miR-17/20–transduced cells (Fig. 2 A).
Figure 2.
miR-17/20 arrests cell cycle at the G1 phase in a cyclin D1–dependent manner. (A) Western blot analysis of the cyclin D1, CDK4, CDK6, and cyclin E expression in miR-17/20–transduced cells. GDI served as loading control. (B) MCF-7 cells were transfected with cyclin D1 siRNA and control siRNA. Western blotting demonstrated the efficient knockdown of cyclin D1 after 72 h of siRNA treatment. (C) Cell cycle analysis indicated the increased population of cells at G0/G1 phase and decreased S and G2/M phase cells in miR-17/20–transduced MCF-7 cells under a cyclin D1 background. This difference was abolished by knockdown of cyclin D1 in cells. The analysis was performed in triplicates (data are equal to mean ± SEM). (D) Northern blot demonstrated the increased expression of miR-17/20 in the miR-17/20–transduced NAFA cells. tRNA served as a loading control. (E) The MTT assay showed the inhibited cell proliferation of NAFA cells by miR-17/20 transduction. The assay was performed in three independent experiments, and the data are presented as the mean ± SEM.
To examine the functional interrelationship between miR-17/20 and cyclin D1, cyclin D1 siRNA was used to transfect miR-17/20–transduced MCF-7 cells (Fig. 2 B). miR-17/20–transduced MCF-7 cells were starved with 5% charcoal-stripped serum for 48 h, and cellular DNA synthesis was stimulated by the addition of 10% normal FBS. Cell cycle analysis was conducted before serum stimulation and upon 16 h of serum treatment. The proportion of cells corresponding to the proportion of cells in the S and G2/M phase was lower in miR-17/20–transduced MCF-7 cells (25.9 ± 0.6% vs. 9.9 ± 0.3%) than that in control cells (31.1 ± 0.4% vs. 14.9 ± 0.8%; Fig. 2 C). The G0/G1 phase was increased in miR-17/20–transduced MCF-7 cells (62.4 ± 0.6%) compared with control cells (51.8 ± 0.1%). The inhibition of S phase by miR-17/20 was dependent on cyclin D1, as cyclin D1 knockdown with siRNA abrogated the G1 phase and S phase differences between miR-17/20 overexpressed MCF-7 cells (G0/G1: 67.5% ± 0.7%; S, 14.8 ± 1.0%) and control cells (G0/G1: 68.8% ± 0.5%; S, 14.4 ± 0.6%; Fig. 2 C).
MCF-7 cells express abundant cyclin D1. In contrast with MCF-7 cells, transduction of MCF-10A and BT-474 cells, which have low abundance of cyclin D1 (Fig. 3), with the miR-17/20 cluster had no demonstratable effect on DNA synthesis induced by FBS (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1). These results indicate that the inhibition of G1/S transition by miR-17/20 is cyclin D1 dependent.
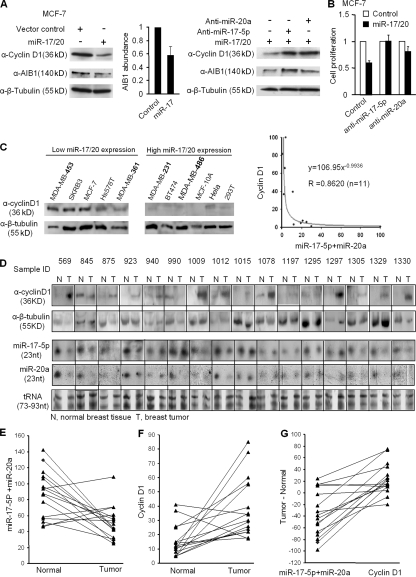
Figure 3.
The inverse correlation between cyclin D1 abundance and miR-17/20 expression in human breast cancer tissues and cell lines. (A) Western blots showed the repression of cyclin D1 and AIB1 abundance by miR-17/20. Both anti–miR-17-5p and anti–miR-20a increased the expression of cyclin D1 and AIB1 in cells. AIB1 is a positive control. β-tubulin was a loading control. (B) Cotransfection of anti–miR-17-5p or anti–miR-20a reversed the antiproliferative function of miR-17/20 in MCF-7 cells. (C) Western blots showed the high cyclin D1 level in low miR-17/20–expressing breast cancer cell lines, and low cyclin D1 in high miR-17/20 cell lines. A Northern blot of miRNA17/20 is shown in Fig. S1 (available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1). (D) The abundances of cyclin D1 and miR-17-5p/miR-20a in 16 human breast tumor tissues and 16 matching normal breast tissues were determined by Western and Northern blots. Tissue sample IDs were provided by the sample provider. Black lines indicate that intervening lanes have been spliced out. N, normal tissue; T, tumor tissue. (E) Statistically significant down-regulation of miR-17/20 expression in breast tumors over matching normal tissue. P = 0.004 by Wilcoxon signed rank test. The grayscale intensity of each band in D was obtained by AlphaImager software. The y axis value stands for the addition of miR-17-5p and miR-20a expression in each sample. (F) Significant up-regulation of cyclin D1 expression in breast tumors over matching normal tissue. P = 0.001. (G) Plotting the paired difference of tumor and normal samples expression for each marker (miR-17/20 vs. cyclin D1). The exact McNemar's test indicates a significant association between the up-regulation of expression in cyclin D1 and the down-regulation of miR-17/20 expression. P = 0.002.
The ErbB2 oncogene is overexpressed or amplified in ∼30% of human breast cancers and is sufficient for mammary tumor generation when targeted to the mammary gland in transgenic mice. The NAFA cell line is derived from MMTV-ErbB2 transgenic tumors. To determine the role of miR-17/20 cluster in ErbB2-mediated tumor growth, the NAFA cell line was transduced with the expression vector encoding an miR-17/20 cluster (Fig. 1 A). The expression of miR-17-5p and miR-20a was induced in NAFA cells transduced with the miRNA17/20 cluster (Fig. 2 D). miR-17/20 expression inhibited proliferation of NAFA cells (Fig. 2 E).
To examine further the specificity with which miR-17/20 inhibits cyclin D1 abundance, chemically modified miRNA inhibitors (anti-mir) were used to block miRNA function. The miR-17/20 locus–transduced MCF-7 cells were transfected with anti–miR-17-5p and anti–miR-20a, which can specifically interact with miR-17-5p and miR-20a, respectively, to block their function. Cyclin D1 abundance is rate-limiting in MCF-7 cells for DNA synthesis and cell proliferation. Both anti–miR-17-5p and anti–miR-20a rescued the inhibition of cyclin D1 abundance by miR-17/20 (Fig. 3 A). Both anti-mirs reversed the antiproliferative function of miR-17/20 (Fig. 3 B), which is consistent with the functional role of cyclin D1. The abundance of AIB1, a target of miR-17-5p repression, was inhibited as previously described by miR-17/20 (Hossain et al., 2006). The down-regulation of cyclin D1 and AIB1 by miR-17/20 could be rescued by either anti–miR-17-5p or anti–miR-20a (Fig. 3 A).
To determine whether endogenous miR-17/20 regulates cyclin D1 abundance and cell proliferation, MCF-7 cells were transfected with anti–miR-17-5p and anti–miR-20a. Inhibition of these endogenous miRNAs' function enhanced cellular proliferation and cyclin D1 abundance (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1).
Cyclin D1 is rate-limiting in tumorigenesis induced by several breast oncogenes (Ras, Src, and ErbB2; Fu et al., 2004). In view of the finding that cyclin D1 level was reduced by miR-17/20 expression, we examined the relative abundance of cyclin D1 and miR-17/20 in 11 different human cell lines and 16 matched normal and tumorous human breast samples. The abundance of cyclin D1 and expression of miR-17/20 were inversely correlated (Fig. 3, C and D). Statistical analysis showed significant down-regulation of miR-17/20 expression in breast tumors over matching normal tissue (Fig. 3 E), and significant up-regulation of cyclin D1 expression in breast tumors over matching normal tissue (Fig. 3 F). There is a significant association between the up-regulation of expression in cyclin D1 and the down-regulation of miR-17/20 expression (Fig. 3 G)
miR-17/20 represses cyclin D1 through a conserved 3′ UTR binding site
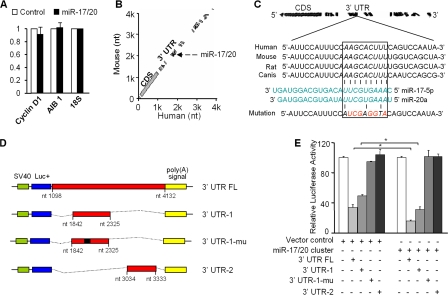
To determine the mechanism by which miR-17/20 inhibited cyclin D1 abundance, quantitative real-time PCR analysis was conducted. No significant difference in cyclin D1 mRNA levels was observed between miR-17/20–transduced cells and control cells (Fig. 4 A). This result suggests that the miR-17/20 cluster regulates cyclin D1 gene expression at the posttranscriptional level. Alignment of the human and mouse cyclin D1 mRNA 3′ UTRs identified a conserved miR-17/20–binding site (Fig. 4 B). The miR-17/20–binding site (nucleotides 2,109–2,117) of human cyclin D1 is well conserved between species (Fig. 4 C). To determine whether miR-17/20 directly regulates cyclin D1, cyclin D1 mRNA 3′ UTR was linked to the firefly luciferase as a reporter gene (Fig. 4 D). Full-length and cyclin D1 3′ UTR fragments containing a wild-type or mutant miR-17/20–binding site were inserted into the luciferase reporter vector (Fig. 4 D). A construct without the miRNA binding sites was used as a negative control. When introduced into MCF-7 cells, the full-length cyclin D1 3′ UTR reporter showed a 90% reduction in luciferase activity in the miR-17/20–transduced cells compared with empty luciferase vector and negative control vector (Fig. 4 E). The reporter vector carrying the cyclin D1 3′ UTR fragment with the miR-17/20 binding site showed reduced luciferase activity in the transduced cells compared with control cells. Mutation of the miR-17/20 binding site abrogated repression of luciferase activity (Fig. 4 E).
Figure 4.
miR-17/20 represses cyclin D1 expression through a conserved 3′ UTR site. (A) Quantitative real-time RT-PCR assay did not show significant difference in mRNA level of cyclin D1 between miR-17/20–transduced cells and control cells. ABI1 was a positive control. 18S served as an internal control for normalization. This experiment was repeated three times in triplicate. Data are mean ± SEM. (B) BLASTN analysis of human and mouse cyclin D1 mRNAs identified a miR-17-5p– and miR-20a–binding site at the conserved 3′ UTR region. (C) Sequence alignment of the miR-17-5p and miR-20a base-paring site in the 3′ UTR of cyclin D1 mRNAs. The region complementary to the 2–10 nt of miR-17/20 is highly conserved among human, mouse, rat, and dog. The “seed” sequence of miR-17/20 that is complementary to cyclin D1 is shown in italics and boxed. The mutant sequence is identical to the wild-type sequence except the mutated nucleotides are shown in red. (D) Luciferase reporter assay constructs. 3′ UTR FL, the full-length 3′ UTR of cyclin D1 inserted to the downstream of the luciferase coding region in the pGL3 vector; 3′ UTR-1, a fragment of cyclin D1 3′ UTR containing the miR-17/20–binding sequence; 3′ UTR-1-mu, the mutated construct identical to 3′UTR-1 but point mutated in the miR-17/20 binding site; 3′ UTR-2, another fragment of cyclin D1 3′ UTR without the miR-17/20–binding sequence. (E) Luciferase reporter assay showed the decreased luciferase activity in miR-17/20 overexpressed cells for both 3′ UTR FL and 3′ UTR-1 constructs, but not for the PGL-3 empty vector, 3′UTR-1-mu and 3′UTR-2 constructs. The luciferase activity was normalized to β-galactosidase. Data are derived from three independent experiments. Values are presented as the mean ± SEM (n = 3). *, P < 0.01
Cyclin D1 is required for induction of miR-17/20 expression
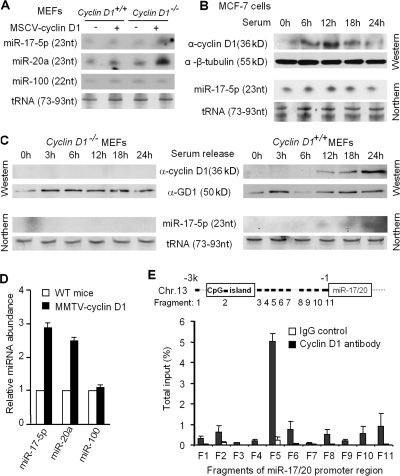
miR-17/20 can be induced by c-myc or E2F1, and miR-17/20 in turn negatively regulates E2F1 translation (O'Donnell et al., 2005; Sylvestre et al., 2007). c-myc, E2F, cyclin D1, and pRb are important regulators of G0/G1–S transition of the cell cycle. The miR-17/20 cluster is activated by c-myc in early G1, and the E2F1 protein accumulates in late G1. Like c-myc, cyclin D1 is activated early in the G1 phase. As c-myc is capable of inducing miR-17/20, we examined the possibility that cyclin D1 may also induce miR-17/20 expression. A retroviral expression vector encoding cyclin D1 was used to transduce wild-type and _cyclin D1_−/− mouse embryonic fibroblasts (MEFs) to determine whether cyclin D1 was sufficient to induce miR-17/20 expression. Cyclin D1 overexpression was confirmed by Western blotting (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1), and miRNA expression was confirmed by Northern blotting (Fig. 5 A). miR-17-5p expression was reduced to undetectable levels in _cyclin D1_−/− cells (Fig. 5 A). Cyclin D1 transduction of _cyclin D1_−/− cells resulted in physiological levels of cyclin D1 and induced miR-17-5p and miR-20a expression (Fig. 5 A, lanes 3 vs. 4). An unrelated miRNA, miR-100, showed no difference in expression in cyclin D1−/− cells by Northern blotting (Fig. 5 A) and miRNA chip analysis (not depicted).
Figure 5.
Cyclin D1 induces miR-17-5p and miR-20a expression. (A) Northern blotting showed the increased expression of miR-17-5p and miR-20a in the MSCV–cyclin D1–infected MEF cells. miR-100 was used as a negative control. tRNA served as loading control. (B) Breast cancer cell line MCF-7. (C) _cyclin D1_−/−, and cyclin D1+/+ MEFs were starved by 5% charcoal-stripped serum for 48 h, followed by 10% FBS stimulation. The time course examination of the cyclin D1 protein level and the miR-17-5p expression level were performed by Western and Northern blots, respectively. The results showed the induction of cyclin D1 and miR-17-5p by serum stimulation in the MCF-7 and cyclin D1+/+ MEF cells, but not in cyclin D1_−/− MEFs. (D) miR-17-5p and miR-20a detection in the MMTV–_cyclin D1 transgene induced mammary tumors (n = 3) compared with normal mice mammary gland. Data from three mammary tumor samples were presented as mean ± SEM. (E) Real-time PCR analysis of cyclin D1 chromatin immunoprecipitated DNA. 11 fragments derived from chromosome 11q31 upstream of miR-17/20 cluster were examined. For each fragment, amplifications on chromatin before immunoprecipitation and chromatin immunoprecipitated with preimmune serum were performed as input and negative control, respectively. This experiment was repeated three times; data are equal to mean ± SEM.
To determine whether miR-17-5p and cyclin D1 are coordinately regulated by physiological stimulation, MCF-7 cells were serum-starved and then stimulated by 10% FBS. The time course of cyclin D1 expression and miR-17-5p induction were detected by Western and Northern blots, respectively (Fig. 5 B). Cyclin D1 protein abundance was increased at 6 h, with maximal abundance at 12 h. miR-17-5p expression increased at 6 h (Fig. 5 B). To determine whether cyclin D1 was required for serum-mediated induction of miR-17-5p, a comparison was made between _cyclin D1_−/− and cyclin D1+/+ MEFs. The induction of miR-17-5p by serum was identified in cyclin D1+/+ but not _cyclin D1_−/− MEFs (Fig. 5 C). To determine if cyclin D1 induced miR-17/20 in vivo, MMTV-cyclin D1 transgenic mice were assessed. RNAs from MMTV–_cyclin D1_–induced mammary tumors, and normal mouse mammary gland were compared and analyzed for miRNA expression using miRNA chips. Consistent with the in vitro data, the miR-17/20 cluster was up-regulated in the _cyclin D1_–induced mammary tumors (Fig. 5 D).
Cyclin D1 binds the promoter of the miR-17/20 cluster
Cyclin D1 induced miR-17/20 expression, and cyclin D1 is known to regulate gene transcription through occupancy of a subset of gene promoters in the context of local chromatin, thereby regulating cellular differentiation and proliferation (Wang et al., 2003; Hulit et al., 2004; Fu et al., 2005). Chromatin immunoprecipitation assays were performed to determine whether cyclin D1 occupied the miR-17/20 promoter in the context of local chromatin in human breast epithelial cells. MCF-10A cells were fixed, and a cell lysate was prepared. Using a cyclin D1 antibody to immunoprecipitate _cyclin D1_–binding DNAs, preimmune serum was used as negative control. DNA was amplified by real-time PCR. 3 kb of the miR-17/20 cluster upstream region was examined by 11 different pairs of primers, which divide the miR-17/20 promoter region into 11 fragments (Fig. 5 E, fragments 1–11). A _cyclin D1_–specific binding site was identified at fragment 5 (Fig. 5 E). Sequence analysis of this 139-bp fragment identified multiple GC boxes upstream of fragment 5 and two TATA boxes downstream of fragment 5. Collectively, these studies demonstrate that cyclin D1 binds to the miR-17/20 promoter and induces miR-17/20 expression at the transcriptional level.
Discussion
Several independent lines of evidence implicate miRNAs as either tumor suppressors or collaborative oncogenes (Calin et al., 2004; Calin and Croce, 2006; Esquela-Kerscher and Slack, 2006; Zhang et al., 2007). miRNAs are differentially expressed in tumors, specific miRNAs are located within regions of chromosomal deletions or rearrangements in tumors, and miRNAs can cooperate with oncogenes in tumorigenesis in animal models (Croce and Calin, 2005). Importantly, the function of individual miRNAs may be cell-type specific. The miR-17/20 cluster has been found to function either as onco-miRNA or as a tumor-suppressor depending on the cell type (He et al., 2005; O'Donnell et al., 2005; Hossain et al., 2006). Expression of the miR-17/20 cluster was increased by 65% in B cell lymphomas, and miR-17/20 cooperates with c-Myc in lymphomagenesis (He et al., 2005). In contrast, the miR-17/20 cluster inhibited proliferation of the P493-6 cell line (O'Donnell et al., 2005). Herein, miR-17/20 inhibited breast cancer cellular proliferation by repression of cyclin D1 translation.
Our findings are of importance to the understanding of breast tumorigenesis, as they directly link the miR-17/20 cluster to the regulation of the cyclin D1 gene. Cyclin D1 plays a key role in regulation of the G1–S phase transition and in tumorigenesis (Fu et al., 2004). The abundance of cyclin D1 is induced by a broad array of oncogenic stimuli and is required for contact-independent growth (Albanese et al., 1995; Fu et al., 2004). Cyclin D1 is required for ErbB2-induced mammary tumorigenesis in vivo (Lee et al., 2000; Yu et al., 2001). Herein, an inverse correlation was found between miR-17/20 and cyclin D1 in human breast cancer samples, in breast cancer cell lines, and in transgenic mice. Our finding that miRNA regulate breast cancer cellular proliferation and contact-independent growth is consistent with recent findings that specific miRNA can regulate breast cancer progression. miR-126 reduces breast tumor growth (Tavazoie et al., 2008), miR-335 inhibits breast cancer metastasis (Tavazoie et al., 2008), and miR-373 and miR-520c stimulate breast cancer cell migration and invasion by suppressing the metastasis suppressor gene CD44 (Huang et al., 2008). The current studies provide evidence for an additional level of fine control in regulation of the G1–S phase transition of the cell cycle. The inhibition of DNA synthesis by miR-17/20 was associated with a reduction in the proportion of cells in the S phase and an increased proportion of cells in G1 phase. Cyclin E, which, like cyclin D1, is capable of enhancing G1–S phase transition in MCF-7 cells, was not regulated by miR-17/20. As the abundance of cyclin D1 is rate-limiting in MCF7 cell growth and G1–S phase progression (Fu et al., 2004), the inhibition of cyclin D1 expression by miR-17/20 likely contributes to the inhibition of DNA synthesis and cellular proliferation in breast cancer cells.
Both cyclin D1 and c-myc are activated early in the G1 phase and are key regulators of the G1–S phase transition. Cyclin D1 activates E2F1 transactivation through phosphorylation of pRb. E2F1 expression is induced by c-myc (Matsumura et al., 2003). miR-17-5p and miR-20a limit _c-myc_–mediated induction of E2F1 translation (O'Donnell et al., 2005), providing a mechanism to dampen reciprocal activation of c-myc to E2F1. Our studies provide evidence for a novel level of fine tuning to regulate the functional interactions between cyclin D1, c-myc, E2F, and miRNAs. Cyclin D1 abundance is increased in ∼50% of human breast cancers. The cyclin D1 gene encodes the labile catalytic subunit of the holoenzyme that phosphorylates and inactivates the pRb protein. The pRb protein serves as a paradigmatic tumor suppressor. The current studies are the first to demonstrate that cyclin D1 abundance is controlled by miRNA. The binding site identified within the cyclin D1 3′ UTR required for repression by miR-17/20 is highly conserved between species. As the relative abundance of cyclin D1 is tightly regulated, multiple distinct mechanisms are involved to control cyclin D1 abundance, including transcriptional induction, posttranslational regulation, and in the current studies, posttranscriptional regulation via miRNA.
The studies herein provide genetic evidence that cyclin D1 binds to an miRNA promoter and regulates a specific miRNA signature. Genetic knockout of cyclin D1 or siRNA treatment demonstrated that the miR-17/20 cluster is induced by cyclin D1. Mammary gland–targeted cyclin D1 expression recapitulated this miRNA signature in the mammary gland tumor. In previous studies, cyclin D1 was identified within chromatin of target endogenous gene promoters in the context of nuclear receptor binding sites (peroxisome proliferator–activated receptor γ) and AP-1/CRE sites (Fu et al., 2005; Wang et al., 2003). Herein, cyclin D1 was identified within the miR-17/20 promoter in the context of local chromatin using chromatin immunoprecipitation assays, extending the potential mechanisms by which cyclin D1 may regulate other miRNAs.
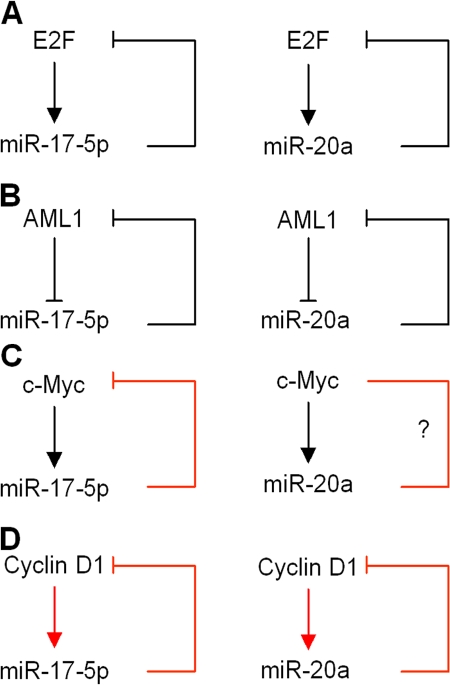
Previous studies have demonstrated the induction of miR-17/20 by c-myc and E2F (O'Donnell et al., 2005; Sylvestre et al., 2007). The E2F family is negatively regulated by miR-17/20 (O'Donnell et al., 2005; Sylvestre et al., 2007). We examined the abundance of c-myc in miR-17/20–overexpressed cells and found the repression of c-myc by miR-17-5p (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1). Thus, four key regulatory proteins—c-Myc, E2F1, AML1 (O'Donnell et al., 2005; Fontana et al., 2007; Sylvestre et al., 2007), and cyclin D1—converge on the miR-17/20 cluster promoter region to create important regulatory feedback loops (Fig. 6). Consistent with this model, translation of the cyclin D1–binding protein AIB1 is repressed by miR-17/20 (Hossain et al., 2006). In turn, miR-17-15p/miR-20a attenuates this activity by inhibiting each of these upstream activators. These attenuating feedback loops provide mechanisms to tightly control proliferative signals. Herein, the expression of miR-17/20 was increased by cyclin D1 in mammary gland–targeted cyclin D1 transgene-induced breast tumors. Thus, the phenotype of cellular growth may be determined by the ability of cyclin D1 to evade translational attenuation by miR-17/20.
Figure 6.
miR-17/20 regulatory loops. (A) E2F-miR-17/20 regulatory loop. (B) AML1-miR-17/20 regulatory loop. (C) c-Myc–miR-17/20 regulatory loop. (D) cyclin D1–miR-17/20 regulatory loop. Red arrows are supported by our data. Black arrows are supported by scientific literature.
Materials and methods
Mice and human breast samples
All animal experiments were done in accordance with the guidelines for the care and use of laboratory animals at Thomas Jefferson University. _Cyclin D1_−/− mice were maintained on a mixed C57B1/6 × 129/SvJ background as described previously (Hulit et al., 2004). Genotyping was done on tail DNA by PCR as described previously (Hulit et al., 2004). Cyclin D1 transgenic mice were made and maintained as described previously (Wang et al., 1994). MMTV–cyclin D1 transgenic mice were obtained from A. Arnold (University of Connecticut, Farmington, CT). Human breast cancer specimens and matching normal breast tissue samples were provided by P. McCue (Thomas Jefferson Hospital, Philadelphia, PA). All the procedures were approved by the Institutional Review Board of Thomas Jefferson University. Expression for miR-17/20 and cyclin D1 in paired tumor and normal samples were analyzed for significant up- or down-regulation using the Wilcoxon signed rank test. The association of down-regulation in miRNA expression and up-regulation in cyclin D1 expression was completed via the exact McNemar's test. AlphaImager (Alpha Innotech) was used for intensity analysis.
Vectors
miR-17/20 expression vector was provided by J. Mendell (Johns Hopkins University School of Medicine, Baltimore, MD) and A. Thomas-Tikhonenko (University of Pennsylvania, Philadelphia, PA). Human cyclin D1 cDNAs were cloned into the mouse stem cell virus (MSCV)-internal ribosome entry site (IRES)-GFP retrovirus vector at the EcoRI site. Different lengths of wild-type and mutated cyclin D1 3′ UTR sequences were inserted into the XbaI–FseI site immediately downstream of the stop codon in the pGL3 firefly luciferase reporter vector. pMSCVpuro vector was used to express miR-17/20 cluster at the EcoRI site as described previously (O'Donnell et al., 2005). A recombinant plasmid that carries the bacterial β-galactosidase cDNA driven by the CMV promoter was used as an internal control for transfection efficiency.
Oligonucleotides
All DNA probes were synthesized by Integrated DNA Technologies. Primer sequences for cyclin D1 3′ UTR cloning were as follows: 3′ UTR FL forward, 5′-GGGCGCCAGGCAG-3′; 3′ UTR FL reverse, 5′-ACAGGACAGACT-3′; 3′ UTR-1 forward, 5′-ACCATCCAGTGGAGG-3′; 3′ UTR-1 reverse, 5′-TGGCAATGTGAGAAT-3′; 3′ UTR-2 forward, 5′-GCCGCAGCTCCATT-3′; and 3′ UTR-2 reverse, 5′-ATGGCTAAGAGAA-3′. The probe sequences for miRNA northern analysis were as follows: miR-17-5p, 5-ACTACCTGCACTGTAAGCACTTTG-3′; miR-18a, 5′-TATCTGCACTAGATGCACCTTA-3′; miR-19a, 5′-TCAGTTTTGCATAGATTTGCACA-3′; miR-19b, 5′-TCAGTTTTGCATGGATTTGCACA-3′; miR-20a, 5′-CTACCTGCACTATAAAGCACTTTA-3′; miR-92, 5′-ACAGGCCGGGGACAAGTGCAATA-3′, and miR-100, 5′- CACAAGTTCGGATCTACGGGTT-3′. siRNAs were obtained from QIAGEN. Target sequence for cyclin D1: 5′-AACAAGCTCAAGTGGAACCTG-3′; target sequence for negative control siRNA: 5′-AATTCTCCGAACGTGTCACGT-3′. Anti-miR miRNA inhibitors are chemically modified, single-stranded nucleic acids antisense to target miRNAs. Anti–miR-17, anti–miR-20, and negative controls were obtained from Ambion.
Cell culture
Cyclin D1+/+ and _cyclin D1_−/− primary MEFs were prepared as described previously (Wang et al., 2003). MCF-10A cells were cultured in DME/F12 medium containing 5% horse serum, 10 μg/ml insulin, 20 ng/ml EGF, 100 ng/ml choleratoxin, 0.5 μg/ml hydrocortisone, and 100 mg/liter of penicillin and streptomycin. 293T, Hela, MCF-7, Hs578T, T47D, BT474, SKBR-3, MDA-MB-231, MDA-MB-361, MDA-MB-453, MDA-MB-486, and the NAFA cell line derived from the MMTV-NeuT mouse cells were cultured in DME containing penicillin and streptomycin (100 mg of each per liter) and supplemented with 10% FBS.
Retrovirus infection, siRNA/plasmid transfection, and luciferase reporter assay
Retroviral production and infection methods have been described previously in detail (Li et al., 2006). The ecotropic packaging vector pSV-Ψ–E-MLV, which provides ecotropic packaging helper function, was used with retrovirus to infect cells. For cellular transfection with plasmid DNA, actively growing cells were seeded on 12-well plates at a density of 1 × 105 cells per well in an antibiotic-free medium. The next day, cells were cotransfected using Lipofectamine 2000 (Invitrogen) with 0.5 μg of pGL3 and 0.1 μg of CMV/β-galactosidase plasmid. 24 h after transfection, luciferase activities were measured by AutoLumat (Berthold Technologies) and normalized with β- galactosidase. For the reverse transfection of siRNA to MCF-7 cells, Lipofectamine RNAiMAX was used according to the manufacturer's instructions (Invitrogen).
miRNA expression profiling and data analysis
miRNA expression arrays were generated at Kimmel Cancer Center at Thomas Jefferson University and probed essentially as described previously (Liu et al., 2004). In brief, 5 μg of total RNA from each sample was biotin-labeled during reverse transcription using random hexanes. Hybridization was performed on an miRNA microarray chip that contained 368 probes in triplicate, corresponding to 245 human and mouse miRNA genes. All probes on these microarrays were 40-mer oligonucleotides spotted by contacting technologies and covalently attached to a polymeric matrix. Hybridization signals were detected by biotin binding of a Streptavidin–Alexa 647 conjugate using a ScanArray XL5K (PerkinElmer). Scanner images were quantified by the Quantarray software (PerkinElmer). Signal intensities were normalized and analyzed using GeneSpring software (Agilent Technologies).
miRNA Northern blot analysis
Northern blot analysis of miRNAs was performed as described previously (Yu et al., 2005). In brief, total RNA (10–20 μg per lane) was loaded on a 15% denaturing polyacrylamide gel and electophoresed at 200 V until the bromophenol blue approached the bottom. The RNA was transferred from the gel to Hybond-N+ membrane using a semidry transfer apparatus (Bio-Rad Laboratories). DNA oligonucleotide probes (20–23 nt) were 5′-end labeled with [γ-32P]ATP, and hybridization was performed using Rapid-Hyb buffer according to the manufacturer's instructions (GE Healthcare).
Real time RT-PCR analysis
Total RNA was extracted with Trizol reagent (Invitrogen). The 7900 HT Sequence Detection System (Applied Biosystems) was used for quantitative RT-PCR assay. Primers were designed using primer Express 1.5/Taqman Primer Design software (Applied Biosystems). The primer sequences were as follows: cyclin D1 forward, 5′-AAGCTGTGCATCTACACCGA-3′; cyclin D1 reverse, 5′-CTTGAGCTTGTTCACCAGGA-3′; AIB-1 forward, 5′-AAACTCCATGGGAAGACCAG-3′; AIB-1 reverse, 5′-GTCTCGCACCTGGTATGCTA-3′; 18S forward, 5′-CTACCACATCCAAGGAAGGCA-3′; and 18S reverse, 5′-TTTTTCGTCACTACCTCCCCG-3′.
Western blot analysis
Whole-cell lysates (50 μg) were separated by 10% SDS-PAGE, and the proteins were transferred to nitrocellulose membrane. The following antibodies were used for Western blotting: anti–cyclin D1 (Thermo Fisher Scientific), anti-AIB1 (BD Biosciences), anti-GDI (RTG Solutions), and anti-Myc (sc-40) and anti–β-tubulin (sc-9104; both from Santa Cruz Biotechnology, Inc.).
Cell proliferation assays
Cells were infected with pMSCVpuro-miR-17/20 cluster or pMSCVpuro empty vector. After puromycin selection, 4 × 104 cells per milliliter were seeded into 6-well plate in triplicate, and the cell number was counted every day for 3–4 d under microscope using a hemocytometer. For the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay, 4 × 103 cells per well were seeded into a 96-well plate in triplicate, and after a 24-h culture, the cell growth was measured by MTT bromide assay.
Colony formation assay
The same amount of miR-17/20 overexpression MCF-7 cells and control cells were plated in triplicate in a 6-well plate in puromycin-containing medium. After incubation at 37°C for 7–10 d, the colonies were stained with Crystal Violet solution (Allied Chemical Corporation) in methanol for 15–30 min. Colonies >50 μm in diameter were counted.
Cell cycle analysis
Cells were starved in DME supplemented with 5% charcoal-stripped serum. After 48 h, medium was changed with DME with 10% normal FBS. Cells were harvested at different time points, and cell cycle parameters were determined using laser scanning cytometry. Cells were processed by standard methods by using propidium iodide staining of cell DNA. 10,000 cells per sample were analyzed by flow cytometry with a FACScan flow cytometer (BD Biosciences). Histograms were analyzed for cell cycle compartments using ModFit version 2.0 (Verity Software House).
Chromatin immunoprecipitation
MCF-10A cells were cross-linked with formaldehyde (final concentration of 1%), and chromatin immunoprecipitation was performed as described previously (Weinmann et al., 2001) using anti–cyclin D1 antibody (SC-450; Santa Cruz Biotechnology, Inc.). Real-time PCR was performed using the SYBR green PCR Master Mix kit (Applied Biosystems) to amplify the immunoprecipitated DNA. Sequences of primers derived from the genomic region upstream of the miR-17/20 cluster are provided in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1).
Online supplemental material
Fig. S1 shows the low miR-17-5p and miR-20a expression in human breast cancer cell lines. Fig. S2 shows the cell cycle analysis of miR-17/20–transduced MCF-10A and BT474 cells. Fig. S3 shows that inhibition of endogenous miR-17/20 by anti–miR-17-5p and anti–miR-20a increased cyclin D1 expression in MCF-7 cells. Fig. S4 shows overexpression of cyclin D1 in the MSCV–cyclin D1–transduced cyclin D1−/− MEFs. Fig. S5 shows the inhibition of c-myc expression by the miR-17/20 cluster in MCF-7 cells. Table S1 shows primer sequences for real-time PCR amplification of cyclin D1 chromatin immunoprecipitation assay. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200801079/DC1.
Supplementary Material
[Supplemental Material Index]
Acknowledgments
The miRNA chip analysis was performed by the Nucleic Acid Facility of Kimmel Cancer Center, Thomas Jefferson University. We thank Dr. Joshua Mendell and Dr. Andrei Thomas-Tikhonenko for providing the miR-17/20 cluster expression vector, Dr. Andrew Arnold for providing MMTV–cyclin D1 transgenic mice, and Dr. Renato Baserga and Dr. Nicole Willmarth for giving good suggestions. We thank the Division of Biostatistics at Thomas Jefferson University for help in analyzing data.
This work was supported in part by National Institutes of Health grants R01CA70896, R01CA75503, R01CA86072, and R01CA107382 (to R.G. Pestell). The Kimmel Cancer Center was supported by the National Institutes of Health Cancer Center Core grant P30CA56036 (to R.G. Pestell). This project is funded in part from the Dr. Ralph and Marian C. Falk Medical Research Trust and a grant from the Pennsylvania Department of Health (to R.G. Pestell and C. Wang). The Pennsylvania Department of Health specifically disclaims responsibility for an analysis, interpretations, or conclusions. There are no conflicts of interest associated with this manuscript.
Abbreviations used in this paper: MEF, mouse embryonic fibroblast; miRNA, microRNA; MMTV, mouse mammary tumor virus; MSCV, mouse stem cell virus; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium; UTR, untranslated region.
References
- Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R.G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 270:23589–23597. [DOI] [PubMed] [Google Scholar]
- Ambros, V. 2004. The functions of animal microRNAs. Nature. 431:350–355. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., and C.M. Croce. 2006. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 6:857–866. [DOI] [PubMed] [Google Scholar]
- Calin, G.A., C.D. Dumitru, M. Shimizu, R. Bichi, S. Zupo, E. Noch, H. Aldler, S. Rattan, M. Keating, K. Rai, et al. 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 99:15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., C. Sevignani, C.D. Dumitru, T. Hyslop, E. Noch, S. Yendamuri, M. Shimizu, S. Rattan, F. Bullrich, M. Negrini, and C.M. Croce. 2004. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA. 101:2999–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin, G.A., M. Ferracin, A. Cimmino, G. Di Leva, M. Shimizu, S.E. Wojcik, M.V. Iorio, R. Visone, N.I. Sever, M. Fabbri, et al. 2005. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 353:1793–1801. [DOI] [PubMed] [Google Scholar]
- Cimmino, A., G.A. Calin, M. Fabbri, M.V. Iorio, M. Ferracin, M. Shimizu, S.E. Wojcik, R.I. Aqeilan, S. Zupo, M. Dono, et al. 2005. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA. 102:13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce, C.M., and G.A. Calin. 2005. miRNAs, cancer, and stem cell division. Cell. 122:6–7. [DOI] [PubMed] [Google Scholar]
- Cullen, B.R. 2004. Transcription and processing of human microRNA precursors. Mol. Cell. 16:861–865. [DOI] [PubMed] [Google Scholar]
- Eiriksdottir, G., G. Johannesdottir, S. Ingvarsson, I.B. Bjornsdottir, J.G. Jonasson, B.A. Agnarsson, J. Hallgrimsson, J. Gudmundsson, V. Egilsson, H. Sigurdsson, and R.B. Barkardottir. 1998. Mapping loss of heterozygosity at chromosome 13q: loss at 13q12-q13 is associated with breast tumour progression and poor prognosis. Eur. J. Cancer. 34:2076–2081. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher, A., and F.J. Slack. 2006. Oncomirs - microRNAs with a role in cancer. Nat. Rev. Cancer. 6:259–269. [DOI] [PubMed] [Google Scholar]
- Fontana, L., E. Pelosi, P. Greco, S. Racanicchi, U. Testa, F. Liuzzi, C.M. Croce, E. Brunetti, F. Grignani, and C. Peschle. 2007. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 9:775–787. [DOI] [PubMed] [Google Scholar]
- Fu, M., C. Wang, Z. Li, T. Sakamaki, and R.G. Pestell. 2004. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology. 145:5439–5447. [DOI] [PubMed] [Google Scholar]
- Fu, M., M. Rao, T. Bouras, C. Wang, K. Wu, X. Zhang, Z. Li, T.-P. Yao, and R.G. Pestell. 2005. Cyclin D1 inhibits PPARgamma -mediated adipogenesis through HDAC recruitment. J. Biol. Chem. 280:16934–16941. [DOI] [PubMed] [Google Scholar]
- Hayashita, Y., H. Osada, Y. Tatematsu, H. Yamada, K. Yanagisawa, S. Tomida, Y. Yatabe, K. Kawahara, Y. Sekido, and T. Takahashi. 2005. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 65:9628–9632. [DOI] [PubMed] [Google Scholar]
- He, L., J.M. Thomson, M.T. Hemann, E. Hernando-Monge, D. Mu, S. Goodson, S. Powers, C. Cordon-Cardo, S.W. Lowe, G.J. Hannon, and S.M. Hammond. 2005. A microRNA polycistron as a potential human oncogene. Nature. 435:828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, L., X. He, L.P. Lim, E. de Stanchina, Z. Xuan, Y. Liang, W. Xue, L. Zender, J. Magnus, D. Ridzon, et al. 2007. A microRNA component of the p53 tumour suppressor network. Nature. 447:1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain, A., M.T. Kuo, and G.F. Saunders. 2006. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 26:8191–8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q., K. Gumireddy, M. Schrier, C. le Sage, R. Nagel, S. Nair, D.A. Egan, A. Li, G. Huang, A.J. Klein-Szanto, et al. 2008. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat. Cell Biol. 10:202–210. [DOI] [PubMed] [Google Scholar]
- Hulit, J., C. Wang, Z. Li, C. Albanese, M. Rao, D. Di Vizio, S. Shah, S.W. Byers, R. Mahmood, L.H. Augenlicht, et al. 2004. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Mol. Cell. Biol. 24:7598–7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, S.M., H. Grosshans, J. Shingara, M. Byrom, R. Jarvis, A. Cheng, E. Labourier, K.L. Reinert, D. Brown, and F.J. Slack. 2005. RAS is regulated by the let-7 microRNA family. Cell. 120:635–647. [DOI] [PubMed] [Google Scholar]
- Krek, A., D. Grun, M.N. Poy, R. Wolf, L. Rosenberg, E.J. Epstein, P. MacMenamin, I. da Piedade, K.C. Gunsalus, M. Stoffel, and N. Rajewsky. 2005. Combinatorial microRNA target predictions. Nat. Genet. 37:495–500. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30:363–364. [DOI] [PubMed] [Google Scholar]
- Lee, R.J., C. Albanese, M. Fu, M. D'Amico, B. Lin, G. Watanabe, G.K. Haines III, P.M. Siegel, M.C. Hung, Y. Yarden, et al. 2000. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Mol. Cell. Biol. 20:672–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., C. Wang, X. Jiao, Y. Lu, M. Fu, A.A. Quong, C. Dye, J. Yang, M. Dai, X. Ju, et al. 2006. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Mol. Cell. Biol. 26:4240–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y.W., J.C. Sheu, L.Y. Liu, C.H. Chen, H.S. Lee, G.T. Huang, J.T. Wang, P.H. Lee, and F.J. Lu. 1999. Loss of heterozygosity at chromosome 13q in hepatocellular carcinoma: identification of three independent regions. Eur. J. Cancer. 35:1730–1734. [DOI] [PubMed] [Google Scholar]
- Liu, C.G., G.A. Calin, B. Meloon, N. Gamliel, C. Sevignani, M. Ferracin, C.D. Dumitru, M. Shimizu, S. Zupo, M. Dono, et al. 2004. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissue. Proc. Natl. Acad. Sci. USA. 101:9740–9744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, E., S. Guttinger, A. Calado, J.E. Dahlberg, and U. Kutay. 2004. Nuclear export of microRNA precursors. Science. 303:95–98. [DOI] [PubMed] [Google Scholar]
- Matsumura, I., H. Tanaka, and Y. Kanakura. 2003. E2F1 and c-Myc in cell growth and death. Cell Cycle. 2:333–338. [PubMed] [Google Scholar]
- O'Donnell, K.A., E.A. Wentzel, K.I. Zeller, C.V. Dang, and J.T. Mendell. 2005. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 435:839–843. [DOI] [PubMed] [Google Scholar]
- Sylvestre, Y., V. De Guire, E. Querido, U.K. Mukhopadhyay, V. Bourdeau, F. Major, G. Ferbeyre, and P. Chartrand. 2007. An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 282:2135–2143. [DOI] [PubMed] [Google Scholar]
- Takamizawa, J., H. Konishi, K. Yanagisawa, S. Tomida, H. Osada, H. Endoh, T. Harano, Y. Yatabe, M. Nagino, Y. Nimura, et al. 2004. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 64:3753–3756. [DOI] [PubMed] [Google Scholar]
- Tavazoie, S.F., C. Alarcon, T. Oskarsson, D. Padua, Q. Wang, P.D. Bos, W.L. Gerald, and J. Massague. 2008. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 451:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., N. Pattabiraman, J.N. Zhou, M. Fu, T. Sakamaki, C. Albanese, Z. Li, K. Wu, J. Hulit, P. Neumeister, et al. 2003. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol. Cell. Biol. 23:6159–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T.C., R.D. Cardiff, L. Zukerberg, E. Lees, A. Arnold, and E.V. Schmidt. 1994. Mammary hyperplasia and carcinoma in MMTV-cyclin D1 transgenic mice. Nature. 369:669–671. [DOI] [PubMed] [Google Scholar]
- Weinmann, A.S., S.M. Bartley, T. Zhang, M.Q. Zhang, and P.J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820–6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, R., Y. Qin, I.G. Macara, and B.R. Cullen. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 17:3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature. 411:1017–1021. [DOI] [PubMed] [Google Scholar]
- Yu, Z., T. Raabe, and N.B. Hecht. 2005. MicroRNA Mirn122a reduces expression of the posttranscriptionally regulated germ cell transition protein 2 (Tnp2) messenger RNA (mRNA) by mRNA cleavage. Biol. Reprod. 73:427–433. [DOI] [PubMed] [Google Scholar]
- Zhang, B., X. Pan, G.P. Cobb, and T.A. Anderson. 2007. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 302:1–12. [DOI] [PubMed] [Google Scholar]
- Zhang, H., F.A. Kolb, L. Jaskiewicz, E. Westhof, and W. Filipowicz. 2004. Single processing center models for human Dicer and bacterial RNase III. Cell. 118:57–68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Material Index]