SUMO modification of PCNA is controlled by DNA (original) (raw)
Abstract
Post-translational modification by the ubiquitin-like protein SUMO is often regulated by cellular signals that restrict the modification to appropriate situations. Nevertheless, many SUMO-specific ligases do not exhibit much target specificity, and—compared with the diversity of sumoylation substrates—their number is limited. This raises the question of how SUMO conjugation is controlled in vivo. We report here an unexpected mechanism by which sumoylation of the replication clamp protein, PCNA, from budding yeast is effectively coupled to S phase. We find that loading of PCNA onto DNA is a prerequisite for sumoylation in vivo and greatly stimulates modification in vitro. To our surprise, however, DNA binding by the ligase Siz1, responsible for PCNA sumoylation, is not strictly required. Instead, the stimulatory effect of DNA on conjugation is mainly attributable to DNA binding of PCNA itself. These findings imply a change in the properties of PCNA upon loading that enhances its capacity to be sumoylated.
Keywords: clamp loader, DNA replication, PCNA, SUMO, SUMO ligase
Introduction
Through its reversible attachment to intracellular proteins the small ubiquitin-related modifier SUMO controls numerous biological processes, ranging from nucleo-cytoplasmic transport to the regulation of transcription and the maintenance of genome stability (Kerscher et al, 2006; Geiss-Friedlander and Melchior, 2007). Modification by SUMO follows a mechanism closely related to that used by the ubiquitin system: the modifier's carboxy terminus is activated by a dedicated activating enzyme (E1), transferred to a conjugating enzyme (E2) and linked to a lysine residue of the substrate protein with the aid of a ligase (E3) that confers selectivity to the reaction. In the SUMO system, however, the single E2, Ubc9, often participates directly in substrate recognition, and some SUMO-specific E3s, such as mammalian RanBP2, stimulate the conjugation reaction in a substrate-independent manner by aligning the SUMO thioester on the Ubc9 active site in a conformation favourable for attack by an incoming lysine (Reverter and Lima, 2005). As a consequence, in vitro sumoylation reactions tend to be highly promiscuous, and even in vivo many substrates can be sumoylated by more than one E3 (Reindle et al, 2006). Selectivity and spatio-temporal control over the modification can sometimes be attributed to signalling cascades resulting in the phosphorylation of the E3 or the substrate, but more often the dynamic regulation of sumoylation is poorly understood (Guo et al, 2007).
In eukaryotic cells, post-translational modifications of the replicative sliding clamp PCNA control the processing of replication intermediates (Ulrich, 2005). In response to DNA damage, ubiquitylation of PCNA promotes the bypass of replication-blocking lesions (Hoege et al, 2002; Stelter and Ulrich, 2003; Kannouche et al, 2004). In budding yeast, PCNA (encoded by POL30) is also subject to damage-independent sumoylation during S phase, which enhances its affinity for an antirecombinogenic helicase, Srs2 (Papouli et al, 2005; Pfander et al, 2005). Recruitment of Srs2 by the modified clamp prevents unscheduled recombination events during replication. When progression of replication forks is stalled by DNA damage, Srs2 thus inhibits resolution by homologous recombination and allows damage bypass via the ubiquitin-dependent pathway. Sumoylation of PCNA occurs on two lysines, predominantly on K164 and to a lesser extent on K127. Modification at K164 in vivo and in vitro requires the E3 Siz1, but Siz1 also stimulates non-selective sumoylation at K127 and the formation of poly-SUMO chains on PCNA (Hoege et al, 2002; Stelter and Ulrich, 2003; Windecker and Ulrich, 2008). During most of the cell cycle, Siz1 is nuclear, with the exception of G2/M phase, when the E3 associates with the bud neck and participates in septin sumoylation (Johnson and Gupta, 2001; Takahashi et al, 2001). A conserved SAP domain, which often binds DNA in other proteins (Okubo et al, 2004; Notenboom et al, 2007), determines nuclear localisation or retention (Takahashi and Kikuchi, 2005) and was found to be required for PCNA modification (Reindle et al, 2006). SUMO is removed from PCNA by the isopeptidase Ulp1, which associates with nuclear pore complexes throughout the cell cycle (Li and Hochstrasser, 1999; Panse et al, 2003; Stelter and Ulrich, 2003).
Under physiological conditions, sumoylation of PCNA is limited to S phase, but it is unclear how this is controlled. We have therefore investigated the signals required for Siz1-dependent modification of PCNA in vivo. To our surprise, we found that SUMO conjugation to PCNA is governed less by the cognate enzymes than by the properties of the clamp itself, in particular its association with DNA. This mechanism of PCNA modification is accurately reproduced by a recombinant in vitro system and suggests a substrate-induced control over sumoylation.
Results
PCNA sumoylation is controlled by conjugation and de-conjugation
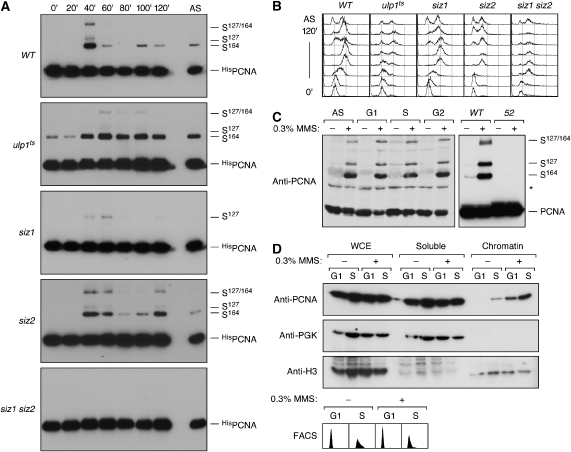
To understand whether regulation of conjugation by Siz1 or of de-conjugation by Ulp1 was primarily responsible for the restriction of PCNA sumoylation to S phase, we analysed the cell cycle dependence of the modification in the relevant mutants (Figure 1A and B). Synchronised cultures of ulp1 ts cells initiated sumoylation in S phase, but due to the slower cell cycle progression, the modification persisted throughout the experiment. The marked upregulation of sumoylation upon entry into S phase even under conditions where de-conjugation is compromised suggested that the cell cycle influences the conjugation reaction. As expected, sumoylation at K164 was abolished in the siz1 mutant. Surprisingly, however, sumoylation at K127 still fluctuated with the cell cycle in this mutant, indicating that Siz1 alone cannot be responsible for upregulating PCNA sumoylation. To assess whether modification at K127 was mediated solely by Ubc9 or depended on a second E3, we deleted SIZ2 in addition to SIZ1, and now conjugation was completely abolished. Therefore, S phase-specific sumoylation of PCNA is not strictly dependent on its cognate E3, Siz1, but can also be performed by the closely related Siz2 enzyme, albeit with lower efficiency and on a different lysine.
Figure 1.
Cell cycle- and DNA damage-dependent sumoylation of PCNA. (A) His POL30 cells of the indicated genotypes were synchronised in G1 and released into the cell cycle. Samples were collected at the indicated times and analysed by Ni-NTA affinity chromatogaphy under denaturing conditions, followed by western blotting with PCNA-specific antibody. Asynchronous cultures (AS) were analysed in parallel. (B) Cell cycle profiles of the cultures shown in (A), determined by flow cytometry. (C) Lethal amounts of DNA damage cause PCNA hyper-sumoylation in WT, but not in pol30-52 cells. Cultures were arrested in G1, S or G2 phase or left asynchronous (AS) and treated where indicated with 0.3% methyl methanesulphonate (MMS) for 90 min. In the right-hand panel, both the WT and the pol30-52 strain (52) were treated during exponential growth. Total cell extracts prepared under denaturing conditions were analysed by western blotting using PCNA-specific antibody. The asterisk indicates a cross-reacting band visible with some batches of the antibody. (D) DNA damage leads to chromatin association of PCNA outside of S phase. G1 and S phase-arrested cells were treated with 0.3% MMS where indicated. Whole cell extracts (WCEs) were prepared by enzymatic lysis, separated into soluble and chromatin-associated fractions and analysed by western blotting for the presence of PCNA. Phosphoglycerate kinase (PGK) and histone H3 served as controls for soluble and chromatin-associated proteins. Arrests were confirmed by flow cytometry (FACS).
These data indicate that both conjugation and de-conjugation contribute to limiting PCNA sumoylation to S phase. However, the cell cycle-dependent fluctuation of the modification is unlikely to be regulated simply by a balance of the respective conjugating and de-conjugating enzymes in the nucleus: first, Siz2—unlike Siz1—remains nuclear in G2 (Takahashi et al, 2003), yet Siz2-dependent sumoylation is lost at this time in siz1 mutants. Second, PCNA is not sumoylated in G1 despite its colocalisation with Siz1 and Siz2 in the nucleus. Therefore, selective modification in S phase appears to require cell cycle-dependent changes in either enzyme activities or substrate properties.
PCNA sumoylation in vivo correlates with its loading onto DNA
In response to lethal concentrations of the alkylating agent methyl methanesulphonate (MMS), PCNA is strongly sumoylated in a Siz1-dependent manner (Hoege et al, 2002; Windecker and Ulrich, 2008). We found that this reaction was independent of the cell cycle stage (Figure 1C). Hence, given the appropriate signal, sumoylation of PCNA is not limited to S phase. We reasoned that the extraordinary levels of DNA damage inflicted by this treatment would likely cause an enhanced engagement of PCNA in repair activities and therefore a significant association with DNA even outside of S phase. Indeed, chromatin-binding assays after 0.3% MMS treatment revealed strongly elevated amounts of PCNA in the chromatin-bound fraction in an S phase-independent manner (Figure 1D). The observed correlation between PCNA hyper-sumoylation and chromatin association therefore suggested that loading onto DNA might exert an effect as a signal for PCNA sumoylation in undamaged cells as well. This notion was also supported by the sumoylation defect of the PCNA mutant encoded by the pol30-52 allele (Figure 1C), which is known for poor loading onto DNA due to reduced trimer stability (Ayyagari et al, 1995).
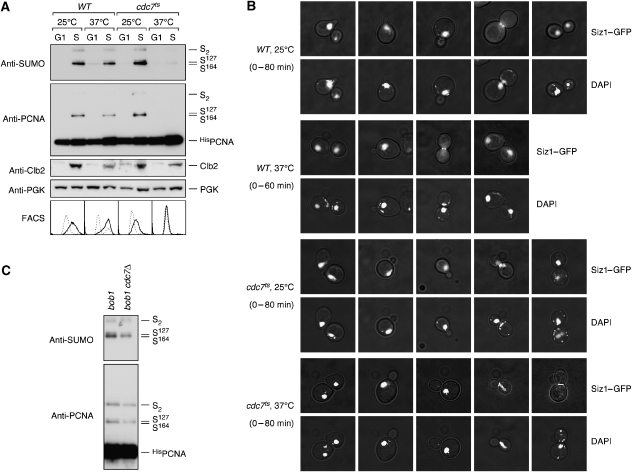
If loading were indeed a prerequisite for sumoylation, preventing PCNA association with DNA during S phase should in turn result in a failure to be modified. We therefore examined PCNA sumoylation in a temperature-sensitive cdc7 ts mutant. CDC7 encodes an essential protein kinase required for the firing of replication origins (Hartwell, 1973). Upon release from G1 arrest at the restrictive temperature, cdc7 ts cells do not initiate DNA replication, although budding pattern and cyclin-dependent kinase activities all resemble a passage through the cell cycle. We found that under these conditions, PCNA was not sumoylated (Figure 2A) at a time when the levels of the mitotic cyclin Clb2 (Figure 2A) and the budding pattern (Figure 2B) indicated an S phase-like state. Siz1 was nuclear at this stage and accumulated at the bud neck only in G2/M phase (Figure 2B), when cdc7 ts cells were arrested due to a failure to undergo mitosis with an unreplicated genome. We excluded the formal possibility that the kinase activity of Cdc7 was required for PCNA sumoylation by confirming the modification in a cdc7 deletion mutant, using a strain background in which CDC7 was rendered non-essential by a mutation in MCM5, a subunit of the replicative helicase (Hardy et al, 1997) (Figure 2C). Therefore, ongoing DNA replication rather than a particular cell cycle regulatory programme appears to bring about PCNA sumoylation, strongly suggesting that the clamp is modified only when it encircles DNA.
Figure 2.
PCNA sumoylation during S phase requires active replication forks. (A) WT and cdc7 ts cells bearing the His POL30 allele, grown at 25°C, were synchronised in G1 and either kept at 25°C or shifted to 37°C for 90 min before releasing them into the cell cycle at the indicated temperatures. Samples were taken before release (G1) and in mid-S phase (S) according to the budding pattern and Clb2 levels (30 min for WT at 25°C and cdc7 ts at both temperatures, 15 min for WT at 37°C). PCNA sumoylation was detected as described in Figure 1, Clb2 and PGK were detected in total cell extracts, and the DNA content was monitored by flow cytometry (FACS; dashed line: G1 arrest; solid line: after release). (B) Subcellular distribution of Siz1 in WT and cdc7 ts cells. Both strains expressing GFP-tagged Siz1 were synchronised in G1 and released into the cell cycle at 25 or 37°C as in (A). Samples were taken at 20-min intervals and analysed by fluorescence microscopy for Siz1–GFP and DNA (DAPI). Representative cells are shown as overlays of fluorescence with interference contrast images. (C) Cdc7 kinase is not required for PCNA sumoylation. Modification of HisPCNA was analysed in asynchronous cultures of bob1 and _bob1 cdc7_Δ mutants. The bob1 mutation affects the MCM5 gene and renders CDC7 non-essential.
PCNA loading stimulates sumoylation in vitro
We have previously demonstrated Siz1-dependent in vitro sumoylation of PCNA in the absence of DNA (Windecker and Ulrich, 2008), but if clamp loading were the main prerequisite for modification in vivo, association with DNA would also stimulate the reaction in vitro. Loading of PCNA is ATP dependent and requires replication factor C (RFC), which opens the PCNA ring and positions the clamp around the DNA at nicks within double-stranded (ds)DNA or 3′ junctions of a primer terminus and single-stranded (ss)DNA (Majka and Burgers, 2004). After loading, PCNA can move freely on DNA, but cannot slide off a circular structure. We therefore examined the effect of purified RFC in the presence of multiply primed circular ssDNA at substrate concentrations that yielded barely detectable levels of Siz1-dependent modification in the absence of DNA. As shown in Figure 3A, addition of DNA and RFC strongly stimulated the reaction. Importantly, neither RFC nor DNA alone enhanced SUMO conjugation, indicating that clamp loading rather than ring opening or the mere presence of DNA was important for stimulation. The same effect was observed with a linear biotinylated DNA containing a 3′ junction, provided that its ends were blocked by the addition of streptavidin (Figure 3B). As RFC-dependent loading occurs in the presence or absence of streptavidin, this result implies that a stable DNA-bound state rather than the process of loading determines the efficiency of PCNA sumoylation. Consistent with the loading defect, modification of the mutant protein encoded by pol30-52 was not stimulated by RFC (Figure 3C), whereas DNA-independent sumoylation at higher protein concentration proceeded with an efficiency comparable to the WT (Figure 3D). This again indicates that PCNA needs to encircle DNA as a trimer to be sumoylated efficiently.
Figure 3.
PCNA sumoylation in vitro is stimulated by loading onto DNA. (A) In vitro sumoylation assays were performed with recombinant Ubc9 and SUMO in the presence or absence of PCNA, E1, RFC, Siz1 and circular, multiply primed ssDNA as indicated. Products were analysed by western blotting with PCNA-specific antibody. (B) In vitro sumoylation reactions were carried out with the complete set of proteins as in (A), but in the presence or absence of streptavidin (SA) and two different linear DNA structures (I and II) derivatised with biotin on both termini. (C) WT HisPCNA and the trimerisation-deficient protein encoded by the pol30-52 allele (52) were compared in sumoylation assays containing E1, Ubc9, Siz1, SUMO and circular primed ssDNA in the presence or absence of RFC. (D) Ubc9- and Siz1-dependent in vitro sumoylation of WT and mutant (52) HisPCNA in the absence of DNA. HisPCNA was used at 3 μM (compared with 50 nM in A–C).
Siz1 binds dsDNA by means of a SAP domain
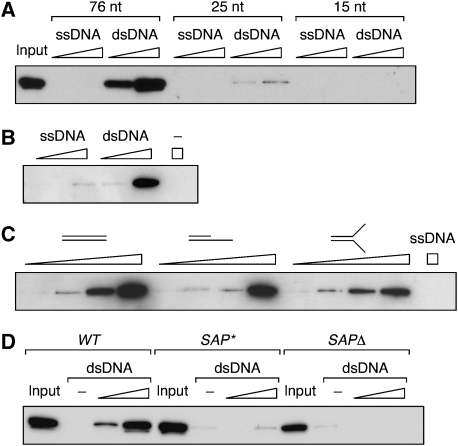
Given that Siz1 contains a SAP domain required for PCNA sumoylation (Reindle et al, 2006), it appeared likely that the enhanced modification of loaded PCNA was attributable to DNA binding of the E3. We therefore asked whether budding yeast Siz1, similar to other members of the PIAS family, was indeed a DNA-binding protein. Recombinant full-length Siz1 was efficiently retained on a biotinylated 76-bp fragment of dsDNA (Figure 4A and B). Its affinity for a 25-bp dsDNA was significantly reduced, and no signal was detected with a 15-bp fragment. Interestingly, binding was strictly limited to dsDNA. Siz1 did not exhibit enhanced affinity for ss–dsDNA junctions or tailed structures when compared with linear dsDNA (Figure 4C), suggesting that the protein primarily recognises ordinary dsDNA. To confirm the importance of the SAP domain for DNA binding, we constructed two Siz1 mutants: SAP*, by mutating three conserved residues within the SAP domain (G55A/K57A/L60A), and _SAP_Δ, by deleting residues 34–68. The purified proteins had no detectable affinity for the 76-bp dsDNA (Figure 4D), indicating that the SAP domain is required for DNA binding.
Figure 4.
The Siz1 SAP domain is required for DNA binding. (A) Siz1 binds to dsDNA, but not ssDNA. Equimolar amounts of biotinylated DNA fragments of the indicated lengths were immobilised on streptavidin Sepharose, and increasing amounts of GSTSiz1FLAGHis were added. Material retained after washing was analysed by western blotting with anti-FLAG antibody. (B) Binding to a 76-nt fragment of ssDNA or dsDNA was analysed as above in the presence of 1 mM EDTA. (C) Equimolar amounts of the indicated DNA structures were immobilised on streptavidin Sepharose, and Siz1 binding was analysed as above. (D) Mutation or deletion of the Siz1 SAP domain results in loss of DNA binding. Equal amounts of Siz1 WT, SAP* and _SAP_Δ were analysed on 76mer dsDNA as above.
The Siz1 SAP domain is dispensable for PCNA sumoylation
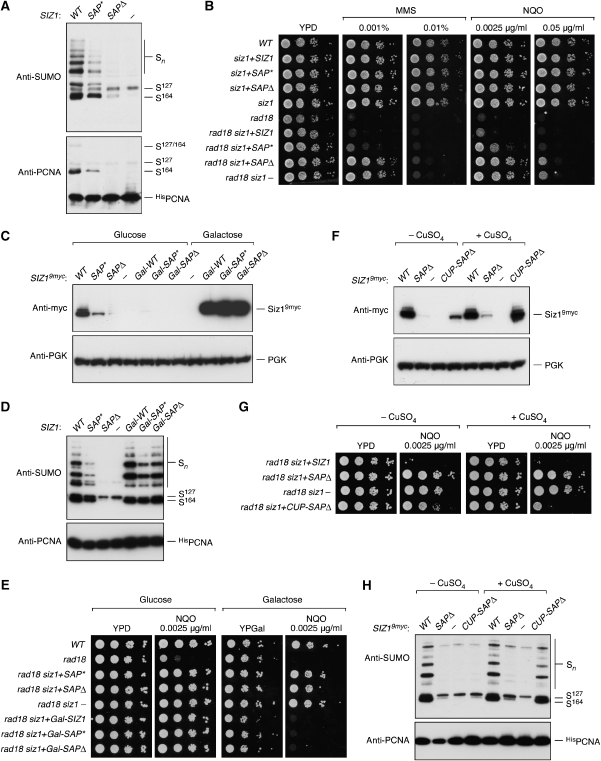
Consistent with previous observations (Reindle et al, 2006), deletion of the SAP domain resulted in loss of sumoylation at K164 in vivo, whereas the SAP* mutation had a partial effect (Figure 5A). This pattern was mirrored in a genetic assay based on suppression of the DNA damage sensitivity of rad18 mutants by loss of SIZ1 function (Stelter and Ulrich, 2003) (Figure 5B). The PCNA sumoylation defect of _siz1(SAP_Δ) had previously been attributed to a defect in nuclear localisation or retention, possibly due to a lack of Siz1 association with the chromatin (Reindle et al, 2006). However, western blot analysis of the mutated Siz1 proteins marked with a 9myc-epitope revealed strongly reduced signals for SAP* and in particular for _SAP_Δ in total cell extracts, implying that the lack of activity towards PCNA in vivo might be due to insufficient protein rather than ineffective nuclear localisation or defective DNA binding (Figure 5C). In fact, overexpression of the mutants from a galactose-inducible promoter completely rescued the sumoylation defect of SAP* and _SAP_Δ (Figure 5D) and fully restored the damage sensitivity of rad18 cells (Figure 5E). To exclude the possibility that overproduction of the mutant Siz1 proteins compensated for a loss in activity, we placed the _SAP_Δ allele under control of the copper-inducible CUP1 promoter, which resulted in protein levels comparable to WT Siz1 under control of its native promoter (Figure 5F). In the presence of copper sulphate, this construct almost completely rescued the phenotype of the siz1 deletion with respect to DNA damage sensitivity (Figure 5G) and PCNA sumoylation (Figure 5H). This indicates that DNA binding of Siz1 might not be a prerequisite for efficient modification of PCNA in vivo.
Figure 5.
The Siz1 SAP domain is dispensable for PCNA sumoylation in vivo. (A) Mutation or deletion of the SAP domain appears to result in partial or complete loss of PCNA sumoylation in vivo. Deletion mutants of siz1 were complemented with integrative plasmids bearing WT, SAP* or _SAP_Δ alleles of SIZ1 or empty vector (−), and modified PCNA was detected in denaturing extracts as described in Figure 1. (B) Mutation or deletion of the SAP domain appears to result in partial or complete loss of SIZ1 function. Sensitivities of the indicated strains to the DNA-damaging agents, methyl methanesulphonate (MMS) and 4-nitroquinoline oxide (NQO) were monitored by growth on plates containing the indicated concentrations of the drugs. Suppression of the damage sensitivity associated with the rad18 deletion indicates a loss of SIZ1 function. (C) Mutation or deletion of the SAP domain results in loss of the Siz1 protein in vivo, which can be rescued by overexpression. The indicated SIZ1 alleles were expressed from integrative plasmids under control of the SIZ1 or the galactose-inducible GAL1 promoter and tagged C-terminally by a 9myc-epitope. An empty plasmid (−) served as a control. Cells were grown in the presence of glucose or galactose, and total extracts were analysed for the presence of Siz19myc by western blotting. Detection of PGK served as a loading control. (D) Overexpression suppresses the sumoylation defects of the SIZ1 SAP domain mutants. The SIZ1 constructs shown in (C) were introduced into the His POL30 siz1 strain, and PCNA modifications were analysed as in Figure 1 after growth in galactose medium. (E) Overexpression of SIZ1 alleles suppresses the loss of function associated with mutation or deletion of the SAP domain. The SIZ1 constructs shown in (C) were introduced into rad18 siz1 strains, and SIZ1 function was analysed as described for (B) on glucose or galactose plates. (F) Expression of the _siz1(SAP_Δ) allele under control of the CUP1 promoter results in near physiological protein levels. The CUP1 promoter was induced by growth in 100 μM CuSO4, and 9myc-tagged versions of the indicated SIZ1 alleles were analysed as in (C). (G) Expression of _siz1(SAP_Δ) under control of the CUP1 promoter suppresses the siz1 phenotype. DNA damage sensitivity assays were carried out with the indicated SIZ1 alleles in rad18 siz1 as in (B, E) in the presence or absence of 100 μM CuSO4. (H) Expression of _siz1(SAP_Δ) under control of the CUP1 promoter restores PCNA sumoylation in vivo. The indicated SIZ1 alleles were analysed in His POL30 siz1 as in (A, D) in the presence or absence of 100 μM CuSO4.
Loading onto DNA changes the properties of PCNA as a sumoylation target
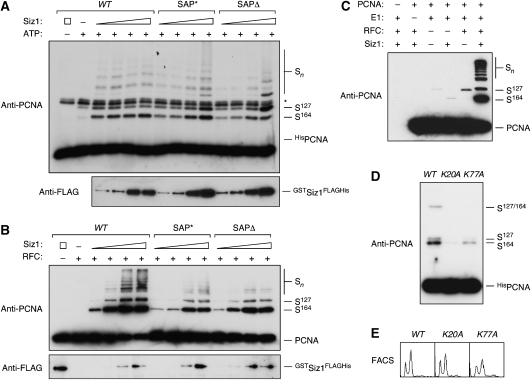
To directly compare the activities of the mutant proteins towards PCNA, we analysed their effects on PCNA sumoylation in vitro at a range of concentrations. In the absence of DNA, the activities of SAP* and _SAP_Δ were slightly lower than those of WT Siz1, and selectivity for K164 was somewhat reduced, possibly indicating some destabilisation of the mutant proteins (Figure 6A). Towards loaded PCNA, both mutants were less active than the WT protein; in particular, formation of poly-SUMO chains was reduced compared with the WT (Figure 6B). Nevertheless, conjugation at K164 by the Siz1 mutants was strongly stimulated by RFC-dependent PCNA loading, indicating that DNA binding of the E3 is not primarily responsible for the effect of PCNA loading on conjugation efficiency. Similar results were obtained with a truncated construct, Siz1(1–508), and its corresponding SAP domain mutants, although the differences in DNA-independent activities between the WT and the SAP domain mutants were even more pronounced (see Supplementary Figure S1).
Figure 6.
PCNA loading stimulates sumoylation by Siz1 SAP mutants and by Ubc9 alone. (A) Mutation or deletion of the Siz1 SAP domain has no effect on PCNA sumoylation in the absence of DNA. In vitro sumoylation assays were performed at high substrate concentration (3 μM) with increasing amounts of Siz1 WT, SAP* or _SAP_Δ protein. GSTSiz1FLAGHis was detected by western blotting with anti-FLAG antibody. (B) PCNA loading stimulates sumoylation by Siz1 SAP mutants. In vitro sumoylation assays in the presence of RFC and circular primed ssDNA were performed at low substrate concentration (50 nM) with increasing amounts of Siz1 WT, SAP* or _SAP_Δ protein (∼1–40 nM). GSTSiz1FLAGHis was detected by western blotting with anti-FLAG antibody. (C) PCNA loading stimulates E3-independent sumoylation. In vitro sumoylation assays in the presence of RFC and circular primed ssDNA were performed with 10-fold elevated concentration of Ubc9 (5 μM). (D) PCNA mutants whose interactions with DNA are altered exhibit reduced sumoylation. PCNA modifications in vivo were analysed in WT, pol30(K20A) and pol30(K77A) as described in Figure 1. (E) Cell cycle distribution of the POL30 alleles shown in (D), determined by flow cytometry (FACS).
The notion that even Siz1 mutants defective in DNA binding preferentially modify PCNA when the clamp encircles DNA raised the question of whether stimulation of the reaction could be ascribed primarily to DNA-induced changes in the substrate rather than a proximity effect mediated by the binding of substrate and E3 to a common stretch of DNA. We therefore examined whether PCNA modification in the absence of Siz1 was also influenced by DNA. Figure 6C shows in vitro sumoylation of PCNA in the presence of primed DNA as before, but at higher Ubc9 concentrations. Surprisingly, RFC stimulated Ubc9-dependent sumoylation at K127 even in the absence of Siz1 (see also Supplementary Figure S2). As we were unable to detect any physical interactions between Ubc9 and either DNA or RFC under our experimental conditions (data not shown), we consider an indirect recruitment of Ubc9 to PCNA unlikely and favour a model in which PCNA itself, when loaded onto DNA, becomes a better substrate for sumoylation.
The inner surface of the PCNA ring is lined by several conserved basic residues, which are likely to directly contact DNA (Fukuda et al, 1995; Lau et al, 2002; Ivanov et al, 2006) and might therefore influence the conformation of loaded PCNA. Indeed, the respective mutants poorly stimulate polymerase δ, although loading and sliding are not affected (Fukuda et al, 1995; Lau et al, 2002). When we examined PCNA sumoylation in two of these alleles, pol30(K20A) and pol30(K77A), we found significant defects, in particular in pol30(K20A), which affects a lysine predicted to interact with bound DNA close to the centre of the minor groove (Ivanov et al, 2006) (Figure 6D). Growth and cell cycle distribution of the corresponding cultures were normal, indicating that the defects in PCNA sumoylation were not due to replication problems (Figure 6E). These observations are consistent with a change in conformation and/or flexibility of PCNA upon DNA binding that is transmitted from the inner surface to the outer rim of the clamp, where it is sensed by the sumoylation system.
Discussion
Control of PCNA sumoylation by Siz1 and Ulp1
Our findings suggest an effective mechanism by which SUMO conjugation can be targeted to S phase. We have shown that the overall levels of SUMO-modified PCNA are influenced both by Siz1-dependent conjugation and by Ulp1-mediated de-conjugation. Yet, a cell cycle-dependent fluctuation of sumoylation is observed even when the relevant enzymes are defective, suggesting that changes in enzyme properties or localisation are not primarily responsible for the temporal control of PCNA modification. Instead, we found that sumoylation of PCNA in vivo prevailed whenever the clamp was associated with DNA. Considering that even in replicating cells a significant part of the cellular pool of PCNA is not bound to DNA (Essers et al, 2005), the total extent of PCNA sumoylation in synchronised cultures (Figure 1A) actually suggests that a major proportion of DNA-loaded PCNA is modified in S phase. Consistent with these observations, we found that the efficiency of PCNA sumoylation in vitro is influenced only to some degree by DNA binding of the E3, but more importantly by the stable loading of the clamp onto DNA. Taken together, S phase-associated sumoylation therefore appears to be triggered mainly by a change in the properties of PCNA induced by RFC-dependent loading.
We cannot exclude a minor contribution of cell cycle-dependent changes in E3 or Ulp1 activity to the regulation of PCNA sumoylation. For example, the re-localisation of Siz1 from the nucleus to the bud neck at mitosis is likely due to its cell cycle-regulated phosphorylation (Johnson and Gupta, 2001) and may well affect the efficiency of PCNA sumoylation at that time. However, given that loading stimulates the reaction with recombinant proteins in vitro, and loaded PCNA can be modified outside of S phase in vivo, a change in substrate properties is sufficient to explain our observations. According to this model, Ulp1-dependent desumoylation of PCNA at the end of S phase could be induced either by a shift in the balance between conjugation and de-conjugation upon unloading or alternatively by an enhanced exposure of ‘soluble' PCNA to the nuclear pore-associated Ulp1. The regulation of PCNA modification thus exemplifies how dynamic control in the SUMO system can be achieved at the substrate level despite the limited number and selectivity of conjugation factors.
Independent signals for PCNA sumoylation and ubiquitylation
In analogy to the system described here, ubiquitylation of PCNA by the E3 Rad18 was shown to be limited to the DNA-bound form both in vivo and in vitro (Garg and Burgers, 2005; Davies et al, 2008). In this case, recruitment of Rad18 by the ssDNA-binding replication protein A (RPA) was found to be required for ubiquitylation in vivo (Davies et al, 2008). In contrast, depletion of RPA does not affect PCNA sumoylation (Davies et al, 2008), and in vitro sumoylation of loaded PCNA proceeds efficiently in the absence of RPA. Hence, although the two modifications affect the same site on PCNA, they are initiated in response to independent signals: whereas ubiquitylation is rendered damage inducible by a dependence on the accumulation of RPA-coated ssDNA at stalled replication intermediates, the sumoylation enzymes react primarily to the loading state of the clamp itself and thereby exert an effect constitutively during S phase.
SUMO conjugation as a probe for the conformation of PCNA
Our observations provide evidence for a conformational change of the clamp upon loading. This concept has been postulated based on molecular dynamics simulations (Ivanov et al, 2006), but has not been demonstrated experimentally due to the difficulties associated with analysing interactions of a topological rather than an affinity-based nature. Interactions between PCNA and several other replication proteins are well known to be affected by DNA. For example, polymerase δ is stimulated only by DNA-bound PCNA, and the productive mode of interaction between PCNA and the flap endonuclease FEN-1 that occurs on DNA differs from that observed in solution (Li et al, 1995; Jonsson et al, 1998; Gomes and Burgers, 2000). However, as both polymerase δ and FEN-1 are DNA-binding proteins themselves, their stimulation by loaded PCNA might be due to their own rearrangement on DNA rather than a conformational change of the clamp. This is unlikely to apply to Ubc9; yet the E2 is able to differentiate between loaded and unloaded PCNA. Hence, the sensitivity of the SUMO conjugation system to the loading state of PCNA demonstrates for the first time that contacts between the DNA and the basic inner surface of the clamp can have an impact on residues situated on the outer edge. Changes in the properties of PCNA may well affect its interactions with other replication- or repair-associated proteins. For example, a contribution of conformational changes within PCNA itself to Rad18-dependent ubiquitylation cannot be excluded until a ligase mutant deficient in DNA binding is examined. A detailed understanding of the nature of these conformational changes will have to await the structural characterisation of PCNA in complex with DNA.
Materials and methods
Yeast strains
Standard procedures were followed for the growth and manipulation of Saccharomyces cerevisiae. Mutants ulp1 ts, siz1, rad18, cdc7 ts, bob1 and _bob1 cdc7_Δ have been described previously (Papouli et al, 2005; Davies et al, 2008; Windecker and Ulrich, 2008). Where required, the His POL30 allele was introduced as described (Stelter and Ulrich, 2003; Windecker and Ulrich, 2008), and His pol30(K20A) and His pol30(K77A) were constructed analogously. SIZ2 was deleted by replacement with a KanMX6 cassette. The pol30-52 allele was introduced on a centromeric plasmid, pBL230-52 (Ayyagari et al, 1995), followed by disruption of endogenous POL30. The SIZ1 alleles were expressed in siz1 deletion strains from integrative plasmids under control of the endogenous or the GAL1 or the CUP1 promoter (see below). For detection by western blotting, these alleles were marked with a 9myc-epitope by integration of a PCR-amplified cassette in place of the stop codon.
Plasmids
The yeast expression vector for His POL30 has been described previously (Papouli et al, 2005; Davies et al, 2008; Windecker and Ulrich, 2008), and mutations K20A and K77A were introduced by site-directed mutagenesis. pBL230-52 was a gift from P Burgers (Ayyagari et al, 1995). Recombinant HisPCNA was expressed in Escherichia coli from pQE-30 (Windecker and Ulrich, 2008), and mutant alleles were constructed in the same vector. pET11c (Novagen) served for expression of recombinant untagged PCNA. Yeast expression vector p416-Siz1-GFP was a gift from E Johnson (Johnson and Gupta, 2001). For expression of native SIZ1 in yeast, the open reading frame with 535 bp of its upstream region was amplified from genomic DNA and inserted into the integrative vector YIplac211, followed by a transcriptional terminator. Mutant alleles were constructed by PCR. The SIZ1 upstream region was replaced by the yeast GAL1 or CUP1 promoter for galactose- or copper-inducible expression, respectively. The E. coli expression vector for GSTSiz1FLAGHis has been described (Windecker and Ulrich, 2008), and the SAP domain mutants were transferred into this construct. An expression vector for yeast RFC was a gift from P Burgers (Franco et al, 2005), those for recombinant HisAos1, Uba2His, Ubc9His and HisSUMO were from E Johnson (Johnson and Gupta, 2001).
Protein purifications
Recombinant HisPCNA, E1 (HisAos1/Uba2His), Ubc9His, HisSUMO, RFC and GSTSiz1FLAGHis were produced as previously described (Franco et al, 2005; Windecker and Ulrich, 2008). The Siz1 SAP domain mutants were expressed and purified by the same procedure as the WT protein. Mutant PCNA proteins were produced as His6-tagged constructs and purified similar to WT HisPCNA.
Untagged PCNA was produced from the expression vector pET11c in E. coli strain BL21-CodonPlus(DE3)-RIL (Stratagene). The bacterial pellet from a 2 l culture was re-suspended in buffer A (25 mM Tris pH 7.5, 1 mM EDTA, 0.5 mM DTT) containing 50 mM NaCl and Complete™ protease inhibitors (Roche) and lysed by sonication. All steps were carried out at 4°C. The lysate was cleared by centrifugation at 40 000 g for 20 min and then at 150 000 g for 45 min. Nucleic acids were removed by Polymin P precipitation, and the lysate was subjected to HiTrap Q chromatography. PCNA-containing fractions were dialysed into buffer A containing 40 mM NaCl and passed through a 5 ml S-Sepharose column. Following MonoQ chromatography (1 ml column), PCNA-containing fractions were pooled and applied to a Superdex 200 gel filtration column equilibrated in buffer A containing 200 mM NaCl and 10% glycerol. The purified protein was stored at −80°C.
Detection of PCNA modifications in vivo
In vivo PCNA modifications were detected by denaturing Ni-NTA affinity chromatography and western blot analysis as described previously, using PCNA- and SUMO-specific antibodies (Papouli et al, 2005; Davies et al, 2008). After treatment with 0.3% MMS, sumoylated PCNA was detected in total cell extracts. Cells were arrested in G1, S or G2 phase with 10 ng/ml α-factor, 100 mM hydroxyurea or 15 μg/ml nocodazole for 1.5–3 h, respectively. Cell cycle stage was monitored by flow cytometry. For induction of the GAL1 promoter, cells were pre-grown in a medium containing 2% raffinose, transferred to 2% galactose medium for overnight growth, and diluted into fresh galactose medium to obtain exponential cultures. Induction of the CUP1 promoter was achieved by overnight growth in 100 μM CuSO4. Control cultures were obtained analogously by transfer and dilution into glucose or copper-free medium.
In vitro PCNA sumoylation assays
In vitro sumoylation assays without DNA were performed as previously described (Windecker and Ulrich, 2008). For reactions in the presence of DNA, 10 oligonucleotides of 28–35 nt length were annealed to ΦX174 virion DNA (New England Biolabs) spaced roughly equally along the sequence (DECAprimed DNA). Unless otherwise noted, sumoylation reactions (20 μl) contained 50 mM HEPES, pH 7.0, 140 mM NaCl, 5 mM MgCl2, 0.1 mM DTT, 1 mM ATP, 2.5 nM DECAprimed DNA, 50 nM PCNA or HisPCNA, 30 nM RFC, 200 nM E1, 500 nM Ubc9His and 8 μM HisSUMO. Siz1 was added at a final concentration of ∼25 nM or titrated in the range of ∼1–40 nM (Figure 6A and B). Reactions in the presence of linear DNA were set up as described above, but contained 25 nM biotinylated primed or unprimed DNA, 18 nM RFC and 1 mM streptavidin where noted. Reactions were incubated at 30°C for 2 h before being terminated by the addition of reducing SDS loading buffer and denaturation at 95°C for 4 min. Samples were analysed by western blotting using an anti-PCNA antibody. PCNA and HisPCNA were modified with equal efficiency in these assays (data not shown).
In vitro DNA-binding assays
A DNA fragment of a given structure, consisting of either a ss 5′-biotinylated oligonucleotide or an annealed pair of oligonucleotides, one of which carried a 5′-biotin label, was immobilised on streptavidin Sepharose in binding buffer (0.1 mg/ml BSA, 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2 or 1 mM EDTA, 1 mM DTT, 0.05% Triton X-100, 150 mM NaCl) for 30 min at room temperature. The beads were washed three times with binding buffer before use. Binding of Siz1 was analysed by incubation of 20 μl beads (∼10 pmol of DNA) with increasing amounts of GSTSiz1FLAGHis (Figure 4A and B: 2 and 8 pmol; Figure 4C: 2, 4, 8 and 15 pmol; Figure 4D: 4 and 12 pmol) for 60 min at 4°C. The beads were washed four times with binding buffer, and bound material was eluted by denaturation in SDS loading buffer and detected by western blotting with anti-FLAG antibody.
Chromatin-binding assays
Total cell extracts prepared by spheroplast lysis were fractionated into soluble and chromatin-bound fractions by centrifugation through a sucrose cushion and analysed by Western blotting as described previously (Davies et al, 2008).
Fluorescence microscopy
WT or cdc7 ts cells harbouring p416-Siz1-GFP (Johnson and Gupta, 2001) were grown to exponential phase in galactose medium at 25°C and treated as described in the legend of Figure 2. Samples were withdrawn at 20-min intervals, and DNA was stained with 0.5 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Cells were analysed by fluorescence microscopy on a DeltaVision Spectris™ system for DAPI and GFP signals. Fluorescence images were processed and overlayed with differential interference contrast images using the Improvision™ software.
Genetic analysis of SIZ1 function
Growth of yeast strains harbouring relevant SIZ1 alleles in a rad18 background was monitored on plates containing various types and concentrations of DNA-damaging agents, using WT, rad18, siz1 and rad18 siz1 as control strains. Loss of SIZ1 function is indicated by a partial suppression of the damage sensitivity associated with the rad18 deletion (Papouli et al, 2005; Windecker and Ulrich, 2008). For analysis of _GAL1_- or _CUP1_-inducible SIZ1 alleles, cultures were pregrown in liquid glucose or galactose medium or in the presence or absence of 100 μM CuSO4 and analysed on plates containing the corresponding carbon source or copper concentration.
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Acknowledgments
We thank Diana Huttner, Angelika Jacobs and Sandra Pahnke for early contributions to the project and Peter Burgers for advice on PCNA loading. We are grateful to Peter Burgers and Erica Johnson for reagents. Funding was provided by Cancer Research UK, an EMBO Young Investigator Award and a Research Training Network grant from the European Commission.
References
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM (1995) A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol 15: 4420–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD (2008) Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell 29: 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers J, Theil AF, Baldeyron C, van Cappellen WA, Houtsmuller AB, Kanaar R, Vermeulen W (2005) Nuclear dynamics of PCNA in DNA replication and repair. Mol Cell Biol 25: 9350–9359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco AA, Lam WM, Burgers PM, Kaufman PD (2005) Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C. Genes Dev 19: 1365–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T (1995) Structure–function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J Biol Chem 270: 22527–22534 [DOI] [PubMed] [Google Scholar]
- Garg P, Burgers PM (2005) Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc Natl Acad Sci USA 102: 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 [DOI] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM (2000) Two modes of FEN1 binding to PCNA regulated by DNA. EMBO J 19: 3811–3821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Yang SH, Witty J, Sharrocks AD (2007) Signalling pathways and the regulation of SUMO modification. Biochem Soc Trans 35: 1414–1418 [DOI] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA (1997) mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA 94: 3151–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH (1973) Three additional genes required for deoxyribonucleic acid synthesis in Saccharomyces cerevisiae. J Bacteriol 115: 966–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) _RAD6_-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Ivanov I, Chapados BR, McCammon JA, Tainer JA (2006) Proliferating cell nuclear antigen loaded onto double-stranded DNA: dynamics, minor groove interactions and functional implications. Nucleic Acids Res 34: 6023–6033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106: 735–744 [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U (1998) Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J 17: 2412–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 [DOI] [PubMed] [Google Scholar]
- Lau PJ, Flores-Rozas H, Kolodner RD (2002) Isolation and characterization of new proliferating cell nuclear antigen (POL30) mutator mutants that are defective in DNA mismatch repair. Mol Cell Biol 22: 6669–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398: 246–251 [DOI] [PubMed] [Google Scholar]
- Li X, Li J, Harrington J, Lieber MR, Burgers PM (1995) Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J Biol Chem 270: 22109–22112 [DOI] [PubMed] [Google Scholar]
- Majka J, Burgers PM (2004) The PCNA-RFC families of DNA clamps and clamp loaders. Prog Nucleic Acid Res Mol Biol 78: 227–260 [DOI] [PubMed] [Google Scholar]
- Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK (2007) Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res 35: 5819–5830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, Yokoyama S, Tanaka H, Yasuda H, Shindo H (2004) NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem 279: 31455–31461 [DOI] [PubMed] [Google Scholar]
- Panse VG, Kuster B, Gerstberger T, Hurt E (2003) Unconventional tethering of Ulp1 to the transport channel of the nuclear pore complex by karyopherins. Nat Cell Biol 5: 21–27 [DOI] [PubMed] [Google Scholar]
- Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell 19: 123–133 [DOI] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436: 428–433 [DOI] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, Johnson ES (2006) Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci 119: 4749–4757 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD (2005) Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425: 188–191 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kikuchi Y (2005) Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J Biol Chem 280: 35822–35828 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Toh EA, Kikuchi Y (2003) Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J Biochem (Tokyo) 133: 415–422 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Toh-e A, Kikuchi Y (2001) A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275: 223–231 [DOI] [PubMed] [Google Scholar]
- Ulrich HD (2005) The RAD6 pathway: control of DNA damage bypass and mutagenesis by ubiquitin and SUMO. Chembiochem 6: 1735–1743 [DOI] [PubMed] [Google Scholar]
- Windecker H, Ulrich HD (2008) Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J Mol Biol 376: 221–231 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2