Mouse Maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 1.
Summary
Tight control of transposon activity is essential for the integrity of the germline. Recently, a germ cell-specific organelle, nuage, was proposed to play a role in transposon repression. To test this hypothesis, we disrupted a murine homolog of a Drosophila nuage protein Maelstrom. Effects on male meiotic chromosome synapsis and derepression of transposable elements (TEs) were observed. In the adult _Mael_−/− testes, LINE-1 (L1) derepression occurred at the onset of meiosis. As a result, _Mael_−/− spermatocytes were flooded with L1 ribonucleoproteins (RNPs) that accumulated in large cytoplasmic enclaves and nuclei. _Mael_−/− spermatocytes with nuclear L1 RNPs exhibited massive DNA damage and severe chromosome asynapsis even in the absence of SPO11-generated meiotic double strand breaks. This study demonstrates that MAEL, a nuage component, is indispensable for the silencing of TEs and identifies the initiation of meiosis as an important step in TE control in the male germline.
Introduction
Germ cells are key to the propagation and evolution of species. Therefore, the ability to prevent and correct the deleterious consequences of endogenous and exogenous damaging agents is critical for the germline function. Genome sequencing efforts over the past decade revealed the enormous impact of transposable elements (TEs) on the evolution and content of genomes throughout the animal kingdom (Babushok and Kazazian, 2007; Kazazian, 2004; Lander et al., 2001; Waterston et al., 2002). Derepression and mobilization of active TEs has been implicated in gene mutation, abrogation or perturbation of gene expression and DNA damage (Girard and Freeling, 1999; Han and Boeke, 2005). The mouse genome contains around 600,000 copies of LINE-1 (L1) non-LTR retrotransposon in various states of decay (Waterston et al., 2002). Only about 3000 of murine L1 elements are thought to be active (DeBerardinis et al., 1998).
In response to the danger posed by active TEs, various species have evolved elegant mechanisms of TE silencing many of which utilize RNA interference (RNAi), small repeat-targeting RNAs and Argonaute proteins (for a recent review see (Slotkin and Martienssen, 2007)). Furthermore, RNAi-related factors often localize to perinuclear electron-dense structures of various shapes and numbers collectively called nuage (for cloud in French) that are present in germ cells from over 80 animal species (Eddy, 1975). These factors include Drosophila Aubergine, AGO3, Armitage, and SPN-E (reviewed in (Klattenhoff and Theurkauf, 2008)), zebrafish ZIWI (Houwing et al., 2007) and mouse MIWI (Kotaja et al., 2006). Based on these observations, nuage has been proposed to function in the repression of TEs (Lim and Kai, 2007). One model for transposon suppression in Drosophila envisions the production of repeat-associated small RNAs in nuage and their subsequent translocation to the nucleus (Klattenhoff and Theurkauf, 2008). Once in the nucleus, small RNAs could guide chromatin modifying complexes to their target TEs to initiate silencing.
In mammalian germ cells, DNA methylation is the primary known mechanism of TE silencing (Bourc'his and Bestor, 2004). Mouse primordial germ cells undergo genome-wide DNA demethylation prior to activation of sex-specific differentiation programs (Hajkova et al., 2002). In males, de novo DNA methylation of TEs is orchestrated by DNMT3l protein in the non-proliferating gonocytes in late gestation (Bourc'his and Bestor, 2004). In addition to the core DNA methylation machinery (Goll and Bestor, 2004), murine PIWI-like proteins MILI (Aravin et al., 2006; Aravin et al., 2007b; Kuramochi-Miyagawa et al., 2008) and MIWI2 (Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008), and their interacting repeat-targeting small RNAs (piRNAs) were recently implicated in DNA methylation-based TE silencing in mice (Aravin et al., 2007a, Aravin and Bourc’his, 2008). Just like in the DNMT3l mutant (Bourc'his and Bestor, 2004), male germ cells lacking either MILI or MIWI2 fail to establish de novo DNA methylation and silence TEs (Kuramochi-Miyagawa et al., 2008). These findings raise the possibility that small RNAs guide de novo DNA methylation machinery to TEs in the mammalian germline (Aravin et al., 2007a; Aravin and Bourc’his, 2008; Girard and Hannon, 2008).
Whether nuage particles participate in TE silencing in the mammalian germline is presently unclear. The best recognized nuage in the mammalian germline is the chromatoid body of round spermatids (Kotaja and Sassone-Corsi, 2007). Given its prominence in haploid germ cells, this specific type of nuage is most commonly associated with translational regulation of germ cell mRNAs possibly by an RNAi-like mechanism as suggested by the presence of MIWI, Dicer and miRNAs (Kotaja et al., 2006). Even though earlier stages lack an equally prominent structure, Eddy (Eddy, 1975) concluded that nuage is present at various stages of germ cell development in rodents. This view is now further supported by studies of other nuage components like MVH (Toyooka et al., 2000), RNF17 (Pan et al., 2005), TDRD1, TDRD6, TDRD7 (Hosokawa et al., 2007) that are present in nuage structures of spermatocytes. However, none of these factors have been reported to be required for TE repression in mice.
To gain further insight into the mechanism of transposon repression in the mouse germline, we focused on a murine homolog of a Drosophila gene maelstrom (mael) (Clegg et al., 1997). Mael belongs to a group of Drosophila genes required for the production of repeat-targeting interfering RNAs, repression of TEs and oocyte axis specification (Findley et al., 2003; Klattenhoff et al., 2007; Lim and Kai, 2007). Mutations in these genes lead to the accumulation of non-meiotic DNA double stand breaks (DSBs) that are thought to trigger a checkpoint response and derail oocyte development (Klattenhoff et al., 2007). In the Drosophila female germline, MAEL is found predominantly in nuage, but its function is unknown (Findley et al., 2003). MAEL contains a domain with weak similarity to a high mobility group (HMG) box DNA-binding domain, but otherwise its amino acid sequence offers little clues about its potential biochemical function. Mouse MAEL is expressed in the male germline where it was reported to localize to the sex body in spermatocytes and to the chromatoid body in round spermatids (Costa et al., 2006). Consistent with its chromatoid body localization, MAEL co-immunoprecipitates with MIWI, MVH and TDRD1 (Costa et al., 2006). In this study we re-examined MAEL localization in the male germline and generated a null allele of Mael. Our studies demonstrate that MAEL is a component of perinuclear nuage and is essential for TE repression in the male germline during early stages of meiosis.
Results
MAEL localizes to perinuclear nuage
Previously, MAEL was reported to localize to the sex body in spermatocytes and to the chromatoid body in round spermatids (Costa et al., 2006). To further clarify MAEL localization during spermatogenesis, we undertook a careful analysis of MAEL localization in male germ cells by immunofluorescence on frozen sections, a three-dimensional preparation technique using fibrin clot-embedded spermatocytes and immunoelectron microscopy (IEM) (Figure 1). The combination of these approaches established that MAEL is a predominantly cytoplasmic protein that localizes to perinuclear nuage. Low MAEL levels were observed in the early stages of meiotic prophase I (leptonema to mid-pachynema) (Figure 1A, 1B, 1I, 1J, 1K). MAEL started to accumulate throughout the cytoplasm and in prominent perinuclear nuage in late pachytene and diplotene spermatocytes (Figure 1C, 1J). Meiotic metaphases and secondary spermatocytes showed a high level of MAEL in the cytoplasm as well as in nuage (Figure 1D, 1K). Round spermatids contained MAEL in the chromatoid body and in a second smaller nuage (Figure 1E and 1I). IEM revealed cytoplasmic (Figure 1M; 1N) and nuclear clusters of MAEL (Figure S1A). These results establish stage-specific recruitment of MAEL to perinuclear nuage during spermatogenesis. However, MAEL was not found in the sex body using our or commercial anti-MAEL antibodies (Figure S4A, C).
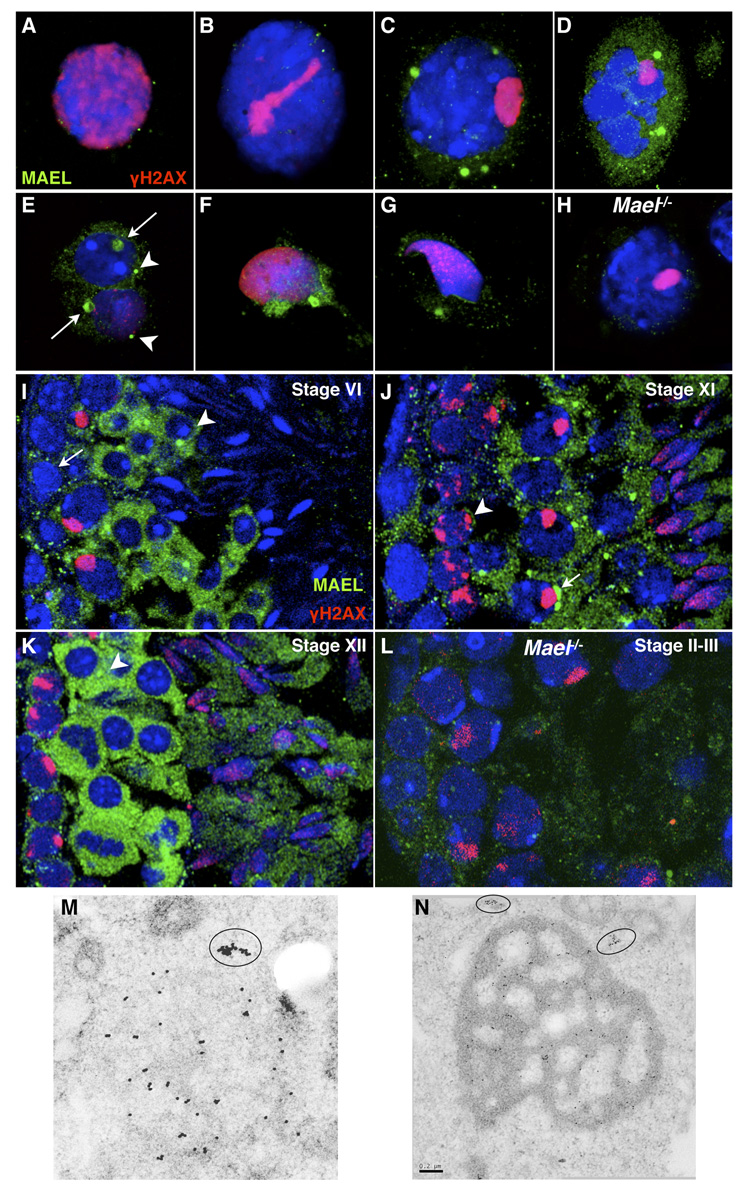
Figure 1. MAEL localizes to perinuclear nuage.
(A–H) Localization of MAEL in spatially preserved fibrin clot-embedded testis cell suspension. This approach underestimates MAEL level (compared to frozen sections below) likely due to a loss of weakly associated MAEL during the fixation step but provides a clearer picture of MAEL localization in nuage. All samples are wild type unless indicated otherwise. Cells are stained with anti-MAEL (green) and anti-γH2AX (red) antibodies. DAPI labels DNA blue.
(A – D) MAEL localization in (A) Leptotene; (B) Late zygotene; (C) Late pachytene; (D) Meiotic metaphase spermatocytes.
(E – G) MAEL localization in (E) round spermatids where arrows point to chromatoid bodies and arrowheads to a smaller nuage; (F and G) elongate spermatids with a single focus in the caudal cytoplasmic lobe also most likely to be the chromatoid body.
(H) _Mael_−/− pachytene spermatocyte subjected to identical labeling with anti-MAEL antibody.
(I–L) Localization of MAEL in germ cells as revealed by immunofluorescence in testicular frozen sections. This approach retains all cellular MAEL and is particularly useful in cells with low or modest MAEL levels. Sections are stained with anti-MAEL (green) and anti-γH2AX (red) antibodies. DAPI labels DNA blue. All samples are wild-type unless indicated otherwise.
(I) Stage VI tubule shows low MAEL signal in the type B spermatogonia (arrow), a modest increase in mid-pachytene spermatocytes (cells with γH2AX-positive sex bodies), and a sharp increase in the cytoplasm and chromatoid body of round spermatids (arrowhead).
(J) Stage XI tubule shows cytoplasmic nuage in zygotene (arrowhead) and diplotene (arrow) spermatocytes. The signal in elongating spermatids (step 10) was largely confined to the cytoplasm.
(K) Stage XII tubule shows strong cytoplasmic MAEL signal in meiotic metaphases and secondary spermatocytes, but cytoplasmic structures are apparent (arrow).
(L) No specific signal was obtained with the anti-MAEL antibody in the _Mael_−/− sections (Stage II-III).
(M) Immunoelectron microscopy localization of MAEL to a single spherical nuage in a wild type spermatocyte and to an adjacent cytoplasmic cluster (circled).
(N) Immunoelectron microscopy localization of MAEL to the chromatoid body in round spermatid and two adjacent cytoplasmic clusters. Scale bar - 0.2 µm.
Targeting of mouse Maelstrom (Mael)
To determine the functional role of MAEL in vivo, we targeted mouse Mael by homologous recombination in mouse embryonic stem (ES) cells and generated Mael null mice. A single-copy Mael locus spans 37 Kb of mouse Chromosome I (chr1:168,038,061 – 168,075,419; Assembly February 2006). The Mael locus contains 12 coding exons that are spliced into a predicted mRNA with a 1,302 nt ORF. The gene does not overlap with any other known or predicted genes, and no known small RNAs map to the Mael locus. Thus, a deletion of the whole or a part of the Mael locus will be expected to exclusively perturb MAEL.
The loss-of-function allele of Mael was generated as outlined in Figure 2A (see Experimental Procedures in the Supplementary Information for details). Matings of Mael+/− animals produced viable wild-type, heterozygous and homozygous mutant offspring in normal Mendelian ratios suggesting that the zygotic function of MAEL is dispensable for viability of embryos, neonates, and adults (Figure 2B). RT-PCR on testicular RNA samples of _Mael_−/− animals revealed no expression of the remaining coding sequence of Mael (Figure 2C). Western blot analysis revealed the presence of a protein of expected molecular weight in the wild-type but not in the _Mael_−/− testes (Figure 2D). Taken together, these results suggest that the generated mutant allele of Mael is null.
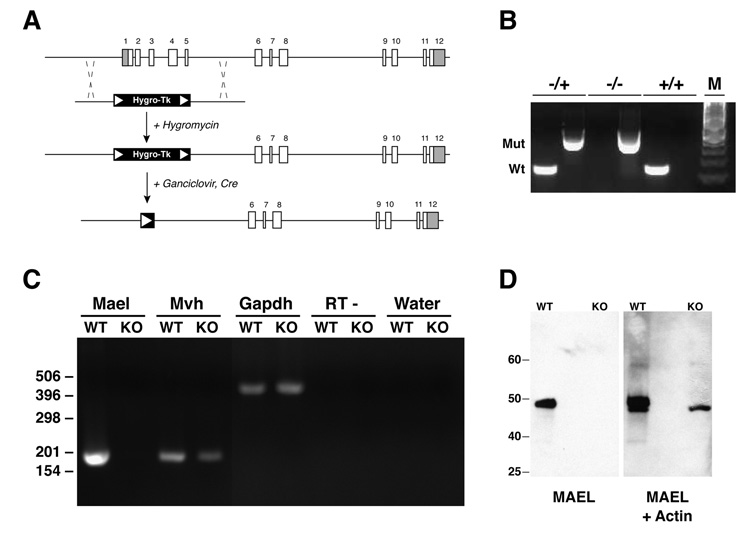
Figure 2. Construction and validation of a Mael null allele.
(A) Exon-intron structure of the Mael locus and knock-out strategy. Shaded areas of exons 1 and 12 represent 5’ and 3’ UTRs respectively. Correctly targeted hygromycin-resistant ES cell clones were transiently transfected with Cre-recombinase and cultured in the presence of ganciclovir to eliminate the selection cassette.
(B) Genotyping by PCR of wild-type and mutant Mael alleles. Tail genomic DNA are assayed independently for the presence of wild-type and mutant alleles. Wt and Mut – amplicons corresponding to the wild-type and mutated allele respectively. Inferred genotypes are shown above. M - 100 bp DNA marker.
(C) Gene expression in the wild-type (WT) and homozygous mutant (KO) testes as determined by RT-PCR using total RNA as templates. Shown are amplicons for Mael, Mvh (germ-cell specific control), and Gapdh (ubiquitous expression control). “RT - ” and “Water” control reactions carried out with Gapdh primers.
(D) Western blot of wild-type and _Mael_−/− testicular lysates probed first with anti-MAEL and then with anti-Actin (Santa Cruz) rabbit polyclonal antibodies.
MAEL is essential for spermatogenesis
Further analysis established that MAEL is essential for spermatogenesis. In contrast to their wild-type and heterozygous siblings, none of tested _Mael_−/− males (n > 20) sired progeny despite normal mating behavior as determined by the presence of mating plugs. Examination of the _Mael_−/− adult reproductive system revealed normal anatomical appearance of their reproductive system with the exception of a smaller size of testes (Figure 3A). Histological analysis of juvenile mutant testes during the first spermatogenic wave revealed a delay in meiotic entry of MAEL-deficient spermatogonia at day 10 postpartum (p10). In our experiments in the C57BL/6 genetic background, 21% of wild-type tubules at p10 and 90% of tubules at p15 contained spermatocytes (Figure 3B). By contrast, only 8% of the p10 and 70% of p20 seminiferous tubules showed evidence of meiosis in _Mael_−/− testes. This analysis also revealed widespread germ cell degeneration leading to a drop in the number of spermatocyte-containing tubules between p20 and p30 (Figure 3B). Consistent with this observation, practically every seminiferous tubule of a 56 day-old _Mael_−/− testis had evidence of a prior or ongoing germ cell degeneration (Figure 3D). Most strikingly, post-meiotic germ cells (round and elongate spermatids) were completely absent suggesting meiotic arrest of spermatogenesis in the absence of MAEL.
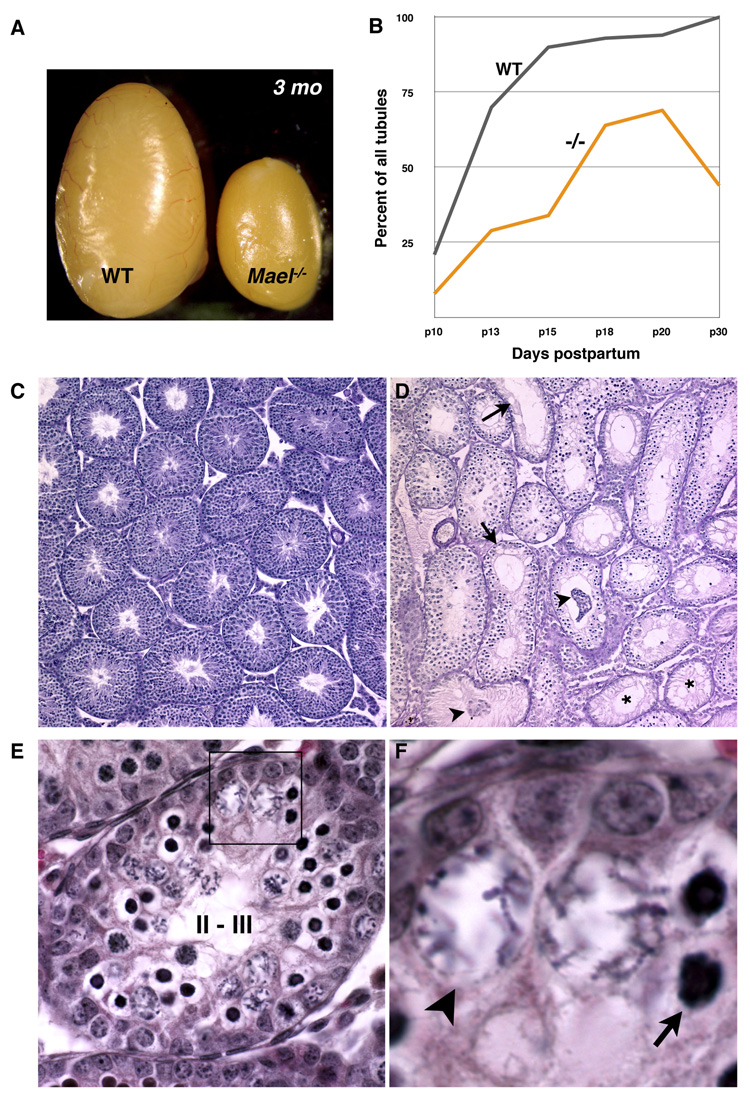
Figure 3. MAEL is essential for spermatogenesis.
(A) Gross anatomical appearance of wild type and _Mael_−/− testes of 3 month-old males.
(B) Share of spermatocyte-containing tubules in wild type and _Mael_−/− testes during the first wave of spermatogenesis (100 tubules counted for each time point).
(C, D) Gross appearance and histological sections of wild-type and _Mael_−/− testes of 56 day old animals. Mutant tubules either entirely lack germ cells (asterisk) or have gaps indicative of germ cell loss (arrows), and sloughing Sertoli and germ cells (arrowheads).
(E, F) Epithelial stage II-III tubule from a p18 _Mael_−/− mutant containing two types of abnormal spermatocytes. Atypical cells with darkly stained nuclei (arrow) appear first among zygotene spermatocytes at stage XI followed by the appearance of atypical spermatocytes with large, nuclei and scattered chromosomes. Additional images are shown in Figure S2.
Further histological analysis revealed that _Mael_−/− spermatocytes failed to complete meiotic prophase I (Figure 3E, F and Figure S2A-D). Specifically, we observed the appearance of atypical spermatocytes of two types: some zygotene – early pachytene spermatocytes began to exhibit compact nuclei (Figure 3E and 3F, arrow) while others developed expanded nuclei with scattered chromatin (Figure 3E and 3F, arrowhead). The two cells types were very similar to abnormal spermatocytes present in Miwi2 mutant testes (Carmell et al., 2007). Abnormal spermatocytes were eliminated by apoptosis by mid-pachynema, around stage IV (Figure S2) consistent with activation of the pachytene checkpoint in response to incomplete synapsis of homologous chromosomes.
MAEL is essential for normal chromosome synapsis in meiosis
To better understand meiotic failure in the absence of MAEL, we examined nuclear spreads of meiotic cells with antibodies against SYCP3 and γH2AX (Figure 4). SYCP3 is an essential component of axial elements (prior to chromosome synapsis) and the lateral elements (after synapsis) of the synaptonemal complex (SC) (Yuan et al., 2000). γH2AX is a marker of double-strand DNA breaks (DSBs) in normal leptotene and zygotene spermatocytes, the sex body and autosomal asynaptic chromatin in pachytene spermatocytes (Celeste et al., 2002; Mahadevaiah et al., 2001; Turner et al., 2005). Hence, in wild-type spermatocytes, SYCP3 and γH2AX exhibit dynamic but characteristic localization patterns during meiotic prophase I (Figure 4B – 4E). Thus, the SYCP3 and γH2AX patterns provide a powerful tool to characterize meiotic progression in the absence of MAEL.
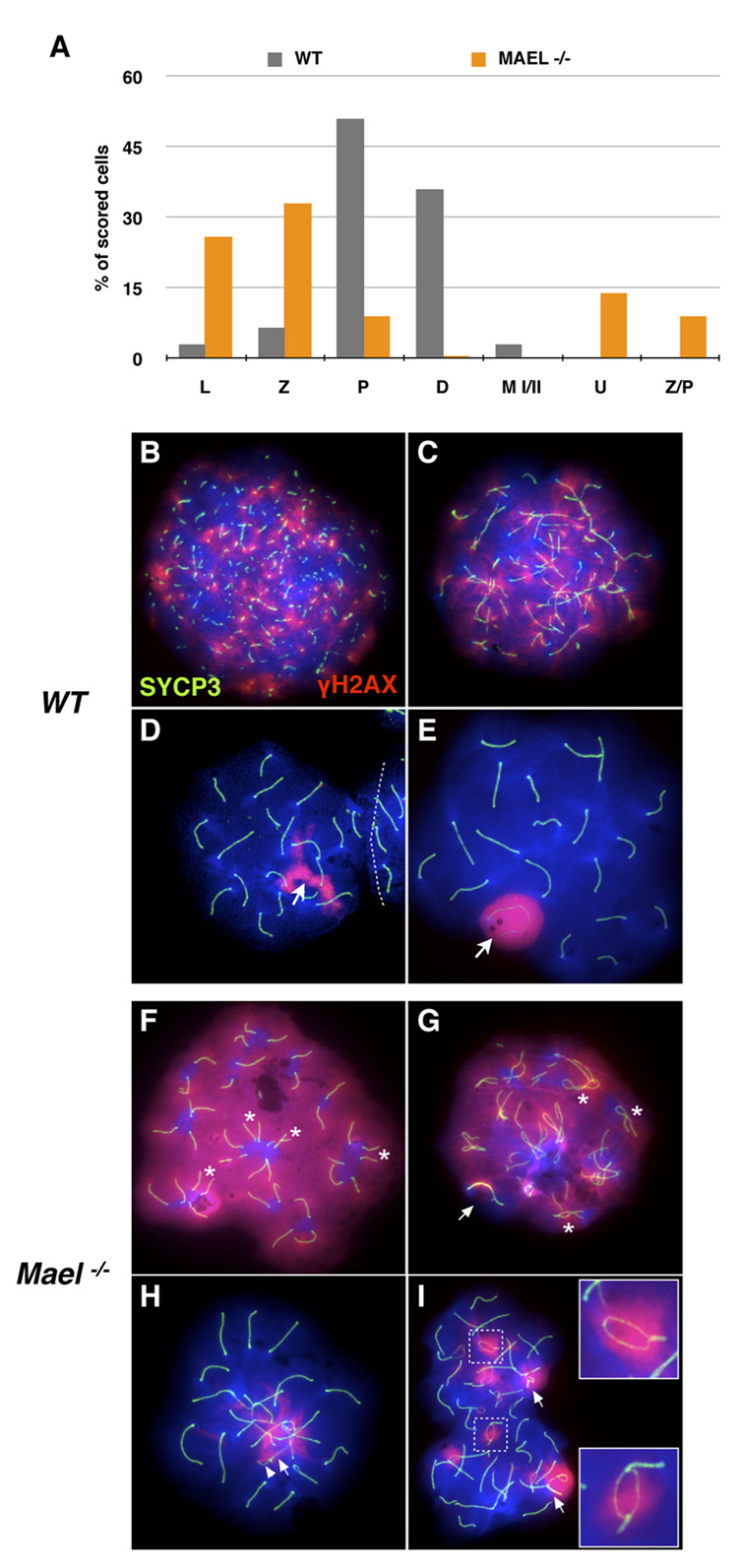
Figure 4. MAEL is required for chromosome synapsis.
Meiotic spreads in (B – I) are stained with antibodies against H2AX phosphorylated on S139 histone (red) and SYCP3 (green). DAPI labels DNA blue.
(A) Distribution of wild-type (gray bars, 400 scored nuclei) and _Mael_−/− (orange bars, 800 scored nuclei) spermatocytes among different substages of the meiotic prophase I (L - leptotene, Z - zygotene, P - pachytene, D - diplotene), metaphase (M I/II) and atypical classes (U − 40 univalents, Z/P - nuclei combining zygotene and pachytene features) of spermatocytes.
(B–E) Leptotene (B), zygotene (C), zygotene-pachytene transition (D) and pachytene (E) stages of the first meiotic prophase in wild type spermatogenesis.
(F–I) Representative examples of univalent (F), Z/P (G, H) and pachytene (I) _Mael_−/− spermatocytes.
(F) A _Mael_−/− spermatocyte with 40 univalent (fully asynaptic) chromosomes and high levels of DNA DSBs as indicated by γH2AX signal. Pairs of equal-sized axial elements in proximity of each other (asterisk) suggest homolog recognition.
(G) A Z/P _Mael_−/− spermatocyte with synapsed sex chromosomes (arrow) but only limited (asterisk) synapsis of autosomes and extensive DNA damage (γH2AX signal).
(H) A Z/P _Mael_−/− spermatocyte with minor autosomal asynapsis and synapsed X (arrow) and Y (arrowhead) chromosomes.
(I) A pachytene _Mael_−/− spermatocyte with asynapsed regions within bivalents (insets) and non-homologous synapsis (arrows).
Careful analysis of 800 meiotic nuclei obtained from two Mael mutant males (2 months of age) uncovered severe impairment of meiosis in the absence of MAEL (summarized in Figure 4A with examples shown in Figure 4F – 4I) (similar results were obtained using meiotic spreads from developmental series of juvenile testes spanning the entire first wave of spermatogenesis (postnatal days p10 – p42) (data not shown)). In contrast to pachytene and diplotene spermatocyte-rich wild-type spreads (Figure 4A), leptotene and zygotene spermatocytes dominated spreads in the Mael mutants (26% and 33%, respectively). Only a minority of spermatocytes (9%) was scored as pachytene (P bar in Figure 4A) and less than 1% of spermatocytes reached diplonema (D bar in Figure 4A). Thus, _Mael_-deficient spermatocytes fail in zygonema of meiotic prophase I.
Closer examination revealed a profound defect in homologous chromosome synapsis in the absence of MAEL. In 14% of _Mael_−/− spermatocytes despite some evidence of homolog interaction, all 40 chromosomes were fully asynaptic (univalent) and contained high levels of γH2AX in euchromatin (U bar in Figure 4A; 4F). Other _Mael_−/− spermatocytes (9% of counted cells) were on the verge of entering pachynema (judging by the extent of sex chromosome synapsis) but were held back by autosomal asynapsis (Z/P bar in Figure 4A; Figure 4G and 4H). Synaptic defects were also prominent in 75% of pachytene _Mael_−/− spermatocytes containing γH2AX-positive sex bodies (P bar in Figure 4A, Figure 4I). Collectively, these findings identify abnormal chromosome synapsis as a common meiotic defect in _Mael_−/− spermatocytes and suggest a role for MAEL downstream of partner recognition but prior or during the initiation of synapsis.
MAEL is not required for meiotic sex chromosome inactivation (MSCI)
To address a previously proposed role of MAEL in MSCI, we examined _Mael_−/− pachytene spermatocytes by fluorescence in situ hybridization with a Cot-1 probe for the presence of nascent transcripts in the γH2AX-positive sex bodies. The sex bodies of wild-type and _Mael_−/− spermatocytes showed a comparably low level of Cot-1 signal (Figure S3). Thus, MAEL is not required for MSCI. Furthermore, we determined that the anti-MAEL antibody used in the original report (Costa et al, 2006) is not specific to MAEL. We carried out Western blot (Figure S4A) and immunofluorescence (Figure S4B) studies on wild type and _Mael_−/− samples with the anti-MAEL antibody used in (Costa et al., 2006). These experiments revealed cross-reactivity of this anti-MAEL antibody with proteins other than MAEL both on Western blots and in the sex body of _Mael_−/− spermatocytes. Collectively, these results rule out a direct role of MAEL in MSCI.
Biogenesis of piRNAs in the absence of MAEL
_Mael_-deficient germ-cell phenotypes were reminiscent of the defects observed in Mili (Kuramochi-Miyagawa et al., 2004) and Miwi2 (Carmell et al., 2007) mutants. Could MAEL be required for biogenesis of MILI/MIWI/MIWI2 associated piRNAs? To address this hypothesis, we examined small RNAs isolated from wild-type and Mael mutant testes. Ethidium bromide staining of the adult wild-type small RNAs revealed a characteristic 30–32 nt piRNA cloud (Figure S5A). In contrast, corresponding RNA species were never observed in adult _Mael_−/− mutant testes. Since repeat-targeting pre-pachytene piRNAs (Aravin et al., 2007b) are present at levels insufficient for their detection by in-gel staining, we turned to Northern blots to evaluate their expression. Using p15 small RNA samples, we found evidence of a pre-pachytene piRNA targeting SINE elements present in _Mael_−/− mutant animals (Figure S5B), demonstrating that mice lacking MAEL remain competent for production of pre-pachytene piRNAs.
However, we found no evidence of pachytene piRNAs in mice lacking MAEL at p18 (Figure S5C) or any later age (Figure S5D). Despite the fact that pachytene piRNAs could never be detected by Northern blot analysis in Mael mutant mice, using long-range RT-PCR we were able to identify, albeit at a lower level, a full-length precursor transcript that gives rise to numerous pachytene piRNAs from a cluster on Chromosome 6 (Figure S5E) (Ro et al., 2007). Given that _Mael_-deficient spermatocytes do not progress past mid-pachynema, lack of pachytene piRNAs could be explained by the failure of _Mael_-deficient spermatocytes to reach a pachytene-piRNA-producing stage of spermatogenesis.
Derepression of transposable elements in the _Mael_−/− male germline
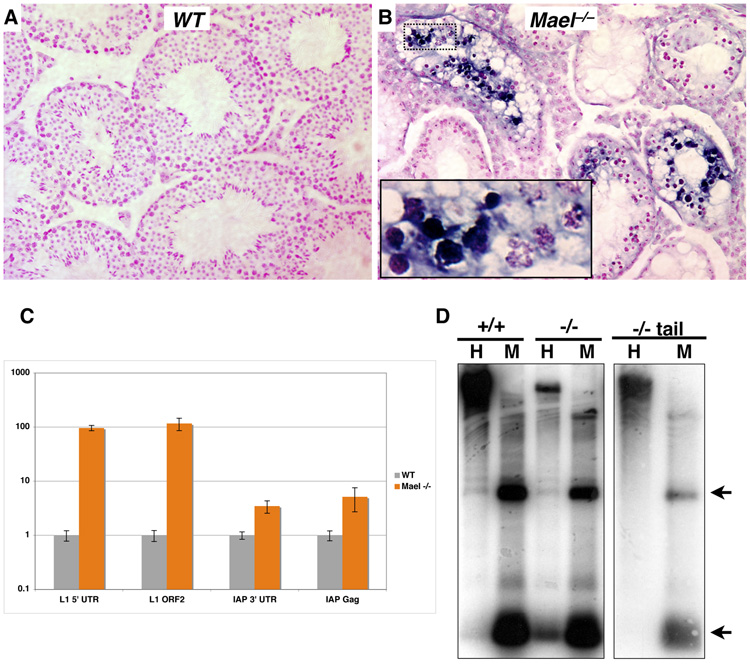
To determine if the loss of mouse MAEL results in TE derepression, we examined expression of L1 element on adult testis sections by in situ RNA hybridization using an anti-sense L1 probe (Figure 5A and 5B). Unlike wild-type, the _Mael_−/− testis had high levels of L1 transcript, particularly in tubules containing zygotene and abnormal early pachytene spermatocytes (inset, Figure 5B). No specific staining was obtained using sense L1 probe (data not shown).
Figure 5. MAEL is essential for TE repression and L1 DNA methylation.
(A, B) In situ RNA hybridization analysis of L1 expression in the testes of adult wild type (A) and _Mael_−/− animals (B). A probe anti-sense to the 5’-UTR of L1 was hybridized to adult testis sections. Sections are counterstained with nuclear fast red.
(C) Quantitative analysis of retroelement expression. qPCR was performed on random-hexamer primed cDNAs made from whole RNA extracted from adult testes. Experiments were performed in triplicate.
(D) Southern blot analysis of L1 DNA methylation. Lanes marked by H contain genomic DNA cut with _Hpa_II, a methylation sensitive restriction enzyme, while M denotes DNA cut with the methylation insensitive enzyme _Msp_I. Blots were probed with the same probe used for in situ analysis. Arrows mark expected bands. Genomic DNA is from testes unless otherwise noted.
To quantify derepression of TEs in the absence of MAEL, we examined L1 and intracisternal A particle (IAP) element expression in adult testis by quantitative RT-PCR (qPCR). In the adult mutant testes, we observed a 100-fold increase in the levels of L1 expression over the wild-type (Figure 5C). We also observed a three- to five-fold increase in IAP expression in adult _Mael_−/− mutants over the wild-type. Thus, a component of mammalian nuage is essential for transcriptional or post-transcriptional silencing of at least two unrelated TEs in the mouse male germline.
To determine if the observed derepression of TEs in Mael mutants was accompanied by reduced CpG methylation, we carried out Southern blot analysis using an L1 probe. Samples of testicular genomic DNA of wild type and _Mael_−/− adult animals were digested individually with isoschizomers of opposite CpG methylation sensitivity. Such analysis demonstrated that the L1 5’UTR was fully methylated in the wild type testes (+/+ sample, Figure 5D). Likewise, genomic DNA extracted from the tail of _Mael_-deficient adult males appeared fully methylated leading us to conclude that MAEL is not required for somatic DNA methylation (−/− tail sample, Figure 5D). However, digests of _Mael_−/− testis samples revealed a loss of DNA methylation in the mutant germ cells (−/− sample, Figure 5D). This result is consistent with the MAEL role in transcriptional repression of TEs by a mechanism that involves DNA methylation.
Loss of MAEL causes a vast accumulation of L1 RNPs
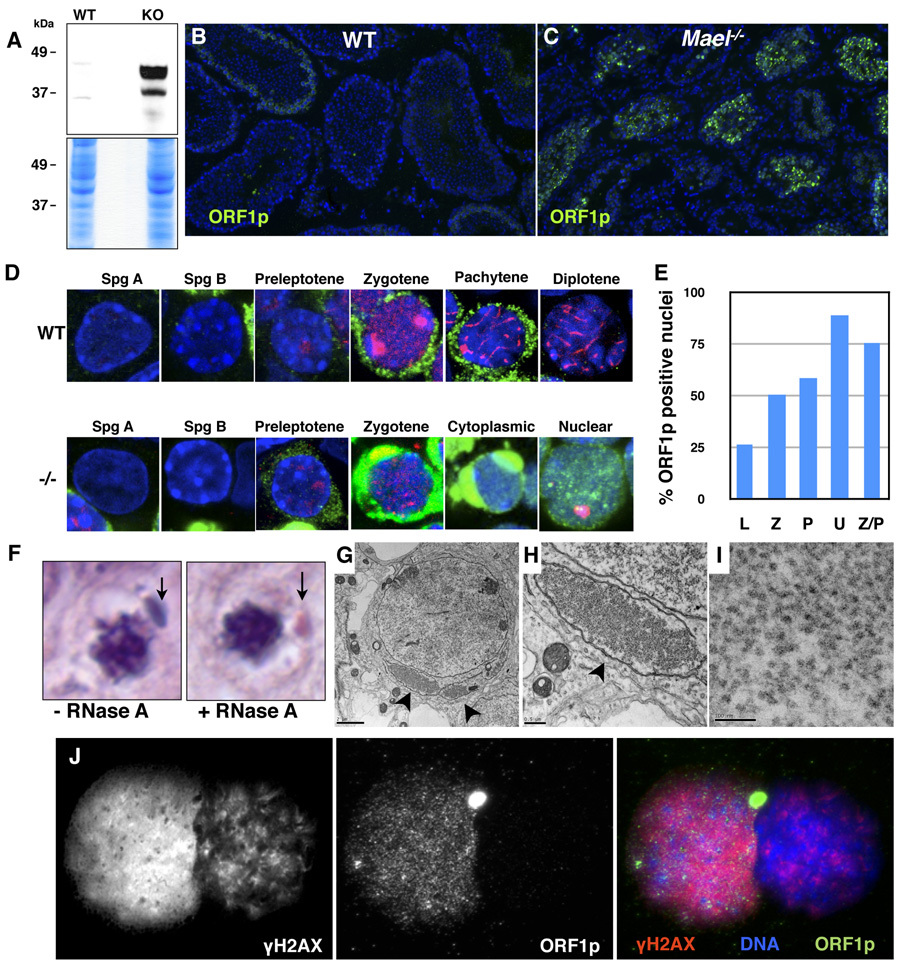
Could L1 derepression be a primary cause of the meiotic failure and excessive DNA damage observed in the Mael mutant testes? L1 transposition depends on the function of both of its encoded proteins, ORF1p and ORF2p (Babushok and Kazazian, 2007; Martin, 2006; Moran et al., 1996). 42 to 44.6 kDa isoforms of ORF1p can be detected in L1-expressing embryonal carcinoma cells (Martin and Branciforte, 1993) and in the testis (Branciforte and Martin, 1994). We probed protein lysates prepared from wild-type and _Mael_−/− adult testes with an antibody to ORF1p (Martin and Branciforte, 1993) and observed a vast increase in the ORF1p levels in the mutant sample (Figure 6A). Consistent with this result, we observed a massive increase in the number of ORF1p-positive germ cells in the _Mael_−/− testis (Figure 6C). Unlike wild-type (Figure 6B) or a different meiotic mutant (_Spo11_−/−, Figure S6) where ORF1p was present in only a subset of tubules at low to moderate levels, _Mael_−/− testes contained a large number of tubules with a high level of ORF1p (Figure 6C). These results demonstrate that transcriptional derepression of L1 elements results in a vast upregulation of ORF1p levels specifically in the _Mael_−/− testis.
Figure 6. L1 ORF1p accumulation in cytoplasmic enclaves and nuclei of _Mael_−/− spermatocytes.
(A) Western blot analysis using an antibody to L1 ORF1p reveals a large increase in the protein levels in _Mael_−/− testis.
(B) Immunofluorescence using anti-ORF1p antibody on frozen sections of wild-type testes.
(C) Immunofluorescence using anti-ORF1p antibody on frozen sections of _Mael_−/− testes. A significant number of _Mael_−/− tubules contain spermatocytes with high levels of ORF1p in the cytoplasm and nuclei.
(D) ORF1p localization in wild type (WT) and _Mael_−/− (−/−) germ cells. Spg A – type A spermatogonia, Spg B – type B spermatogonia, Cytoplasmic – localization of ORF1p to cytoplasmic enclaves. Green - ORF1p, Red - SYCP3, DAPI stains DNA blue.
(E) Percent of ORF1p positive nuclei in various classes of _Mael_−/− spermatocytes described in Figure 4.
(F) Cytoplasmic enclaves (arrows) in a p18 _Mael_−/− spermatocytes. Shown are representative examples of spermatocytes from histological H & E sections either subjected or not to RNase A treatment prior to staining.
(G) Electron microscopy of _Mael_−/− testis reveals perinuclear domains (arrowheads) packed with presumed L1 RNPs (bar - 2 µm).
(H) A close-up view of the cytoplasmic enclave shown in (F) (bar − 0.5 µm).
(I) A close-up view of the presumed L1 RNPs in the cytoplasmic enclave (bar − 100 nm).
(J) Co-localization of ORF1p and γH2AX in the nucleus of a Univalent _Mael_−/− spermatocyte. γH2AX; ORF1p double positive spermatocyte is inferred to be a Univalent spermatocyte based on the pattern of γH2AX staining (high level in euchromatin, very little in heterochromatin; compare to Figure 4F).
The disruption of MAEL altered the timing of ORF1p expression. In the normal testis, detectable ORF1p expression was limited to leptotene, zygotene and early pachytene spermatocytes (Figure 6D and Figure S7). By contrast, in _Mael_−/− testis ORF1p expression was first detected in preleptotene spermatocytes (enhanced cytoplasmic signal in Figure 6D; see Figure S7, _Mael_−/− panel for the original image). Subsequently, all _Mael_−/− leptotene, zygotene, early pachytene and atypical spermatocytes showed a high level of ORF1p. Type A and B spermatogonia, like in the wild type, showed little or no expression of ORF1p. Thus, in the absence of MAEL, L1 ORF1p expression is initiated precociously, in preleptotene rather than in leptotene spermatocytes at the onset of the meiotic program.
High level of L1 expression in the absence of MAEL led to the flooding of the cytoplasm with L1 RNPs. In the cytoplasm, ORF1p was often observed in large perinuclear enclaves (Figure 6D, Cytoplasmic). Similar cytoplasmic, bean-shaped structures were also prominent in our histological samples stained with hematoxylin and eosin (Figure 6F). We posited that these structures are enriched in L1 RNPs. Indeed, a brief pre-treatment of the sections prior to hematoxylin/eosin staining with RNase A reduced detection of these structures by the dye (Figure 6F). In electron micrographs, these enclaves appeared as large cytoplasmic territories almost completely surrounded by membranes and filled with small particles of approximately 25 – 30 nm diameter that could represent L1 RNPs as they were labeled with the anti-ORF1p antibody (Martin and Branciforte, 1993) (immunofluorescence in Figure 6G – 6I, and immunoelectron microscopy Figure S8).
In a dramatic contrast to the wild-type spermatocytes where none of 852 scored ORF1ppositive cells exhibited nuclear localization, 34% of 1098 _Mael_−/− spermatocytes contained nuclear ORF1p (Figure 6D, Nuclear). Double-labeling of _Mael_−/− nuclear spreads with SYCP3 and ORF1p antibodies revealed that 89% of Univalent and 76% of Z/P spermatocytes (described earlier, shown in Figure 4) contained nuclear ORF1p. These results demonstrate that nuclear localization of L1 RNPs correlates with the most prominent representation of chromosome asynapsis in the absence of MAEL.
Overexpression of L1 elements in cultured human cells is accompanied by the accumulation of γH2AX foci suggesting generation of DSBs in the transposition process (Belgnaoui et al., 2006; Gasior et al., 2006). To address the relationship between L1 expression and DNA damage in the mouse germline, we examined 200 _Mael_−/− spermatocytes for nuclear co-localization of ORF1p and γH2AX (Figure 6J). The analysis revealed that 90% of _Mael_−/− spermatocytes with nuclear ORF1p exhibited a high level of γH2AX. Likewise, 90% of γH2AX-positive mutant nuclei contained nuclear ORF1p. Cumulatively, these experiments demonstrated that, in the absence of MAEL, derepression of L1 results in a vast accumulation of ORF1p in cytoplasmic enclaves and in nuclei that show signs of severe DNA damage.
DNA damage in the absence of MAEL is independent from meiotic DNA DSBs
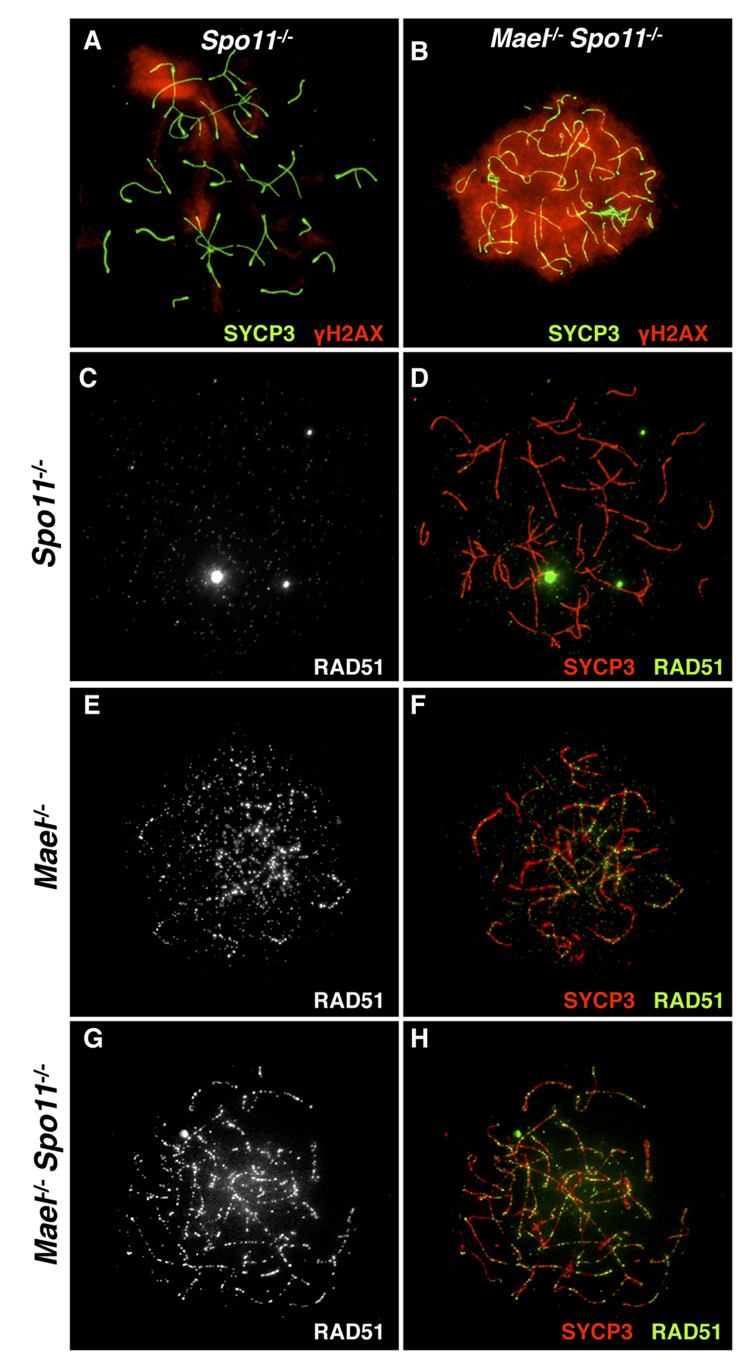
Meiosis is characterized by programmed generation of a large number of DSBs (300 – 400 in the mouse) introduced by SPO11, a type II-like topoisomerase (Bergerat et al., 1997; Keeney et al., 1997). Only around 25 of these DSBs are utilized for the exchange of genetic information between homologous chromosomes, while the remaining DSBs must be repaired to allow further progression of meiosis. To differentiate between the incomplete repair of meiotic DSBs and other DNA damage that might have been accumulating in the absence of MAEL, we created double-mutant animals lacking both MAEL and SPO11 proteins. To reveal DNA DSBs in _Mael_−/− and _Spo11_−/− (Romanienko and Camerini-Otero, 2000) single mutant and _Mael_−/−; _Spo11_−/− double mutant spermatocytes, we used a combination of antibodies to SYCP3 and γH2AX. In agreement with (Baudat et al., 2000; Bellani et al., 2005; Romanienko and Camerini-Otero, 2000), _Spo11_−/− spermatocytes exhibited a zygotene-like arrest and few amorphous areas of γH2AX accumulation (Figure 7A). In contrast, the elimination of MAEL in a _Spo11_-deficient background induced extensive γH2AX signal indicative of the accumulation of a large number of DSBs throughout the genome (Figure 7B).
Figure 7. Accumulation of DNA damage in _Mael_−/−; _Spo11_−/− spermatocytes.
(A, B) SYCP3 (green) and γH2AX (red) localization in _Spo11_−/− (A) and _Mael_−/−;_Spo11_−/− spermatocytes (B).
(C – H) SYCP3 (red) and RAD51 (white in C, E, G and green in D, F, H) localization in _Spo11_−/− (C, D), _Mael_−/− (E, F) and _Mael_−/−; _Spo11_−/− (G, H) spermatocytes.
To confirm the presence of SPO11-independent DSB DNA breaks in the absence of MAEL, we examined the localization of RAD51, a recombinase physically recruited to DSBs (Kawabata et al., 2005). Consistent with the absence of meiotic DSBs, we did not observe RAD51 localization to the axial elements in the _Spo11_−/− spermatocytes (Figure 7C and 7D). Single _Mael_−/− spermatocytes contained large numbers of RAD51 foci (Figure 7E and 7F). In a striking contrast to _Spo11_−/− mutants, _Mael_−/−; _Spo11_−/− late zygotene spermatocytes also contained RAD51 foci on axial elements (Figure 7G and 7H). These results demonstrate that DSBs in _Mael_−/− spermatocytes are created by a SPO11-independent mechanism.
Discussion
To test the hypothesis of nuage participation in transposon repression in the context of the mammalian germline, we studied the localization and functional role of a murine homolog of a Drosophila nuage protein Maelstrom. We demonstrate that mouse MAEL localizes to perinuclear nuage in spermatocytes and round spermatids. Disruption of MAEL results in a profound defect in synapsis of homologous chromosomes in male meiosis, DNA demethylation of L1 elements and a 100-fold increase in L1 expression in the adult testis. These results support the idea of nuage function in transposon silencing in a wide range of animal species.
Our studies of Mael mutant testes suggest close functional relationship of MAEL with MIWI2 and MILI, two of three murine PIWI-like proteins. Mael, Mili and _Miwi2_-deficient spermatocytes fail to complete meiotic prophase and to assemble functional synaptonemal complexes (Kuramochi-Miyagawa et al., 2004; Carmell et al., 2007). Interestingly Mael and _Miwi2_-deficient spermatocytes develop identical atypical morphologies (not reported for the Mili mutant) prior to their elimination by apoptosis (Carmell et al., 2007). Furthermore, Mael and Miwi2 mutant testes exhibit common pattern of TE activation (high levels of L1 expression and only modest derepression of IAP) that contrasts TE expression in the Mili mutant (Aravin et al., 2007b; Carmell et al., 2007; Kuramochi-Miyagawa et al., 2008). These data suggest that MAEL plays a more prominent role in the MIWI2-dependent processes, possibly at a post-piRNA production step (since we did not observe reduction of pre-pachytene piRNAs in the Mael mutant). Given that MAEL was reported to shuttle between the nucleus and cytoplasm in Drosophila (Findley et al., 2003) and MAEL complexes were observed in the nucleus and at nuclear pores in this study, we speculate that MAEL may facilitate trafficking of MIWI2-piRNA complexes to or from nuage.
Of particular interest is our observation of L1 expression at the onset of meiosis in the absence of MAEL. It is generally accepted that de novo DNA methylation of TEs in gonocytes of late gestation male fetuses is crucial for TE repression in the male germline (Bourc'his and Bestor, 2004; La Salle et al., 2007). In addition, recent studies demonstrated essential roles of MILI and MIWI2 in de novo DNA methylation of TEs in fetal gonocytes (Kuramochi-Miyagawa et al., 2008). Once established in the fetal germ cells, repressive DNA methylation of TEs is expected to be maintained in the male germline into the adulthood. One would expect, therefore, L1 derepression to be apparent in both pre-meiotic and meiotic germ cell populations of an adult _Mael_-deficient testis. In our study, however, L1 RNA and ORF1p are found exclusively in the adult _Mael_−/− meiotic germ cells and not in the spermatogonia. L1 ORF1p expression starts in preleptotene _Mael_−/− spermatocytes (characterized by the pre-meiotic S phase) and persists until germ cell elimination. These results suggest a more dynamic control of TE expression at the onset of meiosis and parallel the findings of an earlier study of L1 expression in the testes (Branciforte and Martin, 1994). Taken together, the two studies suggest a tantalizing possibility of existence of an epigenetic reprogramming step at the onset of meiosis that involves transient derepression of TEs.
Our study provides compelling evidence of a highly detrimental effect of TE derepression on the male germline. We demonstrate that L1 transcriptional derepression in the absence of MAEL leads to flooding of spermatocytes with ORF1p found both in the cytoplasm and nuclei. Cytoplasmic ORF1p is often present in large enclaves that contain vast numbers of 25 – 30 nm particles that could be identical to L1 RNPs observed previously (Martin and Branciforte, 1993). Nuclear ORF1p localization strongly correlates with accumulation of severe DNA damage that is mechanistically distinct from developmentally programmed SPO11-generated meiotic DSBs. The presence of numerous RAD51 foci in the absence of SPO11 further supports the idea that _Mael_−/− spermatocytes harbor an endonuclease that is absent or inactive in wild-type and _Spo11_−/− spermatocytes. A leading candidate for such an enzyme is L1-encoded ORF2p whose N-terminus has endonuclease activity (Feng et al., 1996) that is required for retrotransposition and generation of DSBs in cultured human cell lines (Gasior et al., 2006; Moran et al., 1996). Finally, nuclear accumulation of L1 RNPs also strongly correlates with defects in chromosome synapsis. A great majority of _Mael_−/− spermatocytes exhibiting the most severe disruption of synapsis contain nuclear L1 RNPs (89% of cells in the univalent class). The precise mechanism of suppression of synapsis by L1 is yet to be understood, but our results already suggest a strong correlation between the frequency of nuclear L1 RNPs, the severity of DNA damage and asynapsis. Cumulatively, our study vividly illustrates an absolute requirement for efficient TE restraining mechanisms in the mammalian germline.
Experimental Procedures
Generation of Mael mutation
Gene targeting was performed in V6.5 (129SvJae × C57BL/6)F1 ES cells. Following germline transmission of the mutant allele, the Mael mutation was backcrossed to the C57BL/6 genetic background for 10 generations. See Supplemental Information for details.
Anti-MAEL antibody generation
Rabbit polyclonal affinity-purified anti-MAEL antibodies to a peptide corresponding to the C-terminal 22 amino-acids of MAEL were produced at 21st Century Biochemicals (Marlboro, MA).
Histology
Freshly dissected testes were fixed in Bouin’s fixative overnight at room temperature. Testes were processed to paraffin, embedded and sectioned at a thickness of 6 microns. Slides were stained with hematoxylin and eosin.
Cryosections
After dissection of the testis, the tunica was punctured, the testis was fixed (2% PFA in PBS) on ice for 4 hours. Samples were passed through a sucrose gradient (10%, 20%, 30% in water) and embedded in OCT. Sections of 10 µm were used.
Preparation of fibrin-clot embedded samples
Fibrin clots were made as described in (Baart et al., 2000; Hunt et al., 1995).
Cot-1 in situ Hybridization
Cot-1 in situ hybridization was performed as previously described (Turner et al., 2005).
Preparation of surface spreads
Nuclear spreads of testis cell suspension were obtained as described in (Peters et al., 1997), with minor modifications described in Supplemental Information.
Antibodies
Mouse monoclonal anti-γH2AX (1:1000, Upstate; #05-636), rabbit polyclonal anti-SYCP3 (1:750, Abcam; ab15092), guinea pig polyclonal anti-Sycp3 (1:300, R. Benavente), anti-hRAD51 (1:300, R. Kanaar), rabbit polyclonal anti-ORF1p (1:500 for IF, 1:2000 for WB, S. L. Martin), rabbit polyclonal anti-MAEL (1:100 for IF and 1:500 for WB, Abcam; ab28661), rabbit polyclonal anti-MAEL (1:300 for IF, 1:1000 for WB, H. Cooke), rabbit polyclonal anti-Actin (c-11)(1:500, Santa-Cruz; sc-1615). The Zenon Alexa Fluor 594 Rabbit IgG Z25307 labeling kit, was used to directly label Sycp3 antibody according to the manual. Secondary antibodies used were Alexa goat-anti-rabbit 488, Alexa goat-anti-guinea pig 594 (Molecular Probes) and goat-anti-mouse Texas Red (Jackson Immunoresearch). All secondary antibodies were used at a 1:500 dilution.
Supplementary Material
01
Acknowledgements
A.B. is deeply indebted to Dr. David Page for his generous support in the initial phase of the project, discussions and comments on the manuscript. We thank Dr. R. Kanaar and Dr. J. Essers for the gift of the hRAD51 antibody, Dr. R. Benavente for the gift of the SYCP3 antibody, Dr. H. Cooke for the gift of MAEL antibody, Dr. R. D. Camerini-Otero for the generous gift of the Spo11 mice, Dr. Eugenia Dikovsky and the animal facility staff for their invaluable help, Michael Sepanski for assistance with EM studies and Dr. Mary Goll for advice and discussions. We thank Dr. Allan Spradling, Dr. Joseph Gall, Dr. Douglas Koshland, Dr. Marnie Halpern, Dr. Jeff Han, Dr. Safia Malki, Julio Castañeda and Dr. Mary Goll for their comments on the manuscript. This study was initiated when A.B. was a Special Fellow of The Leukemia and Lymphoma Society. S.L.M. research was supported by NIH GM40367. The work was supported by Carnegie Institution of Washington.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. doi: 10.1038/nature04916. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007a;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007b;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Bourc'his D. Small RNA guides for de novo DNA methylation in mammalian germ cells. Genes Dev. 2008;22:970–975. doi: 10.1101/gad.1669408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baart EB, de Rooij DG, Keegan KS, de Boer P. Distribution of Atr protein in primary spermatocytes of a mouse chromosomal mutant: a comparison of preparation techniques. Chromosoma. 2000;109:139–147. doi: 10.1007/s004120050422. [DOI] [PubMed] [Google Scholar]
- Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Human Mutation. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S. Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol. Cell. 2000;6:989–998. doi: 10.1016/s1097-2765(00)00098-8. [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 2006;6:13. doi: 10.1186/1475-2867-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellani MA, Romanienko PJ, Cairatti DA, Camerini-Otero RD. SPO11 is required for sex-body formation, and Spo11 heterozygosity rescues the prophase arrest of Atm−/− spermatocytes. J. Cell Sci. 2005;118:3233–3245. doi: 10.1242/jcs.02466. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cel. Biology. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell. 2007;12:503–514. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg NJ, Frost DM, Larkin MK, Subrahmanyan L, Bryant Z, Ruohola-Baker H. maelstrom is required for an early step in the establishment of Drosophila oocyte polarity: posterior localization of grk mRNA. Development. 1997;124:4661–4671. doi: 10.1242/dev.124.22.4661. [DOI] [PubMed] [Google Scholar]
- Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ. Mouse MAELSTROM: the link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum. Mol. Gen. 2006;15:2324–2334. doi: 10.1093/hmg/ddl158. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Goodier JL, Ostertag EM, Kazazian HH., Jr Rapid amplification of a retrotransposon subfamily is evolving the mouse genome. Nature Gen. 1998;20:288–290. doi: 10.1038/3104. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Germ plasm and the differentiation of the germ cell line. Int. Rev. Cyt. 1975;43:229–280. doi: 10.1016/s0074-7696(08)60070-4. [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development. 2003;130:856–871. doi: 10.1242/dev.00310. [DOI] [PubMed] [Google Scholar]
- Gasior SL, Wakeman TP, Xu B, Deininger PL. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 2006;357:1383–1393. doi: 10.1016/j.jmb.2006.01.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Hannon GJ. Conserved themes in small-RNA-mediated transposon control. Trends Cell Bio. 2008;18:136–148. doi: 10.1016/j.tcb.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard L, Freeling M. Regulatory changes as a consequence of transposon insertion. Dev. Gen. 1999;25:291–296. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2004;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Erhardt S, Lane N, El Maarri O, Reik W, Walter J, Surani MA. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117:15–23. doi: 10.1016/s0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Han JS, Boeke JD. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: domain composition, intracellular localization, and function in male germ cells in mice. Dev. Biol. 2007;301:38–52. doi: 10.1016/j.ydbio.2006.10.046. [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Fillipov DV, Blaser H, Raz E, Moens CB, et al. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell. 2007;129:69–82. doi: 10.1016/j.cell.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Hunt P, LeMaire R, Embury P, Sheean L, Mroz K. Analysis of chromosome behavior in intact mammalian oocytes: monitoring the segregation of a univalent chromosome during female meiosis. Hum. Mol. Gen. 1995;4:2007–2012. doi: 10.1093/hmg/4.11.2007. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Kawabata T, Nishibori M. Role of recA/RAD51 family proteins in mammals. Act. med. Okayama. 2005;59:1–9. doi: 10.18926/AMO/31987. [DOI] [PubMed] [Google Scholar]
- Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev. Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135:3–9. doi: 10.1242/dev.006486. [DOI] [PubMed] [Google Scholar]
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc. Natl. Acad. Sci. USA. 2006;103:2647–2652. doi: 10.1073/pnas.0509333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P. The chromatoid body: a germ-cell-specific RNA-processing centre. Nat. Rev. Mol. Cell Biol. 2007;8:85–90. doi: 10.1038/nrm2081. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Tototki Y, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev. 2008;22:908–917. doi: 10.1101/gad.1640708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Oakes CC, Neaga OR, Bourc'his D, Bestor TH, Trasler JM. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev Biol. 2007;7:104. doi: 10.1186/1471-213X-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lim AK, Kai T. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 2007;104:6714–6719. doi: 10.1073/pnas.0701920104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodriguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat. Genetics. 2001;27:271–276. doi: 10.1038/85830. [DOI] [PubMed] [Google Scholar]
- Martin SL. The ORF1 Protein Encoded by LINE-1: Structure and Function During L1 Retrotransposition. J. Biomed. Biotech. 2006;2006:45621. doi: 10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SL, Branciforte D. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cel. Bio. 1993;13:5383–5392. doi: 10.1128/mcb.13.9.5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development. 2005;132:4029–4039. doi: 10.1242/dev.02003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chrom. Res. 1997;5:66–68. doi: 10.1023/a:1018445520117. [DOI] [PubMed] [Google Scholar]
- Ro S, Park C, Song R, Nguyen D, Jin J, Sanders KM, McCarrey JR, Yan W. Cloning and expression profiling of testis-expressed piRNA-like RNAs. RNA. 2007;13:1693–1702. doi: 10.1261/rna.640307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini-Otero RD. The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol. Cell. 2000;6:975–987. doi: 10.1016/s1097-2765(00)00097-6. [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech. Dev. 2000;93:139–149. doi: 10.1016/s0925-4773(00)00283-5. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Fernandez-Capetillo O, Nussenzweig A, Xu X, Deng CX, Burgoyne PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nature Genetics. 2005;37:41–47. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Yuan L, Liu JG, Zhao J, Brundell E, Daneholt B, Hoog C. The murine SCP3 gene is required for synaptonemal complex assembly, chromosome synapsis, and male fertility. Mol. Cell. 2000;5:73–83. doi: 10.1016/s1097-2765(00)80404-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01