Positive feedback of G1 cyclins ensures coherent cell cycle entry (original) (raw)
. Author manuscript; available in PMC: 2009 Jan 17.
Published in final edited form as: Nature. 2008 Jul 17;454(7202):291–296. doi: 10.1038/nature07118
Abstract
In budding yeast, the Start checkpoint integrates multiple internal and external signals into an all-or-none decision to enter the cell cycle. Here, we show that Start behaves like a switch due to systems-level feedback in the regulatory network. In contrast to current models proposing a linear cascade of Start activation, transcriptional positive feedback of the G1 cyclins Cln1,2 induces the near-simultaneous expression of the ~200-gene G1/S regulon. Nuclear Cln2 drives coherent regulon expression, while cytoplasmic Cln2 drives efficient budding. _cln1,2_-deleted cells frequently arrest as unbudded cells, incurring a large fluctuation-induced fitness penalty due to both the lack of cytoplasmic Cln2 and insufficient G1/S regulon expression. Thus, positive-feedback-amplified expression of Cln1,2 simultaneously drives robust budding and rapid, coherent regulon expression. A similar G1/S regulatory network in mammalian cells, comprised of non-orthologous genes, suggests either the conservation of regulatory architecture or convergent evolution.
Positive feedback in genetic control networks can ensure that cells do not slip back and forth between either cell cycle phases or developmental fates. For example, commitment to sporulation in budding yeast is driven by transcriptional positive feedback of the meiotic inducer IME11–3. In Xenopus laevis, positive feedback underlies the all-or-none characteristics of oocyte maturation4, 5 and mitotic entry6, 7, suggesting the frequent use of positive feedback to regulate cellular transitions.
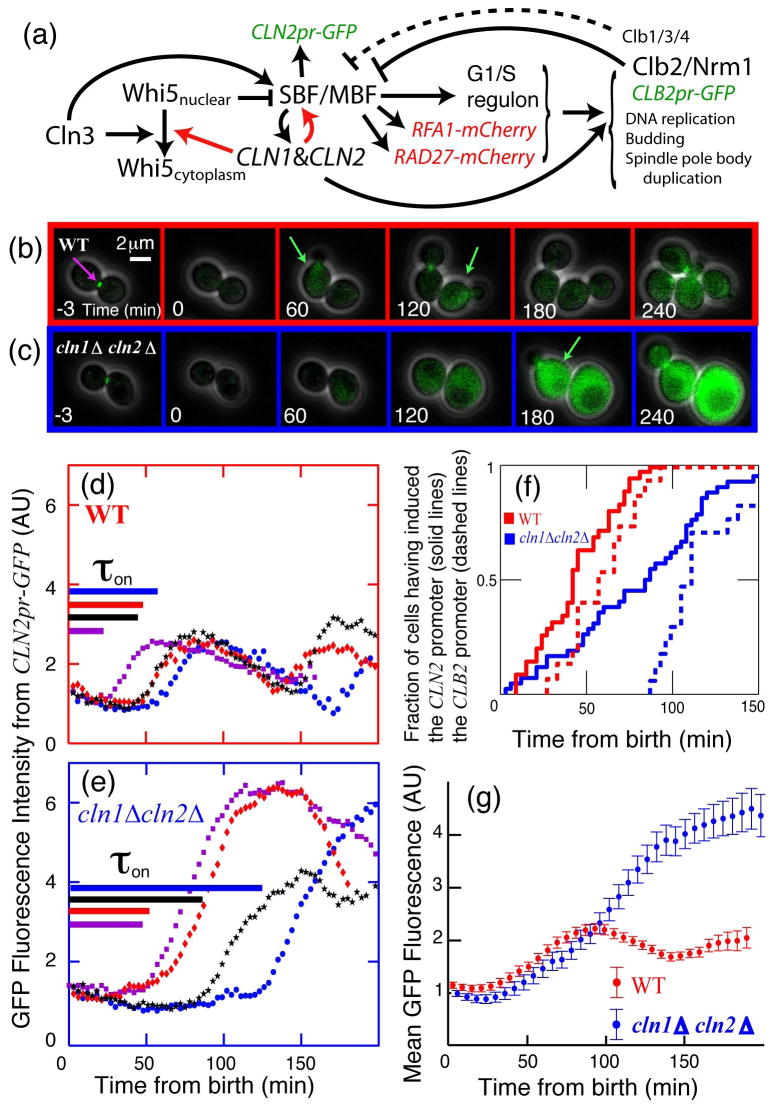
Absent from this list of examples is the well-studied Start checkpoint controlling cell cycle commitment in budding yeast. Nutrient limitation and pheromone exposure arrests cells prior to DNA replication, while size control extends G1 in small daughter cells8–11. Beyond Start, cells proceed through division almost independently of size and environment9. Previous experiments suggested that Start represents a feedback-free cascade of events12 (see schematic in Fig. 1a; omitting red arrows). The transition is initiated by the G1-cyclin Cln313–15, which in complex with Cdc28 activates the transcription of about 200 genes16 by phosphorylating promoter-bound protein complexes that include the transcription factors SBF and MBF17 and the transcriptional inhibitor Whi518, 19. Phosphorylation and inactivation of Whi5 is rate-limiting, and phosphorylated Whi5 rapidly exits the nucleus. The G1/S regulon, which includes two additional G1-cyclins, CLN1,2, contributes to the activation of B-type cyclins, DNA replication, spindle pole body duplication, and bud emergence. Mitotic B-type cyclins then inactivate SBF20 and, with NRM1, inactivate MBF21, thus turning off the G1/S regulon.
Figure 1. Positive feedback drives the Start of the budding yeast cell cycle.
(a) Schematic of the Start transition; novel interactions demonstrated in this paper are shown in red. (b,c): Combined phase and fluorescent images for CLN2pr-GFP MYO1-GFP MET3pr-CLN2 cells, either wild type (b) or _cln1_Δ cln2_Δ (c), grown in -Met (inducing) and plated on +Met (repressing) to normalize initial conditions6 (Fig. S3). Green arrows indicate approximate peak GFP expression from CLN2pr-GFP. (d,e): Single-cell fluorescence intensity for 4 characteristic cells of each genotype; cells are synchronized at birth, marked by the disappearance of a Myo1-GFP ring at the bud-neck (purple arrow in a). Time from birth to CLN2 promoter activation (as defined in Methods),τ_on, for each individual cell is indicated by length of the corresponding line in (d,e). (f) Cumulative distribution of CLN2pr-GFP induction (solid lines) indicates that Cln1,2-dependent positive feedback contributes substantially to the early expression of CLN2; dashed lines indicate induction of CLB2pr-GFP marking the onset of negative regulation of CLN2. (g) Averaging fluorescence intensity for 87 WT and 83 _cln1_Δ _cln2_Δ daughter cells aligned at birth simulates a population study, which would obscure the effect of positive feedback. The results shown are for daughter cells in glucose; changes in cell type or nutrient conditions do not qualitatively influence the results (Table S3).
Any one of the three G1-cyclins suffices to activate the regulon, suggesting the potential for transcriptional positive feedback of CLN1,2 on their own expression22, 23. However, analysis of synchronized populations led to the conclusion that positive feedback, defined as Cln1,2 advancing transcription from the CLN2 promoter, did not occur in WT; rather, Cln3 was the sole activator of firing14, 15.
In sharp contrast to the prevailing linear model, we demonstrate that Cln1,2-dependent positive feedback is central to Start control. We use single-cell time-lapse fluorescent microscopy to show that Cln1,2 advance timing and reduce variability in the activation of CLN2, and of the entire G1/S regulon. We further explore the mechanisms and functional significance of this control.
Positive Feedback of G1-Cyclins
Positive feedback of Cln1,2 on their own transcription should yield faster accumulation of CLN2 mRNA in WT cells than in _cln1_Δ _cln2_Δ cells. Although Cln1,2-dependent positive feedback was clearly demonstrated in the absence of Cln322–24, this does not imply that WT cells function similarly. In synchronized populations, near-identical timing of onset of CLN2 promoter activity was observed in the presence or absence of CLN1,2, leading to the linear model14, 15. Here, we revisit this issue using single cell assays. As a reporter for CLN2 transcription, we use unstable GFP driven by the CLN2 promoter24, 25 (see Methods and Fig S1–2). Birth time was determined using a marker for cytokinesis (disappearance of the Myo1-GFP myosin ring11, which did not influence the CLN2pr-GFP signal). The timing of CLN2 promoter induction in individual cells is sharp and easily quantified computationally (see Methods, Fig 1d,e and Fig. S1–2). Since _cln1_Δ _cln2_Δ cells are larger than WT, we integrated MET3pr-CLN2 in both strains, to conditionally express Cln2 prior to time-lapse imaging so that initially sizes were comparable14 (see methods; Fig. S3&S12 for controls). Thus, we can assay for positive feedback by comparing the time interval from birth to transcriptional activation of CLN2pr-GFP transcription in the first cell cycle after MET3pr-CLN2 turnoff in WT and _cln1_Δ _cln2_Δ cells.
Positive feedback should advance CLN2 promoter activation in WT compared to _cln1_Δ cln2_Δ cells14, 15. Strikingly, in daughter cells, the average time between birth and CLN2 promoter activation (τ_on; Fig. 1d–e,f) was much shorter for WT (41 min) than for _cln1_Δ _cln2_Δ (83 min). Furthermore, CLN2pr-GFP activation was much less variable for WT than for _cln1_Δ _cln2_Δ cells (standard deviation of 21 vs. 47 min). CLN2pr-GFP transcription was Cln3-dependent in _cln1_Δ _cln2_Δ cells. Qualitatively similar results were obtained in mother cells and also in cells growing in glycerol/ethanol instead of glucose. In all cases, the interval from birth to CLN2pr-GFP activation was smaller and less variable in WT than in _cln1_Δ _cln2_Δ, indicating strong positive feedback of Cln1,2 on their own transcription independent of nutrient conditions or cell type (Table S3; P < 10−4).
We explored the potential redundancy of CLN1 and CLN2 in activating the feedback loop. Although budding is slightly delayed in _cln1_Δ CLN2, and _CLN1 cln2_Δ cells compared to WT, the timing of CLN2 promoter activation is similar (Table S3), indicating that CLN1 and CLN2 form redundant conduits for positive feedback.
Our data can be reconciled with previous work14, 15 arguing against positive feedback because measurements averaged over a population of cells necessarily lose information. In addition to delayed onset of transcription, _cln1_Δ _cln2_Δ cells express a more intense and prolonged CLN2pr-GFP signal. The larger peaks are likely due to a delay in the Clb2-mediated repression of SBF/MBF14, 15, 20, 21 (Fig. 1d–e), since the average time between induction of CLN2 and CLB2 was much larger in _cln1_Δ_cln2_Δ strains, (measured using a CLB2pr-GFP cassette) (Fig. 1f; Fig. S13), and Clb2p accumulation is known to be delayed in _cln1_Δ_cln2_Δ strains14.
Therefore, imperfect synchrony11 allows the high and lengthened transcriptional response from the first _cln1_Δ_cln2_Δ cells firing the CLN2 promoter to mask the delayed response of the majority. This effect is reconstituted in Fig. 1g by averaging our measured single-cell data, and explains why positive feedback was not detected in measurements of mRNA levels in populations of synchronized daughter cells14, 15.
Coherent Regulon Expression
Once a cell senses the signal to initiate the cell cycle, it must actuate all the machinery effecting the cell cycle transition. At Start, this requires activating many SBF and MBF regulated genes16 encoding proteins involved in DNA replication and bud site formation. However, noise in protein expression in individual cells26 could interfere with expression of this large regulon. In particular, the delayed and variable induction of the CLN2 promoter in _cln1_Δ _cln2_Δ cells suggested that G1/S regulon expression might be severely disrupted in these feedback-free cells.
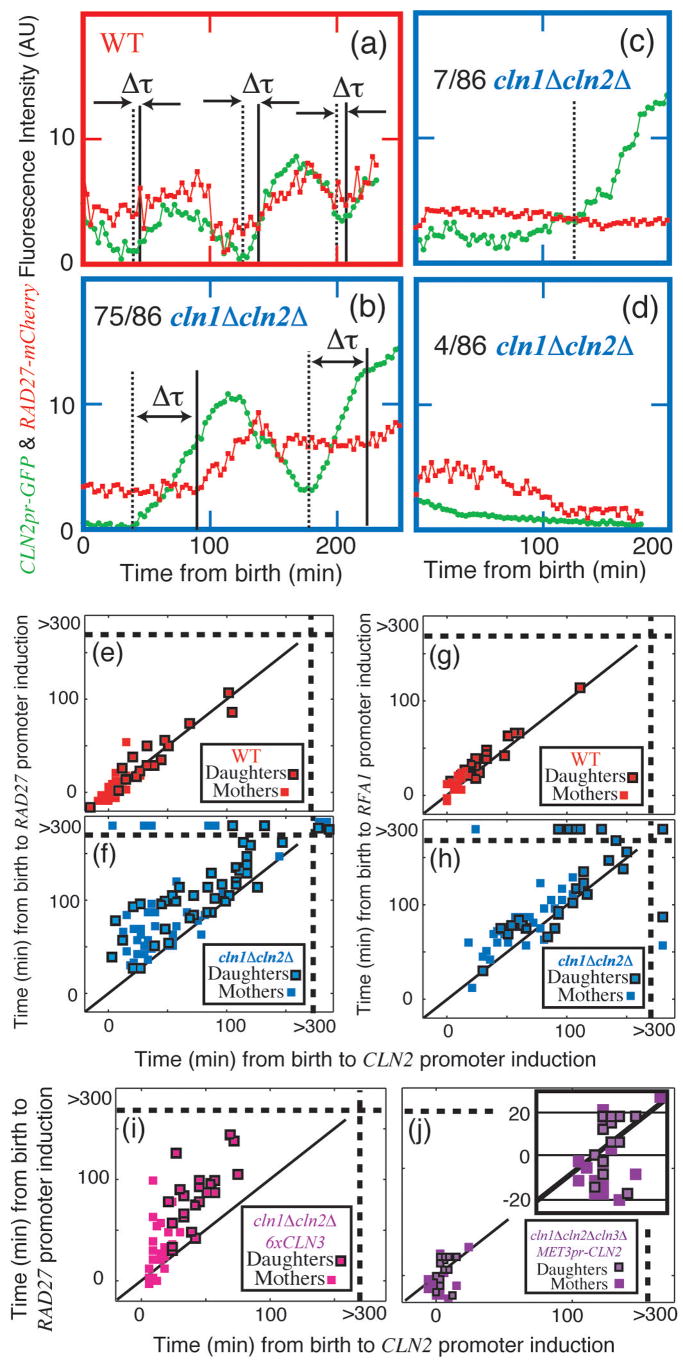
To investigate regulon expression in individual cells, we compared induction of CLN2pr-GFP and RAD27-mCherry, another member of the regulon16 (Fig. 2a–d; Fig. S7–8). RAD27 expression is Cln-dependent (Fig. S11). CLN2 and RAD27 are synchronously induced in WT, while there is a long and variable period of time between the inductions of the two genes in the _cln1_Δ _cln2_Δ mutant (Fig. 2e–f). Indeed, out of the 86 _cln1_Δ _cln2_Δ cells studied, 11 failed to produce a detectable increase in Rad27-mCherry and 4 failed to produce a detectable increase of either marker. We performed identical experiments on strains containing CLN2pr-GFP and RFA1-mCherry, another regulon member16, and obtained similar results (Fig. 2g–h). Our conclusions are valid even after excluding outlying points (P<0.01). Thus, Cln1/2 dependent positive feedback likely promotes coherent and efficient transcription across the SBF/MBF regulon.
Figure 2. Cln1,2 drive coherent expression of the SBF/MBF regulon.
(a–f): Strains containing both CLN2pr-GFP (green) and RAD27-mCherry (red) were examined (see Supplementary Information for detailed methods); τ marks the computed time between CLN2 and RAD27 inductions. WT (a, e): all cells transcribed both markers synchronously. _cln1_Δ _cln2_Δ, (b–d, f): 75 cells transcribed both markers, with variable intervening intervals; 7 cells transcribed CLN2pr-GFP, but not RAD27-mCherry; and 4 cells transcribed neither. Correlation of the initiation of RAD27 and CLN2 transcription in WT (e) and _cln1_Δ _cln2_Δ (f); points beyond the dotted lines in (f) represent no transcription within 300 min (movie limit; see also Table S3). (g–h): Substituting RFA1-mCherry for RAD27-mCherry yielded similar results. (i) _cln1_Δ _cln2_Δ 6xCLN3 cells and (j) _cln1_Δ _cln2_Δ _cln3_Δ cells expressing CLN2 from a MET3 promoter exhibited incoherent regulon expression compared to WT, although expression of both CLN2pr and RAD27pr were faster than in _cln1_Δ _cln2_Δ. P < 10−3 for all comparisons.
Further comparison of these three promoters in _cln1_Δ _cln2_Δ cells reveals that CLN2 is almost always the first of the three to be activated, while the times to subsequent RFA1pr and RAD27pr inductions are significantly different from each other (P=0.004; Table S3). This suggests that the CLN2 promoter is the easiest for Cln3 to induce, followed by the RFA1 promoter, followed by the RAD27 promoter. We note that two MBF targets27–29, RAD27 and RFA1, exhibit different induction timing.
To ask whether lack of coherence in _cln1_Δ _cln2_Δ cells might simply come from low G1 cyclin levels, we analyzed _cln1_Δ _cln2_Δ 6xCLN3 cells. Although expression of both the CLN2 and RAD27 promoters was significantly accelerated by extra CLN3, these cells still exhibited strongly incoherent expression compared to WT (Fig 2i).
To directly short-circuit the proposed positive feedback loop, we examined gene expression in _cln1_Δ _cln2_Δ _cln3_Δ MET3pr-CLN2 cells on methionine-free medium (MET3pr-CLN2 on). Although induction of CLN2pr-GFP and RAD27-mCherry were strongly accelerated by constitutive CLN2 expression, incoherent expression compared to WT was still observed (Fig. 2j). Intriguingly, this incoherence was due to RAD27-mCherry induction prior to CLN2pr-GFP, compared to nearly simultaneous expression in WT (−8±2 min compared to 2±1 min; P<10−3), perhaps due to differential loading of SBF (CLN2) and MBF (RAD27) regulated genes21, 27–30.
Overall, these experiments suggest that the positive feedback architecture is a particularly effective way to promote coherent regulon expression.
Stochastic cell cycle arrest
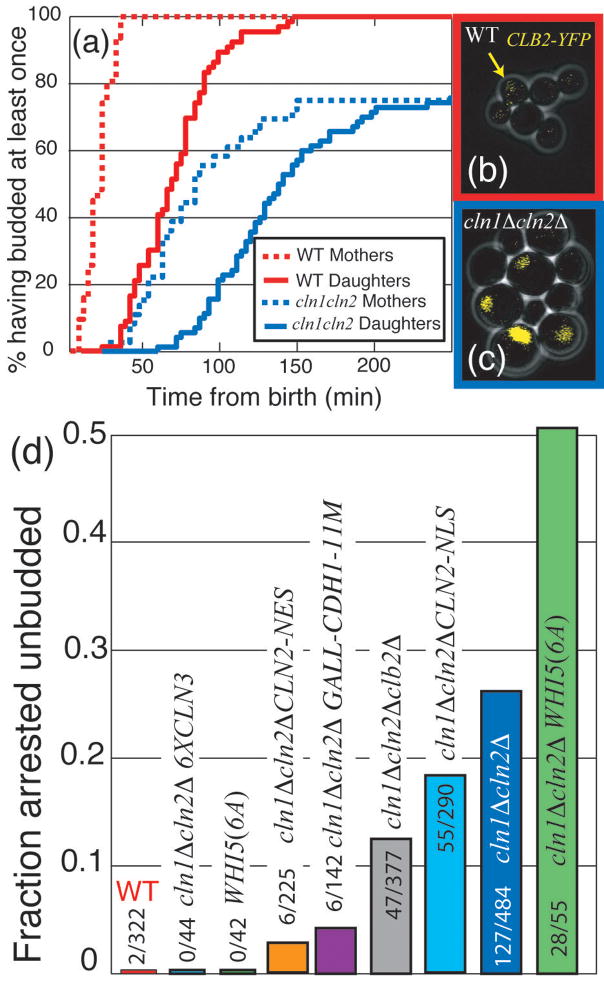
In addition to exhibiting incoherent gene expression, 26% of _cln1_Δ _cln2_Δ cells fail to bud (Fig. 3a). We hypothesized that incoherent gene expression plays a role in this sporadic unbudded arrest. 20 out of 143 assayed _cln1_Δ _cln2_Δ cells were ‘strongly incoherent’: they failed to transcribe one or both of their two transcriptional markers (Fig. 2f,h). 90% of the ‘strongly incoherent’ cells arrested unbudded compared to 26% of all _cln1_Δ _cln2_Δ cells (P<0.003; Fig. 3a). Thus, a lack of coherence in the SBF/MBF regulon is a strong predictor of unbudded arrest within the _cln1_Δ _cln2_Δ population. 6X CLN3 reduced unbudded arrest in _cln1_Δ _cln2_Δ cells, perhaps because of accelerated regulon expression (Fig. 2i). Thus, unbudded arrest in _cln1_Δ _cln2_Δ cells may result from highly delayed expression of some regulon members.
Figure 3. Stochastic unbudded arrest in _cln1_Δ _cln2_Δ cells, and its modulation by mitotic cyclins, Whi5, Cln3, and Cln2.
(a) Cumulative plot of percentage of cells that budded at least once: 26% of _cln1_Δ _cln2_Δ cells arrest unbudded. (b,c): cells with integrated CLB2-YFP fusion protein (endogenous promoter); WT (b) and _cln1_Δ _cln2_Δ (c). Note high nuclear Clb2 specifically in large unbudded (arrested) _cln1_Δ _cln2_Δ cells. (d) Delaying (_cln1_Δ _cln2_Δ _clb2_Δ) or removing (_cln1_Δ _cln2_Δ GALL-CDH1-11M) mitotic cyclin accumulation reduced the fraction of arrested cells; addition of 5 copies of CLN3 eliminated this arrest, while the addition of WHI5(6A) exacerbated the arrest. Unbudded arrest was weakly rescued by nuclear Cln2 (CLN2-NLS), and strongly rescued by cytoplasmic Cln2 (CLN2-NES). Unless stated otherwise in text P < 10−3 for all comparisons.
We hypothesized that in strongly incoherent cells, activation of only some regulon members might lead to activation of mitotic Clbs, which would then inactivate further SBF/MBF regulated expression20 (Fig. 1a; Fig. S9). If genes required for budding in the absence of CLN1,2, such as PCL1,231, had not yet been expressed, unbudded arrest might ensue. Indeed, 20/20 arrested _cln1_Δ _cln2_Δ cells contained large amounts of nuclear Clb2-YFP (Fig. 3b–c).
To further test the role of transcription in unbudded arrest, we deleted the rate-limiting SBF inhibitor CLB2 in a _MET3pr-CLN2 cln1_Δ _cln2_Δ strain and observed a decrease in unbudded arrest from 26% to 13% (Fig. 3d). Additionally, we integrated unphosphorylatable Cdh1 under GAL control (GALL-HA3-CDH1-m11) into a _cln1_Δ _cln2_Δ MET3pr-CLN2 strain to induce rapid degradation of all mitotic cyclins upon galactose induction32. This reduced the unbudded arrested fraction to 4% in the first cell cycle following GAL induction (Fig. 3d). Since the timing of CLB2pr-GFP induction in _cln1_Δ_cln2_Δ cells was similar whether they arrested or not (P = 0.91), the unbudded arrest was not due to unusually early CLB2 induction.
Thus, mitotic cyclins promote unbudded arrest specifically in highly incoherent _cln1_Δ _cln2_Δ cells, probably due to insufficient regulon expression before Clb-dependent SBF/MBF inactivation.
Cln1,2 inactivate the transcriptional inhibitor WHI5
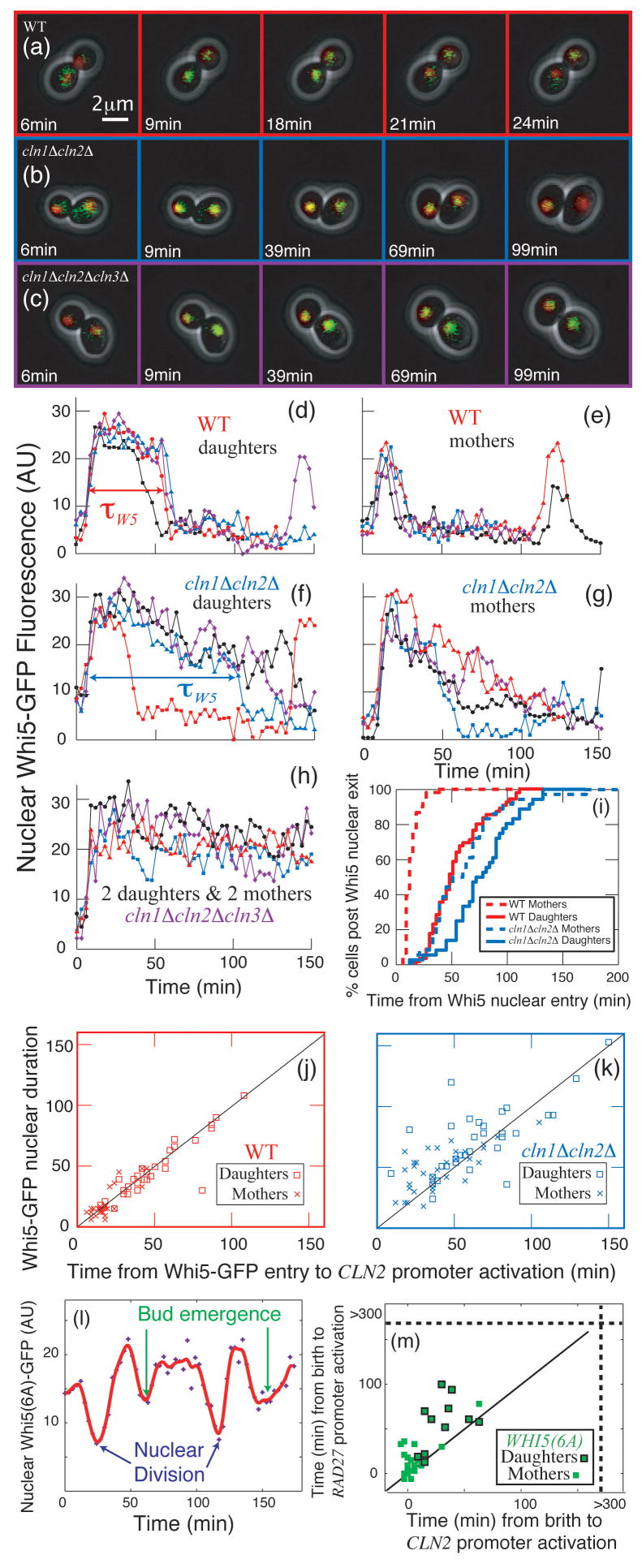
We wanted to determine if Cln1,2-dependent positive feedback operated through Whi5, a transcriptional inhibitor of the G1/S regulon18, 19. Whi5 inactivation is rate-limiting for CLN2 transcription and occurs via Cln-dependent phosphorylation, which leads to nuclear exclusion19.
First, we developed a quantitative assay for nuclear levels of Whi5-GFP by marking the nucleus with HTB2-mCherry (histone H2B) and measuring the difference between nuclear and cytoplasmic GFP fluorescence intensity(Fig. 4a–c). Whi5 entered the nucleus rapidly in both WT and _cln1_Δ _cln2_Δ cells. In WT cells, Whi5 also exited very rapidly. In _cln1_Δ _cln2_Δ cells, Whi5 exited much more slowly (Fig. 4d–g,i) consistent with biochemical data showing that Whi5 remains on the CLN2 promoter longer in _cln1_Δ _cln2_Δ than in WT cells18. Since in _cln1_Δ _cln2_Δ _cln3_Δ cells, Whi5-GFP remained nuclear (Fig. 4h), the slow Whi5 exit in _cln1_Δ _cln2_Δ cells is Cln3-dependent (this also excludes photobleaching artefacts). Thus, Cln3 initiates the slow exit of Whi5 from the nucleus, while Cln1/2 rapidly removes the remainder.
Figure 4. Cln1,2 are required for rapid phosphorylation and inactivation of the rate- limiting repressor Whi5.
(a–c): Combined phase and fluorescent images showing Whi5- GFP and Htb2-mCherry (to mark the nucleus) fusion proteins for (a) WT, (b) _cln1_Δ _cln2_Δ, and (c) _cln1_Δ _cln2_Δ _cln3_Δ cells. The difference between nuclear and non-nuclear fluorescence intensity was used to quantify nuclear Whi5 by automated image analysis. Compared to WT (d,e), in _cln1_Δ _cln2_Δ cells (f,g), Whi5 nuclear exit occurs later and is less sharp. In _cln1_Δ _cln2_Δ _cln3_Δ cells, Whi5 remains nuclear (h). (i) Percent of cells in which Whi5 has left the nucleus (defined as attaining half the maximum level) versus the time from Whi5 nuclear entry. (j,k) Whi5 nuclear exit is tightly correlated with CLN2 promoter activation in WT cells and less correlated in _cln1_Δ _cln2_Δ cells (See also Table S3). (l) WHI5(6A)-GFP19, lacking 6 out of 12 Cln-dependent phosphorylation sites, reproducibly displayed significant, but slower and incomplete, shuttling out of the nucleus at Start and again at nuclear division. In WHI5(6A) strains containing CLN2pr-GFP and RAD27-mCherry (m), CLN2 and RAD27 induction were incoherent, correlating with the poor nuclear transport of Whi5(6A)-GFP.
Since Whi5 exit and CLN2 induction are tightly correlated in WT (Fig. 4j), translocation occurs shortly after Whi5 inactivation and coincides with activation of transcriptional positive feedback. CLN2 promoter activation and Whi5 exit were less tightely correlated in _cln1_Δ _cln2_Δ cells consistent with the gradual exit of Whi5 (Fig. 4k; Fig. S5–6).
To examine the role of Whi5 phosphorylation in positive feedback and regulon coherence, we used a WHI5(6A) allele19 lacking 6 of 12 Cln-dependent phosphorylation sites. Although Whi5(6A) was reported to be constitutively nuclear19, we observed significant, but slower and incomplete, shuttling of Whi5(6A)-GFP out of the nucleus at Start and again at nuclear division (10/10 cells; Fig. 4l). CLN2 and RAD27 induction are less coherent in WHI5(6A) than in WT (Fig. 4m; but more coherent than _cln1_Δ _cln2_Δ), correlating with the poor nuclear transport of Whi5(6A). Thus, interfering with the positive feedback loop by reducing the ability of Cln proteins to phosphorylate Whi5 reduces regulon coherence, even with all three G1 cyclins present.
The addition of WHI5(6A) to _cln1_Δ _cln2_Δ cells increased the frequency of unbudded arrest from 26% to 51%, consistent with the idea that unbudded arrest is a consequence of incoherent regulon expression in _cln1_Δ _cln2_Δ cells.
Overall, these results strongly suggest that Whi5 is a Cln1,2 substrate in WT cells, and that this phosphorylation contributes to positive feedback. To see if Whi5 was the only such substrate, we compared timing of CLN2 promoter activation for _whi5_Δ and _cln1_Δ _cln2_Δ _whi5_Δ cells (Fig. S14; Table S3). Deletion of WHI5 advances CLN2 promoter induction in both WT and _cln1_Δ _cln2_Δ cells. Since _cln1_Δ _cln2_Δ _whi5_Δ cells delayed CLN2pr induction relative to _whi5_Δ cells, Cln1,2 likely act through a Whi5-dependent and a Whi5-independent mechanism to promote positive feedback. Previous results suggested a Whi5-independent Cln3 requirement for SBF activation19, possibly acting through Swi619, 33; a similar mechanism may be employed by Cln1,2.
Separable Cln2 functions
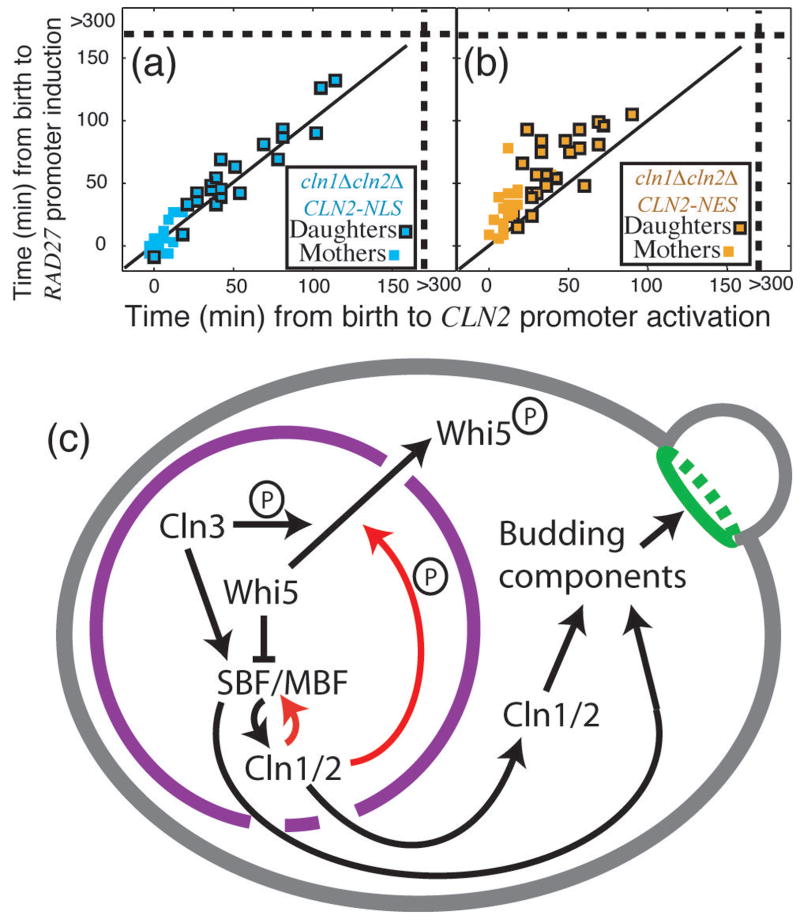
Cln1,2 are pleiotropic effectors of Start with important nuclear and cytoplasmic functions34, 35, complicating interpretation of _cln1_Δ _cln2_Δ phenotypes. Therefore, we tested forced-localization CLN2 alleles, expressed from the wild-type CLN2 promoter, that restrict Cln2 to either the nucleus (CLN2-NLS) or the cytoplasm (CLN2-NES)34. _cln1_Δ _cln2_Δ CLN2-NLS cells exhibit coherent regulon expression (P=0.45 compared to WT), but _cln1_Δ _cln2_Δ CLN2-NES cells are highly incoherent compared to WT (P<10−7), implying that coherent gene expression is primarily a nuclear function of CLN2 (Fig. 6a–b; compare to Fig. 2; Table S3).
Consistent with a role of cytoplasmic Cln2 in budding34, 35, integration of CLN2-NES into _cln1_Δ _cln2_Δ cells strongly reduces arrest (to 3%) in spite of less coherent gene expression. Furthermore, exogenous expression of CLN2 drives cell cycle progression in previously blocked _cln1_Δ _cln2_Δ cells (Fig. S10) and restores viability of _mbp1_Δ _swi4_Δ cells, which lack SBF and MBF and have very low regulon expression36, 37. The localization mutants also have different efficacy for relieving unbudded arrest. Integration of CLN2-NLS into _cln1_Δ _cln2_Δ cells, providing coherent gene expression, leads to a partial but significant reduction of unbudded arrest (from 26% to 19%; P=0.04).
Thus, cell morphogenesis and budding can be driven by two partially redundant pathways: via cytoplasmic Cln1,234, 38 or other genes in the G1/S regulon such as Pcl1,231 (Fig. 6c). Having Cln1,2 coherently activate the G1/S regulon and directly drive bud emergence provides a compact solution to ensure efficient and timely morphogenesis and G1/S regulon expression, before subsequent Clb activation.
Discussion
The regulatory architecture of the G1/S regulon provides an effective design to promote coordinated activation. The promoters are pre-loaded during G1 with a complex of factors that are subsequently rapidly activated by phosphorylation17, 24, 30 removing a potentially rate-limiting step. Furthermore, the upstream cyclin Cln3 is intrinsically more capable of triggering the CLN2 promoter compared to two other randomly selected promoters from the regulon (RFA1 or RAD27; Fig 2e–h). High sensitivity of CLN1/2 to Cln3 means that positive feedback from the initial burst of Cln1,2 will ensure that all other genes fire together. Indeed, in our experiments in WT cells, the genes are expressed too synchronously to evaluate which comes first. We find it likely that positive feedback will be a recurring motif within genetic control networks responsible for the coherent temporal coordination of multiple downstream events.
The sharpness of the Start switch, defined by the rapid exclusion of the transcriptional inhibitor Whi5 and the coherent expression of the G1/S regulon, is principally due to _CLN1,2_-dependent positive feedback (Fig. 6c, red lines) rather than a linear Cln3-Whi5-SBF pathway14, 15, 18, 19. Our data are inconsistent with the sharpness of Start being primarily due to non-linear increases in CLN3 translation39 or nuclear translocation40, or cooperative phosphorylation of Whi5 by Cln319, since these mechanisms all predict a sharp switch in feedback-free _cln1_Δ _cln2_Δ cells.
In budding yeast, Start is a fundamental point of commitment where physiological inputs such as nutrients, mating factor, size and cell type are integrated to produce an all-or-none decision. We show here that positive feedback provides robust switch-like cell cycle entry. Our single-cell data suggest that the point of commitment to the cell cycle, Start, is a very brief interval coinciding with the initiation of positive feedback and Whi5 exclusion. Subsequent Cln-dependent events, such as Sic1 phosphorylation and degradation41 leading to DNA replication, could then be viewed as dependent on, rather than part of, Start.
This work also provides a molecular basis for understanding the modular structure of G111. Two temporally uncorrelated processes in G1 are separated by the molecular event of Whi5 inactivation and nuclear exit. The upstream module is responsible for cell size control, while the downstream size-independent module actuates cell cycle progression11. Here, we showed that rapid Whi5-exit coincides with initiation of Cln1,2-dependent positive feedback. Once feedback is initiated, the rapidly accumulating Cln1,2 likely dominates cellular Cln-kinase activity and Cln3, the rate-limiting upstream activator, is rendered unimportant. In general, we expect modularity, best revealed by single-cell analysis, to be a signature of feedback-driven cellular control networks.
Our systems-level analysis of Start provides a template for further studies of other checkpoints in yeasts or the G1/S transition in mammals. The utility of feedback at Start leads us to expect similar regulatory architecture across eukaryotes, even if the enabling genes are not homologous.
Methods Summary
Strain and plasmid constructions
Standard methods were used throughout. All strains are W303-congenic.
Time-lapse microscopy
Preparation of cells for time-lapse microscopy was performed as previously described24. Since mutant cells are larger than WT, we integrated MET3pr-CLN2 to conditionally express Cln214. On media lacking methionine (MET3pr-CLN2 on), cells bud and divide at comparable sizes (Fig. S3). By pre-growing cells without methionine before plating on media containing methionine (MET3pr-CLN2 off), we are able to begin our time-lapse imaging experiments with similarly sized WT and _cln1_Δ _cln2_Δ cells. We imaged the first Start in cells that were budded at the time of transfer,and that divided least 30 minutes after methionine addition, to allow degradation of Cln213, 42 made before MET3 promoter turnoff.
Image Analysis
Automated image segmentation and fluorescence quantification of yeast grown under time-lapse conditions were performed as previously described11, 24. We added a function to previously described custom software24 to identify nuclei labeled with Htb2-mCherry (histone). The red signal was smoothed, disconnected fragments were eliminated and the cells with nuclei too small, or dim, or oddly shaped (area vs. minimally enclosed rectangle) were eliminated. After background subtraction, the nucleus was defined to be where the fluorescence was greater than 70% of maximum, which controls for cell variability and vertical movement of the nucleus. The nuclear Whi5-GFP signal was the difference between the average nuclear and cytosolic intensities.
Data Analysis
Fluorescence time series were extracted from movies as previously described24. Time-series were fit using smoothing splines (MATLAB) with a smoothing parameter of 0.001. We defined the onset of transcription for a G1/S fluorescent reporter by the maximum in the second derivative that fell between birth and budding (scored separately), which accurately locates rate-changes in spite of noisy data and slow changes in the background fluorescence (Fig. S3–4).
Methods
Strain and plasmid constructions
Standard methods were used throughout. All strains are W303-congenic. In synchronized WT cells, GFP mRNA from the CLN2 promoter and CLN2 mRNA follow similar kinetics, and accumulation of cellular fluorescence follows with a slight delay24. WHI5(6A) and WHI5(6A)-GFP strains with modified WHI5 at the endogenous locus were a gift from M. Tyers. Plasmids for introduction of CLN2-NES and CLN2-NLS under control of the CLN2 promoter were obtained from B. Futcher, and integrated at the ura3 locus in a _cln1_Δ _cln2_Δ background. Histone H2B (HTB2) was C-terminally tagged with mCherry using PCR-mediated tagging, with the template plasmid pKT35543 by J. Bean and B. Timney. RAD27 and RFA1 were tagged similarly. All other alleles were from laboratory stocks described previously.
Time-lapse microscopy
Preparation of cells for time-lapse microscopy was performed as previously described24. Since mutant cells are larger than WT, we integrated MET3pr-CLN2 to conditionally express Cln214. On media lacking methionine (MET3pr-CLN2 on), cells bud and divide at comparable sizes (Fig. S3). By pre-growing cells without methionine before plating on media containing methionine (MET3pr-CLN2 off), we are able to begin our time-lapse imaging experiments with similarly sized WT and _cln1_Δ _cln2_Δ cells. We imaged the first Start in cells that were budded at the time of transfer, and that divided least 30 minutes after methionine addition, to allow degradation of Cln213, 42 made before MET3 promoter turnoff. Briefly, growth of microcolonies was observed with fluorescence time-lapse microscopy at 30ºC using a Leica DMIRE2 inverted microscope with a Ludl motorized XY stage. Images were acquired every 3 minutes for cells grown in glucose and every 6 minutes for cells grown in glycerol/ethanol with a Hamamatsu Orca-ER camera. Custom Visual Basic software integrated with ImagePro Plus was used to automate image acquisition and microscope control.
Image Analysis
Automated image segmentation and fluorescence quantification of yeast grown under time-lapse conditions were performed as previously described24. Budding was scored visually, and cell birth was scored by the disappearance of Myo1-GFP at the bud neck, generally with single frame accuracy. Background was measured as the average fluorescence of unlabelled cells and subtracted from the measured pixel intensities. We added a function to previously described custom software24 to identify nuclei labeled with Htb2-mCherry (histone). The red signal was smoothed, disconnected fragments were eliminated and the cells with nuclei too small, or dim, or oddly shaped (area vs. minimally enclosed rectangle) were eliminated. After background subtraction, the nucleus was defined to be where the fluorescence was greater than 70% of maximum, which controls for cell variability and vertical movement of the nucleus. The nuclear Whi5-GFP signal was the difference between the average nuclear and cytosolic intensities.
Data Analysis
P-values using appropriate tests yielded P<0.001 for all comparisons in the text, except where noted. Fluorescence time series were extracted from movies as previously described24. Time-series were fit using smoothing splines (MATLAB) with a smoothing parameter of 0.001. We defined the onset of transcription for a G1/S fluorescent reporter by the maximum in the second derivative that fell between birth and budding (scored separately). This method was chosen because it accurately locates rate-changes in spite of noisy data and slow changes in the background fluorescence. The onset time was nearly unchanged over a range of 103 in smoothing parameter (Fig. S3–4).
Figure 5. Function of nuclear Cln2 and model for Start regulation by positive feedback.
Comparison of _cln1_Δ _cln2_Δ cells with either a nuclear localized (a) or a nuclear excluded (b) CLN2 allele suggests that nuclear Cln2 is necessary and sufficient for regulon coherence. Strains contained CLN2pr-GFP and RAD27-mCherry. (c) Model for regulon activation and bud emergence: red lines indicate
Acknowledgments
This work was supported by the National Institute of Health (J.M.S., E.D.S., F.R.C.), the Burroughs Wellcome Fund (J.S) and the National Science Foundation (E.D.S.). We thank N. Buchler, G. Charvin, B. Drapkin and J.E. Ferrell for insightful conversations, and J. Widom and C. Wittenberg for thoughtful comments on the manuscript. We thank J.M. Bean, B. Timney and J. Robbins for help with strain/plasmid construction, M. Schwab for the plasmid pWS358, B. Futcher for the CLN2-NES and CLN2-NLS plasmids, E. Bi for the pKT355 mCherry tagging plasmid, and M. Tyers for WHI5 phosphorylation site mutant strains and plasmids. The authors declare no competing financial interests.
References
- 1.Simchen G, Pinon R, Salts Y. Sporulation in Saccharomyces cerevisiae: premeiotic DNA synthesis, readiness and commitment. Exp Cell Res. 1972;75:207–18. doi: 10.1016/0014-4827(72)90538-1. [DOI] [PubMed] [Google Scholar]
- 2.Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–56. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 3.Shenhar G, Kassir Y. A positive regulator of mitosis, Sok2, functions as a negative regulator of meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:1603–12. doi: 10.1128/MCB.21.5.1603-1612.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–8. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 5.Xiong W, Ferrell JE., Jr A positive-feedback-based bistable ‘memory module’ that governs a cell fate decision. Nature. 2003;426:460–5. doi: 10.1038/nature02089. [DOI] [PubMed] [Google Scholar]
- 6.Sha W, et al. Hysteresis drives cell-cycle transitions in Xenopus laevis egg extracts. Proc Natl Acad Sci U S A. 2003;100:975–80. doi: 10.1073/pnas.0235349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pomerening JR, Sontag ED, Ferrell JE., Jr Building a cell cycle oscillator: hysteresis and bistability in the activation of Cdc2. Nat Cell Biol. 2003;5:346–51. doi: 10.1038/ncb954. [DOI] [PubMed] [Google Scholar]
- 8.Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 9.Johnston GC, Pringle JR, Hartwell LH. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 10.Lord PG, Wheals AE. Variability in individual cell cycles of Saccharomyces cerevisiae. J Cell Sci. 1981;50:361–76. doi: 10.1242/jcs.50.1.361. [DOI] [PubMed] [Google Scholar]
- 11.Di Talia S, Skotheim JM, Bean JM, Siggia ED, Cross FR. The effects of molecular noise and size control on variability in the budding yeast cell cycle. Nature. 2007;448:947–51. doi: 10.1038/nature06072. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen P, Tyers M. How cells coordinate growth and division. Curr Biol. 2004;14:R1014–27. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 13.Tyers M, Tokiwa G, Futcher B. Comparison of the Saccharomyces cerevisiae G1 cyclins: Cln3 may be an upstream activator of Cln1, Cln2 and other cyclins. Embo J. 1993;12:1955–68. doi: 10.1002/j.1460-2075.1993.tb05845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dirick L, Bohm T, Nasmyth K. Roles and regulation of Cln-Cdc28 kinases at the start of the cell cycle of Saccharomyces cerevisiae. Embo J. 1995;14:4803–13. doi: 10.1002/j.1460-2075.1995.tb00162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stuart D, Wittenberg C. CLN3, not positive feedback, determines the timing of CLN2 transcription in cycling cells. Genes Dev. 1995;9:2780–94. doi: 10.1101/gad.9.22.2780. [DOI] [PubMed] [Google Scholar]
- 16.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–97. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato M, Hata N, Banerjee N, Futcher B, Zhang MQ. Identifying combinatorial regulation of transcription factors and binding motifs. Genome Biol. 2004;5:R56. doi: 10.1186/gb-2004-5-8-r56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Bruin RA, McDonald WH, Kalashnikova TI, Yates J, 3rd, Wittenberg C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell. 2004;117:887–98. doi: 10.1016/j.cell.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 19.Costanzo M, et al. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell. 2004;117:899–913. doi: 10.1016/j.cell.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 20.Amon A, Tyers M, Futcher B, Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- 21.de Bruin RA, et al. Constraining G1-specific transcription to late G1 phase: the MBF-associated corepressor Nrm1 acts via negative feedback. Mol Cell. 2006;23:483–96. doi: 10.1016/j.molcel.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 22.Cross FR, Tinkelenberg AH. A potential positive feedback loop controlling CLN1 and CLN2 gene expression at the start of the yeast cell cycle. Cell. 1991;65:875–83. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 23.Dirick L, Nasmyth K. Positive feedback in the activation of G1 cyclins in yeast. Nature. 1991;351:754–7. doi: 10.1038/351754a0. [DOI] [PubMed] [Google Scholar]
- 24.Bean JM, Siggia ED, Cross FR. Coherence and timing of cell cycle Start examined at single-cell resolution. Mol Cell. 2006;21:3–14. doi: 10.1016/j.molcel.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 25.Mateus C, Avery SV. Destabilized green fluorescent protein for monitoring dynamic changes in yeast gene expression with flow cytometry. Yeast. 2000;16:1313–23. doi: 10.1002/1097-0061(200010)16:14<1313::AID-YEA626>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 26.Samoilov MS, Price G, Arkin AP. From fluctuations to phenotypes: the physiology of noise. Sci STKE 2006. 2006:re17. doi: 10.1126/stke.3662006re17. [DOI] [PubMed] [Google Scholar]
- 27.Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–8. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- 28.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Simon I, et al. Serial regulation of transcriptional regulators in the yeast cell cycle. Cell. 2001;106:697–708. doi: 10.1016/s0092-8674(01)00494-9. [DOI] [PubMed] [Google Scholar]
- 30.Koch C, Schleiffer A, Ammerer G, Nasmyth K. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 1996;10:129–41. doi: 10.1101/gad.10.2.129. [DOI] [PubMed] [Google Scholar]
- 31.Moffat J, Andrews B. Late-G1 cyclin-CDK activity is essential for control of cell morphogenesis in budding yeast. Nat Cell Biol. 2004;6:59–66. doi: 10.1038/ncb1078. [DOI] [PubMed] [Google Scholar]
- 32.Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–4. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- 33.Wijnen H, Landman A, Futcher B. The G(1) cyclin Cln3 promotes cell cycle entry via the transcription factor Swi6. Mol Cell Biol. 2002;22:4402–18. doi: 10.1128/MCB.22.12.4402-4418.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgington NP, Futcher B. Relationship between the function and the location of G1 cyclins in S. cerevisiae. J Cell Sci. 2001;114:4599–611. doi: 10.1242/jcs.114.24.4599. [DOI] [PubMed] [Google Scholar]
- 35.Miller ME, Cross FR. Distinct subcellular localization patterns contribute to functional specificity of the Cln2 and Cln3 cyclins of Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:542–55. doi: 10.1128/mcb.20.2.542-555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch C, Moll T, Neuberg M, Ahorn H, Nasmyth K. A role for the transcription factors Mbp1 and Swi4 in progression from G1 to S phase. Science. 1993;261:1551–7. doi: 10.1126/science.8372350. [DOI] [PubMed] [Google Scholar]
- 37.Bean JM, Siggia ED, Cross FR. High Functional Overlap Between MBF and SBF in the G1/S Transcriptional Program in Saccharomyces cerevisiae. Genetics. 2005 doi: 10.1534/genetics.105.044560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCusker D, et al. Cdk1 coordinates cell-surface growth with the cell cycle. Nat Cell Biol. 2007;9:506–15. doi: 10.1038/ncb1568. [DOI] [PubMed] [Google Scholar]
- 39.Polymenis M, Schmidt EV. Coupling of cell division to cell growth by translational control of the G1 cyclin CLN3 in yeast. Genes Dev. 1997;11:2522–31. doi: 10.1101/gad.11.19.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Gari E, Verges E, Gallego C, Aldea M. Recruitment of Cdc28 by Whi3 restricts nuclear accumulation of the G1 cyclin-Cdk complex to late G1. Embo J. 2004;23:180–90. doi: 10.1038/sj.emboj.7600022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider BL, Yang QH, Futcher AB. Linkage of replication to start by the Cdk inhibitor Sic1. Science. 1996;272:560–2. doi: 10.1126/science.272.5261.560. [DOI] [PubMed] [Google Scholar]
- 42.Lanker S, Valdivieso MH, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–601. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 43.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–72. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]