Role of JNK1-dependent Bcl-2 Phosphorylation in Ceramide-induced Macroautophagy (original) (raw)
Abstract
Macroautophagy is a vacuolar lysosomal catabolic pathway that is stimulated during periods of nutrient starvation to preserve cell integrity. Ceramide is a bioactive sphingolipid associated with a large range of cell processes. Here we show that short-chain ceramides (C2-ceramide and C6-ceramide) and stimulation of the de novo ceramide synthesis by tamoxifen induce the dissociation of the complex formed between the autophagy protein Beclin 1 and the anti-apoptotic protein Bcl-2. This dissociation is required for macroautophagy to be induced either in response to ceramide or to starvation. Three potential phosphorylation sites, Thr69, Ser70, and Ser87, located in the non-structural N-terminal loop of Bcl-2, play major roles in the dissociation of Bcl-2 from Beclin 1. We further show that activation of c-Jun N-terminal protein kinase 1 by ceramide is required both to phosphorylate Bcl-2 and to stimulate macroautophagy. These findings reveal a new aspect of sphingolipid signaling in up-regulating a major cell process involved in cell adaptation to stress.
Macroautophagy (referred to below as “autophagy”) is a vacuolar, lysosomal degradation pathway for cytoplasmic constituents that is conserved in eukaryotic cells (1–3). Autophagy is initiated by the formation of a multimembrane-bound autophagosome that engulfs cytoplasmic proteins and organelles. The last stage in the process results in fusion with the lysosomal compartments, where the autophagic cargo undergoes degradation. Basal autophagy is important in controlling the quality of the cytoplasm by removing damaged organelles and protein aggregates. Inhibition of basal autophagy in the brain is deleterious, and leads to neurodegeneration in mouse models (4,5). Stimulation of autophagy during periods of nutrient starvation is a physiological response present at birth and has been shown to provide energy in various tissues of newborn pups (6). In cultured cells, starvation-induced autophagy is an autonomous cell survival mechanism, which provides nutrients to maintain a metabolic rate and level of ATP compatible with cell survival (7). In addition, starvation-induced autophagy blocks the induction of apoptosis (8). In other contexts, such as drug treatment and a hypoxic environment, autophagy has also been shown to be cytoprotective in cancer cells (9,10). However, autophagy is also part of cell death pathways in certain situations (11). Autophagy can be a player in apoptosis-independent type-2 cell death (type-1 cell death is apoptosis), also known as autophagic cell death. This situation has been shown to occur when the apoptotic machinery is crippled in mammalian cells (12,13). Autophagy can also be part of the apoptotic program, for instance in tumor necrosis factor-α-induced cell death when NF-κB is inhibited (14), or in human immunodeficiency virus envelope-mediated cell death in bystander naive CD4 T cells (15). Moreover autophagy has recently been shown to be required for the externalization of phosphatidylserine, the eat-me signal for phagocytic cells, at the surface of apoptotic cells (16).
The complex relationship between autophagy and apoptosis reflects the intertwined regulation of these processes (17,18). Many signaling pathways involved in the regulation of autophagy also regulate apoptosis. This intertwining has recently been shown to occur at the level of the molecular machinery of autophagy. In fact the anti-apoptotic protein Bcl-2 has been shown to inhibit starvation-induced autophagy by interacting with the autophagy protein Beclin 1 (19). Beclin 1 is one of the Atg proteins conserved from yeast to humans (it is the mammalian orthologue of yeast Atg6) and is involved in autophagosome formation (20). Beclin 1 is a platform protein that interacts with several different partners, including hVps34 (class III phosphatidylinositol 3-kinase), which is responsible for the synthesis of phosphatidylinositol 3-phosphate. The production of this lipid is important for events associated with the nucleation of the isolation membrane before it elongates and closes to form autophagosomes in response to other Atg proteins, including the Atg12 and LC32 (microtubule-associated protein light chain 3 is the mammalian orthologue of the yeast Atg8) ubiquitin-like conjugation systems (3,21). Various partners associated with the Beclin 1 complex modulate the activity of hVps34. For instance, Bcl-2 inhibits the activity of this enzyme, whereas UVRAG, Ambra-1, and Bif-1 all up-regulate it (22,23).
In view of the intertwining between autophagy and apoptosis, it is noteworthy that Beclin 1 belongs to the BH3-only family of proteins (24–26). However, and unlike most of the proteins in this family, Beclin 1 is not able to trigger apoptosis when its expression is forced in cells (27). A BH3-mimetic drug, ABT-737, is able to dissociate the Beclin 1-Bcl-2 complex, and to trigger autophagy by mirroring the effect of starvation (25).
The sphingolipids constitute a family of bioactive lipids (28–32) of which several members, such as ceramide and sphingosine 1-phosphate, are signaling molecules. These molecules constitute a “sphingolipid rheostat” that determines the fate of the cell, because in many settings ceramide is pro-apoptotic and sphingosine 1-phosphate mitigates this apoptotic effect (31,32). However, ceramide is also engaged in a wide variety of other cell processes, such as the formation of exosomes (33), differentiation, cell proliferation, and senescence (34). Recently we showed that both ceramide and sphingosine 1-phosphate are able to stimulate autophagy (35,36). It has also been shown that ceramide triggers autophagy in a large panel of mammalian cells (37–39). However, elucidation of the mechanism by which ceramide stimulates autophagy is still in its infancy. We have previously demonstrated that ceramide induces autophagy in breast and colon cancer cells by inhibiting the Class I phosphatidylinositol 3-phosphate/mTOR signaling pathway, which plays a central role in inhibiting autophagy (36). Inhibition of mTOR is another hallmark of starvation-induced autophagy (17). This finding led us to investigate the effect of ceramide on the Beclin 1-Bcl-2 complex. The results presented here show that ceramide is more potent than starvation in dissociating the Beclin 1-Bcl-2 complex (see Ref.40). This dissociation is dependent on three phosphorylation sites (Thr69, Ser70, and Ser87) located in a non-structural loop of Bcl-2. Ceramide induces the c-Jun N-terminal kinase 1-dependent phosphorylation of Bcl-2. Expression of a dominant negative form of JNK1 blocks Bcl-2 phosphorylation, and thus the induction of autophagy by ceramide. These findings help to explain how autophagy is regulated by a major lipid second messenger.
MATERIALS AND METHODS
_Reagents_—C2-Cer, C6-Cer, C2-DHCer, and C6-DHCer were from Sigma and were dissolved in ethanol before use. FB1, Myriocin, and TAM were purchased from Biomol. Cell culture medium, Lipofectamine 2000, and fetal bovine serum were from Invitrogen. The radioisotope l-[U-14C]valine (256 mCi/mmol), the ECL™ Western blotting detection kit, and the donkey anti-rabbit antibody were purchased from Amersham Biosciences. Goat anti-mouse and swine anti-goat antibodies were obtained from Bio-Rad and Caltag (Burlingame, CA), respectively. Mouse monoclonal anti-p62 antibody was obtained from BD Biosciences. Mouse monoclonal anti-Bcl-2 and goat polyclonal anti-Beclin 1 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit monoclonal antibodies against p-Bcl-2, p-JNK, and total JNK were obtained from Cell Signaling. Rabbit polyclonal anti-Beclin 1 antibody was obtained from Novus Biologicals. Rabbit polyclonal anti-LC3 antibody was obtained as previously described (41).
_Cell Culture_—Human breast cancer cell line MCF-7 cells stably transfected with beclin 1 (MCF7.beclin 1) were cultured as previously described (42). HeLa cells were obtained from ATCC, and maintained at 37 °C in 10% CO2 in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum and 100 ng/ml each of penicillin and streptomycin. HeLa GFP-LC3 cells, kindly provided by A. M. Tolkovsky (University of Cambridge, UK), were cultured in the presence of 200 μg/ml G418. The human colon cancer cell line HT-29 transfected either with empty vector or vector encoding Bcl-2 were kindly provided by M. T. Dimanche-Boitrel (INSERM U620, France), and cultured at 37 °C in 10% CO2 in Dulbecco's modified Eagle's medium, supplemented with 10% fetal bovine serum and 100 ng/ml each of penicillin and streptomycin plus 200 μg/ml G418. MEF WT and Atg5–/– were kindly given by N. Mizushima (Tokyo Medical and Dental University, Japan), and cultured as previously described (6). The trypan blue exclusion test showed that cell viability was greater than 90% under all the experimental conditions used.
_Quantification of Endogenous Ceramide_—Ceramide was determined using Escherichia coli DAG kinase as previously reported (43). The E. coli strain was kindly provided by Drs. D. K. Perry and Y. A. Hannun (Medical University of South Carolina, Charleston, SC).
_Beclin 1 and Bcl-2 Co-immunoprecipitation_—To immunoprecipitate endogenous Beclin 1 and endogenous Bcl-2 in HeLa cells or stably transfected Beclin 1 in MCF.7 cells, the cells were lysed in CHAPS lysis buffer (20 mm Tris, pH 7.4, 137 mm NaCl, 2 mm EDTA, 10% glycerol, and 2% CHAPS) for 3 h at 4 °C, and immunoprecipitation was performed overnight at 4 °C with a goat polyclonal antibody (1:80 dilution, Santa Cruz Biotechnology). Protein A-Sepharose beads (Amersham Biosciences) were added for 2 h at 4 °C, washed twice with 137 mm NaCl CHAPS wash buffer (20 mm Tris, pH 7.4, 137 mm NaCl, 2 mm EDTA, 10% glycerol, and 0.5% CHAPS), and twice with 274 mm NaCl CHAPS wash buffer. Anti-Beclin 1 immunoprecipitates were subjected to SDS-PAGE, and Bcl-2 was detected by immunoblot analysis (19).
_GFP-LC3 Assay_—The assay was performed in HeLa cells either stably transfected with rat GFP-LC3 (kindly provided by T. Yoshimori, Osaka University, Japan) or in transiently transfected with MCF-7. beclin 1, HT-29, and HeLa cells using Lipofectamine 2000 (19,42). HeLa cells were also transfected either with human GFP-LC3B or the mutant GFP-LC3BΔG (kindly provided by I. Tanida, National Institute of Infectious Disease, Tokyo, Japan). The vector, pcDNA3-FLAG-MKK7-JNK1(APF) (dominant-negative JNK1, dnJNK1), was provided by R. J. Davis (Howard Hughes Medical Institute) (44). When required, co-transfection with a plasmid encoding GFP-LC3 and a vector encoding for dnJNK1, or the empty vector pcDNA3, was performed in MCF-7.beclin 1 cells (ratio GFP-LC3:vector, 1:3). Prior to analysis, the cells were starved for 4 h in Earle's balanced salt solution (EBSS, starvation medium), maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum (control medium), or treated as described in the text. Autophagy was then measured by light microscopic counting of cells with GFP-LC3 puncta as described previously (42). A minimum of 50–100 cells per sample was counted in triplicate samples per condition per experiment.
_Analysis of Protein Degradation_—HT-29 cells were incubated for 24 h at 37 °C with 0.2 μCi/ml l-[14C]valine. Three hours before the end of the radiolabeling period, cells were exposed to increasing concentrations of C2-Cer or C2-DHCer, and when required 100 nm FB1 was added, also 3 h before the end of radiolabeling. At the end of radiolabeling period, the cells were washed three times with PBS, pH 7.4. Cells were then incubated in complete medium supplemented with 10 mm cold valine. After incubating for 1 h, by which time short-lived proteins are degraded, the medium was replaced with fresh nutrient-free medium (EBSS plus 0.1% of bovine serum albumin and 10 mm cold valine), and the incubation was continued for an additional 4 h. Cells and radiolabeled proteins from the 4-h chase medium were precipitated in trichloroacetic acid at a final concentration of 10% (v/v) at 4 °C. The precipitated proteins were separated from the soluble radioactivity by centrifugation at 600 × g for 10 min, and then dissolved in 0.5 ml of 0.2 n NaOH. Radioactivity was determined by liquid scintillation counting. Protein degradation was calculated by dividing the acid-soluble radioactivity recovered from both cells and medium by the radioactivity contained in precipitated proteins from both cells and medium (45).
_Immunoblotting_—After being resolved by SDS-PAGE, proteins were transferred onto a nitrocellulose membrane. The membrane was blocked with 5% non fat dry milk in PBST (PBS and 0.1% Tween 20) for 1 h at room temperature, and then incubated with the appropriate primary antibody overnight at 4 °C in PBST. The antibody dilutions were as follows: anti-LC3 1:10,000; anti-JNK1-P(Thr183/Tyr185), anti-JNK1, anti-Bcl-2-P(Ser70), anti-Bcl-2 1:1000; anti-p62 1:2000; anti-Beclin 1 1:2000; anti-actin 1:5000). After three washes in PBST, the membrane was incubated for 1 h at room temperature with the appropriate horseradish peroxidase-labeled secondary antibody. Bound antibodies were detected using ECL.
_Statistical Analysis_—Statistical analysis of the differences between the groups was performed using Student's t test. p < 0.05 was considered statistically significant.
RESULTS
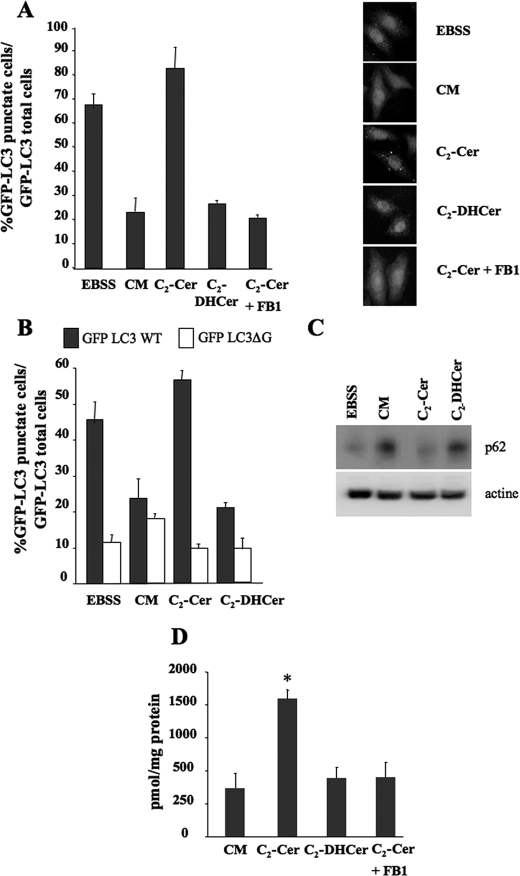
Ceramide Induces Autophagy in Several Cell Lines_—We have previously reported that the permeant short-chain C2-ceramide (C2-Cer) induces autophagy both in MCF-7 breast cancer cells and in HT-29 colon carcinoma cells via the production of endogenous long-chain ceramides (36). Autophagy was monitored by electron microscopy and by measuring the autophagic flux by analyzing the rate of long-lived protein degradationsensitive to 3-methyladenine, an inhibitor of the formation of autophagosomes (46). In a first series of experiments, we extended these findings to HeLa GFP-LC3 cells by analyzing the formation of GFP-LC3 puncta. During autophagy, the LC3 protein is relocated to the autophagosomal membranes as a result of C-terminal conjugation to phosphatidylethanolamine. Thus, the accumulation of GFP-LC3 puncta provides an effective way of detecting autophagosomes (47). C2-Cer treatment induced the accumulation of GFP-LC3 puncta in HeLa GFP-LC3 cells, whereas treatment with C2-dihydroceramide (C2-DHCer), a C2-Cer analogue that does not induce autophagy in MCF-7 and HT-29 cells (36), failed to do so (Fig. 1_A_). To check that the formation of GFP-LC3 puncta in response to C2-Cer was not simply due to clumping of the chimeric protein induced by C2-Cer treatment, we repeated the experiment in cells transfected with the mutant GFP-LC3ΔG, which is unable to support the formation of autophagosomes (48). Under these conditions, the mutant chimeric protein did not form puncta in response to C2-Cer treatment (Fig. 1_B_). Furthermore, C2-Cer was also unable to induce GFP-LC3 puncta in Atg5–/– MEF cells (supplemental Fig. S1_A) that lack the essential autophagy protein Atg5 (49). Unlike C2-Cer, C2-DHCer did not induce autophagy in wt MEF (supplemental Fig. S1_A_). The accumulation of GFP-LC3 puncta was also observed in MCF-7.beclin 1 cells. This cell line was engineered from a MCF-7 cell population with a low level of Beclin 1 expression and provides a convenient tool to for investigating autophagy, because the expression of Beclin 1 is under the control of a Tet-OFF system (50). In the absence of tetracycline we observed that C2-Cer, but not C2-DHCer, induced the formation of GFP-LC3 puncta (supplemental Fig. S2_A_). To confirm that the formation of GFP-LC3 puncta induced by C2-Cer was indeed attributable to stimulation of the autophagic pathway, i.e. to increases in both the formation of autophagosomes and their consumption by the lysosomal compartment, we analyzed the effect of C2-Cer on two independent assays of autophagic flux, the degradation of p62 and that of [14C]valine-labeled long-lived protein. In HeLa GFP-LC3 cells, we observed that protein p62, a substrate for starvation-induced autophagy (51), was just as well degraded in nutrient-free medium (EBSS) as in C2-Cer-treated cells (Fig. 1_C_). In contrast, cells treated with either complete media (CM) or C2-DHCer were unable to stimulate p62 degradation (Fig. 1_C_). In MCF-7.beclin 1 cells, C2-Cer treatment increased the degradation of long-lived proteins sensitive to 3-methyladenine (supplemental Fig. S2_B_).
FIGURE 1.
C2-Cer induces autophagy in HeLa cells. A, HeLa GFP-LC3 cells were cultured for 4 h in either EBSS or complete medium (CM) either alone or supplemented with 100 μm C2-Cer or 100 μm C2-DHCer. Left panel, autophagy was quantified by counting the number of cells with GFP-LC3 puncta. The result shown is representative of three independent experiments. 50–100 cells were analyzed per assay. Right panel, representative images. B, HeLa cells were transfected with a plasmid encoding human GFP-LC3 WT or human GFP-LC3ΔG. 24 h after transfection, the cells were placed for 4 h in either EBSS or complete medium (CM), either alone or supplemented with 100 μm C2-Cer or 100 μm C2-DHCer. Autophagy was quantified by counting the number of cells with GFP-LC3 or GFP-ΔG puncta. The result shown is representative of three independent experiments. 50–100 cells were analyzed per assay. The result shown is representative of three independent experiments. C, HeLa cells were placed for 4 h in EBSS, CM, 100 μm C2-Cer, or 100 μm C2-DHCer. 10 μg of lysates was subjected to immunoblotting using anti-p62 antibody (1:2000) or anti-actin antibody (1:5000). D, determination of endogenous long-chain ceramides by the DAG method described under “Materials and Methods.” Values reported are the mean ± S.D. of three independent experiments. *, p < 0.05_versus_ CM.
Our previous results had shown that the induction of autophagy depends on the elongation of C2-Cer to form long chain ceramides (36). In HeLa GFP-LC3 (Fig. 1_D_), MEF (supplemental Fig. S1_B_) and MCF-7.beclin 1 cells (supplemental Fig. S2_C_) an accumulation of long-chain ceramide was observed after C2-Cer treatment. The elongation step depends on the activity of ceramide synthase is sensitive to FB1 (52). In all the cell lines used, FB1 treatment (100 nm) blocked both the elongation of C2-Cer and C2-Cer-induced autophagy (Fig. 1 and supplemental Figs. S1 and S2). In line with our previous findings (35,36), these results strongly suggest that endogenous long-chain ceramides are potent stimulators of autophagy.
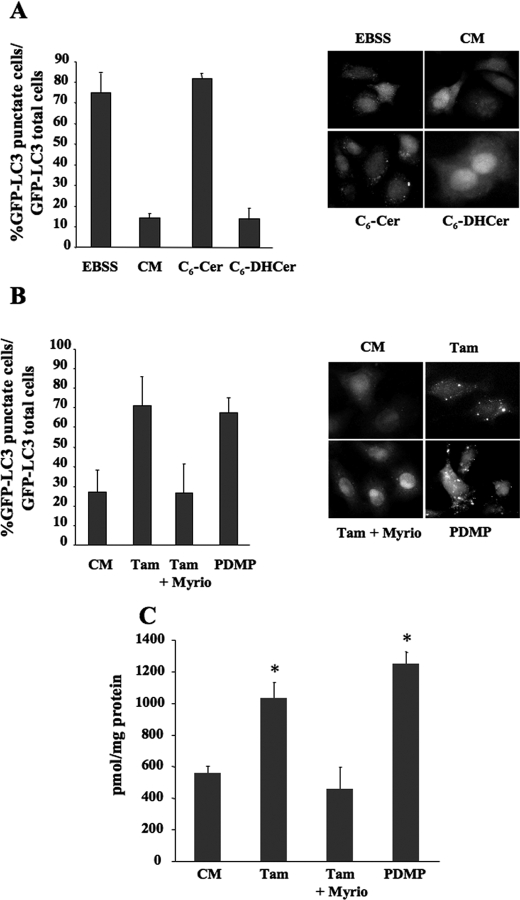
To further substantiate the role of long-chain ceramides in autophagy, we repeated the above experiments using the short chain C6-Cer, which is a good substrate for generating the sphingosine backbone for long-chain ceramides (53). We found that exposing HeLa GFP-LC3 cells to 100 μm of C6-Cer (for 4 h) significantly increased autophagy assessed by counting the number of puncta per cell (Fig. 2_A_). In contrast, its inactive counterpart, C6-DHCer was unable to regulate autophagy (Fig. 2_A_). During the course of these experiments we observed an accumulation of long-chain ceramides in C6-Cer-treated cells. Once again FB1 treatment blocked both the elongation of C6-Cer (Fig. 2_C_) and C6-Cer-induced autophagy (data not shown). Stimulation of autophagy and accumulation of long-chain ceramides were also observed when C6-Cer was used at a lower concentration (40 μm) for different periods of time (supplemental Fig. S3). C6-Cer also increased the autophagic flux as determined by analyzing the rate of long-lived protein degradation (data not shown). We have previously shown that Tamoxifen (Tam) and 1-phenyl-2-decanoylamino-3-morpholino-1-propanol stimulate autophagy in MCF-7 by increasing the level of endogenous long-chain ceramides (36). These treatments both increased the formation of long-chain ceramides presumably either by stimulating the de novo ceramide synthesis in the ER, or by inhibiting the conversion of ceramide to glucosylceramide (36,54). Here we show that Tam and 1-phenyl-2-decanoylamino-3-morpholino-1-propanol stimulated autophagy (Fig. 2_B_) and increased the formation of long-chain ceramides (Fig. 2_C_) in HeLa-GFP-LC3 cells. Moreover, Myriocin, a potent inhibitor of serine palmitoyltransferase the key rate-limiting enzyme of the de novo synthesis of ceramide (52), blocked both autophagy and the accumulation of long-chain ceramides in Tam-treated HeLa GFP-LC3 cells (Fig. 2,B and C).
FIGURE 2.
C6-Cer induces autophagy in HeLa cells. A, HeLa GFP-LC3 cells were cultured for 4 h in either EBSS or complete medium (CM), either alone or supplemented with 100 μm C6-Cer or C6-DHCer. Left panel, autophagy was quantified by counting the number of cells with GFP-LC3 puncta. The result shown is representative of three independent experiments. 50–100 cells were analyzed per assay. Right panel, representative images.B, HeLa GFP-LC3 cells were cultured for 24 h with CM, 1 μm Tam, or 1 μm Tam plus 100 nm Myriocin, or 2.5 μm 1-phenyl-2-decanoylamino-3-morpholino-1-propanol. Left panel, autophagy was quantified by counting the number of cells with GFP-LC3 puncta. The result shown is representative of three independent experiments. 50–100 cells were analyzed per assay. The result shown is representative of three independent experiments. Right panel, representative images.C, quantitation of endogenous long-chain ceramides, by the DAG as described under “Materials and Methods.” Values reported are the mean ± S.D. of three independent experiments. Asterisks indicate p < 0.05 versus CM.
In a previous study, we demonstrated that ceramide stimulates autophagy by interfering with the activation of Akt/PKB upstream of mTOR, a key regulator of autophagy signaling (36). However, we wonder whether ceramide could also stimulate autophagy by directly modulating the activity of the Atg machinery involved in autophagosome formation.
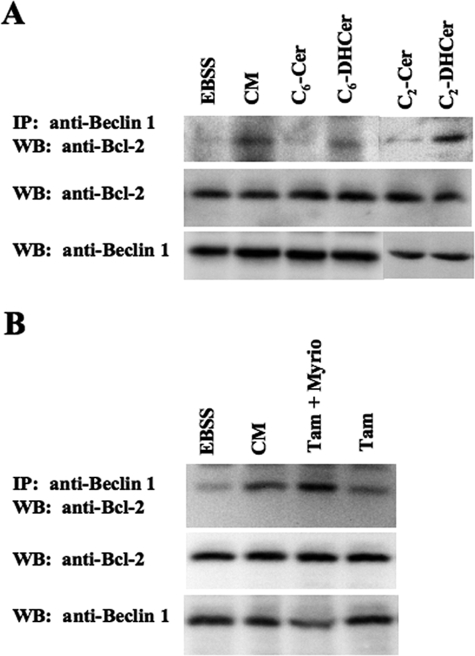
_Ceramide Induces Dissociation of the Beclin 1-Bcl-2 Complex_—Autophagy is tightly regulated by the activity of the Beclin 1 complex in initiating the formation of autophagosomes (22,23). In this complex, the anti-apoptotic protein Bcl-2 represses autophagy. Dissociation of the Beclin 1-Bcl-2 complex stimulates autophagy whether induced by starvation or in response to BH3 mimetic molecules (19,25). However, the role of lipid mediators in the regulation of the Beclin 1-Bcl-2 complex is unknown. We used HeLa cells that express detectable endogenous levels of both Beclin 1 and Bcl-2 to investigate the effect of ceramide on the dissociation of Beclin 1 and Bcl-2. Similarly to starvation (see Ref.19 andFig. 3_A_), co-immunoprecipitation experiments showed that, when C2-Cer or C6-Cer were added to cells, dissociation of the Beclin 1-Bcl-2 complex was observed together with the induction of autophagy. C2-DHCer and C6-DHCer did not stimulate autophagy under the experimental conditions used here, nor did they dissociate the complex (Fig. 3_A_).
FIGURE 3.
Ceramide induces Beclin 1-Bcl-2 complex dissociation in HeLa cells. A, HeLa cells were cultured for 4 h in either EBSS or complete medium (CM), either alone or supplemented with 100 μm C6-Cer, C6-DHCer, C2-Cer, or C2-DHCer. Endogenous Beclin 1 was immunoprecipitated using goat polyclonal antibody (1:80 dilution). Immunoprecipitated proteins were subjected to immunoblotting using a polyclonal anti-Bcl-2 antibody. Lysates were immunoblotted using a polyclonal anti-Bcl-2 or a polyclonal anti-Beclin 1 antibodies. B, HeLa cells were cultured for 24 h in either EBSS or CM, either alone or supplemented with 1 μm Tam, or 1 μm Tam plus 100 nm Myriocin. Endogenous Beclin 1 was immunoprecipitated using goat polyclonal antibody (1:80 dilution). Immunoprecipitated proteins were subjected to immunoblotting using a polyclonal anti-Bcl-2 antibody. Lysates was immunoblotted using a polyclonal anti-Bcl-2 or a polyclonal anti-Beclin 1 antibody. Western blots are representative of three independent experiments.
To confirm that long-chain ceramides are indeed responsible for the dissociation of the Beclin 1-Bcl-2 complex, we next investigated the effects of Tam. We observed a dissociation of the Beclin 1-Bcl-2 complex in Tam-treated cells that correlated with the accumulation of GFP-LC3 puncta (Fig. 3_B_). Moreover, when Myriocin, an inhibitor of serine palmitoyl transferase, a key enzyme in ceramide biosynthesis (52), was present, Tam no longer induced dissociation of the Beclin 1-Bcl-2 complex;i.e. Myriocin reduced the formation of GFP-LC3 puncta by Tam-treated cells (Fig. 3_B_). From these findings, we conclude that the accumulation of long-chain ceramide stimulates autophagy by promoting dissociation of the Beclin 1-Bcl-2 complex.
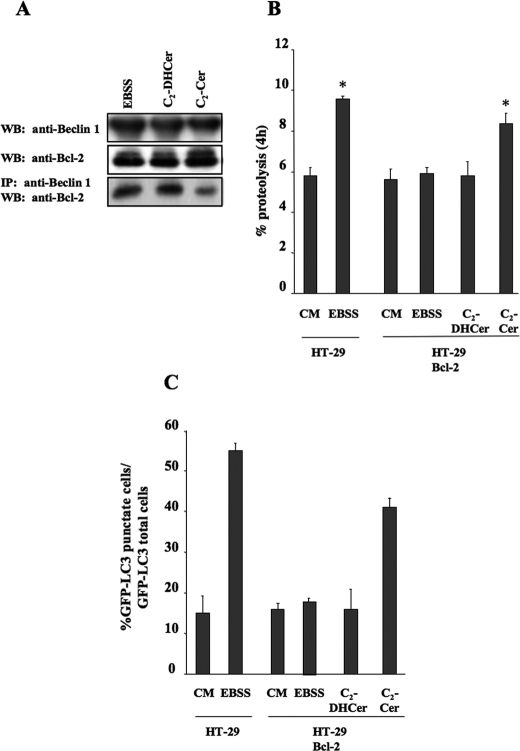
_Effect of Bcl-2 Expression on the Beclin 1-Bcl-2 Complex_—We next wanted to find out whether modulation of Bcl-2 expression mitigates the autophagic response to C2-Cer. To do this, we used the HT-29 colon carcinoma cell line, which we had previously reported to be sensitive to starvation-induced autophagy and to C2-Cer-induced autophagy (36,55). Moreover, this cell line expresses Beclin 1, but no detectable level of Bcl-2 (19). In accordance with previously published results (19), when Bcl-2 was stably expressed in this cell line, a blockade of the Beclin 1-Bcl-2 complex was observed (Fig. 4_A_) as well as inhibition of starvation-induced autophagy, as revealed by the presence of fewer GFP-LC3 puncta (Fig. 4_C_), and no stimulation of long-lived protein degradation (Fig. 4_B_). In contrast, C2-Cer was able to trigger autophagy, as shown by an increase in the number of GFP-LC3 puncta, and stimulated long-lived protein degradation in HT-29-Bcl-2 cells (Fig. 4,B and C). Stimulation of autophagy by C2-Cer in HT-29-Bcl-2 cells was characterized by the dissociation of the Beclin 1-Bcl-2 complex (Fig. 4_A_). In contrast to C2-Cer, C2-DHCer neither induced the dissociation of the Beclin 1-Bcl-2 complex (Fig. 4_A_) nor triggered autophagy in HT-29-Bcl-2 cells (Fig. 4, B and_C_). These results suggest that, under conditions where Bcl-2 blunts starvation-induced autophagy, ceramide is able to overcome this inhibitory effect. These results were confirmed by analyzing the dissociation of the Beclin 1-Bcl-2 complex, and the formation of GFP-LC3 puncta in MCF-7.beclin 1 cells after transfection with a Myc-tagged form of Bcl-2 (Fig. 5). In this setting, starvation did not lead to dissociation of the Beclin 1-Bcl-2 complex but did stimulate the formation of GFP-LC3 puncta when compared with untransfected cells (see Refs.19,40 andFig. 5 (B and_C_)). In contrast, C2-Cer did trigger both autophagy and Beclin 1-Bcl-2 complex dissociation in MCF-7.beclin 1 cells expressing Myc-Bcl-2 (Fig. 5,B and C).
FIGURE 4.
C2-Cer induces both autophagy and Beclin/Bcl-2 complex dissociation in HT-29 cells. A, HT-29 Bcl-2 cells were cultured for 4 h in EBSS, or complete medium (CM) supplemented with 100 μm C2-Cer or 100 μm C2-DH Cer. Endogenous Beclin 1 was immunoprecipitated using a goat polyclonal antibody (1:80 dilution). Immunoprecipitated proteins were subjected to immunoblotting using polyclonal anti-Bcl-2 antibody. Lysates were immunoblotted using polyclonal anti-Bcl-2 or polyclonal anti-Beclin 1 antibodies. B, proteolysis (see “Material and Methods”) was measured in HT-29 cells treated for 4 h with CM or EBSS; or in HT-29 Bcl-2 cells treated with CM, EBSS, 100 μm C2-DHCer, or 100 μm C2-Cer. C, HT-29 and HT-29 Bcl-2 cells were transfected with a plasmid encoding GFP-LC3. 24 h after transfection, HT-29 cells were cultured for 4 h either in EBSS or CM. HT-29 Bcl-2 cells were treated for 4 h with CM, EBSS, 100 μm C2-DHCer, or 100 μm C2-Cer. Autophagy was quantified by counting the number of cells with GFP-LC3 puncta. The result shown is representative of three independent experiments. 50–100 cells were analyzed per assay. Values reported are the mean ± S.D. of three independent experiments.Asterisks indicate p < 0.05 versus CM.
FIGURE 5.
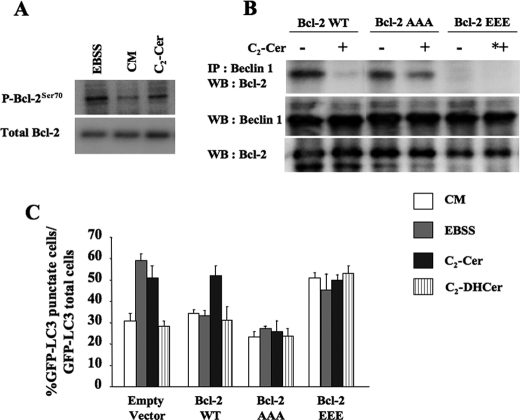
C2-Cer stimulates Bcl-2 phosphorylation in MCF-7.beclin 1 cells. A, MCF-7.beclin 1 cells were cultured for 4 h in EBSS, in complete medium (CM) or in CM supplemented with 100 μm C2-Cer. Lysates were subjected to immunoblotting using a monoclonal antibody against phospho-Bcl-2(Ser70) (1:1000,upper panel), or a mouse monoclonal anti-Bcl-2 antibody (1:1000,lower panel). B, MCF-7.beclin 1 cells were transfected with a plasmid encoding for Bcl-2 WT, Bcl-2 AAA, or Bcl-2 EEE. 24 h after transfection, the cells were placed for 4 h in CM or 100 μm C2-Cer. Beclin 1 was immunoprecipitated using goat polyclonal antibody (1:80 dilution). The immunoprecipitated proteins were subjected to immunoblotting using polyclonal anti-Bcl-2 antibody. Lysates was immunoblotted using a polyclonal anti-Bcl-2 or a polyclonal anti-Beclin 1 antibodies. C, MCF7.beclin 1 cells were cotransfected with a plasmid encoding GFP-LC3, an empty vector, or a plasmid encoding for Bcl-2 WT, Bcl-2 AAA, or Bcl-2 EEE. 24 h after transfection, the cells were cultured for 4 h in EBSS, CM, 100 μm C2-Cer, or 100 μm C2-DHCer. Autophagy was quantified by counting the number of cells with GFP-LC3 puncta. The result shown is representative of four independent experiments ± S.D. 50–100 cells were analyzed per assay.
_Ceramide-induced Autophagy Is Dependent on Bcl-2 Phosphorylation_—We then investigated the mechanism that regulates the ceramide-dependent dissociation of the Beclin 1-Bcl-2 complex. Phosphorylation is one of the post-translational changes known to regulate protein complexes. For example, in the autophagic pathway, the recruitment of partners of Atg1 is phosphorylation-dependent (3). Moreover, ceramide modulates the phosphorylation of Bcl-2 (56), and it has been shown recently that Bcl-2 phosphorylation is essential both for the stimulation of starvation-induced autophagy, and for the regulation of the Beclin 1:Bcl-2 rheostat (40). In a first series of experiments, we analyzed the phosphorylation status of Bcl-2 in MCF-7.beclin 1 cells treated with C2-Cer using a commercial antibody that detects phosphorylation of the Ser residue at position 70. We observed an increase in the phosphorylation at Ser70 after exposure to 100 μm C2-Cer for 4 h (Fig. 5_A_). We also found that starvation induced greater increase of Ser70 phosphorylation than CM (see Ref. (40) andFig. 5_A_). Ser70 is located within a non-structural loop of Bcl-2 characterized by the inclusion of a triad of amino acids Thr69, Ser70, and Ser87, known to be substrates for kinases (57). These same three residues play a major role in Beclin 1-Bcl-2 dissociation in response to starvation (40). To investigate whether these phosphorylation sites are involved in regulating ceramide-induced autophagy and in the dissociation of the Beclin 1-Bcl-2 complex, we transfected MCF-7.beclin 1 cells with the cDNAs that encode wild-type Myc-Bcl-2, Myc-Bcl-2AAA (where Thr69, Ser70, and Ser87 were mutated to Ala by site-directed mutagenesis), and Myc-Bcl-2EEE (where Thr69, Ser70, and Ser87 were mutated to Glu by site-directed mutagenesis).
As illustrated in Fig. 5 (B and_C_), in contrast to starvation (40), C2-Cer stimulated autophagy and induced the dissociation of the Beclin 1-Bcl-2 complex in MCF7.beclin 1 cells expressing the wild-type Myc-Bcl-2. These results are consistent with those obtained in HT-29-Bcl-2 cells (seeFig. 4, A–D). As observed in HT-29-Bcl-2 cells, C2-DHCer did not induce autophagy in MCF-7.beclin 1 cells. C2-Cer treatment did not induce dissociation of the Beclin 1-Bcl-2 complex in MCF-7.beclin 1 cells transfected with the Myc-Bcl-2AAA mutant (Fig. 5_B_ and Ref.40). It was therefore not surprising to find that the autophagic response to C2-Cer treatment and to starvation was blunted in these cells (Fig. 5_C_). In contrast, in MCF-7.beclin 1 cells expressing the Myc-Bcl-2EEE mutant, which mimics the phosphorylated form of Bcl-2, dissociation of the Beclin 1-Bcl-2 complex was observed even when autophagy was not stimulated by either C2-Cer treatment or starvation (Fig. 5_B_ and Ref.40). The autophagy rate under these conditions was similar to that observed after C2-Cer treatment or starvation (Fig. 5_C_). This indicates that autophagy cannot be further stimulated by starvation or C2-Cer once the Beclin 1-Bcl-2 complex has been dissociated. It has been shown recently that simultaneous mutation of the Thr69, Ser70, and Ser87 of Bcl-2 to Ala resulted in a constitutive interaction between Beclin 1 and Bcl-2 during periods of starvation (see Ref.40). In contrast, simultaneous mutation of the Thr69, Ser70, and Ser87 of Bcl-2 to Glu alters the interaction between Beclin 1 and Bcl-2. Accordingly, the effects of ceramide treatment on Beclin 1-Bcl-2 dissociation and the autophagic response shown in Fig. 5 were only observed when all three residues in the non-structural loop of Bcl-2 were mutated to Glu or to Ala (data not shown).
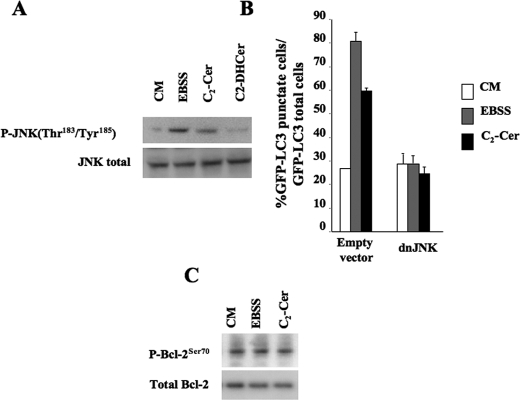
_JNK1 Regulates Ceramide-induced Autophagy and Ceramide-dependent Bcl-2 Phosphorylation_—Residues in the non-structural loop are substrates for various different Ser/Thr protein kinases (57). Among the kinases that phosphorylate the three residues in the non-structural loop, JNK1 has been shown to phosphorylate all three residues (57,58). Moreover, preliminary results showed that inhibitors of PKC and p38 MAPK are not able to mitigate the Beclin 1-Bcl-2 dissociation induced by C2-Cer (data not shown). We therefore first analyzed the phosphorylation of JNK1 in HeLa GFP-LC3 cells. C2-Cer treatment triggered the activation of JNK1 after incubating for 4 h (Fig. 6_A_), by which time the level of long-chain ceramides was significantly increased (seeFig. 1_D_). It is important to note that incubating for 2 h did not modify either Bcl-2 phosphorylation or JNK1 phosphorylation (data not shown), which strengthens the idea that long-chain ceramide synthesis is essential. In addition, we confirmed that starvation stimulated both Bcl-2 phosphorylation (seeFig. 5_A_) and JNK1 phosphorylation (Fig. 6_A_). Following these results, we expressed a dominant-negative form of JNK1. When this mutant was expressed in HeLa GFP-LC3 cells, ceramide-induced autophagy and starvation-induced autophagy were both totally blocked (Fig. 6_B_), suggesting that JNK1 activation is an essential step in the stimulation of autophagy. Moreover, in this context, the increase in Bcl-2 phosphorylation was blocked both in C2-Cer-treated cells and starved cells (Fig. 6_C_).
FIGURE 6.
C2-Cer stimulates autophagy via JNK1 activity in HeLa cells. A, HeLa cells were cultured for 4 h in EBSS, complete medium (CM) alone or supplemented with 100 μm C2-Cer. Lysates were immunoblotted with a rabbit monoclonal anti-Phospho-JNK(Thr183/Tyr185) antibody (1:1000,upper panel) or a rabbit monoclonal anti-JNK antibody (1:1000,lower panel). B, HeLa cells were co-transfected with a plasmid encoding for GFP-LC3, an empty vector or a vector encoding inactive JNK1 (dnJNK1). 24 h after being transfected, the cells were treated for 4 h with EBSS, CM, or 100 μm C2-Cer. Autophagy was quantified by counting the number of cells with GFP-LC3 puncta. The result shown is representative of three independent experiments. 50–100 cells were analyzed per assay. C, HeLa cells were transfected with a plasmid encoding dnJNK1. 4 h after being transfected, the cells were treated for 4 h with EBSS, CM, 100 μm C2-Cer, or 100 μm 100 μm C2-DHCer. The cells were then subjected to electrophoresis and immunoblotting using rabbit monoclonal anti-Phospho-JNK(Thr183/Tyr185) antibody (1:1000), a rabbit monoclonal anti-JNK antibody (1:1000), or a monoclonal antibody raised against phospho-Bcl-2(Ser70) (1:1000, upper panel) or a mouse monoclonal anti-Bcl-2 antibody (1:1000). Values reported are the mean ± S.D. of three independent experiments.
DISCUSSION
Ceramide is a sphingolipid second messenger involved in a wide variety of cell processes (28,31). A better understanding of its role in cellular homeostasis calls for characterization of its intracellular targets. In the work reported here, we identified the Beclin 1-Bcl-2 complex involved in the early stages of autophagosome formation of as a new target for ceramide. In this complex, the anti-apoptotic protein Bcl-2 has an anti-autophagic function (19), and so dissociation of the complex is required to trigger autophagy during starvation to promote cell survival in the absence of nutrients (19). In addition to physiological stimuli, such as starvation or the production of ceramide, drugs relevant to cancer therapy, such as the BH3-mimetic ABT-747, have been shown to dissociate the Beclin 1-Bcl-2 complex by competing with the BH3 domain of Beclin 1 (25).
Based on the present and previous studies (36), we conclude that ceramide stimulates autophagy by interfering with the activities of the mTORC1 complex and of the Beclin1 complex. This situation is reminiscent of that observed during starvation-induced autophagy, where the activity of both these complexes is modulated (23). Interestingly, the Beclin 1 protein is known to act as a scaffold protein that directly or indirectly recruits several partners, including oncoproteins (Bcl-2 and Bcl-XL) and tumor suppressors (UVRAG, Ambra-1, and Bif-1) (22,23). Oncoproteins have an inhibitory effect on the activity of hVps34 (class III phosphatidylinositol 3-phosphate) in the complex, and consequently on autophagy. Tumor suppressors have the opposite effects on both autophagy and hVps34 activity. In this context, it is interesting to note that ceramide, which is a tumor suppressing lipid (31), induces the dissociation of the Beclin 1-Bcl-2 complex and stimulates autophagy.
Ceramide-induced dissociation of the Beclin 1-Bcl-2 complex requires the phosphorylation of Bcl-2. Previous studies have shown that ceramide modulates the anti-apoptotic capacity of Bcl-2 by decreasing its phosphorylation as a result of activating protein phosphatase 2A (56). This would suggest that ceramide triggers autophagy by inhibiting the anti-autophagic function of Bcl-2 via its phosphorylation and apoptosis by dephosphorylating Bcl-2. As far as the anti-autophagic function of Bcl-2 is concerned, the phosphorylation of the non-structural loop alleviates its anti-autophagic capacity, as shown in response to starvation or to ceramide (Ref.40 and the present study). Deletion of the non-structural loop of Bcl-2 blocks both starvation-induced autophagy (see Ref. 40) and ceramide-induced autophagy (data not shown). As discussed elsewhere (59), the absence of the structural loop in viral forms of Bcl-2 is probably a strategy for viruses to block the induction of autophagy during cell infection. Interestingly, deleting this non-structural loop and the absence of phosphorylation of amino acids in this loop both reinforce the anti-apoptotic function of cellular Bcl-2 (58,60).
The subcellular localization of Bcl-2 in ceramide-induced autophagy and ceramide-induced apoptosis is probably an important parameter. Mitochondria have been shown to play a major role in ceramide-induced apoptosis (61). Furthermore, a mitochondrial protein phosphatase 2A has been implicated in the ceramide-dependent dephosphorylation of Bcl-2 that triggers apoptosis (56). These findings suggest that the mitochondrial-targeted Bcl-2 plays a critical role in regulating ceramide-induced apoptosis. In contrast, only the ER-targeted form of Bcl-2 regulates autophagy (19,25). Phosphorylation of ER-located Bcl-2 inhibits its binding to pro-apoptotic family members (62) and induces its dissociation from Beclin 1 (40).
The great plasticity of sphingolipid metabolism means that ceramide can be produced via several different routes (de novo synthesis, hydrolysis of sphingomyelin, and degradation of glycolipids) in different subcellular locations (63). It is interesting to observe that an increase in the de novo synthesis of ceramide in the ER in response to Tam treatment leads to the dissociation of the Beclin 1-Bcl-2 complex. This could suggest that the ER pool of ceramide could probably be involved in modulating autophagy via the Beclin 1-Bcl-2 complex. The transport of ceramide out of the ER is dependent on the CERT protein (64). Inhibition of CERT expression is known to induce an ER stress response (65). On the other hand, in different settings ER stress is known to induce autophagy (66,67). It is tempting to envisage the possibility that ceramide could be a part of the signaling that triggers ER-dependent autophagy. However, it is likely that the ceramide produced by the recycling salvage pathway of sphingosine out of the ER (53) may also interfere with the composition of the Beclin 1-Bcl-2 complex. It would be worth investigating the contributions of the various different pools of ceramide in the induction of autophagy, to see whether this would shed any light on the roles played by the different pools of this lipid in regulating autophagy and apoptosis. Interestingly, a recent reports shows that CD95-dependent ceramide can trigger both pro-survival signals (autophagy) and pro-death signals (apoptosis) in carcinoma cells (68), suggesting that ceramide cannot be considered to be solely a mediator of cell death.
Ceramide mediates the dissociation of the Beclin 1-Bcl-2 complex by stimulating the phosphorylation of Bcl-2 by JNK1. JNKs, which are stress-activated protein kinases, can be activated by ceramide (69,70). One effect of this ceramide-dependent activation of JNKs is the phosphorylation of Bcl-2 family members (71). JNKs are a key regulator of the balance between cell survival and cell death. The duration of activation, as well as the nature of the substrate, are key elements in the equilibrium between the pro- or anti-apoptotic activities of JNK (72). The ceramide-dependent activation of autophagy via JNK is another parameter involved in the pro-survival and pro-death activity of JNK, because of the dual role of autophagy in cell death and survival.
During this work we noticed that ceramide was more potent than starvation in dissociating the Beclin 1-Bcl-2 complex. In fact, ceramide induced dissociation of the complex and stimulated autophagy in a context of the forced expression of Bcl-2 (HT-29-Bcl-2 cells and MCF-7._beclin 1_-Bcl-2 cells), whereas in this context, starvation had no effect on Beclin 1-Bcl-2 dissociation and did not stimulate autophagy (40). This difference between the effects of starvation and ceramide is probably not the consequence of distinct signaling pathways. In fact, both stimuli inhibit the mTOR signaling pathway, which acts upstream of the Beclin 1-Bcl-2 complex during the formation of autophagosomes (17,36). One possible explanation for the robust effect of ceramide on the dissociation of the Beclin 1-Bcl-2 complex is that, in addition to Bcl-2 phosphorylation, ceramide may also promote the dissociation of the complex by other mechanisms. Ceramide has been shown to trigger autophagy via the induction of the BH3-only protein BNIP3 (37). BNIP3 began to accumulate in response to ceramide after 4–5 h of treatment,i.e. on a time scale compatible with the experiments performed here. In addition, several reports indicate that BNIP3 is involved in the induction of autophagy (37,73–75). A recent study has shown that, during hypoxia, BNIP3 is able to displace Bcl-2 from the Beclin 1 complex, probably by competing with the BH3-domain of Beclin 1 (75). This means that we cannot exclude the possibility that ceramide may use two strategies to dissociate Bcl-2 from Beclin1: first, by increasing its phosphorylation of Bcl-2 via JNK1, and second by favoring the BNIP3-Bcl-2 complex rather than the Beclin 1-Bcl-2 complex. One question that remains to be answered is how ceramide leads to an accumulation of Bnip3. Recently, the expression of several autophagy genes, including BNIP3, has been shown to be under the control of the transcription factor FOXO3 (73). FOXO3 is inhibited by Akt/PKB-dependent phosphorylation (76), but this inhibitory effect can be overcome by ceramide, which inhibits Akt/PKB (77). Moreover, as we have reported elsewhere, ceramide increases the level of Beclin 1 mRNA (36). So we hypothesize that the ceramide-induced inhibition of Akt/PKB may stimulate autophagy by two, non-mutually exclusive pathways: by interfering with the activation of the mTORC1 complex and by regulating the expression of autophagy genes via FOXO3.
The present findings suggest that ceramide is a second messenger with a central role in the regulation of autophagy. Elucidation of the targets of ceramide in the autophagic pathway will clarify its role in cell homeostasis. Together with our previous results showing that another sphingolipid second messenger, sphingosine 1-phosphate, also regulates autophagy (35), it is clear that the plasticity of sphingolipid metabolism means that there are a variety of possible mechanisms for triggering autophagy in response to stress situations and to influence cell fate (78).
Supplementary Material
[Supplemental Data]
Acknowledgments
We are grateful to Drs. M.-T. Dimanche-Boitrel, Y. A. Hannun, N. Mizushima, D. K. Perry, I. Tanida, A. M. Tolkovsky, R. J. Davis, and T. Yoshimori for providing us with reagents used in this study.
*
This work was supported, in whole or in part, by National Institutes of Health Grant R01 CA109618 (to B. L.). This work was also supported by institutional funding from INSERM, from University Paris-Sud 11, and from the Association pour la Recherche sur le Cancer (Grant 3503 to P. C. and Grant 4006 to S. P.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “_advertisement_” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
S⃞
The on-line version of this article (available athttp://www.jbc.org) contains supplemental Figs. S1–S3.
Footnotes
2
The abbreviations used are: LC3, Light Chain 3; C2-Cer, C2-ceramide; C2-DHCer, C2-dihydroceramide; C6-Cer, C6-ceramide; C6-DHCer, C6-dihydroceramide; DAG, diacylglycerol; dnJNK1, dominant-negative JNK1; FB1, fumonisin B1; EBSS, Earle's balanced salt solution; Tam, tamoxifen; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; ER, endoplasmic reticulum; PBS, phosphate-buffered saline; CM, complete medium; mTOR, mammalian target of rapamycin.
References
- 1.Levine, B., and Klionsky, D. J. (2004) Dev. Cell 6 463–477 [DOI] [PubMed] [Google Scholar]
- 2.Mizushima, N., Levine, B., Cuervo, A. M., and Klionsky, D. J. (2008) Nature 451 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie, Z., and Klionsky, D. J. (2007) Nat. Cell Biol. 9 1102–1109 [DOI] [PubMed] [Google Scholar]
- 4.Hara, T., Nakamura, K., Matsui, M., Yamamoto, A., Nakahara, Y., Suzuki-Migishima, R., Yokoyama, M., Mishima, K., Saito, I., Okano, H., and Mizushima, N. (2006) Nature 441 885–889 [DOI] [PubMed] [Google Scholar]
- 5.Komatsu, M., Waguri, S., Chiba, T., Murata, S., Iwata, J., Tanida, I., Ueno, T., Koike, M., Uchiyama, Y., Kominami, E., and Tanaka, K. (2006) Nature 441 880–884 [DOI] [PubMed] [Google Scholar]
- 6.Kuma, A., Hatano, M., Matsui, M., Yamamoto, A., Nakaya, H., Yoshimori, T., Ohsumi, Y., Tokuhisa, T., and Mizushima, N. (2004) Nature 432 1032–1036 [DOI] [PubMed] [Google Scholar]
- 7.Lum, J. J., Bauer, D. E., Kong, M., Harris, M. H., Li, C., Lindsten, T., and Thompson, C. B. (2005) Cell 120 237–248 [DOI] [PubMed] [Google Scholar]
- 8.Boya, P., Gonzalez-Polo, R. A., Casares, N., Perfettini, J. L., Dessen, P., Larochette, N., Metivier, D., Meley, D., Souquere, S., Yoshimori, T., Pierron, G., Codogno, P., and Kroemer, G. (2005) Mol. Cell. Biol. 25 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levine, B. (2007) Nature 446 745–747 [DOI] [PubMed] [Google Scholar]
- 10.Mathew, R., Karantza-Wadsworth, V., and White, E. (2007) Nat. Rev. Cancer 7 961–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maiuri, M. C., Zalckvar, E., Kimchi, A., and Kroemer, G. (2007) Nat. Rev. Mol. Cell. Biol. 8 741–752 [DOI] [PubMed] [Google Scholar]
- 12.Shimizu, S., Kanaseki, T., Mizushima, N., Mizuta, T., Arakawa-Kobayashi, S., Thompson, C. B., and Tsujimoto, Y. (2004) Nat. Cell Biol. 6 1221–1228 [DOI] [PubMed] [Google Scholar]
- 13.Yu, L., Alva, A., Su, H., Dutt, P., Freundt, E., Welsh, S., Baehrecke, E. H., and Lenardo, M. J. (2004) Science 304 1500–1502 [DOI] [PubMed] [Google Scholar]
- 14.Djavaheri-Mergny, M., Amelotti, M., Mathieu, J., Besancon, F., Bauvy, C., Souquere, S., Pierron, G., and Codogno, P. (2006) J. Biol. Chem. 281 30373–30382 [DOI] [PubMed] [Google Scholar]
- 15.Espert, L., Denizot, M., Grimaldi, M., Robert-Hebmann, V., Gay, B., Varbanov, M., Codogno, P., and Biard-Piechaczyk, M. (2006) J. Clin. Invest. 116 2161–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qu, X., Zou, Z., Sun, Q., Luby-Phelps, K., Cheng, P., Hogan, R. N., Gilpin, C., and Levine, B. (2007) Cell 128 931–946 [DOI] [PubMed] [Google Scholar]
- 17.Codogno, P., and Meijer, A. J. (2005) Cell Death Differ. 12 Suppl. 2, 1509–1518 [DOI] [PubMed] [Google Scholar]
- 18.Gozuacik, D., and Kimchi, A. (2007) Curr. Top. Dev. Biol. 78 217–245 [DOI] [PubMed] [Google Scholar]
- 19.Pattingre, S., Tassa, A., Qu, X., Garuti, R., Liang, X. H., Mizushima, N., Packer, M., Schneider, M. D., and Levine, B. (2005) Cell 122 927–939 [DOI] [PubMed] [Google Scholar]
- 20.Levine, B., and Yuan, J. (2005) J. Clin. Invest. 115 2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohsumi, Y. (2001) Nat. Rev. Mol. Cell. Biol. 2 211–216 [DOI] [PubMed] [Google Scholar]
- 22.Cao, Y., and Klionsky, D. J. (2007) Cell Res. 17 839–849 [DOI] [PubMed] [Google Scholar]
- 23.Pattingre, S., Espert, L., Biard-Piechaczyk, M., and Codogno, P. (2008) Biochimie (Paris) 90 313–323 [DOI] [PubMed] [Google Scholar]
- 24.Erlich, S., Mizrachy, L., Segev, O., Lindenboim, L., Zmira, O., Adi-Harel, S., Hirsch, J. A., Stein, R., and Pinkas-Kramarski, R. (2007) Autophagy 3 561–568 [DOI] [PubMed] [Google Scholar]
- 25.Maiuri, M. C., Le Toumelin, G., Criollo, A., Rain, J. C., Gautier, F., Juin, P., Tasdemir, E., Pierron, G., Troulinaki, K., Tavernarakis, N., Hickman, J. A., Geneste, O., and Kroemer, G. (2007) EMBO J. 26 2527–2539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oberstein, A., Jeffrey, P. D., and Shi, Y. (2007) J. Biol. Chem. 282 13123–13132 [DOI] [PubMed] [Google Scholar]
- 27.Levine, B., Sinha, S., and Kroemer, G. (2008) Autophagy 4 600–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannun, Y. A., and Obeid, L. M. (2008) Nat. Rev. Mol. Cell. Biol. 9 139–150 [DOI] [PubMed] [Google Scholar]
- 29.Hla, T. (2004) Semin. Cell Dev. Biol. 15 513–520 [DOI] [PubMed] [Google Scholar]
- 30.Kolesnick, R. (2002) J. Clin. Invest. 110 3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogretmen, B., and Hannun, Y. A. (2004) Nat. Rev. Cancer 4 604–616 [DOI] [PubMed] [Google Scholar]
- 32.Spiegel, S., and Milstien, S. (2003) Nat. Rev. Mol. Cell. Biol. 4 397–407 [DOI] [PubMed] [Google Scholar]
- 33.Trajkovic, K., Hsu, C., Chiantia, S., Rajendran, L., Wenzel, D., Wieland, F., Schwille, P., Brügger, B., and Simons, M. (2008) Science 319 1244–1247 [DOI] [PubMed] [Google Scholar]
- 34.Hannun, Y. A. (1996) Science 274 1855–1859 [DOI] [PubMed] [Google Scholar]
- 35.Lavieu, G., Scarlatti, F., Sala, G., Carpentier, S., Levade, T., Ghidoni, R., Botti, J., and Codogno, P. (2006) J. Biol. Chem. 281 8518–8527 [DOI] [PubMed] [Google Scholar]
- 36.Scarlatti, F., Bauvy, C., Ventruti, A., Sala, G., Cluzeaud, F., Vandewalle, A., Ghidoni, R., and Codogno, P. (2004) J. Biol. Chem. 279 18384–18391 [DOI] [PubMed] [Google Scholar]
- 37.Daido, S., Kanzawa, T., Yamamoto, A., Takeuchi, H., Kondo, Y., and Kondo, S. (2004) Cancer Res. 64 4286–4293 [DOI] [PubMed] [Google Scholar]
- 38.Zeng, X., Overmeyer, J. H., and Maltese, W. A. (2006) J. Cell Sci. 119 259–270 [DOI] [PubMed] [Google Scholar]
- 39.Zheng, W., Kollmeyer, J., Symolon, H., Momin, A., Munter, E., Wang, E., Kelly, S., Allegood, J. C., Liu, Y., Peng, Q., Ramaraju, H., Sullards, M. C., Cabot, M., and Merrill, A. H., Jr. (2006) Biochim. Biophys. Acta 1758 1864–1884 [DOI] [PubMed] [Google Scholar]
- 40.Wei, Y., Pattingre, S., Sinha, S., Bassik, M., and Levine, B. (2008) Mol. Cell 30 678–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chaumorcel, M., Souquere, S., Pierron, G., Codogno, P., and Esclatine, A. (2008) Autophagy 4 46–53 [DOI] [PubMed] [Google Scholar]
- 42.Furuya, N., Yu, J., Byfield, M., Pattingre, S., and Levine, B. (2005) Autophagy 1 46–52 [DOI] [PubMed] [Google Scholar]
- 43.Bielawska, A., Perry, D. K., and Hannun, Y. A. (2001) Anal. Biochem. 298 141–150 [DOI] [PubMed] [Google Scholar]
- 44.Lei, K., Nimnual, A., Zong, W. X., Kennedy, N. J., Flavell, R. A., Thompson, C. B., Bar-Sagi, D., and Davis, R. J. (2002) Mol. Cell. Biol. 22 4929–4942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bauvy, C., Meijer, A. J., and Codogno, P. (2008) Methods Enzymol., in press [DOI] [PubMed]
- 46.Seglen, P. O., and Gordon, P. B. (1982) Proc. Natl. Acad. Sci. U. S. A. 79 1889–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kabeya, Y., Mizushima, N., Ueno, T., Yamamoto, A., Kirisako, T., Noda, T., Kominami, E., Ohsumi, Y., and Yoshimori, T. (2000) EMBO J. 19 5720–5728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanida, I., Yamaji, T., Ueno, T., Ishiura, S., Kominami, E., and Hanada, K. (2008) Autophagy 4 131–134 [DOI] [PubMed] [Google Scholar]
- 49.Mizushima, N., Yamamoto, A., Hatano, M., Kobayashi, Y., Kabeya, Y., Suzuki, K., Tokuhisa, T., Ohsumi, Y., and Yoshimori, T. (2001) J. Cell Biol. 152 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang, X. H., Yu, J., Brown, K., and Levine, B. (2001) Cancer Res. 61 3443–3449 [PubMed] [Google Scholar]
- 51.Mizushima, N., and Yoshimori, T. (2007) Autophagy 3 542–545 [DOI] [PubMed] [Google Scholar]
- 52.Merrill, A. H., Jr. (2002) J. Biol. Chem. 277 25843–25846 [DOI] [PubMed] [Google Scholar]
- 53.Kitatani, K., Idkowiak-Baldys, J., and Hannun, Y. A. (2008) Cell Signal. 20 1010–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bleicher, R. J., and Cabot, M. (2002) Biochim. Biophys. Acta 1585 172–178 [DOI] [PubMed] [Google Scholar]
- 55.Ogier-Denis, E., Couvineau, A., Maoret, J. J., Houri, J. J., Bauvy, C., De Stefanis, D., Isidoro, C., Laburthe, M., and Codogno, P. (1995) J. Biol. Chem. 270 13–16 [DOI] [PubMed] [Google Scholar]
- 56.Ruvolo, P. P., Deng, X., Ito, T., Carr, B. K., and May, W. S. (1999) J. Biol. Chem. 274 20296–20300 [DOI] [PubMed] [Google Scholar]
- 57.Blagosklonny, M. V. (2001) Leukemia 15 869–874 [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto, Z., Ichijo, H., and Korsmeyer, S. J. (1999) Mol. Cell. Biol. 19 8469–8478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liang, C., Xiaofie, E., and Jung, J. U. (2008) Autophagy 4 268–272 [DOI] [PubMed] [Google Scholar]
- 60.Maundrell, K., Antonsson, B., Magnenat, E., Camps, M., Muda, M., Chabert, C., Gillieron, C., Boschert, U., Vial-Knecht, E., Martinou, J.-C., and Arkinstall, S. (1997) J. Biol. Chem. 272 25238–25242 [DOI] [PubMed] [Google Scholar]
- 61.Birbes, H., El Bawab, S., Obeid, L. M., and Hannun, Y. A. (2002) Adv. Enzyme Regul. 42 113–129 [DOI] [PubMed] [Google Scholar]
- 62.Bassik, M. C., Scorrano, L., Oakes, S. A., Pozzan, T., and Korsmeyer, S. J. (2004) EMBO J. 23 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Futerman, A. H., and Hannun, Y. A. (2004) EMBO Rep. 5 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanada, K., Kumagai, K., Yasuda, S., Miura, Y., Kawano, M., Fukasawa, M., and Nishijima, M. (2003) Nature 426 803–809 [DOI] [PubMed] [Google Scholar]
- 65.Swanton, C., Marani, M., Pardo, O., Warne, P. H., Kelly, G., Sahai, E., Elustondo, F., Chang, J., Temple, J., Ahmed, A. A., Brenton, J. D., Downward, J., and Nicke, B. (2007) Cancer Cell 11 498–512 [DOI] [PubMed] [Google Scholar]
- 66.Høyer-Hansen, M., and Jäättelä, M. (2007) Cell Death Differ. 14 1576–1582 [DOI] [PubMed] [Google Scholar]
- 67.Yorimitsu, T., and Klionsky, D. J. (2007) Trends Cell Biol. 17 279–285 [DOI] [PubMed] [Google Scholar]
- 68.Park, M. A., Zhang, G., Martin, A. P., Hamed, H., Mitchell, C., Hylemon, P. B., Graf, M., Rahmani, M., Ryan, K., Liu, X., Spiegel, S., Norris, J., Fisher, P. B., Grant, S., and Dent, P. (2008) Cancer Biol. Ther. 7 1648–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Basu, S., and Kolesnick, R. (1998) Oncogene 17 3277–3285 [DOI] [PubMed] [Google Scholar]
- 70.Saslowsky, D. E., Tanaka, N., Reddy, K. P., and Lencer, W. I. (2008) FASEB J., in press [DOI] [PMC free article] [PubMed]
- 71.Kurinna, S., Tsao, C. C., Nica, A. F., Jiffar, T., and Ruvolo, P. P. (2004) Cancer Res. 64 7852–7856 [DOI] [PubMed] [Google Scholar]
- 72.Liu, J., and Lin, A. (2005) Cell Res. 15 36–42 [DOI] [PubMed] [Google Scholar]
- 73.Mammucari, C., Milan, G., Romanello, V., Masiero, E., Rudolf, R., Del Piccolo, P., Burden, S. J., Di Lisi, R., Sandri, C., Zhao, J., Goldberg, A. L., Schiaffino, S., and Sandri, M. (2007) Cell Metabol. 6 458–471 [DOI] [PubMed] [Google Scholar]
- 74.Tracy, K., Dibling, B. C., Spike, B. T., Knabb, J. R., Schumacker, P., and Macleod, K. F. (2007) Mol. Cell. Biol. 27 6229–6242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, H., Bosch-Marce, M., Shimoda, L. A., Tan, Y. S., Baek, J. H., Wesley, J. B., Gonzalez, F. J., and Semenza, G. L. (2008) J. Biol. Chem. 283 10892–10903 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Burgering, B. M., and Medema, R. H. (2003) J. Leukoc. Biol. 73 689–701 [DOI] [PubMed] [Google Scholar]
- 77.Ruvolo, P. P. (2003) Pharmacol. Res. 47 383–392 [DOI] [PubMed] [Google Scholar]
- 78.Lavieu, G., Scarlatti, F., Sala, G., Levade, T., Ghidoni, R., Botti, J., and Codogno, P. (2007) Autophagy 3 45–47 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental Data]