Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2 (original) (raw)
Abstract
The steady-state levels of microRNAs (miRNAs) and their activities are regulated by the post-transcriptional processes. It is known that 3′ ends of several miRNAs undergo post-dicing adenylation or uridylation. We isolated the liver-specific miR-122 from human hepatocytes and mouse livers. Direct analysis by mass spectrometry revealed that one variant of miR-122 has a 3′-terminal adenosine that is introduced after processing by Dicer. We identified GLD-2, which is a regulatory cytoplasmic poly(A) polymerase, as responsible for the 3′-terminal adenylation of miR-122 after unwinding of the miR-122/miR-122* duplex. In livers from GLD-2-null mice, the steady-state level of the mature form of miR-122 was specifically lower than in heterozygous mice, whereas no reduction of pre-miR-122 was observed, demonstrating that 3′-terminal adenylation by GLD-2 is required for the selective stabilization of miR-122 in the liver.
Keywords: miRNA, miR-122, GLD-2, 3′ adenylation, mass spectrometry
MicroRNAs (miRNAs) are a class of small noncoding RNAs (ncRNAs) ∼22 nucleotides (nt) in length. miRNAs repress gene expression post-transcriptionally by partially pairing with the 3′-untranslated region of the target mRNAs. More than 600 species of mature miRNAs have been reported in human (Griffiths-Jones et al. 2008). Primary transcripts of miRNAs (pri-miRNAs), transcribed by RNA polymerase II, are cropped by the Drosha/DGCR8 complex into hairpin-structured pre-miRNAs in the nucleus. Pre-miRNAs are then exported to the cytoplasm, where they are processed into mature miRNAs by the Dicer/TRBP complex (Kim 2005). Post-transcriptional control of miRNA maturation occurs in a tissue-specific and developmentally regulated manner (Mineno et al. 2006; Obernosterer et al. 2006; Thomson et al. 2006; Wulczyn et al. 2007). Several pri-miRNAs accumulate to high levels in human and mouse embryonic stem (ES) cells, mouse embryocarcinoma (EC) cells, and human primary tumors, where their cropping is blocked (Suh et al. 2004; Thomson et al. 2006). Activation of miRNA processing takes place only as development proceeds. Recent studies revealed that the pluripotency factor Lin-28 selectively interacts with pre-let-7 and inhibits its processing by Dicer in ES and EC cells (Rybak et al. 2008). In another case, A-to-I editing by adenosine deaminases acting on RNA (ADARs) blocks the Drosha-mediated cropping of several pri-miRNAs (Yang et al. 2006). These lines of study have increased awareness of the biological significance of the post-transcriptional regulation of miRNA maturation.
Small ncRNAs are known to mature through a series of post-transcriptional processing events after transcription (Chen et al. 2000; Perumal and Reddy 2002; Yu et al. 2005; Ohara et al. 2007). In human, several small ncRNAs, such as 7SL(SRP), 7SK, U2, U6, and 5S RNAs, are post-transcriptionally uridylated or adenylated at their 3′ termini (Perumal and Reddy 2002). A family of noncanonical poly(A) polymerases might be involved in such 3′-terminal adenylation/uridylation of ncRNAs and some deadenylated mRNAs (Kwak and Wickens 2007; Martin and Keller 2007; Rissland et al. 2007). Uridylation and adenylation occur competitively, and a single adenylation prevents further oligo-uridylation (Chen et al. 2000). In addition, 3′-end processing is mediated by 3′–5′ exonuclease activity catalyzed by the exosome or deadenylases (Perumal and Reddy 2002; Goldstrohm and Wickens 2008). Therefore, 3′-end formation of ncRNAs occurs through a delicate balance between the removal and the addition of nucleotides. Recently, large-scale cDNA analyses have identified several miRNAs from mammals (Landgraf et al. 2007; Azuma-Mukai et al. 2008) and Caenorhabditis elegans (Ruby et al. 2006) that have 3′ adenylation or 3′ uridylation. In the plant Arabidopsis, it is known that 3′-terminal methylation protects miRNAs and siRNAs from a 3′ oligo-uridylation (Li et al. 2005). Recently, it has been reported that Lin-28 induces 3′ uridylation of pre-let-7. The 3′-uridylated pre-let-7 fails to be processed by Dicer and undergoes degradation (Heo et al. 2008). These observations imply that 3′ nucleotide addition is a widespread event involved in the process of miRNA maturation.
Results and Discussion
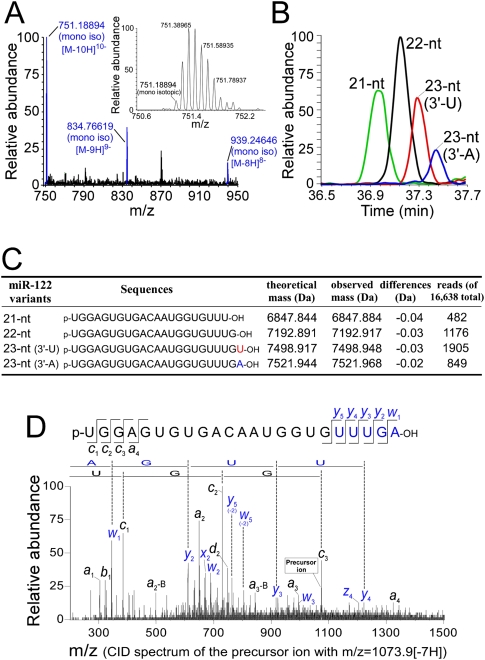
miR-122 is a liver-specific miRNA that plays an important role in the regulation of hepatic function (Chang et al. 2004; Krützfeldt et al. 2005) and is highly expressed in hepatocytes and some hepatoma cells. miR-122 regulates cholesterol and fatty-acid metabolism, and depletion of miR-122 by an antisense oligo lowers the cholesterol level in plasma (Krützfeldt et al. 2005). It was reported that CAT-1 mRNA whose translation was inhibited by miR-122 in the P-body can be released under stress conditions (Bhattacharyya et al. 2006). miR-122 is also required for the replication of hepatitis C virus (Jopling et al. 2005). To characterize the variation in the 3′ termini of miRNAs, we isolated the relatively abundant miR-122 from mouse liver by using the reciprocal circulating chromatography (RCC) method (Miyauchi et al. 2007) and directly analyzed individual molecules using a highly sensitive method of RNA mass spectrometry (capillary liquid chromatography [LC]/nano electrospray ionization mass spectrometry [nanoESI-MS]) (Ohara et al. 2007; Suzuki et al. 2007). We clearly detected multiply charged negative ions derived from deprotonated miR-122 in the mass spectra (Fig. 1A). By measuring the exact mass with Fourier transform mass spectrometry, we identified four species of miR-122 ranging from 21 to 23 nt in length (Fig. 1B,C). Each species was further analyzed in an MS/MS experiment using collision-induced dissociation (CID) to determine its 5′- and 3′-terminal sequences (Fig. 1D; Supplemental Fig. 1). Taking the exact mass and the CID spectra data together, we concluded that all species possessed variations at their 3′ ends (Fig. 1C). Three of the miR-122 variants (21-nt, 22-nt, and 23-nt with a 3′-terminal uridine [3′-U]) exactly coincide with the gene sequence of the pre-miR-122, whereas the 23-nt variant with a 3′-terminal adenosine (3′-A) that is not encoded in the gene appeared to be generated by the post-dicing addition of a single adenosine at the 3′ terminus of the 22-nt variant. The presence of four species of miR-122 was confirmed by the pyrosequencing of the small RNA fraction obtained from mouse liver (Fig. 1C; Supplemental Table 1).
Figure 1.
Mass spectrometric characterization of miR-122 isolated from mouse liver. (A) Mass spectrum of the 23-nt (3′-A) variant of miR-122. The multiply charged negative ions, [M-8H]8−, [M-9H]9−, and [M-10H]10−, of deprotonated miR-122 can be seen. (Inset) A series of isotopic ions of [M-10H]10− are shown in the magnified graph; the monoisotopic ion (m/z 751.19016) is indicated. The mass spectrum was acquired in the range of m/z 750–1500 with mass resolution of 30,000 (FWHM). (B) Mass chromatograms shown by [M-8H]8− and [M-9H]9− of each miR-122 variant, 21-nt (green), 22-nt (black), 23-nt (3′-U), (red) and23-nt (3′-A) (blue). (C) Sequences and exact molecular masses of the miR-122 variants. The read number from the pyrosequencing for each variant is indicated. The total number of reads was 16,638. (D) CID spectrum of the 23-nt (3′-A) variant of miR-122. [M-7H]7− (m/z 1073.9) was selected as a precursor mass for CID. The product ions were assigned according to Mcluckey et al. (1992). Partial sequences from both termini determined by this spectrum are indicated.
We then used a recombinant human Dicer to analyze the dicing patterns of human and mouse pre-miR-122. Human and mouse pre-miR-122s were synthesized by in vitro transcription, then subjected to in vitro processing by using human Dicer. The diced products were analyzed by MALDI-TOF mass spectrometry to definitely identify the cleavage positions of pre-miR-122. Human Dicer only processed a single site on each strand in human and mouse pre-miR-122 to produce a single 22-nt species of mature miR-122 (Supplemental Fig. 2). Thus, the 21-nt variant and the two 23-nt variants (3′-U and 3′-A) of mature miR-122 are likely to be generated by 3′-end processing after dicing.
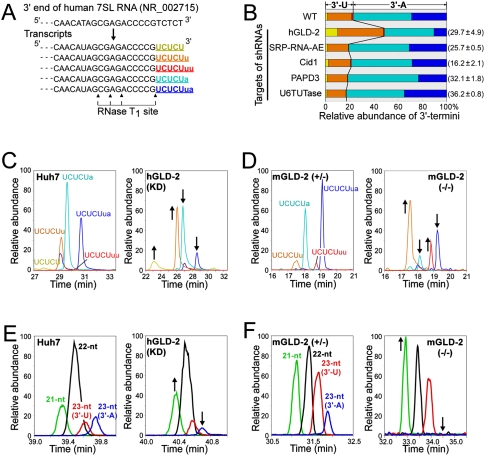
To identify the nucleotidyl transferase that catalyzes uridylation or adenylation at the 3′ end of miR-122, we designed shRNAs (Katoh and Suzuki 2007) that target five candidate RNA-specific nucleotidyl transferases: hGLD-2, SRP-RNA-AE, Cid1, PAPD3, and U6 TUTase. Each shRNA was then introduced into human hepatoma Huh7 cells to knock down each of the candidates. The efficacy of RNAi was estimated by real-time RT–PCR, showing that the steady-state levels of the target mRNAs were decreased to 16%–36% of the levels of untreated cells (Fig. 2B). To readily analyze the 3′ terminus, we used 7SL RNA, which is a more abundant cytoplasmic ncRNA than miR-122. 7SL RNA was isolated from the total RNA of the shRNA-treated cells and then digested by RNase T1. The 7SL RNA digests were analyzed by LC/MS to characterize the 3′-terminal fragments. As shown in Figure 2, A–C, we detected five different 3′-terminal fragments bearing 3′-terminal U or A; three of them had 3′-U residues and the other two had 3′-A. Each fragment was sequenced by CID analyses (Supplemental Fig. 3). In Huh7 cells not treated with shRNAs, 7SL RNAs bearing 3′-A made up 77.2% of the total, and the remaining 22.8% had 3′-U (Fig. 2B). When hGLD-2 was knocked down, 7SL RNAs bearing 3′-A were decreased to 51.5% of the total (Fig. 2B). In fact, the reduction of the 3′-A fragments UCUCUa and UCUCUua correlated well with the increases of UCUCU and UCUCUu, respectively (Fig. 2B,C). Silencing the other candidate transferase genes had no effect on the composition of the 3′ termini of 7SL RNA, although it was reported previously that SRP-RNA-AE is responsible for 3′ adenylation of 7SL RNA in vitro (Perumal et al. 2001). To confirm the effects of reduced GLD-2 expression in mouse, we analyzed 7SL RNA isolated from the livers of mGLD-2 knockout mice (Nakanishi et al. 2007). When compared with the result from heterozygous mGLD-2+/− mice, the proportions of the two 3′-A fragments (UCUCUa and UCUCUua) were substantially lower, and the proportion of UCUCUu was higher in the homozygous mGLD-2−/− (Fig. 2D). Some 3′-A fragments still remained in the mGLD-2−/− mouse (Fig. 2D), suggesting that multiple adenylases, including GLD-2, are redundantly involved in 3′ adenylation of the 7SL RNA in the cytoplasm and/or in the nucleus. Nonetheless, these results clearly demonstrate that GLD-2 is responsible for adenylating the 3′ termini of cytoplasmic ncRNAs.
Figure 2.
GLD-2 is a 3′ adenylase for cytoplasmic ncRNAs. (A) 3′-End sequence of human 7SL RNA gene (NR_002715) and its transcribed sequences with 3′-terminal variations. Cleavage sites of RNase T1 are shown by arrowheads. The five different 3′-terminal fragments are colored yellow (UCUCU), orange (UCUCUu), red (UCUCUuu), light blue (UCUCUa), and blue (UCUCUua). Lowercase letters (u or a) stand for additional nucleosides (uridine or adenosine) attached at the 3′ ends after transcription. 3′-Terminal fragments bearing 2′,3′-cyclic phosphates are not shown. (B) Composition and ratio of 3′ termini of 7SL RNAs isolated from Huh7 cells when each indicated nucleotidyl-transferase was knocked down. The steady-state level of each mRNA in the shRNA treated cells is shown in parentheses. The relative abundance of 3′-terminal fragments was calculated from the intensity of the mass chromatogram for each fragment. The color code of each fragment is the same as in A. (C) Mass chromatograms of doubly charged ions of five different 3′-terminal fragments of 7SL RNAs from Huh7 cells transfected (right panel) or not transfected (left panel) with an shRNA targeting to hGLD-2: UCUCU (m/z 732.00–732.75, yellow), UCUCUu (m/z 885.00–885.75, orange), UCUCUuu (m/z 1038.00–1038.75, red), UCUCUa (m/z 896.50–897.25, light blue), and UCUCUua (m/z 1049.50–1050.25, blue). (D) Mass chromatograms of doubly charged ions of four different 3′-terminal fragments of 7SL RNAs from livers of mGLD-2+/− mice (left panel) or mGLD-2−/− mice (right panel). The color code and m/z value of each fragment are the same as in A–C. (E) Mass chromatograms of the [M-8H]8−, [M-9H]9−, and [M-10H]10− ions of each miR-122 variant [21-nt (MW 6847.84, green), 22-nt (MW 7192.88, black), 23-nt (3′-U) (MW 7498.91, red), and 23-nt (3′-A) (MW 7521.94, blue)] from Huh7 cells transfected (right panel) or not transfected (left panel) with an shRNA targeting hGLD-2. (F) Mass chromatograms of the [M-8H]8−, [M-9H]9−, and [M-10H]10− ions of each miR-122 variant from livers ofmGLD-2+/− mice (left panel) or mGLD-2−/− mice (right panel). The color code and MW value of each variant are the same as in E.
We next analyzed miR-122 isolated from Huh7 cells transfected with the shRNA targeting hGLD-2 and observed the relative abundance of each species of miR-122. The 23-nt variant with 3′-A was present in lower amounts in the treated cells than in the untreated cells (Fig. 2E), whereas the amount of the 21-nt variant was slightly increased in the treated cells (Fig. 2E). The relative amounts of the other two variants (22-nt and 23-nt [3′-U]) remained unchanged. In addition, we analyzed miR-122 isolated from the livers of mGLD-2−/− mice (Fig. 2F). When compared with the profile of miR-122 from the mGLD-2+/− mouse, the 23-nt variant with 3′-A almost disappeared. In contrast, the proportion of the 21-nt variant increased. This observation is consistent with the profile of human miR-122 from the hGLD-2-impaired Huh7 cells (Fig. 2E). Thus, GLD-2 is an enzyme for 3′-terminal adenylation of miR-122, and the adenylation by GLD-2 competes with the shortening of the 3′ ends to control miRNA length.
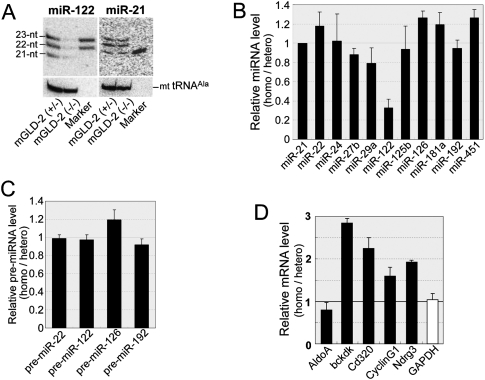
We then used Northern blotting to analyze the steady-state levels of miRNAs. In the livers of mGLD-2−/− mice, the steady-state level of miR-122 decreased significantly when compared with the mGLD-2+/− liver (Fig. 3A), whereas the miR-21 level was unchanged (Fig. 3A). Real-time RT–PCR analysis also demonstrated a specific reduction of miR-122 (∼30%) (Fig. 3B), whereas the steady-state levels of other miRNAs (miR-22, miR-24, miR-27b, miR-29a, miR-125b, miR-126, miR-181a, miR-192, and miR-451) were unchanged (Fig. 3B). In addition, we observed a reduced level of miR-122 in the hGLD-2-impaired Huh7 cells (Supplemental Fig. 4). We also found that the steady-state level of pre-miR-122 was unchanged between mGLD-2−/− and mGLD-2+/− mice (Fig. 3C), suggesting that the transcription of miR-122 was unaffected and that 3′ adenylation by GLD-2 acts specifically to stabilize miR-122 after dicing. We therefore hypothesized that loss of 3′ adenylation by GLD-2 would accelerate the specific degradation of miR-122 by 3′–5′ exonucleases. Indeed, the increased proportion of the 21-nt variant in mGLD-2−/− liver implies that the 3′-exonucleolytic activity had degraded miR-122. We also examined the steady-state levels of five mRNAs that are predicted to be targets of miR-122 (Krützfeldt et al. 2005; Esau et al. 2006; Elmén et al. 2008). Four out of the five genes (bckdk, cd320, CyclinG1, and Ndrg3) were up-regulated in the mGLD-2−/− liver (Fig. 3D). Thus, GLD-2 indirectly controls the levels of various target mRNAs by regulating the cellular stability of miR-122.
Figure 3.
Steady-state level of miR-122 is markedly decreased in mGLD-2 knockout mice. (A) Northern blotting detecting miR-122 (left panel) and miR-21 (right panel) from mGLD-2+/− and mGLD-2−/− mice. As positive controls, 40 fmol each of synthetic miR-122 (22-nt and 23-nt) and miR-21 (21-nt) was run along with the total RNAs. Northern blotting of mitochondrial tRNAAla was used as a loading control for each miRNA. (B) Ratio of the steady-state level of each miRNA from mGLD-2−/− to that from mGLD-2+/−, as measured by real-time RT–PCR. Each ratio was normalized by the ratio of miR-21 (whose value was defined as 1). (C) Ratio of the steady-state level of each pre-miRNA from mGLD-2−/− to that from mGLD-2+/− as measured by real-time RT–PCR. Each ratio was normalized by the ratio of pre-miR-21 (whose value was defined as 1). (D) Expression levels of miR-122-target genes (AldoA, bckdk, cd320, Cyclin G1, and Ndrg3) and GAPDH mRNA in mGLD-2−/− relative to those in mGLD-2+/− were measured by real-time RT–PCR. The relative expression level of each mRNA was normalized by the relative ratio of ACTB mRNA (whose value was defined as 1). The data in B–D are shown as values ± SD and reflect the averages of six independent experiments.
Cytoplasmic polyadenylation regulates the stability and translational activity of mRNAs and is required for early development and synaptic plasticity (Mendez and Richter 2001; Barnard et al. 2004; Rouhana et al. 2005; Richter 2007). GLD-2 was identified as a regulatory cytoplasmic poly(A) polymerase that controls germline progression through meiosis in C. elegans and Schizosaccharomyces pombe (Read et al. 2002; Saitoh et al. 2002; Wang et al. 2002). Recently, it was reported that in Drosophila a GLD-2 homolog is required for long-term memory (Kwak et al. 2008) and oogenesis (Cui et al. 2008). Mammalian GLD-2 is expressed in many tissues and localizes in both the nucleus and cytoplasm (Nakanishi et al. 2006). In the brains of mice and humans, GLD-2 mRNAs are abundant in the regions associated with long-term cognitive and emotional learning, indicating that mammalian GLD-2 is important in synaptic translation (Rouhana et al. 2005). In addition, mouse GLD-2 has been suggested to control the translation of specific mRNAs during oogenesis (Nakanishi et al. 2006). However, disruption of mGLD-2 had no effect on full-term development and oogenesis, implying that another cytoplasmic poly(A) polymerase is redundantly required for oogenesis in mouse (Nakanishi et al. 2007). Thus, the functional roles played by mammalian GLD-2 were unclear. In this study, we show that human and mouse GLD-2 are responsible for 3′ adenylation of miR-122 and 7SL RNA. Disruption of GLD-2 resulted in a specific reduction of the steady-state level of miR-122, suggesting that 3′ adenylation by GLD-2 is required for the cellular stability of miR-122. This is the first report of a mechanism that selectively stabilizes a specific miRNA in mammalian cells.
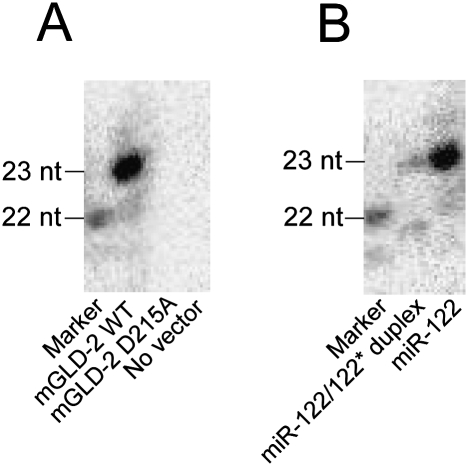
To investigate at which step in the process of miRNA maturation from dicing to RISC loading 3′ adenylation by GLD-2 takes place, we carried out an in vitro 3′ adenylation of miR-122 by Flag-tagged mGLD-2 immunoprecipitated from Huh7 cells. As shown in Figure 4A, a single adenosine was specifically added to the 22-nt variant of miR-122 by the wild-type mGLD-2, not by its inactive mutant (D215A). When we used the miR-122/miR-122* duplex as a substrate, lower activity of 3′ adenylation was observed as compared with the mature form of miR-122 as a substrate (Fig. 4B). The result demonstrated that mGLD-2 preferentially adenylates the single-stranded form of miR-122, indicating that 3′ adenylation occurs after unwinding of the miRNA duplex.
Figure 4.
In vitro 3′ adenylation of miR-122 by immunoprecipitated mGLD-2. (A) The synthetic miR-122 was 3′-adenylated in vitro by the immunoprecipitated mGLD-2 in the presence of [α-32P]ATP. Wild-type mGLD-2 (mGLD-2 WT) or its inactive mutant (mGLD-2 D215A) was immunoprecipitated with anti-Flag M2-agarose beads from the lysate of Huh7 cells transfected with pcDNA3 Flag-HA-mGLD-2 or its inactivated mutant form (pcDNA3 Flag-HA-mGLD-2 D215A). No vector represents a negative control preparation from the same cell lysate untransfected. [α-32P]AMP-labeled miR-122 was analyzed by 20% denaturing PAGE and visualized by an imaging analyzer (BAS5000; FujiFilm). The marker is the 5′-32P-labeled miR-122 (22-nt). (B) The synthetic miR-122 (single strand) and miR-122/mi-R122* duplex were 3′-adenylated in vitro by the immunoprecipitated mGLD-2 in the presence of [α-32P]ATP. The products were analyzed as described in A.
We cannot assume how GLD-2 specifically discriminates miR-122 and 7SL RNA from other nontargeted RNAs by recognizing a common sequence or structure of their 3′ termini, because no consensus sequence was found at the 3′ termini of these RNAs. As GLD-2 is recruited by the phosphorylated CPEB/CPSF complex to the 3′-untranslated region (UTR) of the target mRNAs for cytoplasmic poly(A) addition (Rouhana et al. 2005; Kashiwabara et al. 2008), some as-yet-unidentified protein factors might be involved in recruiting GLD-2 to the 3′ terminus of miR-122. Considering the low specificity of GLD-2, there might be other miRNAs stabilized by 3′ adenylation. GLD-2 probably affects the expression of much broader subsets of mRNAs through the cytoplasmic poly(A) tailing as well as the regulation of miRNA stability.
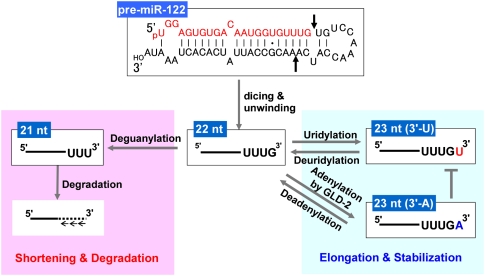
A schematic mechanism of the selective stabilization of miR-122 identified in this study is depicted in Figure 5. Pre-miR-122 is processed by Dicer into a 22-nt mature miR-122. Then the 3′ terminus of the 22-nt variant is elongated with uridylation by a putative poly(U) polymerase or with adenylation by GLD-2. Concomitantly, the elongated 3′ termini of miR-122 are shortened into 21-nt or shorter variants by a 3′–5′ exonuclease or deadenylase. Therefore, a delicate balance of 3′ nucleotide addition and removal might determine the average size of miRNA variants. Because miRNAs of 21 to 23 nt in length are preferentially loaded into Argonaute 2 in the RISC complex, miRNAs shorter than 21 nt or longer than 23 nt might be excluded from the RISC complex and degraded. Thus, only four variants of miR-122, all of 21 to 23 nt, accumulate in the cell. This concept nicely explains why GLD-2 disruption destabilized not only the 23-nt variant with 3′-A, but also the other three variants of miR-122. In general, 3′-oligouridylation stimulates degradation of target RNAs in the cell (Perumal and Reddy 2002; Shen and Goodman 2004). If miR-122 tends to be 3′-oligouridylated, it might therefore be degraded by a putative 3′–5′ exonuclease. In fact, we did observe low copy numbers (33 reads) of the 24-nt variant with 3′-UU using pyrosequencing (Supplemental Table 1). When the 3′ adenylation of miR-122 is impaired by the depletion of GLD-2, 3′ uridylation might be enhanced, and 3′–5′ exonucleolytic degradation relatively increased to decay miR-122 rapidly. To explore the mechanisms of this selective miRNA stabilization in detail, further studies will be necessary to identify the poly(U) polymerase for 3′ uridylation and the 3′–5′ exonuclease that destroys miR-122. A recent study identified a family of the small RNA-degrading nuclease (SDN) that degrades mature miRNAs in Arabidopsis (Ramachandran and Chen 2008). A mammalian homolog of SDN might be a candidate gene involved in destabilization of miR-122.
Figure 5.
Proposed model for the selective stabilization of miR-122 by 3′ nucleotide addition and exonucleolytic degradation. In the cytoplasm, pre-miR-122 is processed by Dicer into mature miR-122 of a uniform 22-nt length (red letters). After unwinding of the miR-122/miR-122* duplex, the 3′ terminus of miR-122 is elongated by the addition of uridine/adenosine by a putative 3′ uridylase or GLD-2 and shortened to 21 nt or less by a putative 3′–5′ exonuclease. Competition between the addition and removal of 3′ nucleotides might determine the steady-state level of miR-122. The average size of miR-122 should be determined by the fact that 21 to 23 nt of miRNAs are preferentially loaded into Argonaute 2 in the RISC machinery.
Materials and methods
RNAi
Five shRNAs that target five candidate RNA-specific nucleotidyl transferases (hGLD-2, SRP-RNA-AE, Cid1, PAPD3, and U6 TUTase) were designed as described in the Supplemental Material. Huh7 cells were transfected with 10 nM shRNAs using Lipofectamine RNAiMAX (Invitrogen). Seventy-two hours after the transfection, cells were treated with ISOGEN (Wako) to isolate total RNAs.
mGLD-2 knockout mouse
Homozygous (−/−) and heterozygous (+/−) mice lacking mGLD-2 were prepared as described previously (Nakanishi et al. 2007). Total RNAs were isolated from homogenized mouse livers from both mGLD-2+/− and mGLD-2−/− mice using Trizol reagent (Invitrogen).
Purification of human and mouse miR-122
Human and mouse miR-122s were isolated by RCC (Miyauchi et al. 2007). The sequences of DNA probes and detailed description of the method are shown in the Supplemental Material.
Capillary LC nano ESI/mass spectrometry
To analyze a limited quantity of digested RNA fragments or intact miRNAs, we used a system of capillary LC coupled with nanoESI-MS (Suzuki et al. 2007). A linear ion trap–orbitrap hybrid mass spectrometer (LTQ Orbitrap XL, Thermo Fisher Scientific) or a tandem-quadrupole time-of-flight mass spectrometer (QSTAR XL, Applied Biosystems) was equipped with a custom-made nanospray ion source, a Nanovolume Valve (Valco Instruments), and a splitless nano HPLC system (DiNa, KYA Technologies). The procedures and conditions for the analyses are described in the Supplemental Material.
3′ Adenylation of miR-122 in vitro by the immunoprecipitated mGLD-2
The detailed conditions are described in the Supplemental Material. Briefly, Huh7 cells (∼2 × 107) cultured in 15-cm dishes were transfected with 50 μg of pcDNA3 Flag-HA-mGLD-2 (wild type or D215A) using Lipofectamine 2000 (Invitrogen). Forty-eight hours after the transfection, the cells were harvested. The cell lysates were mixed with 30 μL of anti-Flag M2-agarose beads (Sigma; 50% slurry) and rotated for 1 h at 4°C. The collected beads were then washed and stocked. For in vitro adenylation assay, the synthetic miR-122 or miR-122/miR-122* duplex (4 pmol each) was incubated for 30 min at 37°C in a 30-μL reaction mixture containing 20 mM Tris-HCl (pH 7.5), 50 mM KCl, 25 mM MnCl2, 50 μg/mL BSA, 0.53 U/μL RNasin ribonuclease inhibitor (Promega), 2.47 kBq/μL [α-32P]ATP, and the immunoprecipitated mGLD-2 (wild type or D215A) on the anti-Flag M2-agarose beads (5 μL of the beads; 50% slurry).
Acknowledgments
We are grateful to the Suzuki laboratory members for technical assistance and fruitful discussions, especially to Hiroki Ueda for his computational support on this study. This work was supported by a grant from the New Energy and Industrial Technology Development Organization (NEDO) to T.S.
Footnotes
References
- Azuma-Mukai A., Oguri H., Mituyama T., Qian Z.R., Asai K., Siomi H., Siomi M.C. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc. Natl. Acad. Sci. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard D.C., Ryan K., Manley J.L., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S.N., Habermacher R., Martine U., Closs E.I., Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Chang J., Nicolas E., Marks D., Sander C., Lerro A., Buendia M.A., Xu C., Mason W.S., Moloshok T., Bort R., et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- Chen Y., Sinha K., Perumal K., Reddy R. Effect of 3′ terminal adenylic acid residue on the uridylation of human small RNAs in vitro and in frog oocytes. RNA. 2000;6:1277–1288. doi: 10.1017/s1355838200000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J., Sackton K.L., Horner V.L., Kumar K.E., Wolfner M.F. Wispy, the Drosophila homolog of GLD-2, is required during oogenesis and egg activation. Genetics. 2008;178:2017–2029. doi: 10.1534/genetics.107.084558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmén J., Lindow M., Silahtaroglu A., Bak M., Christensen M., Lind-Thomsen A., Hedtjärn M., Hansen J.B., Hansen H.F., Straarup E.M., et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esau C., Davis S., Murray S.F., Yu X.X., Pandey S.K., Pear M., Watts L., Booten S.L., Graham M., McKay R., et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I., Joo C., Cho J., Ha M., Han J., Kim V. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- Kashiwabara S., Nakanishi T., Kimura M., Baba T. Non-canonical poly(A) polymerase in mammalian gametogenesis. Biochim. Biophys. Acta. 2008;1779:230–238. doi: 10.1016/j.bbagrm.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Katoh T., Suzuki T. Specific residues at every third position of siRNA shape its efficient RNAi activity. Nucleic Acids Res. 2007;35:e27. doi: 10.1093/nar/gkl1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs.’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- Kwak J.E., Wickens M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak J.E., Drier E., Barbee S.A., Ramaswami M., Yin J.C., Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proc. Natl. Acad. Sci. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G., Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcluckey S.A., Vanberkel G.J., Glish G.L. Tandem mass-spectrometry of small, multiply charged oligonucleotides. J. Am. Soc. Mass Spectrom. 1992;3:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- Mendez R., Richter J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Mineno J., Okamoto S., Ando T., Sato M., Chono H., Izu H., Takayama M., Asada K., Mirochnitchenko O., Inouye M., et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyauchi K., Ohara T., Suzuki T. Automated parallel isolation of multiple species of non-coding RNAs by the reciprocal circulating chromatography method. Nucleic Acids Res. 2007;35:e24. doi: 10.1093/nar/gkl1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T., Kubota H., Ishibashi N., Kumagai S., Watanabe H., Yamashita M., Kashiwabara S., Miyado K., Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Nakanishi T., Kumagai S., Kimura M., Watanabe H., Sakurai T., Kimura M., Kashiwabara S., Baba T. Disruption of mouse poly(A) polymerase mGLD-2 does not alter polyadenylation status in oocytes and somatic cells. Biochem. Biophys. Res. Commun. 2007;364:14–19. doi: 10.1016/j.bbrc.2007.09.096. [DOI] [PubMed] [Google Scholar]
- Obernosterer G., Leuschner P.J., Alenius M., Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T., Sakaguchi Y., Suzuki T., Ueda H., Miyauchi K., Suzuki T. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat. Struct. Mol. Biol. 2007;14:349–350. doi: 10.1038/nsmb1220. [DOI] [PubMed] [Google Scholar]
- Perumal K., Reddy R. The 3′ end formation in small RNAs. Gene Expr. 2002;10:59–78. [PMC free article] [PubMed] [Google Scholar]
- Perumal K., Sinha K., Henning D., Reddy R. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J. Biol. Chem. 2001;276:21791–21796. doi: 10.1074/jbc.M101905200. [DOI] [PubMed] [Google Scholar]
- Ramachandran V., Chen X. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science. 2008;321:1490–1492. doi: 10.1126/science.1163728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R.L., Martinho R.G., Wang S.W., Carr A.M., Norbury C.J. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter J.D. CPEB: A life in translation. Trends Biochem. Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Rissland O.S., Mikulasova A., Norbury C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana L., Wang L., Buter N., Kwak J.E., Schiltz C.A., Gonzalez T., Kelley A.E., Landry C.F., Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Saitoh S., Chabes A., McDonald W.H., Thelander L., Yates J.R., Russell P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- Shen B., Goodman H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Suh M.R., Lee Y., Kim J.Y., Kim S.K., Moon S.H., Lee J.Y., Cha K.Y., Chung H.M., Yoon H.S., Moon S.Y., et al. Human embryonic stem cells express a unique set of microRNAs. Dev. Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Ikeuchi Y., Noma A., Suzuki T., Sakaguchi Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 2007;425:211–229. doi: 10.1016/S0076-6879(07)25009-8. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Wulczyn F.G., Smirnova L., Rybak A., Brandt C., Kwidzinski E., Ninnemann O., Strehle M., Seiler A., Schumacher S., Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yang W., Chendrimada T.P., Wang Q., Higuchi M., Seeburg P.H., Shiekhattar R., Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]