Post-transcriptional control of DGCR8 expression by the Microprocessor (original) (raw)
Abstract
The Microprocessor, comprising the RNase III Drosha and the double-stranded RNA binding protein DGCR8, is essential for microRNA (miRNA) biogenesis. In the miRNA processing pathway certain hairpin structures within primary miRNA (pri-miRNA) transcripts are specifically cleaved by the Microprocessor to release ∼60–70-nucleotide precursor miRNA (pre-miRNA) intermediates. Although both Drosha and DGCR8 are required for Microprocessor activity, the mechanisms regulating the expression of these proteins are unknown. Here we report that the Microprocessor negatively regulates DGCR8 expression. Using in vitro reconstitution and in vivo studies, we demonstrate that a hairpin, localized in the 5′ untranslated region (5′UTR) of DGCR8 mRNA, is cleaved by the Microprocessor. Accordingly, knockdown of Drosha leads to an increase in DGCR8 mRNA and protein levels in cells. Furthermore, we found that the DGCR8 5′UTR confers Microprocessor-dependent repression of a luciferase reporter gene in vivo. Our results uncover a novel feedback loop that regulates DGCR8 levels.
Keywords: microRNA (miRNA), DGCR8, Drosha, Microprocessor, feedback loop
INTRODUCTION
MicroRNAs (miRNAs) are ∼22-nucleotide (nt) small non-coding RNAs that post-transcriptionally regulate gene expression by targeting mRNA for destabilization and translation repression (Filipowicz et al. 2008). miRNAs have been implicated in critical developmental roles, and dysregulation of miRNA expression has been observed in cancer (Calin and Croce 2006; Esquela-Kerscher and Slack 2006; Zhao and Srivastava 2007). The biogenesis of canonical miRNAs relies on two processing events occurring successively in the nucleus and in the cytoplasm of eukaryotic cells (Kim 2005). The first processing step involves a protein complex called the Microprocessor, comprising the type III RNase Drosha and the double-stranded RNA binding protein DGCR8 (Denli et al. 2004; Gregory et al. 2004). This complex recognizes and cleaves primary miRNAs (pri-miRNAs) to generate stem–loop-containing precursor miRNAs (pre-miRNAs) of ∼60–70 nt (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004). The second processing step occurs after export of the hairpin-shaped pre-miRNA by Exportin 5 into the cytoplasm and relies on the processing activity of another type III RNase called Dicer that produces a miRNA duplex of ∼22 nt (Bernstein et al. 2001; Hutvágner et al. 2001). One strand of the duplex, known as the guide miRNA, is recruited by one of the Argonaute (Ago) family members Ago1–4 into the RNA-induced silencing complex (RISC) (Hammond et al. 2001; Liu et al. 2004; Meister et al. 2004; Chendrimada et al. 2005; Gregory et al. 2005). This complex recognizes target messenger RNAs (mRNAs) based on sequence complementarity between the guide miRNA and its targets, resulting in either Ago2-mediated endonucleolytic mRNA cleavage, or translational repression and mRNA destabilization (Filipowicz et al. 2008).
Biochemical studies have provided much insight into the mechanism by which the Microprocessor recognizes and cleaves its pri-miRNA substrates (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004; Yeom et al. 2006; Sohn et al. 2007). Since both subunits are essential for this activity, together with the demonstration that the correct Microprocessor subunit stoichiometry is required for efficient processing, it is likely that there are particular, as yet undefined, regulatory mechanisms to maintain the subunit composition of the complex (Gregory and Shiekhattar 2005). Indeed dysregulation of these pathways may contribute to perturbations of miRNA biogenesis and activity that have been reported in diseases including cancer (Calin and Croce 2006; Esquela-Kerscher and Slack 2006). For example, most miRNA were shown to be down-regulated in primary tumors, and this widespread down-regulation could be a consequence of a block during Microprocessor-mediated processing step (Lu et al. 2005; Thomson et al. 2006). The mechanism involved in this block is not known, and neither is the regulation of the Microprocessor components.
A recent bioinformatics analysis identified two predicted “pre-miRNA-like” hairpin structures contained within the DGCR8 mRNA (Pedersen et al. 2006). This observation prompted us to investigate whether these hairpins could be involved in DGCR8 regulation. We found that DGCR8 is negatively regulated by the Microprocessor through the cleavage of the hairpin that is localized in the DGCR8 5′ untranslated region (5′UTR). This regulatory mechanism likely contributes to maintaining the integrity of the Microprocessor and has implications for miRNA biogenesis and post-transcriptional gene regulation.
RESULTS AND DISCUSSION
Two evolutionarily conserved pre-miRNA-like hairpin structures located in exon 2 of DGCR8 mRNA have been previously computationally predicted (Pedersen et al. 2006; Supplemental Fig. S1A). The first hairpin, which is 88 nt long, is localized in the DGCR8 5′ untranslated region (5′UTR) and will be subsequently referred to as “DGCR8 5′UTR hairpin” (Supplemental Fig. S1B). The second hairpin, which is 85 nt long, is localized 94 nt downstream from the AUG start codon of DGCR8 mRNA (Supplemental Fig. S1C). It will be referred to as the “miR-1306 hairpin,” since a mature miRNA, miR-1306, contained within this hairpin sequence has been recently cloned from human embryonic stem (ES) cells (Griffiths-Jones et al. 2008; Morin et al. 2008).
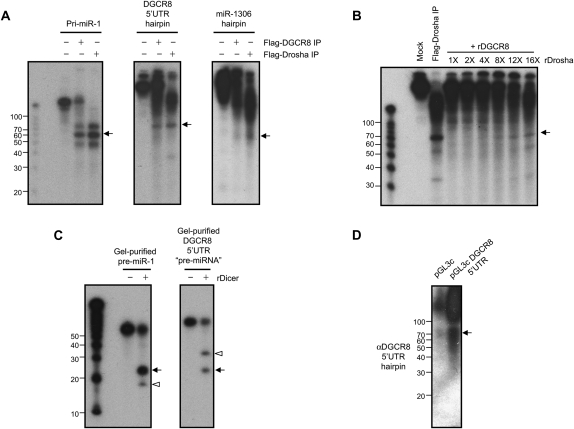
To gain insight into the function of these hairpins, we first asked whether they could be substrates for the Microprocessor in vitro. In vitro transcribed and internally radio-labeled DGCR8 5′UTR RNA or RNA encompassing miR-1306 hairpin was incubated with immunoprecipitated protein complexes associated with Flag-DGCR8 or Flag-Drosha and processing products were analyzed on gel (Fig. 1A). Processing of pri-miR-1 by the Microprocessor, which was used as a positive control, showed accumulation of a product of ∼60 nt that corresponds to pre-miR-1 (Fig. 1A). Interestingly, when the DGCR8 5′UTR hairpin was used as a substrate for in vitro processing a product of about 70 nt was generated. Although we detected robust Microprocessor-mediated cleavage of the DGCR8 5′UTR the processing efficiency was reproducibly lower than that of the pri-miR-1 in multiple independent processing reactions. However, when processing reactions were performed with RNA encompassing miR-1306 hairpin, only a weak band was detected corresponding to a cleavage product of 60 nt (Fig. 1A). Since miR-1306 hairpin appeared to be a poor substrate of the Microprocessor (Fig. 1A; see below), we decided to focus on the DGCR8 5′UTR hairpin. Although it has been demonstrated that the Drosha-DGCR8 (Microprocessor) complex is necessary and sufficient for pri-miRNA processing (Gregory et al. 2004), additional Drosha-associated factors are required for the processing of certain pri-miRNAs (Guil and Cáceres 2007; Michlewski et al. 2008). Therefore, we next asked whether in vitro reconstituted Microprocessor is sufficient to process the DGCR8 5′UTR hairpin. Recombinant Drosha and DGCR8 proteins were produced and incubated together with in vitro transcribed and radio-labeled DGCR8 5′UTR RNA. In processing reactions performed with recombinant Microprocessor, we again observed the accumulation of ∼70 nt processing product that is generated by Flag-Drosha containing complexes isolated from cells (Fig. 1B). Therefore, our results demonstrate that the DGCR8 5′UTR hairpin is a bona fide substrate for the Microprocessor.
FIGURE 1.
Hairpins in the DGCR8 mRNA are cleaved by the Microprocessor. (A) In vitro processing assays performed with pri-miR-1, DGCR8 5′UTR RNA, or RNA encompassing the miR-1306 hairpin. The indicated in vitro transcribed, internally labeled RNA was incubated with or without Flag-immunopurified DGCR8 (Flag-DGCR8 IP) or Drosha (Flag-Drosha IP). Arrows indicate major processing products. (B) Reconstitution of DGCR8 5′UTR RNA processing activity with the recombinant Microprocessor. RNA was incubated with recombinant DGCR8 (rDGCR8) together with an increasing amount of recombinant Drosha (rDrosha). Flag-Drosha IP served as a positive control. Arrow indicates DGCR8 5′UTR “pre-miRNA-like” processing product. (C) In vitro processing of pre-miR-1 and DGCR8 5′UTR pre-miRNA-like hairpin by Dicer. Gel purified pre-miR-1 and DGCR8 5′UTR hairpin from the in vitro processing assays shown in B were incubated with recombinant Dicer (rDicer). Arrows indicate ∼22 nt duplex and arrowheads indicate the terminal loops processed from the hairpin RNA substrates. RNA was resolved on 15% polyacrylamide denaturing gels and visualized by autoradiography (A–C). (D) Detection of the hairpin processed from the DGCR8 5′UTR in vivo. Hela cells were transfected with either the luciferase reporter plasmid containing the DGCR8 5′UTR depicted in Figure 2C (pGL3c DGCR8 5′UTR) or luciferase reporter plasmid without DGCR8 5′UTR (pGL3c). Total RNA was extracted 48 h post-transfection and analyzed by Northern blot using a probe to specifically detect the DGCR8 5′UTR hairpin.
We went on to ask whether the DGCR8 5′UTR hairpin could be processed into mature ∼22 nt miRNA by Dicer in vitro. We incubated the gel-purified ∼70 nt DGCR8 5′UTR hairpin product recovered from the Microprocessor reactions together with recombinant Dicer, and similarly we used gel-purified pre-miR-1 as a positive control. Results showed that both the pre-miR-1 and the DGCR8 5′UTR hairpin were efficiently processed by recombinant Dicer in vitro (Fig. 1C). Processing of pre-miR-1 yielded the expected products of ∼22 and ∼16 nt corresponding to sense/passenger strands and pre-miR-1 loop, respectively. Processing of the DGCR8 5′UTR gel-purified hairpin resulted in the accumulation of ∼22 and ∼32 nt cleavage products (Fig. 1C). These products sizes are consistent with the sense/passenger strands and terminal loop, respectively. These data raise the possibility that the DGCR8 5′UTR encodes for a novel miRNA.
Next we investigated whether the DGCR8 hairpins could yield any mature small RNAs in vivo. We performed Northern blots using a specific probe to detect the DGCR8 5′UTR hairpin. We detected ∼70 nt RNA that likely corresponds to the processing product obtained after Microprocessor cleavage of DGCR8 5′UTR, consistent with that observed in our in vitro processing experiments (Fig. 1D). However, we could not detect any mature ∼22 nt miRNA, even when a luciferase reporter containing DGCR8 5′UTR was used to overexpress this hairpin and long exposure times were used (Fig. 1D). We also performed Northern blot to detect any products processed from the miR-1306 hairpin using probes complementary to both strands of the hairpin. We could not detect any band corresponding either to premature or mature miRNA-1306 by Northern blot (data not shown and see below). Since miR-1306 was originally cloned from ES cells (Morin et al. 2008) and to examine the possibility of cell-type specific processing of this miRNA, we performed 5′ rapid amplification of cDNA ends (5′RACE) on RNA extracted from mouse ES cells and differentiated embryoid bodies. The majority (17/18) of the clones sequenced from both undifferentiated and differentiated cell types correspond to processing products of the DGCR8 5′UTR (Supplemental Fig. S2). Notably, we did not detect a single clone corresponding to the cleavage at the miR-1306 hairpin, consistent with our Northern blots and the low processing efficiency of this hairpin in vitro. Interestingly, all of the clones we sequenced from 7-methylguanosine cap (m7G)-containing mRNAs possessed an additional 44 nt upstream of the annotated cDNA sequence, thus, indicating that we have identified the actual transcriptional start site of the DGCR8 gene (Supplemental Fig. S2). Altogether, these results suggest that DGCR8 mRNA is a target for cleavage by the Microprocessor in vivo. Notably, this cleavage generates a ∼70 nt pre-miRNA-like hairpin that seems not to be processed by Dicer in vivo.
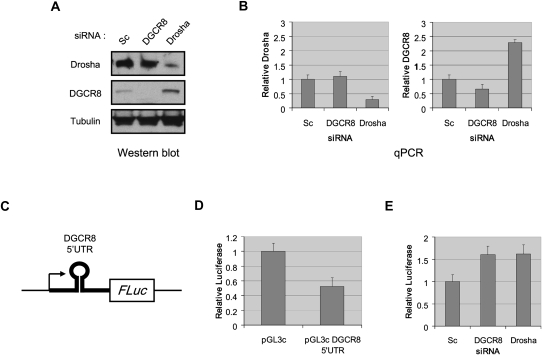
To gain further insights into the regulation of DGCR8 expression and the function of the DGCR8 5′UTR hairpin, we first asked whether Drosha could regulate DGCR8 expression in vivo. We transected Hela cells with siRNA specifically targeting Drosha or DGCR8 mRNA. Scrambled siRNA was used as a negative control. We then analyzed the levels of DGCR8 and Drosha by Western blot and found that RNAi against Drosha resulted in an increase in the level of DGCR8 protein (Fig. 2A). Conversely, the level of Drosha protein was unchanged when DGCR8 was depleted. We also examined DGCR8 and Drosha mRNA levels by qPCR and found that the DGCR8 mRNA level was increased after Drosha knockdown (Fig. 2B). Similar results were obtained in knockdown experiments in mouse embryonic stem cells (using two different siRNA duplexes), thus, demonstrating that this regulatory pathway is conserved in mammals (data not shown). Altogether, these results suggest that Drosha inhibits DGCR8 expression.
FIGURE 2.
DGCR8 expression is regulated by the Microprocessor. Hela cells were transfected with either scrambled (Sc), DGCR8, or Drosha siRNA as indicated. Cells were collected for analysis 60 h post-transfection. (A) Whole cell extracts were analyzed by Western blot using anti-Drosha, DGCR8, and Tubulin antibodies. (B) Total RNA was analyzed by quantitative RT-PCR using primers specific for Drosha, DGCR8, and Actin mRNA. Error bars represent SEM with N = 3. (C) Schematic representation of the firefly luciferase reporter containing DGCR8 5′UTR (pGL3c DGCR8 5′UTR). (D) Hela cells were co-transfected with either pGL3c or pGL3c DGCR8 5′UTR plasmid together with a TK-Renilla luciferase plasmid. Luciferase activity was measured 48 h post-transfection. Firefly luciferase activity was normalized relative to that of Renilla luciferase. Error bars represent SEM with N = 3. (E) Hela cells were transfected with pGL3c DGCR8 5′UTR together with TK-Renilla luciferase vector, and either scrambled, DGCR8, or Drosha siRNA as indicated. Luciferase activity was measured 60 h post-transfection. Renilla luciferase activity was used for normalization. Error bars represent SEM with N = 5.
We hypothesized that the repression of DGCR8 expression by Drosha was a consequence of the processing of the DGCR8 5′UTR hairpin by the Microprocessor. Therefore, we asked whether this hairpin could control the expression of a reporter gene. For this purpose, we inserted DGCR8 5′UTR in the 5′UTR of a luciferase reporter gene (Fig. 2C). The expression of luciferase from the DGCR8 5′UTR-containing reporter construct was only ∼50% that of a control luciferase reporter gene suggesting that processing of the DGCR8 5′UTR hairpin by the Microprocessor could be involved in the repression of the reporter gene (Fig. 2D). To directly test this, we analyzed luciferase activity in cells co-transfected with the DGCR8 5′UTR-containing reporter together with siRNA targeting DGCR8 or Drosha mRNA. Luciferase expression from the DGCR8 5′UTR-containing reporter construct was increased by ∼60% in cells depleted of either DGCR8 or Drosha (Fig. 2E). These results demonstrate that the Microprocessor mediates and regulates DGCR8 expression in vivo and that the majority of this repression is mediated directly through the DGCR8 5′UTR.
We investigated the regulatory role of two “pri-miRNA-like” hairpins localized in the second exon of DGCR8 mRNA. We found that the hairpin localized in the DGCR8 5′UTR is efficiently processed in vitro and in vivo, whereas the other hairpin in the DGCR8 open reading frame is a poor substrate for the Microprocessor. We further showed that DGCR8 expression is regulated by the Microprocessor through the direct cleavage of the DGCR8 5′UTR hairpin. Thus, our data identify a feedback loop that regulates the level of DGCR8 expression (Supplemental Fig. S3).
DGCR8 contains two double-stranded RNA binding domains that endow the Microprocessor with the ability to recognize pri-miRNAs and to guide Drosha-mediated cleavage of the stem of pri-miRNA hairpin. Microprocessor-mediated cleavage of the stem–loop in DGCR8 5′UTR should help to maintain a steady state level of DGCR8 that is physiologically required to achieve efficient processing of pri-miRNA (Supplemental Fig. S3). Though future work will be required to fully understand the physiological significance of maintaining the correct Microprocessor subunit stoichiometry, it was recently reported in a mouse model of 22q11.2 deletion/DiGeorge syndrome that DGCR8 haploinsufficiency leads to altered miRNA levels and behavioral and neuronal deficits characteristic of the 22q11.2 microdeletion (Stark et al. 2008). Deregulation of the DGCR8 steady state level might not only be deleterious to miRNA biogenesis, but could also result in nonspecific targeting and cleavage of other RNA in cells. Altered expression of Drosha/DGCR8 has been described in cancers and shRNA-mediated depletion of Drosha or DGCR8 can lead to increased tumor cell proliferation and tumorigenicity (Lu et al. 2005; Sugito et al. 2006; Blenkiron et al. 2007; Kumar et al. 2007; Muralidhar et al. 2007; Merritt et al. 2008). Moreover, a widespread down-regulation of miRNA expression due to a block at the Microprocessor-mediated processing step has been reported in ES and cancer cells from primary tumors (Thomson et al. 2006). Thus, it should be interesting to investigate the expression level of Microprocessor components and processing of DGCR8 mRNA in these contexts (Ding et al. 2009).
It is notable that we do not find evidence for processing of the miR-1306 hairpin in vivo. Though it is formally possible that this hairpin represents a functional miRNA, based on our observations of inefficient processing in vitro, together with the undetectable expression (by Northern blotting) in different cell types, and the absence of cleaved mRNA detectable by 5′ RACE, suggests that miR-1306 is either expressed at very low levels in cells or it may represent an mRNA degradation product detectable only by cloning and deep sequencing analyses (Griffiths-Jones et al. 2008; Morin et al. 2008).
Recently, two independent groups have also demonstrated Microprocessor regulation of DGCR8 expression in mammals and Drosophila, respectively (Han et al. 2009; Kadener et al. 2009). In the latter study it was similarly demonstrated that repression is mediated through the direct cleavage of the DGCR8 5′UTR hairpin by the Microprocessor leading to DGCR8 mRNA destabilization. Also consistent with our data was their finding that the DGCR8 5′UTR hairpin does not seem to be processed by Dicer in vivo since the corresponding mature miRNA is undetectable in cells (Han et al. 2009). However, our in vitro processing results indicate that the DGCR8 5′UTR processing product generated by the Microprocessor is indeed a suitable substrate for cleavage by recombinant Dicer. This raises the intriguing possibility that additional cellular mechanisms exist to prevent the processing of mature miRNA from the DGCR8 5′UTR. It is possible that it might be achieved through sequestration or degradation of the DGCR8 5′UTR pre-miRNA-like hairpin or degradation of mature product generated from Dicer cleavage. Alternatively, certain cellular factors might also be involved in repression of processing of this hairpin as has been recently demonstrated for Lin28-mediated control of Let-7 processing (Heo et al. 2008; Newman et al. 2008; Piskounova et al. 2008; Rybak et al. 2008; Viswanathan et al. 2008).
MATERIALS AND METHODS
Plasmids and cloning
The DGCR8 5′UTR sequence was cloned by PCR amplification (forward primer: 5′-GGCGGTCGGTCGGTGAGGC-3′; reverse primer: 5′-ATTAAAAGCGCTTAAGACTAGTTTACAAG-3′) into pGEM-T Easy (Promega) for in vitro transcription. For the miR-1306 hairpin, a region encompassing nucleotides 1–900 of DGCR8 coding region was cloned by PCR amplification (forward primer: 5′-TGTAGGTGGGCGGCCACGAC-3′; reverse primer: 5′-ATGTAGGTGGGCGGCCACGAC-3′) into pcDNA 3.1/ CT-GFP-TOPO (Invitrogen). The reporter construct pGL3c DGCR8 5′UTR was generated by cloning the PCR-amplified DGCR8 5′UTR (forward primer: 5′-CCACCAAAGCTTGGCGGTCGGTCGGTGAGGC-3′; reverse primer: 5′-CAACAAAAGCTTATTAAAAGCGCTTAAGACTAGTTTACAAG-3′) into the Hind III site of the pGL3-Control vector (Promega).
Cell culture, transfection, and luciferase assay
Hela cells were maintained in DMEM supplemented with 10% FBS. Flag-DGCR8 and Flag-Drosha stable cell lines were maintained in DMEM supplemented with 10% FBS and 2.5 μg/mL puromycin. For transfection of DNA and RNA, Lipofectamine 2000 (Invitrogen) was used according to manufacturer's protocol. Luciferase assay were realized using Dual Luciferase Reporter Assay System (Promega). Scrambled, Drosha, and DGCR8 siRNA were purchased from Dharmacon (Drosha siRNA: 5′-CGAGUAGGCUUCGUGACUUdTdT-3′; DGCR8 siRNA: 5′-GGAUGUAAAGAUUAGCGUGdTdT-3′). Primers were from IDT.
RNA extraction and Northern blotting
RNA was extracted using Trizol (Invitrogen) according to manufacturer's protocol. Northern blot was performed as previously described using 20 μg of RNA (Gregory et al. 2004). Specific probes for the DGCR8 5′UTR hairpin (5′-CAAGCTGGCCACATTGCTCTTTTCATTAATGTAGACAGC-3′) and for miR-1306 (5′-CACCACCAGAGCCAACGT-3′) were purchased from IDT.
5′ RACE
5′RACE for capped mRNA was performed using RLM-RACE kit (Ambion) according to manufacturer's protocol. 5′RACE for processed RNA was done without calf intestine alkaline phosphatase and tobacco acid pyrophosphatase treatments. Primers were purchased from IDT (3′ outer primer: 5′-GCTGTACACTTGTCTCTCCAT-3′; 3′ inner primer: 5′-TCTAACTCATCGAGCACTGCAT-3′). PCR products were cloned into the PGEM-T Easy vector (Promega) followed by DNA sequencing analysis.
Protein extraction and Western blotting
Protein extracts were prepared in lysis buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10% glycerol, 1.5 mM MgCl2, 0.2 mM PMSF, and 0.5 mM dithiothreitol [DTT]). Proteins were separated on 4%–12% Tris-Glycine gels (Invitrogen) and transferred to PVDF membrane (Millipore) as previously described (Gregory et al. 2006). Antibodies used were; anti-Drosha antibody (a gift from Ramin Shiekhattar), anti-DGCR8 antibody (Proteintech Group), and anti-Tubulin antibody (Abcam).
Reverse transcription and quantitative PCR
Reverse transcription was performed using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to manufacturer's protocol. Quantitative PCR was carried out using iQ SYBR Green Supermix (Bio-Rad) and iCycler iQ Multicolor Real-Time PCR Detection System (Bio-Rad). Specific primers for Drosha (forward primer: 5′-TAGGCTGTGGGAAAGGACCAAG-3′; reverse primer: 5′-GTTCGATGAACCGCTTCTGATG-3′), DGCR8 (forward primer: 5′-CAAGCAGGAGACATCGGACAAG-3′; reverse primer: 5′-CACAATGGACATCTTGGGCTTC-3′), and Actin (forward primer: 5′-TGAAGTGTGACGTGGACATC-3′; reverse primer: 5′-GGAGGAGCAATGATCTTGAT-3′) were purchased from IDT.
In vitro processing assays
Flag-Drosha and Flag-DGCR8 were purified from HEK293 stable cell lines with anti-FLAG M2 affinity gel (Sigma). After twice washing with buffer A (20 mM tris-HCl [pH 7.9], 0.5 M KCl, 10% glycerol, 1 mM EDTA, 5 mM DTT, and 0.2 mM PMSF 0.5% NP40), and once with buffer B (20 mM tris-HCl [pH 7.9], 0.1 M KCl, 10% glycerol, 1 mM EDTA, 5 mM DTT, and 0.2 mM PMSF), the affinity column was eluted with FLAG peptide. Recombinant DGCR8 protein was expressed in bacteria, and recombinant Drosha, and Dicer proteins were expressed in insect cells. Recombinant proteins were isolated as described previously (Gregory et al. 2004; Chendrimada et al. 2005). Processing reactions were performed as previously described within a buffer containing 3.2 mM MgCl2, 1 U/μL RNasin, 20 mM Tris-HCl (pH 7.9), 0.1 M KCl, and 10% glycerol, for 90 min at 37°C (Gregory et al. 2004; Chendrimada et al. 2005; Gregory et al. 2005, 2006).
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to Dr. Ramin Shiekhattar for anti-Drosha antibodies. We thank the Wistar Institute Protein Production Core Facility for help with generating recombinant Drosha and Dicer proteins. R.I.G. is supported by laboratory start-up funds from The Children's Hospital Boston and grants from The Harvard Stem Cell Institute, The March of Dimes Basil O'Conner award, The Charles H. Hood Foundation, and the Emerald Foundation. R.I.G. is a Pew Research Scholar. R.T. and R.I.G. designed the research; R.T., H.-M.C., and R.J.L. performed the research and analyzed the data; and R.T. and R.I.G. wrote the paper.
Footnotes
REFERENCES
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Blenkiron C., Goldstein L.D., Thorne N.P., Spiteri I., Chin S.F., Dunning M.J., Barbosa-Morais N.L., Teschendorff A.E., Green A.R., Ellis I.O., et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chendrimada T.P., Gregory R.I., Kumaraswamy E., Norman J., Cooch N., Nishikura K., Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Ding X.C., Weiler J., Großhans H. Regulating the regulators: Mechanisms controlling the maturation of microRNAs. Trends Biotechnol. 2009;27:27–36. doi: 10.1016/j.tibtech.2008.09.006. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. Oncomirs—MicroRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S.N., Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and post-transcriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Gregory R.I., Chendrimada T.P., Shiekhattar R. MicroRNA biogenesis: Isolation and characterization of the Microprocessor complex. Methods Mol. Biol. 2006;342:33–47. doi: 10.1385/1-59745-123-1:33. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S., Saini H.K., van Dongen S., Enright A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S., Cáceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Boettcher S., Caudy A.A., Kobayashi R., Hannon G.J. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes & Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Pedersen J.S., Kwon S.C., Belair C.D., Kim Y.K., Yeom K.H., Yang W.Y., Haussler D., Blelloch R., Kim V.N. Post-transcriptional cross-regulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Hutvágner G., McLachlan J., Pasquinelli A.E., Bálint E., Tuschl T., Zamore P.D. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Kadener S., Rodriguez J., Abruzzi K.C., Khodor Y.L., Sugino K., Marr M.T., 2nd, Nelson S., Rosbash M. Genome-wide identification of targets of the drosha-pasha/DGCR8 complex. RNA. 2009;15:537–545. doi: 10.1261/rna.1319309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kumar M.S., Lu J., Mercer K.L., Golub T.R., Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Liu J., Carmell M.A., Rivas F.V., Marsden C.G., Thomson J.M., Song J.J., Hammond S.M., Joshua-Tor L., Hannon G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lu J., Getz G., Miska E.A., Alvarez-Saavedra E., Lamb J., Peck D., Sweet-Cordero A., Ebert B.L., Mak R.H., Ferrando A.A., et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Meister G., Landthaler M., Patkaniowska A., Dorsett Y., Teng G., Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Merritt W.M., Lin Y.G., Han L.Y., Kamat A.A., Spannuth W.A., Schmandt R., Urbauer D., Pennacchio L.A., Cheng J.F., Nick A.M., et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N. Engl. J. Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlewski G., Guil S., Semple C.A., Cáceres J.F. Post-transcriptional regulation of miRNAs harboring conserved terminal loops. Mol. Cell. 2008;32:383–393. doi: 10.1016/j.molcel.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.D., O'Connor M.D., Griffith M., Kuchenbauer F., Delaney A., Prabhu A.L., Zhao Y., McDonald H., Zeng T., Hirst M., et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidhar B., Goldstein L.D., Ng G., Winder D.M., Palmer R.D., Gooding E.L., Barbosa-Morais N.L., Mukherjee G., Thorne N.P., Roberts I., et al. Global microRNA profiles in cervical squamous cell carcinoma depend on Drosha expression levels. J. Pathol. 2007;212:368–377. doi: 10.1002/path.2179. [DOI] [PubMed] [Google Scholar]
- Newman M.A., Thomson J.M., Hammond S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen J.S., Bejerano G., Siepel A., Rosenbloom K., Lindblad-Toh K., Lander E.S., Kent J., Miller W., Haussler D. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput. Biol. 2006;2:e33. doi: 10.1371/journal.pcbi.0020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E., Viswanathan S.R., Janas M., LaPierre R.J., Daley G.Q., Sliz P., Gregory R.I. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Sohn S.Y., Bae W.J., Kim J.J., Yeom K.H., Kim V.N., Cho Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- Stark K.L., Xu B., Bagchi A., Lai W.S., Liu H., Hsu R., Wan X., Pavlidis P., Mills A.A., Karayiorgou M., et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- Sugito N., Ishiguro H., Kuwabara Y., Kimura M., Mitsui A., Kurehara H., Ando T., Mori R., Takashima N., Ogawa R., et al. RNASEN regulates cell proliferation and affects survival in esophageal cancer patients. Clin. Cancer Res. 2006;12:7322–7328. doi: 10.1158/1078-0432.CCR-06-0515. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom K.H., Lee Y., Han J., Suh M.R., Kim V.N. Characterization of DGCR8/Pasha, the essential cofactor for Drosha in primary miRNA processing. Nucleic Acids Res. 2006;34:4622–4629. doi: 10.1093/nar/gkl458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Srivastava D. A developmental view of microRNA function. Trends Biochem. Sci. 2007;32:189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]