The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila (original) (raw)
. Author manuscript; available in PMC: 2009 Aug 19.
SUMMARY
The canonical microRNA (miRNA) pathway converts primary hairpin precursor transcripts into ∼22 nucleotide regulatory RNAs via consecutive cleavages by two RNase III enzymes, Drosha and Dicer. In this study, we characterize Drosophila small RNAs that derive from short intronic hairpins termed “mirtrons.” Their nuclear biogenesis appears to bypass Drosha cleavage, which is essential for miRNA biogenesis. Instead, mirtron hairpins are defined by the action of the splicing machinery and lariat-de-branching enzyme, which yield pre-miRNA-like hairpins. The mirtron pathway merges with the canonical miRNA pathway during hairpin export by Exportin-5, and both types of hairpins are subsequently processed by Dicer-1/loqs. This generates small RNAs that can repress perfectly matched and seed-matched targets, and we provide evidence that they function, at least in part, via the RNA-induced silencing complex effector Ago1. These findings reveal that mirtrons are an alternate source of miRNA-type regulatory RNAs.
INTRODUCTION
We now recognize the first microRNA (miRNA) to have been reported in 1993 (Lee et al., 1993; Wightman et al., 1993), with additional examples of miRNA-mediated regulatory phenomena in worms (Moss et al., 1997; Pasquinelli et al., 2000; Reinhart et al., 2000) and Drosophila (Lai et al., 1998; Lai and Posakony, 1997, 1998) arising over the next 7 years. However, it was not until late 2001 that miRNA genes were appreciated to be an abundant feature of eukaryotic genomes (Lagos-Quintana et al., 2001; Lau et al., 2001; Lee and Ambros, 2001). The recent discovery of such an abundant gene class has sparked a torrent of inquiry into their biogenesis and function.
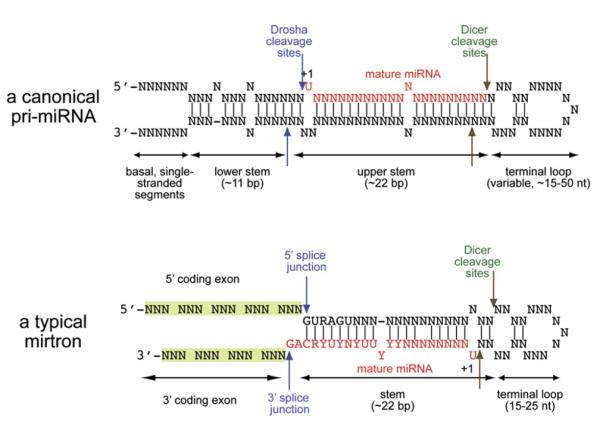
miRNA loci are generally transcribed as long, primary-miRNA (pri-miRNA) transcripts by RNA polymerase II (Lee et al., 2004a), although some are products of RNA polymerase III (Borchert et al., 2006). Most miRNAs derive from the exons or introns of noncoding RNAs, but about one-third are located in the introns of mRNA-encoding host genes (Rodriguez et al., 2004). pri-miRNAs contain an extended hairpin structure that is cleaved near the base by the nuclear RNase III enzyme Drosha, thereby releasing a ∼65 nucleotide (nt) pre-miRNA hairpin (Lee et al., 2003). The pre-miRNA is then exported to the cytoplasm and cleaved near its terminal loop by the RNase III enzyme Dicer, yielding a ∼22 nt miRNA duplex (Grishok et al., 2001; Hutvagner et al., 2001; Ketting et al., 2001). Because RNase III cleavage leaves behind a 2 nt-3′ overhang, miRNA duplexes display these signature overhangs at both ends (Figure 1).
Figure 1. Essential Characteristics of Mirtrons and Pri-miRNAs.
(Top) A typical pri-miRNA transcript contains a “lower stem,” which mediates recognition and cleavage by Drosha (blue arrows). The resulting pre-miRNA hairpin is then cleaved by Dicer-1 (green arrows) to yield a ∼22 nt duplex with ∼2 nt 3′ overhangs. Depending on the miRNA precursor, either the 5′ or the 3′ hairpin product is preferentially transferred into an effector Ago complex; here, it is depicted as the 5′ arm. This diagram was modeled after Han et al. (2006).
(Bottom) A typical mirtron locus lacks the “lower stem” found in pri-miRNAs but is instead constrained by primary nucleotide motifs that mediate their recognition and cleavage by the splicing machinery (i.e., 5′ GURAGU splice donors, 3′ polypyrimidine tracts, and CAG [or much less frequently UAG] splice acceptor sites). Once the spliced mirtron is debranched, it can adopt a pre-miRNA-like hairpin structure and be cleaved by Dicer-1. In contrast to canonical miRNA hairpins, mirtron hairpins are strongly biased to exhibit preferential stability of small RNAs from their 3′ arms.
One strand of the miRNA duplex is preferentially transferred to an active effector complex containing an Argonaute (Ago) protein (reviewed by Du and Zamore [2005]). The other strand, referred to as the miRNA* species, is generally presumed to be a nonfunctional carrier strand that is degraded. The mature miRNA then guides the Ago complex to target transcripts for regulation (reviewed by Valencia-Sanchez et al. [2006]). Experimental and computational approaches showed that Watson-Crick base pairing between positions 2–8 from the 5′ end of an animal miRNA (the miRNA “seed”) and a target transcript is typically necessary and often sufficient for direct regulation (Brennecke et al., 2005; Doench and Sharp, 2004; Lai, 2002; Lewis et al., 2003). Global miRNA target studies suggest that a majority of animal transcripts either are under detectable selective pressure to maintain direct regulation by miRNAs (Grun et al., 2005; Lewis et al., 2003; Stark et al., 2005) or actively avoid the acquisition of miRNA-binding sites (Farh et al., 2005; Stark et al., 2005).
Over a million Drosophila small-RNA sequences were recently generated using 454 pyrosequencing (J.G. Ruby, W. Johnston, D. Bartel, and E.C.L., unpublished data). When analyzing these sequences, the Bartel lab identified 14 short introns with predicted hairpin structure that give rise to novel ∼22 nt RNAs (Ruby et al., 2007). In recognizing their pre-miRNA and intronic features as defining characteristics, they named these introns “mirtrons.” They observed that primary-mirtron precursors composed of mirtronic introns and flanking exonic sequences lack the lower stem of pri-miRNAs (Figure 1), which mediates their recognition and cleavage by the Pasha (DGCR8)/Drosha complex (Han et al., 2006). Instead, their hairpin ends correspond precisely to splice sites (Figures 1 and 2). The “AG” splice acceptor of mirtronic introns typically adopts a 2 nt-3′ overhang to these hairpins, thereby mimicking a Drosha product (Ruby et al., 2007). The processing of these hairpins was further reminiscent of Dicer substrates, since the cloned RNAs derived from mirtrons adopt duplex configurations characteristic of miRNA/miRNA* pairs (Ruby et al., 2007).
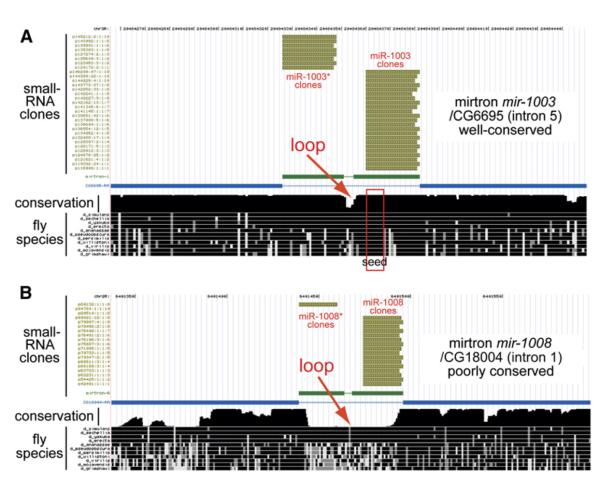
Figure 2. Evolutionary Characteristics of Mirtrons.
(A) Example of a well-conserved mirtron locus, mir-1003. Alignments and conservation data were produced by the UCSC Genome Center (http://genome.ucsc.edu/). Gold tracks depict the positions of a subset of sequenced small RNAs mapping to this locus, the blue track depicts the exon/intron structure of its host CG6695, and the black tracks at bottom depict nucleotide conservation of this region across 12 Drosophilid species. Greater height of the “conservation” track reflects deeper sequence conservation. The mir-1003 mirtron is highly constrained, with perfect conservation of the miR-1003 seed (positions 1–8, red box). Down-stream of the seed, positions 9–13 have undergone significant divergence. Note also that the terminal loop region (red arrow) exhibits accelerated divergence relative to the mirtron hairpin arms.
(B) Example of a poorly conserved mirtron locus, mir-1008. Its sequence is only preserved among melanogaster subgroup species and thus arose sometime in the last <10 million years. Despite its rapid evolution, this mirtron still exhibits accelerated divergence in the terminal loop region (red arrow).
This study reports functional evidence that mirtrons are generated by a nuclear pathway that appears to bypass Drosha but instead involves splicing and intron lariat-debranching enzyme. Debranched mirtron hairpins access the cytoplasm via Exportin-5 and enter the Dicer-1/miRNA biogenesis pathway to yield small regulatory RNAs. Although other effector complexes are not excluded, we provide evidence that mirtron-derived small RNAs associate with and require Ago1 to repress seed-matched targets. Our data support and extend the findings of Ruby, Jan, and Bartel (Ruby et al., 2007) with regard to the fundamental properties of these novel small-RNA genes, whose existence broadens the universe of small regulatory RNAs in animals.
RESULTS
Evolutionary Features of Mirtrons Support Their Status as Regulatory RNAs
Pre-miRNA hairpins collectively yield functional RNAs from both 5′ and 3′ hairpin arms (Figure 1). In contrast, mirtron hairpins yield predominant small RNAs from only their 3′ arms (Ruby et al., 2007). The 5′ splice consensus may intrinsically bias mirtron processing, since miRNAs and mirtron-derived small RNAs exhibit strong preferences to begin with U residues, whereas 5′ mirtron-derived RNAs must begin with a G residue (Ruby et al., 2007). In addition, mirtrons typically exhibit extensive pairing between the 5′ and 3′ splice sequences (Figure 1), a layout that may bias the selection of their 3′ arms as regulatory species (Khvorova et al., 2003; Schwarz et al., 2003). The asymmetry of mirtron processing toward 3′ products is rational from a biological perspective, since the regulatory potential of their 5′ products is perhaps undesirably constrained by the splice donor sequences within their prospective seed regions.
Further support for the regulatory status of mirtrons came from the observation that several mirtronic introns are well conserved among the sequenced Drosophilids (Figures 2A and S1; Ruby et al., 2007). On the other hand, most mirtrons are preserved only within species of the melanogaster subgroup (D. melanogaster, D. simulans/sechellia, and/or D. yakuba/erecta), suggesting that they were born within the last 5–10 million years (Figures 2B and S1; Ruby et al., 2007). Nevertheless, detailed inspection revealed that the evolution of both “old” and “young” mirtrons parallels that of miRNAs in two aspects (reviewed by Lai et al. [2003]). First, as is the case for miRNA hairpins, both young and old mirtrons exhibit accelerated divergence in loop regions relative to the hairpin stems (Figures 2 and S1). Second, mirtron loci display preferential conservation of the 5′ seed region, a key determinant for miRNA target recognition (e.g., Figure 2A). In fact, small RNAs generated by mirtrons resident in CG6695 and CG31772 (miR-1003 and miR-1004, respectively) have the same seed (Figures 2, 6, and S2), indicative of a functional subfamily.
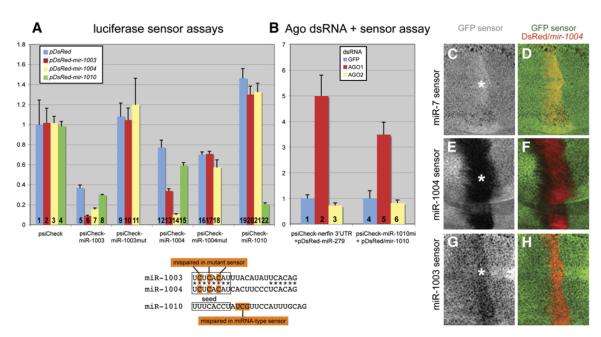
Figure 6. Mirtrons Repress Both Perfect-Match and Seed-Match Targets.
(A) Luciferase sensor assays in S2 cells. Sensors contain two antisense copies of miR-1003, miR-1004, and miR-1010 cloned downstream of renilla luciferase in psiCHECK2; this vector also carries a control firefly luciferase gene. miR-1003 and miR-1004 share the same seed (boxed), and mutant sensors are mispaired at the 3 nt positions highlighted. Sensor plasmids were cotransfected with ub-Gal4 and UAS-DsRed or UAS-DsRed-mirtron plasmids as indicated (see key, inset). Luciferase activity was expressed as the mean ratio of the experimental renilla/firefly luciferase sensor value in the presence of DsRed + mirtron relative to DsRed alone. These data were pooled from quadruplicate transfections, and error bars represent the standard deviations. The following relevant comparisons exhibited p < 0.0001 (equal variance Student’s t test): miR-1003 sensor versus miR-1003 mut sensor (lane 5 versus lane 9), repression of miR-1003 sensor by mir-1003 or mir-1004 (lane 5 versus lane 6 or 7), repression of miR-1004 sensor by mir-1003 or mir-1004 (lane 12 versus lane 13 or 14), and repression of miR-1010 sensor by mir-1010 (lane 19 versus lane 22).
Lanes 1–4: empty sensor. DsRed-mirtron expression had little intrinsic effect on psiCHECK2-luciferase activity.
Lanes 5–8: miR-1003 sensor. This sensor was strongly repressed relative to empty vector (compare lane 5 with lane 1). Expression of mir-1003 (6) and mir-1004 (7) but not mir-1010 (8) mirtron constructs further reduced its activity.
Lanes 9–11: miR-1003 mut sensor. Mutation of its seed-complementary region elevated its basal expression, indicating relief from repression by endogenous miR-1003. Such mutations also abolished its response to mir-1003 and mir-1004 mirtron-expression constructs.
Lanes 12–15: miR-1004 sensor. This sensor was robustly inhibited by mir-1004 (14), mildly suppressed by mir-1003 (13), and largely unaffected by mir-1010 (15) mirtron-expression constructs.
Lanes 16–18: miR-1004 mut sensor. Mutation of its seed-complementary region eliminated its response to ectopic mir-1003 and mir-1004.
Lanes A19–22: miR-1010 sensor. This sensor was specifically repressed by mir-1010 (22) but not mir-1003 (20) or mir-1004 (21) mirtron-expression constructs.
(B) Ago knockdowns in S2R+ cells, followed by transfection with miRNA and mirtron-expression constructs and sensors. The miR-1010 mi sensor contains centrally placed mismatches, as indicated, to render it a miRNA-type sensor. These data were pooled from two independent sets of quadruplicate transfections (n = 8), and error bars represent the standard deviations. The following comparisons exhibited p < 0.00001 (equal variance Student’s t-Test): derepression of nerfin sensor by Ago1 dsRNA (lane 1 versus lane 2) and derepression of miR-1010 mi sensor by Ago1 dsRNA (lane 4 versus lane 5).
Lanes 1–3: Ago1 dsRNA, but not GFP or Ago2 dsRNA, derepressed the nerfin 3′ UTR sensor in the presence of ectopic miR-279.
Lanes 4–6: Ago1 dsRNA, but not GFP or Ago2 dsRNA, derepressed the 4xmiR-1010 mi sensor in the presence of ectopic miR-1010.
(C—H) GFP expression is shown in grayscale in panels (C), (E), and (G) and as a merge (in green) with DsRed/miR-1004 expression (in red) in panels (D) (F), and (H). In (C) and (D), GFP-miR-7 sensor activity, which can be completely abolished by ectopic expression of miR-7 (Lai et al., 2005; Stark et al., 2003), was unaffected by expression of mir-1004. In (E) and (F), GFP-miR-1004 sensor activity was strongly repressed (asterisk) in cells that express the mir-1004 mirtron. In (G) and (H), the GFP-miR-1003, seed-paired, sensor was weakly repressed (asterisk) in cells that express mir-1004.
These observations suggested that mirtrons are RNA genes related to miRNAs. Since experimental evidence presented in this study (see below) and a contemporary study (Ruby et al., 2007) supported their identity as a functional subclass of miRNA genes, we refer to the hairpin introns as mirtrons and their small-RNA products as miRNAs.
Mirtrons Display Distinct Temporal and Spatial Patterns of Expression
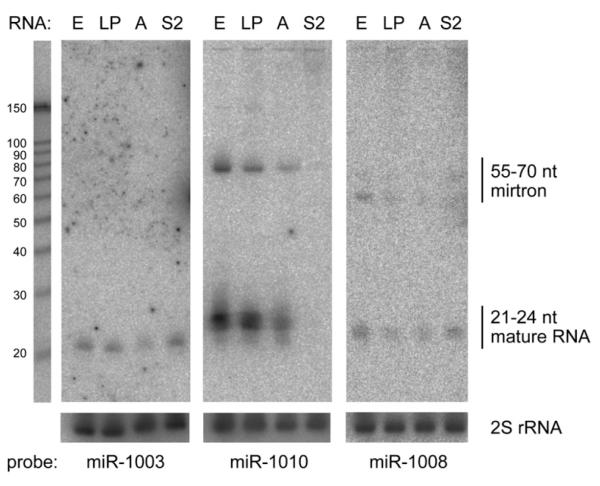
We began our functional studies by using northern analysis to ask whether processed mirtrons could be detected across development or in cultured cells. We probed total RNA from 0–24 hr embryos, third-instar larvae/pupae, adults, and S2 cells to northern analysis with γ-32P-labeled locked nucleic acid (LNA) oligos antisense to the terminal 22–24 nt of mirtron hairpins for mir-1003, mir-1010, and mir-1008. These probes detected mature ∼21–24 nt RNAs and rarer ∼55–70 nt precursors (Figure 3). As with miRNAs, such discretely hybridizing bands reflected precision in mirtron processing and argued against the possibility that the cloned sequences merely represent metabolic intermediates of spliced introns. We also note that mirtrons exhibited variety in their developmental and spatial expression profiles, similar to miRNAs. For example, the small-RNA products of mir-1003 and mir-1008, but not of mir-1010, were detected in S2 cells. mir-1010 also differed in that its expression was much reduced in adults relative to earlier stages (Figure 3).
Figure 3. Distinct Temporal and Spatial Expression of Endogenous Mirtrons.
Northern blots were prepared using RNA from 0–24 hr embryos (E), third-instar larvae, and 0–2 day pupae (LP), adult males and females (A), and S2 cells (S2) and probed with antisense LNA probes to the 3′ ends of several mirtrons. Endogenous ∼21∼24 nt RNAs and ∼55–70 nt precursors were detected in all cases. Blots were stripped and reprobed for 30 nt 2S rRNA as a loading control. RNA sizes were judged with reference to a Decade RNA marker (Ambion) run in parallel. Lengths of the mirtron hairpins inferred from intron boundaries are mir-1003 (56 nt), mir-1010 (71 nt), and mir-1008 (57 nt).
Introns Can Autonomously Dictate Their Entry into the Mirtron Pathway
We next investigated whether mirtrons could be expressed exogenously. To do so, we cloned ∼400 nt primirtron genomic fragments, whose termini lie within the exons flanking the mirtron, and inserted them into the 3′ UTR of a UAS-DsRed vector (Figure 4, construct A). This generic strategy successfully generates mature Drosophila miRNAs from similarly sized pri-miRNA fragments (Lai et al., 2005; Stark et al., 2003). For these studies, we selected both highly conserved (mir-1003 and mir-1010) and newly born (mir-1008 and mir-1004) mirtron loci. When transfected into S2 cells with ub-Gal4, such constructs directed the expression of all four mirtrons and their mature small-RNA products (Figure 4, lanes 2 and 7, and data not shown).
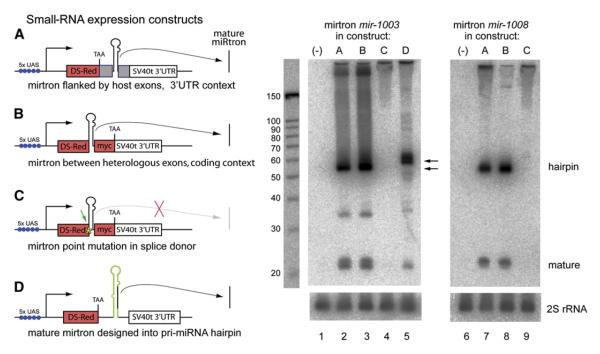
Figure 4. Structure-Function Analysis of Mirtron Biogenesis.
(A)—(D) On the left are four constructs for mirtron expression. In (A), ∼400 nt pri-mirtrons containing their flanking endogenous exons (gray boxes) were cloned into the 3′ UTR of UAS-DsRed. In (B), ∼55-70 nt mirtron hairpins were cloned between coding exons for DsRed and 2xmyc, thereby separating them from endogenous exonic context. In (C), using construct B as a template, a G -> C point mutation in the first base of the 5′ splice donor was introduced (asterisk and arrow). In (D), the mature ∼22 nt mirtron product was substituted into a miRNA precursor based on pri-mir-6-1 and then inserted into the 3′ UTR of UAS-DsRed. On the right, northern analysis of mirtrons cloned into constructs (A)—(D), as depicted on the left, and activated in S2 cells using ub-Gal4. Lanes 2 and 6 show that miR-1003, miR-1008, and their associated mirtron hairpins were efficiently generated from mirtron genomic fragments using construct (A), (compare with lanes 1 and 6 for endogenous mirtron expression; note that the blot has been underexposed relative to blots in Figure 3). miR-1003 and miR-1008 and their mirtron hairpins were also readily expressed from construct B (lanes 3 and 8), demonstrating that endogenous flanking exons are dispensable for entry into the mirtron pathway. Point mutation of the splice donor site demonstrated that splicing is necessary to generate the mirtron hairpin (lanes 4 and 9). Mature miR-1003 could also be expressed by reprogramming a canonical pri-miRNA (lane 5). Since the mir-6-1 hairpin is 63 nt, the hybrid pri-mir-6-1/mir-1003 hairpin is slightly longer than 56-nt-long mirtron for miR-1003 (as indicated by arrows). Blots were stripped and reprobed for 2S rRNA as a loading control.
We then tested the ability of mirtrons to be processed when resident in the coding context of a designed vector. To exclude the potential contribution of specific exonic sequences to mirtron maturation, we designed a host vector in which the mirtronic intron alone is inserted into the coding region of a DsRed-myc transcript (Figure 4, construct B). We found that mature miR-1003, miR-1008, and their corresponding mirtrons were produced from such constructs at levels comparable to their expression from endogenous exonic contexts (Figure 4, lanes 3 and 8; compare with lanes 2 and 7). Therefore, the sequence of a short intron can autonomously dictate its ability to enter the mirtron pathway.
Mirtron Biogenesis Exhibits Little Dependence on Drosha
The concordance between mirtron ends and splice sites (Figures 1 and 2) suggested that their biogenesis might bypass the essential miRNA-producing enzyme Drosha. We tested this by treating S2 cells with dsRNA against Drosha. Western blot analysis confirmed robust knockdown of Drosha protein after 4 days (Figure S2A). As a control, we tested the behavior of a UAS-DsRed-mir-1 construct; S2 cells do not normally express miR-1. As shown in Figure 5A (lanes 1 and 2), the levels of pre-miR-1 hairpin and mature miR-1 were significantly reduced in Drosha dsRNA-treated cells relative to green fluorescent protein (GFP) dsRNA-treated cells. In contrast, endogenous miR-1003 and its mirtron were not demonstrably changed by these treatments (Figure 5A, lanes 6 and 7), and the accumulation of exogenous miR-1003, miR-1010, and their associated mirtrons was also similar between GFP and _Drosha_-knockdown cells (Figure 5A, lanes 11 and 12 and 19 and 20). While these data do not exclude a contribution of Drosha to mirtron processing, they indicate that mirtron biogenesis does not exhibit the strong dependence on Drosha that is characteristic of canonical miRNA pathway substrates.
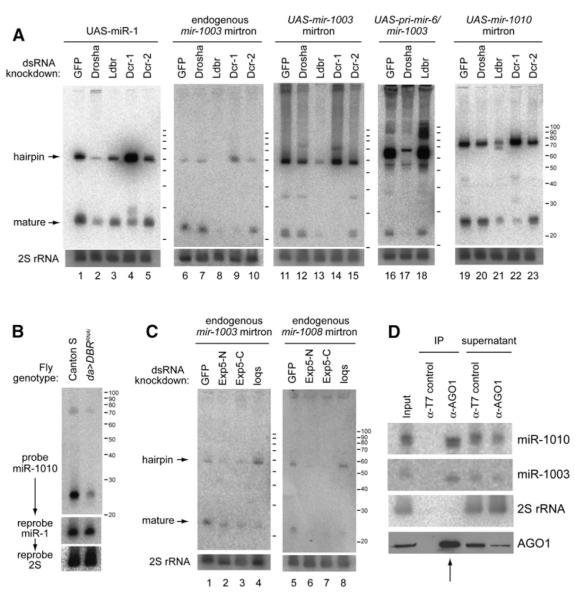
Figure 5. Mirtron Biogenesis Involves a Hybrid Pathway that Couples Splicing and Dicing.
(A) S2 cells were treated with the indicated dsRNAs and transfected with ub-Gal4 and UAS-DsRed-small-RNA plasmids as noted. Total RNA was then extracted and analyzed for small-RNA expression by northern blot; these were stripped and reprobed to detect 2S rRNA as a loading control.
Lanes 1–5: processing of UAS-DsRed-mir-1. Drosha knockdown decreased the amount of hairpin pre-miR-1 and mature miR-1 (lane 2). Ldbr (lariat-debranching enzyme) knockdown had little effect (lane 3). Dicer-1 knockdown caused a profound increase in pre-miR-1 hairpin (lane 4), while Dicer-2 knockdown had little effect (lane 5).
Lanes 6–10: processing of endogenous mir-1003 mirtron. Its accumulation was largely unaffected by Drosha knockdown (lane 7), but Ldbr knockdown abolished the accumulation of mirtron and mature forms of miR-1003 (lane 8). Knockdown of Dicer-1 (lane 9) but not Dicer-2 (lane 10) depleted mature miR-1003 and resulted in the accumulation of its precursor mirtron. Lanes 11–15 depict processing of UAS-DsRed-mir-1003. Its genetic requirements were similar to those of its endogenous counterpart.
Lanes 16–18: processing of UAS-DsRed-pri-mir-6-1/mir-1003. Expression of miR-1003 from a pri-miRNA backbone was similar to that of miR-1, in that it was now strongly dependent on Drosha (lane 17) and appeared not to require Ldbr (lane 18).
Lanes 19–23: processing of a UAS-DsRed-mir-1010 mirtron construct. Similar genetic requirements were seen as for the mir-1003 mirtron.
(B) Mirtron biogenesis requires intron debranching in intact animals. Larvae carrying da-Gal4 and UAS-LdbrRNAi exhibited a marked decrease in the levels of mir-1010 mirtron and mature product compared to wild-type Canton S larvae. The same blot was stripped and reprobed for miR-1, which showed only minor alteration in its steady-state level. The reduction in miR-1010, as normalized to miR-1, was 67 ± 2% SD (see Figure S3).
(C) Mirtron maturation requires Exportin-5 and loqs. Treatment with either of two nonoverlapping dsRNAs against Drosophila Exportin-5 (Exp5-N, Exp5-C) induced substantial loss of mirtron hairpins and small-RNA products for endogenous mir-1003 (lanes 1–3) and mir-1008 (lanes 5–7). Additional Exportin-5 knockdown data are reported in Figure S4. Treatment with loqs dsRNA increased the steady-state level of endogenous mir-1003 mirtron hairpin and decreased its mature form (lane 4); similar results were obtained for endogenous mir-1008 mirtron (lane 8).
(D) Endogenous ∼22 nt small RNAs derived from mirtrons mir-1003 and mir-1010 associate with endogenous Ago1. Coimmunoprecipitation (coIP) assay from 0–10 hr embryos using control mouse α-T7 or mouse α-Ago1. Fifteen percent of the RNA input and supernatants, twenty percent of the protein input and supernatants, and one hundred percent of the IP fractions were loaded. Ago1 lane is a western blot; other lanes are northern blots for the indicated small RNAs. The absence of miRNAs from control IPs, along with absence of 2S rRNA from Ago1 IPs, provided evidence for specific recovery of mirtron-derived miRNAs in the Ago1 IP fraction (an arrow marks the relevant lanes).
Mirtron Biogenesis Requires Intron Splicing and Lariat Debranching
We next tested the alternative hypothesis that splicing might directly initiate mirtron biogenesis. To do so, we made single G -> C substitutions in the 5′ splice sites of otherwise functional mir-1003 and mir-1008 mirtron-expression constructs (Figure 4, construct C). Such mutations completely abolished the accumulation of mirtron hairpins and mature ∼22 mers (Figure 4, lanes 4 and 9), indicating that both forms are obligate splicing products.
Since spliced introns adopt a lariat structure in which the 5′ splice junction is covalently linked to the 3′ branch point, we hypothesized that Drosophila lariat-debranching enzyme (Ldbr) is needed for spliced mirtron lariats to adopt hairpin folds required for their subsequent cleavage by Dicer. We treated cells with Ldbr dsRNA and analyzed their ability to process endogenous mirtrons and products of _UAS-DsRed-_small-RNA plasmids. Quantitative RT-PCR analysis indicated successful knockdown of Ldbr function, as evidenced by a 15-fold accumulation of the actin intron relative to cells that received GFP or Drosha dsRNA (Figure S2B). These cells exhibited only a minor decline in pre-miR-1 hairpin and mature miR-1 (Figure 5A, lane 3). In contrast, the accumulation of hairpin mirtrons and their mature products was strongly decreased or abolished by this treatment, including those of endogenous mir-1003 (Figure 5A, lane 8) and ectopic mir-1003 and mir-1010 (Figure 5A, lanes 13 and 21).
We sought to confirm a positively acting role for Ldbr in mirtron production in the animal using a UAS-LdbrRNAi transgene (Conklin et al., 2005). When activated with da-Gal4, individuals survived well to late larval stages but died during pupation. We therefore selected late third-instar larvae as a compromise time point that was suboptimal for Ldbr knockdown but obviated secondary concerns surrounding the analysis of sickly animals. As with the S2 experiments, RNA from Ldbr knockdown larvae showed increased levels of actin intron (Figure S2B) but decreased levels of endogenous mirtron hairpin and mature product for mir-1010 (Figure 5B). When the same blots were stripped and reprobed for miR-1, we observed little change in the accumulation of this miRNA. We quantified a 67% reduction in miR-1010 as normalized to miR-1 under these conditions (Figure S3).
Finally, to rule out the possibility that specific sequences in mature mirtrons per se influence their choice of nuclear processing pathway, we created a hybrid miRNA/mirtron construct in which the mature miR-1003 sequence was programmed into a pri-mir-6-1 precursor structure (Figure 4, construct D). Such a construct still produced mature miR-1003 (Figure 4, lane 5). However, the hybrid mir-6-1/mir-1003 construct now exhibited behaviors characteristic of a canonical miRNA precursor, in that its processing displayed strong dependence on Drosha but was little affected by Ldbr depletion (Figure 5A, lanes 17 and 18). Thus, we are able to control the choice of RNA substrates to enter the nuclear mirtron or miRNA pathways by manipulating sequence and structural features defined by our biogenesis experiments.
Collectively, these data reveal that mirtron biogenesis, like that of certain snoRNAs (Ooi et al., 1998), positively requires the action of lariat-debranching enzyme. These data do not exclude the possibility that Ldbr is required for the activity or processing of an intermediate factor that in turn mediates the resolution of mirtrons, but they are consistent with the parsimonious explanation that Ldbr acts directly upon mirtron lariats.
The Mirtron Pathway Merges with the Canonical miRNA Pathway during Hairpin Export
The recovery of paired small RNAs from mirtrons that resemble miRNA/miRNA* species suggested that mirtron biogenesis converges at some point with the canonical miRNA pathway (Ruby et al., 2007). We considered that this might occur during nuclear hairpin export, which is believed to be mediated by Exportin-5 (Bohnsack et al., 2004; Lund et al., 2004; Shibata et al., 2006; Yi et al., 2003). Functional studies of Exportin-5 knockdown are complicated since animal mutants have not been described, and only mild effects on miRNA processing can be obtained using RNAi-mediated knockdown (Lund et al., 2004; Shibata et al., 2006).
Consistent with previous results in mammalian and Drosophila cells, we observed modest (∼50%) reduction in endogenous mature miR-2b upon treatment with either of two nonoverlapping Exportin-5 dsRNAs (Figure S4). However, mirtron hairpins proved to be more sensitive to manipulation of Exportin-5. We observed 60%–80% reduction in mirtron hairpins and mature products for endogenous mir-1003 and mir-1008 in cells treated with either Exportin-5 dsRNA (Figure 5C). Similar, although slightly less robust, results were obtained using UAS-DsRed-mir-1003 and UAS-DsRed-mir-1010 mirtron-expression constructs (Figure S4). It is conceivable that mirtron overexpression can partially overcome Exportin-5 knockdown or that there is an alternate mechanism for the nuclear export of pre-miRNAs and mirtrons. However, these data suggest that a considerable proportion of mirtron hairpins transit Drosophila Exportin-5. In addition, the observation that mirtron hairpins decline following Exportin-5 knockdown is consistent with the previous suggestion that nuclear pre-miRNA hairpins are degraded when Exportin-5 is compromised (Yi et al., 2003).
If the mirtron and canonical miRNA pathways merge during hairpin export, one might predict that their cytoplasmic processing should be similar. There are two Drosophila Dicers, with Dicer-1 known to be genetically required for pre-miRNA maturation and Dicer-2 for processing of long dsRNA (Lee et al., 2004b; Saito et al., 2005). We tested their requirements by treating S2 cells with dsRNA against Dicer-1 or Dicer-2 (Figure S1A). As shown previously (Okamura et al., 2004), the maturation of miRNAs exhibited strong dependence on Dicer-1 but not Dicer-2 (Figure 5A, lanes 4 and 5). Dicer-1 was also strongly required for mirtron biogenesis, as its knockdown induced the accumulation of mirtron hairpins and depleted their small-RNA products (Figure 5A; lanes 9, 14, and 22). In contrast, no mirtron tested exhibited substantial sensitivity to Dicer-2 dsRNA (Figure 5A; lanes 10, 15, and 23).
We also analyzed the requirement of loquacious (loqs), a partner of Dicer-1 that is needed for efficient pre-miRNA cleavage (Forstemann et al., 2005; Jiang et al., 2005; Saito et al., 2005). Treatment with loqs dsRNA concomitantly increased the steady-state levels of endogenous mirtron hairpins for mir-1003 and mir-1008 and decreased their mature products (Figure 5C, lanes 4 and 8). Therefore, loqs is also an important cofactor for mirtron cleavage by Dicer-1.
In summary, mirtron biogenesis differs from that of nuclear pre-miRNA biogenesis in that mirtron accumulation appears to bypass Drosha cleavage but, instead, exhibits strong dependence on splicing and intron lariat debranching. However, these pathways converge since both types of hairpins appear to transit Exportin-5 and require Dicer-1/loqs for cleavage into ∼22 nt RNAs.
Mirtrons Generate Active Regulatory RNAs
We assayed the transregulatory activity of mirtrons using renilla luciferase “sensors” bearing sequences antisense to miR-1003, miR-1004, and miR-1010 in psiCHECK2; this vector contains a renilla luciferase “sensor” fused to test sequences and a firefly luciferase gene for normalization. When transfected into S2 cells along with ub-Gal4 and empty UAS-DsRed vector, miR-1003 sensor levels were much lower than those of the empty sensor or miR-1010 sensor (Figure 6A, lane 5; compare with lanes 1 and 19). Since miR-1003, but not miR-1010, is expressed by S2 cells (Figure 3), this suggested that endogenous mirtron-derived miR-1003 directly repressed this sensor. To test this, we mutated the miR-1003 sensor to introduce noncomplementary bases at positions 2, 4, and 6 as measured from its 5′ end. In spite of 16/16 nucleotides of perfect match, the seed mutant miR-1003 sensor was no longer repressed in S2 cells, consistent with its failure to be recognized by endogenous miR-1003 (Figure 6A, lane 9; compare with lane 5).
We next tested the response of these sensors to ectopic mirtrons expressed using ub-Gal4 and UAS-DsRed-mirtron plasmids. We observed that sensors for miR-1003, miR-1004, and miR-1010 were strongly inhibited (5- to 8-fold) in the presence of cognate mirtron-expression constructs (Figure 6A, lanes 6, 14, and 22; compare with lanes 5, 12, 19, respectively). On the other hand, the mir-1010 mirtron construct had little impact on the miR-1003 and miR-1004 sensors (Figure 6A, lanes 8 and 15), while the mir-1003 and mir-1004 mirtron constructs did not repress the miR-1010 sensor (Figure 6A, lanes 20 and 21). The consistent behavior of the different mirtron:sensor pairs demonstrates that mirtrons generate sequence-specific regulatory RNAs.
As is the case with miRNAs, few if any endogenous transcripts are perfectly complementary to mirtron-derived small RNAs. Therefore, we asked whether mirtron-derived small RNAs could recognize seed-matched sites, which constitute the bulk of endogenous miRNA target sites. Since miR-1003 and miR-1004 have the same seed, we performed this test by assaying their mirtron-expression constructs on reciprocal sensors. While weaker than its effect on a perfectly matched sensor, ectopic miR-1004 repressed the miR-1003 sensor by 2-fold (Figure 6A, lane 7); similar repression of the miR-1004 sensor by ectopic miR-1003 was also seen (Figure 6A, lane 13). To test whether the observed regulation was truly mediated by the proposed seed matches, we analyzed the response of seed mutant miR-1003 and miR-1004 sensors. Neither mir-1003 nor mir-1004 mirtron-expression construct could repress either mutant sensor (Figure 6A, lanes 9–11 and 16–18). These data demonstrate that mirtron products can repress targets via seed-matched sites, thereby acting as canonical miRNAs.
Mirtrons Require Ago1 to Repress Seed-Matched Targets
The biogenesis and regulatory properties of mirtrons strongly suggested that their products were incorporated into Ago complexes. We tested whether mirtron products could associate with Ago1, the primary effector of canonical miRNA-mediated regulation in Drosophila (Okamura et al., 2004). We immunoprecipitated (IP-ed) endogenous Ago1 from 0–10 hr embryos and subjected the associated RNAs to northern analysis. As shown in Figure 5D, endogenous mature miR-1003 and miR-1010 co-IPed with endogenous Ago1 protein. Specificity of these interactions was demonstrated by the failure of control T7 antibody to co-IP mirtron-derived small RNAs and the failure of Ago1 to coIP 30 nt 2S rRNA. Since the enrichment of mirtron-derived small RNAs in the IP fraction was less than the observed enrichment of Ago1, however, this left open the possibility that a population of these small RNAs might associate with other partners such as Ago2.
We then examined the functional consequences of Ago knockdown on the ability of mirtrons to regulate seed-matched targets. As a control, we examined the effect of GFP, Ago1, and Ago2 dsRNAs on the ability of miR-279 to regulate a luciferase-nerfin 3′ UTR sensor, which contains at least five miR-279-binding sites (Stark et al., 2003). As seen in Figure 6B, lanes 1–3, knockdown of Ago1, but not Ago2, derepresses the nerfin sensor in the presence of ectopic miR-279. We then analyzed a target bearing four bulged sites for miR-1010 (“miR-1010mi sensor”). We observed that knockdown of Ago1, but not Ago2, also derepresses this sensor in the presence of the mir-1010 mirtron-expression construct (Figure 6B, lanes 4–6). Therefore, while we do not exclude that mirtrons might also function via Ago2, our data provide evidence that small RNAs derived from mirtron hairpins associate with Ago1 to regulate seed-matched targets.
Mirtrons Exhibit Negative Regulatory Activity in Transgenic Drosophila
With these tissue culture data in hand, we challenged mirtrons to regulate target genes in the animal. We used a transgenic assay in which the expression of a ubiquitously expressed GFP “sensor” is tested for modulation by ectopic miRNAs provided in a spatially restricted pattern (Stark et al., 2003). For these tests, we used ptc-Gal4, which is active in a stripe of cells at anterior-posterior compartment boundaries. Specific downregulation of GFP in the ptc > miRNA “stripe” reflects an in vivo miRNA:target relationship.
We created transgenic strains carrying tub-GFP-miR-1003 or tub-GFP-miR-1004 sensors and a UAS-DsRed-mir-1004 mirtron-expression construct. Ectopic mir-1004 had no effect on a functional GFP sensor for miR-7 (Lai et al., 2005; Stark et al., 2003), demonstrating specificity of the assay (Figures 6C and 6D). On the other hand, miR-1004 strongly suppressed its perfect sensor (Figures 6E and 6F) and weakly suppressed the imperfect, seed-matched, miR-1003 sensor (Figures 6G and 6H). These data constitute stringent evidence that mirtrons are processed into functional species that can inhibit both perfectly matched and seed-matched targets in vivo.
DISCUSSION
A New Class of Progenitor Generates miRNA-Type Regulatory RNAs
We have characterized a class of intronic hairpins, termed mirtrons, that generate ∼22 nt regulatory RNAs in Drosophila. The biogenesis of mirtrons is distinct from that of canonical miRNAs. Although alternate mechanisms are not excluded, our data points to a mechanism in which mirtron maturation bypasses cleavage by the pre-miRNA-generating enzyme Drosha but is instead initiated by splicing and intron lariat debranching (Figure 7). This differs explicitly from the processing of canonical intronic miRNA genes, whose cleavage by Drosha occurs prior to host intron splicing (Kim and Kim, 2007). However, the mirtron pathway merges with the canonical miRNA pathway to generate active regulatory RNAs, since debranched mirtrons are productive substrates of Exportin-5 and the Dicer-1/loqs system, yielding small RNAs that can repress target transcripts (Figure 7). We showed specifically that mirtron-derived small RNAs can associate with Ago1 and require Ago1 to regulate seed-matched targets.
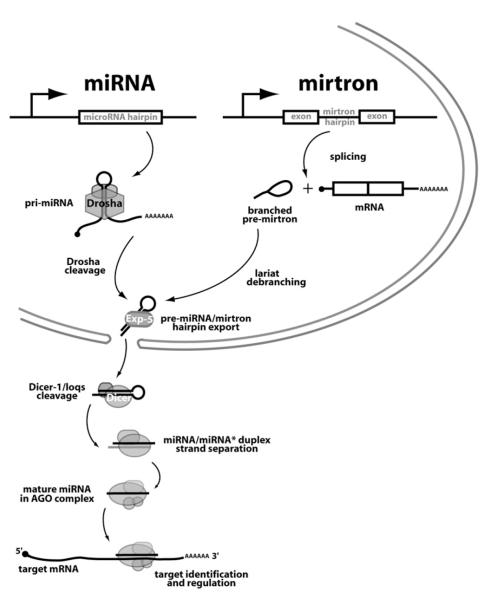
Figure 7. Model for the Convergence of the Mirtron and Canonical miRNA Pathways.
The canonical miRNA pathway initiates with the recognition and cleavage of pri-miRNA transcripts by the Pasha/Drosha complex to yield pre-miRNA hairpins. Our data support the existence of an alternate pathway in which short introns with hairpin potential are spliced and debranched to yield mirtron hairpins. Both pre-miRNA and mirtron hairpins are exported from the nucleus by Exportin-5 and cleaved by Dicer-1/loqs to generate ∼22 nt RNA duplexes. One strand, the active miRNA, is transferred to an Ago complex and guides it to repress fully complementary or seed-matched target transcripts.
The functional similarity between mirtrons and miRNA precursors is bolstered by our observation that miR-10-3p and the small-RNA product of a mirtron hairpin in Vha-SFD are extensively related across their 5′ halves, are derived from the same (right-hand) hairpin arm, are the most abundant products of their respective hairpins, and have the same seed (positions 2–8, AAAUUCG) (Figure S5). In accord with recent nomenclature implemented by the Bartel lab (Ruby et al., 2007), we categorize the small-RNA products of mirtrons as a novel subclass of miRNAs. Our data support and extend their findings with regard to the fundamental properties of mirtrons (Ruby et al., 2007).
Fourteen mirtron loci were identified (Ruby et al., 2007) from a high-throughput sequencing effort that confidently identified 133 canonical miRNA genes (J.G. Ruby, W. Johnston, D. Bartel, and E.C.L., unpublished data); thus, mirtrons constitute a considerable fraction of total miRNA genes in Drosophila. On the other hand, while a majority of canonical miRNA genes are well-conserved among the sequenced Drosophilids, most mirtrons arose recently during evolution. Since newly evolved miRNAs are thought to have fewer targets than highly conserved miRNAs (Chen and Rajewsky, 2007; J.G. Ruby, A. Stark, W. Johnston, M. Kellis, D. Bartel, and E.C.L., unpublished data), the regulatory networks involving mirtrons may be proportionally smaller than those mediated by canonical miRNAs. Still, our findings that both “old” and “young” mirtrons (1) produce miRNAs that associate with Ago1 (Figure 5), (2) can actively repress minimally paired seed targets (Figure 6), and (3) display patterns of divergence on microevolutionary scales that indicate their incorporation into endogenous regulatory networks (Figures 2 and S1) together suggest that mirtrons exert appreciable effects on biological networks. Indeed, the relative ease with which mirtrons have been born and/or lost raises the intriguing possibility that the changing mirtronic content of Drosophila genomes has contributed to fly speciation.
Interpreting the Consequences of Mutations in Small-RNA Processing Enzymes
The existence of mirtrons has implications for the interpretation of miRNA genetics. It is now recognized that the Dicer mutant condition does not solely reflect the loss of miRNAs, since Dicer has additional roles in chromatin dynamics and/or processing of exogenous or other endogenous dsRNA, depending on the organism. Drosha mutant cells do not accurately reflect the loss of miRNAs either; since Drosha processes other ncRNAs, including rRNAs (Wu et al., 2000). More recently, it was suggested that DGCR8/Pasha mutant cells more purely reflect a “miRNA null” state (Wang et al., 2007). This may not be the case either, because the mirtron pathway generates a subclass of miRNAs via a nuclear pathway that is largely, if not completely, distinct from the microprocessor. Therefore, caution should be exercised when using processing-enzyme mutants to assess the contribution of small RNAs to a given biological process.
Do Mirtrons Exist in Other Species?
Our data demonstrate that the Drosophila mirtron pathway merges the splicing/debranching pathway with the dicing pathway to generate functional miRNAs. Since the key parts of this hybrid small-RNA pathway are deeply conserved mechanisms for RNA processing, it seems plausible that mirtrons may exist outside of Drosophila. Since debranched introns are normally quite labile; however, we hypothesize that critical to the operation of the mirtron pathway is a dedicated mechanism to hand-off debranched introns to the hairpin export machinery. Having such a mechanism in place may prove key to the existence of mirtrons in other species.
EXPERIMENTAL PROCEDURES
Mirtron- and miRNA-Expression Constructs
Polymerase chain reaction was used to generate ∼400 nt pri-mirtron and pri-miRNA fragments using Canton S genomic DNA, which we cloned into the 3′ UTR of UAS-DsRed. To express mirtrons from a normal coding exonic context, we created a UAS-DsRed-Asc I-intron-Not I-2xmyc construct. We then cloned appropriate pairs of oligos into this vector to create wild-type and mutant mirtron-expression constructs. To express miR-1003 from a pri-miRNA backbone, we made structurally conservative nucleotide changes to a 160 nt pri-mir-6-1 backbone. Primer sequences and detailed cloning strategy used to generate all of these constructs are available in the Supplemental Data.
Analysis of Mirtron Maturation
To analyze endogenous small RNAs, we isolated small RNAs from staged Canton S animals or cultured S2 cells using Trizol (Life Technologies). To analyze exogenously expressed mirtrons and miRNAs, we transfected 2 × 106 S2 cells with 0.25 μg of ub-Gal4 and 0.5 μg UAS-DsRed-mirtron plasmids using Effectene (QIAGEN) in 6-well plates and extracted total RNA 2 days later. Northern analysis was performed by separating 20 μg of total RNA per lane on 12% polyacrylamide gels, transferring to GeneScreen Plus (Perkin Elmer), and probing with γ-32P -labeled LNA oligonucleotides (Exiqon) antisense to miR-1003 (CTGTGAATATGTAAATGTGAGA), miR-1010 (CTGCAAATGGAACGATAGGTGAAA), and miR-1008 (CTGTAAACACAAAAAGCTGTGA) or DNA oligonucleotide antisense to 2S rRNA (TACAACCCTCAACCATATGTAGTCCAAGCA).
To study the effect of dsRNA knockdowns on endogenous small RNAs, we soaked 2 × 106 S2 cells in 6-well plates with 20 μg/ml dsRNA. GFP, Drosha, Dicer-1, loqs, and Dicer-2 dsRNAs were produced using published pLitmus templates (Forstemann et al., 2005) and T7 Megascript (Ambion). The cloning of Ldbr, Exportin-5, Ago1, and Ago2 pLitmus templates is described in the Supplemental Data; we adopted Ldbr as an abbreviation for lariat-debranching enzyme to avoid confusion with the debra (dbr) gene. To analyze exogenously produced small RNAs, we treated S2 cells with dsRNA for 4 days and then transfected them with ub-Gal4 and UAS-DsRed-mirtron or UAS-DsRed-miRNA plasmids. Cells were then incubated with dsRNA for an additional 2 days before preparing RNA for northern analysis. To inhibit debranching in flies, we crossed a third chromosome insertion of UAS-LdbrRNAi (Conklin et al., 2005)to da-Gal4 and analyzed their transheterozygous progeny. Knockdown of Ldbr activity was assessed by Q-PCR of the actin intron relative to control rp49 sequence in GFP, Drosha, and Ldbr dsRNA-treated cells using SYBR Green (ABI) and a MyiQ Real-Time PCR Detection System (BioRad). Primers are listed in the Supplemental Data. To analyze the association of mirtron products with Ago1, we followed a published protocol (Miyoshi et al., 2005) as described in the Supplemental Data.
Luciferase Assay for Mirtron Regulatory Activity
To generate mirtron targets, we inserted two copies of a sequence that was antisense to miR-1003, miR-1010, or miR-1004 downstream of the renilla luciferase coding region in psiCHECK2 (Promega); this vector contains an internal firefly luciferase gene that serves as an internal control. Control mutant sensors for miR-1003 and miR-1004 contained three point mutations in the seed-match region; a miRNA-type sensor for miR-1010 contained four copies of a bulged target site. Primer sequences are available in the Supplemental Data. We performed quadruplicate transfections of 25 ng target, 12.5 ng ub-Gal4, and 25 ng UAS-DsRed-mirtron plasmids into 1 × 105 S2 cells in 96-well format. Three days later, we lysed the cells and subjected them to dual luciferase assay (Promega) and analyzed these on a Veritas plate luminometer (Turner Biosystems). Statistical analysis was performed using the equal variance Student’s t test.
For RNAi-luciferase assays, 1 × 106 S2R+ cells were seeded per well (12-well plate) in 500 μl of serum-free media. dsRNA was added to a concentration of 15 μg/ml. After 1 hr incubation, an equal volume of media containing 20% FBS was supplemented. After 4 days, cells were seeded 1 × 106 cells per well (96-well plate), and reporter and miRtron overexpression constructs were transfected. After 12 hr, 0.75 μg dsRNA was added to each well, and cells were lysed 2 days after transfection to measure luciferase activity. We performed two replicates of quadruplicate transfections, and these data were analyzed using the equal variance Student’s t test.
Imaginal Disc Sensor Assay for Mirtron Activity
We generated P element-mediated insertions of UAS-DsRed-mirtron and tub-GFP-target transgenes according to standard methods (Best-Gene, Inc.); three to five different insertions were examined for each construct. Previously described transgenes include tub-GFP-miR-7 target (Stark et al., 2003) and ptc-Gal4 (obtained from the Bloomington Stock Center). Sensor assays were performed by dissecting wing imaginal discs from the appropriate genotypes and staining them with rabbit α-GFP (Molecular Probes, 1:1250) followed by Alexa 488- mouse α-rabbit (1:500, Molecular Probes).
Supplementary Material
supp data
ACKNOWLEDGMENTS
We thank Antonio Javier-Lopez for Ldbr reagents, Phil Zamore for dsRNA templates, Greg Hannon for Dcr-1 and Drosha antibodies, Haruhiko and Mikiko Siomi for Ago1 antibody, Stephen Cohen for the miR-7 sensor, and the Bloomington Stock Center for da-Gal4 and ptc-Gal4. Chun-Hong Chen provided advice about the pri-miRNA swap experiment. We are especially grateful to Graham Ruby and David Bartel for communicating unpublished observations on the existence of mirtrons in a collection of 454 Drosophila small-RNA sequences. K.O. was supported by a grant from the Japan Society for the Promotion of Science. J.W.H. was supported by the Mildred and Emil Holland Scholarship, the Margaret and Herman Sokol Fellowship, and NIH MSTP grant GM07739. E.C.L. was supported by the Leukemia and Lymphoma Foundation, the Burroughs Wellcome Foundation, and the V Foundation for Cancer Research.
Footnotes
REFERENCES
- Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of MicroRNA-Target Recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Conklin JF, Goldman A, Lopez AJ. Stabilization and analysis of intron lariats in vivo. Methods. 2005;37:368–375. doi: 10.1016/j.ymeth.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du T, Zamore PD. microPrimer: the biogenesis and function of microRNA. Development. 2005;132:4645–4652. doi: 10.1242/dev.02070. [DOI] [PubMed] [Google Scholar]
- Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli A, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Grun D, Wang Y-L, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comp. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli A, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting R, Fischer S, Bernstein E, Sijen T, Hannon G, Plasterk RH. Dicer functions in RNA interference and in synthesis of small RNAs involved in developmental timing in C. elegans. Genes Dev. 2001;15:2654–2659. doi: 10.1101/gad.927801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J. 2007;26:775–783. doi: 10.1038/sj.emboj.7601512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development. 1997;124:4847–4856. doi: 10.1242/dev.124.23.4847. [DOI] [PubMed] [Google Scholar]
- Lai EC, Posakony JW. Regulation of Drosophila neurogenesis by RNA:RNA duplexes? Cell. 1998;93:1103–1104. doi: 10.1016/s0092-8674(00)81454-3. [DOI] [PubMed] [Google Scholar]
- Lai EC, Burks C, Posakony JW. The K box, a conserved 3′ UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development. 1998;125:4077–4088. doi: 10.1242/dev.125.20.4077. [DOI] [PubMed] [Google Scholar]
- Lai EC, Tomancak P, Williams RW, Rubin GM. Computational identification of Drosophila microRNA genes. Genome Biol. 2003;4:R42. doi: 10.1186/gb-2003-4-7-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N, Lim L, Weinstein E, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004a;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA Silencing Pathways. Cell. 2004b;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss EG, Lee RC, Ambros V. The cold shock domain protein LIN-28 controls developmental timing in C. elegans and is regulated by the lin-4 RNA. Cell. 1997;88:637–646. doi: 10.1016/s0092-8674(00)81906-6. [DOI] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA. 1998;4:1096–1110. doi: 10.1017/s1355838298980785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Müller P, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, Rougvie A, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JG, Jan C, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi MC. Processing of pre-microRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA Targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Bushati N, Russell RB, Cohen SM. Animal MicroRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- Wu H, Xu H, Miraglia LJ, Crooke ST. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 2000;275:36957–36965. doi: 10.1074/jbc.M005494200. [DOI] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supp data