Structure of the Siz/PIAS SUMO E3 ligase Siz1 and determinants required for SUMO modification of PCNA (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 11.
Summary
Siz1 is a founding member of the Siz/PIAS RING family of SUMO E3 ligases. The x-ray structure of an active Siz1 ligase revealed an elongated tripartite architecture comprised of an N-terminal PINIT domain, a central zinc-containing RING-like SP-RING domain, and a C-terminal domain we term the SP-CTD. Structure-based mutational analysis and biochemical studies show that the SP-RING and SP-CTD are required for activation of the E2~SUMO thioester while the PINIT domain is essential for redirecting SUMO conjugation to the proliferating cell nuclear antigen (PCNA) at lysine 164, a non-consensus lysine residue that is not modified by the SUMO E2 in the absence of Siz1. Mutational analysis of Siz1 and PCNA revealed surfaces on both proteins that are required for efficient SUMO modification of PCNA in vitro and in vivo.
Keywords: SUMO, signal transduction, replication, RING E3, PIAS, SIZ, ubiquitin, Ubc9
Introduction
Post-translational modification by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins such as SUMO (small ubiquitin-like modifier) requires the sequential action of E1 activating enzymes, E2 conjugating enzymes and E3 ligases (Hershko and Ciechanover, 1998; Johnson, 2004). The SUMO pathway includes a single E2 that exhibits specificity by directly interacting with substrates that contain the consensus sequence ψ-K-x-D/E where ψ is hydrophobic, K is the lysine conjugated to SUMO, x is any amino acid, and D/E is an acidic residue (Bernier-Villamor et al., 2002; Sampson et al., 2001). While the SUMO E2 can mediate substrate specificity and catalyze SUMO conjugation, SUMO E3 ligases enhance conjugation to SUMO consensus motifs or redirect the specificity of the E2 to promote conjugation to non-consensus lysine residues.
Two distinct classes of SUMO E3 ligases have been identified thus far. The first includes the IR1-M-IR2 domains of the nucleoporin RanBP2/Nup358 (Pichler et al., 2002) and is unique to the SUMO pathway in that it bears no resemblance to either RING or HECT type ubiquitin E3 ligases. Structural and biochemical evidence suggests that the IR1-M domain enhances SUMO E3 activity but makes no contacts to the substrate, so the RanBP2/Nup358 E3 is proposed to enhance SUMO conjugation by aligning the E2~SUMO thioester complex in an optimal configuration to promote catalysis and enable substrate interaction with the active site of the SUMO E2 Ubc9 (Reverter and Lima, 2005). The second class encompasses the SP-RING E3 ligase family which includes human PIAS proteins (PIAS1, PIAS3, PIASxa/β, and PIASy; Kahyo et al., 2001; Sachdev et al., 2001; Schmidt and Muller, 2002) and their yeast homologues (Siz1, Siz2, Mms21 and Zip3; Johnson and Gupta, 2001; Takahashi et al., 2001a; Zhao and Blobel, 2005; Cheng et al., 2006). These E3 ligases contain a conserved SP-RING domain that shares sequence similarity to RING domains of ubiquitin E3 ligases and are thus proposed to function in a similar manner, namely by utilizing the SP-RING domain to recruit the cognate E2 into a complex with the substrate to facilitate conjugation.
The SUMO E3 ligases Siz1 and Siz2 enhance SUMO modification to many substrates in vivo, but they can also exhibit unique substrate specificity (Reindle et al., 2006). Siz1-dependent substrates include Cdc3 and Cdc11 (septin subunits), Prp45 (a splicing factor) and the proliferating cell nuclear antigen (PCNA). While SUMO conjugation to PCNA Lys127 can be detected in cells lacking Siz1, SUMO conjugation to PCNA Lys164 is strictly dependent on Siz1 both in vitro and in vivo (Pfander et al., 2005; Windecker and Ulrich, 2008). A chimera between Siz1 and Siz2 uncovered distinct roles for the N- and C-terminal domains of Siz1 during substrate selection in vivo (Reindle et al., 2006). While a C-terminal Siz1 fragment targeted Siz1 to septins, the N-terminal Siz1 PINIT domain was required for SUMO conjugation to PCNA.
PCNA coordinates DNA replication and DNA repair by integrating signal transduction pathways at the replication fork that include post-translational modification by ubiquitin and SUMO (reviewed in Moldovan et al., 2007). In response to DNA damage, Rad6 (E2) and Rad18 (E3) modify PCNA Lys164 with a single ubiquitin which serves to recruit trans-lesion polymerases such as pol η, pol ζ and pol ί to facilitate error-prone DNA replication. In other situations when error-free replication is favored, PCNA Lys164 is conjugated to a Lys63-linked ubiquitin chain in a process dependent on Rad5 and the E2/UEV complex Ubc13/Mms2. During S-phase, yeast PCNA is preferentially conjugated to SUMO (Smt3 in yeast) on Lys164, the same lysine that is utilized for conjugation to ubiquitin (Hoege et al., 2002). SUMO modification of PCNA facilitates recruitment of the anti-recombinogenic helicase Srs2 (Pfander et al., 2005) which is proposed to inhibit recombination during replication by preventing Rad51 filament formation at the replication fork (Krejci et al., 2003; Veaute et al., 2003). When SUMO modification of PCNA is blocked, DNA lesions can be bypassed through a mechanism that entails recombination. More recently, SUMO modification of PCNA has been implicated in Rad18-Mms2-mediated damage bypass at sister chromatid junctions in a process that is coordinated with Rad51-dependent recombination (Branzei et al., 2008).
To uncover determinants required for SUMO modification by Siz1, we defined a minimal Siz1 ligase that was sufficient to enhance SUMO conjugation to PCNA Lys127 and redirect SUMO conjugation to the non-consensus PCNA Lys164 and determined its structure by x-ray crystallography. The Siz1 structure revealed a modular architecture that includes an N-terminal PINIT domain, a central SP-RING domain, and a unique C-terminal domain (SP-CTD). The SPRING domain shares structural similarity to ubiquitin E3 ligase RING and U-box domains and was sufficient, along with the SP-CTD, to activate the SUMO-E2 thioester for conjugation to substrates containing SUMO consensus motifs. The PINIT domain is juxtaposed to the SPRING domain in an ideal position to recruit substrates to the E2 and we determined that PINIT domain surfaces were required for SUMO conjugation to Lys127 as well as the non-consensus PCNA lysine residue Lys164. Structure-guided mutational analysis of Siz1, PCNA and SUMO coupled with single turnover kinetic assays and in vivo analysis support this model and reveal mechanisms that underlie SP-RING E3 ligase activity.
Results and Discussion
Activities of the Siz1 E3 ligase
A fragment of Siz1 (amino acids 112–465) was reported as sufficient for SUMO conjugation to a septin subunit (Cdc3) and PCNA (Figure 1A and Supplementary Figure 1; Takahashi and Kikuchi, 2005). This fragment included the PINIT and SP-RING domains, but it lacked a potentially important C-terminal SUMO interaction motif (SIM; aa 481–491). We compared SUMO ligase activities for Siz1(112–465; without SIM) and Siz1(112–508; with SIM), however no differences were detected for SUMO conjugation to PCNA at Lys164 under conditions of multiple turnover (not shown). To quantify ligase activity, single turnover assays were conducted to calculate the kinetic constants k2 (a measure of apparent maximum rate) and Kd (a measure of the apparent dissociation constant) (Supplementary Figure 1; Methods). Both constructs promoted SUMO conjugation to PCNA at Lys164 although the SIM-containing construct revealed a ~1.6-fold higher k2 but ~1.5-fold lower Kd (Supplementary Table 1). These data and those presented below suggest that 1) the Siz1 SIM is dispensable for conjugation to Lys164 and 2) Siz1(112–465) encompasses elements necessary for activating the E2 and for redirecting SUMO conjugation to the non-consensus Lys164 acceptor on PCNA.
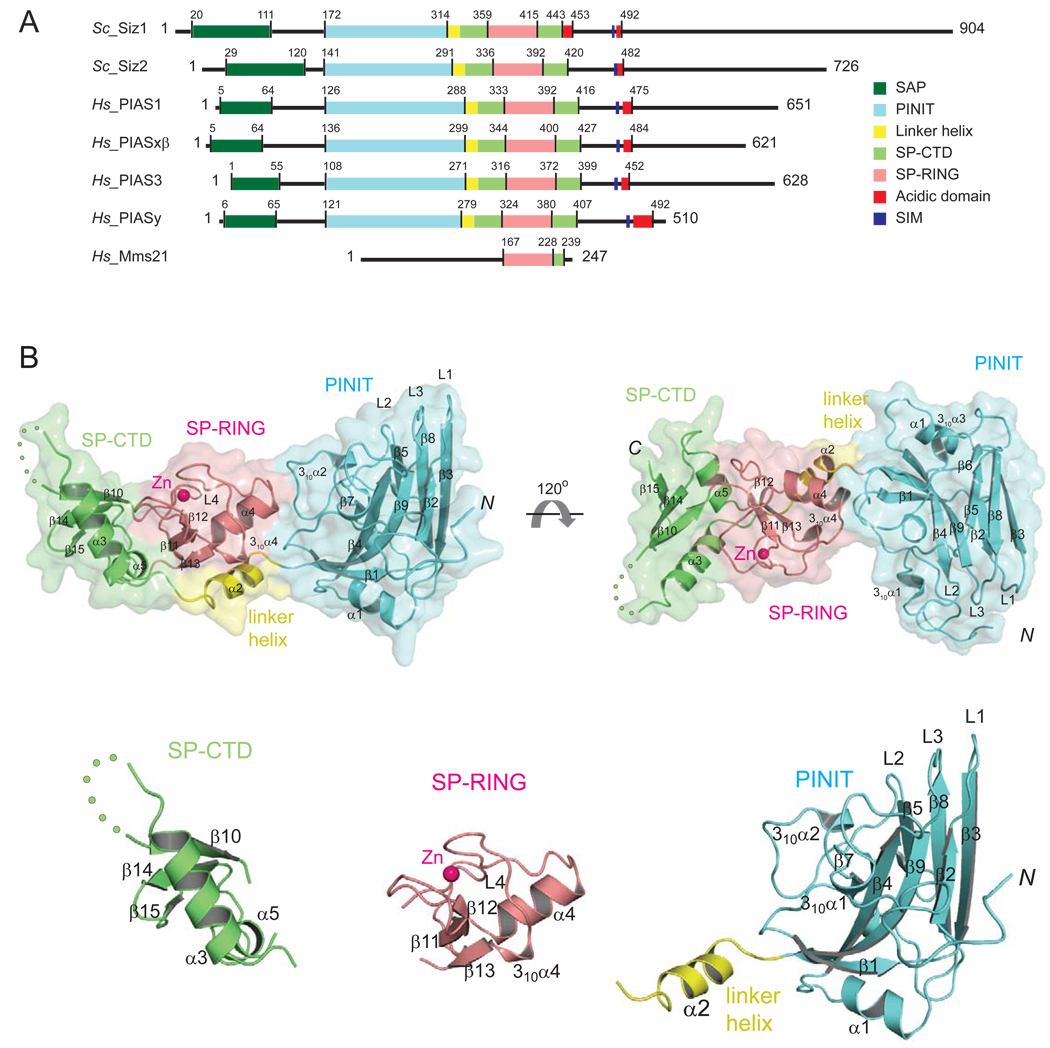
Figure 1. Siz/PIAS E3 ligase family members and structure of the Siz1 catalytic domain.
(A) SP-RING family E3 ligases from S. cerevisiae (Siz1, Siz2), H. sapiens (PIAS1, PIASxβ, PIAS3, and PIASy) and the related SP-RING ligase Mms21. Amino acid numbers and lines indicate boundaries between domains: SAP (dark green), PINIT (cyan), linker helix (yellow), SPRING (salmon), SP-CTD (light green), acidic domain (red) and the SIM motif (blue). (B) The Siz1 structure in ribbon representation enveloped by a transparent molecular surface (top panel) with domains colored as above. Secondary structural elements are labeled and numbered. Zn2+ is depicted as a pink sphere. Each disordered amino acid residue in the SP-CTD is indicated by a green dot. Termini are indicated as N and C, respectively. Bottom panel depicts structures of the separated Siz1 SP-CTD, SP-RING, and PINIT domains.
Structure of a minimal Siz1 E3 ligase
To determine the structural basis for Siz1 ligase activities, we purified and crystallized Siz1(112–465). A single wavelength data set was collected from a crystal containing selenomethionine-substituted Siz1(112–465) (Hendrickson, 1991) and used to calculate phases from the anomalous signal of one zinc and six selenium atoms (Methods). A model including residues 159–347 and 353–444 was built into electron density at 3.2 Å and refined to an Rcrys and Rfree of 21.1 and 25.9, respectively, using data collected from a native crystal that diffracted to 2.6 Å (Methods and Table 1; Supplemental Figure 2A and 2B). Amino acids 112–158, 347–352, and 445–465 were present in the crystal and presumed disordered. Based on a newly defined N-terminal boundary of the globular PINIT domain (aa 167), we designed a minimal construct for Siz1 (aa 167–465). Single turnover kinetic analysis revealed a ~2.7-fold higher apparent rate for Siz1(167–465) when compared to Siz1(112–465) (Supplementary Figure 1 and Supplementary Table 1). The lower activity observed for Siz1(112–465) could be attributed to the higher entropy of the disordered N-terminal amino acids (112–158) as evidenced by the lack of electron density for this segment in the structure. The minimal Siz1 construct (aa 167–465) was used for all subsequent biochemical studies.
Table 1.
Crystallographic Data Statistics
| Data Statistics | Native | SeMet |
|---|---|---|
| Source | APS 24ID | APS 24ID |
| Wavelength (Å) | 0.979 | 0.979 |
| Resolution (Å) | 40 – 2.6 (2.69-2.6) | 50 – 3.20 (3.31-3.20) |
| Space Group | P212121 | P212121 |
| Unit Cell (Å) a, b, c | 44.48, 88.98, 105.59 | 46.53, 89.9, 106.26 |
| # of observations | 69917 | 48529 |
| # of reflections | 13595 | 13835 |
| Completeness (%) | 97.8 (89.2) | 98.1 (94.5) |
| Mean I/σI | 22.0(2.6) | 18.6 (2.9) |
| Rmergea | 7.3(43.4) | 7.0 (28.4) |
| Cut-off criteria I/σI | −0.5 | 0 |
| FOM (SHARP) | 0.29 | |
| FOM (Solomon) | 0.92 | |
| Refinement Statistics | ||
| Resolution Limits (Å) | 34-2.6 (2.76-2.60) | |
| No. of reflections | 13193 | |
| Rcrysb/Rfree | 21.1 (35.1)/25.9 (36.9) | |
| # atoms protein/water | 2236/67 | |
| B-factors protein/water | 67.0/52.9 | |
| Bond r.m.s deviations length (Å)/angles (°) | 0.008/1.5 | |
| Ramachandran plotc | ||
| Most favored | 194 (78.5%) | |
| Additionally allowed | 50 (20.2%) | |
| Generously allowed | 3 (1.2%) | |
| Disallowed region | 0 (0%) |
The Siz1 structure revealed a rod shaped molecule composed of three domains (Figure 1B). The N-terminal PINIT domain (aa 172–315; colored cyan) is formed by two anti-parallel β-sheets, one includes β1, β2, β4, and β9 and the other includes β3, β5 and β8. The β-sheets are connected by protruding loops (L1, L2, and L3) that join strands β2–3, β4–5 and β8–9 at one end of the molecule, while β3–4 and β5–8 are connected by α-helix α1 and a loop, respectively, on the opposite surface. The five amino acid PINIT motif is located near β7 within the hydrophobic core of the domain (Supplemental Figure 5). The signature SP-RING domain is located in the middle of the protein (aa 360–415; colored salmon) between the PINIT and SP-CTD domains. The SP-RING domain includes a β-sheet composed of β11, β12 and β13, and α-helix (α4) while other conserved elements of the SP-RING domain participate in coordination of a single Zn2+ ion (see below). The final element in the Siz1 structure is a domain we term the Siz1 C-terminal domain or SP-CTD (colored green, Figure 1). The SP-CTD includes aa 328–359 and aa 415–443 that form a three-stranded β-sheet (β10, β14 and β15) that is wedged between α-helices α3 and α5 which link the SP-CTD to the linker helix (α2) and SP-RING domain, respectively. The PINIT and SP-CTD are connected by α-helix α2 (colored yellow, Figure 1) which cradles the SP-RING domain. The SP-RING domain shares extensive and hydrophobic contacts with the PINIT and SP-CTD domains which bury a total surface area of 1050 Å2 and 1550 Å2 at the respective interface.
Activities of the Siz1 SP-RING and SP-CTD domains
Siz1 and Siz2 account for the majority of SUMO modification in yeast and most substrates do not exhibit a preference for either Siz1 or Siz2 (Reindle et al., 2006). The absence of E3-mediated substrate specificity suggests that Siz1 and Siz2 can contribute to SUMO conjugation through activation of the E2~SUMO thioester to promote SUMO transfer to substrate lysine residues within SUMO consensus motifs where substrate specificity is dictated by the E2, Ubc9. This activity is perhaps reminiscent of that proposed for the IR1-M-IR2 domains of the Nup358/RanBP2 E3 ligase, domains that enhance SUMO modification but do not alter E2-dependent substrate specificity (Pichler et al., 2004; Reverter and Lima, 2005).
Yeast PCNA is modified by SUMO on Lys127 and Lys164. Lys127 falls within a SUMO consensus motif and can be modified directly by the E2 Ubc9 although modification at Lys127 can be enhanced by Siz1. In contrast, Lys164 is not a substrate for the E2 Ubc9 and its modification is strictly dependent on the Siz1 E3 ligase. These observations suggest that conjugation to Lys164 requires Siz1 to bridge the substrate and E2~SUMO in a complex that bypasses E2 substrate specificity. Although SUMO conjugation to Lys164 still requires the E2, lysine specificity is now dictated by the Siz1 E3 ligase.
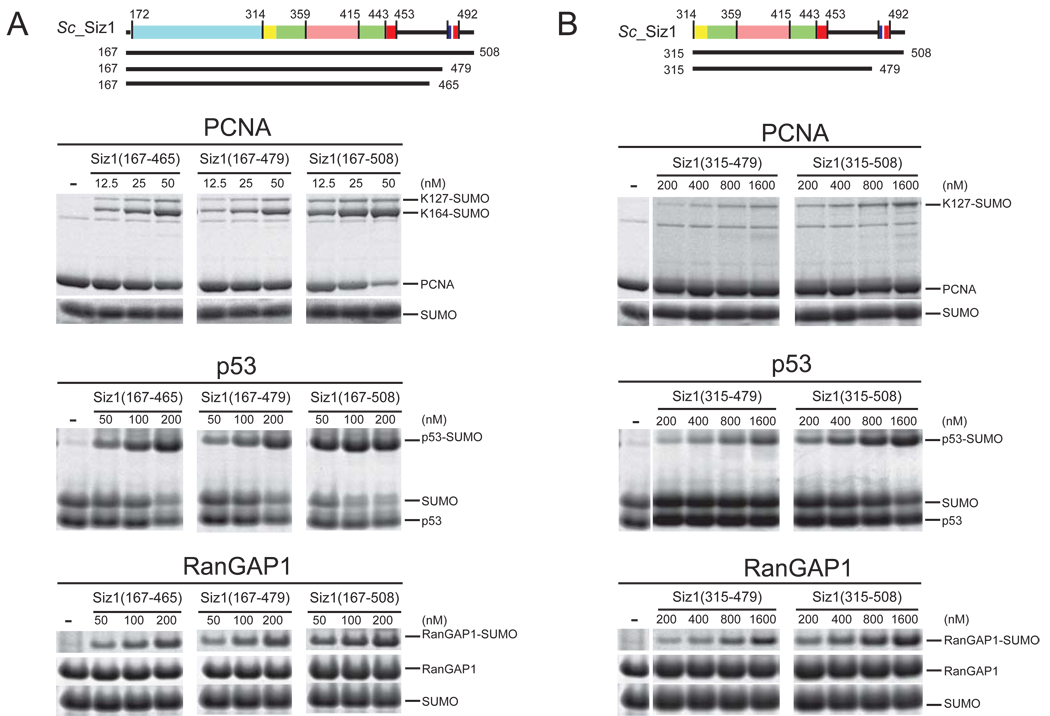
To uncouple these two Siz1 activities, we compared the ligase activities of Siz1(167–508) which contained the PINIT, SP-RING, SP-CTD and SIM domains and Siz1(315–508) which lacked the PINIT domain but retained the SP-RING, SP-CTD, and SIM domains (Figure 2A and 2B). To evaluate the role of the SIM domain, we generated Siz1(167–479) and Siz1(315–479) which lacked the SIM motif. Attempts to generate Siz1(315–465) for comparison to Siz1(167–465) failed as we were unable to obtain Siz1(315–465) as a monodisperse preparation. A comparison between Siz1(167–465) and Siz1(167–479) revealed similar activities suggesting that the 14 additional C-terminal residues do not alter E3 ligase activity (Figure 2A).
Figure 2. Siz1 E3 ligase activities and deletion of the PINIT and SIM domains.
(A) Schematic of Siz1 as in Figure 1A with solid bars indicating termini for proteins used in the assays below. SDS-PAGE for SUMO conjugation to PCNA (top), p53 (middle) and RanGAP1 (bottom) using Siz1(167–465), Siz1(167–479) or Siz1(167–508). (B) Similar to (A) for SUMO conjugation assays using Siz1(315–479) or Siz1(315–508). Each lane depicts the products after 1 hour at 37°C with Siz1 at the indicated concentrations. Proteins detected by SYPRO Ruby. Substrates and products are labeled.
Siz1 and the PINIT-less Siz1 constructs were assessed for ligase activity using three substrates that contain SUMO consensus motifs: yeast PCNA (Lys127), the human p53 C-terminal domain (aa 320–393; Lys386) and the human RanGAP1 C-terminal domain (aa 418–587; F562A/K565A; Lys524). Siz1(315–508) and Siz1(315–479) enhanced SUMO conjugation to p53, RanGAP1 and PCNA (K127) on lysine residues within respective SUMO consensus motifs but failed to promote SUMO conjugation to PCNA Lys164 (Figure 2B). In contrast, Siz1(167–508), Siz1(167–479) and Siz1(167–465) promoted conjugation to all of the substrate lysine residues including PCNA Lys164. It is worth noting that reactions containing Siz1(315–508) or Siz1(315–479) required higher concentrations of the E3 to obtain similar levels of SUMO conjugated product when compared to the respective Siz1 constructs that contained the PINIT domain (compare Figure 2A and 2B). Lower activities observed for Siz1 constructs lacking the PINIT domain could be due to structural destabilization of the SP-RING domain, a distinct possibility given the extensive hydrophobic interface observed between the PINIT and SP-RING domains (see above). These data show that Siz1(315–508) and Siz1(315–479) were sufficient to enhance SUMO conjugation to substrates that contain SUMO consensus motifs.
The activities of Siz1 proteins containing the SIM motif suggest that the SIM motif can stimulate SUMO conjugation by ~2–4 fold independent of the PINIT domain under conditions of multiple turnover (Figure 2). In comparison, we only observed a 1.6-fold stimulation for SUMO conjugation to PCNA under conditions of single turnover (Supplemental Table 1 and Supplemental Figure 1). Since the SIM motif appears less important during single turnover in comparison to multiple turnover, this phenomenon might be explained by the SIM’s ability to recruit the next charged E2~SUMO to the E3 complex. This activity would contribute less to the first round of conjugation (single turnover) but would enhance conjugation under multiple turnover conditions where the E3 is required to recruit multiple E2~SUMO to perform multiple rounds of conjugation.
The Siz/PIAS RING domain shares structural similarity to RING and U-box domains
The SP-RING domain is proposed to bind the SUMO E2 enzyme and is highly conserved (> 36% seq. id.; Figure 3A) in all Siz/PIAS proteins. In support of this function, direct interactions have been detected between the SP-RING domain and Ubc9 by pull-down and yeast-two hybrid analysis (Kahyo et al., 2001; Takahashi and Kikuchi, 2005). A partial match to the signature RING motif CX2CX(9–39)CX(1–3)HX(2–3)C/HX2CX(4–48)CX2C was recognized in sequence alignments; thus the SP-RING domain was postulated to be the structural and functional homologue of RING domains in ubiquitin RING-type E3 ligases (Johnson and Gupta, 2001).
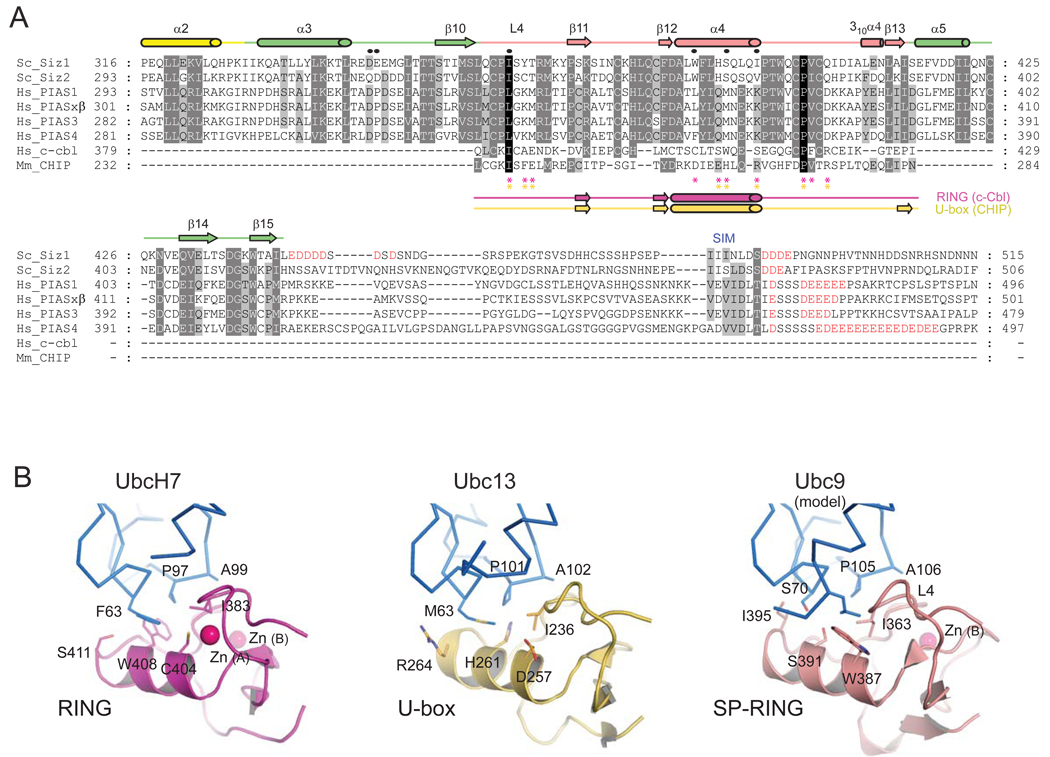
Figure 3. Sequence alignment of SP-RING and SP-CTD and a structural model for SP-RING:Ubc9.
(A) Amino acid alignment for SP-RING and SP-CTD domains from indicated Siz/PIAS family members with acidic residues C-terminal to the SP-CTD colored red. Structure-based sequence alignment for SP-RING, c-Cbl RING, and CHIP U-box (below). Secondary structural elements are labeled and shown above the alignment for Siz1 (color coded as in Figure 1) or below the alignment for c-Cbl RING (magenta) and CHIP U-box (gold). Circles indicate Siz1 residues subjected to mutational analysis. Asterisks indicate residues that contact the E2s in c-Cbl RING (magenta) and CHIP U-box (gold). (B) Structures of the c-Cbl RING:UbcH7 (left; magenta; PDB code 1FBV), CHIP U-box:Ubc13 (middle; gold; PDB code 2C2V) and SP-RING (salmon) with Ubc9 modeled (right) as described in the text. UbcH7, Ubc13 and Ubc9 in stick representation (blue). Residues in the E2:E3 interface in are shown in stick representation and labeled. Zn2+ ions are shown as pink spheres.
The Siz1 SP-RING domain can be aligned to the RING domain from c-Cbl (1.8 Å rmsd over 48 residues with 18% seq. id.; DALI) and the U-box domain from CHIP (1.8 Å rmsd over 52 residues with 17% seq. id.; DALI). A side-by-side comparison of the structures reveals their architectural similarities but it also highlights some important differences (Figure 3B). While RING domains coordinate two Zn2+ ions with a cross-brace architecture (Jackson et al., 2000), the SP-RING domain contains only one Zn2+ ion in site B which is coordinated in a tetrahedral configuration by Cys377, His379, Cys400 and Cys403 (Supplementary Figure 2C and 2D). The structural integrity of the SP-RING domain is likely important for E3 ligase activity because mutation of any side chain that coordinates the Zn2+ ion results in complete loss of activity (Kahyo et al., 2001; Takahashi et al., 2001b).
Three of the four amino acid side chains that coordinate the Zn2+ ion at site A in RING proteins are lacking in Siz/PIAS proteins and are instead substituted to aspartate, serine and tryptophan in Siz1 (Supplementary Figure 2C and 2D). Despite the missing Zn2+ ion, the fold and conformations of the adjacent loops are similar because of a stabilizing network of interactions that include bidendate hydrogen bond contacts between Cys361 Sγ and the backbone amide atoms of Asp384 and Ile363, bidentate hydrogen bond contacts between Asp384 Oδ and backbone amide atoms of Lys369 and Tyr370, and bidentate hydrogen bond contacts between the Ser364 hydroxyl and the Tyr365 and Thr366 backbone amide atoms. Hydrophobic contacts were also observed between the Ser364 Cβ and the aromatic ring of Trp387 (Supplementary Figure 2D). This network of interactions is reminiscent of those in U-box proteins such as CHIP which fold into stable RING-like conformations in the absence of Zn2+ (Zhang et al., 2005).
Mutational analysis and modeling a Siz1-E2~SUMO complex
Analysis of the E3:E2 complexes c-Cbl:UbcH7 (Figure 3B; left panel; Zheng et al., 2000) and CHIP U-box:Ubc13 (Figure 3B; middle panel; Zhang et al., 2005) suggests that RING and RING-like U-box proteins utilize a common surface to recruit their cognate E2 enzymes. E2s share a common mechanism and are highly related at the structural level within the conserved E2 fold. Structural similarities observed for E2 enzymes and SP-RING, RING, and U-box domains enabled superposition of Ubc9 onto Ubc13 and UbcH7 to model SP-RING:Ubc9 interactions (Figure 3B; right panel) indicating that Siz1 Ile363, Trp387, Ser391 and Ile395 were ideally positioned to contact Ubc9. To test this model, Trp387, Ser391, Ile395 and Ile363 were selected for mutational analysis (Figure 3A and 3B). Individual side chain substitution for each residue decreased SUMO conjugation to each lysine residue in p53 and PCNA (Figure 4A and 4B). These data and the structural similarities between E2s and RING, U-box, and SP-RING domains support the hypothesis that Ubc9 interacts with the SP-RING domain in a manner similar to that observed in other E2:RING and E2:U-box complexes (Figure 3B).
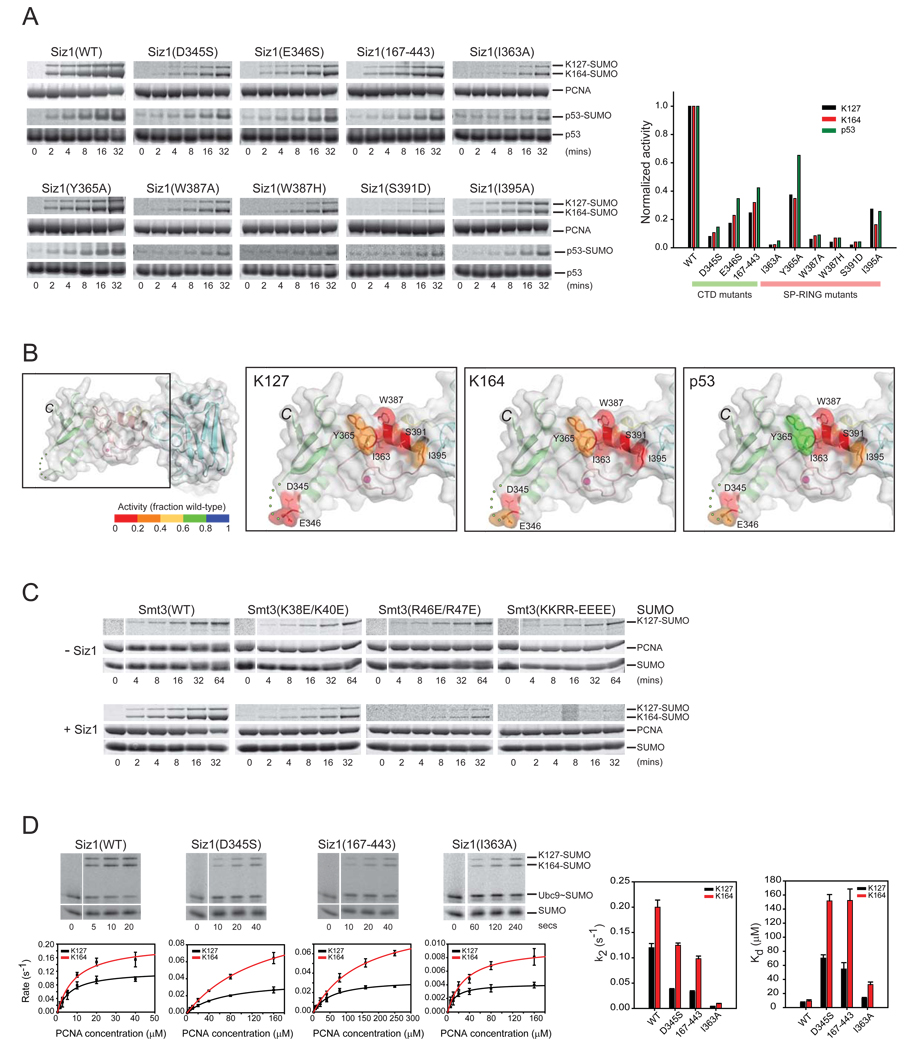
Figure 4. SP-RING, SP-CTD and SUMO surfaces contribute to Siz1-dependent SUMO conjugation of PCNA.
(A) SDS-PAGE and time course (left) and graph depicting rates in the linear range of the assay (right). Rates normalized to one to enable a comparative analysis for SUMO conjugation to p53, PCNA at K127 and K164 with wild-type and mutant Siz1 isoforms. Proteins detected and quantified by SYPRO Ruby (Methods). (B) SP-RING and SP-CTD domains of Siz1 with amino acids subjected to mutational analysis in stick representation and Siz1 surface color-coded according to the level of activity (inset color bar) for SUMO conjugation to PCNA Lys127 (second from left), PCNA Lys164 (second from right), and p53 (right panel). (C) SDS-PAGE and time course for SUMO conjugation to PCNA with wild-type or mutant SUMO isoforms in the absence of Siz1 with 5 µM Ubc9 (top) or presence of Siz1 with 100 nM Ubc9 (bottom). (D) SDS-PAGE and time course for SUMO conjugation to PCNA with wild-type or mutant Siz1 isoforms under conditions of single turnover (left). Proteins detected by fluorescence (Alexa Fluor 488-SUMO; Methods). The initial rate of reaction (s−1) versus PCNA concentration (µM) is shown below each gel. Bar charts (right) for Kd (µM) and k2 (s−1) for indicated Siz1 isoforms. Error bars represent ±1 standard deviation.
We proposed that activation of the E2~SUMO thioester requires an E3 to bind and coordinate both E2 and SUMO to optimally configure the E2~SUMO thioester bond with respect to the incoming nucleophilic lysine substrate (Reverter and Lima, 2005). We posit that the quaternary complex between RanGAP1 (substrate), the E2 Ubc9, SUMO, and the IR1* E3 domain from Nup358/RanBP2 represents a ligated product captured just after conjugation as evidenced by 1) the substrate lysine is in the same position as in substrate complexes (Bernier-Villamor et al., 2002; Yunus and Lima, 2006), 2) the catalytic E2 cysteine is only 3.5 Å from the isopeptide bond, and 3) the asparagine residue that forms the purported oxyanion hole interacts with the SUMO C-terminal carbonyl oxygen. This structure was used to generate a model for E2~SUMO in its activated conformation (Supplementary Figure 3B) and this model was docked onto Ubc9:Siz1 based on structures of other E2:RING complexes (see above).
This model predicted that acidic residues on the SP-CTD surface would be proximal to the conserved basic patch located on the back side of SUMO (Supplementary Figure 3B; right panel; Stehmeier and Muller, 2009). To test if the electrostatic character of the acidic SP-CTD was required for Siz1 E3 ligase activity, we substituted either Asp345 or Glu346 (located on loop between α3 and β10 of Siz1; Figure 3A) to serine and deleted a stretch of acidic residues that were present but disordered in our crystal structure (aa 445-EDDDDSDSD-453) to generate Siz1(167–443). Each of these Siz1 mutant isoforms resulted in diminished SUMO conjugation to PCNA Lys164 and Lys127 as well as to the p53 SUMO consensus motif lysine residue (Figure 4A and 4B). To test if the electrostatic character of the basic SUMO surface contributed to Siz1 E3 ligase activity, we substituted basic residues on the SUMO surface with glutamate to generate Smt3(K38E/K40E), Smt3(R46E/R47E) or Smt3(K38E/K40E/R46E/R47E). These mutations had no detectable effect on the ability of the E1 to charge the E2 (Supplementary Figure 4) or the ability of the E2 enzyme to modify PCNA Lys127 in the absence of Siz1 (Figure 4C; top panel), but each of these mutations decreased the ability of Siz1 to enhance SUMO modification of PCNA. Taken together, these results suggest that the acidic SP-CTD residues and the basic patch on SUMO constitute surfaces important for activation of the E2~SUMO thioester complex during Siz1 mediated SUMO conjugation.
Kinetic and mutational analysis of the SP-RING and SP-CTD
Mutations in the predicted Siz1:E2~SUMO interface were selected for analysis under conditions of single turnover to determine kinetic defects associated with respective mutant Siz1 isoforms. Analysis of Siz1(I363A) revealed a ~30-fold or ~20-fold reduction in apparent catalytic rate (k2) for SUMO conjugation to PCNA Lys127 or Lys164, respectively (Figure 4D; Supplementary Table 1) with only a small decrease in apparent dissociation constant for PCNA (~2-fold and ~3.5-fold defects in Kd for Lys127 and Lys164, respectively; Figure 4D). It is not surprising that rates for SUMO transfer are affected more than PCNA binding because Ile363 lies in the middle of the predicted E2 binding surface and the E2 is required for catalysis.
Siz1(D345S) and Siz1(167–443) were analyzed next because we predicted that acidic residues within the SP-CTD would interact with SUMO to configure the E2~SUMO thioester for catalysis. Kinetic analysis revealed a complex situation in comparison to Siz1(I363A). In these instances, Siz1(D345S) and Siz1(167–443) exhibited slightly lower k2 values (~3-fold for Lys127 and ~1.5-fold for Lys164) but weaker Kd values (~8-fold for Lys127 and ~16-fold for Lys164) when compared to wild-type Siz1 (Figure 4D; right panel; Supplementary Table 1). We believe these mutations could prevent proper coordination of SUMO thus allowing SUMO to sample multiple conformations in the E2~SUMO:Siz1 complex. The inability to coordinate E2~SUMO would 1) misalign the thioester adduct for chemistry (defect in k2) and 2) allow SUMO to interfere with substrate binding (defect in Kd). Similar kinetic effects have been observed during E2~SUMO activation by the Nup358/RanBP2 SUMO E3 ligase (Reverter and Lima, 2005).
These results confirm that basic and acidic surfaces on SUMO and the SP-CTD, respectively, are required for Siz1 function. The basic patch on SUMO is highly conserved across evolution while Siz1, Siz2 and related human PIAS proteins each contain a conserved acidic loop within the Siz1 SP-CTD (located between α3 and β10 of Siz1; Figure 3A). It is important to note that Siz2 and other PIAS proteins lack the C-terminal acidic motif between residues 445–453 (adjacent to β15 of Siz1 and colored red; Figure 3A), suggesting that acidic SIM motifs may play a more important role for other SP-RING E3 ligases than was observed for the Siz1 E3 ligase.
The PINIT domain interacts with PCNA to facilitate SUMO conjugation
Most E3 ligases achieve substrate specificity by recruiting the E2~Ub/Ubl thioester and substrate into a complex to promote conjugation (Pickart, 2001). Siz1 may work in a similar manner by utilizing the SP-RING and SP-CTD domains to coordinate and activate the E2~SUMO thioester while the PINIT domain recognizes Siz1-dependent substrates (Reindle et al., 2006). If true, then mutations on the PINIT surface should decrease conjugation to PCNA while not altering the ability of Siz1 to activate the E2~SUMO thioester for conjugation to substrates containing SUMO consensus motifs.
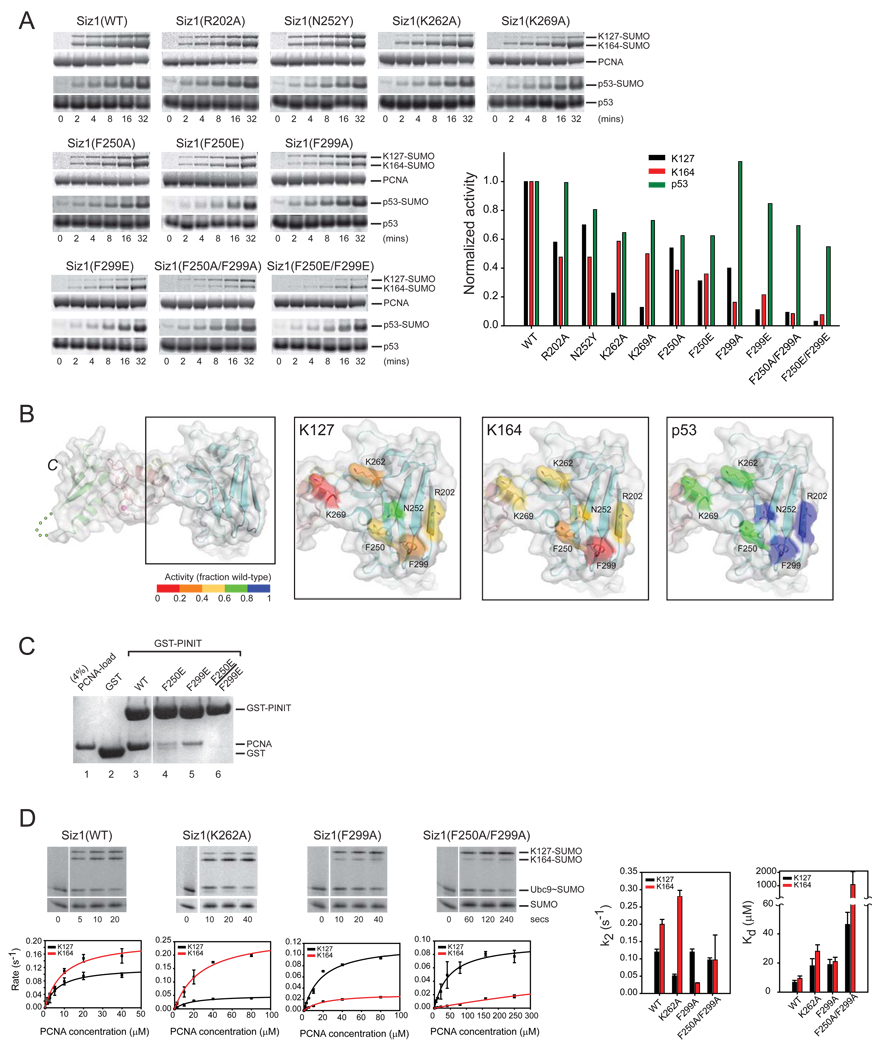
Our model suggested that a unique PINIT domain surface was in an ideal position to coordinate PCNA adjacent to the E2 active site (Supplementary Figure 2A, 2B and 3C). Consistent with this hypothesis, K262A, K269A, F250E, F299A, F299E substitutions and the double substitutions F250A/F299A and F250E/F299E were defective for SUMO conjugation to PCNA on Lys127 or Lys164 relative to wild-type. In contrast, these mutations, especially F250A/F299A or F250E/F299E, had lesser effects on SUMO conjugation to the SUMO consensus motif within p53 (Figure 5A and 5B). Siz1 R202A, F250A, and N252Y also exhibited mild defects for SUMO conjugation to PCNA. Lys262 and Lys269 lie on the loop near 310α2 in the interface with the SP-RING domain while Phe250 and Phe299 lie surface exposed on loops L2 and L3 (Figure 1B).
Figure 5. Surfaces on the Siz1 PINIT domain are important for SUMO conjugation to PCNA.
(A) SDS-PAGE and time course (left) and graph depicting rates in the linear range of the assay (right). Rates normalized to one to enable a comparative analysis for SUMO conjugation to p53, PCNA at K127 and K164 with wild-type and mutant Siz1 isoforms. Proteins detected and quantified by SYPRO Ruby (Methods). (B) PINIT domain of Siz1 with amino acids subjected to mutational analysis in stick representation and Siz1 surface color-coded according to the level of activity (inset color bar) for SUMO conjugation to PCNA Lys127 (second from left), PCNA Lys164 (second from right), and p53 (right panel). (C) SDS-PAGE of pull-down assays to probe interactions between PCNA, GST, or GST fusions to the Siz1 PINIT or indicated mutant isoforms. Protein detected by Coomassie blue (Methods). (D) SDS-PAGE and time course (left) for SUMO conjugation to PCNA with indicated Siz1 isoforms under conditions of single turnover with 10 µM PCNA (near the apparent Kd for Lys164). Proteins detected by fluorescence (Alexa Fluor 488-SUMO; Methods). The initial rate of reaction (s−1) versus PCNA concentration (µM) is shown below each gel. Bar charts (right) for Kd (µM) and k2 (s−1) for indicated Siz1 isoforms. Error bars represent ±1 standard deviation.
We next addressed if the PINIT domain interacted with PCNA directly by a GST-pull down assay using purified PCNA and GST-PINIT fusion proteins (wild-type and mutants). These experiments revealed interactions between PCNA and GST-PINIT (Figure 5C; lane 3) that could be diminished upon substitution of PINIT residues Phe250 or Phe299 to glutamate (Figure 3C; lanes 4 and 5) or abrogated when both Phe250 and Phe299 were mutated to glutamate as evidenced by the inability to detect PCNA (Figure 3C; lane 6). This data is consistent with the defects observed in vitro for the respective Siz1 mutant proteins in E3 ligase assays for SUMO conjugation to PCNA (Figure 5A).
Kinetic analysis of PINIT surface mutations
Siz1 isoforms containing single amino acid substitutions for K262A and F299A and the double amino acid substitution F250A/F299A were selected for kinetic analysis because Siz1(K262A) exhibited a selective defect in conjugation to Lys127, Siz1(F299A) exhibited a selective defect in conjugation to Lys164, and Siz1(F250A/F299A) exhibited a defect in conjugation to both lysine acceptors. For Siz1(K262A), kinetic analysis revealed ~3-fold weaker Kd for conjugation of PCNA at Lys127 and Lys164 (Figure 5D; Supplementary Table 1). Interestingly, k2 values decreased by ~2.5-fold for conjugation to PCNA at Lys127 and increased by ~1.5-fold for conjugation to Lys164. This suggests that Siz1(K262A) maintains the ability to interact with PCNA in a conformation suitable for modification at Lys164 while disfavoring interactions that promote modification at Lys127.
Consistent with results obtained under multiple turnover, Siz1(F299A) exhibited a 2 to 3-fold weaker Kd for conjugation to both Lys127 and Lys164 and while k2 remained unaffected for conjugation to Lys127, the apparent rate was diminished by ~7-fold for conjugation to Lys164. Defects were exacerbated for the Siz1(F250/F299A) double mutant for both lysine acceptors revealing a ~7-fold and ~115-fold weaker Kd for SUMO conjugation to PCNA Lys127 and Lys164, respectively (Figure 5D; Supplementary Table 1). In this case, the apparent rate of SUMO conjugation decreased slightly for Lys127 and by ~2-fold for Lys164.
A PCNA surface contributes to Siz1-dependent SUMO modification
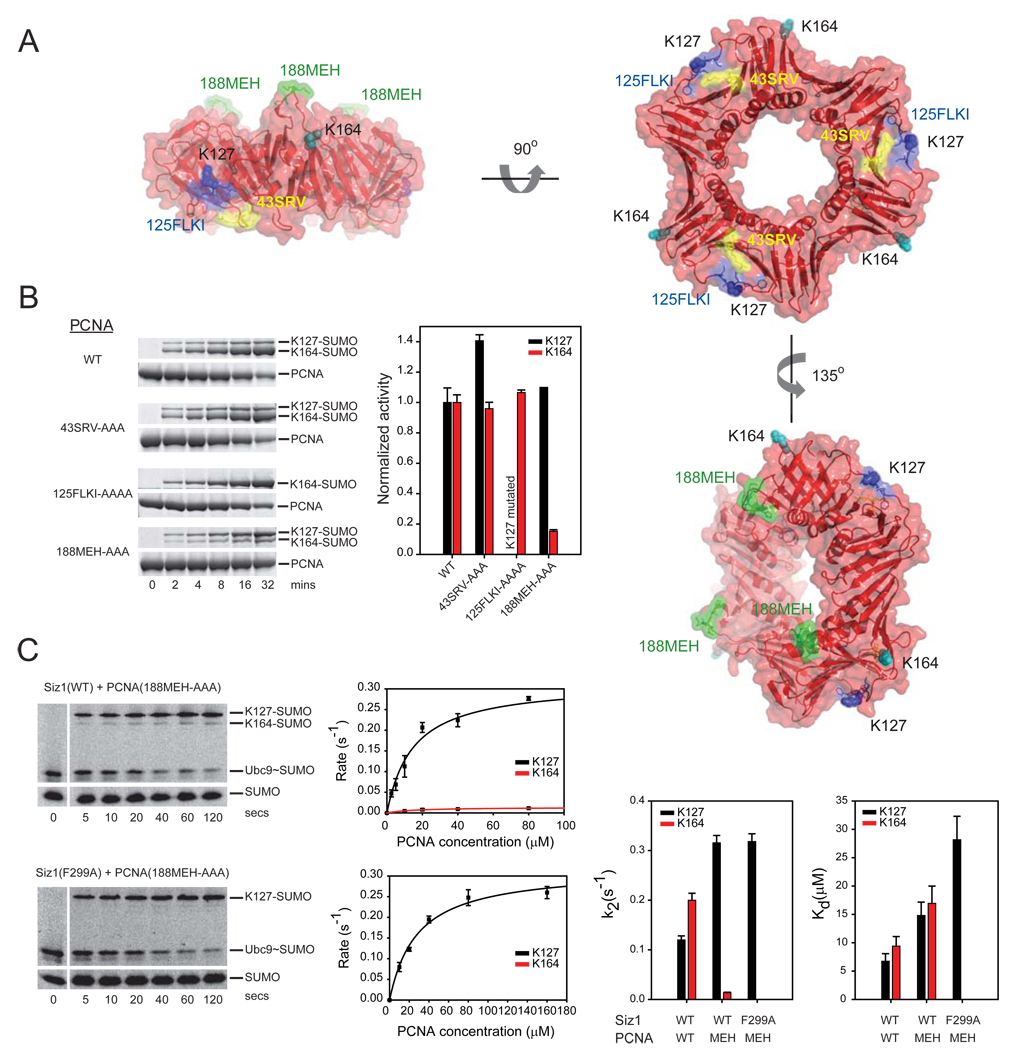
Mutations on the PINIT domain surface that disrupted conjugation to PCNA Lys164 and Lys127 are located approximately 30 Å from the proposed E2 active site in our model (Supplemental Figure 3). We explored the importance of three surface exposed loops in yeast PCNA for Siz1-mediated SUMO conjugation (Figure 6A). Each of these PCNA surface loops were previously subjected to mutational analysis in human PCNA (Jonsson et al., 1998). These included the FLKI inter-domain connector loop (125-FLKI-128; colored blue), the SRV loop (43-SRV-45; colored yellow), and the MEH loop (188-MEH-190; colored green) (Figure 6A). The FLKI inter-domain connector loop is a major site of interaction for DNA polymerases (Reynolds et al., 2000), the FEN1 endonuclease (Sakurai et al., 2005), DNA ligase and p21 (Montecucco et al., 1998; Warbrick et al., 1995), factors important for processes that include DNA replication, DNA repair, and the cell cycle. The SRV loop has been implicated in interactions with Pol δ, Pol ε and RF-C (Ayyagari et al., 1995). The MEH loop protrudes from the PCNA surface on the opposite surface relative to the two other loops. Previous studies have not identified a function for this PCNA surface (Jonsson et al., 1998).
Figure 6. A PCNA surface is important for Siz1-mediated SUMO conjugation.
(A) Surface representations of PCNA depicting loops 43-SRV (yellow), 125-FLKI (blue) and 188-MEH (green). PCNA Lys127 and Lys164 colored blue and cyan, respectively. (B) SDS-PAGE and time course (left) and graph depicting rates in the linear range of the assay (right) for SUMO conjugation to PCNA Lys127 and Lys164 with wild-type Siz1. Rates normalized to one for wild-type PCNA to enable a comparative analysis to PCNA mutant isoforms. Proteins detected by SYPRO Ruby (Methods). (C) SDS-PAGE and time course (left) for SUMO conjugation to PCNA(188-MEH-AAA) with Siz1(WT) or Siz1(F299A) under conditions of single turnover with 10 µM PCNA. Proteins detected by fluorescence (Alexa Fluor 488-SUMO; Methods). The initial rate of reaction (s−1) versus PCNA concentration (µM) is shown below each gel. Bar charts (right) for Kd (µM) and k2 (s−1) for indicated PCNA and Siz1 isoforms. Error bars represent ±1 standard deviation.
Loop residues were substituted with alanine and respective PCNA isoforms were expressed, purified and assayed for SUMO conjugation using wild-type Siz1(167–465) under conditions of multiple turnover (Figure 6B). Alanine substitution of the FLKI loop had no adverse effect on SUMO conjugation at Lys164 but resulted in mutation of Lys127. Alanine substitution of the SRV loop had no detectable effect on SUMO conjugation at Lys164 or Lys127. In contrast, alanine substitution of the MEH loop led to a selective reduction of SUMO conjugation at Lys164 while conjugation at Lys127 was maintained (Figure 6B; see graph). It is important to note that 1) mutations of the analogous loops in human PCNA did not affect the stability of the PCNA trimer as previously shown by native-gel electrophoresis (Jonsson et. al. 1998) and 2) loop mutations in yeast PCNA were able to fully complement the essential functions for PCNA as evidenced by plasmid shuffle and complementation of a Δpcna yeast strain (Methods; see below). These data suggest that the respective PCNA mutations do not affect the overall structure of PCNA.
Single turnover kinetic analysis was employed to uncover the basis for defects associated with SUMO conjugation to PCNA(188-MEH-AAA) using wild-type Siz1. The apparent Kd weakened by 2-fold for conjugation to PCNA Lys164 and Lys127 while k2 decreased by ~14-fold for conjugation at Lys164 and increased by ~3-fold for conjugation at Lys127. Defects in conjugation were further exacerbated for both lysine acceptors when Siz1(F299A) was combined with PCNA(188-MEH-AAA). In this case the apparent rate decreased by ~2.5-fold and the apparent Kd weakened by ~4-fold for conjugation to Lys127 and conjugation could no longer be detected at Lys164 (Figure 6C; Supplementary Table 1). These results are consistent with the 188-MEH loop participating in a Siz1-PCNA interface that is essential for directing SUMO conjugation to the non-consensus Lys164 side chain and for enhancing modification at Lys127.
Analysis of PINIT and PCNA mutations in vivo
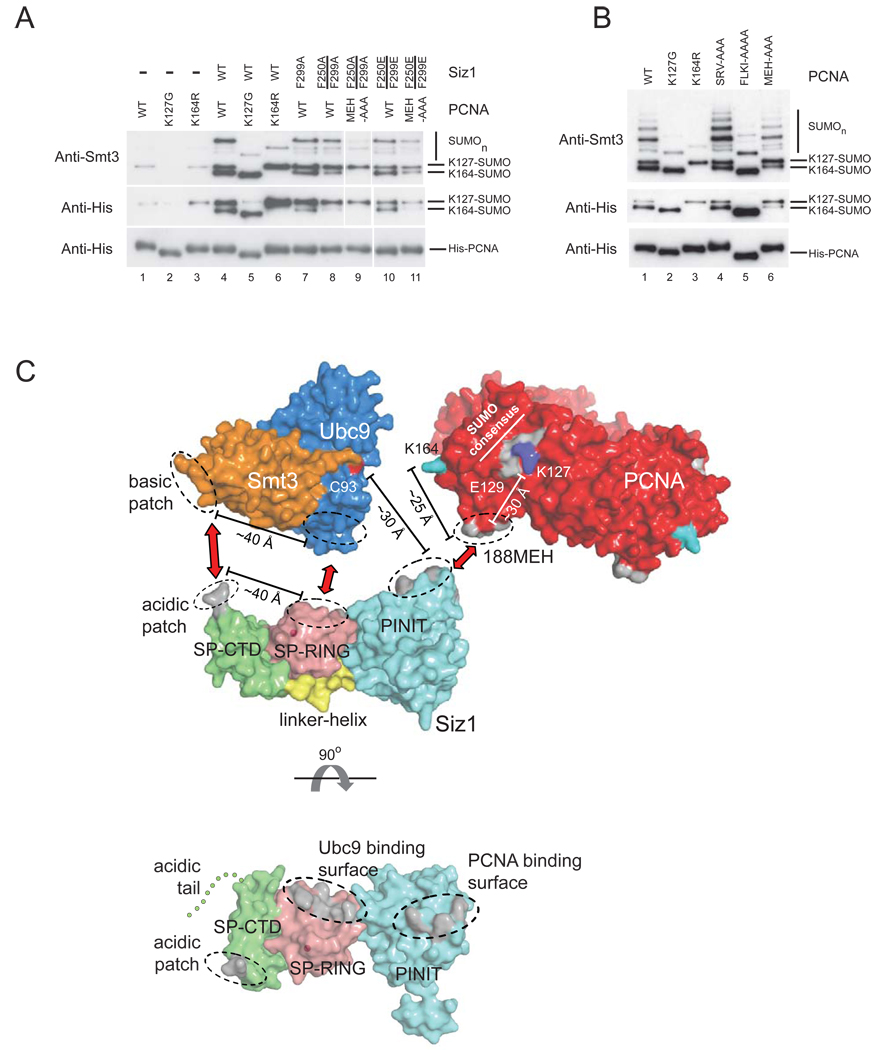
The preceding in vitro binding and activity assays identified Siz1 residues Phe250 and Phe299 on the PINIT domain surface as important determinants for SUMO conjugation to PCNA because F299A and F250A/F299A decreased SUMO conjugation at Lys164 and Lys127 under conditions of single (Figure 5D) and multiple turnover (Figure 5A) while F250E, F299E and F250E/F299E decreased SUMO conjugation to Lys164 and Lys127 under conditions of multiple turnover (Figure 5A). To test if these mutations had an effect on SUMO modification of PCNA in vivo, we introduced full-length wild-type and mutant siz1 alleles into a siz1Δ strain in which N-terminal His8-tagged PCNA isoforms were expressed (Pfander et al., 2005; Methods). Strains lacking Siz1 (siz1Δ) were unable to maintain SUMO conjugation at PCNA Lys164 and while SUMO conjugation was detected at PCNA Lys127, it was diminished in comparison to strains harboring wild-type Siz1 (Figure 7A; lane 1). Introduction of a plasmid encoding wild-type Siz1 under control of its endogenous promoter restored SUMO conjugation at Lys164 and Lys127 (Figure 7A; compare lanes 1 and 4). Plasmids harboring Siz1(F299A) could not fully complement the siz1Δ strain with respect to SUMO conjugation at PCNA Lys164 while the double mutants Siz1(F250A/F299A) and Siz1(F250E/F299E) could not complement the siz1Δ strain with respect to SUMO conjugation at either PCNA Lys164 or Lys127 when compared to wild-type Siz1 (Figure 7A; compare lanes 1, 4, 8 and 10).
Figure 7. Surfaces on Siz1 and PCNA important for SUMO conjugation in vitro are important in vivo and a model for E2~SUMO activation and selection of PCNA Lys164.
(A) Western blot to detect His-tagged PCNA and SUMO conjugated PCNA isolated from yeast cultures treated with MMS (Methods). Siz1 and PCNA alleles are indicated above each lane. (B) Same as (A) except that the yeast strain (pol30Δ) expressed wild-type Siz1 from the genomic locus and was complemented with PCNA (WT, K127G, K164R, 43SRV-AAA, 125FLKI-AAAA or 188MEH-AAA). Anti-His antibody used to detect PCNA and SUMO conjugates (middle and bottom). An anti-Smt3 antibody was used to confirm the identity of SUMO conjugated PCNA (top). (C) Model for an activated E3:E2~SUMO:PCNA complex with the Siz1 E3 ligase (colored as in Figure 1A), Ubc9 (blue), Smt3 (orange) and PCNA (red) (top). The Ubc9 catalytic cysteine (Cys93) is colored red, Lys164 of PCNA is colored cyan and SUMO consensus motif is colored gray with Lys127 in blue and Glu129 labeled. Surfaces identified in this study as important for SUMO conjugation by mutational analysis of Siz1 and PCNA are colored gray. Red arrows and dashed ellipses indicate putative pair-wise interactions. Distances between protein surfaces are indicated. (Bottom panel) Siz1 shown in an orthogonal orientation to reveal surfaces proposed to interact with Smt3, Ubc9 and PCNA (colored gray indicated by ellipses).
The importance of the PCNA 188-MEH loop for SUMO conjugation to PCNA was evaluated in vivo by complementing a pol30Δ strain with plasmids harboring N-terminal His8-tagged wild-type PCNA or mutant pcna alleles. Similar to data obtained in vitro, PCNA(125FLKI-AAAA) had no effect on SUMO conjugation to Lys164 and PCNA(43SRV-AAA) had no effect on SUMO conjugation to Lys164 or Lys127 (Figure 7B; compare lanes 1, 4 and 5). In contrast, SUMO conjugation of PCNA(188MEH-AAA) at Lys164 was selectively diminished in comparison to Lys127 (Figure 7B; compare lanes 1 and 6). Furthermore, SUMO conjugation at Lys164 was abrogated in siz1Δ strains expressing His8-PCNA(188MEH-AAA) and either Siz1(F250A/F299A) or Siz1(F250E/299E) while SUMO conjugation to Lys127 was diminished in comparison to wild-type (Figure 7A; lanes 9 and 11). Results obtained in vivo are in accordance with our structure, mutational analysis, and data obtained from SUMO conjugation assays in vitro (Figure 6C).
Models for interaction of PCNA with Siz1 and Ubc9
We identified surfaces on the Siz1 PINIT domain and PCNA that are required for SUMO modification at PCNA Lys164 and contribute to SUMO conjugation at PCNA Lys127. We propose a structural model where surfaces of the PINIT domain and PCNA shown to be important for SUMO conjugation activity can be juxtaposed to position Lys164 proximal to the E2 active site (Supplementary Figure 6A). This model is plausible given that the distance between the PINIT surface and Cys93 of Ubc9 (~30Å) is comparable to the distance between the PCNA MEH loop and Lys164 (~25 Å) (Figure 7C and Supplementary Figure 6A). This model is also consistent with observations that PCNA is competent for SUMO modification when loaded on DNA (Parker et al., 2008) because the central PCNA channel that encircles DNA is not occluded by interactions with Siz1 (Supplemental Figure 6).
Mutation of the SUMO consensus motif glutamate (Glu129) can disrupt SUMO modification at Lys127 in the presence of Siz1 (Windecker and Ulrich 2008). The importance of this residue suggests that the PCNA SUMO consensus motif binds to Ubc9 in a manner similar to that observed for other SUMO consensus motifs in structures of Ubc9:substrate complexes (Bernier-Villamor et al., 2002; Lin et al., 2002). Based on these structures, we modeled the PCNA SUMO consensus motif into the E2 substrate binding pocket (Supplementary Figure 6B). This model revealed a steric clash between the Siz1 PINIT domain and PCNA that could be alleviated if the Siz1 PINIT domain swings away from the SP-RING domain. Although PINIT and SP-RING domains share a hydrophobic interface that buries 1050 Å2 of total surface area, this conformational change is possible because the PINIT and SP-RING domains are connected via a single linker helix (Figure 1B). Interestingly, the two mutations (K262A and K269A) we isolated that diminish conjugation to PCNA Lys127 in comparison to PCNA Lys164 (Figure 5A and 5D) lie near the interface between the SP-RING and PINIT domains.
Other models can be envisioned to explain how the PINIT domain enhances SUMO conjugation to Lys127. Both Lys127 and Lys164 acceptors occupy positions along the midline of PCNA nearly equidistant from the MEH loop (Figure 6 and Figure 7). If interactions between the Siz1 PINIT domain and PCNA MEH surfaces were dynamic this might allow PCNA to swivel about the MEH:PINIT interaction to facilitate conjugation to Lys127. Support for alternate conformations is supported by the same Siz1 mutations (K262A and K269A) which led to a selective loss for SUMO conjugation at Lys127 in comparison to Lys164. In another model, interactions between PCNA and Siz1 would increase the probability of Lys127 modification by bringing three SUMO consensus motifs in the PCNA trimer within striking distance of the activated E2~SUMO thioester.
Conclusions
The Siz1 structure exemplifies conserved elements in the Siz/PIAS E3 ligase family and reveals a tripartite domain organization formed by an N-terminal PINIT domain, a central SPRING domain and a conserved SP-CTD domain. The central SP-RING domain is structurally similar to ubiquitin E3 ligase RING and U-box domains and likely interacts with the E2 Ubc9 in a manner similar to that observed in other ubiquitin E2:E3 complexes because mutations predicted to disrupt the E2:E3 interaction failed to support SUMO conjugation to any substrate tested. Interestingly, Siz1 E3 ligase activity also required acidic elements within the SP-CTD domain and we propose that the SP-CTD domain is required for interactions with SUMO since basic residues on the SUMO surface were not required for E2-mediated conjugation but were required for Siz1-mediated SUMO conjugation. Thus our model suggests that the SP-RING and SP-CTD domains are both required to properly configure the E2~SUMO thioester to facilitate catalysis and interactions between the substrate and E2 active site (Figure 7C).
The N-terminal PINIT domain plays a unique role in substrate recognition. The PINIT domain is essential for modification at PCNA Lys164 because this lysine is a non-consensus SUMO acceptor and is not a substrate for the SUMO E2 even at high E2 concentrations. Thus, the PINIT domain is required to bind PCNA and position Lys164 proximal to the E2 active site. Binding interactions between the PINIT and PCNA can also enhance modification at Lys127 in a manner that remains dependent on the SUMO consensus motif, a natural substrate for the SUMO E2. Our studies suggest that the PINIT domain serves as a scaffold to coordinate PCNA to promote SUMO conjugation to both consensus and non-consensus lysine side chains because mutations on the PINIT domain surface disrupted conjugation to both lysine acceptors. PINIT domains are unique to and conserved in all Siz/PIAS family members although amino acid residues that constitute PINIT domain surfaces deviate greatly between SP-RING family members. This latter feature is consistent with its role in dictating substrate specificity. Our studies have revealed amino acid residues on the surface of Siz1, PCNA, and SUMO that are required for efficient modification of PCNA in vitro and in vivo and provide useful insights to guide future studies aimed at resolving functions for SUMO modification of PCNA during replication and DNA damage response.
Experimental Procedures
Cloning, expression and purification of recombinant proteins
Procedures for cloning, expression and purification of recombinant proteins utilized for crystallization trials and in vitro biochemistry are described in Supplementary materials or have been described previously (Mossessova and Lima, 2000; Bernier-Villamor et al., 2002; Lois and Lima, 2005; Reverter and Lima, 2005; Yunus and Lima, 2009).
Crystallographic analysis
Procedures for crystallographic analysis are provided in Supplementary material. Refinement and data statistics are reported in Table I. All graphic representations of structure were generated with Pymol (DeLano, 2002).
Plasmids and yeast strains for detection of SUMO conjugation to PCNA in vivo
Plasmids and yeast strains are described in Supplemental material. Assays for detecting SUMO conjugates of yeast PCNA have been described (Hoege et al., 2002; Johnson and Blobel, 1999). Briefly, strains were grown to an A600 of 1, treated with 0.3% MMS for 90 mins or 250 mins and cells were harvested by centrifugation. Cell pellets were lysed with 1.85 N NaOH, 7.5% βME. Protein was precipitated with an equal volume of 50% TCA, suspended in 6 M guanidinium-HCl, 100 mM Na2HPO4, 20 mM Tris pH 8.0 and His8-PCNA was isolated by metal affinity chromatography. Samples were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using 12% Bis-Tris gels and MOPS buffer (Invitrogen). Smt3 and PCNA were detected by western blotting using anti-Smt3 or anti-His antibodies (Invitrogen), respectively.
Biochemical assays
Procedures for SUMO conjugation assays under conditions of multiple or single turnover have been described (Yunus and Lima, 2005; 2009) or are provided in Supplemental material. Single turnover assays to assess Smt3 conjugation to PCNA and estimate kinetic constants (k2 and Kd) were carried out at 4°C because rates were too fast to measure reliably at higher temperatures.
E2-SUMO thioester assays were carried out at 37°C. Reaction mix included 50 mM NaCl, 20 mM HEPES pH 7.5, 1 mM ATP, 5 mM MgCl2, 1 mM DTT, 1 µM Ubc9(K153R), 100 nM E1, and 1 µM Smt3 (wild-type or mutant isoforms). Ubc9(K153R) was used to suppress Smt3 conjugation to the E2 at Lys153. Reactions were initiated by addition of ATP. Aliquots were removed at indicated time points and denatured in 50 mM Tris-HCl, pH 6.8, 2% SDS, 4 M Urea, and 10% glycerol. Samples were separated by SDS-PAGE and analyzed by western-blot analysis using an anti-Smt3 antibody.
In vitro binding assay
We mixed 300 µg of GST or wild-type and mutant isoforms of GST-PINIT (Siz1 aa 172–315 as a C-terminal fusion to GST) in 400 µl binding buffer (50 mM NaCl, 20 mM Tris 8.0, and 1 mM BME) with 50 µl of 50% (v/v) Glutathione-Sepharose beads (GE Healthcare) for 30 minutes at 23ºC to saturate beads with the respective proteins. Unbound protein was removed by three washes with binding buffer. Beads were then mixed with 400 µl of 10 µM PCNA in binding buffer, incubated 60 minutes at 23ºC, and washed three times with binding buffer. The bound fraction was eluted with 100 µl of 350 mM NaCl, 20 mM Tris 8.0, 1 mM BME and 20 mM glutathione, analyzed by SDS-PAGE (4–12% Bis-Tris; Invitrogen) and detected by Coomassie Blue staining.
Structure factors and coordinates are deposited in the RCSB Protein Data Base with accession code 3I2D.
Supplementary Material
01
Acknowledgements
We thank Jaclyn Gareau and Ryan Kniewel for assistance with yeast strains, Firaz Mohideen for suggestions, and Anthony Armstrong and Shaun Olsen for input on this manuscript. This work is based in part upon research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by award RR-15301 from the NCRR and NIH. Use of the Advanced Photon Source is supported by the U.S. Dept. of Energy, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357. This work was supported by NIH GM065872 (C.D.L.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol Cell Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier-Villamor V, Sampson DA, Matunis MJ, Lima CD. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell. 2002;108:345–356. doi: 10.1016/s0092-8674(02)00630-x. [DOI] [PubMed] [Google Scholar]
- Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Hendrickson WA. Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science. 1991;254:51–58. doi: 10.1126/science.1925561. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Jackson PK, Eldridge AG, Freed E, Furstenthal L, Hsu JY, Kaiser BK, Reimann JD. The lore of the RINGs: substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 2000;10:429–439. doi: 10.1016/s0962-8924(00)01834-1. [DOI] [PubMed] [Google Scholar]
- Johnson ES. Protein modification by SUMO. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. EMBO J. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- Lin D, Tatham MH, Yu B, Kim S, Hay RT, Chen Y. Identification of a substrate recognition site on Ubc9. J Biol Chem. 277:21740–21748. doi: 10.1074/jbc.M108418200. [DOI] [PubMed] [Google Scholar]
- Lois LM, Lima CD. Structures of the SUMO E1 provide mechanistic insights into SUMO activation and E2 recruitment to E1. EMBO J. 2005;24:439–451. doi: 10.1038/sj.emboj.7600552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Montecucco A, Rossi R, Levin DS, Gary R, Park MS, Motycka TA, Ciarrocchi G, Villa A, Biamonti G, Tomkinson AE. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigen: identification of a common targeting mechanism for the assembly of replication factories. EMBO J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Lima CD. Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol Cell. 2000;5:865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- Parker JL, Bucceri A, Davies AA, Heidrich K, Windecker H, Ulrich HD. SUMO modification of PCNA is controlled by DNA. EMBO J. 2008;27:2422–2431. doi: 10.1038/emboj.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- Pichler A, Knipscheer P, Saitoh H, Sixma TK, Melchior F. The RanBP2 SUMO E3 ligase is neither HECT- nor RING-type. Nat Struct Mol Biol. 2004;11:984–991. doi: 10.1038/nsmb834. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Reindle A, Belichenko I, Bylebyl GR, Chen XL, Gandhi N, Johnson ES. Multiple domains in Siz SUMO ligases contribute to substrate selectivity. J Cell Sci. 2006;119:4749–4757. doi: 10.1242/jcs.03243. [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–692. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Warbrick E, Fantes PA, MacNeill SA. Essential interaction between the fission yeast DNA polymerase delta subunit Cdc27 and Pcn1 (PCNA) mediated through a C-terminal p21(Cip1)-like PCNA binding motif. EMBO J. 2000;19:1108–1118. doi: 10.1093/emboj/19.5.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, Grosschedl R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001;15:3088–3103. doi: 10.1101/gad.944801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai S, Kitano K, Yamaguchi H, Hamada K, Okada K, Fukuda K, Uchida M, Ohtsuka E, Morioka H, Hakoshima T. Structural basis for recruitment of human flap endonuclease 1 to PCNA. EMBO J. 2005;24:683–693. doi: 10.1038/sj.emboj.7600519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehmeier P, Muller S. Phospho-regulated SUMO interaction modules connect the SUMO system to CK2 signaling. Mol Cell. 2009;33:400–409. doi: 10.1016/j.molcel.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kahyo T, Toh EA, Yasuda H, Kikuchi Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem. 2001a;276:48973–48977. doi: 10.1074/jbc.M109295200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Kikuchi Y. Yeast PIAS-type Ull1/Siz1 is composed of SUMO ligase and regulatory domains. J Biol Chem. 2005;280:35822–35828. doi: 10.1074/jbc.M506794200. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Toh-e A, Kikuchi Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene. 2001b;275:223–231. doi: 10.1016/s0378-1119(01)00662-x. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- Warbrick E, Lane DP, Glover DM, Cox LS. A small peptide inhibitor of DNA replication defines the site of interaction between the cyclin-dependent kinase inhibitor p21WAF1 and proliferating cell nuclear antigen. Curr Biol. 1995;5:275–282. doi: 10.1016/s0960-9822(95)00058-3. [DOI] [PubMed] [Google Scholar]
- Windecker H, Ulrich HD. Architecture and assembly of poly-SUMO chains on PCNA in Saccharomyces cerevisiae. J Mol Biol. 2008;376:221–231. doi: 10.1016/j.jmb.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol Biol. 2009;497:167–186. doi: 10.1007/978-1-59745-566-4_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01