Dynamin2 GTPase and Cortactin Remodel Actin Filaments (original) (raw)
Abstract
The large GTPase dynamin, best known for its activities that remodel membranes during endocytosis, also regulates F-actin-rich structures, including podosomes, phagocytic cups, actin comet tails, subcortical ruffles, and stress fibers. The mechanisms by which dynamin regulates actin filaments are not known, but an emerging view is that dynamin influences F-actin via its interactions with proteins that interact directly or indirectly with actin filaments. We show here that dynamin2 GTPase activity remodels actin filaments in vitro via a mechanism that depends on the binding partner and F-actin-binding protein, cortactin. Tightly associated actin filaments cross-linked by dynamin2 and cortactin became loosely associated after GTP addition when viewed by transmission electron microscopy. Actin filaments were dynamically unraveled and fragmented after GTP addition when viewed in real time using total internal reflection fluorescence microscopy. Cortactin stimulated the intrinsic GTPase activity of dynamin2 and maintained a stable link between actin filaments and dynamin2, even in the presence of GTP. Filaments remodeled by dynamin2 GTPase in vitro exhibit enhanced sensitivity to severing by the actin depolymerizing factor, cofilin, suggesting that GTPase-dependent remodeling influences the interactions of actin regulatory proteins and F-actin. The global organization of the actomyosin cytoskeleton was perturbed in U2-OS cells depleted of dynamin2, implicating dynamin2 in remodeling actin filaments that comprise supramolecular F-actin arrays in vivo. We conclude that dynamin2 GTPase remodels actin filaments and plays a role in orchestrating the global actomyosin cytoskeleton.
Controlled assembly and disassembly of actin filaments underlies movement, shape, division, trafficking of lipids and proteins of the cell and pathogenesis by infectious bacteria and viruses. Several proteins and signaling circuits modulate actin filament dynamics, including proteins that nucleate formation of new filaments, filament cross-linking proteins that stabilize branched and bundled filament arrays, and depolymerizing factors that promote filament disassembly (1). Studies with reconstituted systems show that a single actin nucleating factor, such as the Arp2/3 complex together with a nucleation-promoting factor, a barbed end capping protein to preserve the actin monomer pool and promote nucleation, and a filament disassembly factor, such as ADF/cofilin, are sufficient to establish a dynamic dendritic actin network in vitro that mimics many properties of actin networks at the leading edge of migrating cells (2–4). However, the mechanisms for coordinating the organization and dynamics of actin filaments associated with higher-order cellular structures such as the subcortical F-actin network, F-actin at focal adhesions, and actomyosin arrays are not as well understood.
Considerable evidence indicates that the large GTPase dynamin, a key mediator of membrane remodeling and fission, also influences actin filaments (reviewed in Refs. 5–7). Although the mechanisms are unknown, dynamin could influence actin filaments via its interactions with a number of proteins that directly or indirectly regulate actin filament assembly, filament stability, or filament organization. For example, several protein scaffolds biochemically link dynamin and the Arp2/3 complex activating factor, N-WASP, suggesting that the machinery for de novo actin assembly may be targeted or activated by dynamin (6, 8, 9). Dynamin2 is associated with several dynamic F-actin-containing structures in vivo, including podosomes, F-actin comet tails, phagocytic cups, dynamic cortical ruffles, and pedestal structures elaborated by enteropathogenic Escherichia coli (10–20). Cortactin, which directly binds both dynamin and actin filaments, is associated with many of the same dynamic actin structures as dynamin (5, 7) and is required for both clathrin-dependent and -independent endocytosis (21, 22). Thus, dynamin-cortactin interaction may be an important link between actin filaments and dynamin during formation or turnover of F-actin-rich structures.
Considerable evidence supports the notion that GTP hydrolysis by dynamin catalyzes membrane fission activity via GTPase-dependent changes in conformation (23, 24) or via GTPase-dependent cycles of assembly and disassembly (25, 26). We hypothesize that GTPase-dependent changes in dynamin linked via its interacting proteins to actin filaments or actin regulators could similarly influence actin filaments. Overexpressed, dominant negative dynamin mutant proteins impaired in binding or hydrolyzing GTP (most often the dynamin-K44A mutation) perturb a variety of F-actin-rich cellular structures, including stress fibers and focal adhesions (27, 28), dendritic spines of neurons (29), podosomes (12, 30), actin comet tails (13, 14), phagocytic cups and bacteria-induced pedestal structures (16, 19), and dynamic cortical ruffles (15, 17). In addition, F-actin of stress fibers and overall cell morphology were perturbed in Clone9 cells expressing a mutant dynamin2 protein lacking the C-terminal proline-rich domain, the domain through which dynamin2 interacts with actin regulatory factors (11). Whereas existing data indicates that the specific effects of dynamin GTPase activity on F-actin structures are cell type- and structure-specific, a general conclusion is that dynamin GTPase activity influences the organization or turnover of a subset of actin filaments.
To determine the mechanisms by which dynamin2 GTPase activity influences actin filaments, we developed biochemical and microscopic approaches to quantitatively assess and observe GTPase-dependent effects on actin filaments formed in vitro with Arp2/3 complex, cortactin, and dynamin2. The activities of dynamin2 on actin filaments in vivo were examined in cells with disrupted dynamin2 function using siRNA2-mediated suppression or pharmacologic inhibition. We report that dynamin2 GTPase, together with cortactin, functions as a dynamic actin filament remodeling complex that influences the global organization of the actomyosin cytoskeleton.
EXPERIMENTAL PROCEDURES
Proteins, Plasmids, Reagents, and Antibodies
Actin was purified from acetone powder of rabbit muscle (31) and gel-filtered on a Sephacryl S-200 HR column (GE Healthcare). Pyrene-labeled actin (32) and Alexa 488-labeled actin (33) were prepared using reactive fluorophores purchased from Molecular Probes. Arp2/3 complex was purified from bovine calf thymus (34). Recombinant glutathione _S_-transferase (GST)-cortactin and GST-cortactin-W525K were expressed in bacteria and purified as described (35); cortactin proteins were cleaved from GST using Tev protease and purified on a HiTrapQ column (GE Healthcare). His-tagged rat dynamin2 was expressed in Hi5 insect cells and purified using Talon affinity resin (36). Baculovirus to express mutant rat dynamin2-R399A was prepared after site-directed mutagenesis of pFastBac-dynamin2 using the QuikChange Mutagenesis kit (Stratagene). The sequence of dynamin2-R399A in pFastBac1 was confirmed prior to generating baculovirus stocks using the Bac-to-Bac Baculovirus Expression system (Invitrogen). Recombinant dynamin2-R399A was expressed in Hi5 insect cells and purified by affinity chromatography using amphiphysin II-SH3 conjugated to GST (37). Recombinant GST-cofilin was expressed in bacteria and purified on glutathione-Sepharose resin; cofilin was cleaved from GST using thrombin. The plasmid for expressing GST-cofilin was obtained from Pekka Lappalainen (University of Helsinki, Helsinki, Finland).
Purified proteins were dialyzed in the following buffers before use: actin in G-buffer (2 mm Tris-HCl, pH 8.0, 0.2 mm ATP, 0.1 mm dithiothreitol (DTT), 0.005% NaN3, and 0.2 mm CaCl2); Arp2/3 complex (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 0.2 mm EGTA, 0.2 mm MgCl2, 1 mm NaN3, 0.1 mm ATP, 0.5 mm DTT); cortactin and cortactin-W525K (20 mm Tris-HCl, pH 8.0, 50 mm KCl, 1 mm EDTA, 1 mm EGTA, 0.5 mm DTT, and 0.1 mm ATP); dynamin2 and dynamin2-R399A (20 mm HEPES, pH 7.5, 150 mm KCl, 1 mm EDTA, 1 mm EGTA, 0.5 mm DTT); and cofilin (20 mm Tris-HCl, pH 7.5, 50 mm KCl, 0.5 mm DTT). Proteins were quantified from absorbance at 280 nm (or 290 nm for actin) using the following molar extinction coefficients: actin, 26,600 m−1 cm−1; dynamin2, 53,490 m−1 cm−1; cortactin, 69,060 m−1 cm−1; cortactin-W525K, 67,855 m−1 cm−1; and cofilin, 13,850 m−1 cm−1.
GTP, GDP, and GTPγS (Sigma) were prepared as stock solutions at 50 mm in 10 mm Tris-HCl, pH 7.5, 50 mm MgCl2; the pH as adjusted to approximately pH 7.8 using 1 m Tris base; aliquots were stored at −80 °C. GMP-PCP was used for some experiments and was obtained from Jena Bioscience (Jena, Germany). Rhodamine-phalloidin was purchased from Molecular Probes. Antibodies used in this work include: goat anti-dynamin2 (C-18, Santa Cruz Biotechnology), goat anti-dynamin1 (C-16, Santa Cruz Biotechnology), mouse monoclonal anti-actin (C-4, Chemicon), mouse monoclonal anti-α-actinin (H-2, Santa Cruz Biotechnology), rabbit anti-non-muscle myosin heavy chain IIA (Covance), and mouse anti-cortactin (4F11, Upstate). Secondary antibodies were obtained from Jackson ImmunoResearch (horseradish peroxidase conjugated) or from Molecular Probes (Alexa Fluor conjugated). Dynasore (Sigma) was prepared at 20 mm in DMSO.
TIRF Microscopy
Glass coverslips were cleaned 1–2 h in Piranha solution (3:1, H2SO4:H2O2), rinsed in water followed by 100% ethanol, and stored in 100% ethanol. Reactions containing 1.5 μm actin (30% Alexa labeled), 50 nm Arp2/3 complex, 500 nm cortactin, and 500 nm dynamin2 in imaging buffer (IB50, 20 mm imidazole, pH 7.0, 100 mm KCl, 1 mm EGTA, 2 mm MgCl2, 0.3% methylcellulose, 3 mg/ml glucose, 0.2 mm ATP, 20 μg/ml catalase, 100 μg/ml glucose oxidase, and 100 nm dithiothreitol) were incubated 10 min at room temperature prior to observation. A 15-μl aliquot of the reaction mixture was placed directly on the coverslip surface; subsequent reagents (1 mm GTP, 1 mm GTPγS, or buffer) were added directly to the drop. TIRFM was performed on an Olympus X71 inverted microscope equipped with a ×60, 1.45 N.A. oil objective lens, an Argon laser, and a Cool-Snap ES CCD camera (Photometrics). Images (0.5 s exposure) were collected at 2- or 5-s intervals over 5–10 min using Isee imaging software (Innovision).
Electron Microscopy
Reactions containing 1.5 μm actin, 50 nm Arp2/3 complex, 500 nm cortactin, and 500 nm dynamin2 were incubated for 10 min in MKEI-50 (20 mm imidazole, pH 7.0, 100 mm KCl, 1 mm EGTA, 2 mm MgCl2, 0.4 mm ATP, and 0.2 mm DTT) at room temperature. Guanine nucleotides were added to reactions 2 min prior to application to the EM grid. Copper mesh grids with a Formvar support film (Electron Microscopy Sciences) were pre-coated with ∼6 Å of carbon prior to applying the samples. Samples were stained with 2% uranyl acetate (Electron Microscopy Sciences). Electron micrographs were collected on a Jeol 1010 transmission electron microscope (Peabody, MA) operating at 60 kV and equipped with a 16-megapixel CCD camera (SIA-12C; Scientific Instruments and Applications, Inc.). Images were collected at a magnification of ×20,000 using Maxim DL software (Diffraction Limited). Measurements of filament bundle width were obtained every 250 nm along the length of each bundle using Image J (NIH).
Low Speed Sedimentation Assay
Reactions containing 1.5 μm actin, 50 nm Arp2/3 complex, 500 nm cortactin, and varying amounts of dynamin2 were incubated for 30 min at room temperature in MKEI-50. GTP or buffer was added for 5 min and samples were centrifuged in a Microfuge 18 centrifuge (Beckman) at 14,000 rpm (18,000 × g) at 4 °C for 15 min to pellet bundled filaments. Aliquots of each reaction, and corresponding supernatant and pellet fractions, were prepared for SDS-PAGE on a 10% gel. The amounts of protein in supernatant and pellet fractions were quantified from the Coomassie Blue-stained gels using gel analysis tools in Image J.
GTPase Assay
Dynamin2 GTPase activity was measured using a colorimetric assay as described (38) with the following modifications. Dynamin2 and cortactin were dialyzed in assay buffer (20 mm HEPES, pH 7.5, 150 mm KCl, 2 mm MgCl2, and 1 mm DTT) prior to each experiment; reactions contained 0.25 μm dynamin2 (or dynamin2-R399A), 0.25 mm GTP, and varying concentrations of cortactin or cortactin-W525K. Proteins were incubated at 37 °C for 5 min prior to the addition of GTP and reactions were carried out at 37 °C. Aliquots (10 μl) were removed at each time point and mixed with 100 mm EDTA (3 μl of 0.5 m EDTA, pH 8.0) to stop the reaction. When all reactions were complete, 75 μl of malachite green solution (1 mm malachite green, 10 mm ammonium molybdate tetrahydrate in 1 n HCl) was added and the absorbance at 650 nm was measured using a microplate reader. The amount of phosphate ion released was determined from a standard curve. Rates of GTP hydrolysis were determined from the initial linear portion of each time course and plotted versus the concentration of cortactin; data were fit to a hyperbolic function. Each experiment contained duplicate samples and was carried out at least 4 times.
Actin Polymerization Assay
Bundled filament “seeds” were prepared in reactions containing 2 μm actin, 50 nm Arp2/3 complex, 500 nm cortactin, and 500 nm dynamin2 incubated in MKEI-50 buffer for 1 h. Pre-formed seeds were treated with various concentrations of guanine nucleotide or buffer for 45 s, followed by a 1:3 dilution into 2 μm actin (10% pyrene-labeled) in MKEI-50; the fluorescence of pyrene-actin was monitored over time (excitation at 365 nm; emission at 386 nm). GTP/GTPγS stock solutions were diluted into MKEI-50 containing 7 mm MgCl2 prior to use. For reactions containing cofilin, bundled filament seeds were treated with or without 0.6 mm GTP or buffer for 15 s, followed by addition of 0.5 μm cofilin for an additional 30 s prior to 1:3 dilution into 2 μm actin (10% pyrene-labeled).
Cell Culture, Transfection, and Labeling
U2-OS cells (ATCC HTB-96) were plated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and glutamine on coverslips coated with 5 μg/ml fibronectin. Dynamin2 was depleted using siGenome siRNAs (Dharmacon) targeting human dynamin2: D2-02, GACAUGAUCCUGCAGUUCA (catalog number D-004007-02) and D2-18, AGUCCUACAUCAACACGAA (catalog number D-004007-18); a non-targeting siRNA was used as a control (catalog number D-001210-05). siRNAs (2 μg) were delivered into U2-OS cells using an Amaxa Nucleofector II, program X-001, and nucleofection kit V (Amaxa Biosystems, Lonza) according to the manufacture's protocols. Cells were used 48–72 h after nucleofection.
Wild type rat dynamin2, which is not targeted by the siRNAs used to target human dynamin2, was expressed in dynamin2-depleted cells after nuclear microinjection of a plasmid driving its expression (4 μg/ml in injection buffer (75 mm KCl, 10 mm KH2PO4, pH 7.0)) together with a plasmid driving expression of GFP-paxillin (a gift of Dr. Rick Horwitz) to identify injected cells. Cells were fixed 4 h after injection and stained as described below. For time-lapse movies, cells were transfected with plasmid (2 μg) to express GFP-actin (a gift of Dr. Beat Imhof) together with the siRNAs. For experiments with dynasore-treated cells, cultures were preincubated for 30 min in Opti-MEM followed by incubation for 20 min in Opti-MEM containing 80 μm dynasore; control cells were incubated with 0.4% (v/v) DMSO. Total cell extracts were prepared in SDS-urea sample buffer (2% SDS, 8 m urea, 2 m thiourea, 10% glycerol, 0.1 m Tris, pH 7, 2 mm EDTA, 0.08 m DTT, 0.1 mg/ml bromphenol blue, 0.1 mg/ml pyronin Y) and analyzed by SDS-PAGE and Western blotting using chemiluminescent detection.
For immunofluorescence detection, cells were fixed for 10 min at 37 °C with 2% paraformaldehyde in IF buffer (127 mm NaCl, 5 mm KCl, 1.1 mm NaH2PO4, 0.4 mm KH2PO4, 2 mm MgCl2, 5.5 mm glucose, 1 mm EGTA, and 20 mm Pipes, pH 7.3), permeabilized 10 min in IF buffer containing 0.05–0.5% (v/v) Triton X-100, and stained with antibodies and/or 7.5 nm rhodamine-phalloidin. Coverslips were mounted in 1% _n_-propyl gallate in 50 mm Tris-Cl, pH 8.5, containing 50% glycerol. Micrographs from focal planes spaced 0.4-μm apart were collected using a Zeiss LSM510 confocal microscope equipped with a ×63, 1.4 N.A. objective lens; identical laser intensity and photo-detector gain were applied for all image acquisition. Images from all focal planes were rendered as a single maximum intensity, using projected image software available from Zeiss; images were processed for final figures using Adobe Photoshop. The extent of immunostaining was quantified from the micrographs by tracing each cell and obtaining the integrated fluorescence intensity/cell using analysis tools available in NIH Image J; the integrated intensity/cell area was used as a measure of immunostaining. Time-lapse imaging of live cells expressing GFP-actin was collected using a wide-field epifluorescence microscopy and a Photometrics Cool-Snap HQ camera (Roper Scientific).
RESULTS
Dynamin2 GTPase Activity Remodels Actin Filaments in Vitro
To determine how GTP hydrolysis by dynamin2 influences actin filaments, we analyzed actin filaments in reactions with dynamin2 and cortactin. We previously observed GTP-dependent changes in the morphology of bundled actin filaments assembled by Arp2/3 complex, cortactin, and dynamin2 in the presence of phosphatidylinositol 4,5-bisphosphate-containing liposomes in vitro, but the arrangement of actin filaments within bundles and the mechanisms for GTPase-dependent reorganization were not apparent from static, light-level micrographs of phalloidin-stained preparations (15). Therefore, to observe filament organization within bundles and to glean insight into the mechanisms for GTPase-dependent filament remodeling, we observed filaments using negative-stained transmission electron microscopy (TEM) and in real time using TIRFM.
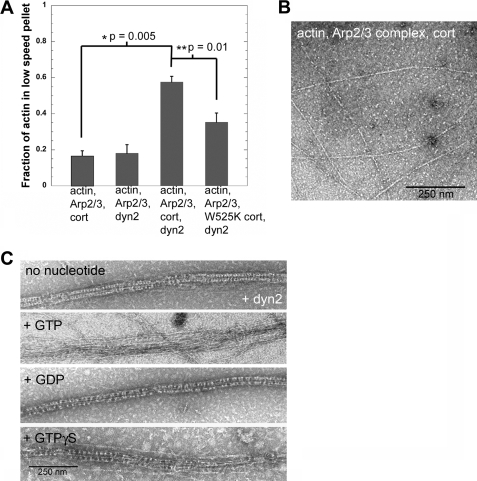
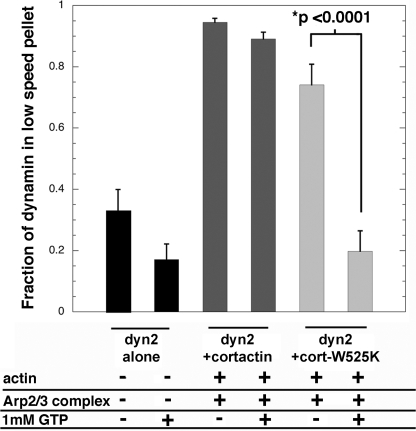
Actin filaments formed as bundles in reactions containing Arp2/3 complex, cortactin, and dynamin2 and the extent of filament bundling depended on the concentration of dynamin2 (Fig. 1 and supplemental Fig. S1, A and B). Approximately 60% of the actin in these reactions sedimented at low speed in the presence of 500 nm dynamin2, whereas <20% sedimented at low speed in the absence of dynamin2. Omitting cortactin, or substituting a mutant form of cortactin (cortactin-W525K) with reduced affinity for dynamin2 (15), decreased the amount of actin in bundles 4- and 1.7-fold, respectively (Fig. 1A). Thus, we conclude that a complex of dynamin2 and cortactin cross-links actin filaments and forms bundled arrays in vitro.
FIGURE 1.
Dynamin2 and cortactin remodel bundled actin filaments in a GTPase-dependent manner. A, F-actin cross-linking by dynamin2 (dyn2) depends on cortactin. Plotted is the fraction of actin pelleted after low speed centrifugation (18,000 × g for 30 min) in reactions containing 1.5 μm actin, 50 nm Arp2/3 complex, 500 nm cortactin (cort), or 500 nm cortactin-W525K and 500 nm dynamin2, as indicated. Data plotted are the mean from three to four experiments; error bars indicate the S.E. Representative SDS gels are shown in supplemental Fig. S1_B_. B, transmission electron micrograph of negatively stained actin filaments formed after 10 min by 1.5 μm actin, 50 nm Arp2/3 complex, and 500 nm cortactin. Scale bar is 250 nm. C, transmission electron micrographs of negatively stained actin filaments formed after 10 min by 1.5 μm actin, 50 nm Arp2/3 complex, 500 nm cortactin, and 500 nm dynamin2. Filaments were assembled in the absence of guanine nucleotide for 10 min, followed by a 2-min incubation with 0.4 mm GTP, 1 mm GDP, 0.4 mm GTPγS, or buffer, as indicated. Scale bar is 250 nm.
When viewed in negative-stained preparations by TEM, most of the actin filaments formed in reactions containing Arp2/3 complex, cortactin, and dynamin2 were associated in tight bundles with an average width of 47.5 ± 0.6 nm (Fig. 1C, Table 1). Bundles were decorated along their length with regularly spaced, transverse striations (Fig. 1C). Actin filaments formed in reactions lacking dynamin2 were not bundled (Fig. 1B). A few bundles without striated decorations formed in reactions lacking cortactin (supplemental Fig. 1_C_).
TABLE 1.
Nucleotide-dependent properties of dynamin2-induced filament bundles
Reported is the mean ± S.E. obtained from 6 to 21 bundles for each parameter at each condition.
| Treatment | Bundle width |
|---|---|
| nm | |
| No nucleotide | 47.5 ± 0.6 (n = 12) |
| 0.4 mm GTP | 80.8 ± 2.8a (n = 17) |
| 1.0 mm GDP | 50.1 ± 0.8 (n = 21) |
| 0.4 mm GTPγS | 56.0 ± 1.5a (n = 17) |
| 0.4 mm GMP-PCP | 54.2 ± 0.1a (n = 11) |
GTP hydrolysis by dynamin2 changed the organization of actin filaments within bundles. Although actin filaments remained bundled in the presence of GTP, individual filaments within bundles became loosely associated and the striated decorations were lost (Fig. 1C). The overall width of GTP-treated bundles nearly doubled to 80.8 ± 2.8 nm (Table 1). Filaments in bundles treated with GTPγS or GMP-PCP remained tightly associated, however, overall bundle width increased slightly to 56.0 ± 1.5 and 54.2 ± 0.1, respectively (Fig. 1C, Table 1). Binding of non-hydrolysable GTP analogs to dynamin2 may induce slight changes in filament organization within bundles that increase overall bundle width. A few filaments also appeared loosely associated with some bundles treated with GTPγS; this could result from slow hydrolysis of GTPγS by dynamin2 (39). Bundles in reactions treated with GDP were similar in width (50.1 ± 0.8 nm) to those formed in the absence of nucleotide (Fig. 1C, Table 1). GTPase-dependent reorganization of filaments within bundles depended on cortactin because no changes in filament packing or patterning were observed when GTP was added to the few bundles formed in the absence of cortactin (supplemental Fig. S1_D_). Taken together, TEM views of actin filaments cross-linked by dynamin2 and cortactin reveal that GTP hydrolysis by dynamin2 changes the organization of filaments within bundled arrays.
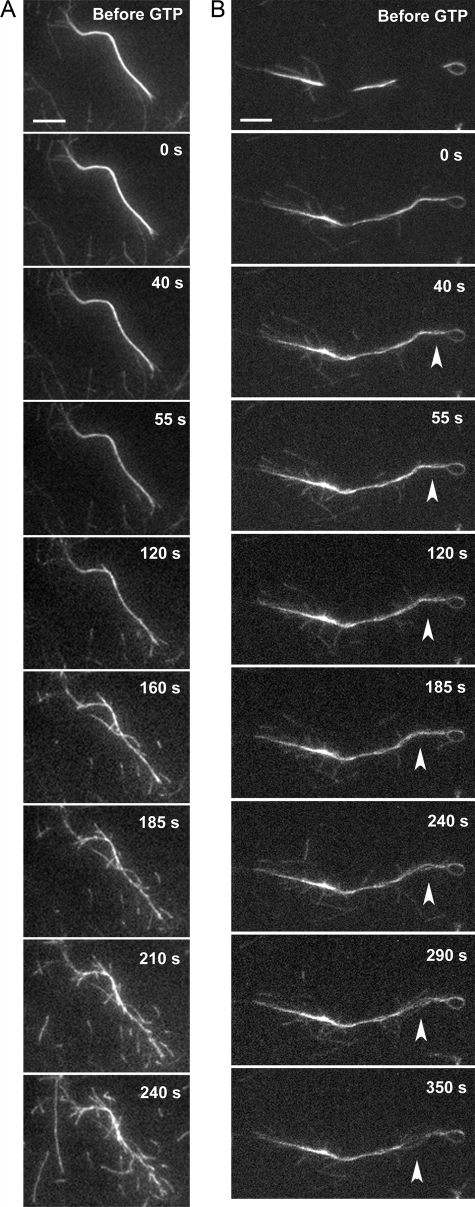
To directly observe reorganization of actin filaments resulting from dynamin2 GTPase activity, we monitored the effects of GTP on bundled actin filaments in real time using TIRFM (Fig. 2). Bundles labeled with Alexa 488-actin were uniform in width along most of their length and tapered slightly at each end. Actin filaments were elongated from both ends of the bundles indicating that filaments within bundles were of mixed polarity. Occasionally, segments of bundles were situated above the evanescent field and were not visible, however, those segments usually moved into the evanescent field when additional reagents, such as guanine nucleotide/buffer, were applied (Fig. 2B, compare the first two panels of time-lapse sequence).
FIGURE 2.
Observing dynamin2 GTPase-dependent filament remodeling in real time. Time-lapse series shows Alexa 488-labeled actin filaments formed by Arp2/3 complex, cortactin, and dynamin2 following addition of 1 mm GTP to the reaction. The numbers indicate the time in seconds following GTP addition. A, actin filaments grow from the sides of uniformly bundled filaments ∼120 s after addition of 1 mm GTP, resulting in a “frayed” appearance along the length of the bundled filaments. At later times short filaments fall onto the coverslip surface from above. This sequence of images corresponds to supplemental Movie S1. Scale bar is 5 μm. B, GTPase-dependent unraveling of actin filaments in a region along a bundle that is not attached to the coverslip surface (arrowhead). This sequence of images corresponds to supplemental Movie S2. Scale bar is 5 μm.
Actin filaments of bundles were remodeled in several ways after GTP addition. First, actin filaments began to elongate from the sides of the bundled arrays 15–45 s after GTP was added, causing the bundles to appear frayed along their length (Fig. 2A and supplemental Movie S1). Second, in regions of bundles that were not attached to the coverslip surface, bundled filaments unraveled and the filaments waved about wildly (Fig. 2B, supplemental Movie S2). Third, short actin filaments appeared to break off the bundles and short filaments fell onto the coverslip surface, suggesting that filaments in bundled arrays above the evanescent field were also fragmented (Fig. 2A, supplemental Movie S1). Filaments within bundles did not appear to slide with respect to one another along the bundle axis; however, small changes in the relative position of filaments within bundles would be difficult to detect. Over time, other bundles fell onto the coverslip and most appeared frayed with filaments emanating along their length (not shown).
No changes in the organization of actin filaments within bundles were observed when either GTPγS or buffer containing no guanine nucleotide was added (supplemental Movies S3 and S4). The few bundles formed in the absence of cortactin also were not altered after GTP addition (supplemental Movie S5). Taken together, the results of the TEM and TIRFM experiments indicate that dynamin2 and cortactin cross-link actin filaments and that GTPase-dependent changes within dynamin2 are transduced via cortactin to dynamically remodel actin filaments.
Filament Remodeling by Dynamin2 GTPase Activity Reveals Actin Filament-barbed Ends and Targets Actin-binding Proteins to Cross-linked Filaments
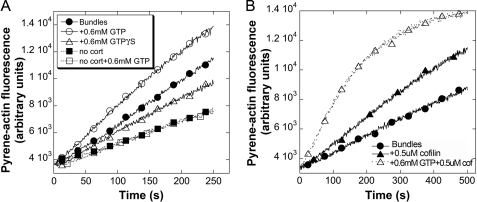
Our observations of filament remodeling using TIRFM suggested several mechanisms by which dynamin2 GTPase activity could influence actin filaments. First, the frayed appearance resulting from filaments elongating from the sides of bundles suggested that dynamin2 GTPase activity exposes filament-barbed ends that become accessible to polymerize actin. To determine whether dynamin2 GTPase activity creates or reveals actin filament-barbed ends within bundled arrays, we quantified free barbed ends associated with bundled filaments using actin assembly assays. Bundled actin filament seeds pre-formed from Arp2/3 complex, cortactin, and dynamin2 were incubated for 30 s with GTP, GTPγS, or buffer prior to dilution into 2 μm G-actin (pyrene-labeled) and the rate of actin assembly from treated seeds was monitored. GTP treatment increased the initial rate of actin assembly from bundled seeds ∼1.3-fold over that obtained from buffer-treated seeds or seeds treated with GTPγS (Fig. 3A). Adding GMP-PCP also did not alter the rate of actin assembly from seeds (data not shown). Cortactin was required for the dynamin2 GTPase-dependent increase in barbed ends.
FIGURE 3.
GTP hydrolysis by dynamin2 reveals actin filament-barbed ends and increases the sensitivity of filaments in bundles to cofilin. A, actin filaments were polymerized 1 h in the presence of 50 nm Arp2/3 complex and 500 nm dynamin2, with (circles and triangles) or without 500 nm cortactin (squares), creating bundled filament seeds. Seeds were treated for 45 s with 0.6 mm GTP (open circles and squares), 0.6 mm GTPγS (open triangles), or with buffer (filled circles and squares) prior to dilution into 2.0 μm G-actin (10% pyrene-labeled). Plotted is the fluorescence of pyrene-actin versus time after dilution. B, bundled filament seeds pre-formed for 1 h were treated with 0.6 mm GTP (open triangles) or with buffer (closed triangles) for 15 s prior to addition of 0.5 μm cofilin for 30 s. Seeds were diluted into 2 μm G-actin (10% pyrene-labeled) and the fluorescence of pyrene-actin was recorded versus time after dilution.
A second mechanism by which dynamin2 GTPase-dependent filament remodeling could influence actin filaments is by modulating binding of other actin-binding proteins to filaments within bundled arrays. For example, bundled actin filaments are weakly sensitive to actin depolymerizing factors (40). Turnover of filaments within bundled arrays could be enhanced if dynamin2 GTPase activity unraveled filaments, making them more accessible to targeting by actin depolymerizing factors. To test the hypothesis that dynamin2 GTPase-dependent filament remodeling promotes targeting of actin-binding proteins to filaments in bundles, we assessed the ability of cofilin to sever filaments in bundled arrays in the presence and absence of GTP. Bundled actin filament seeds were assembled from Arp2/3 complex, cortactin, and dynamin2 and incubated with buffer, cofilin, or cofilin and GTP before addition into assembly reactions containing 2 μm G-actin (pyrene-labeled). The rate of actin assembly from seeds increased 1.5-fold after the seeds were incubated for 30 s in 0.5 μm cofilin (Fig. 3B); this increased rate of actin polymerization likely results from cofilin-mediated severing of filaments in the seed preparations that were not in bundles or of individual filaments emanating from the ends of bundles. However, the rate of actin assembly from seeds increased nearly 4-fold when bundles were treated for 15 s with GTP prior to addition of cofilin (Fig. 3B). Treatment with the non-hydrolysable GTP analogs, GTPγS or GMP-PCP, had no effect on the rate of assembly from cofilin-treated bundled seeds (data not shown). Taken together, our results suggest that GTPase-dependent remodeling by dynamin2 reorganizes filaments within bundles to expose or create new filament-barbed ends and generate loosely bundled actin filaments that become sensitive to severing by actin depolymerizing factors.
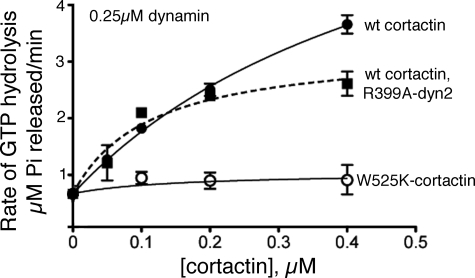
Cortactin Stimulates the Intrinsic GTPase Activity of Dynamin2 and Stabilizes the Association of Dynamin2 and Actin Filaments
Dynamins exhibit an intrinsic GTPase activity that is stimulated by conditions that promote its self-assembly such as low ionic strength buffers or anionic lipids, and by interactions with proteins that bind the C-terminal proline-rich domain (8, 41–44). To determine whether cortactin also influences dynamin2 GTPase activity, we measured GTPase activity in the presence of increasing concentrations of cortactin. Cortactin, but not cortactin-W525K, which binds dynamin2 poorly, stimulated the basal GTPase activity of dynamin2 (Fig. 4). Stimulation by cortactin did not result from enhanced dynamin2 self-assembly because the GTPase activity of a mutant form of dynamin2 (dynamin2-R399A) defective in self-assembly (45) was also stimulated by cortactin (Fig. 4). Thus, cortactin enhances the intrinsic GTPase activity of unassembled dynamin2 via an interaction that depends on the cortactin SH3 domain.
FIGURE 4.
Cortactin stimulates the intrinsic GTPase activity of dynamin2. Plotted are the initial rates of GTP hydrolysis catalyzed by 0.25 μm dynamin2 (circles) or 0.25 μm dynamin2-R399A (squares) with increasing concentrations of wild type (WT) cortactin (filled circles and squares) or cortactin-W525K (open circles). Reactions contained 0.25 mm GTP. Data are presented as the mean initial rate ± S.E. obtained from four to seven experiments. Lines show the fit of the data to a hyperbolic function after correction for the rate of GTP hydrolysis in the absence of cortactin.
Cortactin also stabilized the association of dynamin2 and actin filaments in the presence of GTP. When subjected to low speed centrifugation in a buffer containing 50 mm KCl but without actin filaments, ∼30% of dynamin2 was recovered in the pellet fraction, presumably due to its self-assembly at these ionic conditions. As expected from experiments showing the GTP hydrolysis-dependent release of dynamin2 from lipid tubules (26, 44, 46), approximately half of the assembled dynamin2 was released to the supernatant by GTP (Fig. 5, black bars). In contrast, in the presence of cortactin and actin filaments, nearly all of the dynamin2 sedimented at low speed with bundled filaments with only a small fraction (∼5%) released to the supernatant by GTP (Fig. 5, dark gray bars). Approximately 70% of the dynamin2 sedimented with bundles in reactions containing cortactin-W525K, but nearly all was released from bundles with GTP (Fig. 5, light gray bars). Thus, cortactin stabilized the association of dynamin2 with actin filaments in the presence of GTP, via interactions that depend on Trp-525 of cortactin. Taken together with the ability of cortactin to stimulate the intrinsic dynamin2 GTPase activity, these findings suggest that some GTPase-dependent functions of dynamin2 are executed in synergy with cortactin.
FIGURE 5.
Cortactin stabilizes the interaction of dynamin2 and actin filaments in the presence of GTP. Plotted is the fraction of dynamin2 associated with bundled actin filaments obtained after low speed centrifugation in reactions treated, or not, with GTP. Actin filaments were pre-formed from 1.5 μm actin, 50 nm Arp2/3 complex, 500 nm cortactin (or 500 nm cortactin-W525K), and 500 nm dynamin2 for 30 min, then treated with either 1 mm GTP or buffer for 5 min before low speed centrifugation at 18,000 × g for 10 min. A control reaction with dynamin2 alone (black bars, n = 4) showed that ∼30% of dynamin2 sedimented independent of the other components, presumably due to self-assembly at the ionic conditions used; other reactions contained actin, dynamin2, and either wild type cortactin (dark gray bars, n = 4) or mutant cortactin-W525K (light gray bars, n = 3) as indicated. Data are presented as the mean ± S.E. obtained from three to four experiments. Error bars indicate the S.E.
Dynamin2 Influences the Global Organization of the Actomyosin Cytoskeleton of U2-OS Cells
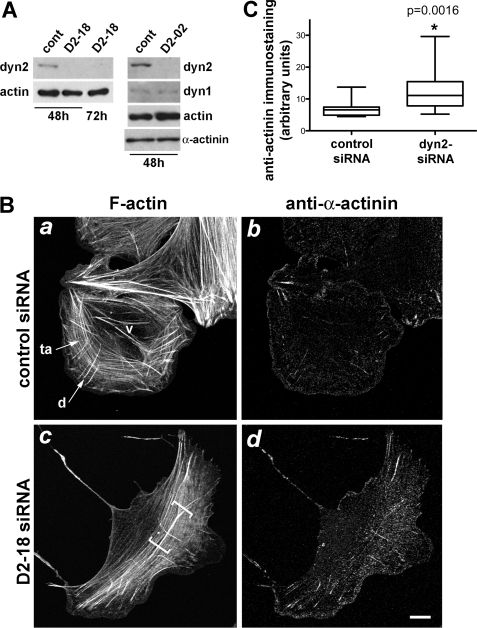
To determine how dynamin2 influences actin filaments in vivo, we observed actin filaments and the distributions of α-actinin and myosin IIA in U2-OS cells in which dynamin2 was depleted using an RNA interference-based approach. Two different siRNAs targeting human dynamin2 reduced [dynamin2] in U2-OS cells by >90% in 48 h, as determined by quantitative immunoblotting of extracts from dynamin2-depleted cells and dilutions of extracts from control cells treated with a non-targeting siRNA (Fig. 6A and data not shown). Efficient depletion of dynamin2 was confirmed using immunolocalization with anti-dynamin2 (supplemental Fig. S2). Dynamin1, which is expressed at low amounts by U2-OS cells, was unaltered in cells treated with siRNAs targeting dynamin2 (Fig. 6A).
FIGURE 6.
Dynamin2 influences the distributions of F-actin and α-actinin in U2-OS cells. A, dynamin2 was efficiently depleted from U2-OS cells using two different siRNAs targeting human dynamin2. Cell extracts obtained 48–72 h after treatment with control siRNA (cont) and siRNAs targeting dynamin2 (D2-02 and D2-18) were analyzed for dynamin2, dynamin1, actin, and α-actinin. Extracts were prepared from equal numbers of cells in all samples and equal volumes were loaded in each lane. B, cells were fixed and stained with rhodamine-phalloidin (a and c) and anti-α-actinin (b and d) 48 h after transfection with control (a and b) or dynamin2-specific (D2-18) (c and d) siRNAs. The predominant types of stress fibers elaborated by U2-OS cells are labeled in panel a: ta, transverse arc; d, dorsal stress fiber; v, ventral stress fiber; the diffuse transverse arcs in dynamin2-depleted cells are indicated with brackets in panel c and correspond to regions where staining for α-actinin is enhanced. Representative cells are shown for each condition; additional images of control and dynamin2-depleted cells stained to reveal F-actin and α-actinin are provided in supplemental Fig. S3. Scale bar is 10 μm. C, box and whisker plot shows the integrated fluorescence intensity/area for anti-α-actinin immunostaining in control siRNA-treated and dynamin2 siRNA-treated U2-OS cells. Data were collected from 17 cells in each group; boxes indicate the median value and the upper and lower quartile values, and whiskers indicate the minimum and maximum values of the data. * indicates p = 0.0016.
Actomyosin-containing stress fibers are the predominant F-actin structures in U2-OS cells and three major classes of stress fibers have been described (Fig. 6B, panel a): 1) distinct transverse arcs arrayed circumferentially and parallel to the cell edge; 2) shorter dorsal stress fibers oriented perpendicular to the cell edge and anchored at one end via a peripheral focal adhesion and intersecting transverse arcs more centrally; and 3) ventral stress fibers in which both ends are anchored to focal adhesions on the ventral surface (47–49).
The organization of F-actin in transverse arcs was perturbed in most dynamin2-deficient cells as visualized in fixed cells stained with fluorescent-phalloidin (Fig. 6B, panel c; supplemental Fig. S3, A and B). Notably, actin filaments comprising transverse arcs appeared diffuse in dynamin2-depleted cells compared with the distinct filament bundles that comprise transverse arcs of control siRNA-treated cells. Although it was difficult to quantify the organizational state of F-actin visualized in fixed, phalloidin-stained cells, transverse arcs were scored by blinded observers as diffuse/disorganized in ∼60% of dynamin2-depleted cells compared with ∼20% of control siRNA-treated cells. In addition, when present in dynamin2-depleted cells, dorsal stress fibers were frequently long and poorly integrated with transverse arcs, intersecting arcs at varying angles or curved, rather than straight. Time-lapse observations of control and dynamin2-depleted cells expressing GFP-actin confirmed that well defined, distinct stress fibers comprise the transverse arcs of control cells, whereas diffusely arrayed transverse arcs that were difficult to distinguish from the soluble pool of GFP-actin, formed predominantly in dynamin2-depleted cells (supplemental Movies S6 and S7).
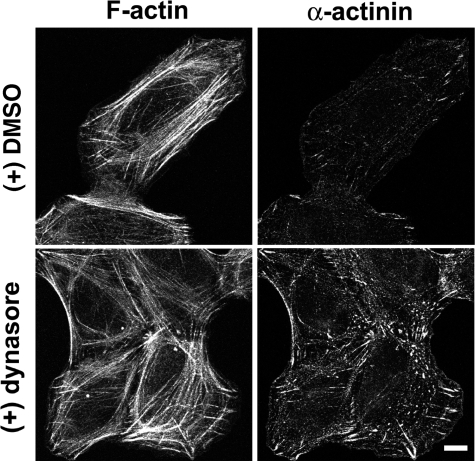
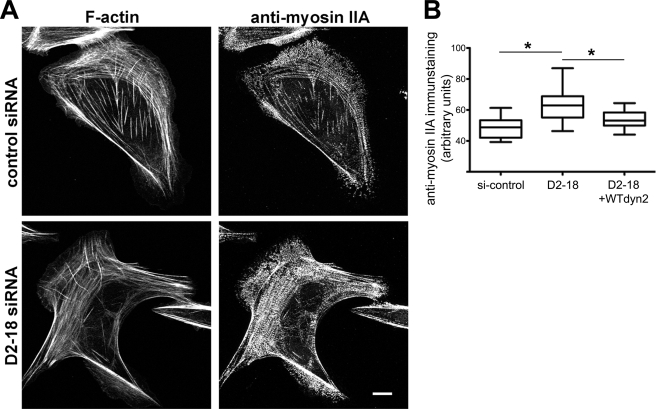
The effects of dynamin2 depletion on the actin cytoskeleton were also reflected in distributions of two other proteins: α-actinin and non-muscle myosin IIA. α-Actinin, which is enriched at the base of dorsal fibers and distributed in a finely stippled pattern in control U2-OS cells (Fig. 6B, panel b), exhibited an enhanced association with the diffusely arrayed transverse arcs in dynamin2-depleted cells (Fig. 6B, panel d, and supplemental Fig. S3, A and B). Quantitative analysis of the intensity of the anti-α-actinin immunostaining/cell area in control and dynamin2-depleted cells showed that α-actinin immunostaining was increased in dynamin2-depleted cells without changes in the cellular level of α-actinin, suggesting that more α-actinin was associated with the cytoskeleton (Fig. 6, A and C). Interestingly, α-actinin immunostaining was also enhanced in U2-OS cells treated for 20 min with the dynamin inhibitor, dynasore, however, no dramatic changes in the organization of F-actin were apparent (Fig. 7). Similarly, myosin IIA appeared poorly organized, particularly in regions where transverse arcs were diffuse, and the intensity of anti-myosin IIA immunostaining was increased in dynamin2-depleted cells (Fig. 8, A and B). Expression in dynamin2-depleted cells of rat wild type dynamin2 resistant to the D2-18 siRNA targeting human dynamin2 restored anti-myosin IIA immunostaining to a level close to that observed in control siRNA-treated cells (Fig. 8B). Thus, disrupting dynamin2 function by decreasing the amount of dynamin2 protein perturbs the global organization of actomyosin. Moreover, acute, pharmacologic perturbation of dynamin function rapidly alters the distribution of α-actinin, suggesting that changes in actin filament cross-linking may be immediate effects of perturbing dynamin function.
FIGURE 7.
Acute perturbation of dynamin with dynasore influences α-actinin distribution but not the overall F-actin organization. U2-OS cells were treated with 80 μm dynasore or DMSO (0.4%) for 20 min at 37 °C before fixing and immunostaining with anti-α-actinin (right panels) and rhodamine-phalloidin (left panels). Scale bar is 10 μm.
FIGURE 8.
Dynamin2 influences the distributions of F-actin and non-muscle myosin IIA; exogenous rat dynamin2 restores the myosin IIA distribution in U2-OS cells. A, cells were fixed and stained with rhodamine-phalloidin (left panels) or anti-myosin IIA (right panels) 48 h after transfection with control (upper panels) or dynamin2-specific (D2-18) siRNAs. Representative cells are shown for each condition; additional images of actomyosin distributions in control and dynamin2-depleted cells are shown in supplemental Fig. S4. Scale bar is 10 μm. B, box and whisker plot shows the integrated fluorescence intensity/area of anti-α-actinin immunostaining in control siRNA-treated, D2-18 siRNA-treated and D2-18 siRNA-treated cells injected with a plasmid driving expression of rat wild type dynamin2 for 6 h before fixing. Data were collected from 19 to 26 cells in each group; boxes indicate the median value and upper and lower quartile values, and whiskers indicate the minimum to maximum values of each data set. * indicates p < 0.0001.
DISCUSSION
We investigated the mechanisms by which dynamin2 and cortactin influence actin filaments in vitro and in vivo. In a simple biochemical system composed of actin, Arp2/3 complex, cortactin, and dynamin2, actin filaments were cross-linked into bundles and GTP hydrolysis by dynamin2 remodeled actin filaments within the bundled arrays. Most intriguing were real time observations by TIRF microscopy that filaments cross-linked by dynamin2 and cortactin unraveled and waved about in the presence of GTP. This GTP hydrolysis-dependent activity of dynamin2, which we call filament remodeling, required cortactin. Cortactin also stimulated the GTPase activity of unassembled dynamin2. We suggest that dynamin2, together with cortactin, comprise a dynamic actin filament remodeling complex that functions during formation of higher-order cytoskeletal structures. Consistent with this notion, actomyosin was perturbed in U2-OS cells depleted of dynamin2, as reflected in the distributions of F-actin, α-actinin, and myosin IIA. Two of these components, α-actinin and myosin IIA, cross-link actin filaments, suggesting that dynamin2 regulates the extent of actin filament cross-linking.
The simplest model of the dynamin-cortactin actin filament remodeling complex is a dynamin dimer/tetramer interacting with actin filaments via cortactin bridges (supplemental Fig. S5). An alternate, not mutually exclusive, arrangement is that a dynamin-cortactin complex associates with a single actin filament via interactions of each cortactin-F-actin binding site with the same filament (not shown). Cortactin, which is essential for remodeling in vitro and stimulates the basal GTPase activity of unassembled dynamin, is likely an essential linkage in vivo. Dynamic remodeling of actin filaments could result if GTP hydrolysis-dependent conformational changes within dynamin2 are transduced via cortactin to actin filaments, resulting in changes in the relative orientations of the filaments. By changing the relative orientations of actin filaments, dynamin2 GTPase activity could influence associations to filaments of actin-binding proteins, such as α-actinin or cofilin. In addition, if both cortactin-F-actin binding sites of the complex were associated with the same actin filament, GTPase-induced remodeling could sever the filament and thereby contribute to filament turnover. Thus, actin filament remodeling by dynamin2 could influence the actin cytoskeleton via two mechanisms: 1) by organizing filaments and modulating their interactions with actin-binding proteins, and 2) by promoting filament turnover. Although not yet supported by our experiments, the GTPase cycle of dynamin2 could also act as a timer of filament remodeling/turnover to govern local changes in cytoskeletal organization in response to signals from cell surface receptors, cell-generated tension, or other stimuli.
The poorly organized actomyosin arrays observed in U2-OS cells depleted of dynamin2 suggests that actin filament remodeling by dynamin2 contributes to maintaining the global actin cytoskeleton. Although neither dynamin2 nor cortactin are prominently associated with stress fibers or transverse arcs, both proteins are enriched within dendritic actin networks that comprise the lamellipodia at the cell periphery (11, 15, 35, 50). Transverse arcs of U2-OS cells are proposed to originate from Arp2/3 complex-dependent, dendritic actin filaments (48) and studies in migrating fibroblasts suggest that lamellipodial actin filaments give rise to actomyosin filaments of the cell body (51, 52). We speculate that dynamin2 and cortactin influence the assembly and integration of actomyosin arrays by remodeling a subset of lamellipodial dendritic actin filaments that ultimately comprise the transverse arcs of the lamella. Similarly, dynamin2 could remodel dendritic actin networks to generate other specialized F-actin-rich structures, such as phagocytic cups (10, 16), F-actin pedestal-like structures induced by enteropathogenic E. coli (19), or podosomes (12, 30). Moreover, GTPase-dependent filament remodeling by dynamin2 could establish actin filaments that contribute to early and later steps of clathrin-mediated endocytosis (53–56).
In addition to organizing filaments, another possible consequence of dynamin2-dependent filament remodeling activity is regulating filament turnover. In reactions with purified components, GTP hydrolysis-dependent filament remodeling increased the susceptibility of bundled filaments to severing by cofilin, which selectively severs single versus bundled actin filaments (40, 57). Furthermore, some actin filaments of GTP-treated bundles appeared to break when viewed in real time. Thus, dynamin2 GTPase activity could promote actin filament turnover in vivo by severing filaments directly and by increasing the sensitivity of actin filaments to actin depolymerizing factors. It is unlikely that dynamin2 functions as a general regulator of filament turnover, because proteins of the ADF/cofilin family efficiently provide that function. Rather, we suggest that dynamin2-dependent remodeling may fine-tune the turnover of a subset of actin filaments. In this mode, filament remodeling encompasses filament “trimming” as well as orienting filaments. Experiments that examine the dynamics and organization of specialized cortical actin structures, such as phagocytic cups or podosomes, in live cells will pinpoint which F-actin structures are influenced by the actin-related activities of dynamin2.
It is possible that the perturbations in cytoskeletal organization observed in dynamin2-depleted U2-OS cells result, in part, from impaired endocytosis and signaling. Depleting dynamin2 was recently reported to slow internalization of activated β1-integrins (58), which could indirectly influence the organization of actomyosin filaments by altering focal adhesion turnover. Overexpression of mutant dynamin2-K44A that potently blocks endocytosis resulted in robustly distinct stress fibers (Ref. 27, and data not shown), unlike the diffuse transverse arcs elaborated by dynamin2-depleted U2-OS cells. Whether or not dynamin2 GTPase regulates actin filaments independently of its role in endocytosis remains to be determined.
In summary, we conclude that dynamin2 and cortactin comprise a dynamic, GTPase-dependent actin filament remodeling complex that may influence actin filaments via two mechanisms: modulating filament organization and the extent of filament cross-linking and filament turnover. Each of these processes could be involved in orchestrating actomyosin organization. Whether or not actin filament remodeling by dynamin2 acts coordinately with dynamin2-dependent membrane remodeling during endocytosis is an intriguing idea that remains to be investigated. Both dynamin2 and cortactin are involved in processes in which membranes are remodeled (22, 59). Dynamin2 could remodel actin filaments during early rate-limiting steps of clathrin-mediated endocytosis as cargo receptors cluster and nascent coated pits begin to form. Filament remodeling could also orient actin filaments during the late steps of clathrin-mediated endocytosis that are thought to involve dynamic actin (55, 60,1–62). The abilities of dynamin2 to simultaneously remodel both membranes and actin filaments could optimally promote coated vesicle formation.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Pekka Lappalainen and Rick Horwitz for plasmids used in this work, Ammasi Periasamy of the W.M. Keck Center for Cellular Imaging at the University of Virginia, Suthirta Datta for assistance with TIRF microscopy, and John Cooper for stimulating discussions.
*
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM067222 (to D. A. S.) from the NIGMS and Ruth L. Kirschstein National Research Service Award F32 GM083538 (to O. L. M.). This work was also supported by National Institutes of Health R01 GM038542 (awarded to J. A. Cooper, Washington University School of Medicine).
2
The abbreviations used are:
siRNA
small interfering RNA
GST
glutathione _S_-transferase
GMP-PCP
guanosine 5′-(β,γ)-methylenetriphosphate
TIRFM
total internal reflection fluorescence microscopy
DTT
dithiothreitol
SH3
Src homology domain 3
GFP
green fluorescent protein
TEM
transmission electron microscopy
Pipes
1,4-piperazinediethanesulfonic acid
GTPγS
guanosine 5′-3-_O_-(thio)triphosphate.
REFERENCES
- 1.Pollard T. D., Borisy G. G. (2003) Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 2.Loisel T. P., Boujemaa R., Pantaloni D., Carlier M. F. (1999) Nature 401, 613–616 [DOI] [PubMed] [Google Scholar]
- 3.Upadhyaya A., van Oudenaarden A. (2003) Curr. Biol. 13, R734–R744 [DOI] [PubMed] [Google Scholar]
- 4.Akin O., Mullins R. D. (2008) Cell 133, 841–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orth J. D., McNiven M. A. (2003) Curr. Opin. Cell Biol. 15, 31–39 [DOI] [PubMed] [Google Scholar]
- 6.Kruchten A. E., McNiven M. A. (2006) J. Cell Sci. 119, 1683–1690 [DOI] [PubMed] [Google Scholar]
- 7.Schafer D. A. (2004) Traffic 5, 463–469 [DOI] [PubMed] [Google Scholar]
- 8.Soulet F., Yarar D., Leonard M., Schmid S. L. (2005) Mol. Biol. Cell 16, 2058–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yarar D., Waterman-Storer C. M., Schmid S. L. (2007) Dev. Cell 13, 43–56 [DOI] [PubMed] [Google Scholar]
- 10.Gold E. S., Underhill D. M., Morrissette N. S., Guo J., McNiven M. A., Aderem A. (1999) J. Exp. Med. 190, 1849–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McNiven M. A., Kim L., Krueger E. W., Orth J. D., Cao H., Wong T. W. (2000) J. Cell Biol. 151, 187–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ochoa G. C., Slepnev V. I., Neff L., Ringstad N., Takei K., Daniell L., Kim W., Cao H., McNiven M., Baron R., De Camilli P. (2000) J. Cell Biol. 150, 377–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee E., De Camilli P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 161–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orth J. D., Krueger E. W., Cao H., McNiven M. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 167–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schafer D. A., Weed S. A., Binns D., Karginov A. V., Parsons J. T., Cooper J. A. (2002) Curr. Biol. 12, 1852–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Di A., Nelson D. J., Bindokas V., Brown M. E., Libunao F., Palfrey H. C. (2003) Mol. Biol. Cell 14, 2016–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krueger E. W., Orth J. D., Cao H., McNiven M. A. (2003) Mol. Biol. Cell 14, 1085–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNiven M. A., Baldassarre M., Buccione R. (2004) Front. Biosci. 9, 1944–1953 [DOI] [PubMed] [Google Scholar]
- 19.Unsworth K. E., Mazurkiewicz P., Senf F., Zettl M., McNiven M., Way M., Holden D. W. (2007) Cell Microbiol. 9, 438–449 [DOI] [PubMed] [Google Scholar]
- 20.Liu Y. W., Surka M. C., Schroeter T., Lukiyanchuk V., Schmid S. L. (2008) Mol. Biol. Cell 19, 5347–5359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao H., Orth J. D., Chen J., Weller S. G., Heuser J. E., McNiven M. A. (2003) Mol. Cell. Biol. 23, 2162–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sauvonnet N., Dujeancourt A., Dautry-Varsat A. (2005) J. Cell Biol. 168, 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinshaw J. E. (2000) Annu. Rev. Cell Dev. Biol. 16, 483–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mears J. A., Ray P., Hinshaw J. E. (2007) Structure 15, 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bashkirov P. V., Akimov S. A., Evseev A. I., Schmid S. L., Zimmerberg J., Frolov V. A. (2008) Cell 135, 1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pucadyil T. J., Schmid S. L. (2008) Cell 135, 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Damke H., Baba T., Warnock D. E., Schmid S. L. (1994) J. Cell Biol. 127, 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ezratty E. J., Partridge M. A., Gundersen G. G. (2005) Nat. Cell Biol. 7, 581–590 [DOI] [PubMed] [Google Scholar]
- 29.Gray N. W., Kruchten A. E., Chen J., McNiven M. A. (2005) J. Cell Sci. 118, 1279–1290 [DOI] [PubMed] [Google Scholar]
- 30.Bruzzaniti A., Neff L., Sanjay A., Horne W. C., De Camilli P., Baron R. (2005) Mol. Biol. Cell 16, 3301–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spudich J. A., Watt S. (1971) J. Biol. Chem. 246, 4866–4871 [PubMed] [Google Scholar]
- 32.Bryan J. (1986) Methods Enzymol. 134, 13–23 [DOI] [PubMed] [Google Scholar]
- 33.Mahaffy R. E., Pollard T. D. (2006) Biophys. J. 91, 3519–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgs H. N., Blanchoin L., Pollard T. D. (1999) Biochemistry 38, 15212–15222 [DOI] [PubMed] [Google Scholar]
- 35.Weed S. A., Karginov A. V., Schafer D. A., Weaver A. M., Kinley A. W., Cooper J. A., Parsons J. T. (2000) J. Cell Biol. 151, 29–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin H. C., Barylko B., Achiriloaie M., Albanesi J. P. (1997) J. Biol. Chem. 272, 25999–26004 [DOI] [PubMed] [Google Scholar]
- 37.Marks B., Stowell M. H., Vallis Y., Mills I. G., Gibson A., Hopkins C. R., McMahon H. T. (2001) Nature 410, 231–235 [DOI] [PubMed] [Google Scholar]
- 38.Leonard M., Song B. D., Ramachandran R., Schmid S. L. (2005) Methods Enzymol. 404, 490–503 [DOI] [PubMed] [Google Scholar]
- 39.Eccleston J. F., Binns D. D., Davis C. T., Albanesi J. P., Jameson D. M. (2002) Eur. Biophys. J. 31, 275–282 [DOI] [PubMed] [Google Scholar]
- 40.Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C. J., Martiel J. L., Blanchoin L. (2007) Curr. Biol. 17, 825–833 [DOI] [PubMed] [Google Scholar]
- 41.Warnock D. E., Hinshaw J. E., Schmid S. L. (1996) J. Biol. Chem. 271, 22310–22314 [DOI] [PubMed] [Google Scholar]
- 42.Sweitzer S. M., Hinshaw J. E. (1998) Cell 93, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 43.Barylko B., Binns D., Lin K. M., Atkinson M. A., Jameson D. M., Yin H. L., Albanesi J. P. (1998) J. Biol. Chem. 273, 3791–3797 [DOI] [PubMed] [Google Scholar]
- 44.Danino D., Moon K. H., Hinshaw J. E. (2004) J. Struct. Biol. 147, 259–267 [DOI] [PubMed] [Google Scholar]
- 45.Ramachandran R., Surka M., Chappie J. S., Fowler D. M., Foss T. R., Song B. D., Schmid S. L. (2007) EMBO J. 26, 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stowell M. H., Marks B., Wigge P., McMahon H. T. (1999) Nat. Cell Biol. 1, 27–32 [DOI] [PubMed] [Google Scholar]
- 47.Small J. V., Rottner K., Kaverina I., Anderson K. I. (1998) Biochim. Biophys. Acta 1404, 271–281 [DOI] [PubMed] [Google Scholar]
- 48.Hotulainen P., Lappalainen P. (2006) J. Cell Biol. 173, 383–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naumanen P., Lappalainen P., Hotulainen P. (2008) J. Microsc. 231, 446–454 [DOI] [PubMed] [Google Scholar]
- 50.Wu H., Parsons J. T. (1993) J. Cell Biol. 120, 1417–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Small J. V., Resch G. P. (2005) Curr. Opin. Cell Biol. 17, 517–523 [DOI] [PubMed] [Google Scholar]
- 52.Anderson T. W., Vaughan A. N., Cramer L. P. (2008) Mol. Biol. Cell 19, 5006–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loerke D., Mettlen M., Yarar D., Jaqaman K., Jaqaman H., Danuser G., Schmid S. L. (2009) PLoS Biol. 7, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sever S., Damke H., Schmid S. L. (2000) J. Cell Biol. 150, 1137–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itoh T., Erdmann K. S., Roux A., Habermann B., Werner H., De Camilli P. (2005) Dev. Cell 9, 791–804 [DOI] [PubMed] [Google Scholar]
- 56.Roux A., Uyhazi K., Frost A., De Camilli P. (2006) Nature 441, 528–531 [DOI] [PubMed] [Google Scholar]
- 57.Huang S., Robinson R. C., Gao L. Y., Matsumoto T., Brunet A., Blanchoin L., Staiger C. J. (2005) Plant Cell 17, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chao W. T., Kunz J. (2009) FEBS Lett. 583, 1337–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salvarezza S. B., Deborde S., Schreiner R., Campagne F., Kessels M. M., Qualmann B., Caceres A., Kreitzer G., Rodriguez-Boulan E. (2009) Mol. Biol. Cell 20, 438–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Merrifield C. J., Feldman M. E., Wan L., Almers W. (2002) Nat. Cell Biol. 4, 691–698 [DOI] [PubMed] [Google Scholar]
- 61.Merrifield C. J., Perrais D., Zenisek D. (2005) Cell 121, 593–606 [DOI] [PubMed] [Google Scholar]
- 62.Yarar D., Waterman-Storer C. M., Schmid S. L. (2005) Mol. Biol. Cell 16, 964–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data