Genome-Wide Screen Identifies Localized RNAs Anchored At Cell Protrusions Through Microtubules And APC (original) (raw)
. Author manuscript; available in PMC: 2009 Nov 25.
Published in final edited form as: Nature. 2008 May 1;453(7191):115–119. doi: 10.1038/nature06888
Abstract
RNA localization is important for establishment and maintenance of polarity in multiple cell types. Localized RNAs are usually transported along microtubules or actin filaments1, and become anchored at their destination to some underlying subcellular structure. Retention commonly involves actin or actin-associated proteins2–7, although cytokeratin filaments and dynein anchor certain RNAs8,9. RNA polarization is important for diverse processes ranging from cell fate determination to synaptic plasticity. However, there has been to date no comprehensive identification of localized RNAs in any mammalian cell type. Here we have addressed this issue, focusing on migrating fibroblasts, which polarize to form a leading edge and a tail, in a process that involves asymmetric distribution of RNAs10–14. We used a fractionation scheme15 combined with microarrays to identify for the first time, on a genome-wide scale, localized RNAs in mammalian cells. We find that a diverse group of RNAs accumulates in pseudopodial protrusions during cell migration. Through their 3’UTRs these transcripts are anchored in granules concentrated at the plus ends of detyrosinated microtubules. RNAs in the granules associate with the APC tumor suppressor and the Fragile X Mental Retardation protein (FMRP). APC is required for accumulation of transcripts in protrusions. Our results reveal a new type of RNA anchoring mechanism as well as a novel, unanticipated function for APC in localizing RNAs.
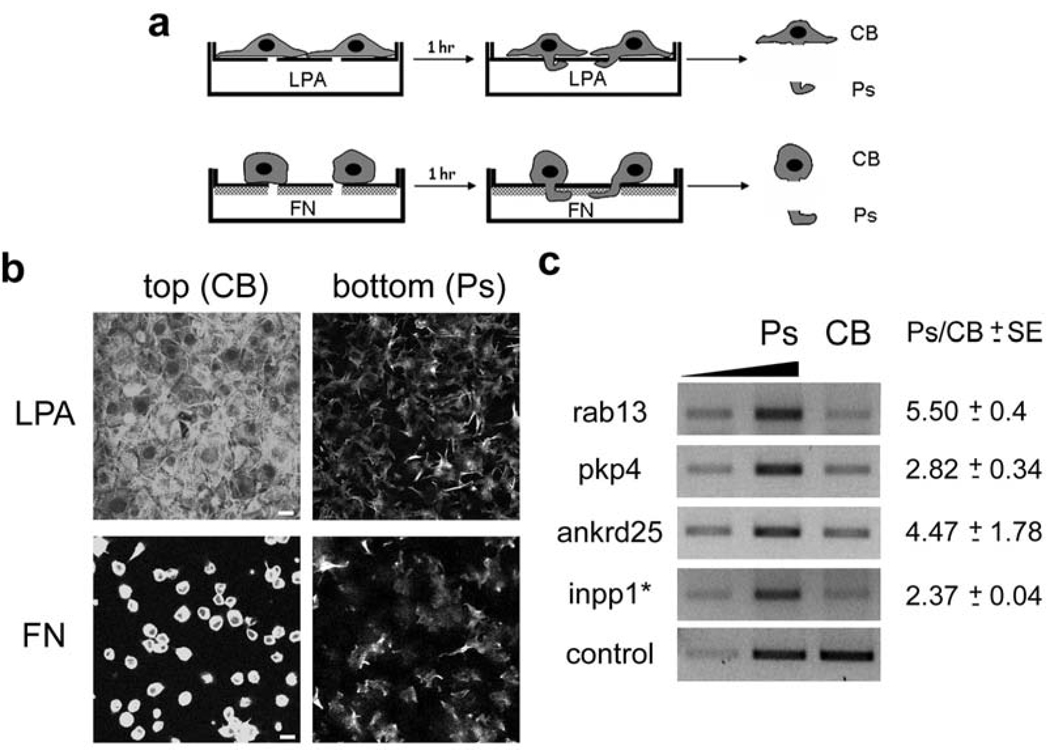
To identify on a genome-wide scale RNAs that are enriched at the leading edge of migrating cells, we employed a fractionation method in which cells are plated on a microporous filter and are induced to polarize and extend pseudopodial protrusions in response to a migratory stimulus. Pseudopodia and cell bodies are then physically isolated and their contents compared15. We isolated pseudopodia (Ps) and cell body (CB) fractions from NIH/3T3 cells migrating towards a chemotactic (lysophosphatidic acid, LPA) or a haptotactic (fibronectin, FN) stimulus (Fig. 1a, b and S1a). The quality of the fractionation was assessed by immunoblotting for activated FAK (phosphorylated at Y397), which is concentrated at the leading edge during migration15,16. Activated FAK was enriched in the pseudopodial fraction (Fig. S1b), whereas total FAK and Ran were not. Total RNA from Ps and CB fractions was hybridized on Affymetrix GeneChip arrays, which analyze the expression level of over 39,000 transcripts. The resulting signals were processed to identify transcripts that show an asymmetric distribution.
Figure 1. Several RNAs are enriched in protruding pseudopodia of migrating cells.
a, Schematic depicting strategies used for isolation of pseudopodia (Ps) and cell bodies (CB). NIH/3T3 cells were allowed to attach onto FN-coated microporous filters and were induced to extend protrusions by adding LPA in the bottom chamber. Alternatively, only the underside of microporous filters was coated with FN and cells plated on top extended protrusions towards the FN-coated surface. CB and Ps fractions were physically isolated after 1 hr of induction. b, Cells extending protrusions in response to LPA or FN, as described in a, were fixed and stained with FITC-phalloidin. Confocal images of the top and bottom side of the filter are shown. Scale bar: 15µm. c, Total RNA was isolated from Ps and CB fractions of cells extending protrusions in response to LPA. The mRNAs indicated on the left were detected by quantitative RT-PCR. Increasing amounts of the Ps sample were amplified (lanes 1 and 2) to ensure linearity of the amplification. RNA levels were normalized to the control, arpc3, mRNA and the Ps/CB ratio was calculated. Values on the right indicate mean ± SEM of 3 independent experiments.
About 50 RNAs were significantly enriched in pseudopodia in response to both migratory stimuli (table 1 and S1). This enrichment was verified by quantitative RT-PCR (Fig. 1c) and real-time RT-PCR (Fig. S1c) of representative RNAs. The majority of transcripts enriched in pseudopodia encode proteins with various functions, ranging from membrane traffic to cytoskeletal organization, signaling, microtubule-based transport and RNA metabolism, as well as a number of uncharacterized proteins (see table 1 and S1). However, comparison of the primary sequences of these transcripts did not reveal any readily identifiable motifs shared among them, suggesting that potential localization signals are likely defined by a combination of primary sequence and higher order structure, as is common for most localized RNAs17.
Table 1. Partial list of RNAs significantly enriched in pseudopodia in response to both LPA and fibronectin (FN) stimulation.
Fold change indicates the average fold enrichment in pseudopodia from two independent experiments for each stimulus.
| Accession | LPAfold change | FN fold change | Gene name |
|---|---|---|---|
| vesicle/membrane trafficking | |||
| BC027214 | 5.1 | 3.58 | RAB13, member RAS oncogene family |
| AK013287 | 3.19 | 3.26 | Mak3 homolog (S. cerevisiae) |
| BM222792 | 3.26 | 1.67 | Liprin a1 |
| BC003764 | 2.3 | 2.98 | synaptobrevin like 1 |
| motors | |||
| BC016221 | 4.46 | 2.29 | kinesin family member 1C |
| NM_133796 | 4.01 | 1.6 | dynein light chain LC8 type 2 |
| BI328541 | 2.84 | 2.05 | kinesin family member 5B |
| AV059518 | 2.39 | 2.91 | dynein light chain Tctex-type 3 |
| cytoskeleton organization/signalling | |||
| BQ266693 | 6.46 | 6.92 | inositole polyphosphate 1-phosphatase* (Inpp1*)a |
| AV286396 | 4.14 | 1.99 | plakophilin 4 |
| BB795075 | 4.01 | 2.97 | discoidin domain receptor member 2 |
| AW541674 | 3.81 | 1.93 | mitogen activated protein kinase kinase 7 |
| BG071905 | 3.16 | 2.24 | Palladin |
| AW049055 | 2.94 | 1.59 | GRB2-associated binding protein 2, Gab2 |
| AV219419 | 2.73 | 1.91 | septin 7 |
| BM124893 | 2.47 | 2.92 | adenomatosis polyposis coli |
| NM_007462 | 2.39 | 1.85 | adenomatosis polyposis coli |
| BB794177 | 2.28 | 2.37 | regulator of G-protein signaling 20 |
| RNA metabolism | |||
| AW543705 | 2.97 | 3.63 | HIV TAT specific factor 1 |
| BB381966 | 2.55 | 1.77 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26 |
| AK011930 | 2.55 | 1.59 | heterogeneous nuclear ribonucleoprotein A3 |
| BG797460 | 2.47 | 2.82 | eukaryotic translation initiation factor 2a |
| BG796845 | 2.47 | 3.19 | La protein |
| BB032885 | 2.22 | 1.74 | eukaryotic translation initiation factor 3, subunit 6 |
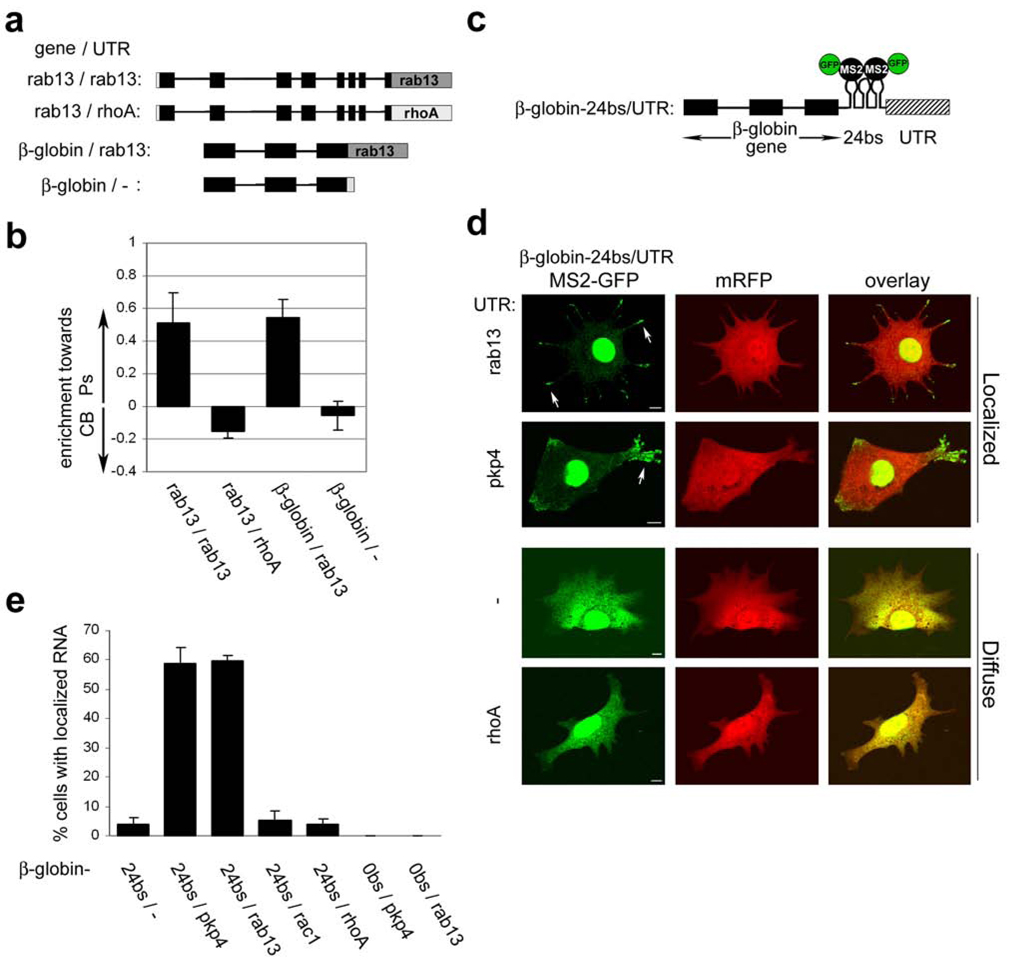
To dissect the localization mechanisms, we focused on the rab13 and plakophilin 4 (pkp4) mRNAs, which both showed robust localization. First, we expressed in NIH/3T3 cells the rab13 gene encompassing the whole open reading frame and 3’UTR (Fig. 2a). A sequence encoding the FLAG tag was introduced at the 5’end of the first coding exon to distinguish the exogenous mRNA from endogenous transcript. Transfected cells plated on filters were induced to migrate by addition of LPA to the bottom chamber, and Ps and CB fractions were isolated. RT-PCR analysis revealed that, like the endogenous rab13 mRNA, the reporter transcript was enriched in pseudopodia (Fig. 2b). Pseudopodial enrichment was abolished by replacement of the 3’ UTR of the rab13 gene with the corresponding region from the non-localized rhoA gene. Moreover, the non-localized β-globin mRNA was recruited to pseudopodia when attached to the rab13 3’ UTR (Fig. 2a, b). All exogenous RNAs were expressed at similar levels, close to the expression level of the endogenous rab13 mRNA (Fig. S2a,b), and in all experiments the distribution of the endogenous rab13 mRNA was determined in parallel, to ensure the reproducibility of the fractionation (data not shown). We conclude that the rab13 3’UTR is necessary and sufficient to direct RNA accumulation in pseudopodia.
Figure 2. The 3’UTRs direct RNAs to accumulate in granules at tips of protrusions.
a, Schematic of transfected constructs. Black boxes: exons, black lines: introns, grey boxes: 3’UTR, white box: FLAG tag. b, Cells transfected with the constructs depicted in a, were fractionated into Ps and CB fractions after induction with LPA. The enrichment of each RNA towards the Ps or CB fraction was calculated as described in Methods. Error bars: SEM c, Schematic depicting the general structure of β-globin constructs used in d, and e,. 24bs: 24 MS2-binding sites d, Cells were co-transfected with plasmids encoding mRFP, MS2-GFP and β-globin constructs with various 3’UTRs. Live cells were imaged during spreading on fibronectin. Shown are representative examples of the localization patterns observed when β-globin mRNA carried the UTRs indicated on the left; localized in granules at the tips of protrusions (arrows) or diffuse in the cytoplasm. Scale bar: 10µm e, Quantification of the percentage of cells exhibiting localized RNA distribution when transfected as in d, with constructs carrying the indicated 3’UTRs.
To visualize the localization pattern conferred by the rab13 3’UTR, we used the MS2 system18. The MS2 bacteriophage coat protein binds with high affinity to RNA elements, multiple repeats of which are introduced into a reporter transcript. MS2 is fused to green fluorescent protein (GFP) and carries a nuclear localization signal to force accumulation in the nucleus. When co-expressed, the MS2-GFP binds to the reporter RNA and accompanies it to the cytoplasm, thus providing indirect detection of reporter RNA distribution in the cytoplasm of live cells.
Twenty-four repeats of the MS2-binding site were introduced into the β-globin gene downstream of the coding region, followed by different 3’UTR sequences (Fig. 2c). These constructs were co-expressed with MS2-GFP and fluorescence was monitored in live cells during spreading on a fibronectin-coated surface. Cells expressing β-globin mRNA attached to control 3’UTRs derived from the vector or from two non-localized mRNAs, rac1 and rhoA, exhibited diffuse fluorescence throughout the cytoplasm. The GFP signal overlapped entirely with the fluorescence from co-transfected mRFP protein, used as a diffuse cytosolic marker (Fig. 2d, bottom panels, and 2e, constructs β-globin-24bs/-, -24bs/rac1 and -24bs/rhoA). By contrast, in a large proportion of the cells that expressed a β-globin mRNA carrying the rab13 3’UTR, GFP was concentrated in granules at the tips of protrusions (Fig. 2d, arrows, top panels, and 2e, construct β-globin-24bs/rab13). Granular accumulation did not result from differences in expression levels, as all RNAs were expressed at similar amounts (Fig. S2c). Furthermore, the signal was dependent on the presence of the 24 MS2-binding sites in the mRNA (Fig. 2e, construct β-globin-0bs/rab13), confirming that it reflects the distribution of the transfected RNA. Significantly, a similar localization was conferred by the 3’UTR of pkp4 (Fig. 2d, e, construct β-globin-24bs/pkp4). Therefore, certain 3’UTRs can direct RNAs to granules at the tips of protrusions, and this property is shared among mRNAs enriched in pseudopodia.
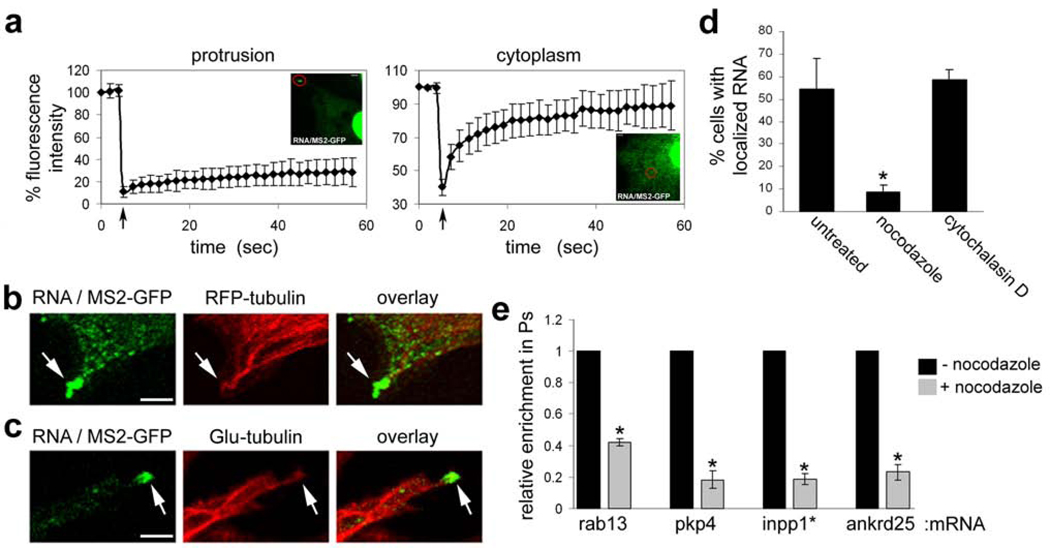
When imaged over time, the localized RNA granules are relatively stationary (suppl. video 1) and appear and disappear with surprisingly slow kinetics (Fig. S3). This stable accumulation of RNAs could represent either the steady-state of a dynamic movement of RNA molecules to and from the granules, or result from sequestration into some cellular structure. To distinguish between these possibilities, we performed fluorescence recovery after photobleaching (FRAP). Since the RNA granules vary in size (see Fig. 2d and S3), we focused, for these experiments, on the smaller granules of ca 1–2 µm in diameter. While bleaching of the fluorescence signal in internal cytoplasmic areas was followed by rapid recovery (Fig. 3a), bleaching of the localized RNA granules was followed by very slow fluorescence recovery (Fig. 3a and suppl. videos 2 and 3), indicating that the transcripts present in these granules are stably associated/anchored in these structures and do not exchange rapidly with free cytoplasmic RNA molecules.
Figure 3. Localized RNA granules are anchored at the plus ends of detyrosinated microtubules.
a, Cells were co-tranfected with constructs encoding MS2-GFP and the β-globin-24bs/pkp4 RNA. Fluorescence intensity in granules at protrusive areas (left panel, red circle) or within the cytoplasm (right panel, red circle) was monitored before and after photobleaching. Arrow indicates time of bleach. Curves represent average values of 10 and 5 independent experiments, respectively. Error bars: SD. b,c, Confocal fluorescence images of cells expressing MS2-GFP, the β-globin-24bs/pkp4 RNA and either RFP-tubulin (b) or fixed and stained with anti-Glu-tubulin antibody (c). Panels show edges of protrusive areas. Scale bar: 3µm. d, Cells expressing mRFP, MS2-GFP and the β-globin-24bs/pkp4 RNA were treated for 30 min with 10µM nocodazole or 1µM cytochalasin D. The percentage of cells exhibiting localized RNA distribution was quantified as in figures 2b and 2c. *: p-value<0.005 by two-tailed t-test versus untreated control. e, Cells on microporous filters were induced to extend protrusions in response to LPA in the absence or presence of 10µM nocodazole (added during the last 25 min of the assay). Enrichment of the indicated mRNAs in pseudopodia was determined as described in Methods and expressed relative to that of the untreated control cells. Values are averages of 3 independent experiments. Error bars: SEM. *: p-value<0.005 by paired, two-tailed t-test versus untreated control.
To gain insight into the identity of the structures that anchor transcripts at the tips of protrusions, the localized RNAs were expressed together with fluorescently-tagged markers of various cellular structures. The RNA granules did not co-localize significantly with Dcp1-containing P-bodies, focal adhesions (Fig. S4a, c, d), cortical actin or actin stress fibers (Fig. S4b). Significantly, however, the granules appeared to be associated with microtubules (MTs), visualized through expression of RFP-tubulin, and were specifically concentrated at their +ends (Fig. 3b and S5). Consistent with an MT association, the RNA granules largely disappeared when cells were treated with the MT-depolymerizing agent, nocodazole (Fig. 3d and S6b), at concentrations that do not affect the actin cytoskeleton (Fig. S6a). By contrast, the presence of RNA granules was not affected when the actin cytoskeleton was disrupted by cytochalasin D (Fig. 3d and S6a,b). Importantly, brief treatment with nocodazole significantly reduced the pseudopodial enrichment of four different endogenous localized RNAs (Fig. 3e), without affecting the overall integrity and number of protrusions (Fig S6c). We conclude that association with MT +ends is a property shared by multiple RNAs found in pseudopodia.
The fact that the RNA granules remain stationary over several minutes (suppl video 1) suggested that the MTs with which they associate do not exhibit dynamic instability. Indeed, the RNA granules did not co-localize significantly with EB1-RFP or RFP-CLIP170 (Fig. S7a, b), two +end tracking proteins (+TIPs) known to associate with +ends of dynamic MTs 19,20. Furthermore, FRAP analysis on protrusions of EB1-GFP-expressing cells showed that, in contrast to the RNA granules, EB1 is highly dynamic with fluorescence recovering after a few seconds (Fig. S8a and suppl video 4). Therefore, we tested whether the RNA granules associate specifically with the +ends of stable MTs, which are marked by post-translational modifications of tubulin. Interestingly, while the RNA granules were not attached to acetylated-MTs (Fig. S9a), they did associate with the +ends of detyrosinated-MTs (Glu-MTs) (Fig. 3c and S9b). Glu-MTs do not significantly grow or shrink over a period of minutes21, in agreement with the dynamics of the RNA granules we observe.
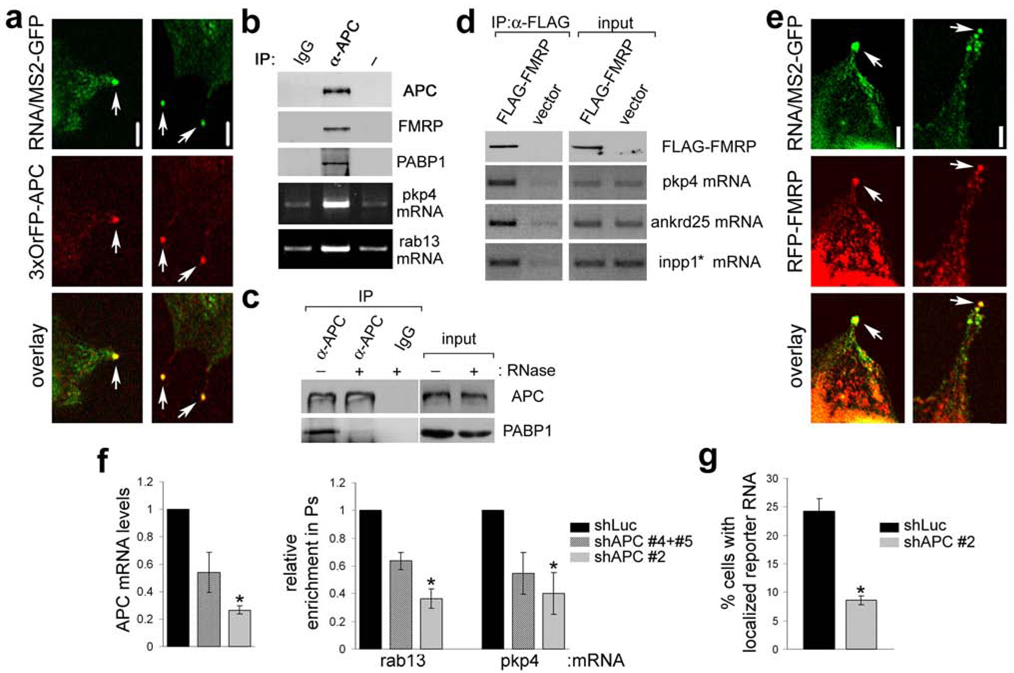
APC is an unusual +TIP that associates with only a minority of MTs, and has been observed in particular at the +ends of Glu-MTs24. In migrating cells APC is attached to a subset of the MTs growing into protrusions towards the leading edge22,23. When NIH/3T3 cells were induced to spread on a fibronectin-coated surface, we found that GFP-APC was mainly concentrated in granules at the tips of protrusions, reminiscent of the distribution exhibited by the localized RNAs (Fig. S10a, arrows). Indeed, co-expression of a localized reporter RNA with APC tagged with 3 copies of orange fluorescent protein (3xOFP-APC) revealed that >90% (n=80) of the RNA granules at the tips of protrusive areas co-localize with APC (Fig. 4a, arrows and S10b). Furthermore, FRAP analysis showed that APC present in granules is stably anchored there, exhibiting a very slow exchange rate (Fig. S8b and suppl video 5). Taken together these data strongly suggest that APC and the localized RNAs are part of the same complex.
Figure 4. APC associates with RNP complexes containing FMRP and is required for localization of RNAs in protrusions.
a, Confocal fluorescence images of cells expressing MS2-GFP, the β-globin-24bs/pkp4 RNA and 3xOrFP-APC. Panels show edges of protrusive areas. b, Lysates of NIH/3T3 cells were immunoprecipitated (IP) with control antibody (IgG), anti-APC antibody (α-APC) or no antibody (-) and analyzed to detect the indicated proteins and mRNAs. c, same as in b, except that prior to IP lysates were treated (+) or not (−) with RNase. d, Lysates of cells transfected with FLAG-FMRP or vector were immunoprecipitated with anti-FLAG antibody and analyzed to detect the indicated proteins and mRNAs. e, Confocal fluorescence images of cells expressing MS2-GFP, the β-globin-24bs/pkp4 RNA and RFP-FMRP. Panels show edges of protrusive areas. f, Cells expressing shRNAs against Luciferase (shLuc) or APC (shAPC), individually or in combination as indicated, were analyzed to determine APC mRNA levels. Shown are average values normalized to β-actin mRNA and expressed relative to that of the shLuc control (left panel). In parallel, cells were plated on filters and induced to extend protrusions in response to LPA. Enrichment of the indicated mRNAs in pseudopodia was determined as described in Methods and expressed relative to that of the shLuc control (right panel). Values are averages of 2–3 independent experiments. *: p-value<0.05 by paired, two-tailed t-test versus shLuc control. g, shLuc and shAPC-expressing cells were transfected with MS2-GFP and the β-globin-24bs/pkp4 RNA. The percentage of cells exhibiting localized RNA distribution, as described in Fig. 2d, was quantified. Values are averages of 7 independent experiments. *: p-value<0.0001 by two-tailed t-test. Error bars: SEM. Scale bar: 5µm.
To test this hypothesis, we asked if these RNAs bind specifically to endogenous APC. Indeed, APC co-immunoprecipitated with both rab13 and pkp4 mRNAs (Fig. 4b). Moreover, APC also bound the cytoplasmic poly(A)-binding protein (PABP1) and this interaction was disrupted by pre-treatment with RNase, indicating that it is mediated through RNA (Fig. 4b,c). Interestingly, we also found that APC associates with the Fragile X Mental Retardation protein (FMRP) (Fig. 4b), a known translational regulator of localized RNAs in other systems25,26. FMRP co-localized with the RNA granules at tips of protrusions and co-IPs with endogenous localized mRNAs (Fig. 4d,e). We conclude that APC is a component of RNP complexes that contain localized RNAs, PABP1 and FMRP.
To directly test whether APC mediates anchoring of the localized RNAs, we knocked-down APC expression using short-hairpin RNAs (shRNAs) (Fig. 4f and S11a,b). Knockdown of APC did not significantly affect the ability of cells to extend protrusions (see Fig S12 and S13 and data not shown), but reduced both the enrichment of the endogenous rab13 and pkp4 transcripts in pseudopodia as well as the localization of the MS2 reporter RNA in protrusions (Fig. 4f,g). It is unlikely that this effect of APC on RNA localization is mediated indirectly through effects of APC on MT dynamics. Consistent with previous reports27,28, APC knockdown does not affect the overall MT organization or the presence of Glu-MTs (Fig S12). APC knockdown did affect the organization of acetylated-MTs (Ac-MTs), causing more cells to exhibit short Ac-MTs (Fig. S13 and ref.27). However, RNA granules are not associated with Ac-MTs (Fig S9a) and can form in cells with short Ac-MTs (Fig S9a, middle panels). Thus, we conclude that APC is directly required for accumulation and anchoring of RNAs in protrusions.
This study provides the first genome-wide identification of asymmetrically distributed RNAs in mammalian cells. We show that, in response to migratory stimuli, >50 mRNAs accumulate in cellular protrusions of fibroblasts, revealing that even in less differentiated polarized cells, RNA localization mechanisms are widely utilized. These RNAs do not include β-actin mRNA and the mRNAs for subunits of the arp2/3 complex, which have been previously observed at lamellipodial regions11,13,14, possibly because in some cell types β-actin mRNA accumulates at the leading edge in only a small percentage of cells29. Significantly, the RNAs identified here are anchored at protrusions through a novel mechanism, which includes accumulation in granules at the plus ends of Glu-MTs. The tumor suppressor protein APC, a protein whose disruption or loss of function affects polarization and cell migration27,30,31, is part of the anchoring mechanism. APC associates both with RNA-binding proteins and RNAs and is required for RNA accumulation in protrusions, raising the possibility that at least some of the effects of APC are mediated through its ability to anchor mRNAs at the tips of cellular protrusions.
Methods
Detailed methods are provided in the Supplementary Information
Supplementary Material
01
Acknowledgements
We thank Yongde Bao for the microarray and real-time PCR analysis. Jens Lykke-Andersen, Inke Nathke, J.Thomas Parsons, Robert Darnell and Gregg Gundersen for plasmids and antibodies. S.M. is a fellow of the Leukemia and Lymphoma Society. This work was supported by grant GM070902 from the NIH to I.G.M.
Footnotes
Author contributions
S.M. performed the experiments. S.M., K.M. and I.G.M. designed experiments and analyzed the data. K.M. provided reagents. S.M. and I.G.M. wrote the manuscript.
References
- 1.St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 2.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 3.Babu K, Cai Y, Bahri S, Yang X, Chia W. Roles of Bifocal, Homer, and F-actin in anchoring Oskar to the posterior cortex of Drosophila oocytes. Genes Dev. 2004;18:138–143. doi: 10.1101/gad.282604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erdelyi M, Michon AM, Guichet A, Glotzer JB, Ephrussi A. Requirement for Drosophila cytoplasmic tropomyosin in oskar mRNA localization. Nature. 1995;377:524–527. doi: 10.1038/377524a0. [DOI] [PubMed] [Google Scholar]
- 5.Jankovics F, Sinka R, Lukacsovich T, Erdelyi M. MOESIN crosslinks actin and cell membrane in Drosophila oocytes and is required for OSKAR anchoring. Curr Biol. 2002;12:2060–2065. doi: 10.1016/s0960-9822(02)01256-3. [DOI] [PubMed] [Google Scholar]
- 6.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu G, et al. Interactions of elongation factor 1alpha with F-actin and beta-actin mRNA: implications for anchoring mRNA in cell protrusions. Mol Biol Cell. 2002;13:579–592. doi: 10.1091/mbc.01-03-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alarcon VB, Elinson RP. RNA anchoring in the vegetal cortex of the Xenopus oocyte. J Cell Sci. 2001;114:1731–1741. doi: 10.1242/jcs.114.9.1731. [DOI] [PubMed] [Google Scholar]
- 9.Delanoue R, Davis I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- 10.Chicurel ME, Singer RH, Meyer CJ, Ingber DE. Integrin binding and mechanical tension induce movement of mRNA and ribosomes to focal adhesions. Nature. 1998;392:730–733. doi: 10.1038/33719. [DOI] [PubMed] [Google Scholar]
- 11.Kislauskis EH, Zhu X, Singer RH. beta-Actin messenger RNA localization and protein synthesis augment cell motility. J Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leung KM, et al. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mingle LA, et al. Localization of all seven messenger RNAs for the actinpolymerization nucleator Arp2/3 complex in the protrusions of fibroblasts. J Cell Sci. 2005;118:2425–2433. doi: 10.1242/jcs.02371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shestakova EA, Singer RH, Condeelis J. The physiological significance of beta -actin mRNA localization in determining cell polarity and directional motility. Proc Natl Acad Sci U S A. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho SY, Klemke RL. Purification of pseudopodia from polarized cells reveals redistribution and activation of Rac through assembly of a CAS/Crk scaffold. J Cell Biol. 2002;156:725–736. doi: 10.1083/jcb.200111032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moissoglu K, Schwartz MA. Integrin signalling in directed cell migration. Biol Cell. 2006;98:547–55. doi: 10.1042/BC20060025. [DOI] [PubMed] [Google Scholar]
- 17.Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. Rna. 2007;13:625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertrand E, et al. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 19.Galjart N. CLIPs and CLASPs and cellular dynamics. Nat Rev Mol Cell Biol. 2005;6:487–498. doi: 10.1038/nrm1664. [DOI] [PubMed] [Google Scholar]
- 20.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Infante AS, Stein MS, Zhai Y, Borisy GG, Gundersen GG. Detyrosinated (Glu) microtubules are stabilized by an ATP-sensitive plus-end cap. J Cell Sci. 2000;113(Pt 22):3907–3919. doi: 10.1242/jcs.113.22.3907. [DOI] [PubMed] [Google Scholar]
- 22.Mimori-Kiyosue Y, Shiina N, Tsukita S. Adenomatous polyposis coli (APC) protein moves along microtubules and concentrates at their growing ends in epithelial cells. J Cell Biol. 2000;148:505–518. doi: 10.1083/jcb.148.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nathke IS, Adams CL, Polakis P, Sellin JH, Nelson WJ. The adenomatous polyposis coli tumor suppressor protein localizes to plasma membrane sites involved in active cell migration. J Cell Biol. 1996;134:165–179. doi: 10.1083/jcb.134.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wen Y, et al. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. doi: 10.1038/ncb1160. [DOI] [PubMed] [Google Scholar]
- 25.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6:376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 26.Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- 27.Kroboth K, et al. Lack of adenomatous polyposis coli protein correlates with a decrease in cell migration and overall changes in microtubule stability. Mol Biol Cell. 2007;18:910–918. doi: 10.1091/mbc.E06-03-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kita K, Wittmann T, Nathke IS, Waterman-Storer CM. Adenomatous polyposis coli on microtubule plus ends in cell extensions can promote microtubule net growth with or without EB1. Mol Biol Cell. 2006;17:2331–2345. doi: 10.1091/mbc.E05-06-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vikesaa J, et al. RNA-binding IMPs promote cell adhesion and invadopodia formation. Embo J. 2006;25:1456–1468. doi: 10.1038/sj.emboj.7601039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watanabe T, et al. Interaction with IQGAP1 links APC to Rac1, Cdc42, and actin filaments during cell polarization and migration. Dev Cell. 2004;7:871–883. doi: 10.1016/j.devcel.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Etienne-Manneville S, Hall A. Cdc42 regulates GSK-3beta and adenomatous polyposis coli to control cell polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01