Targeted chromosomal deletions in human cells using zinc finger nucleases (original) (raw)
Abstract
We present a novel approach for generating targeted deletions of genomic segments in human and other eukaryotic cells using engineered zinc finger nucleases (ZFNs). We found that ZFNs designed to target two different sites in a human chromosome could introduce two concurrent DNA double-strand breaks (DSBs) in the chromosome and give rise to targeted deletions of the genomic segment between the two sites. Using this method in human cells, we were able to delete predetermined genomic DNA segments in the range of several-hundred base pairs (bp) to 15 mega-bp at frequencies of 10−3 to 10−1. These high frequencies allowed us to isolate clonal populations of cells, in which the target chromosomal segments were deleted, by limiting dilution. Sequence analysis revealed that many of the deletion junctions contained small insertions or deletions and microhomologies, indicative of DNA repair via nonhomologous end-joining. Unlike other genome engineering tools such as recombinases and meganucleases, ZFNs do not require preinsertion of target sites into the genome and allow precise manipulation of endogenous genomic scripts in animal and plant cells. Thus, ZFN-induced genomic deletions should be broadly useful as a novel method in biomedical research, biotechnology, and gene therapy.
The ability to generate targeted deletions of genomic DNA greater than 10 kilobase pairs (kbp) in length could expand genetic and genomic studies in new dimensions by allowing the selective removal of gene clusters, intergenic regions, exons, and introns from a genome and may have broad applications in research, biotechnology, and gene therapy, but it has been difficult, if not impossible, to achieve this aim in higher eukaryotic cells and organisms. Recombinase systems such as Flp/FRT (Ryder et al. 2007) and Cre/loxP (Ramirez-Solis et al. 1995) and bacterial artificial chromosome (BAC)-based gene targeting (Valenzuela et al. 2003) have been used to delete large genomic DNA segments; however, practically, these approaches are limited to murine embryonic stem (ES) cells, which are more amenable to genetic manipulation via homologous recombination (HR) than are other cells. Furthermore, recombinase systems require two rounds of FRT or loxP insertion into the genome via HR, isolation of cells in which two target sites are inserted in the same chromosome but not in different homologous chromosomes, and subsequent treatment with Flp or Cre recombinases, respectively, to delete the intervening DNA segment, a process that still leaves a single FRT or loxP site behind in the genome. BAC-based gene targeting also has limitations associated with the preparation of BAC vectors and the screening of recombinant clones because of the huge size of these vectors. In addition, false positive clones are often isolated, which results from the breakage and partial integration of BAC vectors (Gomez-Rodriguez et al. 2008). Thus, these approaches are highly laborious and time-consuming even in murine ES cells, and, to our knowledge, have never been used to delete predetermined genomic DNA segments in other higher eukaryotic cells or organisms.
Here we introduce a new approach using engineered zinc finger nucleases (ZFNs) to generate efficient genomic deletions in higher eukaryotic cells and organisms. ZFNs are artificial restriction enzymes made by fusing tailor-made DNA-binding ZF arrays to the FokI nuclease domain. ZFNs have been used to introduce site-specific, local mutations in various cell lines and organisms but not to induce large genomic deletions in a targeted manner thus far. We now provide a platform for ZFN-induced chromosomal deletions, which employ ZFNs that target two separate sites in a chromosome and introduce two concurrent DNA double-strand breaks (DSBs) to delete the genomic segment between the two sites.
Results
Targeted genomic deletions at the human CCR2 and CCR5 loci
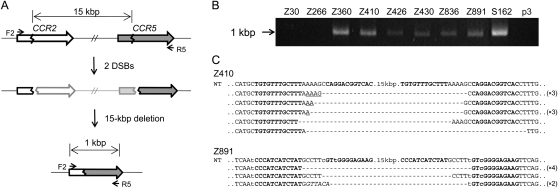
First, we used previously characterized ZFNs that target the genes encoding the human chemokine receptors 5 (CCR5) and 2 (CCR2) to demonstrate targeted deletions of genomic DNA segments in human cells. These two genes are adjacent to each other on human chromosome 3 and are highly homologous. We and others have produced ZFNs that target several different sites at the CCR5 locus and efficiently introduce mutations at these sites (Perez et al. 2008; Kim et al. 2009). Many but not all of these _CCR5_-targeting ZFNs also show site-specific genome editing activities at the corresponding, homologous sites at the CCR2 locus. We investigated whether these ZFNs could induce large genomic deletions in addition to site-specific point mutations at each locus. To this end, we isolated genomic DNA from human embryonic kidney (HEK) 293 cells transfected with ZFN expression plasmids and performed PCR analysis to detect genomic deletions. We used two primers, whose sequences correspond to the CCR2 region or to the CCR5 region and are separated by 16 kbp (Fig. 1A). No PCR product was observed from cells transfected with the control empty plasmid. Apparently, the intervening DNA segment was too long to be amplified under conventional PCR conditions. To our surprise, we observed amplified DNA segments from cells expressing each of the seven different ZFNs whose target sites are conserved between the CCR5 and CCR2 loci (Fig. 1B). The size of the PCR products was about 1 kbp, which was as expected if the DNA segments between the two ZFN target sites were deleted from the chromosome. In contrast, we did not observe amplified DNA segments from cells expressing ZFNs Z30 and Z266 (the numbers indicate the position of the ZFN cleavage sites relative to the initiator codon), whose recognition sites are not conserved at the CCR2 locus and thus show genome editing activity only at the CCR5 locus, suggesting no genomic deletion occurred in these cells. Apparently, ZFNs generating two DSBs in a chromosome but not those generating only one DSB could give rise to large genomic deletions in human cells.
Figure 1.
ZFN-induced genome deletions at the CCR2 and CCR5 loci. (A) Schematic representation of ZFN-mediated genome deletions. Zigzag lines indicate ZFN target sites. F2 and R5 are PCR primers (arrows) used for the detection of genome deletion events. (B) PCR products corresponding to the 15-kbp genomic DNA deletions in cells treated with ZFNs. p3 is the empty plasmid used as a negative control. (C) DNA sequences of PCR products. PCR products were cloned and sequenced. ZFN target sites are shown in boldface letters. Microhomologies are underlined and inserted bases are shown in italics. Dashes indicate deleted bases. Nonconserved bases at the CCR2 and CCR5 loci are shown in lowercase letters. In cases in which a deletion sequence was detected more than once, the number of occurrences is shown in parentheses. (WT) Wild-type DNA sequence.
We cloned the PCR products and determined their DNA sequences, which revealed that, indeed, the CCR2 and CCR5 sites were joined and the intervening 15-kbp DNA segments were deleted (Fig. 1C; Supplemental Fig. 1). The DNA sequences of the deletion junctions were consistent with the DNA cleavage patterns of ZFNs. ZFNs function as dimers, and each monomer recognizes one of two 9- or 12-bp half-sites, which are separated by a 5- or 6-bp spacer. ZFNs cleave DNA at the spacer and generate 5′ 4- or 5-base overhangs (Smith et al. 2000). Sequence analysis of PCR amplicons showed that a ZFN half-site at the CCR2 locus was directly linked to a ZFN half-site at the CCR5 locus and that the entire DNA segment spanning from the other half-site at the CCR2 locus to the other half-site at the CCR5 locus was gone. The breakpoint junction sequences often showed small (2–19 bp) deletions (in addition to 15-kbp deletions) and insertions (indels) (shown as dashes and italicized letters, respectively, in Fig. 1C and Supplemental Fig. 1). These mutagenic patterns are characteristic of DSB DNA repair via nonhomologous end-joining (NHEJ) (Morton et al. 2006). In addition, microhomologies (shown as underlined letters in Fig. 1C and Supplemental Fig. 1) of 1–5 bases often were observed at the junctions, which also strongly supports that the DSB repair was mediated by the NHEJ system.
Although the indels and microhomologies frequently observed at the deletion junctions strongly suggest the involvement of NHEJ in the DNA repair, it remains possible that the large deletions might have resulted from an alternative DNA repair process, i.e., nonallelic homologous recombination (NAHR) (Shaw and Lupski 2004). This possibility should not be neglected, if one considers that the two loci, CCR5 and CCR2, are highly homologous and that all of the ZFNs that gave rise to deletions target two conserved sites. The CCR5 region might be used as a template for NAHR when a DSB was formed at the CCR2 locus or vice versa. We reasoned that the use of two different ZFNs, targeting nonhomologous sites, could resolve this issue, as shown below.
Deletions with two ZFN pairs
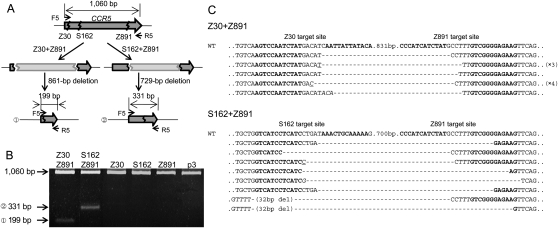
We tested whether two ZFN pairs, each of which targets one of two different sites in a chromosome, could induce deletions of the intervening DNA segments. We chose two sets of _CCR5_-targeting ZFNs for this analysis: Z30 + Z891 and S162 + Z891. (S162 had been termed ZFN-224 by Perez et al. [2008] but, to avoid confusion, is herein denoted as S162, using our numbering scheme.) Genomic DNA was prepared from cells transfected with plasmids that encode two ZFN pairs, and PCR was performed using primers spanning the CCR5 coding sequence. Cells transfected with the Z30 + Z891 set and the S162 + Z891 set are expected to give rise to the amplification of a 199-bp (1060 − 891 + 30) DNA band and a 331-bp (1060 − 891 + 162) band, respectively, in addition to a 1060-bp band corresponding to the entire CCR5 coding region, if specific deletions occur (Fig. 2A). Electrophoretic analysis showed that, indeed, PCR products whose sizes match those predicted were amplified from cells expressing two ZFN pairs but not from cells expressing only one ZFN pair (Fig. 2B). We cloned and sequenced these PCR products, which confirmed specific deletions of ∼861-bp (891 − 30) DNA segments in the cells expressing Z30 + Z891 and of ∼729-bp (891 − 162) DNA segments in the cells expressing S162 + Z891 (Fig. 2C). As noted above, small indels in addition to the expected deletions were observed at the joints. These results strongly support the idea that ZFN-induced genomic deletions were mediated via NHEJ. Because we used two ZFN pairs—each of which targets one of two different sites at the CCR5 locus—and still observed targeted deletions, we ruled out the possibility that NAHR is responsible for the deletion events.
Figure 2.
Use of two ZFNs for targeted genome deletions within the CCR5 locus. (A) Schematic representation of two different deletion events within the CCR5 locus. F5 and R5 are PCR primers (arrows) used for the detection of genome deletion events. (B) PCR products validating ZFN-induced genomic deletions. Approximate sizes of PCR products corresponding to deletion events (199 bp and 331 bp) and to the intact wild-type sequence (1060 bp) are indicated as predicted in A. p3 is the empty plasmid used as a negative control. (C) DNA sequences of PCR products corresponding to deletion events. Symbols are as in Figure 1.
Importantly, the different spacer sequences of two ZFN target sites did not preclude genomic deletions. For example, Z30 and Z891 would generate 5′-ACAT and 5′-CCTT overhangs, respectively, at their cognate target sites but still gave rise to deletions of ∼861-bp DNA segments at the CCR5 locus. Interestingly, one of the PCR amplicon sequences showed that these two overhangs were filled in and the two ends were rejoined to generate the 5′-ACATCCTT-3′ sequence at the breakpoint junction (Fig. 2C). Other clones showed several base deletions of the overhang sequences and the ZFN binding sites. Frequently observed indels at breakpoint junctions suggest that the spacer sequences of the two ZFN target sites do not need to be identical to promote genomic deletions.
Large nested deletions
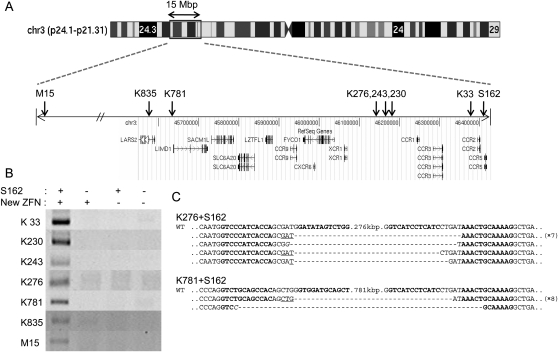
We investigated whether it is possible to delete very long stretches of DNA from the human genome using two ZFN pairs. To this end, we synthesized a series of ZFN pairs, whose target sites lie far upstream of the CCR5 locus. We used 17 naturally occurring ZFs encoded in the human or Drosophila genome as modules to assemble four-finger ZFNs (that is, ZFNs that consist of tandem arrays of four ZF modules). Our previous work, which involved characterization of 315 ZFN pairs at 33 endogenous sites in human cells, showed that only 26% of four-finger ZFNs displayed genome editing activities and that naturally occurring ZFs were more reliable than others for use as modules (Kim et al. 2009). Each of the 17 ZFs recognizes different 3-bp subsites and, collectively, they cover 21 out of 64 triplet subsites. Because ZFNs function as dimers, two four-finger ZFN monomers were prepared per site. Four-finger ZFNs recognize two 12-bp half-sites or 24-bp full sites. A total of 30 ZFN pairs were synthesized, and these newly prepared ZFNs recognized sites 30 kbp to 46 Mbp upstream of the CCR5 locus.
Each ZFN pair was coexpressed in HEK 293 cells with the S162 pair that targets the CCR5 locus. Seven of the 30 ZFNs coexpressed with S162 yielded PCR products corresponding to gross deletions (Fig. 3A,B). (Thus, the success rate was 23%, which is on par with the 26% rate we had observed previously with four-finger ZFNs [Kim et al. 2009]. We note, however, that “successful ZFNs” in our previous study had been defined as those that give rise to site-specific mutations at the CCR5 locus, whereas successful ZFNs in this report were those that give rise to genomic deletions when coexpressed with S162.) Sequence analysis of the PCR products unequivocally corroborated large deletions of 33-kbp, 230-kbp, 243-kbp, 276-kbp, 781-kbp, 835-kbp, and 15.1-Mbp genomic DNA segments, respectively (Fig. 3C; Supplemental Fig. 2). As observed with deletion events between the CCR2 and CCR5 loci and within the CCR5 locus, many sequences of PCR products showed small indels and microhomologies at the breakpoint junctions. None of the ZFN pairs (including the S162 pair) used alone resulted in the production of any PCR product corresponding to the gross genomic deletions.
Figure 3.
Large nested genomic deletions. (A) ZFN target sites on a chromosomal ideogram. Arrows indicate the locations of ZFN target sites on the amplified view of the relevant chromosome 3 region. (B) PCR products validating large nested deletions. S162 and each of the seven new ZFNs were coexpressed in HEK 293 cells. DNA sequences of PCR primers used in this analysis are listed in Supplemental Table 3. (C) DNA sequences of PCR products corresponding to deletion events. Symbols are as in Figure 1.
We also investigated whether the use of two new ZFNs in the absence of S162 could give rise to corresponding genomic deletions at loci other than CCR2 and CCR5. We tested various combinations of two ZFNs among the seven active ZFNs and were able to confirm deletions in each case (Supplemental Fig. 3). For example, the ZFNs termed K230 and M15 gave rise to 230-kbp and 15.1-Mbp deletions, respectively, when coexpressed with S162. When these two ZFNs were coexpressed without S162 in human cells, 14.9-Mbp (15.1 − 0.23) genomic DNA segments between the two target sites were deleted. Sequence analysis of the breakpoint junctions also revealed microhomologies and small indels in addition to the gross deletions.
In summary, we observed, with various combinations of ZFNs, two (∼729-bp and ∼861-bp) deletions within the CCR5 locus, seven different 15-kbp deletions between the CCR2 and CCR5 loci, seven (33-kbp, 230-kbp, 243-kbp, 276-kbp, 781-kbp, 835-kbp, and 15.1-Mbp) deletions between the CCR5 locus and loci upstream of CCR5, and three (538-kbp, 551-kbp, and 14.9-Mbp) deletions at loci other than CCR2 and CCR5 in human cells. Although we were not able to test all possible combinations of the 15 active ZFNs (eight previously characterized ZFNs targeting the CCR5 locus and seven new ZFNs targeting loci upstream of CCR5) used in this study, in every combination of two ZFNs we tested, specific genomic deletions were detected by PCR and confirmed by DNA sequence analyses. These results strongly suggest that use of any two active ZFNs targeting different sites in a chromosome could give rise to specific genome deletions in human cells.
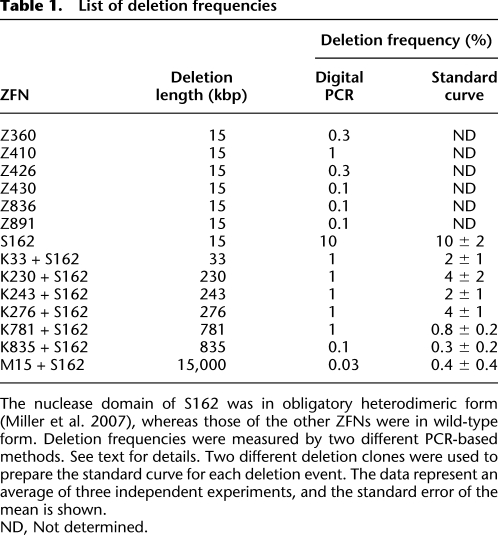
Frequencies of genomic deletions
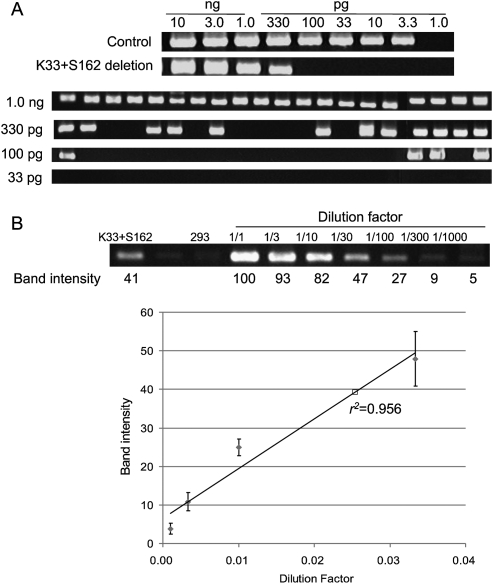
We next investigated the frequencies of ZFN-induced genomic deletions. Two different PCR analyses were used to estimate the frequencies of each deletion event. First, genomic DNA samples were serially diluted in distilled water, and these diluted samples were subjected to “digital” PCR analysis(Fig. 4A) (Flores et al. 2007). In this assay, PCR products corresponding to deletion events would be observed from samples with no or limited dilutions but undetectable from highly diluted samples. The most diluted sample that still yielded positive PCR products was assumed to contain a single chromosomal DNA molecule with a genomic deletion. As a reference point, 3.3 pg ([3.0 × 109 bp × 650 g/mol/bp]/6.0 × 1023) of genomic DNA per reaction was considered to be equivalent to “a haploid genome.” Control PCR using appropriate primers to detect wild-type genomic sequences was performed to confirm that this amount of genomic DNA was indeed equivalent to a haploid genome (Fig. 4A). To account for stochastic amplifications of only a few template molecules in diluted samples, 20 reactions were performed in parallel at each critical dilution point. For example, 12 out of 20 aliquots of 330 pg of genomic DNA purified from cells expressing K33 + S162 (giving rise to 33-kbp deletions) were PCR positive, and only four out of 20 aliquots of 100 pg were positive (Fig. 4A). By contrast, none of 20 aliquots of 33 pg was PCR positive. The least amount of genomic DNA (in this case, 330 pg) that gave rise to PCR products in at least 10 aliquots (that is, >50%) was determined and used to estimate deletion frequencies, which were calculated by dividing 3.3 pg (which is equivalent to a haploid genome) with this amount in picograms. Thus, in the case of the K33 + 162 treatment, the deletion frequency was estimated to be 1%.
Figure 4.
Frequencies of genomic deletions induced by ZFNs. (A) “Digital” PCR analysis. (Upper panel) Genomic DNA isolated from cells cotransfected with plasmids encoding K33 and S162 were serially diluted in a buffer and subjected to PCR analysis in a reaction volume of 10 μL. (Lower panel) At critical dilution points, PCR was performed in 20 aliquots. (B) A standard curve for the estimation of deletion frequencies. A plasmid containing a PCR product corresponding to the 33-kbp genomic deletion induced by ZFNs was serially diluted in a solution containing genomic DNA isolated from HEK 293 cells and the diluted samples were subjected to PCR analysis. Intensities of DNA bands corresponding to the deletion event were measured and plotted against dilution factors. At high values, band intensities reached plateau and thus were excluded when the standard curve was plotted. The open square corresponds to the PCR product from cells treated with ZFNs.
Next, we measured deletion frequencies in a different setting to confirm our results from digital PCR analysis. An individual PCR product (corresponding to a deletion event in question) cloned in a plasmid (0.01 pg, 3 kbp) was mixed with genomic DNA (10 ng, 3 Gbp) isolated from HEK 293 cells such that the mixture contained one deletion clone per haploid genome. This mixture was assumed to be equivalent to genomic DNA with 100% deletion frequency and was serially diluted in the solution containing HEK 293 genomic DNA. A standard curve was generated by plotting dilution factors vs. the amounts of PCR products, measured on an agarose gel, corresponding to the deletion event (Fig. 4B). We used two different plasmid clones to prepare the standard curve for each deletion event and found that there was no significant clone-to-clone variation. Next, the genomic DNA in question was subjected to PCR, and the amount of PCR product was used to calculate the deletion frequency in the sample. The deletion frequency with K33+S162 was estimated to be 2 ± 1% using this method, which was in line with our estimation obtained using the digital PCR analysis described above. These two different methods in general yielded comparable results, which are presented in Table 1.
Table 1.
List of deletion frequencies
Frequencies of various ZFN-mediated genomic deletions ranged from 0.1% to 10%. We found that there was no significant correlation between the deletion size and the frequency (_r_2 = 0.02). Rather, the activity of ZFNs appeared to determine the deletion frequency. For example, the seven ZFNs that induced 15-kbp genomic deletions between the CCR2 and CCR5 loci resulted in deletion frequencies that ranged from 0.1% to 10%. The highest deletion frequency (10%) was observed with S162, which had caused higher site-specific mutation efficiencies (53%) at the CCR5 locus than did other ZFNs. Larger genomic segments also were deleted efficiently. For example, coexpression of S162 and K781 gave rise to 781-kbp genomic deletions with a frequency of 1%. These high frequencies suggest that one should be able to isolate clonal populations of cells, in which a genomic segment is deleted, by dilution cloning.
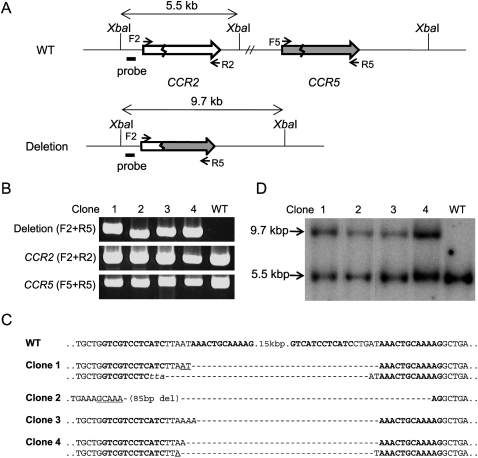
Analysis of clonal populations of cells
Next, we screened for clones derived from single cells in which the target regions were deleted. We chose S162 for this analysis because this ZFN resulted in the highest deletion frequency. Cells expressing S162 were plated at limiting dilution (0.7 cell/well in a 96-well plate) and cultured for 15 to 21 d. Expanded populations of clonal cells were then analyzed by PCR to detect genomic deletions. Out of 87 clones, four yielded amplified DNA segments with the expected size (Fig. 5B). Sequence analysis of these PCR products confirmed specific deletions of 15-kbp DNA segments (Fig. 5C). Interestingly, Clones 1 and 4 each showed two different DNA sequences commensurate with the 15-kbp DNA deletions. This result suggests that genomic deletions occurred at two homologous chromosomes in these cells. HEK 293 is a multiploid cell line that appears to contain at least three copies of chromosome 3 (Bylund et al. 2004). Indeed, we were able to detect the presence of intact chromosomal segments in each of these two clones using PCR analysis (Fig. 5B). Clones 2 and 3 each showed only one DNA sequence commensurate with 15-kbp deletions. As observed above, indels and microhomologies were detected at breakpoint junctions.
Figure 5.
Analyses of clonal populations of cells whose genomic segments were deleted by ZFNs. (A) Schematic of CCR2 and CCR5 loci in wild-type cells and in clonal cells whose genomic segments were deleted. The probe (black bar) and the restriction enzyme, XbaI, used for Southern blot analysis are indicated. F2, R2, F5, and R5 are PCR primers (arrows) used for the detection of genome deletion events or the intact CCR2 and CCR5 loci. (B) PCR products validating genomic deletions and the intact CCR2 and CCR5 region. PCR primers are shown in parentheses. Clones 1 to 4 are clonal cells isolated by limiting dilution. (WT) Wild-type cells. (C) DNA sequences of deletion junctions in clonal cells. Symbols are as in Figure 1. (D) Southern blot analysis of clonal cells.
These clonal cells were further analyzed by Southern blotting to confirm the presence of genomic deletions. Genomic DNA prepared from each clone was treated with XbaI and subjected to agarose gel electrophoresis. A radiolabeled DNA segment near the CCR2 locus was used as a probe. As shown in Figure 5, A and D, a 9.7-kb band, which corresponds to the genomic deletion, was detected in addition to a 5.5-kb band corresponding to the region in the wild-type chromosome from all four clones but not from parental HEK 293 cells.
Because we were able to detect six different deletion events in four out of 87 clones, the deletion frequency with S162 was 4.6% (4/87) per cell or 2.3% (6/261) per allele (assuming that HEK 293 is a triploid). These values are somewhat smaller than our estimation (10%) obtained by the two different PCR-based methods described above. This discrepancy may arise from the difference in the time points used in the analyses. PCR-based estimations used cells at day 3 post-transfection, whereas clonal analysis used cells at day 15 to 21 post-transfection. Not all cells in which the target genomic segments are deleted may survive because of the cytotoxicity of ZFN expression. ZFN cytotoxicity has been reported previously and may arise from its off-target effects (Szczepek et al. 2007).
Discussion
Recent analyses of individual human genome sequences revealed that a variety of genomic rearrangements including deletions, insertions, and duplications contribute to genetic diversity in humans (Iafrate et al. 2004; Redon et al. 2006). Some of these copy number variations are associated with diverse phenotypes and genetic diseases. Sequence analyses of these variations suggest the involvement of both NAHR and NHEJ in their generation (Lupski and Stankiewicz 2005). Because DSBs greatly enhance the rate of both NAHR and NHEJ, it is assumed that most, if not all, chromosomal rearrangements arise, through NAHR or NHEJ, from the repair of DSBs that occur accidentally in the genome (van Gent et al. 2001). Unfortunately, a direct test of this hypothesis has not been possible because experimental systems to study the etiology of these chromosomal aberrations have been lacking. A rare-cutting endonuclease, I-SceI, was used to induce the deletion of a small DNA segment between two I-SceI recognition sites that had been incorporated into the mammalian genome (Guirouilh-Barbat et al. 2004; Honma et al. 2007). However, it is not known whether two randomly occurring DSBs at endogenous sites in a chromosome could give rise to large genomic deletions in mammalian cells, and, if they do, how frequently the events would occur. Our ZFN experiments address these questions and reveal that two DSBs in a chromosome are sufficient to promote frequent genomic deletions of endogenous DNA segments.
It is remarkable to find that the endpoints of two distantly occurring DSBs are joined together to give rise to large genomic deletions. To explain how these events occur in cells, two different models can be considered, which were first proposed to explain the etiology of chromosomal translocations that are frequently observed in cancer (Nikiforova et al. 2000; Aten et al. 2004). The “contact-first” model assumes that the sites where DSBs occur in a chromosome must be in close physical proximity before the formation of DSBs to give rise to deletions (or translocations). By contrast, the “breakage-first” model postulates that DSBs occur first and that distantly located broken ends are brought together and joined to give rise to deletions of intervening DNA segments. Note that our ZFNs target many different sites, separated by up to 15 Mbp, in chromosome 3 and that ZFNs generating two DSBs always give rise to deletions. Because it is unlikely that all the ZFN target sites in chromosome 3 make prior contact with each other, our results strongly support the breakage-first model to explain how genomic deletions occur.
Recently, Brunet et al. (2009) reported on chromosomal translocations artificially induced by I-SceI and ZFNs in human cells. They found that coexpression of these enzymes, which generates concurrent DSBs in two chromosomes, gave rise to translocations at frequencies of 10−6 to 10−5. These frequencies are significantly smaller than the deletion frequencies we observed, which were in the range of 10−3 to 10−1 (P < 0.01, Student's _t_-test). It is possible that this discrepancy may arise from the use of different enzymes targeting different chromosomes in the study by Brunet et al. (2009) and this study. Alternately, DSB-induced chromosomal deletions could occur much more frequently than do translocations in human cells. In this regard, it is of note that each interphase chromosome occupies a separate region in a nucleus, referred to as a chromosome territory (Meaburn and Misteli 2007). Perhaps the endpoints derived from two DSBs occurring in a single chromosome within a territory are ligated much more readily to give rise to deletions than are those in two different chromosomes, each occupying a separate territory, to give rise to translocations. Yet another possibility is that cells might tolerate DSB-induced deletions better than translocations. Thus, cells that harbor translocations would tend to die out more frequently than do those that harbor chromosomal deletions.
To our knowledge, this is the first report that demonstrates genomic deletions of large, predetermined endogenous DNA segments in a targeted fashion in human cells. The sizes of deleted DNA segments range from several-hundred bp to 15 Mbp and thus span greater than four orders of magnitude. We envision that ZFN-induced chromosomal deletions described herein can be applied in virtually any organism or cell whose genome has been sequenced. The microhomologies and indels that are often observed with genomic deletions induced by ZFNs suggest that NHEJ but not HR is involved in these events. Because NHEJ rather than HR is the dominant DSB repair system in both animals and plants (Ray and Langer 2002), ZFN-induced chromosomal deletions should allow targeted genome engineering in diverse organisms and cells.
It is of note that ZFNs, unlike other genome engineering tools such as recombinases or meganucleases, do not require prior manipulation of genomic sequences. Thus, ZFNs induce targeted deletions of endogenous genomic segments and do not leave any footprints. By contrast, insertion of target elements must precede the use of recombinases or meganucleases for genome engineering, and those elements remain in the genome after the genetic engineering is complete. Also note that ZFNs do not require selection markers or gene targeting vectors, whose preparation is time-consuming. These features make ZFNs versatile tools in genome engineering.
Targeted genome deletions via ZFNs could be broadly useful in many areas of life science, biotechnology, and medicine. Geneticists and molecular biologists could use ZFNs to delete gene clusters in a genome of interest. It often is the case that a single gene knockout does not result in any discernable phenotypic changes in animal studies or in vitro experiments. More often than not, this phenotypic masking is caused by the presence of a homologous gene(s). Interestingly, homologous genes tend to cluster in the genome. For example, CCR2 and CCR5 are right next to each other at chromosome 3p21. The 230-kbp genomic DNA segment that was deleted with two ZFNs in our study also contains two additional genes, CCR1 and CCR3, that encode chemokine (C–C motif) receptors. ZFNs can be used to delete clusters of homologous genes as a unit. In addition, ZFNs could enable scientists to selectively delete intergenic regions and introns. These noncoding regions represent greater than 98% of the human and other mammalian genomes (Venter et al. 2001) and contain critical information such as regulatory elements and noncoding RNAs but are largely unexplored. Recent studies revealed that noncoding regions are pervasively transcribed in mammalian cells (The ENCODE Project Consortium 2007), implying physiological roles associated with these regions. Conventional gene targeting approaches alone may not be sufficient for the systemic study of these vast regions in human and other higher eukaryotic genomes.
The use of ZFNs for generating genome deletions could also lead to important advances in stem cell research and gene therapy. Genes associated with diseases could be selectively removed from stem cells or somatic cells using ZFNs. Perez et al. (2008) elegantly demonstrated that S162 could be used to induce frameshift mutations at the CCR5 locus and to produce T cells deficient in wild-type CCR5 protein and resistant to HIV-1 infection in animal models. A derivative of S162 is now under clinical investigation for the treatment of AIDS patients. Unlike the immunodeficient NOG mice used in Perez et al. (2008), patients with AIDS are still immune-competent and could respond to the novel CCR5 epitopes generated by ZFN-induced frameshift mutations at the CCR5 locus. As a result, the very T cells that are devoid of wild-type CCR5 but express mutant forms of CCR5 could be eliminated by the host immune system. As we have shown here, S162 alone, which targets both the CCR2 and CCR5 sites, can induce genomic deletions of 15-kbp DNA segments, which encompass not only the 5′ part of CCR5 but also the promoter of CCR5, and could give rise to T cells that do not express functional CCR5 at all. It will be interesting to see whether T cells in which this region is deleted would be maintained in vivo and provide therapeutic benefits to patients. (It still is possible that CCR2–CCR5 fusion epitopes would be generated by S162-mediated genome deletion.) In this regard, deleting a promoter as well as a coding region by targeting two sites might be advantageous to avoid opportunistic immune responses against novel epitopes that could be generated by a ZFN targeting the coding region only. S162-mediated chromosomal deletions also highlight the possibility that undesired chromosomal rearrangements in addition to targeted genome editing could be induced by ZFNs when homologous target sites are present in the genome.
ZFN-induced chromosomal deletions could broaden the utility and applicability of ZFN research as well. Thus far, ZFNs have been used to introduce site-specific mutations in the coding regions of many genes in diverse organisms (Bibikova et al. 2002, 2003; Doyon et al. 2008; Meng et al. 2008) and cell lines (Urnov et al. 2005; Maeder et al. 2008; Santiago et al. 2008; Kim et al. 2009; Zou et al. 2009) but not to generate large genomic deletions as described in this report. To knock out a gene of interest (GOI), ZFN users have searched for potential target sites in coding regions but not in noncoding regions. As we have demonstrated previously, not all sites are equally “targetable” with ZFNs and not all ZFs are equally effective and reliable as modules for making ZFNs (Kim et al. 2009). For example, naturally occurring ZFs (preselected in a yeast-based one-hybrid system) are more reliable building blocks for making functional ZFNs than are engineered ZFs (selected via phage display or produced by site-directed mutagenesis). GNN repeat sites [e.g., 5′-(NNC)3N6(GNN)3-3′] can be more efficiently targeted with ZFNs than other sites (Ramirez et al. 2008). However, these preferable sites occur rarely and may not exist at the coding regions of a GOI. Considering that coding regions represent less than 2% of the human genome, whereas noncoding regions represent greater than 98%, our genome deletion approach significantly expands targetable regions for ZFN-mediated gene knockout experiments. A promoter or an exon could be deleted by targeting two sites in intergenic regions or introns. Furthermore, as discussed above, targeted deletions of DNA segments including the promoter region would completely knock out a GOI and give rise to a 100% null phenotype. By contrast, conventional targeted mutagenesis in coding regions using a single ZFN pair could generate truncated proteins or proteins fused to novel amino acid sequences that arise from frameshift mutations. It certainly is possible that these mutated proteins could be partially active or, in rare cases, display gain-of-function phenotypes. In this regard, it is of note that the naturally occurring 32-base deletion in CCR5 may be a gain-of-function mutation (Chelli and Alizon 2001; Agrawal et al. 2004).
It remains a challenge for potential ZFN users to prepare efficient ZFNs for their experiments, but the current progress in this field should make them routine and accessible tools in the near future. Three different platforms for ZFN synthesis currently are available to the science community. Each platform has its own pros and cons. Sigma-Aldrich in collaboration with Sangamo Biosciences offers a custom ZFN synthesis service to academic scientists. Thus far, four mammalian genes, two zebrafish genes, and three plant genes have been successfully mutated with ZFNs produced by Sangamo Biosciences. Many more genes in diverse organisms will certainly follow. But the high price tag attached to this service may make this system unaffordable to most academic users (Pearson 2008). Do-it-yourself kits also are available from two sources. The Zinc Finger Consortium has developed an _Escherichia coli_-based semi-selection method for making ZFNs (termed the OPEN system) (Maeder et al. 2008). In this system, ZF arrays are first selected in E. coli from pools of ZFs with identical DNA-binding specificities and then attached to the FokI nuclease domain to produce ZFNs. The Consortium has reported successful targeted mutagenesis using OPEN ZFNs of one plant gene, four human genes, and five zebrafish genes. Although OPEN ZFNs may take the context effect of ZF modules into account, the entire process is time-consuming and labor-intensive.
A faster, more convenient, and easy-to-practice approach is modular assembly of precharacterized ZFs using standard recombinant DNA technology. Unfortunately, this method had not been successfully used to yield ZFNs that are functional in mammalian cells. For example, an extensive survey of a large number of modularly assembled ZFNs showed high failure rates (Ramirez et al. 2008). We found, however, that success and failure rates depend on the choice of ZF modules and that naturally occurring ZFs are more reliable building blocks for making functional ZF arrays than are engineered ZFs (Bae et al. 2003). Indeed, we were able to make dozens of modularly assembled ZFNs that target eight different endogenous sites in human cells using a carefully chosen set of ZFs (Kim et al. 2009). In this report, we showed that modularly assembled ZFNs could also induce genomic deletions in addition to site-specific mutations in human cells. The plasmids that encode these modules are available from Addgene (http://www.addgene.org/zfc) or ToolGen.
Methods
Plasmids encoding ZFNs
Plasmids that encode ZFNs targeting far upstream of the CCR5 locus were constructed as described (Kim et al. 2009). Briefly, potential ZFN target sites 30 kbp to 46 Mbp upstream of the CCR5 locus were identified using a computer algorithm (http://www.toolgen.com/ZFNfinder/). Thirty four-finger ZFN pairs were assembled using 17 naturally occurring ZFs (Supplemental Table 1). Successful ZFNs that gave rise to genomic deletions and their target sequences are listed in Supplemental Table 2.
Cell culture and transfection
Human embryonic kidney 293T/17 (ATCC, CRL-11268) cells were maintained in Dulbecco's modified Eagle medium (Welgene Biotech) supplemented with penicillin (100 units/mL), streptomycin (100 μg/mL), and fetal bovine serum (10%). Cells were seeded at 1 × 106 cells/well in a six-well plate, incubated for 24 h, and then transfected with ZFN-encoding plasmids (total 4 μg) using polyethylenimine (Aldrich). At 3 d post-transfection, genomic DNA was isolated from cells using the AccuPrep Genomic DNA Extraction Kit (Bioneer), as described by the manufacturer's protocol, and used for PCR analysis.
PCR analysis of genomic deletions and DNA sequencing of breakpoint junctions
To detect genomic deletions, genomic DNA (50 ng per reaction) was subjected to PCR analysis using Taq DNA polymerase (GeneAll Biotech) and appropriate primers (Supplemental Table 3). PCR products were analyzed by agarose gel electrophoresis. For sequencing analysis, PCR products corresponding to genomic deletions were purified using QIAquick Gel Extraction Kit (QIAGEN) and cloned into the T-Blunt vector using the T-Blunt PCR Cloning Kit (SolGent). Cloned plasmids were sequenced using M13 primers or primers used for PCR amplification.
Deletion frequencies estimated by two different PCR-based methods
For digital PCR analysis, genomic DNA samples isolated from cells transfected with plasmids encoding ZFNs were serially diluted in distilled water, and diluted samples were subjected to hemi-nested PCR using appropriate primers (Supplemental Table 3). For standard curve analysis, a cloned PCR product (0.01 pg, 3 kbp) was first added to a solution containing genomic DNA (10 ng, 3 Gbp) isolated from HEK 293T17 cells, which was then serially diluted in the genomic DNA solution. PCR was performed using appropriate primers and PCR product concentration was quantified using the software LabWorks ver.4.6 (UVP).
Isolation of clonal populations of cells
293T/17 cells transfected with plasmids encoding S162 were incubated for 6 h and then seeded in a 96-well plate at limiting dilution (0.7 cell per well on average). After 15 to 21 d of culture, 87 clones were grown to confluency. A small portion of cells was maintained in culture and the rest was used for the isolation of genomic DNA, which was then subjected to PCR analysis to detect genomic deletions. Methods for genomic DNA extraction, PCR, and sequencing analyses were described above.
Southern blot analysis
Genomic DNA (30 μg) was treated with XbaI (New England Biolabs), electrophoresed on agarose gels (0.8%), and transferred to Hybond-N+ membrane (Amersham). The DNA probe was prepared by PCR amplification of a cloned human CCR2 DNA template using primers F2f and R2u (Supplemental Table 3) and subsequent labeling with [α-32P]dCTP using the Megaprime DNA Labeling System (Amersham). After hybridization at 65°C in Church buffer for 24 h, the membrane was washed and exposed to an imaging plate (Fuji Photo Film Co.), which was then scanned using the BAS-2500 system (FUJIFILM Life Science). Band intensities were quantified using Multi Gauge Ver3.0 (Fuji Photo Film Co.) software.
Acknowledgments
We thank members of Genome Engineering Laboratory for helpful discussions. J.-S.K. is supported in part by a grant (R17-2007-019-01001-0) from the Korea Science and Engineering Foundation.
Footnotes
References
- Agrawal L, Lu X, Qingwen J, VanHorn-Ali Z, Nicolescu IV, McDermott DH, Murphy PM, Alkhatib G. Role for CCR5Δ32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J Virol. 2004;78:2277–2287. doi: 10.1128/JVI.78.5.2277-2287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aten JA, Stap J, Krawczyk PM, van Oven CH, Hoebe RA, Essers J, Kanaar R. Dynamics of DNA double-strand breaks revealed by clustering of damaged chromosome domains. Science. 2004;303:92–95. doi: 10.1126/science.1088845. [DOI] [PubMed] [Google Scholar]
- Bae KH, Kwon YD, Shin HC, Hwang MS, Ryu EH, Park KS, Yang HY, Lee DK, Lee Y, Park J, et al. Human zinc fingers as building blocks in the construction of artificial transcription factors. Nat Biotechnol. 2003;21:275–280. doi: 10.1038/nbt796. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylund L, Kytola S, Lui WO, Larsson C, Weber G. Analysis of the cytogenetic stability of the human embryonal kidney cell line 293 by cytogenetic and STR profiling approaches. Cytogenet Genome Res. 2004;106:28–32. doi: 10.1159/000078556. [DOI] [PubMed] [Google Scholar]
- Chelli M, Alizon M. Determinants of the trans-dominant negative effect of truncated forms of the CCR5 chemokine receptor. J Biol Chem. 2001;276:46975–46982. doi: 10.1074/jbc.M106432200. [DOI] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores M, Morales L, Gonzaga-Jauregui C, Dominguez-Vidana R, Zepeda C, Yanez O, Gutierrez M, Lemus T, Valle D, Avila MC, et al. Recurrent DNA inversion rearrangements in the human genome. Proc Natl Acad Sci. 2007;104:6099–6106. doi: 10.1073/pnas.0701631104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Rodriguez J, Washington V, Cheng J, Dutra A, Pak E, Liu P, McVicar DW, Schwartzberg PL. Advantages of q-PCR as a method of screening for gene targeting in mammalian cells using conventional and whole BAC-based constructs. Nucleic Acids Res. 2008;36:e117. doi: 10.1093/nar/gkn523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Honma M, Sakuraba M, Koizumi T, Takashima Y, Sakamoto H, Hayashi M. Non-homologous end-joining for repairing I-SceI-induced DNA double strand breaks in human cells. DNA Repair. 2007;6:781–788. doi: 10.1016/j.dnarep.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, Stankiewicz P. Genomic disorders: Molecular mechanisms for rearrangements and conveyed phenotypes. PLoS Genet. 2005;1:e49. doi: 10.1371/journal.pgen.0010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T. Cell biology: Chromosome territories. Nature. 2007;445:379–381. doi: 10.1038/445379a. [DOI] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Morton J, Davis MW, Jorgensen EM, Carroll D. Induction and repair of zinc-finger nuclease-targeted double-strand breaks in Caenorhabditis elegans somatic cells. Proc Natl Acad Sci. 2006;103:16370–16375. doi: 10.1073/pnas.0605633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Stringer JR, Blough R, Medvedovic M, Fagin JA, Nikiforov YE. Proximity of chromosomal loci that participate in radiation-induced rearrangements in human cells. Science. 2000;290:138–141. doi: 10.1126/science.290.5489.138. [DOI] [PubMed] [Google Scholar]
- Pearson H. Protein engineering: The fate of fingers. Nature. 2008;455:160–164. doi: 10.1038/455160a. [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, Winfrey RJ, Sander JD, Fu F, Townsend JA, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- Ray A, Langer M. Homologous recombination: Ends as the means. Trends Plant Sci. 2002;7:435–440. doi: 10.1016/s1360-1385(02)02327-0. [DOI] [PubMed] [Google Scholar]
- Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, et al. The DrosDel deletion collection: A Drosophila genomewide chromosomal deficiency resource. Genetics. 2007;177:615–629. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum Mol Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Valenzuela DM, Murphy AJ, Frendewey D, Gale NW, Economides AN, Auerbach W, Poueymirou WT, Adams NC, Rojas J, Yasenchak J, et al. High-throughput engineering of the mouse genome coupled with high-resolution expression analysis. Nat Biotechnol. 2003;21:652–659. doi: 10.1038/nbt822. [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams EW, Myers PW, Li RJ, Mural GG, Sutton HO, Smith M, Yandell CA, Evans RA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell. 2009;5:97–110. doi: 10.1016/j.stem.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]