High-resolution nucleosome mapping reveals transcription-dependent promoter packaging (original) (raw)
Abstract
Genome-wide mapping of nucleosomes has revealed a great deal about the relationships between chromatin structure and control of gene expression, and has led to mechanistic hypotheses regarding the rules by which chromatin structure is established. High-throughput sequencing has recently become the technology of choice for chromatin mapping studies, yet analysis of these experiments is still in its infancy. Here, we introduce a pipeline for analyzing deep sequencing maps of chromatin structure and apply it to data from S. cerevisiae. We analyze a digestion series where nucleosomes are isolated from under- and overdigested chromatin. We find that certain classes of nucleosomes are unusually susceptible or resistant to overdigestion, with promoter nucleosomes easily digested and mid-coding region nucleosomes being quite stable. We find evidence for highly sensitive nucleosomes located within “nucleosome-free regions,” suggesting that these regions are not always completely naked but instead are likely associated with easily digested nucleosomes. Finally, since RNA polymerase is the dominant energy-consuming machine that operates on the chromatin template, we analyze changes in chromatin structure when RNA polymerase is inactivated via a temperature-sensitive mutation. We find evidence that RNA polymerase plays a role in nucleosome eviction at promoters and is also responsible for retrograde shifts in nucleosomes during transcription. Loss of RNA polymerase results in a relaxation of chromatin structure to more closely match in vitro nucleosome positioning preferences. Together, these results provide analytical tools and experimental guidance for nucleosome mapping experiments, and help disentangle the interlinked processes of transcription and chromatin packaging.
Eukaryotic DNA is packaged in nucleosomes, composed of 147 bp of DNA wrapped ∼1.7 turns around an octamer of histone proteins (Luger et al. 1997; Kornberg and Lorch 1999). Nucleosomes influence the expression of a huge fraction of yeast genes (Wyrick et al. 1999) and the precise positioning of nucleosomes relative to underlying DNA, controls access to protein-binding sites, and thereby affects regulatory programs (Stunkel et al. 1997; Lomvardas and Thanos 2002; Lam et al. 2008; Radman-Livaja and Rando 2009). In yeast, transcription start sites (TSSs) are generally positioned just within the +1 nucleosome, downstream of a nucleosome-depleted region generally referred to as the nucleosome-free region (NFR) (Yuan et al. 2005; Albert et al. 2007; Mavrich et al. 2008a). Nucleosome positioning is different between promoter types; TATA-less promoters tend to be “housekeeping” genes and are characterized by a canonical NFR upstream of their TSS; conversely, TATA-containing genes tend to be stress-responsive, noisily expressed, are more responsive to genetic perturbation of chromatin remodeling complexes, and their promoters are often at least partly occupied by nucleosomes (Basehoar et al. 2004; Ioshikhes et al. 2006; Newman et al. 2006; Field et al. 2008; Tirosh and Barkai 2008; Choi and Kim 2009).
While nucleosomes can package almost any sequence, work over the past few decades has established sequence rules that influence this packaging. Most importantly, rigid poly(A) tracts in DNA are unfavorable for nucleosome assembly and can direct formation of NFRs in vitro (Kunkel and Martinson 1981; Drew and Travers 1985; Iyer and Struhl 1995; Sekinger et al. 2005; Yuan et al. 2005; Ioshikhes et al. 2006; Kaplan et al. 2008; Yuan and Liu 2008; Segal and Widom 2009). This was recently confirmed globally in an in vitro reconstitution study, emphasizing a major role for nucleosome-excluding sequences in “programming” promoter architecture (Kaplan et al. 2008; Zhang et al. 2009). Much of the remainder of in vivo chromatin structure is proposed to result from “statistical positioning,” the idea that packing as many nucleosomes as possible in a short stretch of the genome will result in positioned nucleosomes (Kornberg and Stryer 1988; Yuan et al. 2005; Mavrich et al. 2008a). While some aspects of yeast chromatin structure can, in principle, be predicted based on sequence and packing rules, a number of protein machines, such as the ATP-dependent chromatin remodeling complexes, regulate transcription by moving nucleosomes (Workman and Kingston 1998; Clapier and Cairns 2009). Perhaps the most widespread chromatin-perturbing complex is RNA polymerase, whose passage disrupts histone–DNA contacts, and at very high transcription rates results in nucleosome eviction (Studitsky et al. 1994, 1997; Lee et al. 2004; Schwabish and Struhl 2004; Bondarenko et al. 2006; Dion et al. 2007; Field et al. 2008).
The advent of high-throughput sequencing technology has enabled rapid genome-wide analysis of nucleosome positioning at high resolution and has been used to map nucleosomes in yeast, worms, flies, medaka, and humans (Field et al. 2008; Mavrich et al. 2008a,b; Schones et al. 2008; Shivaswamy et al. 2008; Valouev et al. 2008; Sasaki et al. 2009). Yet, analytical and experimental methods for deep-sequencing analysis of chromatin are still in their infancy and mostly focus on averaged nucleosome occupancy levels at genomic loci, effectively transforming deep-sequencing data into an analog of tiling array data.
Here, we describe a novel analytical method for identifying nucleosome positions from Illumina sequencing data and automatically estimate nucleosome position, occupancy, and length from S. cerevisiae data. Using parameters extracted from these nucleosome calls allows us to identify groups of functionally related genes with significantly high or low values of a given parameter. Experimentally, we examine the impact of nuclease titration levels on this assay and identify properties of yeast chromatin that are influenced by the level of digestion as well as invariant properties. Finally, we explore the role of RNA polymerase in chromatin structure through the use of a temperature-sensitive mutant in the gene encoding the large subunit of RNA polymerase II (Pol II), RPO21 (also known as RPB1). Comparing nucleosome positions and properties before and after Pol II inactivation, we confirm the role of RNA polymerase in nucleosome eviction at promoters, and find a surprising role in retrograde movement of nucleosomes over genes. By quantitatively analyzing changes in nucleosome positioning after Pol II shutoff we find that different classes of genes are subject to distinct perturbations by RNA polymerase. Finally, we confirm a role for RNA polymerase in perturbing nucleosomes from their thermodynamically favored positions.
Results
Identifying nucleosome positions by template filtering
The application of deep sequencing to mononucleosomal DNA results in million of reads from both ends of the mononucleosomal DNA segments. Published methods for calling nucleosome positions from Illumina data typically involve extending each single-end short sequenced read to the expected segment length of ∼140 bp and then examining the coverage of different genomic loci by the accumulated extended segments. These methods clearly highlight nucleosome-depleted vs. nucleosome-occupied regions, for example, in averaged gene alignments (Albert et al. 2007; Field et al. 2008; Shivaswamy et al. 2008). More elaborate approaches identify nucleosome positions by identifying the center of these inferred segments and estimating the occupancy at different center locations (Shivaswamy et al. 2008). These methods do not account for the possibility that nucleosomes might occupy different positions in subpopulations of cells and assume uniform nucleosome lengths (i.e., no variability in digestion level at different nucleosomes).
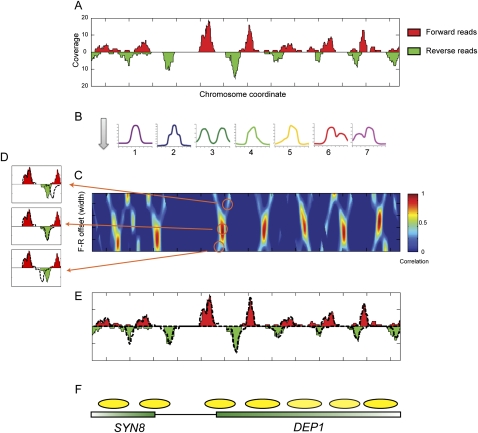
To overcome these issues, we developed a method for calling nucleosome positions, occupancy, and length using template filtering (Turin 1960). This method is based on the observation that sequencing the ends of a nucleosome will result in an expected pattern of offset forward (F) and reverse (R) strand reads at the two ends (Fig. 1A). Due to variability in exact nucleosome position from cell to cell and variability in the extent of MNase digestion at each end of a nucleosome, the peak of reads at each nucleosome end will form a distribution of variable width. Our method is based on identifying occupancy templates of forward and reverse reads that are typical of nucleosomes. The method then uses a fast procedure to identify locations where the forward and reverse read distributions correlate with a series of model templates (Fig. 1B; Supplemental Fig. S1). The method examines a range of distances between forward and reverse templates to capture over- and underdigestion of the ∼147-bp nucleosomal DNA, and determines nucleosome length by choosing an F-to-R offset that maximizes correlation to F and R templates (Fig. 1C,D).
Figure 1.
Template filtering overview. (A) Deep sequencing data for a typical stretch of the yeast genome. Coverage by forward-strand sequencing reads are shown as red peaks, whereas coverage by reverse-strand sequencing reads are shown as inverted green peaks. (B) Templates. Forward and reverse-strand read distributions are cross-correlated with each of the seven templates shown. (C) Correlation coefficient heat map of template 1 for forward and reverse templates at varying center positions (_x_-axis) and distances (_y_-axis). (D) Examples of templates spaced too far apart (top), at the optimal distance (middle), or too close together (bottom). Dotted lines indicate template outlines being compared with the underlying data. (E) Read distributions explained by the optimal template matches are shown as dotted lines for the region in A. (F) Schematic of nucleosome calls and underlying gene annotations.
To further elaborate the method, we aimed to identify the common read distributions from the data. In other words, what are the typical distributions of end reads for the nucleosomes in an experimental data set? We first applied our method using a Gaussian-shaped template to create a preliminary map of nucleosome positions. We next examined read distributions at the ends of these initial nucleosome predictions, clustered read patterns to identify typical behaviors, and selected seven representative templates for use in our scans (Fig. 1B; Supplemental Fig. S1). Six of the templates exhibited several peaks, potentially indicating nucleosomes with variable ends resulting from subpopulations and/or variable nuclease digestion. For example, nucleosomes with ends that match template 2 have sets of ends separated by 10 bp (Supplemental Fig. S1), suggesting that the first stage in overdigestion of nucleosomes by micrococcal nuclease is to cut one helical turn further into the nucleosomal DNA.
Our method fits the sequencing data to the best-correlated templates with varying distances between F and R templates. Local maxima in the correlation spectrum (Fig. 1C) are identified as potential nucleosome calls (Fig. 1E,F). To assemble the final set of nucleosomes, we use a greedy approach to choose the best-correlating template and distance per nucleosome (Methods). Occupancy is determined for each nucleosome by the number of reads contributing to a given nucleosome call. To account for different sequencing yields, we normalize nucleosome occupancies to a mean value of 1.
To evaluate the quality of our method's nucleosome calls, we used the nucleosome positions, templates, and occupancy calls to regenerate the sequenced reads that account for our called nucleosomes. This simulated data set is sampled with the same number of reads as the original sequencing run. These simulated reads closely match the original sequencing data, and the small difference (residual) between the measured and reconstructed data indicate that our nucleosome calls account for 88% of the sequencing reads, indicating that the small number of extracted parameters capture the majority of the experimental sequencing data (Supplemental Fig. S2).
We also examined data from seven additional previously published deep-sequencing datasets (Kaplan et al. 2008; Shivaswamy et al. 2008). We find that the same templates are common in all datasets (Supplemental Fig. S3A), confirming the generality of our approach. However, we did find that the occurrence frequency of different templates differed between datasets, which we ascribe to differences in numbers of reads as well as differences in digestion between different MNase preparations (Supplemental Fig. S3B; see below). Positions of nucleosome calls were generally concordant between datasets (Supplemental Figs. S4, S5), with the major difference occurring in −1 nucleosome calls, which we also identify below as being dependent on digestion levels.
Finally, we compared our nucleosome calling method to an alternative calling algorithm based on identifying peaks in data where forward and reverse reads have been shifted a half-nucleosome width toward one another (Shivaswamy et al. 2008). Nucleosome calls were highly concordant for both methods (Supplemental Fig. S5A), particularly for clearly well-positioned nucleosomes (those that match Template 1 in our approach). Examination of regions where nucleosome calls differed revealed that our method fails in regions where only one end of a nucleosome generates reads, whereas our method better captures data from “fuzzy” regions such as mid-coding regions (Supplemental Fig. S5B,C), thereby better capturing nucleosome occupancy over such regions.
By examining distributions of end reads, we found that nucleosomes at different locations vary in their digestion patterns. Over- and underdigestion of DNA can be due to many possible factors, including properties of the DNA sequence (e.g., sequence composition, bendability, etc.) and properties of the nucleosome (e.g., histone modification state). To investigate these two factors we tested whether the digestion template at nucleosome ends is associated with specific sequence properties and/or specific modification annotation. Consistent with previous reports of MNase sequence preference (Dingwall et al. 1981; Horz and Altenburger 1981), we find that different templates are associated with a clear sequence preference for location of A/T dinucleotides (Supplemental Fig. S6B). On the other hand, we also observed that multimodal templates such as template 3 were enriched at locations previously described as “fuzzy” or delocalized, such as mid-coding regions and over promoters of stress-responsive genes (Supplemental Fig. S6C,D). These results suggest that both nucleosomal subpopulations (i.e., delocalization) as well as sequence composition are significant sources of variability in patterns of nucleosome reads. While these two factors cannot be completely disentangled when using MNase to map nucleosomes, we note that the use of several templates provides an automated means for taking MNase sequence biases into account.
Nucleosome positioning in growing yeast
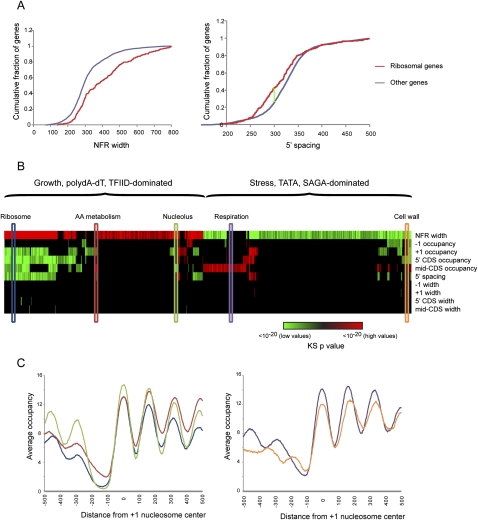
Genome-wide maps of nucleosomes in actively growing yeast have been the subject of a rage of recent studies (Yuan et al. 2005; Albert et al. 2007; Lee et al. 2007; Field et al. 2008; Mavrich et al. 2008a; Shivaswamy et al. 2008). To further evaluate our methodology, we carried out deep sequencing of mononucleosomal DNA from actively growing yeast, and applied our method to generate a map of nucleosome locations (Supplemental Table S1). Our method automatically extracts features of interest, such as NFR width, nucleosome length (which is not available using previous methods), nucleosome spacing, and occupancy. To systematically examine these features we used a compendium of experimental gene annotations that we previously collected (Dion et al. 2007; Wapinski et al. 2007) and compared these against multiple nucleosome attributes associated with each gene (e.g., +1 nucleosome occupancy, spacing between the +1 and +3 nucleosome, etc.). Using the Kolmogorov-Smirnov (KS) test (Fig. 2A) we discovered attributes whose distribution in specific gene groups was significantly (false discovery rate [FDR] < 0.05) different from the background (Fig. 2B; Supplemental Table S2).
Figure 2.
Different promoter types are differently packaged. (A) Cumulative distribution function (CDF) plots for two significant Kolmogorov-Smirnov (KS) enrichments. The gene set of 270 ribosomal genes is enriched for long NFRs (left), and close +1 to +3 nucleosome spacing (right). For example, 45% of ribosomal genes have 5′ nucleosome spacing of <300 bp (green line), whereas only 25% of all genes have this spacing. (B) Enrichment of high or low values of various parameters for a set of promoter types. Various parameters such as +1 nucleosome occupancy (listed at right) were extracted for all yeast promoters. Each of the gene sets previously gathered (Dion et al. 2007; Wapinski et al. 2007) was tested (using KS test) for significantly high or low values of the various parameters. Significant (FDR < 0.05) high values are shown in red, and significant low values are in green. Colors represent log10 of the _P_-value of the KS enrichment (saturated at P < 1 × 10−20). Colored elongated boxes indicate gene classes evaluated in C. (C) Averages for five promoter types as indicated in B, aligned according to +1 nucleosome center. The _y_-axis represents average normalized nucleosome occupancy, in number of reads per million mappable reads.
This analysis highlights previously described features of yeast chromatin as well as novel ones. We recapitulate the dichotomy between promoter packaging of growth (TFIID-regulated, TATA-less, high-expression growth genes) and stress (SAGA-regulated, TATA, noisy expression, rapid histone replacement) genes (Fig. 2B) (Ioshikhes et al. 2006; Newman et al. 2006; Albert et al. 2007; Dion et al. 2007; Choi and Kim 2008, 2009; Field et al. 2008; Tirosh and Barkai 2008). For example, canonical growth genes, such as those encoding ribosome proteins, are enriched with wider NFRs and often also exhibit lower nucleosomal occupancy of coding regions, whereas stress genes, such as heat shock-induced genes, show the opposite behavior. This dichotomy is clearly the major feature that separates classes of genes in yeast, but our analysis also shows finer distinctions beyond this dichotomy. In general, the second distinguishing axis beyond NFR width is coding region nucleosome occupancy, which further can be separated into occupancy of 5′ nucleosomes such as the +1 nucleosome, and occupancy of mid-CDS nucleosomes located distal from either end of the gene. For example, within the “growth” class of genes, ribosomal and metabolic genes can be roughly distinguished by the occupancy of coding-region nucleosomes (Fig. 2B,C). While these distinctions appear subtle, it is important to note that they are of similar magnitude to physiologically relevant (Ihmels et al. 2005) changes observed in the chromatin packaging of mitochondrial ribosomal genes between S. cerevisiae and C. albicans (Field et al. 2009), suggesting that such small differences can, indeed, play roles in transcriptional control.
To explore the role of transcription in shaping chromatin architecture we analyzed the relationship between Pol II enrichment (Methods) at a given gene and the various chromatin parameters described above (Supplemental Fig. S7). NFR width was positively correlated with transcription rate, while +1 occupancy, mid-CDS occupancy, and +1 to +3 spacing were slightly anticorrelated with transcription rate. To determine the extent to which gene set enrichments from Figure 2A were driven by transcription level, we corrected each gene's chromatin parameters to account for transcription rate (Methods) and repeated the KS enrichment analysis from Figure 2A (Supplemental Fig. S7D). The majority of enrichments from Figure 2B repeated after correcting for polymerase abundance (699 of 1001 retained, 302 lost, 42 gained; see Supplemental Table S2), indicating that different regulatory mechanisms are linked to different chromatin architecture of gene sets.
Analysis of nuclease titration levels
A striking aspect of our analysis is that the majority of nucleosomes were matched using templates with multiple ends, suggesting that the majority of nucleosome ends are partially trimmed during a typical MNase digestion. Furthermore, in comparing our data to published datasets we found global variation in the distribution of nucleosome widths between datasets (Supplemental Fig. S3B), as well as in the relative occupancy of various nucleosome classes (i.e., genome-wide averages of +1 occupancy vs. mid-CDS occupancy differs between our data and that of Shivaswamy et al. (2008) (data not shown). Indeed, a recent study using MNase titrations followed by q-PCR identified variation in quantitative MNase susceptibility across nucleosomes associated with GAL genes (Bryant et al. 2008). We therefore sought to more thoroughly explore the influence of digestion levels in nucleosome positioning and occupancy. We have previously reported little change in nucleosome maps as measured by tiling microarray when mononucleosomal DNA is isolated from an early digestion step with only ∼40% mononucleosomal DNA (Yuan et al. 2005). However, dynamic range compression by microarrays might hide changes in a relative abundance of nucleosomes, and we were unable to obtain enough DNA for microarray analysis from less-digested (<40% mononucleosome) titration steps. Moreover, small changes in nucleosome segment lengths are difficult to detect with tiling microarrays.
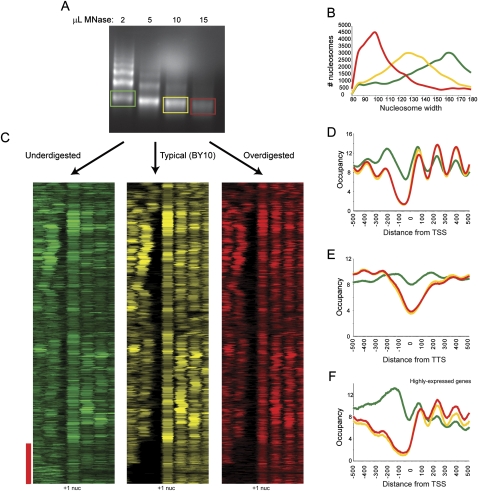
Since limited digestion with trypsin has proven a valuable structural probe for proteins, we were also interested in whether the same might be true of limited nuclease digestion of chromatin. We therefore carried out a titration of micrococcal nuclease, and gel-purified mononucleosomal DNA from three different titration levels—underdigested (∼15% mononucleosomes, “BY2”), typical digestion (∼80% mononucleosomes, “BY10”), and overdigested (only mononucleosomal DNA, “BY15”) (Fig. 3A). We used Template Filtering to call nucleosome positions in our titration data. As expected, nucleosome length was correlated with digestion level (Fig. 3B), with increasing digestion leading to shorter and shorter nucleosomes, presumably due to “chewing” of nucleosome ends by MNase.
Figure 3.
Effects of MNase level on chromatin structure. (A) Mononucleosomal DNA was isolated from ladders from three different MNase titration levels, and sequenced by Illumina sequencing. (B) Data from titration series was subjected to template filtering to generate nucleosome calls. Width distributions for nucleosomes from the three titration steps are plotted. Green, yellow, and red correspond to under-, mid-, and overdigested chromatin, respectively. (C) Data for under- (green), mid- (yellow), and over- (red) digested chromatin is shown in cluster view. Genes are aligned using BY10 +1 nucleosome center (indicated); all three clusters have genes ordered by clustering for BY10 data. Red bar indicates genes with wide NFRs in mid- and overdigested chromatin (largely highly expressed genes such as ribosomal genes), which are partially filled in underdigested chromatin. (D) TSS-aligned nucleosome occupancy data for all genes. (E) Stop-codon-aligned nucleosome occupancy for all genes. (F) As in D, but only for genes with Pol II ChIP occupancy >1, top 7% of genes.
Inspection of nucleosome maps for the three digestion levels revealed extensive similarities between the three maps. However, notable changes occur, particularly between underdigested and typical digestion (Fig. 3C). To globally assess differences between the different titration steps, we aligned genes by transcriptional start site (Fig. 3D) or stop codon (Fig. 3E) and averaged data from all genes at the three different titration levels. At the 5′ ends of genes we found an anticorrelation between +1 nucleosome occupancy and digestion level as expected. +1 nucleosomes are most abundant in underdigested chromatin and least abundant in overdigested chromatin. This is seen both in TSS-aligned averages of all genes (Fig. 3D), as well as in systematic analysis of changes in relative nucleosome occupancy calls (Supplemental Fig. S8).
The loss of promoter-proximal nucleosomes during digestion was even more pronounced for −1 nucleosomes, where underdigested chromatin (BY2) showed high levels of the −1 (and a resulting decrease in the width of the average NFR), whereas this nucleosome was less abundant or completely missing in BY10 and BY15 (Fig. 3C, red bar). A similar effect was observed at the 3′ NFR (Fig. 3E). The presence of an easily digested nucleosome over the NFR was most commonly observed at the long NFRs associated with highly expressed genes (red bar), and can easily be seen in averaged data for genes with the highest levels of RNA polymerase (Fig. 3F). Thus, consistent with recent studies in Drosophila (Henikoff et al. 2009) and human (Jin et al. 2009) cells, we find that at least some of the “nucleosome-free” region seen in typical nucleosome mapping studies corresponds to a loosely bound (as determined by salt extraction in those studies), easily digested (seen here) nucleosome. Overall, nucleosome occupancy levels are most even in BY10, which is the digestion level we typically use for nucleosome mapping (Yuan et al. 2005) and histone modification mapping (Liu et al. 2005).
Together, the results of the titration series suggest the presence of easily digested nucleosomes or other protein complexes at the promoters of highly expressed genes and point toward the necessity of knowing the extent of digestion when comparing nucleosome maps from different labs or different experiments.
The role of RNA polymerase in chromatin structure
A number of recent studies have claimed that intrinsic affinity of various genomic sequences for the histone octamer accounts for much, or most, of the chromatin structure observed in vivo in yeast (Ioshikhes et al. 2006; Segal et al. 2006; Peckham et al. 2007; Field et al. 2008; Kaplan et al. 2008; Yuan and Liu 2008). However, experimental determination of nucleosome positioning after in vitro reconstitution revealed that intrinsic preferences can almost entirely be ascribed to the role of poly(dA/dT) in excluding nucleosomes, with little additional translational positioning information encoded in the genome (Kaplan et al. 2008; Zhang et al. 2009). The huge discrepancy between in vitro sequence preferences and in vivo nucleosome positioning is likely to result from the action of numerous factors, most notably protein complexes in vivo that move nucleosomes from their preferred positions, such as the ATP-dependent chromatin remodeler Isw2 (Whitehouse and Tsukiyama 2006; Whitehouse et al. 2007). Almost certainly the most widespread of these _trans_-acting factors is Pol II, as ∼2/3 of the yeast genome codes for proteins, and combining distributions of RNA abundance (Yassour et al. 2009) with typical absolute mRNA abundances (Iyer and Struhl 1996) and half-lives (Wang et al. 2002) indicates that well over half of the yeast genome is likely transcribed at least once during a given cell cycle. The passage of RNA polymerase disrupts DNA–histone contacts (Wasylyk and Chambon 1980), leading us to ask how RNA polymerase globally affects chromatin structure in vivo.
We examined the effects of RNA polymerase on chromatin structure by using the rpb1-1 yeast strain, which contains a temperature-sensitive allele of the gene encoding the large subunit of Pol II (Nonet et al. 1987). To identify both the early, as well as longer term effects of RNA polymerase deactivation, we performed MNase-seq at 0, 20, and 120 min after shifting these cells from 25°C to 37°C. Interestingly, we found that despite minimal change in cell density, increasing amounts of MNase were required to generate similar nucleosome ladders (Supplemental Fig. S9A–C) as the time course progressed, indicating a global role for RNA polymerase in increasing overall chromatin accessibility. Consistent with the increased MNase required, we found that nucleosome length decreased over the time course (Supplemental Fig. S9D). Importantly, none of our conclusions below are affected by the titration level, as the inferred effects of polymerase loss do not match effects of overdigestion (see below).
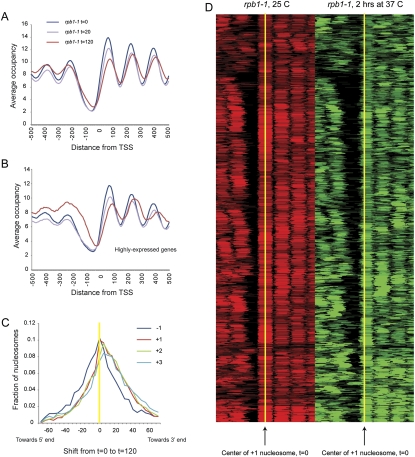
Averaging genes by TSS for the three time points reveals two effects of polymerase loss on nucleosome positioning (Fig. 4A,B; Supplemental Fig. S10). First, NFR width decreases over time, largely because of gains in nucleosome occupancy at the −1 position (Venters and Pugh 2009). This was not a digestion artifact at later time points, as the nucleosome length distribution at 120 min was consistent with MNase overdigestion (Supplemental Fig. S9D), whereas increased −1 nucleosome occupancy is a general property of underdigested chromatin (Fig. 2). Second, coding-region nucleosomes shift downstream over time. This latter observation is interesting, as it is consistent with predictions from biochemical studies: The process of RNA polymerase transiting a nucleosome in vitro results in transient dissociation of 5′ DNA from the octamer, followed by recapture of upstream DNA on the octamer surface, resulting in a predicted retrograde shift of the octamer relative to polymerase movement (Studitsky et al. 1994, 1997).
Figure 4.
Effects of RNA polymerase on chromatin structure. (A) Nucleosomes were isolated from rpb1-1 yeast grown at 25°C, and shifted to 37°C for 20 or 120 min. Data are presented in TSS-aligned average. (B) As in A, but for highly expressed genes. (C) Nucleosomes over genes shift downstream upon Pol II loss. For each indicated nucleosome type (−1, +1, +2, +3) we plot the distribution of center-to-center distances between the nucleosome calls at 0 and 120 min after Pol II inactivation. We find that 43% of −1 nucleosomes, 59% of +1 nucleosomes, 61% of +2 nucleosomes, and 60% of +3 nucleosomes shift away from the NFR. (D) Global view of +1 nucleosome shifts during Pol II inactivation. Nucleosome calls for all promoters with a downstream +1 nucleosome shift are shown as a heatmap, aligned by the center of the +1 nucleosome (yellow) before Pol II inactivation (red). After 2 h of Pol II inactivation (green), downstream shifts of these 59% of +1 nucleosomes are apparent.
We investigated nucleosome position shifts by plotting the direction of nucleosome shift between pairs of time points during the temperature shift (Fig. 4C,D). Nucleosome shifts over coding regions were clearly biased in the downstream direction, consistent with the averaged view in Figure 4A. Importantly, −1 nucleosome shifts were not biased in either direction. This small (∼10 bp on average) downstream shift is specific, as comparisons between other pairs of chromatin datasets (such as pre- and post-heat shock) show distributions of nucleosome shifts centered on zero (Supplemental Fig. S11). To determine whether nucleosome shifts preferentially occurred over particular classes of genes, we tested gene classes for significant deviation from the average shift of the −1, +1, +2, or +3 nucleosome by the KS statistic. Again, very highly expressed gene classes, such as those encoding ribosomal proteins or amino acid metabolism genes, exhibited more dramatic nucleosome shifts from 20 to 120 min (data not shown). Highly expressed genes are generally down-regulated during heat stress (Gasch et al. 2000), but nucleosome shifts at ribosomal genes are unlikely to be a consequence of heat shock-induced changes—reanalysis of the heat shock data from Shivaswamy et al. (2008) do not recapitulate the shifts we observe here (Supplemental Fig. S11A).
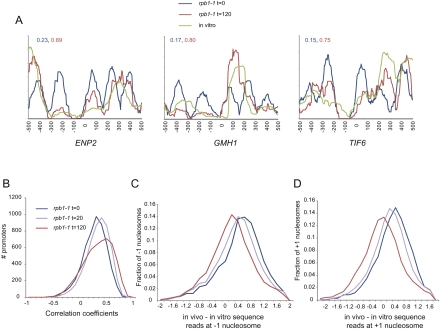
Why do nucleosomes shift after loss of polymerase? As noted above, RNA polymerase is a major factor in nucleosome movement and eviction in vivo, and the in vivo nucleosome positions over highly expressed genes deviate from in vitro nucleosome preferences to a greater extent than they do over poorly expressed genes. Thus, we compared our data with the in vitro data from Kaplan et al. (2008), reasoning that loss of RNA polymerase might allow chromatin to relax to more closely match local thermodynamic minima. We noticed that nucleosomes at many promoters, indeed, more closely match in vitro preferences after 2 h of polymerase inactivation (Fig. 5A).
Figure 5.
Nucleosomes relax toward in vitro preferred locations after Pol II loss. (A) Three examples of promoters where data from Pol II inactivation matches in vitro nucleosome assembly data better than data from before Pol II inactivation. Shown are extended read coverage along 1000 bp centered on TSS. Numbers shown in the inset are correlations between in vitro coverage and t = 0 (blue) and t = 120 (red) in vivo coverage. (B) Promoter chromatin architecture globally shifts toward in vitro preferences as polymerase is inactivated. Extended read coverage along the 1 kb centered on the TSS was extracted for all genes, and correlation coefficients were calculated to equivalent data for in vitro nucleosome reconstitution experiments (Kaplan et al. 2008). Histograms show a global shift of promoters toward the in vitro nucleosome pattern. (C) Normalized occupancy of −1 nucleosome better matches in vitro data after polymerase loss. For all −1 nucleosomes (called at t = 0 or at t = 120), the difference between in vivo normalized occupancy and in vitro normalized occupancy at the center of the in vivo nucleosome were calculated and presented as a histogram. (D) As in C, but for all +1 nucleosomes.
To generalize this result, we calculated the correlation between in vitro nucleosome assembly data (Kaplan et al. 2008) and in vivo nucleosome positioning for 1-kb windows at the 5′ ends of genes (Fig. 5B). At t = 0, correlation coefficients centered around 0.3, consistent with the ability of in vitro assembly to highlight NFRs (Sekinger et al. 2005; Kaplan et al. 2008; Zhang et al. 2009). After 2 h of polymerase inactivation, the distribution of correlations shifted to a higher value of ∼0.5, indicating that RNA polymerase does help maintain nucleosomes in thermodynamically unfavored locations in vivo. Of course, much of this is due to the above-mentioned role of transcription in −1 nucleosome eviction (Fig. 5C). We also asked whether lateral repositioning of nucleosomes upon polymerase loss resulted in relaxation to thermodynamically preferred positions. Strikingly, distances between +1 nucleosomes and the nearest in vitro occupancy peak decreased after polymerase inactivation (Fig. 5D; Supplemental Fig. S12), consistent with the hypothesis that both nucleosome eviction and sliding by RNA polymerase antagonize the thermodynamically preferred chromatin state.
Discussion
Genome-wide mapping of nucleosome positions in S. cerevisiae has been a tremendously productive method for illuminating the principles underlying chromatin structure and function. Deep sequencing methods have multiple advantages over tiling microarrays for genomic localization studies such as nucleosome mapping studies, including single-nucleotide resolution, expanded dynamic range, and nearly whole-genome coverage. Here, we present a novel method for analyzing deep sequencing data for chromatin maps. Our method automatically extracts nucleosome position, occupancy, and width, and accounts for variability in end digestion by MNase.
Analysis of chromatin packaging across the yeast genome confirmed previously described aspects of yeast chromatin structure, including widespread 5′ and 3′ nucleosome-depleted regions, a dichotomy between stress and growth genes reflected in NFR width, increased nucleosome fuzziness distal to the NFR, and a subtle anticorrelation between coding-region nucleosome occupancy and transcription. We also identified finer distinctions between certain classes within the major stress/growth branches. Most interestingly, we found that +1 to +3 nucleosome spacing was significantly shorter over ribosomal genes than over other gene types (see below).
We also analyzed data from an MNase titration series, as different laboratories isolate nucleosomes from different MNase digestion levels (see, for example, Shivaswamy et al. 2008). Analysis of data from underdigested chromatin revealed the abundant presence of nucleosome-sized peaks in the “NFR.” A recent analysis of Drosophila chromatin also identified nucleosomes in NFRs in a low-salt extraction from underdigested chromatin (Henikoff et al. 2009), and similar results hold in human cells (Jin et al. 2009). Does this material correspond to easily digested nucleosomes or to DNA protected from MNase by other proteins such as transcription factors? Two lines of evidence support the former hypothesis. One, the equivalent material in Drosophila is associated with the histone variant H3.3, indicating the presence of histones at these locations (Henikoff et al. 2009). Second, we only find nucleosomes filling in NFRs that are larger than 140 bp, suggesting that these are bona fide nucleosomes. Interestingly, analysis of sequence motifs that exhibit occupancy differences include the CGCG motif recently shown to be bound by the Rsc3/30 subunits of the RSC chromatin remodeling complex (Badis et al. 2008), suggesting that the easily digested −1 peak might correspond to a nuclease-accessible RSC-remodeled nucleosome state.
As many features of yeast chromatin correlate with transcription rate, we mapped nucleosomes before and after inactivation of Pol II. We find that NFRs become shorter and shallower upon loss of Pol II, particularly at highly expressed genes, consistent with a previously described role for RNA polymerase in eviction of −1 nucleosomes (Venters and Pugh 2009). We also found a surprising general role for RNA polymerase in nucleosome sliding—nucleosomes over coding regions generally shifted away from the NFR upon loss of polymerase (Fig. 4). These results are consistent with the predictions from biochemical studies—in vitro, RNA polymerase is capable of transiting a nucleosome without evicting histones, apparently by invading a nucleosome edge and then propagating a bubble of DNA around the octamer surface (Studitsky et al. 1994, 1997; Hodges et al. 2009). This retrograde nucleosome movement may play a role in the stereotyped +1 nucleosome positioning in vivo, which is not explained by intrinsic thermodynamic preferences as measured by in vitro nucleosome assembly (Kaplan et al. 2008; Zhang et al. 2009). We speculate that after assembly of a newly replicated DNA into nucleosomes, polymerase passage could be responsible for nucleosome shifts toward the NFR until the +1 nucleosome is as close to either poly(A)s or to the preinitiation complex as physically possible. This retrograde nucleosome movement may also play a role in the surprisingly tight packing of nucleosomes over highly transcribed ribosomal genes and, indeed, loss of polymerase results of relaxation of the +1 to +3 spacing at these genes (Supplemental Figs. S10, S13).
This interpretation must be tempered by dynamic studies in yeast, which indicate that some nucleosomes (particularly +1 nucleosomes) are rapidly exchanged during G1 (Dion et al. 2007; Jamai et al. 2007; Rufiange et al. 2007)—how is it that translational effects of transcription are observed on nucleosomes, given that nucleosomes are often rapidly exchanged (in some cases many times per cell cycle)? We consider two of many possible ways to reconcile these results. First, given that current locus-specific dynamic exchange measurements rely on transcriptional activation of tagged histones, there is a lower bound (∼15–30 min) for the fastest exchange rates measurable. However, if the fastest exchange rates are indeed on the order of ∼15 min, then for many genes a high proportion of cells in a population will have had polymerase pass through the gene since the last histone turnover cycle, resulting in a detectable polymerase-driven retrograde shift in the population measurement despite ongoing nucleosome replacement. Second, we do not currently know the extent of correlation in the dynamics of adjacent nucleosomes at the single-gene level. In other words, at a highly transcribed gene with high levels of histone replacement throughout the gene body, does +1 eviction affect +2 eviction? If these do not always co-occur, then retrograde shifts of surrounding nucleosomes could provide a local “memory” of prior polymerase passage, such that a leftward-shifted +2 would constrain the replacement location for a replaced +1 nucleosome.
Finally, our results bear on the relationship between thermodynamic sequence preferences and in vivo chromatin structure. Dramatic claims have been made regarding the extent to which genomic sequence dictates the positioning of nucleosomes in the cell (Segal et al. 2006; Kaplan et al. 2008), although most studies find the effects of sequence on chromatin architecture to be modest (Ioshikhes et al. 2006; Peckham et al. 2007; Yuan and Liu 2008). While in vitro chromatin assembly correlates well with in vivo nucleosome positions (Kaplan et al. 2008; Zhang et al. 2009), this almost entirely results from the depletion of nucleosomes over poly(A) and related sequences (Kunkel and Martinson 1981; Drew and Travers 1985; Iyer and Struhl 1995; Sekinger et al. 2005). Our results confirm the expected role for RNA polymerase in movement of nucleosomes away from thermodynamically preferred positions.
Together, these results further emphasize the role for RNA polymerase in shaping the chromatin landscape of the genome and point toward the difficulty in disentangling cause and effect in the relationship between chromatin and transcription.
Methods
Nucleosome isolation
Yeast culture, fixation, and MNase titrations were carried out as previously described (Yuan et al. 2005). For rpb1-1 temperature shifts, cells were grown to an OD of 0.6 in YPD at 25°C, then culture aliquots were immediately shifted to 37°C by addition of an equal volume of YPD at 49°C. After recovery of digested DNA, mononucleosomal was gel-purified and subjected to Illumina sequencing as described in Shivaswamy et al. (2008).
Template filtering algorithm
Using a sliding window across the genome, we cross-correlate each position with a pair of templates, one matching the forward reads and one the reverse reads. We enumerate all 7 × 7 possible combinations of forward and reverse templates. We repeat this scan with different spacing between both templates to capture over- and underdigestion of the ∼146-bp nucleosomal DNA fragments. As a result, we obtain a correlation “heat map” for each pair of templates containing the correlation coefficient for each center position and width. Next, we search for local maxima points within this “heat map”; each maxima point is a potential nucleosome at a given position with a specific width. Finally, to assemble the final set of nucleosomes, we are using a greedy approach to select the best assignment of nucleosomes under overlapping constrains. Potential nucleosomes are sorted according to correlation score and occupancy, and are then selected to the final set allowing maximum overlap of 40% between adjacent nucleosomes.
Selecting representative templates
To generate a variety of templates that represents the prototypical distributions of reads at nucleosome ends, we first applied our method using a Gaussian-shaped template and obtained a preliminary map of nucleosome predictions. We aligned all predicted nucleosome ends and created a matrix of read patterns using a window of 80 bp flanking the edges. Next, we clustered this matrix using k-mean clustering and selected seven representative templates that capture the range of template shapes observed.
Correcting chromatin parameters to account for Pol II enrichment in KS tests
We represent each gene as a vector of chromatin properties (i.e., +1 nucleosome occupancy, NFR width, mid-CDS occupancy, etc). Using a compendium of experimental gene annotations we previously collected (Dion et al. 2007; Wapinski et al. 2007), we compared the distribution of each chromatin parameter for each gene set vs. the background. We discovered 1001 gene sets with at least one enriched chromatin property. To correct the genes' properties vectors for RNA polymerase levels, we plotted RNA polymerase occupancy (measured by microarray as in Steinmetz et al. 2006; T Kim, S Buratowski, A Novogrodski, M Yassour, N Friedman, and O Rando, in prep.) vs. each chromatin property and calculated the LOESS curve for each property (Supplemental Fig. S7A–C). We then subtracted the smoothed LOESS curves from each gene, obtaining new chromatin properties vectors that represent the distance of each property from the LOESS curve. We repeated the K-S enrichment with these new vectors.
Source code
Source code is available at http://compbio.cs.huji.ac.il/NucPosition.
Acknowledgments
We thank F. Winston for the rpb1-1 strain. We thank current and past members of the Friedman and Rando labs for comments and discussions. O.J.R. is supported in part by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund. This research was supported by grants to O.J.R. and N.F. from the NIGMS, and to N.F. from the US-Israel Bi-National Foundation and a European Union FP7 “Model-In” collaborative grant.
Author contributions: O.J.R. designed the experiments in discussion with N.F., and A.H. carried them out. A.W. and M.Y. developed the analysis methodology and analyzed the data. O.J.R., A.W., and N.F. wrote the manuscript.
Footnotes
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Badis G, Chan ET, van Bakel H, Pena-Castillo L, Tillo D, Tsui K, Carlson CD, Gossett AJ, Hasinoff MJ, Warren CL, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basehoar AD, Zanton SJ, Pugh BF. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004;116:699–709. doi: 10.1016/s0092-8674(04)00205-3. [DOI] [PubMed] [Google Scholar]
- Bondarenko VA, Steele LM, Ujvari A, Gaykalova DA, Kulaeva OI, Polikanov YS, Luse DS, Studitsky VM. Nucleosomes can form a polar barrier to transcript elongation by RNA polymerase II. Mol Cell. 2006;24:469–479. doi: 10.1016/j.molcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- Bryant GO, Prabhu V, Floer M, Wang X, Spagna D, Schreiber D, Ptashne M. Activator control of nucleosome occupancy in activation and repression of transcription. PLoS Biol. 2008;6:2928–2939. doi: 10.1371/journal.pbio.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JK, Kim YJ. Epigenetic regulation and the variability of gene expression. Nat Genet. 2008;40:141–147. doi: 10.1038/ng.2007.58. [DOI] [PubMed] [Google Scholar]
- Choi JK, Kim YJ. Intrinsic variability of gene expression encoded in nucleosome positioning sequences. Nat Genet. 2009;41:498–503. doi: 10.1038/ng.319. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Lomonossoff GP, Laskey RA. High sequence specificity of micrococcal nuclease. Nucleic Acids Res. 1981;9:2659–2673. doi: 10.1093/nar/9.12.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Kaplan T, Kim M, Buratowski S, Friedman N, Rando OJ. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges C, Bintu L, Lubkowska L, Kashlev M, Bustamante C. Nucleosomal fluctuations govern the transcription dynamics of RNA polymerase II. Science. 2009;325:626–628. doi: 10.1126/science.1172926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Ioshikhes IP, Albert I, Zanton SJ, Pugh BF. Nucleosome positions predicted through comparative genomics. Nat Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Struhl K. Absolute mRNA levels and transcriptional initiation rates in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1996;93:5208–5212. doi: 10.1073/pnas.93.11.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2008;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- Kornberg RD, Stryer L. Statistical distributions of nucleosomes: Nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel GR, Martinson HG. Nucleosomes will not form on double-stranded RNa or over poly(dA).poly(dT) tracts in recombinant DNA. Nucleic Acids Res. 1981;9:6869–6888. doi: 10.1093/nar/9.24.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam FH, Steger DJ, O'Shea EK. Chromatin decouples promoter threshold from dynamic range. Nature. 2008;453:246–250. doi: 10.1038/nature06867.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D. Modifying gene expression programs by altering core promoter chromatin architecture. Cell. 2002;110:261–271. doi: 10.1016/s0092-8674(02)00822-x. [DOI] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008a;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008b;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Rando OJ. Nucleosome positioning: How is it established, and why does it matter? Dev Biol. 2009 doi: 10.1016/j.ydbio.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Mello CC, Shimada A, Nakatani Y, Hashimoto S, Ogawa M, Matsushima K, Gu SG, Kasahara M, Ahsan B, et al. Chromatin-associated periodicity in genetic variation downstream of transcriptional start sites. Science. 2009;323:401–404. doi: 10.1126/science.1163183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004;24:10111–10117. doi: 10.1128/MCB.24.23.10111-10117.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol. 2009;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, Iyer VR. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ, Warren CL, Kuehner JN, Panbehi B, Ansari AZ, Brow DA. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol Cell. 2006;24:735–746. doi: 10.1016/j.molcel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Studitsky VM, Kassavetis GA, Geiduschek EP, Felsenfeld G. Mechanism of transcription through the nucleosome by eukaryotic RNA polymerase. Science. 1997;278:1960–1963. doi: 10.1126/science.278.5345.1960. [DOI] [PubMed] [Google Scholar]
- Stunkel W, Kober I, Seifart KH. A nucleosome positioned in the distal promoter region activates transcription of the human U6 gene. Mol Cell Biol. 1997;17:4397–4405. doi: 10.1128/mcb.17.8.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turin G. An introduction to matched filters. IRE Trans Inf Theory. 1960;6:311–329. [Google Scholar]
- Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venters BJ, Pugh BF. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 2009;19:360–371. doi: 10.1101/gr.084970.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci. 2002;99:5860–5865. doi: 10.1073/pnas.092538799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Wasylyk B, Chambon P. Studies on the mechanism of transcription of nucleosomal complexes. Eur J Biochem. 1980;103:219–226. doi: 10.1111/j.1432-1033.1980.tb04306.x. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Workman JL, Kingston RE. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:545–579. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- Wyrick JJ, Holstege FC, Jennings EG, Causton HC, Shore D, Grunstein M, Lander ES, Young RA. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature. 1999;402:418–421. doi: 10.1038/46567. [DOI] [PubMed] [Google Scholar]
- Yassour M, Kaplan T, Fraser HB, Levin JZ, Pfiffner J, Adiconis X, Schroth G, Luo S, Khrebtukova I, Gnirke A, et al. Ab initio construction of a eukaryotic transcriptome by massively parallel mRNA sequencing. Proc Natl Acad Sci. 2009;106:3264–3269. doi: 10.1073/pnas.0812841106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Moqtaderi Z, Rattner BP, Euskirchen G, Snyder M, Kadonaga JT, Liu XS, Struhl K. Intrinsic histone-DNA interactions are not the major determinant of nucleosome positions in vivo. Nat Struct Mol Biol. 2009;16:847–852. doi: 10.1038/nsmb.1636. [DOI] [PMC free article] [PubMed] [Google Scholar]