The MIRA method for DNA methylation analysis (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 18.
Abstract
DNA methylation patterns are often altered in human cancer and aberrant methylation is considered a hallmark of malignant transformation. Several methods have been developed for the characterization of gene-specific and genome-wide DNA methylation patterns. In this chapter, we describe the methylated-CpG island recovery assay (MIRA), which is based on the high affinity of the MBD2b/MBD3L1 complex for double-stranded CpG-methylated DNA. MIRA has been used in combination with microarray platforms to map DNA methylation patterns across the human genome.
Keywords: DNA methylation analysis, methyl-CpG binding protein, microarrays, CpG islands, tiling arrays
1. Introduction
Methylation of DNA at the 5-position of cytosines within CpG dinucleotides is an important component of epigenetic gene silencing systems. Aberrations in DNA cytosine methylation may underlie human disease. In particular, the genome of cancer cells is known to undergo substantial changes in DNA methylation (1). Most notable are genome-wide hypomethylation events that specifically target repetitive DNA elements, and gene-specific hypermethylation of CpG islands. CpG islands are sequences with greater than normal G+C DNA content (2). They are usually between 0.5 and 2 kb long and contain a relatively high frequency of CpG dinucleotides. CpG sequences normally are underrepresented in mammalian genomes owing to mutational pressure and/or lack of efficient DNA repair at methylated CpGs (3). However, in most normal tissues and in the germ line, CpG islands are unmethylated. Accordingly, they are not subject to erosion by mutational events and retain a close to expected frequency of CpG dinucleotides. In cancer tissue, CpG islands often are found methylated and each individual tumor may contain several hundred methylated CpG islands. The exact extent of DNA methylation changes and the mechanism that elicits these events are unknown and are subject to intense investigation. Therefore, it is of great importance to have technologies available that can interrogate the methylation status of normal and diseased tissues or cell types at a genome-wide level and at high resolution.
Several different techniques have been developed to analyze DNA methylation patterns on a genome-wide scale (4). These methods include several restriction enzyme-based techniques, such as restriction landmark genomic scanning (5, Chapter 11), methylation-sensitive representational difference analysis (6, Chapter 10), and differential methylation hybridization (7, Chapter 7). Huang and colleagues were the first to apply microarrays for the analysis of DNA methylation thus providing a significant advancement to researchers in this field (7). The methods using methylation-sensitive restriction endonucleases are naturally limited by the occurrence of the respective sequences within a CpG island or any other target sequence. Another commonly used approach to identify methylated genes is based on mRNA expression arrays to identify genes reactivated by treatment with DNA methylation inhibitors such as 5-aza-deoxycytidine (8-11, Chapter 13). This approach can only be used effectively with cell lines and some genes may be refractory to demethylation-coupled reactivation due to additionally imposed chromatin modifications that are independent of DNA methylation. Antibodies against 5-methylcytosine have been used in immunoprecipitation experiments combined with microarrays in analogy with chromatin immunoprecipitation (ChIP on chip) assays (12, Chapter 5). Another variation of current methylation microarray approaches is the use of the methylation-dependent restriction enzyme _Mcr_BC to cleave methylated DNA (13, 14). Finally, high-throughput direct sequencing of bisulfite-converted genomes can be used to derive high resolution and precise DNA methylation maps (15, Chapter 14). However, for complete genome-wide analysis, considerable bioinformatics challenges will need to be overcome since, with the exception of the rare 5mC bases, the genome consists of only three DNA bases (U or T, A, G) after sodium bisulfite conversion of cytosine.
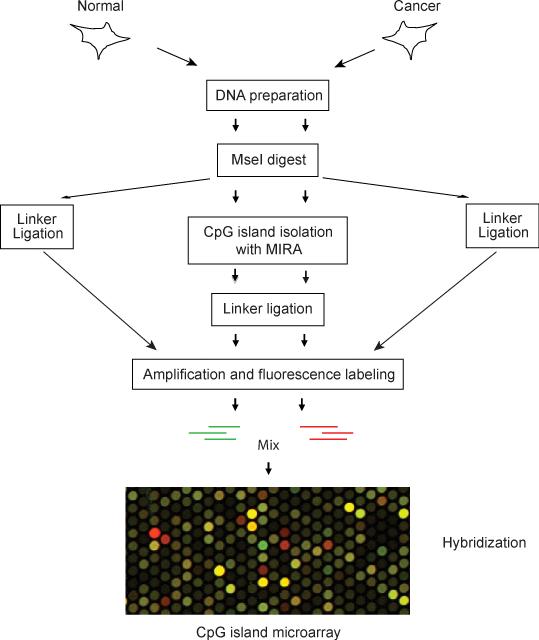
Among the methods suitable for genome-wide mapping of DNA methylation, the methylated CpG island recovery assay (MIRA) represents an approach that is based neither on restriction endonucleases, antibodies or sodium bisulfite treatment of the genomic DNA. MIRA depends on the facts that the methyl-CpG binding protein MBD2b specifically recognizes methylated CpG dinucleotides (16) and that this interaction is strongly enhanced by the MBD3L1 protein (17-19), a heterodimerization partner of MBD2 (20). Among all methyl-CpG binding proteins known, MBD2b has the highest affinity for methylated DNA and displays the greatest capacity to differentiate between methylated and unmethylated DNA. It recognizes a wide range of methylated CpG sequences with little sequence specificity (21). In our lab, lack of a defined sequence specificity of the MBD2b/MBD3L1 complex was confirmed by cloning and random sequencing of MIRA-enriched DNA fragments. Pulldown of methylated fragments is most efficient when at least two methylated CpG sites are present (17). In the MIRA procedure, fragmentized genomic DNA is incubated with the MBD2b/MBD3L1 high affinity protein complex. Unlike the anti-5-methylcytosine antibody precipitation technique, which requires single-stranded DNA for antibody recognition, MIRA works on normal double-stranded DNA; in fact the complex does not bind to single-stranded DNA. The CpG-methylated DNA is easily captured from the binding reaction via the GST-tagged MBD2b and glutathione beads. The isolated CpG-methylated fraction is linker ligated and then PCR amplified. The MIRA-enrichment method has been proven to be compatible with several types of microarray platforms. Fig. 1 outlines the procedure of the MIRA technology.
Figure 1.
Schematic diagram of the MIRA microarray method. Input and MIRA-enriched fractions are prepared, labeled with different dyes, mixed, and hybridized to the microarray slides. In a simplified version, MIRA-enriched DNA from normal and tumor cells can be mixed and hybridized directly.
2. Materials
Buffer components and other reagents used in the MIRA procedure must be molecular biology grade fine chemicals.
2.1. Expression and purification of recombinant proteins
- LB liquid media and agarose plates for bacterial work can be prepared according to standard bacterial protocols (22).
- BL21 (DE) Epicurian Coli Competent Cells (Agilent/Stratagene, Santa Clara, CA)
- Lysozyme (Sigma; St. Louis, MO).
- STE (Sodium-Tris-EDTA) buffer for GST-tagged protein purification (SGPP buffer): 10 m_M_ Tris-HCl, pH 7.8, 150 m_M_ NaCl, 1 m_M_ EDTA.
- PMSF (phenylmethylsulfonylfluoride) solution: 100 m_M_ PMSF (Sigma) is dissolved in isobutanol and stored at -20°C. Add PMSF to buffers just before use.
- Lysis solution: 10 % (w/v) N-lauroylsarcosine in water.
- Triton X-100 solution: 10 % (v/v) Triton X-100 in water.
- Glutathione Sepharose 4B beads (GE Healthcare; Uppsala, Sweden).
- Glutathione bead wash buffer: Phosphate buffered saline (137 m_M_ NaCl, 2.7 m_M_ KCl, 10 m_M_ Na2HPO4, 1.8 m_M_ KH2PO4) and 0.1% (v/v) Triton X-100.
- GST-tagged protein elution buffer: 50 m_M_ Tris-HCl, pH 8.5, 150 m_M_ NaCl, 20 m_M_ reduced glutathion (Sigma) and 0.1 % (v/v) Triton X-100.
- Protein dialysis buffer: 50 m_M_ Hepes, pH 7.4, 150 m_M_ NaCl, 5m_M_ β-mercaptoethanol and 50 % (v/v) glycerol.
- STE (Sodium-Tris-EDTA) buffer for His-tagged protein purification (SHPP buffer): 10 m_M_ Tris-HCl, pH 7.8, 150 m_M_ NaCl, 0.1 m_M_ EDTA.
- Ni-NTA agarose beads (Novagen/EMG; Darmstadt, Germany).
- Ni-NTA agarose beads wash buffer: 50 m_M_ NaH2PO4, pH to 8.0, 300 m_M_ NaCl, 20 m_M_ imidazole. The pH is adjusted with 1 M NaOH.
- His-tagged protein elution buffer: 50 m_M_ NaH2PO4, pH to 8.0, 300 m_M_ NaCl, 250 m_M_ imidazole). The pH is adjusted with 1 M NaOH.
2.2. MIRA procedure and amplicon generation
- 1
MseI enzyme, NEB 2 buffer and 1 mg/mL BSA (New England Biolabs; Ipswich, MA). - 2
QIAquick PCR purification kit (Qiagen; Valencia, CA). - 3
Sonicated JM110 bacterial DNA (see Note 1). The JM110 bacterial strain can be purchased from Agilent/Stratagene (Santa Clara, CA). The JM110 strain can grow in antibiotic free LB medium and chromosomal DNA is prepared from bacteria according to a standard protocol (23). Purified JM110 DNA is sonicated to ~ 500 bp long fragments, ethanol precipitated and redissolved in TE buffer. - 4
TE buffer: 10 m_M_ Tris-HCl, pH 8.0 and 1 m_M_ EDTA. - 5
10 × MIRA binding buffer: 100 m_M_ Tris-HCl (pH 7.9), 500 m_M_ NaCl, 10 m_M_ DTT, 100 m_M_ MgCl2, 1.0% (v/v) Triton X-100. - 6
MagneGST beads and magnetic stand (Promega; Madison, WI). Magnetic beads should be washed before use to remove preservatives. Take 2.5 μL of MagneGST beads and wash 3 times with 1 mL of PBS containing 0.1 % (v/v) Triton X-100. To reduce non-specific binding to the surface of the beads, set up a blocking reaction similar to the MIRA reaction but containing only MIRA binding buffer and sonicated JM110 DNA. Incubate it at 4°C for 20 min on a rotating platform. Capture the beads by using the magnetic stand and carefully remove the supernatant letting ~10 μL buffer remain on the beads. - 7
MIRA wash buffer: 10 m_M_ Tris-HCl (pH 7.5), 700 m_M_ NaCl, 1 m_M_ EDTA, 3 m_M_ MgCl2, 0.1 % (v/v) Triton X-100. - 4
Long-linker 23-mer: 5’-AGCAACTGTGCTATCCGAGGGAT-3’ and Mse-linker 12-mer: 5’-TAATCCCTCGGA-3’. Two unidirectional linkers are annealed by combining 50 μL of 100 μ_M_ Long- and 50 μL of 100 μ_M_ Mse-linkers. This mixture is boiled for 1-2 min in a water bath, then allowed to slowly cool to room temperature. The annealed double stranded linker can be stored indefinitely at -20°C. - 5
T4 DNA ligase and 10 × ligase buffer (New England Biolabs; Ipswich, MA). - 6
Taq DNA polymerase, 10 × PCR buffer, and 5 × Q solution (Qiagen; Valencia, CA).
3. Methods
3.1 Preparation of GST-MBD2b and His-MBD3L1 proteins for MIRA
3.1.1 Expression of GST-tagged MBD2b protein
- Recombinant plasmids for bacterial expression of GST-MBD2b and His-MBD3L1 proteins are available upon request.
- Transform BL21 (DE3) competent cells with GST-MBD2b protein expressing plasmid and plate them on ampicillin containing LB plates.
- Inoculate 50 mL LB (amp) with 20 well-developed bacterial colonies and grow at 37°C until OD reaches 0.6 at fixed wavelength A 600.
- Add 50 μL of 100 m_M_ IPTG to induce expression of GST-tagged MBD2b protein.
- Allow the cells to grow for an additional 4-6 hours at 37°C.
- Transfer the induced bacterial culture into a 50 mL tube and centrifuge at 3500_g_ for 10 min at 4°C, pour off supernatant. Bacterial cells can be stored at -80°C for several months, or proceed with protein purification.
3.1.2. Purification of GST-tagged MBD2b protein
- Resuspend bacterial pellet in 10 mL of SGPP buffer containing 100 μg/mL lysozyme.
- Add 100 μL of PMSF solution and incubate on ice for 10 min.
- Lyse bacterial cells by addition of 1 mL of lysis solution.
- Sonicate bacterial lysate until it clears up and is not viscous anymore.
- Add 1 mL Triton X-100 solution to the lysate and vortex it for 20 s.
- Centrifuge the lysate at 3500_g_ for 10 min.
- Transfer supernatant into a new tube.
- Add 0.1 mL of Glutathione Sepharose 4B beads (50% slurry - see Note 2) to 12 mL cleared lysate and mix gently by shaking at 4°C for 45 min.
- Pellet the beads at 1000_g_ for 1 min.
- Add 10 mL of glutathione bead wash buffer and invert the tube several times.
- Collect the beads by centrifugation at 1000_g_ for 1 min.
- Repeat the previous two steps two more times.
- Elute the GST-tagged MBD2b protein from the beads with 1 mL of GST-elution buffer at 4°C for 4 hrs on a rotating platform.
- The eluted GST-MBD2b protein should be dialyzed against 2 liters of PBS in the cold-room for 5 hrs and then overnight against protein dialysis buffer. After dialysis, MBD2b can be kept at -20°C for 6 months.
- Check the protein concentration on a 10 % SDS-PAGE gel using BSA controls.
3.1.3. Expression of His-tagged MBD3L1 protein
- Transform BL21 (DE3) competent cells with His-MBD3L1 protein expressing plasmid and plate them on kanamycin-containing LB plates.
- Inoculate 50 mL LB (Kan) with 20 well-developed bacterial colonies and grow at 37°C until OD reaches 0.6 at fixed wavelength A 600.
- Add 50 μL of 100 m_M_ IPTG to induce expression of His-tagged MBD3L1 protein.
- Allow the cells to grow for an additional 4-6 hrs at 37°C.
- Transfer the induced bacterial culture into a 50 mL tube and centrifuge at 3500_g_ for 10 min at 4°C, pour off supernatant. Bacterial cells can be stored at -80°C for several months, or proceed with protein purification.
3.1.4. Purification of His-tagged MBD3L1 protein
- Resuspend bacterial pellet in 10 mL of SHPP buffer containing 100 μg/mL lysozyme.
- Add 100 μL of PMSF solution and incubate on ice for 10 min.
- Lyse bacterial cells by addition of 1 mL of lysis solution.
- Sonicate bacterial lysate until it clears up and is not viscous anymore.
- Add 1 mL of Triton X-100 solution to the lysate and vortex it for 20 sec.
- Centrifuge the lysate at 3500_g_ for 10 min.
- Save supernatant into a new tube.
- Add 0.1 mL of Ni-NTA Agarose beads to 12 mL of cleared lysate and mix gently by shaking at 4°C for 30-45 min.
- Pellet the beads at 1000_g_ for 1 min.
- Add 10 mL of Ni-NTA agarose bead wash buffer and invert tube several times (see Note 3).
- Collect the beads by centrifugation at 1000_g_ for 1 min.
- Repeat the previous two steps two more times.
- MBD3L1 can be eluted from the beads with His-elution buffer at 4°C for 30 min on a rotating platform.
- The eluted MBD3L1 protein should be dialyzed against 2 liters of PBS for 5 hrs in the cold-room and then overnight against protein dialysis buffer. After dialysis, MBD3L1 can be kept at -20°C. for 6 months.
- Check the protein concentration on a 10 % SDS-PAGE gel using BSA controls.
3.2. Genomic DNA preparation for MIRA
3.2.1. Genomic DNA purification
Genomic DNA can be isolated from cells or tissue samples by any standard proteinase K and phenol/chloroform extraction protocol.
3.2.2. Genomic DNA fragmentation with _Mse_I endonuclease
Purified high molecular weight genomic DNA must be fragmentized for MIRA. This can be achieved by restriction endonuclease digestion. We suggest using _Mse_I enzyme for fragmentation which cuts 5’-TTAA-3’ sequences and leaves most CpG islands intact (see Note 4).
- Set up the following reaction: 2-3 μg of genomic DNA; 5.0 μL of 10 × NEBuffer 2 buffer; 5.0 μL of 1 μg/μL BSA; 2 μL of _Mse_I (10 U/μL) and add H2O to 50 μL.
- Incubate the reaction at 37°C overnight.
- Check whether the digestion is complete by running the sample on a 1.5 % agarose gel.
- Purify digested genomic DNA with Qiagen PCR purification kits according to the company's recommendations.
- Measure the concentration of MseI cut genomic DNA by using a spectrophotometer.
3.3. MIRA binding reaction
- Set up the following binding reaction in a 1.5 mL Eppendorf tube: 40 μL of 10 × MIRA buffer; 10 μL of 50 ng/μL of JM110 DNA; 1 μg of purified GST-MBD2b; 1 μg of purified His-MBD3L1 and add H2O to a final volume of 350 μL.
- Mix by pipetting and pre-incubate at 4°C for 20 min on a rotating platform.
- Add 250-500 ng of _Mse_I cut genomic DNA in 50 μL. (The final volume is now 400 μL).
- Incubate the binding reaction at 4°C at least for 4 hours (or overnight) on a rotating platform.
- Add 10.0 μL of pre-blocked MagneGST beads.
- Incubate it at 4°C for 45 min on a rotating platform.
- Retrieve MagneGST beads carrying the enriched methylated DNA fraction. Use the magnetic stand to capture the beads, and carefully remove the supernatant with a pipette.
- Add 800 μL of MIRA wash buffer into the tube and invert 4-5 times.
- Retrieve beads (methyl-CpG rich fraction) by using the magnetic stand and carefully decant supernatant.
- Repeat steps 8 and 9 two more times.
- Elute and purify the mCpG-enriched fraction from the MagneGST beads with QIAquick PCR purification kit according to the company's protocol.
- Elute methyl-CpG rich fraction from the column with 50 μL of H2O.
- Reduce the volume of the eluted fraction to 5 μL in a Speed Vac concentrator.
3.4. Linker-ligation and amplification
- Set up the following reaction: 5.0 μL of MIRA-enriched fraction; 1.0 μL of 10 × ligase buffer; 3.0 μL of 50 μ_M_ double stranded linker; 1.0 μL of T4 DNA ligase.For the ligation of “input” samples, add 10 ng of the _Mse_I-digested genomic DNA into the reaction.
- Incubate at 4°C overnight.
- Set up the following PCR by adding to the previous ligation mix: 10.0 μL of 10 × PCR buffer; 20.0 μL of 5 × Q solution; 4.8 μL of 25 m_M_ MgCl2; 14.0 μL of 2.5 m_M_ dNTPs; 2.0 μL of Taq polymerase (2.5 U/μL) and 39.2 μL of H2O.
- Before the amplification, let Taq polymerase work at 72°C for 7 min to fill in the 3’ ends of the ligated double stranded linkers. The two strands of the unligated double-stranded linkers are separated at this temperature and can serve as primers in the subsequent PCR.
- Cycling parameters: denaturation: 94°C for 20 s; annealing: 68°C for 30 s; and elongation: 72°C for 2 min. Repeat the previous cycle 10-12 times (see Note 5).
- Purify the amplicon by using a Qiagen PCR purification kit according to the company's recommendations.
- Measure the DNA content of the amplicon by using either a Nanodrop or another spectrophotometer. If the yield is lower than expected (less than 0.5 to 1 μg), take 10 ng of the first amplicon and repeat the amplification once more (see Note 6).
- MIRA-enriched and input amplicons, for example from control and tumor tissue, can be labeled with Cy3 and Cy5 dyes, respectively, and hybridized to commercially available CpG island or promoter arrays according to the manufacturer's instructions (see Note 7).
4. Notes
- JM110 is a Dam and Dcm methylation minus bacterial strain. Sonicated JM110 DNA is used to reduce the background by blocking non-specific binding of the mammalian DNA to the beads and proteins.
- A 50% slurry of Glutathione Sepharose 4B beads is prepared according to the company's protocol.
- Ni-NTA Agarose beads can strongly bind to some plastic tubes; you may add 0.1% (v/v) Triton X-100 into the Ni-NTA agarose beads wash buffer to minimize the loss.
- As an alternative to restriction enzyme digestion, careful sonication to produce fragments 300-500 bp in length can be used. In this case T4 DNA polymerase treatment is essential and blunt-ended double-stranded linkers must be used for creation of amplicons. MIRA can also be used with un-amplified DNA but much more DNA is needed as starting material. Approximately 25 μg of input DNA have been used successfully (19).
- PCR should be performed in such a way that cycling is stopped right after the linear phase of amplification. The easiest way of monitoring the amplification is to perform the PCR in a real-time thermocycler. Adding SYBR green dye into the PCR does not interfere with any of the subsequent procedures.
- You have to add 0.5 μM long-linker oligonucleotide into the 2nd PCR.
- We have successfully used microarrays from the UHN Microarray Centre, University of Toronto, Canada (17), Agilent (19), Affymetrix (unpublished data), and NimbleGen (19). We followed the companies’ protocols for array hybridization. For NimbleGen arrays, the labeling of amplicons, microarray hybridization, and scanning were performed by the NimbleGen Service Laboratory as previously described (24). Data was extracted from scanned images using NimbleScan 2.3 extraction software (NimbleGen Systems Inc.; Madison, WI) (19). For Agilent CpG island microarrays, which contain 237,000 oligonucleotide probes covering 27,800 CpG islands, two micrograms each of the amplicons from MIRA-enriched DNA and control samples were labeled with the BioPrime Array CGH Genomic Labeling kit (Invitrogen; Carlsbad, CA) with either Cy5-dCTP (e.g. tumor) or Cy3-dCTP (e.g. control) following the manufacturer's instructions. The purified labeled samples were then mixed and microarray hybridization was performed according to the Agilent ChIP-on-chip protocol (v.9.0). The hybridized arrays were scanned on an Axon 4000B microarray scanner and the images were analyzed with Axon GenePix software v.5.1. Image and data analysis were done as described (17). When screening for methylated CpG islands in cancer tissue, it is important to define a reliable cut-off value for methylation-positive CpG islands. We found good concordance of the array data with bisulfite-based methylation assays by considering individual CpG islands as methylation-positive when at least two adjacent probes within the CpG island scored a fold-difference factor of >3.0 when comparing tumor and normal tissue DNA.
Acknowledgements
This work was supported by NIH grants CA104967 and CA128495 to GPP.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 3.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr. Top. Microbiol. Immunol. 2006;301:259–281. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 5.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 6.Ushijima T, Morimura K, Hosoya Y, et al. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc. Natl. Acad. Sci. USA. 1997;94:2284–2289. doi: 10.1073/pnas.94.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan PS, Chen CM, Shi H, et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001;61:8375–8380. [PubMed] [Google Scholar]
- 8.Sato N, Fukushima N, Maitra A, et al. Discovery of novel targets for aberrant methylation in pancreatic carcinoma using high-throughput microarrays. Cancer Res. 2003;63:3735–3742. [PubMed] [Google Scholar]
- 9.Shi H, Wei SH, Leu YW, et al. Triple analysis of the cancer epigenome: an integrated microarray system for assessing gene expression, DNA methylation, and histone acetylation. Cancer Res. 2003;63:2164–2171. [PubMed] [Google Scholar]
- 10.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 11.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 12.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 13.Lippman Z, Gendrel AV, Colot V, Martienssen R. Profiling DNA methylation patterns using genomic tiling microarrays. Nat. Methods. 2005;2:219–224. doi: 10.1038/nmeth0305-219. [DOI] [PubMed] [Google Scholar]
- 14.Nouzova M, Holtan N, Oshiro MM, et al. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J. Pharmacol. Exp. Ther. 2004;311:968–981. doi: 10.1124/jpet.104.072488. [DOI] [PubMed] [Google Scholar]
- 15.Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res. 2007;67:8511–8618. doi: 10.1158/0008-5472.CAN-07-1016. [DOI] [PubMed] [Google Scholar]
- 16.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell. Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rauch T, Li H, Wu X, Pfeifer GP. MIRA-assisted microarray analysis, a new technology for the determination of genome-wide DNA methylation patterns, identifies frequent methylation of homeodomain containing genes in lung cancer cells. Cancer Res. 2006;66:7939–7947. doi: 10.1158/0008-5472.CAN-06-1888. [DOI] [PubMed] [Google Scholar]
- 18.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab. Invest. 2005;85:172–180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 19.Rauch T, Wang Z, Zhang X, et al. Homeobox gene methylation in lung cancer studied by genome-wide analysis with a microarray-based methylated CpG island recovery assay. Proc. Natl. Acad. Sci. USA. 2007;104:5527–5532. doi: 10.1073/pnas.0701059104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang CL, Jin SG, Pfeifer GP. MBD3L1 is a transcriptional repressor that interacts with MBD2 and components of the NuRD complex. J. Biol. Chem. 2004;279:52456–52464. doi: 10.1074/jbc.M409149200. [DOI] [PubMed] [Google Scholar]
- 21.Fraga MF, Ballestar E, Montoya G, Taysavang P, Wade PA, Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swords WE. Chemical transformation of E.. coli. Methods Mol. Biol. 2003;235:49–55. doi: 10.1385/1-59259-409-3:49. [DOI] [PubMed] [Google Scholar]
- 23.Bickley J, Owen RJ. Preparation of bacterial genomic DNA. Methods Mol. Biol. 1995;46:141–148. doi: 10.1385/0-89603-297-3:141. [DOI] [PubMed] [Google Scholar]
- 24.Selzer RR, Richmond T,A, Pofahl NJ, et al. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer. 2005;44:305–319. doi: 10.1002/gcc.20243. [DOI] [PubMed] [Google Scholar]