Direct conversion of fibroblasts to functional neurons by defined factors (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 25.
Published in final edited form as: Nature. 2010 Jan 27;463(7284):1035–1041. doi: 10.1038/nature08797
Abstract
Cellular differentiation and lineage commitment are considered robust and irreversible processes during development. Recent work has shown that mouse and human fibroblasts can be reprogrammed to a pluripotent state with a combination of four transcription factors. This raised the question of whether transcription factors could directly induce other defined somatic cell fates, and not only an undifferentiated state. We hypothesized that combinatorial expression of neural lineage-specific transcription factors could directly convert fibroblasts into neurons. Starting from a pool of nineteen candidate genes, we identified a combination of only three factors, Ascl1, Brn2, and Myt1l, that suffice to rapidly and efficiently convert mouse embryonic and postnatal fibroblasts into functional neurons in vitro. These induced neuronal (iN) cells express multiple neuron-specific proteins, generate action potentials, and form functional synapses. Generation of iN cells from non-neural lineages could have important implications for studies of neural development, neurological disease modeling, and regenerative medicine.
The diverse cell types present in the adult organism are produced during development by lineage-specific transcription factors that define and reinforce cell type specific gene expression patterns. Cellular phenotypes are further stabilized by epigenetic modifications that allow faithful transmission of cell-type specific gene expression patterns over the lifetime of an organism1,2. Recent work by Yamanaka and colleagues showing that four transcription factors are sufficient to induce pluripotency in primary fibroblasts demonstrated that fully differentiated cells can be induced to undergo dramatic cell fate changes 3. Similarly, the transfer of somatic cell nuclei into oocytes, as well as cell fusion of pluripotent cells with differentiated cells have proven to be capable of inducing pluripotency 4–9. This remarkable transformation has been interpreted as a reversion of mature into more primitive developmental states, with a concomitant erasure of developmentally relevant epigenetic information 10. Therefore, direct reprogramming between divergent lineages could be unique to reprogramming into an embryonic state, and might not be possible between different somatic cell states. However, cell fusion or forced expression of lineage-specific genes in somatic cells can induce traits of other cell types 11,12. For example, the basic helix-loop-helix (bHLH) transcription factor MyoD can induce muscle-specific properties in fibroblasts but not hepatocytes 13,14; ectopic expression of IL2 and GM-CSF receptors can lead to myeloid conversion in committed lymphoid progenitor cells15; expression of Cebpα in B-cells or Pu.1 and Cebpα in fibroblasts induces characteristics of macrophages 16–18; deletion of Pax5 can induce B-cells to de-differentiate toward a common lymphoid progenitor 19; and the (bHLH) transcription factor neurogenin3, in combination with Pdx1 and MafA, can efficiently convert pancreatic exocrine cells into functional β-cells in vivo 20. Here, we set out to determine whether specific transcription factors could directly convert fibroblasts into functional neurons.
A screen for neuronal fate-inducing factors
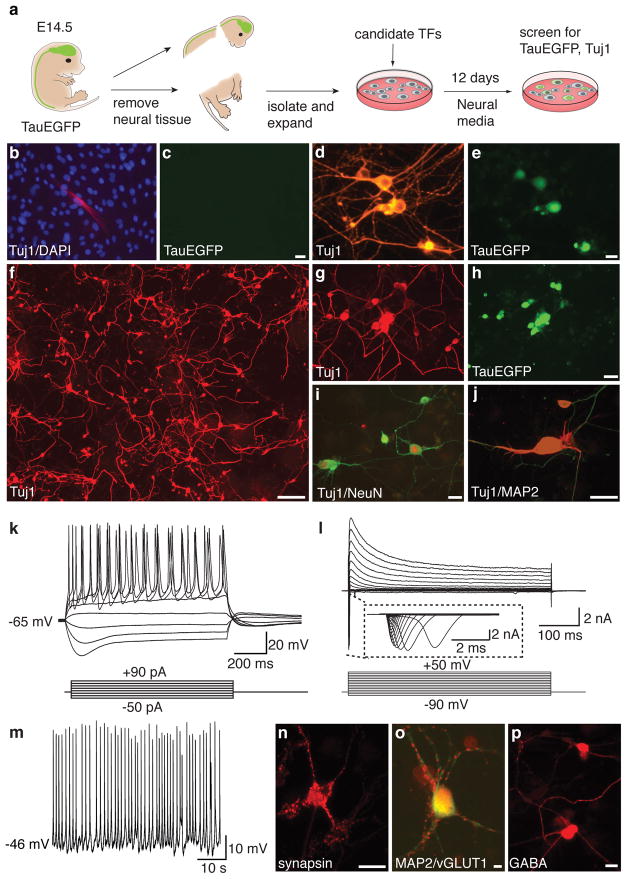
Reasoning that multiple transcription factors would likely be required to reprogram fibroblasts to a neuronal fate, we cloned a total of nineteen genes that are specifically expressed in neural tissues, play important roles in neural development, or have been implicated in epigenetic reprogramming (Supplementary Table 1). A pool of lentiviruses containing all nineteen genes (19F pool) was prepared to infect mouse embryonic fibroblasts (MEFs) from TauEGFP knock-in mice, which express EGFP specifically in neurons 21,22 (see Fig. 1a for experimental outline). Great care was taken to exclude neural tissue for the MEF isolation, and we were unable to detect evidence for the presence of neurons or neural progenitor cells in these cultures using immunofluorescence, fluorescence activated cell sorting (FACS), and RT-PCR analyses (Supplementary Fig. 1). However, uninfected MEFs did contain rare Tuj1-positive, TauEGFP-negative cells with fibroblast-like morphology, indicative of weak Tuj1 (i.e. β-III-tubulin) expression in non-neuronal cells (Fig. 1b,c; Supplementary Figure 1a). In contrast, 32 days after infection with the 19F pool, we detected Tuj1-positive cells with typical neuronal morphologies and bright TauEGFP fluorescence (Fig. 1d,e). Thus, some combination(s) of the genes in the 19F pool was capable of converting MEFs into induced neuronal (iN) cells.
Figure 1. A screen for neuronal fate inducing factors and characterization of MEF-derived iN cells.
a, Experimental rationale. b, Uninfected, p3 TauEGFP MEFs contained rare Tuj1-positive cells (red) with flat morphology. Blue: DAPI counterstain. c, Tuj1-positive fibroblasts do not express visible TauEGFP. d–e, MEF-iN cells express Tuj1 (red) and TauEGFP (green) and display complex neuronal morphologies 32 days after infection with the 19-factor (19F) pool. f, Tuj1 expression in MEFs 13 days after infection with the 5F pool. g–j, MEF-derived Tuj1-positive iN cells co-express the pan-neuronal markers TauEGFP (h), NeuN (red,i) and MAP2 (red,j). k, Representative traces of membrane potential responding to step depolarization by current injection (lower panel). Membrane potential was current-clamped at around −65 mV. l, Representative traces of whole-cell currents in voltage-clamp mode, cell was held at −70 mV, step depolarization from −90 mV to 60 mV at 10 mV interval were delivered (lower panel). Insert showing Na+currents. m, Spontaneous action potentials (AP) recorded from a 5F MEF-iN cell 8 days post infection. No current injection was applied. n–p, 22 days post-infection 5F MEF-iN cells expressed synapsin (red,n) and vesicular glutamate transporter 1 (vGLUT1) (red,o) or GABA (p). Scale bars = 5 μm (o), 10 μm (e,n,p) 20 μm (c,h,i), and 200 μm (f).
We next set out to narrow down the number of transcription factors required for generation of iN cells. Given their important roles in neuronal cell fate determination23–26 we first tested the bHLH transcription factors Ascl1, (also known as Mash1) and Neurod1 individually. Surprisingly, we observed occasional Tuj1-, TauEGFP-positive cells exhibiting a simple mono- or bipolar morphology after infection with only Ascl1 (Supplementary Fig. 2b). However, 19F-iN cells exhibited more complex morphologies, which indicated that the activity of Ascl1 alone was not sufficient to recapitulate the full activity of the 19F pool (compare to Fig. 1d,e). We therefore tested the neuron-inducing activity of Ascl1 in combination with each of the remaining eighteen candidate genes (Supplementary Fig. 2a). Five genes (Brn2, Brn4, Myt1l, Zic1, and Olig2) substantially potentiated the neuron-inducing activity of Ascl1 (Supplementary Fig. 2a–b). Importantly, none of these five genes generated iN cells when tested individually (data not shown). Next, we tested whether combinatorial expression of these factors with Ascl1 could further increase the induction of neuron-like cells by infecting TauEGFP MEFs with a pool of Brn2, Myt1l, Zic1, Olig2, and Ascl1 viruses (5F pool). Given its close similarity to Brn2, we did not include Brn4 in the 5F pool. Twelve days after infection, we detected a frequent Tuj1-positive iN cells with highly complex morphologies (Fig. 1f). These 5F-iN cells also expressed the pan-neuronal markers MAP2, NeuN, and synapsin (Fig. 1i–j, n). Similar results were obtained with iN cells derived from Balb/c MEFs (Supplementary Fig. 3a).
Characterization of 5-factor iN cells
To explore whether iN cells have functional membrane properties similar to neurons, we performed patch-clamp recordings of TauEGFP-positive cells on days 8, 12, and 20 after infection. Action potentials could be elicited by depolarizing the membrane in current clamp mode the majority of the iN cells analyzed (85.1%, n=47) (Fig. 1k–l). Six cells (14.2%, n=42) exhibited spontaneous action potentials, some as early as eight days after transduction (Fig. 1m). These action potentials could be blocked by tetradotoxin (TTX), a specific inhibitor of Na+ ion channels (Supplementary Fig. 3e). Moreover, in voltage-clamp mode we observed both fast, inactivating inward and outward currents, which likely correspond to opening of voltage-dependent K+- and Na+-channels, respectively, with a possible contribution of Ca2+-channels to the whole cell currents (Fig. 1l, Supplementary Fig. 3f). The resting membrane potentials (RMP) ranged between −30 and −69 mV with an average of ~−55 mV on day 20 (n=12, Fig. 3c, Supplementary Table 2,3). Additionally, we asked whether these cells possessed functional ligand-gated ion channels. iN cells responded to exogenous application of GABA, and this response could be blocked by the GABA receptor antagonist picrotoxin (Supplementary Fig. 3g). Thus, MEF-derived iN cells appear to exhibit the functional membrane properties of neurons and possess ligand-gated GABA-receptors.
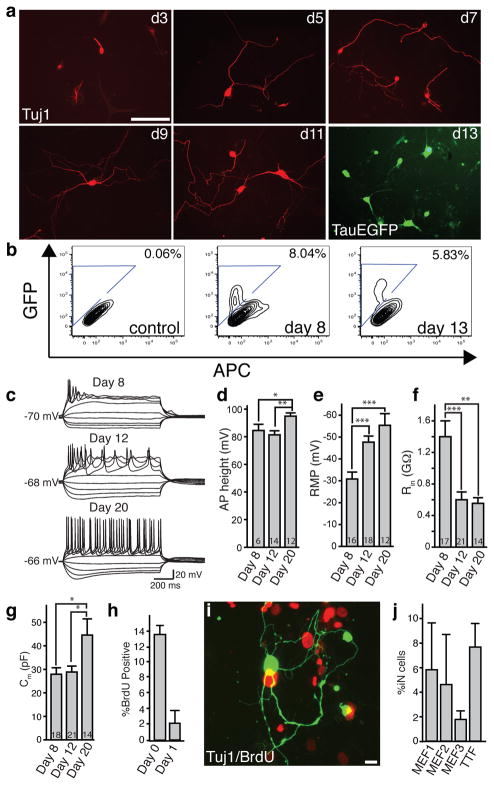
Figure 3. The 5F pool induced conversion is rapid and efficient.
a, Tuj1-positive iN cells (red) exhibit morphological maturation over time after viral infections. At day 13, TauEGFP expression outlines neuronal processes. b, FACS analysis of TauEGFP expression 8 and 13 days post infection. Control = Uninfected TauEGFP MEFs. c, Representative traces showing action potentials elicited from MEF-iN cells at days 8, 12, and 20 post infection. Cells were maintained at a potential of ~ −65 to −70mV. Step current injection protocols were used from −50 to +70 pA. Scale bars apply to all traces. d–g, Quantification of membrane properties in MEF-iN cells at 8, 12, and 20 days post infection. Numbers in the bars represent the numbers of recorded cells. Data are presented as mean±S.E.M. * p<0.05; **p<0.01; *** p<0.001 (Student’s _t_-test). AP: Action Potentials; RMP: Resting Membrane Potentials; Rin: Membrane input resistances; Cm: Membrane Capacitance. AP heights were measured from the baseline. h, BrdU-positive iN cells following BrdU treatment from day 0–13 or day 1–13 after transgene induction. i, Example of a Tuj1 (green) positive cell not labeled with BrdU (red) when added at day 0 after addition of doxycycline. Data are presented as mean ±S.D. j, Efficiency estimates for iN cell generation 13 days after infection (see methods). Every bar represents an independent experiment. Doxycycline was added to 48 hours after plating in MEF experiment #1 and after 24 hours in MEF experiments #2, #3. Error bars = ±1 S.D. of cell counts. Scale Bars = 10 μm (j), 100 μm (a).
We then sought to characterize the neurotransmitter phenotype of iN cells. After 21 days of culture in minimal neuronal media, we detected vGLUT1-positive puncta outlining MAP2-positive neurites of some cells, indicating the presence of excitatory, glutamatergic neurons (Fig. 1o). In addition, we found iN cells labeled with antibodies against GABA, the major inhibitory neurotransmitter in brain (Fig. 1p). Some iN cells (9 out of ~500) contained the Ca2+-binding protein calretinin, a marker for cortical interneurons and other neuronal subtypes (Supplementary Fig. 3c). No expression of tyrosine hydroxylase (TH), choline acetyltransferase (ChAT) or serotonin (5HT) was detected. The vast majority of iN cells were negative for peripherin, an intermediate filament characteristic of peripheral neurons (data not shown)27.
Functional neurons from tail fibroblasts
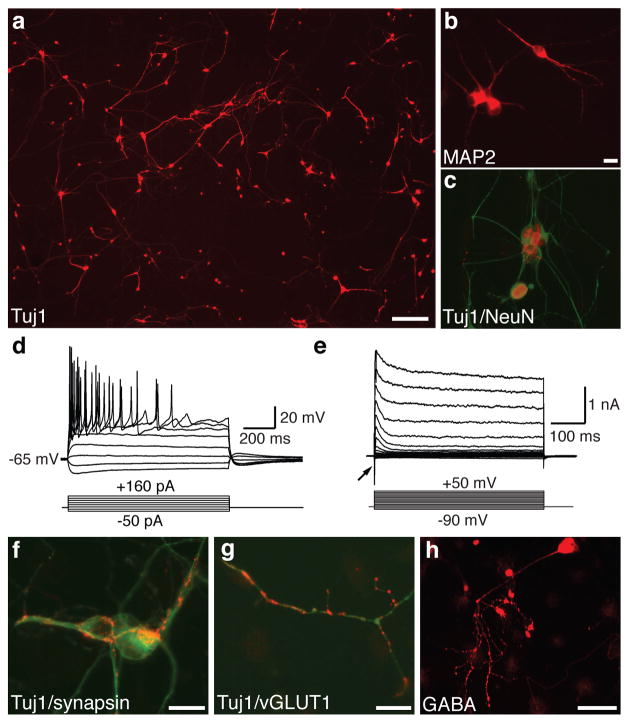
To evaluate whether iN cells could also be derived from postnatal cells, we isolated tail-tip fibroblasts (TTFs) from three-day-old TauEGFP and Rosa26-rtTA mice 28. Similar to our MEF cultures, we could not detect preexisting neurons, glia, or neural progenitor cells (Supplementary Fig. 1a). Twelve days after infecting TTFs with the 5F pool, Tuj1-positive iN cells with a complex, neuronal morphology could be readily detected (Fig. 2a). TTF-iN cells expressed the pan-neuronal markers NeuN, MAP2, and synapsin (Fig. 2b–c, f). Electrophysiological recordings twelve days after infection demonstrated an average RMP of ~ −57 mV (range: −35 to −70mV, n=11), firing of APs (81.8%, n=11) (Fig 2d), and expression of functional voltage-gated membrane channel proteins (Fig. 2e, Supplemental Table 2,3). We were also able to detect vGLUT1- as well as GABA-positive cells (Fig. 2g–h). Despite extensive screening (>1,000 iN cells analyzed), we were unable to detect tyrosine hydroxylase, choline-acetyltransferase, or serotonin expression. Individual iN cells exhibited peripherin-positive filaments (Supplementary Fig. 3d).
Figure 2. Efficient induction of neurons from perinatal tail-tip fibroblasts.
a, Tuj1-stained tail-tip fibroblast 13 days after infection 5F pool. b–c, TTF-iNs express the pan-neuronal markers MAP2 (b) and NeuN (c). d, Representative traces showing action potentials elicited at day 13 post infection. Nine of eleven cells recorded exhibited APs. e, Whole cell currents recorded in voltage-clamp mode. Inward fast inactivating sodium currents (arrow) and outward currents can be observed. f–h, 21 days after infection TTF-iN cells express synapsin (red, f), vGLUT1 (red g) and GABA (h). c, f, and g are overlay images with the indicated marker (red) and Tuj1 (green). Scale bars = 20 μm (b,f,g), 100 μm (h), 200 μm (a).
Neuronal induction is fast and efficient
Next, we assessed the kinetics and efficiency of 5F-induced neuronal conversion. In MEFs, Tuj1-positive cells with immature neuron-like morphology were found as early as three days after infection (Fig. 3a). After five days, neuronal cells with long, branching processes were readily detected, and over time increasingly complex morphologies were evident, suggesting an active process of maturation in newly formed iN cells (Fig. 3a). Similarly, we detected TauEGFP expression as early as day five (Supplementary Fig. 3h). The fraction of TauEGFP-positive cells remained similar at eight and thirteen days post-infection, as determined by FACS analysis suggesting no de-novo generation of iN cells after day 8 (Fig. 3b). Electrophysiological parameters such as action potential height, resting membrane potential, membrane input resistance, and membrane capacitance also showed signs of maturation over time (Fig. 3c–g, Supplementary Tables 2,3).
To estimate the conversion efficiency, we first determined how many of the MEF-derived iN cells divided after induction of the viral transgenes by treating the cells with BrdU throughout the duration of the culture period and beginning one day after gene induction. The results showed that the vast majority of iN cells became postmitotic by 24 hours after transgene activation (Fig. 3h–i). This allowed us to roughly estimate the conversion efficiency of our method by quantifying the total number of Tuj1-positive iN cells in the entire dish on day twelve, and dividing this number by the number of plated cells (see methods). Using this method, the efficiency ranged from 1.8–7.7% in MEF and TTF-iN cells (Fig. 3j) presumably due to slight variations in titers of the viruses. These calculations might be an underestimation of the true conversion rate because not all cells receive the necessary complement of viral transgenes.
iN cells form functional synapses
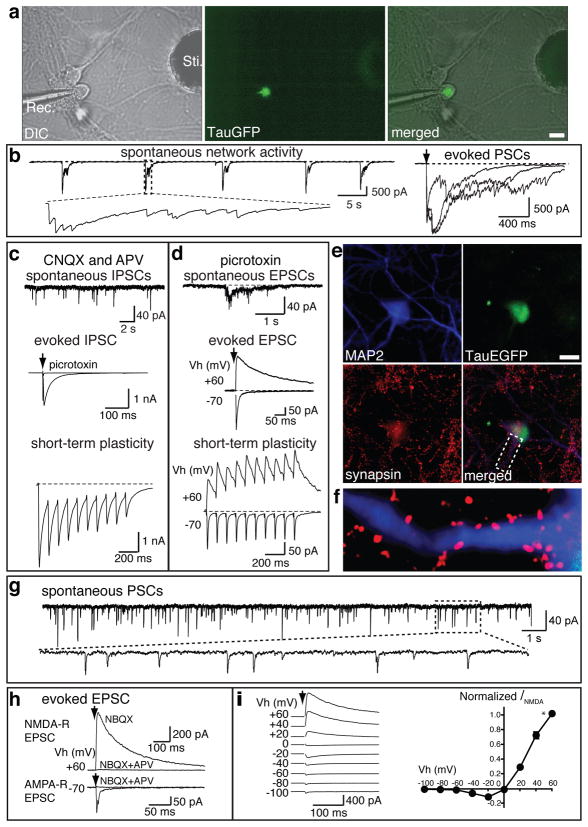
Since iN cells exhibit the membrane properties of neurons, we next wanted to assess whether iN cells have the capacity to form functional synapses. To accomplish this we used two independent methods. First, we determined whether iN cells were capable of synaptically integrating into preexisting neural networks. We employed FACS to purify TauEGFP-positive iN cells seven days after infection and re-plated the 5F-iN cells onto neonatal cortical neurons that had been cultured for seven days in vitro. One week after re-plating, we performed patch-clamp recordings from TauEGFP-positive iN cells and observed spontaneous and rhythmic network activity typical of cortical neurons in culture (Fig. 4a–b). Both excitatory and inhibitory postsynaptic currents (EPSCs and IPSCs) could be evoked following electrical stimulation delivered from a concentric electrode placed 100–150 μm away from the patched iN cells, (Fig. 4b–d). In the presence of the AMPA and NMDA receptor channel blockers CNQX and D-APV, spontaneous IPSCs were reliably detected (Fig. 4c, upper panel). Evoked IPSCs could be blocked by further addition of picrotoxin (Fig. 4c, middle panel). Similarly, at a holding potential of −70 mV and in the presence of picrotoxin, fast-decaying EPSCs mediated by AMPA-receptors could be evoced (Fig. 4d, middle panel). Conversely, at a holding potential of +60 mV (which relieves the voltage-dependent blockade of Mg2+ to NMDA-receptors), slow-decaying NMDA-receptor mediated EPSCs could be recorded (Fig. 4d, middle panel).
Figure 4. MEF-derived iN cells exhibit functional synaptic properties.
TauEGFP-positive iN cells were FACS purified 7–8 days post infection of MEFs and plated on cortical neuronal cultures (7 days in vitro, a–f) or on monolayer glial cultures (g–i). Electrophysiological recordings were performed 7–10 days after sorting. a, Recording electrode (Rec.) patched onto an TauEGFP-positive cell (middle panel) with a stimulation electrode (Sti.). right panel, merged picture of DIC and fluorescence images showing the recorded cell is TauEGFP positive. b, Representative traces of spontaneous synaptic network activities and representative evoked postsynaptic currents (PSCs) following stimulation. c, In the presence of 20 μM CNQX and 50 μM D-APV, upper panel shows a representative trace of spontaneous IPSCs. Evoked IPSC could be elicited (middle panel) and blocked by the addition of picrotoxin. When a train of 10 stimulations was applied at 10 Hz, evoked IPSCs exhibit depression (lower panel). d, In the presence of 30 μM picrotoxin, excitatory synaptic activities from EGFP-positive cells were observed. Spontaneous-(upper panel), and evoked-(middle panel) EPSCs. At a holding potential of −70 mV, AMPA receptor (R) -mediated EPSCs were monitored. When holding potential were set at +60 mV, both AMPA R- and NMDA R-mediated EPSCs could be recorded. Lower panel shows the short-term synaptic plasticity of both AMPA R- and NMDA R- mediated synaptic activities. e, Example of a TauEGFP-positive iN cell expressing MAP2 among cortical neurons. f, High magnification of area marked with dotted lines in e. g, Representative spontaneous postsynaptic currents (PSCs) recorded from MEF-iN cells co-cultured with glia. h, Representative traces of evoked EPSCs. NMDA R-mediated EPSCs in the presence of 10 μM NBQX were recorded at holding potential (Vh) of +60 mV. Application of D-APV blocked the response. AMPA R-mediated EPSCs were recorded at Vh of −70 mV. AMPA R-evoked response is blocked by NBQX and APV. i, Current-voltage (I-V) relationship of NMDA R-mediated EPSCs, left panel; representative traces of evoked EPSCs at different Vh as indicated. Right panel shows the summarized I-V relationship. NMDA-R EPSC amplitudes (_I_NMDA) are normalized to EPSCs at Vh of +60 mV (indicated by *, n=5). NMDA-R EPSCs show ratifications at negative holding potentials, presumably because of the blockade of NMDA-R by Mg2+. Scale bars = 10 μm (a,d).
Moreover, synaptic responses recorded from iN cells showed signs of short-term synaptic plasticity, such as depression of IPSCs and facilitation of EPSCs during a high frequency stimulus train (Fig. 4c–d, lower panels). The presence of synaptic contacts between iN cells and cortical neurons was independently corroborated by the immunocytochemical detection of synapsin-positive puncta surrounding MAP2-positive dendrites originating from EGFP-positive cells (Fig. 4e–f). We were also able to observe synaptic responses in similar experiments performed with iN cells derived from TTFs, (Supplementary Fig. 4). These data demonstrate that iN cells can form functional postsynaptic compartments and receive synaptic inputs from cortical neurons.
Next we asked whether iN cells were capable of forming synapses with each other. To address this question we plated FACS-sorted TauEGFP-positive, MEF-derived 5F-iN cells eight days after infection onto a monolayer culture of primary astrocytes, which are thought to play an essential role in synaptogenesis29,30. Importantly, we confirmed that these cultures were free of preexisting Tuj1 or MAP2-positive neurons (data not shown). Patch clamp recordings at 12–17 days after sorting indicated the presence of spontaneous post synaptic currents in (5/11 cells) (Fig. 4g). Upon extracellular stimulation, evoked EPSCs could be elicited in a majority of the cells (9/11 cells, Fig 4h). Similar to iN cells cultured with primary cortical neurons, we were able to record both NMDA receptor mediated (9/11 cells) and AMPA receptor-mediated EPSCs (8/11 cells; Fig. 4h–i). Interestingly, we were unable to detect obvious IPSCs in a total of fifteen recorded 5F-iN cells. These data indicate that iN cells are capable of forming functional synapses with each other, and that the majority of iN cells exhibit an excitatory phenotype.
Genes sufficient for neuronal conversion
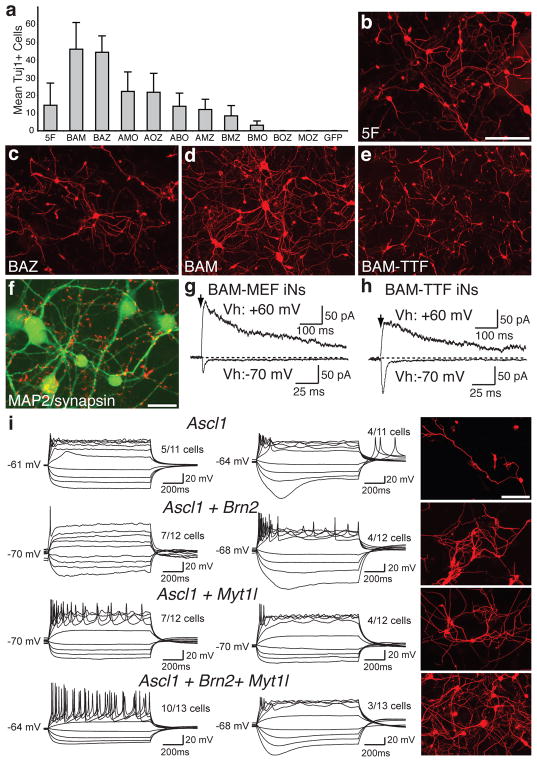
As stated earlier, Ascl1 was the only gene from the 5F pool that was sufficient to induce neuron-like cells in MEFs. We next attempted to determine the relative contribution of each of the five genes by removing each gene from the pool and assessing the efficiency of iN cell generation. Surprisingly, only the omission of Ascl1 had a dramatic effect on induction efficiency (Supplementary Fig. 6a). Thus, we tested the effects of removing two genes at a time by evaluating all possible three gene combinations. Our results indicated that either Ascl1 or both Brn2 and Myt1l must be present in order to generate iN cells (Fig. 5a). The most efficient conversions were achieved when Ascl1 and Brn2 were combined with either Myt1l (BAM pool) or Zic1 (BAZ pool). The efficiencies in these conditions were two to threefold higher than the 5F pool when the total amount of virus was kept constant (Fig. 5a–d). In this experiment the BAM-iN cells appeared to have a more complex morphology than the BAZ cells (Fig. 5c–d, Supplementary Fig. 7). Therefore, we focused our further analysis on the BAM pool.
Figure 5. Defining a minimal pool for efficient induction of functional iN cells.
a, Quantification of Tuj1-positive iN cells from TauEGFP MEFs infected with different 3-factor combinations of the five genes. Each gene is represented by the first letter in its name. Averages from 30 randomly selected visual fields are shown (error bars= ± S.D.) b–d, Representative images of Tuj1 staining of MEFs infected with the 5F (b), Ascl1+Brn2+Zic1 (ABZ) (c) and Ascl1+Brn2+Myt1L (BAM) (d) pools. e, Tuj1 staining of perinatal TTF-iN cells 13 days after infection with the BAM pool. f, BAM-induced MEF-iN cells express MAP2 (green) and synapsin (red) 22 days after infection. g, Representative traces of synaptic responses recorded from MEF-derived BAM (3F)-iN cells co-cultured with glia after isolation by FACs. Vh: holding potential. At Vh of −70 mV, AMPA R-mediated EPSCs were recorded; at Vh of +60 mV, NMDA R-mediated EPSCs were revealed. h, Synaptic responses recorded from TTF-derived 3F-iN cells. Scale bars in (h) apply to traces in (g). i, Representative traces of action potentials elicited from MEF-derived iN cells transduced with the indicated gene combinations, recorded 12 days after infection. Cells were maintained at a resting membrane potential of ~−65 to −70mV. Step current injection protocols were used from −50 to +70 pA. Traces in each subgroup (left or right panels) represent subpopulations of neurons with similar responses. Numbers indicate the fraction of cells from each group that were qualitatively similar to the traces shown. Right panels: representative images of Tuj1 staining after recordings from each condition. Scale bars = 20 μm (f) and 100 μm (b,i).
MEF-derived BAM-iN cells expressed the pan-neuronal markers MAP2 and synapsin (Fig. 5f). The BAM-pool was capable of efficiently generating iN cells from perinatal tail tip fibroblasts (Fig. 5e, Supplementary Fig. 8a–e). After infecting tail tip fibroblasts from adult mice with these three factors, we could detect neuronal cells expressing TauEGFP, Tuj1, NeuN and MAP2 (Supplementary Fig. 9). Importantly, when co-cultured with astrocytes, both MEF and perinatal tail-tip fibroblast-derived BAM-iN cells were capable of forming functional synapses as determined by the presence of both NMDA- and AMPA-receptor mediated EPSCs (Fig. 5g–h). Similar to 5F-iN cells, no IPSCs were detected in MEF-derived (n=16) or tail-derived (n=12) BAM-iN cells. This functional evidence suggests that a majority of BAM-iN cells are excitatory. Indeed, 53% (111/211 cells) of MEF BAM-iN cells expressed Tbr1, a marker of excitatory cortical neurons, whereas less than 1% (3/~500 cells) were GAD –positive (Supplementary Fig. 8f).
Our results left open the possibility that one or two factors might be able to induce functional neuronal properties in MEFs. Thus, we tested smaller subsets of the BAM pool to determine their functionality. In many _Ascl1_-induced cells, current injection elicited action potentials, but their properties appeared to be immature, consistent with their simple neurite morphology (Fig. 5i, Supplementary Fig. 2b). MEFs infected with Ascl1 and Brn2 or Myt1l generated more mature action potentials and displayed more complex neuronal morphologies. In contrast, the majority of BAM-iN cells exhibited repetitive action potentials with more mature characteristics, and displayed the most complex neuronal morphologies. Thus, it appears likely that Ascl1 alone is sufficient to induce some neuronal traits, such as expression of functional voltage-dependent channel proteins that are necessary for the generation of APs, however, co-infection of additional factors is necessary to facilitate neuronal conversion and maturation.
Discussion
Here we show that expression of three transcription factors can rapidly and efficiently convert mouse fibroblasts into functional neurons (iN cells). While the single factor Ascl1 was sufficient to induce immature neuronal features, the additional expression of Brn2 and Myt1l generated mature iN cells with efficiencies of up to 19.5% (Supplementary Fig. 6b). Three factor iN cells displayed functional neuronal properties such as the generation of trains of action potentials and synapse formation. These transcription factors were identified from a total of nineteen candidates that we selected because of their specific expression in neural cell types or their roles in reprogramming to pluripotency (see Methods).
Despite the heterogeneity of embryonic and tail-tip fibroblast cultures, the highly efficient nature of this process effectively rules out the possibility that directed differentiation of rare stem or precursor cells with neurogenic potential can explain our observations. Future studies will have to be performed to unequivocally demonstrate that terminally differentiated cells such as mature B- or T-lymphocytes can be directly converted into neurons using this approach 31,32.
It will now be of great interest to decipher the molecular mechanism of this fibroblast to neuron conversion. We assume that high expression levels of strong neural cell-fate determining transcription factors can activate salient features of the neuronal transcriptional program. Auto-regulatory feed-back and feed-forward activation of downstream transcriptional regulators could then reinforce the expression of important cell fate determining genes and help to further stabilize the induced transcriptional program. Robust changes in transcriptional activity could also lead to genome-wide adjustments of repressive and active epigenetic features such as DNA methylation, histone modifications, and changes of chromatin remodeling complexes that further stabilize the new transcriptional network. 12,33 It is possible that certain subpopulations of cells are “primed” to respond to these factors, depending on their pre-existing transcriptional or epigenetic states34.
The majority of iN cells described in this report are excitatory and express markers of cortical identity. One of the next important steps will be to generate iN cells of other specific neuronal subtypes and from human cells. A low proportion of iN cells expressed markers of GABAergic neurons, but no other neurotransmitter phenotypes were detected. Our data suggest the intriguing possibility that additional combinations of neural transcription factors might also be able to generate functional neurons whose phenotypes remain to be explored.
Future studies will be necessary to determine whether iN cells could represent an alternative to generate patient-specific neurons. The generation of iN cells is fast, efficient, and devoid of tumorigenic pluripotent stem cells, a key complication of iPS cell approaches in regenerative medicine. Therefore, iN cells could provide a novel and powerful system for studying cellular identity and plasticity, neurological disease-modeling, drug discovery, and regenerative medicine.
Methods Summary
Fibroblast isolation, cell culture, and molecular cloning
Homozygous TauEGFP knock-in mice 21 were purchased from the Jackson Laboratories and bred with C57Bl6 mice (Taconic) to generate TauEGFP heterozygous embryos. MEFs were isolated from E14.5 embryos using a dissection microscope (Leica). Tail tips were sliced into small pieces, trypsinized and plated to derive fibroblast cultures. All fibroblasts were expanded for three passages before being used for experiments. Complementary DNAs for candidate genes were cloned into doxycycline-inducible lentiviral vectors, as described previously35. MEFs were infected overnight and cultured in MEF media with doxycycline for 48 hours before being transferred into N3 media with doxycycline.
FACS analysis, cortical cultures, glial cultures
For glial and cortical co-cultures cells were trypsinized eight days after infection and EGFP-positive cells were isolated using a FACS Aria II (Becton Dickinson). EGFP-positive cells were replated on seven days in vitro cortical cultures from wild type p0 mice, or alternatively, passage three primary astrocytes from isolated from p5 pups (as described previously 30,36,37).
Electrophysiology and Immunofluorescence Analysis
Cells were analysed at indicated times after infection. Action potentials (APs) were recorded with current-clamp whole-cell configuration. The pipette solution for current clamp experiments contained (in mM) 123 K-gluconate, 10 KCl, 1 MgCl2, 10 HEPES, 1 EGTA, 0.1 CaCl2, 1 K2ATP, 0.2 Na4GTP, and 4 glucose, pH adjusted to 7.2 with KOH. Membrane potentials were kept around −65 to −70 mV, and step currents were injected to elicit action potentials. Whole-cell currents including sodium currents, potassium currents were recorded at a holding potential of −70 mV, voltage steps ranging from −80 mV to +90 mV were delivered at 10 mV increments. Synaptic responses were measured as described previously36,37. For immunofluorescence experiments, cells were fixed in 4% paraformaldehyde in PBS for 10 minutes. Antibodies were diluted to indicated concentrations (see supplementary methods).
FULL METHODS (for online PDF)
Embryonic fibroblast isolation
Homozygous TauEGFP knock-in mice 21 were purchased from the Jackson Laboratories and bred with C57Bl6 mice (Taconic) to generate TauEGFP heterozygous embryos. Balb/c mice were purchased from Taconic. Rosa26-rtTA mice were obtained from Rudolf Jaenisch28. MEFs were isolated from E14.5 embryos under a dissection microscope (Leica). The head, vertebral column (containing the spinal cord), dorsal root ganglia, and all internal organs were removed and discarded to ensure the removal of all cells with neurogenic potential from the cultures. The remaining tissue was manually dissociated and incubated in 0.25% Trypsin (Sigma) for 10–15 minutes to create a single cell suspension. The cells from each embryo were plated onto a 15cm tissue culture dish in MEF media (Dulbecco’s Modified Eagle Medium (Invitrogen) containing 10% Fetal Bovine Serum (FBS) (Hyclone), beta-mercaptoethanol (Sigma-Aldrich), non-essential amino acids, sodium pyruvate, and penicillin/streptomycin (all from Invitrogen). Cells were grown at 37oC for 4–7 days until confluent, and then split once before being frozen. After thawing, cells were cultured on 15cm plates and allowed to become confluent before being split onto plates for infections using 0.25% Trypsin. Postnatal tail tip fibroblasts were prepared by removing the bottom third of tail from 3-day-old pups using surgical scissors. Cells were rinsed in ethanol, washed with HBSS (Sigma), and then dissociated using scissors and 0.25% Trypsin. Tail tip fibroblasts were cultured in MEF media until confluent and passaged once before being pooled together and frozen down for further use.
Cell culture, molecular cloning, and viral infections
We had three criteria for identifying candidates with neuron-inducing activity: (i) we reasoned that cell-fate inducing factors should be enriched in the gene category of transcriptional regulators. (ii) We included factors previously involved in reprogramming to pluripotency (Klf4, c-Myc, and Sox2). (iii) We searched for genes specifically expressed in neural tissues. Those) were selected based on published expression arrays of MEFs, ES cells and neural progenitor cells retrieved from the Gene Expression Omnibus database (GSE8024, http://www.ncbi.nlm.nih.gov/gds) and the EST Profile function of NCBI’s Unigene database (http://www.ncbi.nlm.nih.gov/unigene). cDNAs for the factors included in the nineteen factor pool were cloned into lentiviral constructs under the control of the tetracycline operator 35. Replication-incompetent, VSVg-coated lentiviral particles were packaged in 293T cells as described35. Passage three TauEGFP and Balb/c MEFs were infected in MEF media containing polybrene (8 μg/mL). After 16–20 hours in media containing lentivirus, the cells were switched into fresh MEF media containing doxycycline (2 μg/mL) to activate expression of the transduced genes. After 48 hours in MEF media with doxycycline (Sigma), the media was replaced with N3 media 22 containing doxycycline. The media was changed every 2–3 days for the duration of the culture period. For BrdU experiments, 10μM BrdU was added to the culture media and was maintained throughout media changes until the cells were fixed.
Immunofluorescence, RT-PCR, and flow cytometry
Neuronal cells were defined as cells, which stained positive for Tuj1and had a process at least 3 times longer than the cell body. For immunofluorescence staining, cells were washed with PBS and then fixed with 4% paraformaldehyde for 10 minutes at room temperature (RT). Cells were then incubated in 0.2% Triton X-100 (Sigma) in PBS for 5 minutes at RT. After washing twice with PBS, cells were blocked in a solution of PBS containing 4% BSA, 1% FBS for 30 minutes at RT. Primary and secondary antibodies were diluted in a solution of PBS containing 4% BSA and 1% FBS. Fields of cells for staining were outlined with a PAP pen (DAKO). Primary and secondary antibodies were typically applied for 1 hour and 30 minutes, respectively. Cells were washed three times with PBS between primary and secondary staining. For anti-BrdU staining, cells were treated with 2N HCl in PBS for 10 minutes and washed twice with PBS before permeablization with TritonX-100 (Sigma). The following antibodies were used for our analysis: goat anti-ChAT (Millipore, 1:100), rabbit anti-GABA (Sigma, 1:4000), rabbit-GFAP (DAKO, 1:4000), mouse anti-MAP2 (Sigma, 1:500), mouse anti-NeuN (Millipore, 1:100), mouse anti-Peripherin (Sigma, 1:100), mouse anti-Sox2 (R&D Systems, 1:50), rabbit anti-Serotonin (Biogenesis, 1:1000), rabbit anti-Tuj1 (Covance, 1:1000), mouse anti-Tuj1 (Covance, 1:1000), goat anti-Brn2 (Santa Cruz Biotechnology, 1:100), mouse anti-BrdU (Becton-Dickinson, 1:3.5), mouse anti-Calretinin (DAKO, 1:100), sheep anti-Tyrosine Hydroxylase (Pel-Freez, 1:1000), E028 rabbit anti-synapsin (gift from Thomas Südhof, 1:500), guinea pig anti-vGLUT1 (Millipore, 1:2000), mouse anti-GAD6 (Developmental Studies Hybridoma Bank (DSHB), 1:500), mouse anti-Pax3 (DSHB, 1:250), mouse anti-Pax6 (DSHB, 1:50), mouse anti-Pax7 (DSHB, 1:250), mouse anti-Nkx2.2 (DSHB, 1:100), mouse anti-Olig1 (NeuroMab, 1:100). Fitc-, and Cy3-conjugated secondary antibodies were obtained from Jackson Immunoresearch. Alexa-488, Alexa-546 and Alexa-633-conjugated secondary antibodies were obtained from Invitrogen. TauEGFP expressing cells were analyzed and sorted on a FACS Aria II (Becton Dickinson). Flow cytometry data was analyzed using Flowjo (Tree Star). After sorting, cells were plated on cortical cultures or glial cultures derived from neonatal brains. Cells were kept in 50% N3 media and 50% growth media (see media composition below) and 2 μg/ml doxycycline for one week before being switched to growth media without doxycycline until electrophysiological analysis was completed. For RT-PCR analysis, RNA was isolated using Trizol (Invitrogen) following the manufacturer’s instructions, treated with DNAse (NEB) and 1.5 μg was reverse-transcribed with Superscript II (Invitrogen). PCR was performed using the following primers Sox1 (F- TCGAGCCCTTCTCACTTGTT, R-TTGATGCATTTTGGGGGTAT), Sox10 (F- GAACTGGGCAAGGTCAAGAA, R-CGCTTGTCACTTTCGTTCAG), β-Actin (F-CGTGGGCCGCCCTAGGCACCA, R-CTTAGGGTTCAGGGGGGC). PCR products were analyzed on a 1% gel.
Efficiency Calculation
The following method was used to calculate the efficiency of neuronal induction. The total number of Tuj1+ cells with a neuronal morphology, defined as cells having a circular, three dimensional appearance that extend a thin process at least three times longer than their cell body, were quantified twelve days after infection. This estimate was based on the average number of iN cells present in 30 randomly selected 20× visual fields. The area of a 20× visual field was then measured, and we used this estimated density of iN cells to determine the total number of neurons present in the entire dish. We then divided this number by the number of cells plated before infection to get a percentage of the starting population of cells that adopted neuron-like characteristics.
Cortical cultures
Primary cortical neurons were isolated from newborn wildtype mice as described 36 with modifications. Briefly, cortices were dissociated by papain (10 U/ml, with 1 μM Ca2+, and 0.5 μM EGTA) digestions and plated on Matrigel coated circle glass coverslips (Ø 11 mm). The neurons were cultured in vitro in growth media consisting of: MEM (Invitrogen) supplemented with B27 (Invitrogen), glucose, transferrin, FBS and Ara-C (Sigma).
Glial cell isolation
Forebrains were dissected from postnatal day five wild-type mice and were manually dissociated into ~.5 mm2 pieces in a total of 2 mL of HBSS. 500 μL of 2.5% Trypsin and 1% DNase were added and dissociated tissue was incubated at 37° for 15 minutes. Solution was mixed every 5 minutes. The supernatant was then transferred into 1.5 mL of FBS. 4 mL of HBSS, 500 μL 2.5% Trypsin, and 500 μL DNase were again added to the remaining dissociated tissue and incubated at 37° for 15 minutes, mixing every 5 minutes. The supernatant was again removed and added to the FBS-containing solution. Using a pipette, the remaining tissue was further dissociated and passed through a 70 μM nylon mesh filter (BD Biosciences) into the FBS-containing solution. The cell mixture was then spun at 1000 rpm for 5 minutes and resuspended in MEF media. Glial cells were passaged three times before culturing with MEF or TTF-derived iN cells. Contaminating neurons in p3 glial cell cultures could not be detected when stained for either Tuj1 or MAP2.
Electrophysiology
Recordings were performed from MEF- and tail cell-derived iN cells at 8, 12 and 20 days after viral infection, or 7–13 days after co-culturing with cortical neurons. Spontaneous or evoked synaptic responses were recorded in the whole-cell voltage-clamp mode. Evoked synaptic responses were triggered by 1 ms current injection through a local extracellular electrode (FHC concentric bipolar electrode, Catalogue No. CBAEC75) with a Model 2100 Isolated Pulse Stimulator (A-M Systems, Inc.), and recorded in whole-cell mode using a Multiclamp 700B amplifier (Molecular Devices, Inc.) 37. Data were digitized at 10 kHz with a 2 kHz low-pass filter. The whole-cell pipette solution for synaptic current recordings contained (in mM): CsCl 135, HEPES 10, EGTA 1, Mg-ATP 4, Na4GTP 0.4, and QX-314 10, pH 7.4. The bath solution contained (in mM): NaCl 140, KCl 5, CaCl2 2, MgCl2 0.8, HEPES 10, and glucose 10, pH 7.4. IPSCs were pharmacologically isolated by addition of 50 μM D-AP5 and 20 μM CNQX to the bath solution. EPSCs were pharmacologically isolated by addition of 30 μM picrotoxin and 50 μM D-APV. Data were analyzed using Clampfit 10.02 (Axon Instruments, Inc). Action potentials (APs) were recorded with current-clamp whole-cell configuration. The pipette solution for current clamp experiments contained (in mM) 123 K-gluconate, 10 KCl, 1 MgCl2, 10 HEPES, 1 EGTA, 0.1 CaCl2, 1 K2ATP, 0.2 Na4GTP, and 4 glucose, pH adjusted to 7.2 with KOH. Membrane potentials were kept around −65 to −70 mV, and step currents were injected to elicit action potential. Whole-cell currents including sodium currents, potassium currents were recorded at a holding potential of −70 mV, voltage steps ranging from −80 mV to +90 mV were delivered at 10 mV increments.
Supplementary Material
1
2
Acknowledgments
We would like to thank Samuele Marro and Patty Lovelace for help with FACS sorting, Sana Hafeez and Yul Huh for assistance with molecular cloning and mouse husbandry, and Karen Jann for assistance with the diagram in Figure 1. We would also like to thank Isabella Graef, Ruchi Bajpai, Joanna Wysocka, Jia-Ren Lin, and Jia-Yun Chen for contributing reagents and help with analysis. This work was supported by start-up funds from the Institute for Stem Cell Biology and Regenerative Medicine at Stanford (M.W.), the Donald E. and Delia B. Baxter Foundation (M.W.), an award from William Stinehart, Jr. and the Reed Foundation (M.W.), the National Institute of Health Training Grant 1018438-142-PABCA (A.O) and the Ruth and Robert Halperin Stanford Graduate Fellowship (T.V.). Z.P. Pang is partly supported by NARSAD Young Investigator Award and NIH/NINDS Epilepsy Training Grant 5T32NS007280.
Footnotes
Author contributions
T.V., A.O., and M.W. designed and conceived the experiments. T.V., Y.K., and M.W. produced the lentiviral vectors. T.V. and A.O. performed the lentiviral infections, isolated the fibroblasts, and completed the molecular characterization of the iN cells. Z.P. and T.S. designed, performed, and analysed the electrophysiological assays. T.V., A.O., Z.P., T.S. and M.W. wrote and edited the manuscript and produced the figures.
Author Information
The authors declare no competing financial interests.
References
- 1.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–81. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proc Natl Acad Sci U S A. 1952;38:455–63. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–5. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- 6.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380:64–6. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 7.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–8. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 8.Do JT, Scholer HR. Nuclei of embryonic stem cells reprogram somatic cells. Stem Cells. 2004;22:941–9. doi: 10.1634/stemcells.22-6-941. [DOI] [PubMed] [Google Scholar]
- 9.Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–73. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- 10.Silva J, Smith A. Capturing pluripotency. Cell. 2008;132:532–6. doi: 10.1016/j.cell.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blau HM. How fixed is the differentiated state? Lessons from heterokaryons. Trends Genet. 1989;5:268–72. doi: 10.1016/0168-9525(89)90100-5. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Q, Melton DA. Extreme makeover: converting one cell into another. Cell Stem Cell. 2008;3:382–8. doi: 10.1016/j.stem.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 13.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 14.Schafer BW, Blakely BT, Darlington GJ, Blau HM. Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature. 1990;344:454–8. doi: 10.1038/344454a0. [DOI] [PubMed] [Google Scholar]
- 15.Kondo M, et al. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–6. doi: 10.1038/35030112. [DOI] [PubMed] [Google Scholar]
- 16.Bussmann LH, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–66. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 17.Feng R, et al. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–62. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–76. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 19.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–7. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–32. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tucker KL, Meyer M, Barde YA. Neurotrophins are required for nerve growth during development. Nat Neurosci. 2001;4:29–37. doi: 10.1038/82868. [DOI] [PubMed] [Google Scholar]
- 22.Wernig M, et al. Tau EGFP embryonic stem cells: an efficient tool for neuronal lineage selection and transplantation. J Neurosci Res. 2002;69:918–24. doi: 10.1002/jnr.10395. [DOI] [PubMed] [Google Scholar]
- 23.Lee JE, et al. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–44. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 24.Guillemot F, et al. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–76. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- 25.Farah MH, et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 26.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17:639–47. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Escurat M, Djabali K, Gumpel M, Gros F, Portier MM. Differential expression of two neuronal intermediate-filament proteins, peripherin and the low-molecular-mass neurofilament protein (NF-L), during the development of the rat. J Neurosci. 1990;10:764–84. doi: 10.1523/JNEUROSCI.10-03-00764.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–8. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 29.Christopherson KS, et al. Thrombospondins are astrocyte-secreted proteins that promote CNS synaptogenesis. Cell. 2005;120:421–33. doi: 10.1016/j.cell.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 30.Wu H, et al. Integrative genomic and functional analyses reveal neuronal subtype differentiation bias in human embryonic stem cell lines. Proc Natl Acad Sci U S A. 2007;104:13821–6. doi: 10.1073/pnas.0706199104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–8. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- 32.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–64. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–82. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature. 2009;460:49–52. doi: 10.1038/nature08180. [DOI] [PubMed] [Google Scholar]
- 35.Wernig M, et al. A drug-inducible transgenic system for direct reprogramming of multiple somatic cell types. Nat Biotechnol. 2008;26:916–24. doi: 10.1038/nbt1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maximov A, Pang ZP, Tervo DG, Sudhof TC. Monitoring synaptic transmission in primary neuronal cultures using local extracellular stimulation. J Neurosci Methods. 2007;161:75–87. doi: 10.1016/j.jneumeth.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 37.Maximov A, Sudhof TC. Autonomous function of synaptotagmin 1 in triggering synchronous release independent of asynchronous release. Neuron. 2005;48:547–54. doi: 10.1016/j.neuron.2005.09.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2