Targeted Capture and Massively Parallel Sequencing of Twelve Human Exomes (original) (raw)
. Author manuscript; available in PMC: 2010 Mar 23.
Published in final edited form as: Nature. 2009 Aug 16;461(7261):272–276. doi: 10.1038/nature08250
Abstract
Genome-wide association studies suggest that common genetic variants explain only a small fraction of heritable risk for common diseases, raising the question of whether rare variants account for a significant fraction of unexplained heritability1,2. While DNA sequencing costs have fallen dramatically3, they remain far from what is necessary for rare and novel variants to be routinely identified at a genome-wide scale in large cohorts. We have therefore sought to develop second-generation methods for targeted sequencing of all protein-coding regions (`exomes'), to reduce costs while enriching for discovery of highly penetrant variants. Here we report on the targeted capture and massively parallel sequencing of the exomes of twelve humans. These include eight HapMap individuals representing three populations4, and four unrelated individuals with a rare dominantly inherited disorder, Freeman-Sheldon syndrome (FSS)5. We demonstrate the sensitive and specific identification of rare and common variants in over 300 megabases (Mb) of coding sequence. Using FSS as a proof-of-concept, we show that candidate genes for monogenic disorders can be identified by exome sequencing of a small number of unrelated, affected individuals. This strategy may be extendable to diseases with more complex genetics through larger sample sizes and appropriate weighting of nonsynonymous variants by predicted functional impact.
Protein coding regions constitute ~1% of the human genome or ~30 Mb, split across ~180,000 exons. A brute-force approach to exome sequencing with conventional technology6 is expensive relative to what may be possible with second-generation platforms3. However, the efficient isolation of this fragmentary genomic subset is technically challenging7. Hodges et al. (2007) described enrichment of an exome by hybridization of shotgun libraries constructed from 140 micrograms of genomic DNA to seven microarrays8. To improve the practicality of hybridization capture, we developed a protocol to enrich for coding sequences at a genome-wide scale starting with 10 micrograms of DNA and using two microarrays. Our initial target was 27.9 Mb of coding sequence defined by CCDS (the NCBI Consensus CDS database)9. This curated set avoids the inclusion of spurious hypothetical genes that contaminate broader exome definitions10. The target is reduced to 26.6 Mb upon exclusion of regions that are poorly mapped to with our anticipated read length due to paralogous sequences elsewhere in the genome (Supplementary Data 1).
We captured and sequenced the exomes of eight individuals previously characterized by the HapMap4 and Human Genome Structural Variation11 projects. We also analyzed four unrelated individuals affected with Freeman-Sheldon syndrome (FSS; OMIM #193700) or distal arthrogryposis type 2A, a rare autosomal dominant disorder caused by mutations in MYH35. Unpaired, 76 base-pair (bp) reads12 from post-enrichment shotgun libraries were aligned to the reference genome13. On average, 6.4 gigabases (Gb) of mappable sequence was generated per individual (20-fold less than whole genome sequencing with the same platform12), and 49% of reads mapped to targets (Supplementary Table 1). After removing duplicate reads that represent potential PCR artifacts14, the average fold-coverage of each exome was 51× (Supplementary Fig. 1). On average per exome, 99.7% of targeted bases were covered at least once, and 96.3% (25.6 Mb) were covered sufficiently for variant calling (>=8× coverage and _phred_-like15 consensus quality>=30). This corresponded to 78% of genes having >95% of their coding bases called (Supplementary Fig. 2, Supplementary Data 2). The average pairwise correlation coefficient between individuals for gene-by-gene coverage was 0.87, consistent with systematic bias in coverage between individual exomes.
False positives and false negatives are critical issues in genomic resequencing. We assessed the quality of our exome data in four ways. First, comparing sequence-based calls for the eight HapMap exomes to array-based genotyping, we observed a high concordance with both homozygous (99.94%; n = 219,077) and heterozygous (99.57%; n = 43,070) genotypes (Table 1). Second, we compared our coding single-nucleotide polymorphism (cSNP) catalogue to ~1 megabase of coding sequence determined in each of the eight HapMap individuals by molecular inversion probe (MIP) capture and direct resequencing16. At coordinates called in both datasets, 99.9% of all cSNPs (n = 4,620) and 100% of novel cSNPs (n = 334) identified here were concordant, consistent with a low false discovery rate (FDR). Third, we compared the NA18507 cSNPs identified here to those called by recent whole genome sequencing of this individual12, and found substantial overlap (Supplementary Fig. 3). The relative numbers of cSNPs called by only one approach, and the proportions of these represented in dbSNP, indicate that exome sequencing has equivalent sensitivity for cSNP detection as compared to whole genome sequencing. Fourth, we compared our data to cSNPs in high quality Sanger sequence of single haplotype regions from fosmid clones of the same HapMap individuals17. 38 of 40 fosmid-defined cSNPs were at coordinates with sufficient coverage in our data for variant calling. Of these, 38 of 38 were correctly identified as variant.
Table 1. Sequence coverage and array-based validation.
The number of coding bases covered at least 1× and with sufficient coverage to variant call (>=8× and consensus quality >=30) are listed for each exome, with the fraction of the aggregate target (26.6 Mb) that this represents in parentheses. For the eight HapMap individuals, concordance with array genotyping (Illumina Human1 M-Duo) is listed for positions that are homozygous for the reference allele, heterozygous, or homozygous for the non-reference allele (according to the array genotype). YRI = Yoruba HapMap; CHB = Chinese HapMap; JPT = Japanese HapMap; CEU = CEPH HapMap; Eur = European-American ancestry (non-HapMap).
| Concordance with Illumina Human1 M-Duo calls | |||||
|---|---|---|---|---|---|
| Individual | Covered >=1× | Sequence called | Homozygous reference | Heterozygous | Homozygous non-reference |
| NA18507 (YRI) | 26,477,161 (99.7%) | 25,795,189 (97.1%) | 23757/23762 (99.98%) | 5553/5583 (99.46%) | 3582/3592 (99.72%) |
| NA18517 (YRI) | 26,476,761 (99.7%) | 25,748,289 (97.0%) | 23701/23705 (99.98%) | 5575/5601 (99.54%) | 3568/3579 (99.69%) |
| NA19129 (YRI) | 26,491,035 (99.8%) | 25,733,587 (96.9%) | 23701/23708 (99.97%) | 5482/5510 (99.49%) | 3681/3690 (99.76%) |
| NA19240 (YRI) | 26,486,481 (99.7%) | 25,576,517 (96.3%) | 23546/23551 (99.98%) | 5600/5634 (99.40%) | 3542/3549 (99.80%) |
| NA18555 (CHB) | 26,475,665 (99.7%) | 25,529,861 (96.1%) | 23980/23984 (99.98%) | 4877/4893 (99.67%) | 3776/3786 (99.74%) |
| NA18956 (JPT) | 26,454,942 (99.6%) | 25,683,248 (96.7%) | 24217/24221 (99.98%) | 4890/4910 (99.59%) | 3751/3760 (99.76%) |
| NA12156 (CEU) | 26,476,155 (99.7%) | 25,360,704 (95.5%) | 23789/23794 (99.98%) | 5493/5514 (99.62%) | 3206/3213 (99.78%) |
| NA12878 (CEU) | 26,439,953 (99.6%) | 25,399,572 (95.6%) | 23885/23891 (99.97%) | 5413/5425 (99.78%) | 3274/3292 (99.45%) |
| FSS10066 (Eur) | 26,467,140 (99.7%) | 25,546,738 (96.2%) | n.a. | n.a. | n.a. |
| FSS10208 (Eur) | 26,461,768 (99.6%) | 25,576,256 (96.3%) | n.a. | n.a. | n.a. |
| FSS22194 (Eur) | 26,426,401 (99.5%) | 25,454,551 (95.9%) | n.a. | n.a. | n.a. |
| FSS24895 (Eur) | 26,478,775 (99.7%) | 25,602,677 (96.4%) | n.a. | n.a. | n.a. |
A comparison of our data to past reports on exonic18 or exomic8 array-based capture revealed roughly equivalent capture specificity, but greater completeness in terms of coverage and variant calling (Supplementary Table 2). These improvements likely arise from a combination of greater sequencing depth, differences in array designs and in experimental conditions for capture. Within the set of called positions, the high concordance with heterozygous array-based genotypes (>99%) provides an estimate of our sensitivity for rare variant detection, as rare variants are overwhelmingly expected to be heterozygous. However, sensitivity was limited in that ~4% of known heterozygous genotypes were at coordinates where there was insufficient coverage to make a confident call.
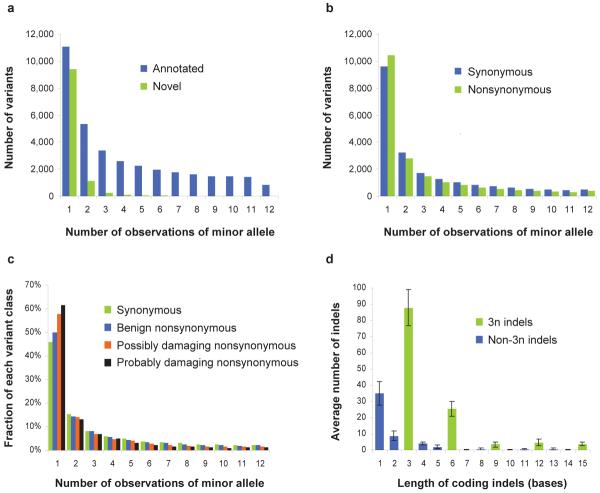
56,240 cSNPs were called in one or more individuals, of which 13,347 were novel. On average, 17,272 cSNPs were called per individual, of which 92% were already annotated in a public database (dbSNP v129) (Table 2a). The proportion of previously annotated cSNPs was consistent by population, and higher for European (94%; n = 6) and Asian (93%; n = 2) than Yoruba (88%; n = 4) ancestry. These confirmation rates are ~10% higher than recent whole genome analyses12,19–22. The most likely explanation is that coding sequences have historically been more heavily ascertained than non-coding sequences, although other factors such as dbSNP version, prior ascertainment of HapMap individuals, and different FDRs may contribute as well. For the subset of cSNPs at coordinates with sufficient coverage for variant calling in all 12 individuals (n = 47,079), 32% of annotated variants and 86% of novel variants were singleton observations across 24 chromosomes (Fig. 1a).
Table 2. Coding variation across 12 human exomes.
(a) cSNPs called in each individual, relative to the reference genome, are broken down by the fraction in dbSNP and by genotype; (b) Extrapolation of observed numbers of cSNPs in each individual to an exactly 30 Mb exome YRI = Yoruba HapMap; CHB = Chinese HapMap; JPT = Japanese HapMap; CEU = CEPH HapMap; Eur = European-American ancestry (non-HapMap).
| (a) | |||||
|---|---|---|---|---|---|
| Individual | cSNP calls | # in dbSNP | % in dbSNP | # heterozygous | # homozygous |
| NA18507 (YRI) | 19720 | 17577 | 89.1% | 12896 | 6824 |
| NA18517 (YRI) | 19737 | 17326 | 87.8% | 13039 | 6698 |
| NA19129 (YRI) | 19761 | 17298 | 87.5% | 12845 | 6916 |
| NA19240 (YRI) | 19517 | 17168 | 88.0% | 12866 | 6651 |
| NA18555 (CHB) | 16047 | 14894 | 92.8% | 9181 | 6866 |
| NA18956 (JPT) | 16011 | 14848 | 92.7% | 9132 | 6879 |
| NA12156 (CEU) | 16119 | 15250 | 94.6% | 10179 | 5940 |
| NA12878 (CEU) | 15970 | 15051 | 94.2% | 9928 | 6042 |
| FSS10066 (Eur) | 16229 | 15144 | 93.3% | 10240 | 5989 |
| FSS10208 (Eur) | 16073 | 15018 | 93.4% | 9966 | 6107 |
| FSS22194 (Eur) | 16094 | 15128 | 94.0% | 10005 | 6089 |
| FSS24895 (Eur) | 15986 | 15027 | 94.0% | 9920 | 6066 |
| (b) | |||||
|---|---|---|---|---|---|
| Individual | est. total cSNPs | est. total heterozygous | est. total homozygous | est. total synonymous | est. total nonsynonymous |
| NA18507 (YRI) | 22727 | 14876 | 7851 | 12466 | 10261 |
| NA18517 (YRI) | 22841 | 15135 | 7706 | 12550 | 10291 |
| NA19129 (YRI) | 22907 | 14906 | 8001 | 12693 | 10214 |
| NA19240 (YRI) | 22814 | 15063 | 7751 | 12565 | 10249 |
| NA18555 (CHB) | 18722 | 10677 | 8045 | 10275 | 8447 |
| NA18956 (JPT) | 18523 | 10585 | 7938 | 10072 | 8451 |
| NA12156 (CEU) | 18825 | 11818 | 7007 | 10220 | 8605 |
| NA12878 (CEU) | 18544 | 11455 | 7089 | 10110 | 8434 |
| FSS10066 (Eur) | 18836 | 11795 | 7041 | 10240 | 8596 |
| FSS10208 (Eur) | 18591 | 11444 | 7147 | 10075 | 8516 |
| FSS22194 (Eur) | 18667 | 11539 | 7128 | 10144 | 8523 |
| FSS24895 (Eur) | 18508 | 11466 | 7042 | 10169 | 8339 |
Figure 1. Minor allele frequency and coding indel length distributions.
(a) The distribution of minor allele frequencies is shown for previously annotated versus novel cSNPs. (b) The distribution of minor allele frequencies is shown for synonymous versus nonsynonymous cSNPs. (c) The distribution of minor allele frequencies (by proportion, rather than count) is shown for synonymous cSNPs (n = 21,201) versus nonsynonymous cSNPs predicted to be benign (n = 13,295), possibly damaging (n = 3,368), or probably damaging (n = 2,227) by PolyPhen24. (d) The distribution of lengths of coding insertion-deletion variants is shown (average numbers per exome). Error bars indicate s.d.
We also estimated the total number of cSNPs in each individual relative to the reference genome (Table 2b). As the precise and comprehensive definition of the human exome remains incomplete, we extrapolated our data to an estimated exome size of exactly 30 Mb. The results were remarkably consistent by population. As expected, a higher number of nonsynonymous cSNPs were estimated for Yoruba (avg. 10,254; n = 4) than non-Africans (avg. 8,489; n = 8). More heterozygous cSNPs were estimated for four Yoruba (avg. 14,995) than six European Americans (avg. 11,586) and two Asians (avg. 10,631). The ratio of synonymous to nonsynonymous cSNPs was 1.2 within any single individual, and 1.1 when calculated for a non-redundant list of variants identified across all individuals. The difference results from the slightly shifted allele frequency distribution of nonsynonymous variants (Fig. 1b). Consistent with expectation23, the trend is more pronounced for nonsynonymous variants predicted to be damaging (by PolyPhen24) (Fig. 1c).
Nonsense mutations (NMs) and splice-site disruptions (SSDs) are often assumed to be deleterious, but have a broad range of potential fitness effects25–27. Our non-redundant cSNP catalogue included 225 NMs (112 novel) and 102 SSDs (49 novel). Excluding 86 nonsense alleles that are common in this dataset (2+ observations) or in a recent study by Yngvadottir et al.25 (>5% allele frequency), our genome-wide estimate (projected to 30 Mb) for the average number of relatively rare mutations introducing premature nonsense codons in an individual genome was 10 for non-Africans (n = 8) and 20 for Yoruba (n = 4). However, these are likely overestimates, given that our catalogue of common nonsense mutations remains incomplete.
Short insertion-deletions (indels) in coding sequence are likely to be functionally important when they cause frameshifts but are difficult to detect with short reads. We developed and applied an approach for identifying indels from our unpaired 76 bp reads. In total, 664 coding indels were called in 1+ individuals. On average, 166 coding indels were called per individual, of which 63% were previously annotated in dbSNP (Supplementary Table 3). To assess our sensitivity, we compared our data for NA18507 to Bentley et al.12. 73% of their coding indels were also observed in our data (136 of 187). To assess specificity, we attempted PCR and Sanger sequencing of 28 novel coding indels chosen at random. Of 21 successful assays, 20 coding indels were verified, and 1 was a false positive. We anticipate that future use of paired-end reads will improve detection of coding indels.
The shape of the distribution of coding indel lengths was consistent with other studies10,20 as well as across the 12 exomes (Fig. 1d), demonstrating a preference for multiples of 3 (“3n”). Of the 664 coding indels observed here, 65% were 3n in length. The allele frequency distribution for novel indels relative to annotated indels was markedly shifted towards rarer variants (Supplementary Fig. 4). However, the length histogram for novel versus annotated coding indels were similar (Supplementary Fig. 5), reinforcing that our set of novel coding indels is not excessively contaminated with false positives (as these would not be expected to have the observed 3n bias). Excluding indels that were common in this dataset (2+ observations), the average number of relatively rare frameshifting indels identified per individual was 8 for non-Africans (n = 8) and 17 for Yoruba (n = 4).
The number of synonymous, missense, nonsense, splice site, frameshifting indel, and non-frameshifting indel variants observed in each individual (as well as the size of the subsets that are novel and singleton observations) are presented in Supplementary Table 4. Also shown are the average numbers of variants of each class for non-Africans and Yoruba.
Phenotypes inherited in an apparently Mendelian pattern often lack sufficiently sized pedigrees to pinpoint the causal locus. We evaluated whether exome sequencing could be applied to directly identify the causative gene underlying a monogenic human disease (FSS), i.e. with neither linkage data nor candidate gene analysis. Even in this simple scenario for “whole exome/genome genetics”, the key challenge that arises immediately is that the large number of apparently private mutations present by chance in any single human genome makes it difficult to identify which variant is causal, even when only considering nonsynonymous variants. Jones et al. recently overcame this in the context of hereditary pancreatic cancer by restricting focus to only nonsense mutations and also resequencing tumor DNA from the same individual, but this approach greatly limits sensitivity and is only relevant to a subset of mechanisms within one disease class28.
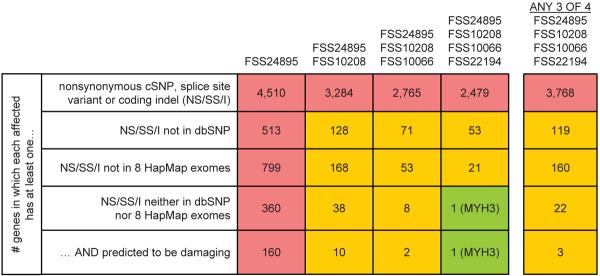
To quantify this background of non-causal variants in our exome data, we first asked how many genes had one or more nonsynonymous cSNPs, splice site disruptions, or coding indels in one or several FSS exomes (Fig. 2, row 1). Simply requiring that a gene contain variants in multiple affected individuals was clearly insufficient, as over 2,000 candidate genes remained even after intersecting four FSS exomes. We then applied filters to remove presumably common variants, as these are unlikely to be causative. Removing dbSNP catalogued variants from consideration reduced the number of candidates considerably (Fig. 2, row 2). Remarkably, the 8 HapMap exomes provided a filter nearly equivalent to dbSNP (Fig 2, row 3). Combining the two catalogues had a synergistic effect (Fig. 2, row 4), such that the candidate list could be narrowed to a single gene (MYH3, previously identified by a candidate gene approach as causative for FSS5). Specifically, MYH3 is the only gene where: (a) at least one (but not necessarily the same) nonsynonymous cSNP, splice-site disruption, or coding indel is observed in all four individuals with FSS; (b) the mutations are not in dbSNP, nor in the eight HapMap exomes. Taking the predicted deleteriousness of individual mutations into account served as an effective filter as well (Fig. 2, row 5), but was not required to identify MYH3. Ranges of candidate list sizes when other permutations of individuals are used are shown in Supplementary Fig. 6.
Figure 2. Direct identification of the causal gene for a monogenic disorder by exome sequencing.
Boxes list the number of genes with 1+ nonsynonymous cSNP, splice-site SNP, or coding indel (“NS/SS/I”) meeting specified filters. Columns show the effect of requiring that 1+ NS/SS/I variants be observed in each of 1 to 4 affected individuals. Rows show the effect of excluding from consideration variants found in dbSNP, the 8 HapMap exomes, or both. Column 5 models limited genetic heterogeneity or data incompleteness by relaxing criteria such that variants need only be observed in any 3 of 4 exomes for a gene to qualify.
MYH3 was well-covered in our data. To assess our sensitivity more globally, we calculated the probability that a mutation would have been identified in all four FSS-affected individuals for each gene, based on our overall coverage of that gene in each individual (Supplementary Data 2). The average probability across all genes was 86%. This is likely still an overestimate of sensitivity, as functional non-coding or structural mutations would be missed. It also remains challenging to detect mutations in segmentally duplicated regions of the genome with short read sequencing.
Nevertheless, our analysis suggests that direct sequencing of exomes of small numbers of unrelated individuals (but more than one) with a shared monogenic disorder can serve as a genome-wide scan for the causative gene. The availability of the 8 HapMap exomes was clearly helpful, suggesting that the power of this approach will improve as the 1000 Genomes Project29 generates a catalogue of common variation that is more complete and evenly ascertained than dbSNP. Also, FSS is inherited in an autosomal dominant pattern so the presence of only one mutant allele is sufficient to cause disease. Applying this strategy to a recessive disease would likely be easier, because there are far fewer genes in each exome that are homozygous or compound heterozygous for rare nonsynonymous variants. We also note that modeling of even a modest degree of genetic heterogeneity or data incompleteness is observed to significantly impact performance (Fig. 2, column offset to right). Moving along the spectrum from rare monogenic disorders to complex common diseases, it is likely that the increasing extent of genetic heterogeneity will need to be matched by increasingly large sample sizes30, and/or more sophisticated weighting of predicted mutational impact.
A clear limitation of exome sequencing is that it does not identify the structural and non-coding variants found by whole genome sequencing. At the same time, it allows a given amount of sequencing to be extended across at least 20 times as many samples as compared to whole genome sequencing. In studies focused on identifying rare variants or somatic mutations with medical relevance, sample size and the interpretability of functional impact may be critical to achieving meaningful success. It is the context of such studies that exome sequencing may be most valuable.
In summary, we demonstrate that targeted capture and massively parallel sequencing represents a cost-effective, reproducible, and robust strategy for the sensitive and specific identification of variants causing protein-coding changes in individual human genomes. The 307 megabases determined here across 12 individuals is the largest dataset reported to date of human coding sequence ascertained by second-generation sequencing methods. Finally, our successful demonstration that the causative gene for a monogenic disorder can be identified directly by exome sequencing of several unrelated individuals provides increasing context to the possibility that exome or genome sequencing may represent a new approach for identifying gene-disease relationships.
Methods Summary
DNA samples, targeted capture, and massively parallel sequencing
DNA samples were obtained from Coriell Repositories (HapMap) or by M.B. (FSS). Each shotgun library was hybridized to two Agilent 244K microarrays for target enrichment, followed by washing, elution and additional amplification. The first array targeted CCDS (2007), while the second was designed against targets poorly captured by the first array plus updates to CCDS in 2008. All sequencing was performed on the Illumina GA2 platform. Oligonucleotides used are listed in Supplementary Table 5.
Read mapping and variant analysis
Reads were mapped to the reference human genome (UCSC hg18), initially with ELAND (Illumina) for quality recalibration, and then again with Maq13. Sequence calls were also performed by Maq, and filtered to coordinates with >= 8× coverage and a _phred_-like15 consensus quality >= 30. Sequence calls for HapMap individuals were compared against Illumina Human1M-Duo genotypes. NA18507 SNPs from whole genome data12 were obtained from Illumina, Inc. Annotations of cSNPs were based on NCBI and UCSC databases, supplemented with PolyPhen Grid Gateway24 predictions for nonsynonymous SNPs. Identification of coding indels. This involved: a) gapped alignment of unmapped reads to the genome to generate a set of candidate indels using cross_match; b) ungapped alignment of all reads to the reference and alternative alleles for all candidate indels using Maq; c) filtering by coverage and allelic ratio.
Data access
Sequencing reads for HapMap individuals are available from the NCBI Short Read Archive under center name `UWGS-JS'. Variants identified in HapMap individuals have been submitted to NCBI dbSNP under handle `SEATTLESEQ'. Variants identified in FSS individuals are available to approved investigators through NCBI dbGaP, accession phs000204. Individual genotypes for variants identified in HapMap individuals, as well as the collapsed CCDS 2008 definition (prior to masking of coordinates listed in Supplementary Data 1), are available at http://krishna.gs.washington.edu/12_exomes.
Methods
Genomic DNA Samples
Targeted capture was performed on genomic DNA from 8 HapMap individuals (4 Yoruba (NA18507, NA18517, NA19129, NA19240), 2 East Asians (NA18555, NA18956), and 2 European-Americans (NA12156, NA12878)) and 4 European-American individuals affected by Freeman-Sheldon syndrome (FSS10066, FSS10208, FSS22194, FSS24895). Genomic DNA for HapMap individuals was obtained from Coriell Cell Repositories (Camden, NJ). Genomic DNA for Freeman-Sheldon syndrome individuals was obtained by M.B.
Oligonucleotides and adaptors
All oligonucleotides were synthesized by Integrated DNA Technologies (IDT) and resuspended in nuclease-free water to a stock concentration of 100 uM. Sequences are provided in Supplementary Table 5. Double-stranded library adaptors SLXA_1 and SLXA_2 were prepared to a final concentration of 50 uM by incubating equimolar amounts of SLXA_1_HI and SLXA_1_LO together and SLXA_2_HI and SLXA_2_LO together at 95°C for 3 mins and then leaving the adaptors to cool to room temperature in the heat block.
Shotgun library construction
Shotgun libraries were generated from 10ug of genomic DNA (gDNA) using protocols modified from the standard Illumina protocol12. Each library provided sufficient material for hybridization to two microarrays. For each sample, gDNA in 300ul 1× Tris-EDTA was first sonicated for 30min using a Bioruptor (Diogenode) set at high, then end-repaired for 45 mins in a 100ul reaction volume with using 1× End-It Buffer, 10ul dNTP mix and 10ul ATP as supplied in the End-It DNA End-Repair Kit (Epicentre). The fragments were then A-tailed for 20 mins at 70°C in a 100ul reaction volume with 1× PCR buffer (Applied Biosystems), 1.5mM MgCl2, 1mM dATP and 5U AmpliTaq DNA polymerase (Applied Biosystems). Next, library adaptors SLXA_1 and SLXA_2 were ligated to the A-tailed sample in a 90ul reaction volume with 1× Quick Ligation Buffer (New England Biolabs) with 5ul Quick T4 DNA Ligase (New England Biolabs) and each adaptor in 10× molar excess of sample. Samples were purified on QIAquick columns (Qiagen) after each of these four steps and DNA concentration determined on a Nanodrop-1000 (Thermo Scientific) when necessary.
Each sample was subsequently size selected for fragments of size 150–250bp using gel electrophoresis on a 6% TBE-polyacrylamide gel (Invitrogen). A gel slice containing the fragments of interest was then excised and transferred to a siliconized 0.5ml microfuge tube (Ambion) with a 20G needle-punched hole in the bottom. This tube was placed in a 1.5ml siliconized microfuge tube (Ambion), and centrifuged at 13.2rpm for 5mins to create a gel slurry that was then resuspended in 200ul 1× Tris-EDTA and incubated at 65°C for 2hrs, with periodic vortexing. This allowed for passive elution of DNA, and the aqueous phase was then separated from gel fragments by centrifugation through 0.2um NanoSep columns (Pall Life Sciences) and the DNA recovered using a standard ethanol precipitation.
Recovered DNA was resuspended in EB buffer (10mM Tris-Cl, pH8.5, Qiagen) and the entire volume used in a 1ml bulk PCR reaction volume with 1× iProof High-Fidelity Master Mix (Bio-Rad) and 0.5uM each of primers SLXA_FOR_AMP and SLXA_REV_AMP in the following conditions – 98°C for 30s; 20 cycles at 98°C for 30s, 65°C for 10s and 72°C for 30s; and finally 72°C for 5 min. PCR products were purified across 4 QIAquick columns (Qiagen) and all the eluants pooled.
Design of exome capture arrays
We targeted all well-annotated protein coding regions as defined by the CCDS (version 20080902, http://www.ncbi.nlm.nih.gov/projects/CCDS/). Coordinates were extracted from entries with “public” status, and regions with overlapping coordinates were merged. This resulted in a target with 164,007 discontiguous regions summing to 27,931,548 bp. By comparison, coding sequence defined by all of RefSeq (NCBI 36.3) comprises 31.9 Mb (14% larger). Hybridization probes against the target were designed primarily such that they were evenly spaced across each region. Probes were also constrained a) to be relatively unique, such that the average occurrence of each 15-mer in the probe sequence is less than 1008, b) to be between 20–60 bases in length, with preference for longer probes, and c) to have a calculated melting temperature (Tm) ≤ 69°C, with preference for higher Tms. Tm was calculated by 64.9 + 41 * (number of G+Cs − 16.4) / length of probe.
Two arrays (Agilent, 244K format) were designed and used per individual. The first array was common to all individuals, and contained 241,071 probes designed mainly against the subset of the target that was also found in a previous version of the CCDS (CCDS20070227). For most exomes, the second array was custom-designed specifically against target regions that had not been adequately represented after capture on the first array and subsequent sequencing. For two individuals (FSS10066, FSS10208), the matching was to a different individual's first-array data. However, this did not appear to significantly impact performance, likely because features capturing poorly on the first array largely did so consistently. Additionally, all of the second arrays also targeted sequences found in CCDS20080902 that were not in CCDS20070227 and hence not targeted by the first array. A subset of arrays used lacked control grids.
Targeted capture by hybridization to DNA microarrays
Hybridizations to Agilent 244K arrays were performed per manufacturer's instructions with modifications. For each enrichment, a 520ul hybridization solution containing 20ug of the bulk amplified gDNA library, 1× aCGH Hybridization Buffer (Agilent), 1× Blocking Agent (Agilent), 50ug Human CotI DNA (Invitrogen) and 0.92nmol each of the blocking oligos SLXA_FOR_AMP, SLXA_REV_AMP, SLXA_FOR_AMP_rev, SLXA_REV_AMP_rev was incubated at 95°C for 3 min and then at 37°C for at least 30mins. The hybridization solution was then loaded and the hybridization chamber assembled as per manufacturer's instructions. Incubation was done at 65°C for at least 66hrs with rotation at 20rpm in a hybridization oven (Agilent).
After hybridization, the slide-gasket sandwich was removed from the chamber and placed in a 50ml conical tube filled with aCGH Wash Buffer 1 (Agilent). The slide was separated from the gasket while in the buffer and then washed, first with fresh aCGH Wash Buffer 1 at room temperature for 10mins on an orbital shaker (VWR) set on low speed, and then in pre-warmed aCGH Wash Buffer 2 (Agilent) at 37°C for 5mins. Both washes were also done in 50ml conical tubes.
A Secure-Seal (SA2260, Grace Biolabs) was then affixed firmly over the active area of the washed slide and heated briefly according to manufacturer's instructions. One port was sealed with a seal tab and the seal chamber completely filled with approximately 1ml of hot EB (95°C). The other port was sealed and the slide incubated at 95°C on a heat block. After 5min, one port was unsealed and the solution recovered. DNA was purified from the solution using a standard ethanol precipitation.
Precipitated DNA was resuspended in EB and the entire volume used in a 50ul PCR volume comprising of 1× iTaq SYBR Green Supermix with ROX (Bio-Rad) and 0.2uM each of primers SLXA_FOR_AMP and SLXA_REV_AMP. Thermal cycling was done in a MiniOpticon Real-time PCR system (Bio-rad) with the following program: 95°C for 5min, then 30 cycles of 95°C for 30sec, 55°C for 2min, and 72°C for 2min. Each sample was monitored and extracted from the PCR machine when fluorescence began to plateau. Samples were then purified on a QIAQuick column (Qiagen) and sequenced.
Sequencing
All sequencing of post-enrichment shotgun libraries was carried out on an Illumina Genome Analyzer II as single-end 76 bp reads, following the manufacturer's protocols and using the standard sequencing primer. Image analysis and base-calling was performed by the Genome Analyzer Pipeline version 1.0 or 1.3 with default parameters but no pre-filtering of reads by quality. Quality values were recalibrated by alignment to the reference human genome with the Eland module.
Read mapping
The reference human genome used in these analyses was UCSC assembly hg18 (NCBI build 36.1), including unordered sequence (chrN_random.fa) but not including alternate haplotypes. For each lane, reads with calibrated qualities were extracted from the Eland export output. Base qualities were rescaled and reads mapped to the human reference genome using Maq (version 0.7.1)13. Unmapped reads were dumped using the −u option and subsequently used for indel mapping. Mapped reads that overlapped target regions (“target reads”) were used for all other analyses.
Target masking
All possible 76-bp reads that overlapped the aggregate target were simulated, mapped using Maq and consensus called using maq assemble with parameters −q 1 −r 0.2 −t 0.9. Target coordinates that had read depth < 76 (i.e. half of the expected depth), reflecting poor mappability (Supplementary Data 1), were removed from consideration for downstream analyses, leaving a 26,553,795 bp target.
Variant calling
All reads with a map score > 0 from each individual were merged and filtered for duplicates such that only the read with the highest aggregate base quality at any given start position and orientation was retained. Sequence calls were obtained using maq assemble with parameters −r 0.2 −t 0.9, and only coordinates with at least 8× coverage and an estimated _phred-_like consensus quality value of at least 30 were used for downstream variant analyses.
Comparison of sequence calls to array genotypes, dbSNP, and whole genome sequencing
For the 8 HapMap individuals, sequence calls were compared to array-based genotyping data (Illumina Human1M-Duo) provided by Illumina, Inc. We excluded from consideration genotyping assays where all 8 individuals were called by the arrays as homozygous non-reference as well as the MHC locus at chr6:32500001–33300000, as both sets are likely to be error-enriched in the genotyping data. We downloaded dbSNP(v129) from ftp://ftp.ncbi.nih.gov/snp/organisms/human_9606/chr_rpts on 08-May-13. ~14.2 million non-redundant coordinates were defined by this file-set. For comparison of NA18507 cSNPs to whole genome data, variant lists from Bentley et al. (2008) were obtained from Illumina, Inc.
Identification of coding indels
Reads for which Maq was unsuccessful in identifying an ungapped alignment were converted to fasta format and mapped to the human reference genome with cross_match (v1.080812, http://www.phrap.org), using parameters –gap_ext -1 –bandwidth 10 -minmatch 20 –maxmatch 24. Output options –tags –discrep_lists –alignments –score_hist were also set. Alignments with an indel were then filtered for those that a) had a score at least 40 more than the next best alignment, b) mapped at least 75 bases of the read, c) had no substitutions in addition to the indel, and d) overlapped a target region. Reads from filtered alignments that mapped to the negative strand were then reverse complemented and, together with the rest of the filtered reads, re-mapped with cross_match using the same parameters. This was to reduce ambiguity in called indel positions due to different read orientations. After the second mapping, alignments were re-filtered using the same criteria a) through d). For each sample, a putative indel event was called if at least 2 filtered reads covered the same event. A fasta file containing the sequences of all called events +/− 75 bp, as well as the reference sequence at the same positions was then generated for each individual. All the reads from each individual were then mapped to its “indel reference” with Maq using default parameters. Reads that mapped multiple times (map score 0) or had redundant start sites were removed, after which the number of reads mapping to either the reference or the non-reference allele was counted for each individual and indel. An indel was called if there were at least 8 non-reference allele reads making up at least 30% of all reads at that genomic position. Indels were called as heterozygous if non-reference alleles were 30–70% of reads at that position, and homozygous non-reference if >70%.
Variant annotation
For cSNP annotation, we constructed a local server that integrates data from NCBI (including dbSNP and Consensus CDS files) and from UCSC Genome Bioinformatics. We also generated PolyPhen predictions24 for all cSNPs identified here, using the PolyPhen Grid Gateway and Perl scripts supplied by Dr. Ivan Adzhubey. The server reads files with SNP locations and alleles, and produces annotation files available for download. Annotation includes dbSNP rs IDs, overlapping-gene accession numbers, SNP function (e.g. whether coding missense), conservation scores, HapMap minor-allele frequencies, and various protein annotations (sequence, position, amino acid changes with physicochemical properties, and PolyPhen classification). Indels were considered annotated by dbSNP if an entry was found with the same allele (or reverse complemented) within 1 bp of the variant position. This was to allow for ambiguities in calling the indel position.
Calculation of genome-wide estimates
Extrapolated estimates for the genome-wide number of cSNPs of various classes (Table 2b) were calculated based on the number of cSNP calls in that individual, the estimated sensitivity for making a variant call in that individual at any given position within the aggregate target (based on the fraction of array-based genotypes of that class that were successfully called; calculated separately for heterozygous and homozygous non-reference variants), and extrapolation to an estimated exome size of exactly 30 Mb (i.e. multiplying by 30/26.6 = 1.13). A similar approach was taken to estimate the genome-wide number of uncommon cSNPs introducing nonsense codons, starting with the number observed in each individual and extrapolating based on estimated sensitivity for heterozygote detection and an estimated exome size of exactly 30 Mb.
Freeman-Sheldon Syndrome Mutations
For FSS10066, FSS22194, and FSS24895, the identified mutation was a C>T at chr17:10485359, and the corresponding amino acid change was R672H. For FSS10208, the mutation was C>T at chr17:10485360, and the corresponding amino acid change was R672C.
Supplementary Material
1
2
Acknowledgements
For helpful discussions or assistance with genotyping data, we thank P. Green, J. Akey, R. Patwardhan, G. Cooper, J. Kidd, D. Gordon, J. Smith, I. Stanaway, and M. Rieder. For assistance with project management, computation, data management and submission, we thank E. Torskey, S. Thompson, T. Amburg, B. McNally, S. Hearsey, M. Shumway, and L. Hillier. For Human1M-Duo genotype data on HapMap samples, we thank Illumina, Inc. Our work was supported in part by grants from the National Institutes of Health / National Heart Lung and Blood Institute, the National Institutes of Health / National Human Genome Research Institute, and National Institutes of Health / National Institute of Child Health and Human Development. S.B.N. is supported by the Agency for Science, Technology and Research, Singapore. E.H.T. and A.W.B. are supported by a training fellowship from the National Institutes of Health / National Human Genome Research Institute. E.E.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
References
- 1.Cohen JC, et al. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 2.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nature reviews. 2009;10(4):241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 3.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 4.IHC A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toydemir RM, et al. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman-Sheldon syndrome and Sheldon-Hall syndrome. Nature genetics. 2006;38(5):561–565. doi: 10.1038/ng1775. [DOI] [PubMed] [Google Scholar]
- 6.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314(5797):268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 7.Olson M. Enrichment of super-sized resequencing targets from the human genome. Nat Methods. 2007;4(11):891–892. doi: 10.1038/nmeth1107-891. [DOI] [PubMed] [Google Scholar]
- 8.Hodges E, et al. Genome-wide in situ exon capture for selective resequencing. Nature genetics. 2007;39(12):1522–1527. doi: 10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 10.Ng PC, et al. Genetic variation in an individual human exome. PLoS Genet. 2008;4(8):e1000160. doi: 10.1371/journal.pgen.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kidd JM, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453(7191):56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456(7218):53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008 doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature genetics. 2008;40(6):722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 1998;8(3):186–194. [PubMed] [Google Scholar]
- 16.Turner EH, Lee C, Ng SB, Shendure J. Massively parallel exon capture and library-free resequencing across 16 individuals. Nat Methods. 2009 Apr 6; doi: 10.1038/nmeth.f.248. Advanced Online Publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kidd JM, et al. Haplotype sorting using human fosmid clone end-sequence pairs. Genome Res. 2008;18(12):2016–2023. doi: 10.1101/gr.081786.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albert TJ, et al. Direct selection of human genomic loci by microarray hybridization. Nat Methods. 2007;4(11):903–905. doi: 10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler DA, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452(7189):872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5(10):e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456(7218):66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boyko AR, et al. Assessing the evolutionary impact of amino acid mutations in the human genome. PLoS Genet. 2008;4(5):e1000083. doi: 10.1371/journal.pgen.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunyaev S, et al. Prediction of deleterious human alleles. Hum Mol Genet. 2001;10(6):591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
- 25.Yngvadottir B, et al. A genome-wide survey of the prevalence and evolutionary forces acting on human nonsense SNPs. Am J Hum Genet. 2009;84(2):224–234. doi: 10.1016/j.ajhg.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson MV. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 1999;64(1):18–23. doi: 10.1086/302219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cohen J, et al. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nature genetics. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 28.Jones S, et al. Exomic Sequencing Identifies PALB2 as a Pancreatic Cancer Susceptibility Gene. Science. 2009 doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siva N. 1000 Genomes project. Nat Biotechnol. 2008;26(3):256. doi: 10.1038/nbt0308-256b. [DOI] [PubMed] [Google Scholar]
- 30.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2