Genome beginnings: rooting the tree of life (original) (raw)
Abstract
A rooted tree of life provides a framework to answer central questions about the evolution of life. Here we review progress on rooting the tree of life and introduce a new root of life obtained through the analysis of indels, insertions and deletions, found within paralogous gene sets. Through the analysis of indels in eight paralogous gene sets, the root is localized to the branch between the clade consisting of the Actinobacteria and the double-membrane (Gram-negative) prokaryotes and one consisting of the archaebacteria and the firmicutes. This root provides a new perspective on the habitats of early life, including the evolution of methanogenesis, membranes and hyperthermophily, and the speciation of major prokaryotic taxa. Our analyses exclude methanogenesis as a primitive metabolism, in contrast to previous findings. They parsimoniously imply that the ether archaebacterial lipids are not primitive and that the cenancestral prokaryotic population consisted of organisms enclosed by a single, ester-linked lipid membrane, covered by a peptidoglycan layer. These results explain the similarities previously noted by others between the lipid synthesis pathways in eubacteria and archaebacteria. The new root also implies that the last common ancestor was not hyperthermophilic, although moderate thermophily cannot be excluded.
Keywords: tree of life, root, indels, eubacteria, firmicutes, archaebacteria, eukaryotes
1. Introduction
Today, there is enormous interest in reconstructing the rooted tree of prokaryotic life, but there is little or no agreement about the topology of the net, the web, the ring, the tree or the non-tree of life (Hilario & Gogarten 1993; Doolittle 1999; Jain et al. 2003; Rivera & Lake 2004; Konstantinidis & Tiedje 2005; Dagan & Martin 2006; McInerney & Pisani 2007; Sorek et al. 2007), and there is even less agreement about the location of the root.
Rooting is important because rooted ‘trees’ provide a framework for answering fundamental questions about the evolution of life. An accurate representation of a rooted tree, or graph, of life's history allows one to test theories for novel innovations in the evolution of life. With rooting information, it becomes possible to relate genetic, biochemical, ultrastructural and behavioural innovations to geological, paleontological and climatological events, thereby allowing one to trace the interdependent histories of the Earth and its microbiota and to test theories for the order of appearance of novel biological innovations.
Rooted trees allow central assumptions to be tested, such as: did the cenancestor, the last common population ancestral to all extant life, live in a hot environment? Many think so, but others argue for moderate temperatures (Miller & Bada 1988), and even the concept of a knowable cenancestor (Gogarten et al. 2002) has been questioned. Did the archaebacteria evolve in a hot environment? Again many think so, but the guanine–cytosine (GC) compositions of ribosomal RNAs suggest that the most recent common ancestor was not an extreme thermophile (Galtier et al. 1999). Did carbon heterotrophy evolve before autotrophy, or vice versa? Did prokaryotes with single membranes precede (Gupta 1998) or follow (Cavalier-Smith 2002) prokaryotes with double membranes? Did early organisms respire, or was substrate level phosphorylation used, and was methanogenesis early or late (Russell & Hall 1997; Grassineau et al. 2001; Bada & Lazcano 2002; Martin & Russell 2003; Purdy et al. 2003; Chistoserdova et al. 2004; Russell & Martin 2004; Ferry & House 2006; Gribaldo & Brochier-Armanet 2006)? Are phylogenetic signals sufficiently well preserved in eukaryotic genomes to permit the founding prokaryotic partners in eukaryogenesis to be identified, as some have claimed (Lake 1988; Woese 2002), or has all signal been overwritten by the passage of time and subsequent gene transfers (GTs)? All of these questions are controversial, and obtaining a workable representation of prokaryotic evolution may help answer them.
2. Four rooted trees of life
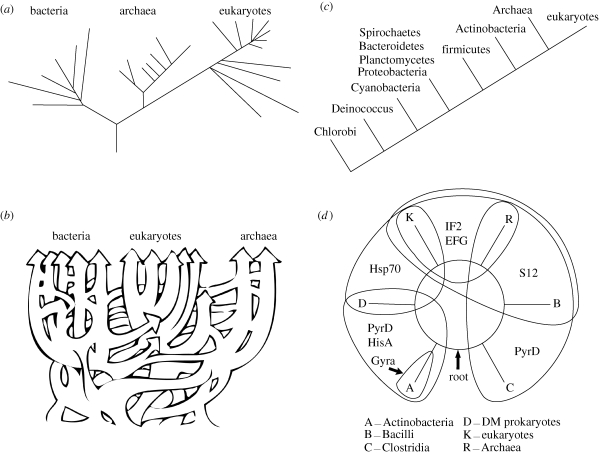
Four different rooted trees of life are illustrated in figure 1. The root, or cenancestor (Fitch & Upper 1987), of the traditional tree based on the pioneering work of Gogarten et al. (1989), Iwabe et al. (1989) and their collaborators, is placed between the archaebacteria and the bacteria, as shown in figure 1a. The non-tree, tree of life (Doolittle 1999), in figure 1b, illustrates the extensive lateral/horizontal GTs that may have erased most, if not all, of the phylogenetic signal from the early evolution of life on Earth, making it unlikely that the topology of the tree, and even less likely that the root, can be determined. A tree rooted within the double-membrane prokaryotes, based on transition analyses (Cavalier-Smith 2006), is illustrated in figure 1c. Figure 1d illustrates the indel-based rooted tree/ring of life recently developed in a series of papers from our laboratory (Lake et al. 2008).
Figure 1.
A summary of four prominent rooted trees of life. The trees are: (a) the traditional, three kingdom rooted tree (Gogarten et al. 1989; Iwabe 1989); (b) the lateral GT rooted tree (Doolittle 1999); (c) the transition analysis rooted tree (Cavalier-Smith 2006); and (d) the indel-based rooted ring of life (Lake et al. 2008).
As the variety of differing rooted trees suggests, determining roots is a complex problem that is sensitive to various sources of error. Here we discuss some of the difficulties encountered in rooting the tree of life and describe some recent progress made in this area.
Based on the proximity of the archaebacteria to the traditional root (figure 1a), it is thought that some archaebacterial metabolisms and adaptations, such as methanogenesis and hyperthermophily, may be indicators of the energy sources used and the environments that were present at the time of the cenancestor. Thus there is uncertainty about whether the archaebacteria are phylogenetically ancient, or whether they simply evolved rapidly after diverging from eubacterial ancestors. Furthermore, the most deeply branching bacterial phylum, the Aquificae, is adjacent to the root, adding support for a hyperthermophilic cenancestor. Since a hyperthermophilic cenancestor is inconsistent with molecular stability arguments (Miller & Lazcano 1995) and with correlations between phylogenetic analyses and ribosomal RNA GC compositions (Galtier et al. 1999), this suggests that life might have started at mesophilic temperatures. These findings call the traditional root of the tree of life into question and make it important to examine other types of rooting data.
3. Long branch attraction
There is also a growing awareness that the traditional root might be an artefact of phylogenetic reconstruction resulting from long branch attraction (Felsenstein 1978; Lake 1991) and other sequence analysis artefacts (Philippe & Forterre 1999; Zhaxybayeva et al. 2005). Because the root is located at the bottom of the tree, the phylogenetic signal generated by this earliest bifurcation has had the longest time to decay. This makes the location of the root extremely sensitive to artefacts of phylogenetic reconstruction, arguably making the root of the tree of life one of the most difficult phylogenetic signals to reconstruct. As the name ‘long branch attraction’ implies, this artefact can erroneously connect the longest branches in trees together. Thus when one uses paralogous gene sets to locate the root between the sets, long branch attraction can connect the fast-evolving, i.e. the longest-branched, taxa in each of the sets and thereby incorrectly root the tree (Lake 1991; Philippe & Forterre 1999).
One way to reduce the harmful effects of long branch attraction is to use phylogenetic characters that evolve more slowly than the nucleotide and amino acid sequences traditionally used. Potentially useful slower evolving characters include using the absences/presences of genes and the absences/presences of indels (insertions and deletions within genes). Indels are named indels because it is not always possible to determine whether they are _in_serts or _del_etions, and calling them indels acknowledges this inherent uncertainty. Since it is known that indels can persist over long time scales without changing length or position, as evidenced by the fact that numerous amino acid replacements occur within indels, they are potentially less affected by long branch attraction and other artefacts. Hence, in principle, they permit one to delve deeper into the rooted tree of life.
4. Gene transfers
Lateral/horizontal GTs represent a second confounding artefact. When genes are transferred between taxa, they misplace branches. There are three principal mechanisms that facilitate GT in prokaryotes, reviewed in Syvanen & Kado (1998). Genes can be transferred by means of transformation, conjugation and transduction. Transformation is the process whereby prokaryotes take up free DNA from their surroundings. Conjugation, also known as bacterial sex, occurs when an organism builds a tube-like structure known as the pilus, joins it to its ‘mate’ and transfers a plasmid through the tube. This process can transfer genes between very different taxa. Most astounding is the demonstration of Escherichia coli conjugating with the eukaryote Saccharomyces cerevisiae (Heinemann & Sprague 1989). Finally, transduction is a process for moving genes from one prokaryotic species to another via viruses (Hendrix et al. 1999).
GTs directly affect the calculation of the root by moving branches, thereby turning trees into tangles of webs, or nets, and making prokaryotic evolution less clonal and more population driven (Doolittle 1999; Gogarten et al. 1999; Ochman et al. 2000; Zhaxybayeva & Gogarten 2004; Lake et al. 2005) (also see the articles by Gogarten et al., Dagan & Martin, Beiko & Ragan and Sørensen et al. this volume). However, the effects of GT can be reduced by the careful choice of taxa used for analyses.
5. The diversity of life on earth
Because of the pervasiveness of GTs, one cannot realistically expect to reconstruct the root of a perfectly bifurcating, high-resolution tree. However, since GTs are less likely to occur between phylogenetically and functionally distant groups (Jain et al. 1999; Jain 2003), their effects may be reduced by choosing taxonomic groups that are phylogenetically well separated, thereby minimizing intergroup gene/indel transfers. These phylogenetically well-separated and relatively homogeneous prokaryotic taxa (Skophammer et al. 2006, 2007; Lake et al. 2007) are: the archaebacteria, the Bacilli and relatives, the Clostridia and relatives, the Actinobacteria and the double-membrane, Gram-negative, prokaryotes (Cyanobacteria, Proteobacteria, Spirochaetes, Chlorobi, Chloroflexi and 17 additional phyla) (for more detailed definitions of these groups, including some recent taxonomic changes, see Ohno et al. (2000), Garrity & Holt (2001), Wu et al. (2005) and Lake et al. (2008)). Together these five prokaryotic super-taxa, plus the eukaryotes, include all-known life (Boone & Castenholz 2001).
The Actinobacteria, characterized by high GC genomic compositions, are morphologically diverse and include many human pathogens, including those that cause leprosy and tuberculosis. The archaebacteria, a presumably paraphyletic taxon (Gouy & Li 1989; Archibald 2008; Cox et al. 2008), contain extreme halophiles, methanogens, hyperthermophiles and other unique phenotypes. The Clostridia and the Bacilli are firmicutes, characterized by their low GC compositions, although not exclusively (Ueda et al. 2004). The Clostridia are unique among the single-membrane prokaryotes for including photosynthetic as well as fermenting organisms. The double-membrane prokaryotes constitute an inordinately speciose, probably primitively photosynthetic, taxon. Some of their novelty is associated with the double-membrane system that surrounds them and encloses the periplasmic space.
6. A rooting example
The Actinobacteria are an extremely diverse prokaryotic taxon and have properties that suggested that this taxon might contain the root of life. They are among the most morphologically diverse prokaryotes and are widely distributed in both terrestrial and aquatic ecosystems (Embley & Stackebrandt 1994). Actinobacteria employ varied metabolic mechanisms, although no photosynthetic members are known. They are primarily chemoheterotrophs, which either respire or ferment. Their oxygen tolerances vary from strictly aerobic, to facultatively anaerobic, to microaerophilic, or strictly anaerobic. In addition to using some unique biochemical pathways not found in other prokaryotes, they also synthesize many macromolecules absent from other organisms, such as unique cell wall peptidoglycans (Gokhale et al. 2007). Given their diverse morphological and biochemical repertoires (Embley & Stackebrandt 1994; Boone & Castenholz 2001; Garrity & Holt 2001), properties that might indicate a deep placement in the tree of life, we were anxious to learn whether the root of life was contained within the Actinobacteria.
The GyrA protein contains an indel (Gao & Gupta 2005) that is ideal for determining whether the root is contained within the Actinobacteria, provided a suitable paralogous gene can be found. GyrA is a DNA topoisomerase, an essential protein that is found in all-known life. Topoisomerases serve to relieve the topological strains encountered by a cell during replication, transcription, recombination and chromatin remodelling. Type II DNA topoisomerases introduce double-strand breaks and are ATP dependent. Type II DNA topoisomerases are further subdivided into type IIA found in all domains of life and type IIB topoisomerases found only in archaebacteria. Gyrase and topoIV are well-documented paralogues in the Topo IIA family and exhibit extensive sequence similarity (Champoux 2001). Thus we use the indel in GyrA and in the paralogously related topoIV, ParC, to determine whether the root could be located within the Actinobacteria.
Table 1 summarizes the distribution of indels in approximately 1000 GyrA and ParC protein sequences. Sequences preceded by an A, D, F or R correspond to those from Actinobacteria, double-membrane prokaryotes, firmicutes and archaebacteria, respectively. Note that the GyrA indel sequence is present in all-known GyrA proteins from Actinobacteria, shown at the top of the table, and is absent in GyrA protein sequences from all other prokaryotic taxa. In the paralogous ParC sequences shown at the bottom of the table, the indel is absent in all sequences (Servin et al. 2008).
Table 1.
Summary of the GyrA/ParC indel. A summary of GyrA and ParC alignments within the NGSSG/GPDFPT region, corresponding to Escherichia coli residues 167–217 in the outgroup ParC sequence.
 |
|---|
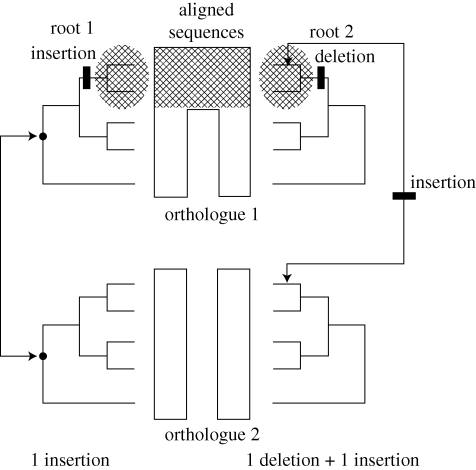
The process of indel rooting (Rivera & Lake 1992; Baldauf & Palmer 1993; Gupta 1998) is illustrated in figure 2 using the GyrA/ParC indels from table 1. The top two taxa in the trees, shown by shading at the left and the right of the figure, correspond to the actinobacterial taxa shown at the top of table 1. The sequences corresponding to the actinobacterial sequences, also shown by shading, are at the top of the centre panel and contain the four amino acid insert. The bottom two-thirds of the GyrA (orthologue 1) sequences and all of the ParC (orthologue 2) sequences lack the four amino acid insert and are not shaded. The tree on the left side of the figure is rooted outside the actinobacterial clade, as illustrated. This root requires only a single change (an insertion at the top of the tree) to produce the observed indel pattern. By contrast, the tree on the right side of the figure is rooted within the actinobacterial clade and requires two changes. Four amino acids must be inserted somewhere between the orthologue 1 and the orthologue 2 sequences, and this sequence must be subsequently deleted on the branch leading from the Actinobacteria to the remainder of the orthologue 1 tree. We conclude most parsimoniously that the root of the tree of life cannot be located within the shaded regions corresponding to the Actinobacteria. In this way, analyses of indels in orthologous/paralogous gene pairs can be used to exclude the root from regions of the tree.
Figure 2.
The process of indel rooting illustrated for two alternative rootings. In the centre of the figure, the two top sequences within orthologue 1 contain an insertion (shaded), whereas the bottom two-thirds of orthologue 1 and all of orthologue 2 lack the insertion. The trees on the left and the right sides of the figure represent two different rooted trees that relate orthologues 1 and 2. The tree on the right is rooted through the shaded region corresponding to those sequences that contain the insert, and the tree on the left is rooted outside the shaded region. The right tree is less parsimonious than the left tree, indicating that the root of the tree cannot lie within the shaded region.
In earlier studies (Rivera & Lake 1992; Gupta & Singh 1994; Philippe et al. 1999), it was assumed that incomplete gene sets could not be used for rooting, since ubiquitous gene sets were required for sequence-based rooting (Gogarten et al. 1989; Iwabe 1989; Brown & Doolittle 1995; Boucher et al. 2003; Zhaxybayeva et al. 2005). We now know that ubiquitous genes are not necessarily required for indel rooting since indel- and sequence-based rooting studies use different types of information. Although the ParC sequence is missing from the archaebacteria, this complication was not considered in the simplified analysis in figure 2. However, when insertions and deletions of genes and insertions and deletions of indels were simultaneously analysed, it was shown that the GyrA/ParC data exclude the root within the Actinobacteria (Servin et al. 2008). In fact, one frequently finds indel sets that exclude some roots, even though genes are missing from some taxa.
Recently, solutions have been found for some technical problems that previously prevented the analyses of indel data. For example, in the past, it was generally thought that a single indel could not be statistically significant. Now, however, methods have been developed for determining the statistical significance of indels by analysing the large amounts of sequence data that are available at the margins of indels. The statistical significance of an indel is a function of the length of the indel (longer indels are better), the proportion of exceptions to the indel pattern (fewer exceptions are better) and the evolutionary histories of the flanking sequences (Lake et al. 2008). Solutions have also been found for other problems related to indel rooting. For example, improved methods for identifying paralogous gene sets are now available (Skophammer et al. 2007), further increasing the number of useful indels.
Having workable solutions to some of the technical problems associated with indels rooting has greatly increased the number of indel sets available for rooting studies. In the following sections, we discuss the current state of indel-based rooting studies and some of the resulting biological implications.
7. A new root
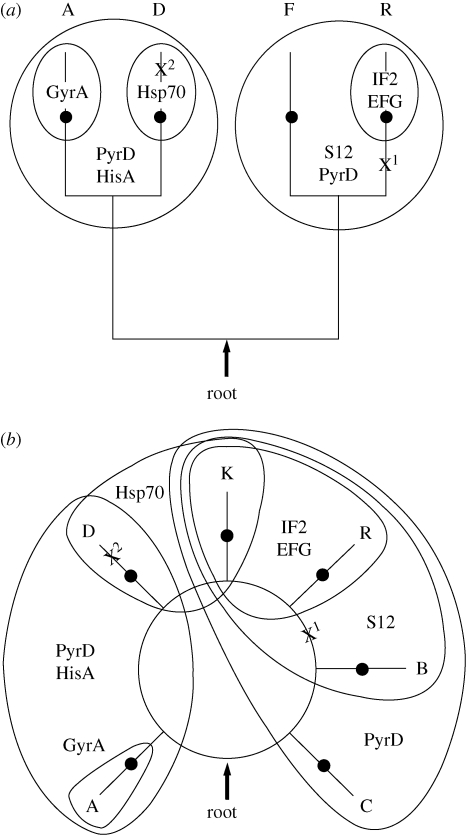
To date, indel-based rooting has excluded the root from the tree/graph of life, except for the site marked ‘root’ in the rooted tree and graph in figure 3. The prokaryotic tree of life is shown in figure 3a, and the graph of life, including the eukaryotes, is shown in figure 3b. The figure summarizes the results of eight indel analyses that exclude the root from the tree/ring of life consisting of the Actinobacteria, A; the double-membrane prokaryotes, D; the firmicutes, F; the archaebacteria, R and the eukaryotes, K. The excluded regions are indicated by the surrounding balloons, and the names of the indel-containing proteins that exclude these roots are given within the balloons. All eight indels significantly, p > 0.95, exclude their respective clades.
Figure 3.
(a) A summary of the new root of the tree of life and (b) for the ring of life. The relevant four taxa representing known prokaryotic diversity are the double-membrane prokaryotes (D), the firmicutes (F), the Actinobacteria (A) and the archaebacteria (R). The eukaryotes (K) are present in the ring of life (b), and the Bacilli (B) and the Clostridia (C) form a paraphyletic grouping within the ring. The regions from which the root is excluded are circled and labelled with the name(s) of the relevant indel(s) that exclude(s) them. The dots present on the distal portions of the leaves represent the last common ancestors of each crown group. For reference, the root based on ground-breaking analyses of anciently duplicated gene paralogues (Gogarten et al. 1989; Iwabe 1989), marked by an ‘X1’, is located between the archaebacteria and (b) the Archaea contain extreme halophiles, methanogens, hyperthermophiles and other unique phenotypes, bacteria, and the root based on transition analysis (Cavalier-Smith 2006), marked by an ‘X2’, is within the double-membrane prokaryotes.
In addition to providing a new root, indel-based rooting provides a non-tree-based view of evolution. For example, the tree of life becomes a graph of life when the eukaryotes are included because indel-based analyses provide data, directed quartets (Lake 2008), that allow one to distinguish between trees and graphs. To understand how this is possible, consider the phylogenetic relationships between eukaryotes and archaebacteria and the eukaryotes and the double-membrane prokaryotes. Based on indels present in protein synthesis initiation factor IF2 and in protein synthesis elongation factor EFG, the eukaryotes and the archaebacteria are sister taxa, as shown by the relevant excluded region in figure 3b. By contrast, based on the indel present in heat shock protein Hsp70, the eukaryotes and the double-membrane prokaryotes are sister taxa (figure 3b). Since trees do not allow a taxon to be simultaneously the sister taxon to two different taxa, i.e. the archaebacteria and the double-membrane prokaryotes, these results can no longer be represented by a tree. In other words, it is impossible for a tree to join the eukaryotes to the archaebacteria and simultaneously to the double-membrane prokaryotes. This relationship can only be represented by a ring, as shown in figure 3b. This ability of indel-based rootings to distinguish graphs from trees may become important in future studies designed to reconstruct a low-resolution graph of life.
As previously discussed, an important property of indel-based rooting is that indels can sometimes exclude roots, even though genes may be missing from some taxa. For example, the Hsp70/MreB sequences that exclude the root from the double membrane/eukaryotic clade shown in figure 3b were suitable for analysis, despite the possibility that some of these genes had been transferred from eubacterial taxa. The reason that archaebacterial sequences were not needed is related to the fact that indel analyses exclude roots from regions, rather than provide positive evidence that a particular root exists. This novel property is highly useful.
Indel-based rooting also provides a more dynamic view of what a root actually represents. Imagine, for a moment, that the archaebacteria had not yet been discovered, even though they were alive on the Earth. In that case, in order to root the tree of life, one could only search for a root within the eubacteria because, in this example, no other prokaryotes would have been available. Once a unique root was found by exclusion that cenacestor would have been useful for understanding the evolution of known life on Earth. Consider now what would happen if the archaebacteria were then discovered! For one thing, all of the previous work done to exclude the root from the eubacteria would still be useful and valid. But the discovery of the archaebacteria would create new potential root locations. Thus each newly discovered, fundamentally different organism potentially represents a new rooting alternative. Because roots are found by exclusion, if a newly discovered organism generates new rooting alternatives, then these alternatives and only these new ones would have to be excluded in order to again obtain a unique root. In this new view, the studies described here do not find a ‘universal root of life’, but only find the root of all (currently known) life. Thus as new organisms are discovered, they can potentially generate new, even more ancient, roots. This is an exciting outcome, because every time a deeper branching organism is discovered, we have a new opportunity to learn about even earlier events in the evolution of life.
8. Discussion
The new root localized in figure 3b provides a new view of the early evolution of life. For one thing, it excludes the root from the archaebacteria. Furthermore, since four separate indels independently exclude the root from the archaebacteria, this is a robust finding. This is perhaps not surprising, since literature searches provide little support for a root within the archaebacteria. Note that a root within the archaebacteria is not supported by the traditional root located between the archaebacteria and the eubacteria, marked by an X1 in figure 3 (Gogarten et al. 1989; Iwabe 1989). For example, a recent phylogenetic analysis of ancient orthologue/paralogue sets found only a single gene set that placed the root within the archaebacteria, whereas nine sets supported the traditional root between the archaebacteria and the eubacteria and seven sets supported a root in the eubacteria (Zhaxybayeva & Gogarten 2004), but see Philippe & Forterre (1999).
Three independent indels, Hsp70, PyrD and HisA, exclude the root from the double-membrane prokaryotes and directly argue against the root based on transition analyses shown in figure 3b (Cavalier-Smith 2006). Again this fits well with other data and is consistent with the derivation of double-membrane prokaryotes from simpler single-membrane prokaryotes. The double-membrane arrangement is known to greatly complicate many processes that are much simpler in single-membrane prokaryotes. For example, complex design changes are needed to accommodate transport across the double-membrane arrangement. Specifically, special ABC transporters differing considerably from those present in single-membrane prokaryotes are required to facilitate the uptake of vitamin B12 in double-membrane prokaryotes (Locher et al. 2002), and the process of flagellar assembly is considerably more complex in double-membrane prokaryotes than in single-membrane prokaryotes. In double-membrane prokaryotes, the process requires the construction of novel flagellar rings, the L and P ring assemblies, in addition to the M and S rings found in the cytoplasmic membrane of single- and double-membrane prokaryotes (Macnab 2003).
Parsimonious reasoning applied to this root also indicates that members of the cenancestral population were enclosed by ester-linked lipid membranes and surrounded by a peptidoglycan layer, because the Actinobacteria and the double-membrane prokaryotes on the left side of the root in figure 3b and the Clostridia and the Bacilli on the right side of the root in figure 3b, all share these character states. The location of the root also implies that during the transition from the root to the archaebacteria, through the Clostridia and the Bacilli (figure 3b), the peptidoglycan layer was lost and the ester-linked membrane lipids were replaced with ether-linked lipid membranes. If this occurred, then it is possible that remnants of this transition still exist in the lipid biosynthetic pathways found in the firmicutes today.
This possibility prompted us to re-examine the taxonomic distributions of genes coding for archaebacterial lipid pathways. With the appearance of the first archaebacterial and firmicute genomes, detailed phylogenetic comparisons of lipid biosynthetic pathways pointed towards some novel lipid-based archaebacterial–firmicute connections (Lange et al. 2000; Smit & Mushegian 2000; Boucher et al. 2003, 2004; Daiyasu et al. 2005). These simply did not make sense in light of the then current ideas concerning the uniqueness and the early evolution of the archaebacteria. However, those genomic findings make sense when viewed in the light of the new rooted tree of life. The findings reported in those prior genomic analyses suggest that parts of the mevalonate (MVA) synthesis pathway, central to archaebacterial lipid synthesis, may have been present in the common population immediately ancestral to the Bacilli and the archaebacteria. In addition, they imply that one of the enzymes thought to be responsible for the unique archaebacterial _sn_-1 stereochemistry was present in this ancestral population.
Many bacilli have complete, or nearly complete, MVA lipid synthesis pathways, similar to those found in the archaebacteria (Smit & Mushegian 2000; Boucher et al. 2004), whereas most eubacteria use the DXPS lipid synthesis pathway (Lange et al. 2000). In detailed phylogenetic analyses of the genes present in the prokaryotic MVA pathways (Boucher et al. 2004), few double-membrane prokaryotes contain MVA genes. Furthermore, the MVA genes that are present in double-membrane prokaryotes are non-parsimoniously distributed throughout the trees in no particular phylogenetic order, suggesting that they arose from multiple GTs. In contrast, the Bacilli form a single clade in most MVA gene trees (Boucher et al. 2004). The phylogenetic distribution of MVA biosynthetic genes within the Bacilli parsimoniously suggests that many genes in the pathway were vertically inherited from the last common ancestral population, relating the Bacilli and the archaebacteria.
Similar evidence suggests that at least one antecedent of the enzymes contributing the unique sn-1 stereochemistry existed in the ancestral population common to the Bacilli and the archaebacteria. Geranylgeranylglyceryl phosphate (GGGP) synthase, a terminal member of the MVA pathway, is one of the enzymes thought to be responsible for the unique _sn_-1 stereochemistry of the archaebacterial glycerol phosphate backbone (Boucher et al. 2004). GGGP synthase appears to have been vertically inherited from the ancestral population common to the Bacilli and the archaebacteria, since, except for a single Cytophaga species (Boucher et al. 2004), only the Bacilli and the archaebacteria contain genes for GGGP synthase. Furthermore, the Bacilli and the archaebacteria are not intermixed in the GGGP synthase tree, as would be expected if extensive GT from either group had been the source of GGGP synthase. Instead, they are resolved into their respective groups. Although the GGGP synthase genes cannot polarize the root, they are consistent with our indel evidence, excluding the root from the firmicute/archaebacterial clade. We parsimoniously infer that the ancestral population contained a nearly complete archaebacterial-like MVA pathway and one of the genes necessary for producing the unique archael lipid backbone stereochemistry. Thus this new root offers a simple explanation for a previously puzzling observation.
Almost no proposal regarding the evolution of life has provided more thought provoking discussion than the possibility of a hyperthermal origin of life. The new root discussed here does not support the proposition that the cenancestral population was hyperthermophilic, because the taxa adjacent to the root are not hyperthermophilic. The Actinobacteria, adjacent to the left side of the root, are primitively mesophilic, and the Clostridia on the right of the root are not hyperthermophilic (figure 3b). However, neither a moderately thermophilic nor a mesophilic cenancestor can be excluded for this root, since two recently discovered thermophilic Clostridia occupy a position near the root in concatenated protein sequence trees (Wu et al. 2005). Symbiobacterium thermophilum (Ohno et al. 2000) grows optimally at 60°C, and Carboxydothemus hydrogenoformans (Wu et al. 2005) grows optimally at 78°C. Thus this new root parsimoniously places constraints on the growth temperature of the cenancestor. These results, together with other evidence and arguments for mesophilic origins (Miller & Lazcano 1995; Galtier et al. 1999; Philippe & Forterre 1999), make lower temperature hydrothermal sites such as the Lost City field (Russell & Martin 2004; Kelly et al. 2005), increasingly attractive for the cenancestral evolution of life. We hope that this new root will form a basis for synthesizing the rapidly growing database of genomic- and Earth science-based information becoming available on the early evolution of life.
Acknowledgements
Supported by grants from NSF and the UCLA NASA Astrobiology Institute to J.A.L. R.G.S., J.A.S. and C.W.H. were supported by an IGERT training grant from NSF, a Cell and Molecular Biology Training Grant from NIH and a Genomic Interpretation and Analysis Training Grant from NIH, respectively.
Footnotes
References
- Archibald J. M.2008The eocyte hypothesis and the origin of eukaryotic cells. Proc. Natl Acad. Sci. USA 105, 20 049–20 050 (doi:10.1073/pnas.0811118106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bada J. L., Lazcano A.2002Origin of life. Some like it hot, but not the first biomolecules. Science 296, 1982–1983 (doi:10.1126/science.1069487) [DOI] [PubMed] [Google Scholar]
- Baldauf S. L., Palmer J. D.1993Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl Acad. Sci. USA 90, 11 558–11 562 (doi:10.1073/pnas.90.24.11558) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone D., Castenholz R. W.2001The Archaea and the deep branching and phototrophic Bacteria New York, NY: Springer [Google Scholar]
- Boucher Y., Douady C. J., Papke R. T., Walsch D. A., Boudreau M. E. R., Nesbo C. L., Case R. J., Doolittle W. F.2003Lateral gene transfer and the origins of prokaryotic groups. Annu. Rev. Genet. 37, 283–328 (doi:10.1146/annurev.genet.37.050503.084247) [DOI] [PubMed] [Google Scholar]
- Boucher Y., Kamekura M., Doolittle W. F.2004Origins and evolution of isoprenoid lipid biosynthesis in Archaea. Mol. Microbiol. 52, 515–527 (doi:10.1111/j.1365-2958.2004.03992.x) [DOI] [PubMed] [Google Scholar]
- Brown J. R., Doolittle W. F.1995Root of the universal tree of life based on ancient aminoacyl-transfer RNA synthetase gene duplications. Proc. Natl Acad. Sci. USA 92, 2441–2445 (doi:10.1073/pnas.92.7.2441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T.2002The neomuran origin of archaebacteria, the negibacterial root of the universal tree and bacterial megaclassification. Int. J. Syst. Evol. Microbiol. 52, 7–76 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T.2006Rooting the tree of life by transition analyses. Biol. Direct 1, 1–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J.2001DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (doi:10.1146/annurev.biochem.70.1.369) [DOI] [PubMed] [Google Scholar]
- Chistoserdova L., Jenkins C., Kalyuzhnaya M. G., Marx C. J., Lapidus A., Vorholt J. A., Staley J. T., Lidstrom M. E.2004The enigmatic planctomycetes may hold a key to the origins of methanogenesis and methylotrophy. Mol. Biol. Evol. 21, 1234–1241 (doi:10.1093/molbev/msh113) [DOI] [PubMed] [Google Scholar]
- Cox C. J., Foster P. G., Hirt R. P., Harris S. R., Embley T. M.2008The archaebacterial origin of eukaryotes. Proc. Natl Acad. Sci. USA 105, 20 356–20 361 (doi:10.1073/pnas.0810647105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T., Martin W.2006The tree of one percent. Genome Biol. 7, 118 (doi:10.1186/gb-2006-7-10-118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daiyasu H., Kuma K.-I., Yokoi T., Morii H., Koga Y., Toh H.2005A study of archaeal enzymes involved in polar lipid synthesis linking amino acid sequence information, genomic contexts and lipid composition. Archaea 1, 399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle W. F.1999Phylogenetic classification and the universal tree. Science 284, 2124–2128 (doi:10.1126/science.284.5423.2124) [DOI] [PubMed] [Google Scholar]
- Embley T. M., Stackebrandt E.1994The molecular phylogeny and systematics of the actinomycetes. Annu. Rev. Microbiol. 48, 257–289 [DOI] [PubMed] [Google Scholar]
- Felsenstein J.1978Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 (doi:10.2307/2412923) [Google Scholar]
- Ferry J. G., House C. H.2006The stepwise evolution of early life driven by energy conservation. Mol. Biol. Evol. 23, 1286–1292 (doi:10.1093/molbev/msk014) [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Upper K.1987The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harb. Symp. Quant. Biol. 52, 759–767 [DOI] [PubMed] [Google Scholar]
- Galtier N., Tourasse N., Gouy M.1999A nonhyperthermophilic common ancestor to extant life forms. Science 283, 220–221 (doi:10.1126/science.283.5399.220) [DOI] [PubMed] [Google Scholar]
- Gao B., Gupta R. S.2005Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 55, 2401–2412 (doi:10.1099/ijs.0.63785-0) [DOI] [PubMed] [Google Scholar]
- Garrity G., Holt J. G.2001The road map to the manual. In Bergey's manual of systematic bacteriology (eds Boone D., Castenholz R. W.). New York, NY: Springer [Google Scholar]
- Gogarten J. P., et al. 1989Evolution of the vacuolar H+-ATPase—implications for the origin of eukaryotes. Proc. Natl Acad. Sci. USA 86, 6661–6665 (doi:10.1073/pnas.86.17.6661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogarten J. P., Murphey R. D., Olendzenski L.1999Horizontal gene transfer: pitfalls and promises. Biol. Bull. 196, 359–361 (doi:10.2307/1542970) [DOI] [PubMed] [Google Scholar]
- Gogarten J. P., Doolittle W. F., Lawrence J. G.2002Prokaryotic evolution in light of gene transfer. Mol. Biol. Evol. 19, 2226–2238 [DOI] [PubMed] [Google Scholar]
- Gokhale R. S., Saxena P., Chopra T., Mohanty D.2007Versatile polyketide enzymatic machinery for the biosynthesis of complex mycobacterial lipids. Nat. Prod. Rep. 24, 267–277 (doi:10.1039/b616817p) [DOI] [PubMed] [Google Scholar]
- Gouy M., Li W. H.1989Phylogenetic analysis based on ribosomal-RNA sequences supports the archaebacterial rather than the eocyte tree. Nature 339, 145–147 (doi:10.1038/339145a0) [DOI] [PubMed] [Google Scholar]
- Grassineau N. V., Nisbet E. G., Bickle M. J., Fowler C. M. R., Lowry D., Mattey D. P., Abell P., Martin A.2001Antiquity of the biological sulphur cycle: evidence from sulphur and carbon isotopes in 2700 million-year-old rocks of the Belingwe Belt, Zimbabwe. Proc. R. Soc. B 268, 113–119 (doi:10.1098/rspb.2000.1338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaldo S., Brochier-Armanet C.2006The origin and evolution of Archaea: a state of the art. Phil. Trans. R. Soc. B 361, 1077–1022 (doi:10.1098/rstb.2006.1841) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S.1998Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R. S., Singh B.1994Phylogenetic analysis of 70-kD heat-shock protein sequences suggests a chimeric origin for the eukaryotic cell-nucleus. Curr. Biol. 4, 1104–1114 (doi:10.1016/S0960-9822(00)00249-9) [DOI] [PubMed] [Google Scholar]
- Heinemann J. A., Sprague G. F., Jr1989Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature 340, 205–209 (doi:10.1038/340205a0) [DOI] [PubMed] [Google Scholar]
- Hendrix R. W., Smith M. C., Burns R. N., Ford M. E., Hatfull G. F.1999Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl Acad. Sci. USA 96, 2192–2197 (doi:10.1073/pnas.96.5.2192) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilario E., Gogarten J. P.1993Horizontal transfer of ATPase genes—the tree of life becomes a net of life. Biosystems 31, 111–119 (doi:10.1016/0303-2647(93)90038-E) [DOI] [PubMed] [Google Scholar]
- Iwabe N., Kuma K., Hasegawa M., Osawa S., Miyata T.1989Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl Acad. Sci. USA 86, 9355–9359 (doi:10.1073/pnas.86.23.9355) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Rivera M. C., Lake J. A.1999Horizontal gene transfer among genomes: the complexity hypothesis. Proc. Natl Acad. Sci. USA 96, 3801–3806 (doi:10.1073/pnas.96.7.3801) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Rivera M. C., Moore J. E., Lake J. A.2003Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20, 1598–1602 (doi:10.1093/molbev/msg154) [DOI] [PubMed] [Google Scholar]
- Kelly D., et al. 2005A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307, 1428–1434 (doi:10.1126/science.1102556) [DOI] [PubMed] [Google Scholar]
- Konstantinidis K. T., Tiedje J. M.2005Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264 (doi:10.1128/JB.187.18.6258-6264.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. A.1988Origin of the eukaryotic nucleus determined by rate-invariant analysis of ribosomal RNA sequences. Nature 331, 184–186 (doi:10.1038/331184a0) [DOI] [PubMed] [Google Scholar]
- Lake J. A.1991Tracing origins with molecular sequences: metazoan and eukaryotic beginnings. Trends Biochem. Sci. 16, 46–50 (doi:10.1016/0968-0004(91)90020-V) [DOI] [PubMed] [Google Scholar]
- Lake J. A.2008Reconstructing evolutionary graphs: 3D parsimony. Mol. Biol. Evol. 25, 1677–1682 (doi:10.1093/molbev/msn117) [DOI] [PubMed] [Google Scholar]
- Lake J. A., Moore J. E., Simonson A. B., Rivera M. C.2005Fulfilling Darwin's dream. In Microbial evolution: concepts and controversies, (ed. Sapp J.), pp. 184–206 New York, NY: Oxford University Press [Google Scholar]
- Lake J. A., Herbold C. W., Rivera M. C., Servin J. A., Skophammer R. G.2007Rooting the tree of life using non-ubiquitous genes. Mol. Biol. Evol. 24, 130–136 (doi:10.1093/molbev/msl140) [DOI] [PubMed] [Google Scholar]
- Lake J. A., Servin J. A., Herbold C. W., Skophammer R. G.2008Evidence for a new root of the tree of life. Syst. Biol. 57, 835–843 (doi:10.1080/10635150802555933) [DOI] [PubMed] [Google Scholar]
- Lange B. M., Rujan T., Martin W., Croteau R.2000Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes. Proc. Natl Acad. Sci. USA 97, 13 172–13 177 (doi:10.1073/pnas.240454797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locher K. P., Allen T. L., Rees D. C.2002The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science 296, 1091–1097 (doi:10.1126/science.1071142) [DOI] [PubMed] [Google Scholar]
- Macnab R. M.2003How bacteria assemble flagella. Annu. Rev. Microbiol. 57, 77–100 (doi:10.1146/annurev.micro.57.030502.090832) [DOI] [PubMed] [Google Scholar]
- Martin W., Russell M. J.2003On the origins of cells: a hypothesis for the evolutionary transitions from abiotic geochemistry to chemoautotrophic prokaryotes, and from prokaryotes to nucleated cells. Phil. Trans. R. Soc. Lond. B 358, 59–85 (doi:10.1098/rstb.2002.1183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney J. O., Pisani D.2007Paradigm for life. Science 318, 1390–1391 (doi:10.1126/science.1151657) [DOI] [PubMed] [Google Scholar]
- Miller S. L., Bada J. L.1988Submarine hot springs and the origin of life. Nature 334, 609–611 (doi:10.1038/334609a0) [DOI] [PubMed] [Google Scholar]
- Miller S. L., Lazcano A.1995The origin of life—did it occur at high temperatures? J. Mol. Evol. 41, 689–692 [DOI] [PubMed] [Google Scholar]
- Ochman H., Lawrence J. G., Groisman E. A.2000Lateral gene transfer and the nature of bacterial innovation. Nature 405, 299–304 (doi:10.1038/35012500) [DOI] [PubMed] [Google Scholar]
- Ohno M., et al. 2000_Symbiobacterium thermophilum_ gen. nov., sp. nov., a symbiotic thermophile that depends on co-culture with a Bacillus strain for growth. Int. J. Syst. Evol. Microbiol. 50, 1829–1832 [DOI] [PubMed] [Google Scholar]
- Philippe H., Forterre P.1999The rooting of the universal tree of life is not reliable. J. Mol. Evol. 49, 509–523 (doi:10.1007/PL00006573) [DOI] [PubMed] [Google Scholar]
- Philippe H., Budin K., Moreira D.1999Horizontal transfers confuse the prokaryotic phylogeny based on the HSP70 protein family. Mol. Microbiol. 31, 1007–1012 (doi:10.1046/j.1365-2958.1999.01185.x) [DOI] [PubMed] [Google Scholar]
- Purdy K. J., Nedwell D. B., Embley T. M.2003Analysis of the sulfate-reducing bacterial and methanogenic archaeal populations in contrasting Antarctic sediments. Appl. Environ. Microbiol. 69, 4501 (doi:10.1128/AEM.69.6.3181-3191.2003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A.1992Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science 257, 74–76 (doi:10.1126/science.1621096) [DOI] [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A.2004The ring of life: evidence for a genome fusion origin of eukaryotes. Nature 431, 152–155 (doi:10.1038/nature02848) [DOI] [PubMed] [Google Scholar]
- Russell M. J., Hall A. J.1997The emergence of life from iron monosulphide bubbles at a submarine hydrothermal redox and pH front. J. Geol. Soc. Lond. 154, 377–402 (doi:10.1144/gsjgs.154.3.0377) [DOI] [PubMed] [Google Scholar]
- Russell M. J., Martin W.2004The rocky roots of the acetyl–CoA pathway. Trends Biochem. Sci. 29, 358–363 (doi:10.1016/j.tibs.2004.05.007) [DOI] [PubMed] [Google Scholar]
- Servin J. A., Herbold C. W., Skophammer R. G., Lake J. A.2008Evidence excluding the root of the tree of life from the Actinobacteria. Mol. Biol. Evol. 25, 1–4 (doi:10.1093/molbev/msm249) [DOI] [PubMed] [Google Scholar]
- Skophammer R. G., Herbold C. W., Rivera M., Servin J. A., Lake J. A.2006Evidence that the root of the tree of life is not within the Archaea. Mol. Biol. Evol. 23, 1648–1651 (doi:10.1093/molbev/msl046) [DOI] [PubMed] [Google Scholar]
- Skophammer R. G., Servin J. A., Herbold C. W., Lake J. A.2007Evidence for a Gram positive, eubacterial root of the tree of life. Mol. Biol. Evol. 24, 1761–1768 (doi:10.1093/molbev/msm096) [DOI] [PubMed] [Google Scholar]
- Smit A., Mushegian A. R.2000Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 10, 1468–1484 (doi:10.1101/gr.145600) [DOI] [PubMed] [Google Scholar]
- Sorek R., Zhu Y., Creevery C. J., Francino M. P., Bork P., Rubin E. M.2007Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318, 1449–1452 (doi:10.1126/science.1147112) [DOI] [PubMed] [Google Scholar]
- Syvanen M., Kado C. I. (eds) 1998Horizontal gene transfer. London, UK: Chapman & Hall [Google Scholar]
- Ueda K., Yamashita A., Ishikawa J., Shimada M., Watsuji T., Morimura K., Ikeda H., Hattori M., Beppu T.2004Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res. 32, 4937–4944 (doi:10.1093/nar/gkh830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R.2002On the evolution of cells. Proc. Natl Acad. Sci. USA 99, 2742–2747 (doi:10.1073/pnas.132266999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., et al. 2005Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 1, 563–574 (doi:10.1371/journal.pgen.0010065) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O., Gogarten J. P.2004Cladogenesis, coalescence and the evolution of the three domains of life. Trends Genet. 20, 182–187 (doi:10.1016/j.tig.2004.02.004) [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O., Lapierre P., Gogarten J. P.2005Ancient gene duplications and the root(s) of the tree of life. Protoplasma 227, 53–64 (doi:10.1007/s00709-005-0135-1) [DOI] [PubMed] [Google Scholar]