Establishment of HIV Latency in Primary CD4+ Cells Is due to Epigenetic Transcriptional Silencing and P-TEFb Restriction (original) (raw)
Abstract
The development of suitable experimental systems for studying HIV latency in primary cells that permit detailed biochemical analyses and the screening of drugs is a critical step in the effort to develop viral eradication strategies. Primary CD4+ T cells were isolated from peripheral blood and amplified by antibodies to the T-cell receptor (TCR). The cells were then infected by lentiviral vectors carrying fluorescent reporters and either the wild-type Tat gene or the attenuated H13L Tat gene. After sorting for the positive cells and reamplification, the infected cells were allowed to spontaneously enter latency by long-term cultivation on the H80 feeder cell line in the absence of TCR stimulation. By 6 weeks almost all of the cells lost fluorescent protein marker expression; however, more than 95% of these latently infected cells could be reactivated after stimulation of the TCR by α-CD3/CD28 antibodies. Chromatin immunoprecipitation assays showed that, analogously to Jurkat T cells, latent proviruses in primary CD4+ T cells are enriched in heterochromatic markers, including high levels of CBF-1, histone deacetylases, and methylated histones. Upon TCR activation, there was recruitment of NF-κB to the promoter and conversion of heterochromatin structures present on the latent provirus to active euchromatin structures containing acetylated histones. Surprisingly, latently infected primary cells cannot be induced by tumor necrosis factor alpha because of a restriction in P-TEFb levels, which can be overcome by activation of the TCR. Thus, a combination of restrictive chromatin structures at the HIV long terminal repeat and limiting P-TEFb levels contribute to transcriptional silencing leading to latency in primary CD4+ T cells.
The introduction of highly active antiretroviral therapy (HAART) in the mid 1990s led to a dramatic increase in patient longevity due to the ability of antiretroviral drugs to suppress HIV replication to below threshold detection levels (<50 copies HIV RNA/ml) (23, 52). Unfortunately, despite the intensive therapy, there is continuing viral replication at levels below the limits of detection of most clinical assays due to inefficient antiviral pharmacodynamics that create environments where drug potency is reduced (12, 13, 41, 43). For example, there is recent evidence for ongoing HIV replication in gut-associated lymphoid tissue during long-term antiretroviral therapy (7). A second cause of HIV treatment failure is the creation of a subpopulation of HIV-infected CD4+ T lymphocytes that harbors latent replication-competent proviruses. Since no viral proteins are produced, the latently infected cells cannot be recognized by the antiviral immune response and are highly resistant to antiretroviral therapy. The development of these latent and slowly replicating viral reservoirs during HIV infections has immense practical consequences for treatment of HIV infections because it provides a mechanism that allows the virus to evade immune clearance and the effects of antiviral drugs while still retaining an ability to quickly revert to the productive state upon interruption of drug therapy or in response to cellular activation signals (6, 17).
Multiple complementary mechanisms are required to silence HIV transcription and permit its entry into latency. Although HIV silencing can readily occur in transformed cell lines, several features of the metabolism of resting CD4 cells ensure that latent proviruses remain transcriptionally inactive for long periods. First, a key factor contributing to the restricted transcriptional initiation that is characteristic of HIV transcriptional silencing is the sequestration of the cellular initiation factors NF-κB and NFAT in the cytoplasm of quiescent T cells (28, 37). The second major transcriptional block seen in latently infected cells is the incorporation of the P-TEFb elongation factor into an inactive complex containing HEXIM and 7SK RNA (8, 56). This restricts P-TEFb levels in the cell and creates a block to efficient transcription elongation from the HIV promoter. In addition, posttranscriptional restrictions further reduce HIV gene expression. For example, limiting nuclear levels of the PTB splicing factor in quiescent cells leads to a block to the export of HIV-specific RNA transcripts (32). Finally, miRNAs that inhibit translation of HIV mRNAs may also play an important role in maintaining HIV latency (24, 38).
Entry into latency is also strongly correlated with the recruitment of histone deacetylases (HDACs) to the HIV long terminal repeat (LTR) (9, 50). For example, we have recently demonstrated that CBF-1 (for latency C-promoter binding factor 1), a DNA-binding protein that plays a central role in the Notch signaling pathway, can direct transcriptional silencing of the HIV LTR through recruitment of HDAC-1 (49). The HDACs help to establish restrictive chromatin structures that limit HIV transcriptional initiation and elongation. Additional chromatin restrictions due to histone H3 methylation by histone methyltransferase Suv39H1, lead to the accumulation of HP1 proteins on transcriptionally inactive proviruses (14, 35). We have also been able to monitor the progressive HIV silencing in isolated Jurkat T-cell clones and showed that latency is associated with these chromatin modifications (40).
Although all silenced HIV proviruses appear to acquire restrictive chromatin structures near the viral promoter, the cellular integration site can have a profound influence on the extent of proviral silencing. Viral integration into actively transcribed host genes can led to transcriptional interference caused by the elongating RNA polymerase II (RNAPII) transcribing through the viral promoter (15, 20, 33). Similarly, integration into heterochromatic regions can accelerate proviral silencing (25, 34).
Whereas there is compelling evidence that silencing through histone remodeling is a key feature mediating the establishment of HIV latency, the involvement of DNA methylation is more controversial. Recent studies have shown that proviruses that are poorly responsive to T-cell activation signals also tend to be hypermethylated (26). However, in many silenced HIV clones proviral expression does not correlate with DNA methylation (42). Thus, it seems likely that although DNA methylation is a powerful silencing mechanism for retroviruses, in the natural setting, DNA methylation is not inevitably imposed and partially methylated promoters can still be reactivated (26).
Molecular studies of HIV latency have been severely hampered by the absence of reliable cellular models. The rarity of latently infected cells in patients (less than 1 in 106 resting CD4+ T cells in the peripheral circulation) makes it almost impossible to isolate them in sufficient numbers for biochemical studies (41). As a result, virtually all molecular investigations of HIV latency have involved the use of transformed cell lines, such as the popular Jurkat T-cell line which carries a functional T-cell receptor (TCR) signaling apparatus (25, 30, 40). However, because the quiescent phenotype of the latently infected CD4+ T cells found in vivo is drastically different from the replicating and constitutively activated Jurkat T cells, many laboratories are working to develop more suitable experimental models for HIV latency using primary cells. A particularly informative model system has been developed by Zack and coworkers, who have effectively used the HIV SCID-hu (Thy/Liv) mouse model to recapitulate the generation of latently infected naive T cells during thymopoiesis both in vivo (4) and in vitro (5). Similarly, Cloyd and coworkers have reported that latently infected, quiescent CD4+ T cells can be obtained by cultivating HIV-infected, activated normal CD4+ T lymphocytes on feeder cell layers (45).
In a significant recent set of advances, sophisticated cell culturing techniques have been used to recapitulate T-cell differentiation events in vitro and generate latently infected cells. First, Bosque and coworkers (26) have taken advantage of polarizing conditions to force activated T cells to differentiate and enter quiescence. Similarly, Marini et al. (36) used low doses of interleukin-7 (IL-7) to generate and maintain latently infected memory CD4+ T cells in vitro. Although promising, both methods are of limited use for biochemical analyses of latent proviruses because the yields of viable latently infected cells are low. Greater quantities of latently infected cells have recently been obtained by Yang et al. (53), who used Bcl2 to partially transform primary CD4+ T cells isolated from peripheral blood mononuclear cells (PBMC). We report here a novel ex vivo method that permits, for the first time, the generation of large populations of latently infected primary CD4+ T cells. We used this system to demonstrate that both creation of heterochromatic structures on the HIV provirus and restrictions in P-TEFb levels contribute to the establishment of HIV latency in primary cells.
MATERIALS AND METHODS
Cell culture and lentiviral vectors.
CD4+ primary T cells and H80 cells were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum with or without IL-2 (20 U of recombinant human IL-2 [R&D Systems, Inc.]/ml). The lentiviral vectors pHR′P-PNL-mCherry or pHR′P-PNL-d2EGFP vectors were constructed and pseudotyped HIV particles were packaged as described previously (49).
Isolation, stimulation, and culture of CD4+ T lymphocytes.
The CD4+ cells were either isolated from the blood drawn from healthy donors or were isolated from discarded tonsils. CD4+ T cells were purified by negative selection method using a MACS kit (Miltenyi Biotechnology, Auburn, CA). Purified CD4+ T cells (>98% pure) were stimulated for 4 days in RPMI 1640-10% fetal bovine serum with 25 μl of α-CD3/CD28 antibodies conjugated to magnetic beads (Dynal Biotech) per million cells, along with 20 U of IL-2/ml. One million cells were infected with VSV-G-pseudotyped HIV viruses. After 2 days, the fluorescent cells were purified by fluorescence-activated cell sorting (FACS) and again propagated in the presence of α-CD3/CD28 antibody-conjugated Dynal beads (25 μl/106 cells) and 20 U of IL-2/ml for 2 to 3 weeks. Fresh medium was added every 4 days, and the cultures were maintained at a density of 1.5 × 106 to 2.0 × 106 cells/ml. Once 0.5 × 108 to 1 × 108 cells were obtained they were placed on 30 to 40% confluent H80 cell layers (45). Every 2 to 3 days, half of the culture medium was replaced by fresh IL-2-containing medium, and every 2 weeks the T lymphocytes were transferred to the fresh flasks of H80 feeder cells.
Flow cytometry.
Cells were analyzed for fluorescent reporter gene expression FACSCalibur flow cytometer (Becton Dickinson, Franklin Lakes, NJ). Multicolor flow analysis of cell surface marker expression was performed by using a LSRII flow cytometer. Antibodies were conjugated to the fluorophores indicated in the figure legends. Between 20,000 and 80,000 events were acquired for each antibody and its appropriate isotype control. The data was analyzed with the FlowJo program.
Staining of cells with BrdU and Ki67.
Cells were labeled for 18 h with bromodeoxyuridine (BrdU) using a commercial BrdU incorporation assay protocol (BD Pharmingen). Approximately 3 × 105 labeled cells were permeabilized using 500 μl of Cytoperm (BD Biosciences) for 10 min in the dark at 37°C. After a washing step, the cells were incubated for 30 to 60 min with antibodies to BrdU and α-Ki67 or control antibodies. The unbound antibodies were then removed by washing, and the cells were fixed and resuspended in 100 μl of 1% paraformaldehyde in phosphate-buffered saline plus 200 μl of wash buffer prior to FACS analysis.
ChIP assays.
Chromatin immunoprecipitation (ChIP) was performed as previously described (49). To activate cells, we used either 10 ng of tumor necrosis factor alpha (TNF-α)/ml or 25 μl of α-CD3/CD28 antibodies bound to Dynal beads (Dynal Biotech) per 106 cells. Most antibodies were purchased from Santa Cruz, including anti-RNAPII (N-20), CBF-1(H-50), CIR (C-19), mSIN3A (AK-11), HDAC-1 (H-51), HDAC-2 (H-54), and p65 (C-20). Anti-acetylated histone-3 and histone-4 antibodies were obtained from Upstate.
Western blot analysis.
Western blotting was performed according to standard protocols. Anti-NF-κB, anti-CycT1, and anti-CDK-9 antibodies were obtained from Santa-Cruz. Secondary horseradish peroxidase-conjugated anti-rabbit or anti-mouse antibodies were from Dako.
RESULTS
Efficient generation of pure populations of latently infected CD4+ memory T cells.
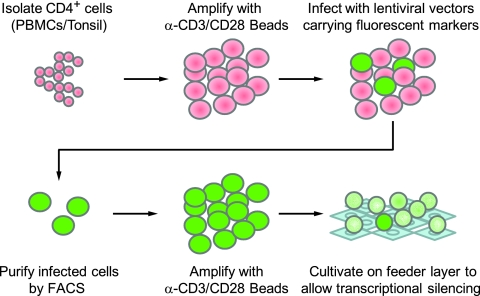
We developed a novel ex vivo model system to study HIV latency in primary CD4+ T cells. As outlined in Fig. 1, pure populations (∼97%) of primary CD4+ T cells were isolated from PBMC by using a negative selection MACS kit from Miltenyi Biotech. The cells were then expanded using α-CD3/CD28 antibody-coated beads in the presence of IL-2. After 4 days, the cells were infected with HIV-derived vectors (Fig. 2A) that express fluorescent protein reporter genes (either the short-lived d2EGFP or mCherry) in place of the nef gene, as previously described (27, 49). Like HIV itself, the viruses included the regulatory proteins Tat and Rev, which provide a positive feedback circuit that enhances HIV transcriptional elongation and export of mRNA from the nucleus. In the majority of our experiments, in order to increase the frequency of latently infected cells in the population, we utilized Tat carrying the H13L mutation. This partially attenuated Tat variant was originally identified in the U1 latently infected cell line (16, 44) and was earlier shown by us to expedite HIV entry into latency (40). The use of an attenuated Tat mutant is consistent with recent studies which have shown that latently infected cells accumulate Tat variants with reduced transactivation potential (55). However, as described below, latently infected cells can also be readily generated by using wild-type Tat.
FIG. 1.
Method for obtaining large populations of latently infected primary CD4+ T cells.
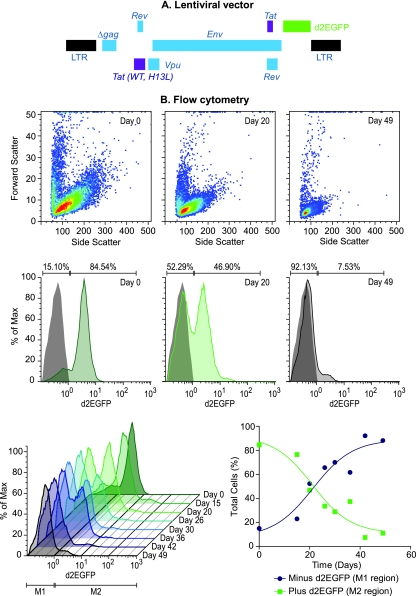
FIG. 2.
Progressive silencing of HIV expression in infected CD4+ T cells. (A) Structure of lentiviral vectors. In some experiments, mCherry was used in place of the d2EGFP fluorescent reporter depicted in this diagram. (B) Flow cytometric analysis of infected cells placed on H80 feeder cells. The top panels show a light scatter analysis of the cell size. During proviral silencing, the activated T cells become quiescent and become reduced in size, as indicated by reductions in both forward and side light scatter. The middle panels display selected histograms showing the fraction of cells expressing d2EGFP at day 0, day 20, and day 49 after cultivation of sorted cells on the H80 feeder cell line. A gray histogram shows uninfected control cells used to set the gates indicated by the bars. The percentage of cells in each gate for the various silenced cell populations is given above the bars. The bottom panels show the time course of proviral silencing during cultivation of CD4+ T cells on H80 feeder cells. Histograms are shown on the left. The graph on the right shows the percent d2EGFP+ cells (green line; >2 × 10°) and the percent d2EGFP− cells (black line; >2 × 10°) for each time point (C).
To obtain a homogeneous population of HIV-infected cells, cells expressing the fluorescent reporters were purified by cell sorting. The purified cells were further expanded with magnetic beads with α-CD3/CD28 antibodies. After 4 to 6 weeks, the cell population reached between 50 × 106 and 100 × 106 cells; the magnetic beads were removed, and the cells were placed on H80 feeder cells in the presence of IL-2, as originally described by Cloyd and coworkers (45).
As shown in Fig. 2B, cultivation of primary T cells infected with viruses carrying the H13L mutation in Tat on the feeder layers led to the progressive loss of HIV gene expression and the entry of the cells into a largely quiescent state characterized by a dramatic reduction in cell size. After 6 weeks, 92.13% of the cells lost virtually all d2EGFP reporter gene expression (Fig. 2B). As shown in Fig. 2B and 8B, comparison of the d2EGFP profiles in the silenced cell population to uninfected control cells shows that the silenced cells display a very low level of fluorescence, suggesting that there are only minimal levels of continuing transcription. Very few of the silenced cells have lost the provirus, since most of them can be efficiently reactivated by stimulation of the TCR (see Fig. 8B). Importantly, the quiescent T cells can be maintained on the H80 feeder cells for more than 6 months without any noticeable loss of viability or reactivation capability.
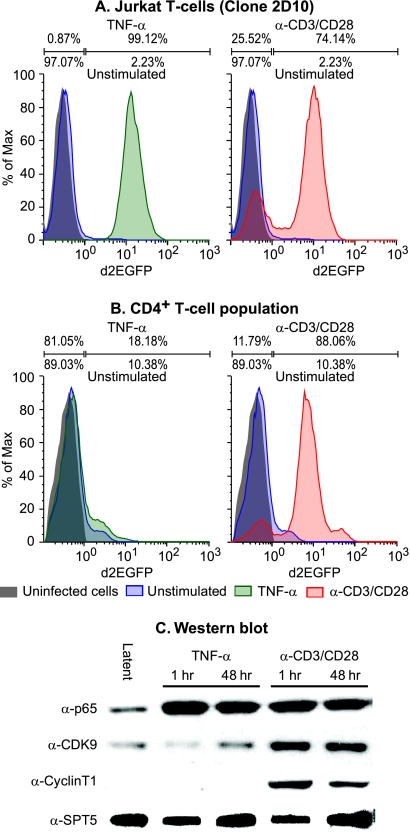
FIG. 8.
Latently infected primary CD4+ T cells have restricted nuclear P-TEFb levels. (A) Latently infected Jurkat T cells (clone 2D10 [40]) were stimulated for 18 h by TNF-α (left) or by α-CD3/CD28 antibodies (right) and analyzed for d2EGFP expression by flow cytometry. The gray histogram shows uninfected control cells used to set the gates, indicated by the bars above the histograms. The percentage of cells in each gate for the activated cell population is given above the bars, and the percentage of cells in each gate for the control unstimulated cell population is given below the bar. (B) CD4+ T cells from PBMC were infected with pHR-p-d2EGFP vector carrying H13L Tat and allowed to enter latency by culturing on H80 feeder cells. The latently infected cell population was then stimulated for 18 h by TNF-α (left) or by α-CD3/CD28 antibodies (right) and analyzed for d2EGFP expression by flow cytometry. The gray histogram shows uninfected control cells used to set the gates, indicated by the bars above the histograms. The percentage of cells in each gate for the activated cell population is given above the bars, and the percentage of cells in each gate for the control unstimulated cell population is given below the bar. (C) Western blotting was performed on nuclear extracts obtained from latently infected cells before and after activating them with either TNF-α or with antibodies against TCR. Antibodies used were: α-NFκB p65, α-CDK9, α-CyclinT1 (CycT1), and α-SPT5. Note that NF-κB p65 is efficiently induced by the both TNF-α and TCR stimulation. In contrast, P-TEFb levels (CDK-9, CycT1) in the nucleus are strongly stimulated by TCR activation but not by TNF-α treatment.
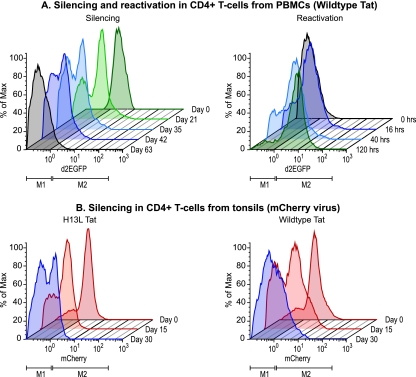
The progressive silencing of HIV gene expression as cells enter quiescence can also be observed with viruses carrying wild-type Tat (Fig. 3A). In this experiment, 86.15% of the cells carrying proviruses became silenced by day 63. After stimulation of the TCR by antibodies to CD3 and CD28, 90.99% of the cells were reactivated. As previously observed in our studies with Jurkat T cells (40), the silencing of proviruses carrying the wild-type Tat is generally slower and less efficient than the silencing of proviruses carrying attenuated Tat genes.
FIG. 3.
Epigenetic silencing of HIV expression CD4+ T cells obtained from PBMC and tonsil tissue. (A) Silencing and reactivation in CD4+ T cells from PBMC. (Left panel) CD4+ T cells isolated from PBMC were infected with viruses carrying wild-type Tat and the d2EGFP reporter. After sorting for d2EGFP+ cells the population was cultivated on H80 feeder cells for up to 63 days. (Right panel) At day 63, the latently cells were reactivated by stimulation of the TCR with α-CD3/CD28 antibodies. During the next 5 days there was a gradual reactivation of the entire latently infected cell population. (B) Silencing in CD4+ T cells from tonsils. CD4+ T cells were isolated from discard tonsils and infected with viruses carrying either H13L Tat (left) or wild-type Tat (right) and the mCherry reporter. After sorting for mCherry+ cells, the populations were cultivated on H80 feeder cells for the next 30 days. During this period there was progressive silencing of the proviruses.
The silencing of HIV proviruses was also observed in CD4+ T cells derived from tonsil tissues infected with viruses carrying either H13L Tat or wild-type Tat and the mCherry fluorescent reporter (Fig. 3B). After 30 days, 52.35% of cells infected with viruses carrying the H13L Tat mutation and 46.79% of cells carrying wild-type Tat were silenced. Because cells derived from tonsil tissues show variable degrees of activation and are slower to enter quiescence than CD4+ T cells isolated from PBMC, the remaining experiments in the present study were performed with PBMC.
Latently infected cells have resting central memory cell phenotype.
In HIV patients undergoing HAART, the vast majority of HIV-infected cells (over 89%) in the peripheral circulation are resting memory CD4+ T cells, although a small but significant fraction of infected cells (3%) have a naive CD4+ T-cell phenotype (2). To document the changes that occur within the population during the expansion and subsequent establishment of the latent virus, the cells were stained with multiple antibodies to cell surface markers and examined using multicolor FACS. The most striking feature of this analysis is that the expansion and culturing on H80 cells results in a more homogeneous population than the CD4+ cells originally isolated from PBMC (Fig. 4).
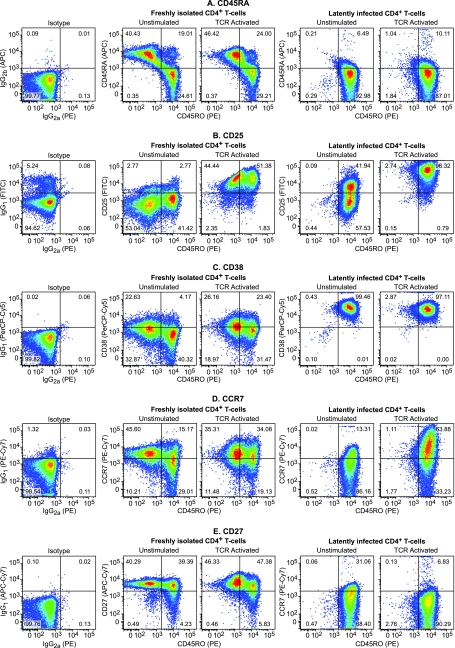
FIG. 4.
Latently infected cells are central resting memory cells. Flow cytometric analysis of CD4+ PBMC and quiescent CD4+ T cells was performed. (A) CD45RA versus CD45RO. Note that the PBMC population contains a mixture of CD45RA+ CD45RO− naive T cells and CD45RA− CD45RO+ memory T cells. (B) CD25 versus CD45RO. (C) CD38 versus CD45RO. (D) CCR7 versus CD45RO. (E) CD27 versus CD45RO.
As shown in Fig. 4A, freshly isolated T cells contain a mixture of naive T cells (40.43%, CD45RA+ CD45RO−), memory cells (24.61%, CD45RO+ CD45RA−), and a small population (19.01%) of dual-positive cells which represent cells that remain in the transitional phase between naive and memory T cells. In contrast, latently infected cells that have been cultured on H80 cells for 6 weeks display a uniform CD45RA− CD45RO+ phenotype (92.98%), indicating that they are primarily resting memory cells (Fig. 4A and Fig. 5B).
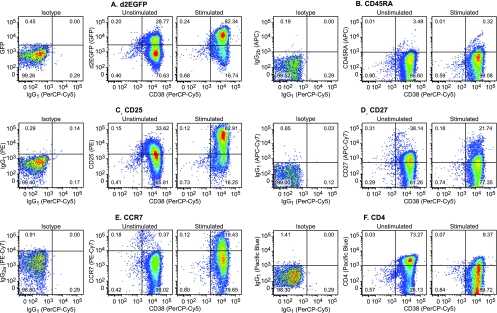
FIG. 5.
Changes in surface marker expression during the activation of cells latently infected with viruses carrying H13L Tat. Primary CD4+ cells harboring latent provirus which had been maintained on the H80 feeder cells were analyzed by multicolor flow cytometry before and after activation for 18 h with α-CD3/CD28 antibodies. (A) d2EGFP (GFP) versus CD38 (PerCP-Cy5). (B) CD45RA (APC) versus CD38 (PerCP-Cy5). (C) CD25 (PE) versus CD38 (PerCP-Cy5). (D) CD27 (APC-Cy7) versus CD38 (PerCP-Cy5). (E) CCR7 (PE-Cy7) versus CD38 (PerCP-Cy5) (F) CD4 (Pacific Blue) versus CD38 (PerCP-Cy5). Note that CD4 is downregulated while d2EGFP, CD25, and CCR7 are upregulated after proviral activation.
To further define the phenotypes of the latently infected cells, we performed multicolor flow cytometric analysis (FACS) using antibodies for a wide range of different cell surface protein markers (Fig. 4, 5, and 6). The cells shown in Fig. 4 and 5 were infected with viruses carrying the H13L Tat gene.
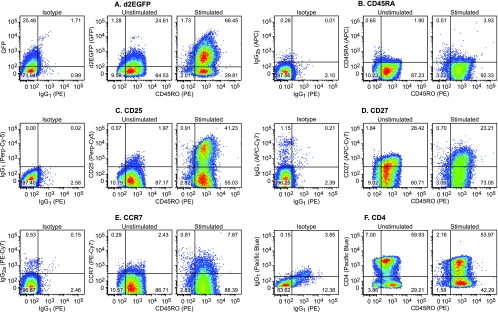
FIG. 6.
Changes in surface marker expression during the activation of cells latently infected with viruses carrying wild-type Tat. Primary CD4+ cells infected with viruses carrying wild-type Tat were allowed to enter latency by cultivation on H80 feeder cells for over 60 days. Cells harboring latent provirus were analyzed by multicolor flow cytometry before and after activation for 16 h with α-CD3/CD28 antibodies. (A) d2EGFP (GFP) versus CD45RO (PE). (B) CD45RA (APC) versus CD45RO (PE). (C) CD25 (Perp-Cy5) versus CD45RO (PE). (D) CD27 (APC-Cy7) versus CD45RO (PE). (E) CCR7 (PE-Cy7) versus CD45RO (PE). (F) CD4 (Pacific Blue) versus CD45RO (PE). Note that the latently infected cells constitutively express CD45RO, CD38, and CD27. The cells used in this experiment were from a different donor than the cells used in the experiment in Fig. 5.
As shown in Fig. 5A, 70.63% of the latently infected cells used in these experiments did not express the d2EGFP marker and the remaining 28.77% expressed only low levels of d2EGFP. However, 82.34% of the cells resumed HIV transcription and expressed high levels of d2EGFP after stimulation of the TCR.
CD38 is only expressed at very low levels on naive T cells and unactivated memory T cells, but is present at high levels on both activated T cells and mature thymocytes (11). Consistent with these observations, we observed that freshly isolated naive and memory cells express only low levels of CD38 (Fig. 4C). In contrast, the latently infected memory (CD45RA− CD45RO+) positive cells showed uniformly high levels of CD38 (Fig. 4C, and horizontal axis, Fig. 5A to F).
Activation of the cells through the TCR also resulted in the upregulation of the IL-2 receptor (CD25). In the experimental results shown in Fig. 4B, 96.32% of the cells upregulated CD25, whereas 82.91% of the cells were activated in the experimental results shown in Fig. 5C.
CD27 (Fig. 4E and 5D) is an important T-cell activation and expansion marker, which is constitutively expressed on central memory T cells and strongly upregulated after TCR activation of naive cells (10). Upon acquisition of effector functions, the levels of CD27 on central memory cells declines as CD27 becomes soluble and detaches from the cell surface (18). CD27 has also been linked with T-cell survival during expansion (21). In the latently infected population of cells, there is a low level of CD27 on the cells surface that is modestly reduced after TCR activation (Fig. 4E and 5D). This is consistent with the identification of the latently infected cells as resting central memory cells.
The CCR7 receptors are the hallmark of activated central memory cells since CCR7 enhances the retention of these cells by lymph nodes (46). In contrast, effector memory cells do not express CCR7 but do express other receptors that stimulate migration to inflamed tissues (46). As shown in Fig. 4D and 5E, CCR7 is upregulated on the latently infected cells, which is again consistent with their identification as a resting central memory cell population.
Finally, as d2EGFP levels increase (Fig. 5A), there is a concomitant decrease in levels of CD4 (Fig. 5F). The downregulation of CD4 is probably due to the increased expression of HIV proteins (Vpu and Env) from the latent proviruses.
Very similar patterns of surface marker expression were observed in cells obtained from a second donor that were infected with viruses carrying the wild-type Tat gene (Fig. 6). In this cell population, 96.38% of the infected cells did not express the d2EGFP marker, and the remainder only expressed low levels of d2EGFP. However, 66.45% of the cells became strongly activated after stimulation of the TCR (Fig. 6A). As in the previous experiment, the latently infected cells are all CD45RA− CD45RO+ (Fig. 6B) and expressed low levels of CD27 (Fig. 6D) and CCR7 (Fig. 6E), both of which became somewhat upregulated after TCR activation. The cells also showed strong upregulation of CD25 (Fig. 6C) and strong downregulation of CD4 upon stimulation of the TCR (Fig. 6F).
Latently infected cells show reduced DNA synthesis and cell proliferation.
To analyze the proliferation capability of the quiescent cells, we compared the incorporation of BrdU into cellular DNA before and after activating them with anti-CD3/CD28 antibodies (Fig. 7A) (29). For control cells we utilized freshly isolated, but not previously activated, CD4+ T cells isolated from PBMC. In parallel, we also performed intracellular staining for the nuclear antigen Ki67 (Fig. 7B) and CCR7 (Fig. 7C). The IL-2 receptor chain, CD25, was included as an additional dimension for the flow cytometry. Mock-infected cells were used for these experiments to avoid interference of the d2EGFP signal expressed by the lentiviral vectors with the fluorescein isothiocyanate (FITC)-labeled antibodies utilized in the BrdU incorporation assay.
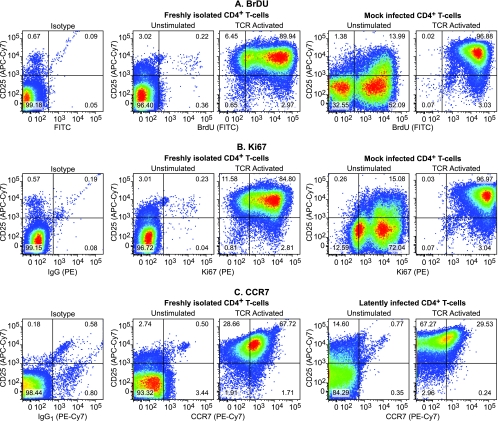
FIG. 7.
CD4+ cells show restricted DNA synthesis after cultivation on H80 feeder cell lines. (A) The proliferation capability of freshly isolated CD4+ T cells from PBMC and mock-infected cells that had been cultivated on the H80 feeder cell line for more than 60 days was analyzed by measuring BrdU nucleotide incorporation into cellular DNA (FITC) before and after activating them with α-CD3/CD28 antibodies. Labeling with BrdU was done for 18 h. The data are displayed against a second cell activation marker, CD25 (APC-Cy7). (B) Ki67 (PE) expression after activation of quiescent T cells. (C) CCR7 expression after activation of the quiescent T cells.
As shown in Fig. 7, during cultivation on the H80 feeder cells, the majority of the HIV-infected cells show strongly reduced CD25 expression. In order to look for cells in the population that were undergoing low levels of DNA synthesis, we labeled the cells for 18 h with BrdU, an unusually long labeling period compared to the more typical 2-h “pulse” that is used in cell cycle studies. Even under these conditions, in the latently infected cell population, 32.55% of the cells failed to incorporate any BrdU (Fig. 7A). The remaining cells showed measurable levels of BrdU incorporation. As expected, after TCR activation 96.88% of the latently infected cells incorporated BrdU. Furthermore, the level of BrdU incorporation increased 10-fold (Fig. 7A). As shown in Fig. 7B, the latently infected cell population also had moderate levels of Ki67 expression compared to freshly isolated and unstimulated CD4+ T-cell controls. Activation of latently infected cells through the TCR resulted in the strong upregulation of both Ki67 and CD25 in over 96% of the cells (Fig. 7B). Thus, the latently infected cells appear to be replicating slowly, but they can be readily reactivated. As previously noted, CCR7 expression is also upregulated when these cells were activated (Fig. 7C).
NF-κB is necessary but not sufficient to activate latently infected primary CD4+ T cells.
Work by Zack and coworkers has shown that, in contrast to Jurkat cells, induction of NF-κB is necessary but not sufficient to induce latent proviruses in naive primary T cells (3). Similarly, we have found that TNF-α is unable to induce latent proviruses in central memory T cells (Fig. 8B), although it is a potent inducer of proviral expression in latently infected Jurkat T cells (Fig. 8A). In contrast, as described above, activation of the TCR strongly activates HIV transcription in both Jurkat T cells (Fig. 8A) and latently infected primary CD4+ T cells (Fig. 8B). To confirm that NF-κB was activated in the primary cells by both TNF-α and activation of the TCR, we performed Western blots to measure the nuclear levels of the NF-κB p65 subunit. As shown in Fig. 8C, nuclear levels of p65 increase >10-fold after exposure of the quiescent cells to either activator.
We next examined whether P-TEFb levels are limiting in these cells, as was originally reported by Rice and coworkers for freshly isolated PMBC (22). As shown in Fig. 8C, nuclear extracts of the latent cells show very low levels of CDK-9 and CycT1. There is no increase in the nuclear levels of CDK-9 and CycT1 after exposure to TNF-α. In contrast, activation of the TCR leads to a >5-fold increase in CDK-9 and a >50-fold increase in CycT1. This strongly suggests that P-TEFb levels are limiting in the latently infected memory cells and that both NF-κB and P-TEFb have to be activated in order to induce proviral transcription at both initiation and elongation steps of transcription.
Latent proviruses in primary CD4+ cells have heterochromatic structures.
Extensive studies in transformed cell systems have shown that latent HIV proviruses typically contain high levels of histone deacetylases and heterochromatic structures (9, 14, 35, 49, 50). It is believed that these chromatin modifications are used to restrict transcription initiation and thus promote viral entry into latency (49). In primary cells, entry into latency is associated with differentiation events, leading to entry of cells into a quiescent state (31). Because it is not known whether some, or all, of the chromatin silencing mechanisms seen in the transformed cells are also observed in primary cells, we used ChIP assays to study the chromatin of latent HIV proviruses in our primary CD4+ T-cell model.
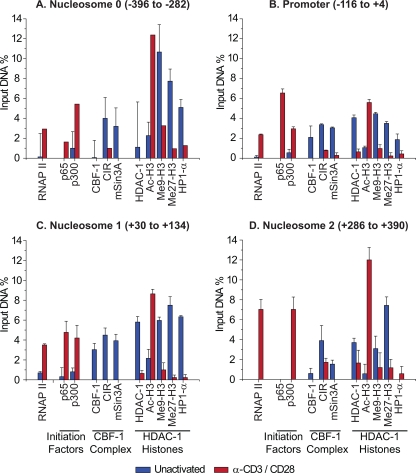
As shown in Fig. 9B, the latently infected CD4+ T cells have low, but detectable, levels of RNAP II at the promoter region of the LTR (−116 to +4). Similarly to Jurkat T cells, HDAC-1 is also present at high levels, whereas the levels of acetylated histone H3 were very low. We have previously reported that CBF-1 acts as an effective recruiter of HDACs to the HIV LTR (49). As shown in Fig. 9B, CBF-1 and its cofactors CIR and mSin3A are all present on the latent proviruses in primary cells. Induction of the latently infected cells by treating them with α-CD3/CD28 antibodies increased RNAP II levels at the promoter by >7-fold. In contrast, the levels of the repressors CBF-1, CIR, mSIN3A, and HDAC-1 decreased substantially. As expected, treatment of latent primary CD4+ lymphocytes with α-CD3/CD28 antibodies resulted in NF-κB p65 and histone acetyltransferase p300 recruitment to the promoter. After TCR activation there was a 4- to 7-fold increase in acetylated histone H3 present at the provirus. The latent proviruses also carry heterochromatic markers at the promoter region of the LTR, including trimethylated histone H3 at positions K9 and K27 (Fig. 9B). HP1-α, a heterochromatic protein that binds exclusively to trimethyl-K9-H3 histones, is also present at the HIV LTR (14). After TCR activation, there were significant decreases in the levels of trimethylated histone H3 (K9 and K27) and in the HP1-α repressor protein, whereas the levels of acetylated histones rise at the HIV LTR.
FIG. 9.
Fluctuations in the levels of different transcription and chromatin-associated factors, before and after activation of latently infected primary CD4+ T cells with α-CD3/CD28 antibodies. ChIP analysis was performed on primary cells latently infected with proviruses carrying H13L Tat and the d2EGFP marker. Antibodies used for the analysis included RNAP II (N20), transcription initiation factors (p65 and p300), CBF-1 repressor complex (CBF-1, CIR, and Sin3A), and histones and chromatin-modifying proteins (HDAC-1, acetylated histone H3, trimethyl-lysine-9-histone H3, trimethyl-lysine-27-histone H3, and HP-1α). (A) Primers directed to the nucleosome 0 region (−396 to −282). (B) Promoter region (−116 to +4). (C) Nucleosome 1 (+30 to +134). (D) Nucleosome 2 (+286 to +390). Blue bars, latently infected cells; red bars, cells after 30 min of treatment with α-CD3/CD28 antibodies.
Similar results were obtained by using primers directed to the upstream nucleosome 0 region of the LTR (Fig. 9A) and the downstream nucleosome 1 (Fig. 9C) and nucleosome 2 (Fig. 9D) regions. Because the resolution of the ChIP assay is between 500 and 700 bp, there is some signal overlap between each of these adjacent regions. Nonetheless, some important differences in protein distribution can be observed. The most notable difference is that there are much higher levels of histones present in the nucleosome 0 and 1 regions than at the promoter of the latent proviruses. These observations are consistent with previous studies with transformed cell lines that demonstrated that the HIV promoter region is relatively devoid of histones (51). The data also demonstrate that modified histones are present both upstream and downstream of the transcription start site. In contrast, as expected, p65 is largely restricted to the promoter region (Fig. 9D).
DISCUSSION
The recognition that an extremely stable HAART-resistant reservoir of latent HIV is present in most patients has focused attention on the molecular mechanisms governing HIV latency. Unfortunately, both because it is difficult to isolate large numbers of latently infected cells from patient samples and because there have been problems in developing culture conditions that allow study of HIV latency in primary cells, virtually all molecular investigations of HIV latency have involved the use of transformed cell lines, such as Jurkat T cells. These studies have suggested that there are many close parallels in the behavior of the latent proviruses in transformed cells and primary cells. For example, in both cell types, latent proviruses are preferentially integrated into actively transcribed genes (19, 34) and the addition of HDAC inhibitors induces latent proviruses, suggesting that chromatin restrictions play a universal role in maintaining latency (54). However, because the quiescent phenotype of the infected CD4+ T cells that make up the bulk of the latent reservoir is drastically different from the constitutively activated Jurkat T cells, important differences in the viral reactivation and shutdown pathways are also likely to exist. Thus, there was a pressing need to develop reliable and biochemically tractable model systems to study HIV latency in primary CD4+ T cells.
Recent reports have demonstrated that it is possible to generate latent HIV infection in primary human CD4+ T cells using in vitro maturation (5), coculture (45), spinoculation (48), or manipulation of the interleukin levels (26, 36). Although these are promising developments, none of these methods yield sufficiently large quantities of latently infected cells to perform extensive biochemical analyses.
A frequently encountered problem in attempts to generate large homogeneous populations of latently infected primary T cells is that infected T cells rapidly enter apoptosis once TCR signaling is interrupted. This leads to massive cell loss during in vitro T-cell differentiation by specific cytokine exposure regimes (26, 36). Consequently, the recent ex vivo systems used to generate latently infected primary cells typically only yield less than 20% viable cells at the end of resting phase. Furthermore, in both the method of Bosque and coworkers (26) and the method of Marini (36) a low frequency of HIV infection results in less than 10% of the recovered cell populations carrying proviruses. The Siliciano laboratory has been exploring a potential solution to this problem through the transduction of Bcl-2, an antiapoptotic factor that allows the primary CD4+ T cells to survive in a quiescent state in vitro (53). Although promising, this strategy raises the concern that overexpression of Bcl-2 may alter the normal physiology of primary resting CD4+ T cells. Similarly, the use of IL-7 in the method of Marini et al. (36) raises concerns that these cells may remain partially activated, since IL-7 is known to be a potent inducer of latent HIV (47).
We have described here a novel primary CD4+ T-cell-based model system that allows us to obtain large populations of pure, latently infected, cells. The model is based on our previous observation that lentiviral vectors carrying an attenuated Tat mutation (H13L) rapidly and efficiently enter latency. In adopting this strategy for use with primary cells, a key step was to utilize the feeder layer system established by Sahu et al. (45) to maintain infected CD4+ T cells in the absence of TCR stimulation.
Our method has proven to be highly reproducible and robust during the last 2 years. During the course of this work, we have produced latently infected cells from six different donors using four different viral vectors carrying either wild-type or H13L Tat and either the d2EGFP or mCherry fluorescent reporters. In general, we have found that it was useful to include the attenuated H13L Tat mutant in our experiments, since viruses carrying attenuated Tat enter latency more efficiently than viruses carrying the wild-type Tat. It is therefore not surprising that recent viral isolation studies have shown that latently infected cells obtained from patients tend to accumulate Tat variants with reduced transactivation potential (55).
Extensive multicolor flow cytometric analysis of the latently infected T cells shows that they represent a very homogeneous population of cells that is CD45RA−, CD45RO+, CD38+, CD25−, CD27+, and CCR7+. This combination of surface markers suggests that these cells represent a central memory T-cell population (39). The cells are largely, but not completely, quiescent since although they do not divide, they are still able to incorporate BrdU and express moderate levels of Ki67. This partially activated phenotype may be a consequence of the IL-2 present in the culture medium, which is needed to maintain cell viability.
A striking result from these studies is that, just as we have previously observed in Jurkat T cells, there is progressive epigenetic shutdown of HIV transcription as the virus enters latency. Using ChIP assays we have shown that latent proviruses recruit HDACs via CBF-1 and its cofactors CIR and mSIN3A. We were also able to demonstrate that the latent HIV LTR contains stable heterochromatic structures. Histone H3 trimethylated at positions K9 and K27 and the HP-1α protein are all present. These results are consistent with the hypothesis that establishment of restrictive heterochromatic structures on the latent HIV provirus involves a series of sequential events starting with the recruitment of HDACs to the promoter via CBF-1, followed by methylation of histones by Suv39H1 and binding of the HP proteins to the methylated histones (14, 35, 49). Since the ChIP experiments were performed with mixed populations of cells that contained thousands of separate integration events, rather than clones, we can conclude that the epigenetic silencing we observed is due to recognition of specific features of the HIV proviral genome rather than a consequence of integration into a specific site.
As in the Jurkat T-cell models, activation of latently infected primary CD4+ lymphocyte by stimulation of the T-cell receptor using α-CD3/CD28 antibodies reverses these chromatin modifications. After TCR activation, there are significant reductions in the levels of trimethylated histone H3 (K9 and K27) and a concomitant fall in the levels of HP1 repressor protein. TCR activation also resulted in the reversal of the acetylation status of histones represented by the higher presence of acetylated histone 3 at the LTR. However, the reactivation of latent proviruses in primary cells is more complex than proviral reactivation in Jurkat T cells. Most importantly, although TNF-α is an extremely potent inducer of latent proviruses in Jurkat T cells, it is totally ineffective in primary cells. The failure of TNF-α to activate latent proviruses in primary cells appears to be due to limiting quantities of free P-TEFb, which we have measured by examining the levels of CDK-9 and CycT1 present in nuclear extracts. Under the extraction conditions we have used, the large, inactive form of P-TEFb is found in the cytoplasmic fraction (1). Consistent with observations made in freshly isolated PMBCs (22), activation of the cells through the TCR produces a dramatic upregulation of P-TEFb. In our experiments, this activation of P-TEFb is demonstrated by the >20-fold increase in the levels of CDK-9 and CycT1 in the nuclear extracts after activation of latently infected cells by antibodies to CD3 and CD28.
In summary, the model system we have described presents exciting opportunities to study specific molecular aspects of both the entry and exit from HIV latency. Using our method, latently infected cells carrying informative mutations in the viral LTR and regulatory genes can be readily produced. Furthermore, since we have been able to generate pure populations of latently infected primary T cells in amounts suitable for biochemical studies, our model should facilitate the screening of drug candidates as part of the effort to develop new therapeutic approaches designed to eliminate latent HIV reservoirs.
Acknowledgments
We thank our coworkers Scott Sieg and Douglas A. Bazdar for assistance in the multicolor flow cytometry, and we thank laboratory members Joseph Hokello, Uri Mbonye, Julian Wong, Julia Friedman, Julie Jadlowsky, Amzie Pavlisin, Kara Lassen, and Zafeiria Athanasiou for their help and fruitful discussions. Darell Bigner (Duke University) and Miles Cloyd (UT Galveston) kindly provided the H80 cells. We also thank the CWRU/UH Center for AIDS Research (P30-AI036219) for the provision of flow cytometry services.
This study was supported by NIH grants R01-AI067093 and DP1-DA028869 to J.K.
Footnotes
▿
Published ahead of print on 21 April 2010.
REFERENCES
- 1.Biglione, S., S. A. Byers, J. P. Price, V. T. Nguyen, O. Bensaude, D. H. Price, and W. Maury. 2007. Inhibition of HIV-1 replication by P-TEFb inhibitors DRB, seliciclib, and flavopiridol correlates with release of free P-TEFb from the large, inactive form of the complex. Retrovirology 4**:**47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenchley, J. M., B. J. Hill, D. R. Ambrozak, D. A. Price, F. J. Guenaga, J. P. Casazza, J. Kuruppu, J. Yazdani, S. A. Migueles, M. Connors, M. Roederer, D. C. Douek, and R. A. Koup. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J. Virol. 78**:**1160-1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks, D. G., P. A. Arlen, L. Gao, C. M. Kitchen, and J. A. Zack. 2003. Identification of T cell-signaling pathways that stimulate latent HIV in primary cells. Proc. Natl. Acad. Sci. U. S. A. 100**:**12955-12960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks, D. G., S. G. Kitchen, C. M. Kitchen, D. D. Scripture-Adams, and J. A. Zack. 2001. Generation of HIV latency during thymopoiesis. Nat. Med. 7**:**459-464. [DOI] [PubMed] [Google Scholar]
- 5.Burke, B., H. J. Brown, M. D. Marsden, G. Bristol, D. N. Vatakis, and J. A. Zack. 2007. Primary cell model for activation-inducible human immunodeficiency virus. J. Virol. 81**:**7424-7434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chun, T. W., R. T. Davey, Jr., D. Engel, H. C. Lane, and A. S. Fauci. 1999. Re-emergence of HIV after stopping therapy. Nature 401**:**874-875. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T. W., D. C. Nickle, J. S. Justement, J. H. Meyers, G. Roby, C. W. Hallahan, S. Kottilil, S. Moir, J. M. Mican, J. I. Mullins, D. J. Ward, J. A. Kovacs, P. J. Mannon, and A. S. Fauci. 2008. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 197**:**714-720. [DOI] [PubMed] [Google Scholar]
- 8.Contreras, X., M. Barboric, T. Lenasi, and B. M. Peterlin. 2007. HMBA releases P-TEFb from HEXIM1 and 7SK snRNA via PI3K/Akt and activates HIV transcription. PLoS Pathog. 3**:**1459-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coull, J. J., F. Romerio, J. M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74**:**6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Jong, R., W. A. Loenen, M. Brouwer, L. van Emmerik, E. F. de Vries, J. Borst, and R. A. van Lier. 1991. Regulation of expression of CD27, a T cell-specific member of a novel family of membrane receptors. J. Immunol. 146**:**2488-2494. [PubMed] [Google Scholar]
- 11.Dianzani, U., A. Funaro, D. DiFranco, G. Garbarino, M. Bragardo, V. Redoglia, D. Buonfiglio, L. B. De Monte, A. Pileri, and F. Malavasi. 1994. Interaction between endothelium and CD4+ CD45RA+ lymphocytes: role of the human CD38 molecule. J. Immunol. 153**:**952-959. [PubMed] [Google Scholar]
- 12.Dinoso, J. B., S. Y. Kim, A. M. Wiegand, S. E. Palmer, S. J. Gange, L. Cranmer, A. O'Shea, M. Callender, A. Spivak, T. Brennan, M. F. Kearney, M. A. Proschan, J. M. Mican, C. A. Rehm, J. M. Coffin, J. W. Mellors, R. F. Siliciano, and F. Maldarelli. 2009. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc. Natl. Acad. Sci. U. S. A. 106**:**9403-9408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornadula, G., H. Zhang, B. VanUitert, J. Stern, L. Livornese, Jr., M. J. Ingerman, J. Witek, J. R. Kedanis, J. Natkin, J. DeSimone, and R. J. Pomerantz. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282**:**1627-1632. [DOI] [PubMed] [Google Scholar]
- 14.du Chene, I., E. Basyuk, Y. L. Lin, R. Triboulet, A. Knezevich, C. Chable-Bessia, C. Mettling, V. Baillat, J. Reynes, P. Corbeau, E. Bertrand, A. Marcello, S. Emiliani, R. Kiernan, and M. Benkirane. 2007. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 26**:**424-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duverger, A., J. Jones, J. May, F. Bibollet-Ruche, F. A. Wagner, R. Q. Cron, and O. Kutsch. 2009. Determinants of the establishment of human immunodeficiency virus type 1 latency. J. Virol. 83**:**3078-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emiliani, S., W. Fischle, M. Ott, C. van Lint, C. A. Amella, and E. Verdin. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72**:**1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5**:**512-517. [DOI] [PubMed] [Google Scholar]
- 18.Hamann, D., P. A. Baars, M. H. Rep, B. Hooibrink, S. R. Kerkhof-Garde, M. R. Klein, and R. A. van Lier. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186**:**1407-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, X. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78**:**6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han, Y., Y. B. Lin, W. An, J. Xu, H. C. Yang, K. O'Connell, D. Dordai, J. D. Boeke, J. D. Siliciano, and R. F. Siliciano. 2008. Orientation-dependent regulation of integrated HIV-1 expression by host gene transcriptional readthrough. Cell Host Microbe 4**:**134-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hendriks, J., L. A. Gravestein, K. Tesselaar, R. A. van Lier, T. N. Schumacher, and J. Borst. 2000. CD27 is required for generation and long-term maintenance of T-cell immunity. Nat. Immunol. 1**:**433-440. [DOI] [PubMed] [Google Scholar]
- 22.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase, TAK, activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72**:**9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373**:**123-126. [DOI] [PubMed] [Google Scholar]
- 24.Huang, J., F. Wang, E. Argyris, K. Chen, Z. Liang, H. Tian, W. Huang, K. Squires, G. Verlinghieri, and H. Zhang. 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat. Med. 13**:**1241-1247. [DOI] [PubMed] [Google Scholar]
- 25.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22**:**1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kauder, S. E., A. Bosque, A. Lindqvist, V. Planelles, and E. Verdin. 2009. Epigenetic regulation of HIV-1 latency by cytosine methylation. PLoS Pathog. 5**:**e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, Y. K., C. F. Bourgeois, R. Pearson, M. Tyagi, M. J. West, J. Wong, S. Y. Wu, C. M. Chiang, and J. Karn. 2006. Recruitment of TFIIH to the HIV LTR is a rate-limiting step in the emergence of HIV from latency. EMBO J. 25**:**3596-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kinoshita, S., L. Su, M. Amano, L. A. Timmerman, H. Kaneshima, and G. P. Nolan. 1997. The T-cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity 6**:**235-244. [DOI] [PubMed] [Google Scholar]
- 29.Kovacs, J. A., R. A. Lempicki, I. A. Sidorov, J. W. Adelsberger, B. Herpin, J. A. Metcalf, I. Sereti, M. A. Polis, R. T. Davey, J. Tavel, J. Falloon, R. Stevens, L. Lambert, R. Dewar, D. J. Schwartzentruber, M. R. Anver, M. W. Baseler, H. Masur, D. S. Dimitrov, and H. C. Lane. 2001. Identification of dynamically distinct subpopulations of T lymphocytes that are differentially affected by HIV. J. Exp. Med. 194**:**1731-1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76**:**8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lassen, K., Y. Han, Y. Zhou, J. Siliciano, and R. F. Siliciano. 2004. The multifactorial nature of HIV-1 latency. Trends Mol. Med. 10**:**525-531. [DOI] [PubMed] [Google Scholar]
- 32.Lassen, K. G., K. X. Ramyar, J. R. Bailey, Y. Zhou, and R. F. Siliciano. 2006. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2**:**e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lenasi, T., X. Contreras, and B. M. Peterlin. 2008. Transcriptional interference antagonizes proviral gene expression to promote HIV latency. Cell Host Microbe 4**:**123-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewinski, M. K., D. Bisgrove, P. Shinn, H. Chen, C. Hoffmann, S. Hannenhalli, E. Verdin, C. C. Berry, J. R. Ecker, and F. D. Bushman. 2005. Genome-wide analysis of chromosomal features repressing human immunodeficiency virus transcription. J. Virol. 79**:**6610-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marban, C., S. Suzanne, F. Dequiedt, S. de Walque, L. Redel, C. Van Lint, D. Aunis, and O. Rohr. 2007. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 26**:**412-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marini, A., J. M. Harper, and F. Romerio. 2008. An in vitro system to model the establishment and reactivation of HIV-1 latency. J. Immunol. 181**:**7713-7720. [DOI] [PubMed] [Google Scholar]
- 37.Nabel, G., and D. A. Baltimore. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326**:**711-713. [DOI] [PubMed] [Google Scholar]
- 38.Nathans, R., C. Y. Chu, A. K. Serquina, C. C. Lu, H. Cao, and T. M. Rana. 2009. Cellular microRNA and P bodies modulate host-HIV-1 interactions. Mol. Cell 34**:**696-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Okada, R., T. Kondo, F. Matsuki, H. Takata, and M. Takiguchi. 2008. Phenotypic classification of human CD4+ T-cell subsets and their differentiation. Int. Immunol. 20**:**1189-1199. [DOI] [PubMed] [Google Scholar]
- 40.Pearson, R., Y. K. Kim, J. Hokello, K. Lassen, J. Friedman, M. Tyagi, and J. Karn. 2008. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 82**:**12291-12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18**:**665-708. [DOI] [PubMed] [Google Scholar]
- 42.Pion, M., A. Jordan, A. Biancotto, F. Dequiedt, F. Gondois-Rey, S. Rondeau, R. Vigne, J. Hejnar, E. Verdin, and I. Hirsch. 2003. Transcriptional suppression of in vitro-integrated human immunodeficiency virus type 1 does not correlate with proviral DNA methylation. J. Virol. 77**:**4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6**:**82-85. [DOI] [PubMed] [Google Scholar]
- 44.Reza, S. M., L.-M. Shen, R. Mukhopadhyay, M. Rosetti, T. Pe'ery, and M. B. Mathews. 2003. A naturally occurring substitution in human immunodeficiency virus Tat increases expression of the viral genome. J. Virol. 77**:**8602-8606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sahu, G. K., K. Lee, J. Ji, V. Braciale, S. Baron, and M. W. Cloyd. 2006. A novel in vitro system to generate and study latently HIV-infected long-lived normal CD4+ T lymphocytes. Virology 355**:**127-137. [DOI] [PubMed] [Google Scholar]
- 46.Sallusto, F., D. Lenig, R. Forster, M. Lipp, and A. Lanzavecchia. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 401**:**708-712. [DOI] [PubMed] [Google Scholar]
- 47.Scripture-Adams, D. D., D. G. Brooks, Y. D. Korin, and J. A. Zack. 2002. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J. Virol. 76**:**13077-13082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swiggard, W. J., C. Baytop, J. J. Yu, J. Dai, C. Li, R. Schretzenmair, T. Theodosopoulos, and U. O'Doherty. 2005. Human immunodeficiency virus type 1 can establish latent infection in resting CD4+ T cells in the absence of activating stimuli. J. Virol. 79**:**14179-14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyagi, M., and J. Karn. 2007. CBF-1 promotes transcriptional silencing during the establishment of HIV-1 latency. EMBO J. 26**:**4985-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15**:**1112-1120. [PMC free article] [PubMed] [Google Scholar]
- 51.Verdin, E., P. J. Paras, and C. Van Lint. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12**:**3249-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373**:**117-122. [DOI] [PubMed] [Google Scholar]
- 53.Yang, H. C., S. Xing, L. Shan, K. O'Connell, J. Dinoso, A. Shen, Y. Zhou, C. K. Shrum, Y. Han, J. O. Liu, H. Zhang, J. B. Margolick, and R. F. Siliciano. 2009. Small-molecule screening using a human primary cell model of HIV latency identifies compounds that reverse latency without cellular activation. J. Clin. Invest. 119**:**3473-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ylisastigui, L., N. M. Archin, G. Lehrman, R. J. Bosch, and D. M. Margolis. 2004. Coaxing HIV-1 from resting CD4 T cells: histone deacetylase inhibition allows latent viral expression. AIDS 18**:**1101-1108. [DOI] [PubMed] [Google Scholar]
- 55.Yukl, S., S. Pillai, P. Li, K. Chang, W. Pasutti, C. Ahlgren, D. Havlir, M. Strain, H. Gunthard, D. Richman, A. P. Rice, E. Daar, S. Little, and J. K. Wong. 2009. Latently infected CD4+ T cells are enriched for HIV-1 Tat variants with impaired transactivation activity. Virology 387**:**98-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, Q., and J. H. Yik. 2006. The Yin and Yang of P-TEFb regulation: implications for human immunodeficiency virus gene expression and global control of cell growth and differentiation. Microbiol. Mol. Biol. Rev. 70**:**646-659. [DOI] [PMC free article] [PubMed] [Google Scholar]