Two Nedd4-binding Motifs Underlie Modulation of Sodium Channel Nav1.6 by p38 MAPK (original) (raw)
Abstract
Sodium channel Nav1.6 is essential for neuronal excitability in central and peripheral nervous systems. Loss-of-function mutations in Nav1.6 underlie motor disorders, with homozygous-null mutations causing juvenile lethality. Phosphorylation of Nav1.6 by the stress-induced p38 MAPK at a Pro-Gly-Ser553-Pro motif in its intracellular loop L1 reduces Nav1.6 current density in a dorsal root ganglion-derived cell line, without changing its gating properties. Phosphorylated Pro-Gly-Ser553-Pro motif is a putative binding site to Nedd4 ubiquitin ligases, and we hypothesized that Nedd4-like ubiquitin ligases may contribute to channel ubiquitination and internalization. We report here that p38 activation in hippocampal neurons from wild-type mice, but not from Scn8amedtg mice that lack Nav1.6, reduces tetrodotoxin-S sodium currents, suggesting isoform-specific modulation of Nav1.6 by p38 in these neurons. Pharmacological block of endocytosis completely abolishes p38-mediated Nav1.6 current reduction, supporting our hypothesis that channel internalization underlies current reduction. We also report that the ubiquitin ligase Nedd4-2 interacts with Nav1.6 via a Pro-Ser-Tyr1945 motif in the C terminus of the channel and reduces Nav1.6 current density, and we show that this regulation requires both the Pro-Gly-Ser-Pro motif in L1 and the Pro-Ser-Tyr motif in the C terminus. Similarly, both motifs are necessary for p38-mediated reduction of Nav1.6 current, whereas abrogating binding of the ubiquitin ligase Nedd4-2 to the Pro-Ser-Tyr motif results in stress-mediated increase in Nav1.6 current density. Thus, phosphorylation of the Pro-Gly-Ser-Pro motif within L1 of Nav1.6 is necessary for stress-induced current modulation, with positive or negative regulation depending upon the availability of the C-terminal Pro-Ser-Tyr motif to bind Nedd4-2.
Keywords: Ion Channels, MAPKs, Membrane Proteins, Sodium Channels, Ubiquitin Ligase, Hippocampal Neurons, Stress, Voltage-gated
Introduction
The voltage-gated sodium channel (VGSC)3 Nav1.6 is abundant at axon initial segments (AIS), at mature nodes of Ranvier in myelinated fibers, and along nonmyelinated axons within the central and peripheral nervous system (1–4). Loss-of-function mutations of Nav1.6 underlie motor disorders, and the null phenotype is lethal in mice (5, 6). Nav1.6 has been linked to axonal degeneration in multiple sclerosis (7, 8) and to neuronal death following traumatic brain injury (9). Furthermore, Nav1.6 regulates motility and phagocytosis in activated microglia and macrophages (10–12). Thus, modulation of Nav1.6 may impact diverse functions of neurons and glial cells.
Recently, we have shown that activation of the stress-induced p38 MAPK (activated p38, pp38, phosphorylated at Thr180/Tyr182) reduces Nav1.6 peak currents in a dorsal root ganglion-derived cell line (13). Phosphorylation of Nav1.6 at a single serine residue (Ser553) within the sequence motif Pro-Gly-Ser553-Pro in loop 1 (Nav1.6/L1) by pp38 is necessary for current reduction (13). This suggests that neuronal stress responses may involve phosphorylation of Nav1.6 and reduction of its current, resulting in attenuated neuronal excitability. However, the mechanism of pp38-mediated Nav1.6 current reduction is not yet known.
Ubiquitin ligases, for example Nedd4-like proteins, can induce internalization of ion channels (14). Nedd4 family members carry different types and numbers of WW domains (amino acid segments delineated by two tryptophan residues), which determine their target specificity (15). Type I WW domains bind to P_X_Y motifs, whereas type IV WW domains interact with phosphorylated P_X_(pS/T)P motifs (16). Furthermore, Nedd4 family members have been shown to bind to VGSCs via P_X_Y motifs in the C termini of the channels (17, 18), thereby reducing peak currents in different heterologous expression systems (17–20). The ubiquitin ligase Nedd4-2 contains both type I and type IV WW domains (15) and interacts with the Pro-Ser-Tyr1945 motif in the C terminus of Nav1.6 (Nav1.6/C) (17). Notably, phosphorylation of Ser553 by pp38 could convert the Pro-Gly-Ser-Pro motif in Nav1.6/L1 into a type IV WW-binding site. Thus, we hypothesized that the WW type IV-binding motif in Nav1.6/L1, alone or together with the WW type I-binding motif in Nav1.6/C, might recruit Nedd4-2 to the channel, thereby inducing internalization and reduction of Nav1.6 current.
Here, we show that stress-induced activation of p38 specifically reduces endogenous Nav1.6 current in hippocampal neurons, an effect that is abolished by a pharmacological block of endocytosis. We also show that both the Pro-Gly-Ser553-Pro motif in Nav1.6/L1 and the Pro-Ser-Tyr1945 motif in Nav1.6/C are necessary to induce internalization of a reporter protein. We also demonstrate that co-expression of Nedd4-2 and Nav1.6 reduces sodium currents, which is dependent upon both binding motifs being functional. Surprisingly, unlike its effect on wild-type channels, p38 activation increases the peak current of the Nav1.6Y1945A mutant channel. Taken together, these data suggest that a cooperative interaction of the two Nedd4-binding motifs in the L1 and C terminus is necessary for pp38-mediated regulation of Nav1.6 currents.
EXPERIMENTAL PROCEDURES
Antibodies
Mouse monoclonal p38 antibody, rabbit polyclonal p38 antibody, and rabbit polyclonal pp38 antibody were purchased from Cell Signaling (Danvers, MA). Mouse monoclonal HA antibody was from Covance (Princeton, NJ), mouse monoclonal CD4 antibody was from Beckman Coulter (Fullerton, CA), and chicken polyclonal MAP2 antibody was from Encor Biotechnology (Gainesville, FL). Rabbit polyclonal Nav1.6 antibody was from Sigma, and goat polyclonal p38β antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Secondary antibody-HRP conjugates were purchased from Dako (Carpinteria, CA). Secondary antibodies for immunolabeling were donkey anti-chicken Cy5 (Chemicon, Temecula, CA), donkey anti-rabbit Alexa 488 (Invitrogen), donkey anti-goat Alexa 546 (Invitrogen), and donkey anti-rabbit Alexa 546 (Invitrogen).
Plasmid Constructs
The plasmid pcDNA3-Nav1.6R encoding full-length mouse Nav1.6 has been described previously (21) and permits Nav1.6 currents to be recorded in isolation from neurons after other sodium currents are blocked with tetrodotoxin (13, 22, 23). It contains the substitution Y371S, which converts the tetrodotoxin sensitivity of Nav1.6 from nanomolar to micromolar concentrations upon expression in mammalian cell lines or in DRG neurons (13, 21, 24–26); the tetrodotoxin-R version of Nav1.6 will be referred to as WT channel in this study. The mutation Y1945A was introduced using QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). For generation of CD4 chimera proteins, a human CD4 amplicon (1254 bp), lacking the C-terminal domain (123 bp) of CD4, was cloned into the BglII and EcoRI sites of pEGFP-N1 to create a CD4-EGFP fusion protein. The L1 region or the C terminus of murine Nav1.6 was then cloned into the EcoRI site, which was retained in the CD4-EGFP construct. To generate the CD4-L1-C-EGFP construct, a second EcoRI site was introduced during amplification of the L1 fragment from CD4-L1-EGFP and cloned into the EcoRI site of CD4-C-EGFP. Sequence analyses verified that all inserts retained intact open reading frames. Plasmids encoding glutathione _S_-transferase (GST) fusion proteins with intracellular Nav1.6 fragments have been described previously (13). The mutation Y1945A was introduced into the GST-Nav1.6/C terminus fusion protein using QuikChange II XL site-directed mutagenesis kit. The plasmid pSwick-Nedd4-2-HA was a kind gift from Dr. Peter Snyder (University of Iowa, Iowa City). Recombinant human Nedd4-2 encoded by this plasmid includes a double HA epitope at its C terminus.
Cell Culture
HEK293 cells were maintained in Dulbecco's modified Eagle's medium/F-12 (1:1) (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), penicillin (100 units/ml), and streptomycin (100 μg/ml) (Invitrogen). The DRG-derived cell line ND7/23 (27) was kept in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, penicillin, and streptomycin (concentrations as above). To generate HEK293 cell lines stably expressing reporter proteins, transfected cells were selected with 600 μg/ml G418 (Invitrogen), and clonal lines were derived from single cells.
Hippocampal Neuronal Culture
Animal protocols complied with National Institutes of Health guidelines and were approved by the Veterans Affairs Connecticut Healthcare System Animal Use Committee. Hippocampal neurons were isolated from P14 to P16 WT C57/BL6 mice (Harlan, Indianapolis, IN) or from Scn8amedtg mice as described previously (28). Briefly, mice were deeply anesthetized (ketamine/xylazine, 100/10 mg/kg, intraperitoneal) and sacrificed by decapitation. Brains were removed and hippocampi dissected out in ice-cold HABG (HA medium (Brain Bits, Springfield, IL) supplemented with 2% B-27 (Invitrogen) and 0.5 mm Glutamax (Invitrogen)). Tissue was cut into small pieces and digested for 30 min at 30 °C with papain (30 units/ml; Worthington) in calcium-free HA medium (Brain Bits) with 0.5 mm Glutamax. The tissue was resuspended in HABG and carefully triturated 10 times through a plastic pipette tip. The supernatant was centrifuged at 800 × g for 2 min at room temperature and resuspended in culture medium (Neurobasal-A, supplemented with 2% B-27, 0.5 mm Glutamax, 10 μg/ml gentamycin, 5 ng/ml recombinant mouse FGF2, and 5 ng/ml recombinant mouse PDGFbb (all components from Invitrogen)). Cells were plated on 12-mm glass coverslips (Fisher), which had previously been coated with poly-d-lysine (70–150 kDa, Sigma) at 37 °C overnight. After allowing cells to adhere for 30 min, coverslips were washed once with HABG and incubated in 0.5 ml of culture medium at 37 °C.
Immunocytochemistry
To obtain brain sections for immunohistochemistry, adult WT C57/BL6 mice were perfused through the heart with PBS and then with 4% paraformaldehyde in 0.14 m Sorensen's phosphate buffer. Brains were excised, cryoprotected with 30% glucose in PBS, and frozen, and 12-μm sections were cut with a cryostat. Staining was then done as detailed below for hippocampal neurons, except that the blocking solution contained 0.3% Triton X-100.
Hippocampal neurons were immunostained 24 h after isolation and in some experiments after treatment for 30 min at 37 °C with 10 μg/ml anisomycin or 0.05% DMSO (vehicle control). Cells were washed once with PBS and fixed for 10 min with 4% paraformaldehyde in 0.14 m Sorensen's phosphate buffer. After another wash with PBS, cells were sequentially incubated in the following solutions: 1) blocking solution (PBS, 10% donkey serum, 2% bovine serum albumin, 0.1% Triton X-100) for 30 min at room temperature; 2) primary antibodies (rabbit anti-Nav1.6, 1:100, chicken anti-MAP2, 9.5 μg/ml; goat anti-p38β, 8 μg/ml; rabbit anti-p38, 1:100, or rabbit anti-pp38, 1:100) in blocking solution at 4 °C overnight; 3) 6× washes in PBS for 5 min each; 4) secondary antibodies in blocking solution for 2 h at room temperature (donkey anti-chicken Cy5, 15 μg/ml; donkey anti-rabbit Alexa 488, 2 μg/ml; donkey anti-goat Alexa 546, 2 μg/ml, donkey anti-rabbit Alexa 546, 2 μg/ml); 5) 6× washes in PBS for 5 min each. Coverslips were mounted with Aqua-Poly mount (Polysciences, Warrington, PA), and images were acquired with a Nikon (Tokyo, Japan) E600 microscope equipped with confocal optics.
Immunostaining of HEK293 cells transiently expressing the CD4-EGFP reporter protein derivatives was performed as above except for the blocking solution, which did not contain Triton X-100. After blocking for 30 min, cells were incubated with mouse anti-CD4 (0.8 μg/ml in blocking solution) at room temperature for 2 h. The secondary antibody (donkey anti-mouse Alexa 546, 2 μg/ml) was added to the coverslips and incubated for 2 h at room temperature. Coverslips were mounted with Aqua-Poly mount (Polysciences, Warrington, PA), and images were acquired with a Nikon E600 microscope equipped with confocal optics.
Reporter Protein Internalization Assay
HEK293 cells stably or transiently transfected with different CD4-EGFP reporter proteins were seeded on 12-mm glass coverslips coated with poly-d-lysine/laminin (BD Biosciences). 24 h after transfection or seeding, cells were treated with 10 μg/ml anisomycin or 0.05% DMSO (vehicle control) for 30 min at 37 °C. After washing once with PBS, cells were fixed for 10 min with ice-cold 4% paraformaldehyde in 0.14 m Sorensen's phosphate buffer, washed once with PBS, and incubated in blocking solution (PBS, 10% goat serum, 2% bovine serum albumin, 0.1% gelatin) for 30 min at room temperature. Extracellular CD4 was reacted with mouse anti-CD4 (0.8 μg/ml in blocking solution) for 2 h at room temperature. After washing six times for 5 min each with PBS, goat anti-mouse HRP conjugate (1:5000 in blocking solution) was added for 2 h at room temperature. Coverslips were washed six times for 5 min each with PBS, and luminescence was induced with SuperSignal ELISA femto maximum sensitivity substrate (Pierce) and measured within 5 min using a multiwell reader (Synergy-HT, BioTek, Winooski, VT).
Pulldown of Nedd4-2 with GST-Nav1.6 Fusion Proteins
HEK293 cells were transfected with pSwick-Nedd4-2-HA using Lipofectamine 2000 (Invitrogen). After 24 h, cells were resuspended in lysis buffer containing 150 mm NaCl, 50 mm Tris/HCl, pH 7.4, 2 mm EDTA, Complete protease inhibitor mixture, and 1% Triton X-100. Cells were lysed by sonication on ice, and proteins were solubilized by head-over mixing for 30 min at 4 °C. Samples were centrifuged at 18,000 × g for 20 min at 4 °C, and supernatants containing recombinant Nedd4-2 were collected and used for pulldown experiments. Fragments of murine Nav1.6 were produced as GST fusion proteins in Escherichia coli Rosetta 2 (DE3) cells and purified on glutathione beads as described previously (13). Loading of the beads was quantified by SDS-PAGE (4–12% BisTris, NuPAGE gels, Invitrogen) and Coomassie Blue staining. Equal amounts of GST-Nav1.6 fusion proteins were incubated with lysates of HEK293 cells expressing Nedd4-2-HA, incubated for 2 h at 4 °C, and extensively washed with PBS. Proteins were eluted by heating with sample buffer (NuPAGE, Invitrogen) for 10 min at 70 °C, separated by 4–12% BisTris NuPAGE gels, and blotted onto nitrocellulose membranes (NuPAGE, Invitrogen). After blocking overnight at 4 °C with 10% skim milk in TBST (Tris-buffered saline with 0.1% Tween 20), mouse anti-HA antibody (1 μg/ml in 5% bovine serum albumin in TBST) was added for 2 h at room temperature, followed by incubation with goat anti-mouse HRP conjugate (0.11 μg/ml in 1.25% bovine serum albumin in TBST) for 2 h at room temperature. Luminescence was induced by Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences) and detected by x-ray film exposure (BioMax XAR, Kodak, Rochester, NY).
Western Blot of Total p38 and pp38
HEK293 cells were incubated with 10 μg/ml anisomycin or 0.05% DMSO (vehicle control) for 30 min at 37 °C and resuspended in the same lysis buffer as above. After lysis by sonication, proteins were solubilized for 30 min at 4 °C. Samples were centrifuged at 18,000 × g for 20 min at 4 °C, and supernatants were collected and used for Western blotting as described above. Primary antibodies were mouse anti-p38, 1:1000, or rabbit anti-pp38, 1:1000. Secondary antibodies were goat anti-mouse HRP conjugate (0.11 μg/ml) or goat anti-rabbit HRP conjugate (0.03 μg/ml).
Kinase Assay
HEK293 cells were transiently transfected with the reporter proteins CD4-L1-EGFP or CD4-L1/SA-EGFP which carries the mutation S553A, using Lipofectamine 2000. 24 h after transfection, cells were lysed as described above, and CD4 fusion proteins were pulled down with a mouse monoclonal anti-CD4 antibody (Beckman) and protein A/G beads (Pierce). Immunoprecipitated proteins bound to beads were used as substrates in a kinase reaction containing 20 mm MOPS, pH 7.2, 25 mm β-glycerophosphate, 5 mm EGTA, 1 mm Na3VO4, 1 mm dithiothreitol, 0.5% DMSO, 10 mm MgCl2, 25 μm ATP, 10 μCi [γ-32P]ATP, and 10 ng/μl recombinant activated p38α (Cell Signaling). The active p38 MAPK inhibitor SB203580 (10 μm) was added to one reaction tube to confirm that the incorporation of radioactivity was not due to a contaminating kinase that may have co-precipitated with the CD4 fusion proteins. The reactions were carried out for 5 min at 30 °C in a 100-μl volume, and then stopped with 1 ml of PBS, 20 mm EDTA. After extensive washing of the beads, proteins were eluted with sample buffer by heating for 10 min at 80 °C and resolved by SDS-PAGE as above. The gels were dried and exposed to x-ray film (BioMax XAR) for 2–5 days.
Electrophysiology
Whole-cell voltage clamp recordings were performed with an Axopatch 200B amplifier (Axon Instruments, Foster City, CA) at room temperature (20–22 °C). Fire-polished electrodes were fabricated from 1.6-mm outer diameter borosilicate glass micropipettes (World Precision Instruments, Sarasota, FL). Pipette potential was adjusted to zero before seal formation, and liquid junction potential was not corrected. Capacity transients were cancelled, and series resistance was compensated by 80–90%. Leakage current was subtracted digitally on line using hyperpolarizing potentials applied after the test pulse (P/4 procedure). Currents were acquired with Clampex 9.2 5 min after establishing whole-cell configuration, sampled at a rate of 50 or 100 kHz, and filtered at 5 kHz.
Hippocampal neurons were patched 20–36 h after isolation, using only cells with well defined neurites. Pipette resistances were 1.1–2.6 megohms. In some experiments, neurons were pretreated with 10 μm SB203580 or 10 μm SB202474 for 30 min at 37 °C, followed by 10 μg/ml anisomycin for 30 min at 37 °C in the continued presence of SB203580 or SB202474. After break-in, neurons were held at −80 mV. For current-voltage relationships, a 500-ms prepulse at −120 mV was introduced before a range of step depolarizations (from −80 to +40 mV for 100 ms in 5-mV increments) to relieve inactivation of sodium channels. Peak inward currents (I) were plotted as a function of depolarization potential to generate I-V curves. Neurons generating more than 20 nA currents or exhibiting large voltage errors were not included for analysis. Activation curves were obtained by converting I to conductance (G) at each voltage (V) using the equation G = I/(V − _V_rev), where _V_rev is the reversal potential that was determined for each neuron individually. Activation curves were fit with a Boltzmann function in the form of G = _G_max/{1 +exp((_V_½,act − V)/k)}, where _G_max is the maximal sodium conductance; _V_½,act is the potential at which activation is half-maximal; V is the test potential, and k is the slope factor. Steady-state fast inactivation was achieved with a series of 500-ms prepulses (from −10 to −150 mV in 10-mV increments), and noninactivated channels were measured by a 40-ms test pulse to −10 mV. Peak inward currents obtained from steady-state fast inactivations were normalized with the maximal peak current (_I_max) and fit with a Boltzmann function, I/_I_max = A + (1 − A)/{1 + exp((V − _V_½,inact)/k)}, where V represents the inactivating prepulse potential, _V_½,inact represents the midpoint of inactivation curve, and A is the offset.
In some experiments, neurons were pretreated with 80 μm Dynasore (Tocris Bioscience, Ellisville, MI), an endocytosis inhibitor (29, 30), in complete medium lacking B-27 but supplemented with 10% NuSerum (BD Biosciences) for 15 min at 37 °C. Then 10 μg/ml anisomycin or a vehicle control was added for 30 min at 37 °C in the continued presence of Dynasore. Whole-cell recording was done as detailed above, except that neurons were held at −100 mV after break-in, and sodium currents were measured in the presence of 80 μm Dynasore in the bath solution.
To investigate the effects of pp38 or Nedd4-2 on Nav1.6, we used the DRG-derived cell line ND7/23 cultured on 12-mm glass coverslips coated with poly-d-lysine/laminin (BD Biosciences). Pipette resistances were 0.7–1.3 megohms. DNA constructs (0.8 μg per well) were co-transfected with pEGFP-C1 (0.2 μg per well; Clontech) using Lipofectamine 2000 (Invitrogen) as described previously (13, 22, 23). Cells with robust green fluorescence were selected 24 h after transfection for recordings. For current-voltage relationships, cells were held at −120 mV and stepped to a range of potentials (−80 to +60 mV in 10-mV increments) for 100 ms each. Peak inward currents (I) were plotted as a function of depolarizing potential to generate I-V curves. Steady-state fast-inactivation was achieved with a series of 500-ms prepulses (−140 to −10 mV in 10-mV increments), and the noninactivated channels were measured by a 40-ms test pulse to −10 mV. Properties of activation and steady-state fast-inactivation were analyzed the same way as for hippocampal neurons.
Pipette solution for both hippocampal neurons and ND7/23 cells contained (in mm) the following: 140 CsF, 10 NaCl, 1 EGTA, and 10 HEPES, pH 7.30 (with CsOH), and osmolarity was adjusted to 314 mosmol/liter with dextrose. For voltage clamp recordings in hippocampal neurons, the extracellular bath solution contained (in mm) the following: 30 NaCl, 110 choline-Cl, 3 KCl, 20 tetraethylammonium, 1 MgCl2, 1 CaCl2, 10 HEPES, 5 CsCl, 0.1 CdCl2, pH 7.32 (with NaOH), and the osmolarity was 322 mosmol/liter. For measurements on ND7/23 cells, the extracellular solution contained (in mm) the following: 140 NaCl, 3 KCl, 20 tetraethylammonium, 1 MgCl2, 1 CaCl2, 10 HEPES, 5 CsCl, 0.1 CdCl2, pH 7.32 (with NaOH), and the osmolarity was 328 mosmol/liter. For experiments with ND7/23 cells, 300 nm tetrodotoxin was added to the extracellular bath solution to block endogenous voltage-gated sodium currents (13, 22, 23).
Data Analysis
Data were analyzed using Clampfit 9.2 (Molecular Devices, Sunnyvale, CA) and OriginPro 8 (Microcal Software, Northampton, MA) and presented as means ± S.E. Unpaired Student's t test was used for experiments with hippocampal neurons and for analysis of biophysical properties of Nav1.6 and Nav1.6Y1945A. Significance of reporter protein internalization was tested by one-way analysis of variance with Tukey post hoc analysis. Kruskal-Wallis nonparametric test was used to compare Nav1.6 current densities obtained from transfected ND7/23 cells.
RESULTS
Co-expression of Nav1.6 and p38 in Mouse Hippocampal Neurons
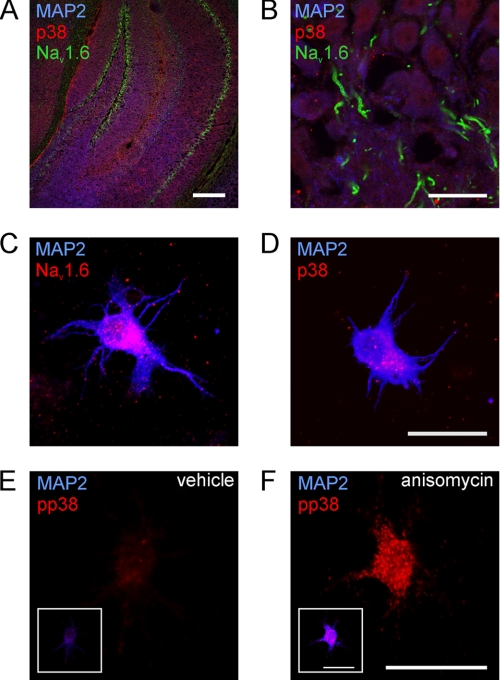
We investigated the expression of Nav1.6 and p38 MAPK in hippocampal sections from adult C57/BL6 mice and in cultured hippocampal neurons isolated from C57/BL6 mice at postnatal days 14–16 (P14–P16). In hippocampal sections, we found expression of p38 in neurons in the CA1, CA3, and DG regions (Fig. 1, A and B). As demonstrated by co-localization with the somatodendritic marker MAP2, p38 was mainly localized in neuronal cell bodies, whereas Nav1.6 immunoreactivity was detected at the AIS of hippocampal neurons (Fig. 1B).
FIGURE 1.
Expression of Nav1.6 and p38 MAPK in mouse hippocampal neurons. A and B, adult C57/BL6 mouse hippocampal sections were stained with specific antibodies against p38 MAPK (red), Nav1.6 (green), and the somatodendritic neuronal marker protein MAP2 (blue). Shown are total hippocampus (A) and the CA1 region (B). Scale bars represent 200 μm (A) or 20 μm (B). C–F, co-immunostainings of cultured hippocampal neurons. Neurons were isolated from P14 to P15 C57/BL6 mice and cultured for 24 h. Co-immunostaining of Nav1.6 and MAP2 (C), total p38, and MAP2. D, activated p38 (pp38) in native neurons (E) and pp38 after stimulation with anisomycin (F); insets in E and F show co-staining of the same neuron with MAP2. Scale bars represent 20 μm.
Adult central nervous system neurons express multiple VGSC isoforms, including Nav1.1, Nav1.2, and Nav1.6 (32). To investigate the effect of pp38 on neuronal Nav1.6, we isolated hippocampal neurons from mice at P14 or P15, because Nav1.6 expression starts after birth and reaches a plateau by P14 (33). Neurons were kept in culture for only 24 h to minimize space clamp artifacts during patch clamp recordings of sodium currents (34). Immunostaining of hippocampal neurons 24 h after cell isolation confirmed expression of Nav1.6 under these culture conditions (Fig. 1C). In contrast to Nav1.6 immunostaining in hippocampal sections (Fig. 1B), Nav1.6 immunoreactivity was localized within the somata of cultured hippocampal neurons at this early time point after isolation, in agreement with previous reports (35). Co-immunostaining of p38 and MAP2 (Fig. 1D) in hippocampal neurons after 1 day in culture showed expression of p38 in these cells. Similar to Nav1.6, p38 was mostly localized in the somatodendritic compartment of the neurons. This co-expression of p38 and VGSCs in hippocampal neurons in vivo and in cultured neurons suggests that pp38 may regulate VGSCs in hippocampal neurons.
Co-immunostaining of hippocampal neurons in culture for pp38 and MAP2 (Fig. 1E) showed the presence of low levels of pp38 in these neurons, possibly due to the culture procedure itself. However, p38 has been shown to be activated in cultured cells by incubation with the protein synthesis inhibitor anisomycin (36). Thus, cultured hippocampal neurons were treated with anisomycin, and enhanced activation of p38 was ascertained by immunocytochemistry. Treatment of these neurons with anisomycin for 30 min clearly increased the level of pp38 (compare Fig. 1, E and F).
Reduction of Nav1.6 Currents by pp38 in Hippocampal Neurons
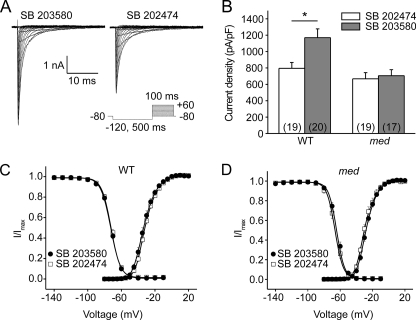
We have previously demonstrated that pp38 reduces Nav1.6 current density in the DRG-derived cell line ND7/23 (13). Nav1.6 currents have been shown to be modulated in a cell type-specific manner (25, 37). Therefore, we assessed pp38-mediated regulation of Nav1.6 in native hippocampal neurons. Although central nervous system neurons express TTX-S sodium channels other than Nav1.6, e.g. Nav1.1 and Nav1.2 (32), which carry potential MAPK phosphorylation sites (SP or TP), it is notable that the Pro-Gly-Ser-Pro motif in L1 is unique in Nav1.6. A functional significance of this motif may be inferred from its conservation in Nav1.6 orthologues from all vertebrate species reported thus far in databases (Fig. 2). To investigate if other TTX-S channels in hippocampal neurons are modulated by pp38, we compared the effect of pp38 on sodium currents in hippocampal neurons from wild-type C57/BL6 and Scn8amedtg mice, which lack Nav1.6 (6). To avoid interference of endogenous basal pp38 activity (see Fig. 1E), we pretreated the cultured neurons with either the pp38 inhibitor SB203580 or the inactive control compound SB202474 for 30 min and then added anisomycin and continued incubation for 30 min before recording of sodium currents. Sodium currents were measured by whole-cell voltage clamp recordings (representative traces shown in Fig. 3A). Activation of p38 in hippocampal neurons from WT C57/BL6 mice (Fig. 3B, left) significantly reduced the peak current density (SB203580, 1170 ± 141 pA/pF, n = 19, versus SB202474, 796 ± 71 pA/pF, n = 20; p < 0.05). In contrast, p38 activation had no effect on the peak current density in _Scn8amedtg_ neurons (Fig. 3_B_, _right_), which lack Nav1.6 (SB203580, 705 ± 74 pA/pF, _n_ = 17, _versus_ SB202474, 667 ± 76 pA/pF, _n_ = 19; _p_ > 0.05). The reduced total sodium current observed in neurons from Scn8amedtg mice is consistent with reports indicating a significant contribution of Nav1.6 to the total sodium current in central nervous system neurons (38, 39). Lack of an effect of pp38 on hippocampal neurons from Scn8amedtg mice suggests that Nav1.6 is the primary target for modulation by pp38 among sodium channels in these native neurons.
FIGURE 2.
Conservation of Pro-Gly-Ser-Pro motif in Nav1.6 orthologues from vertebrate species. Alignment of peptide sequence straddling the Pro-Gly-Ser-Pro motif from Nav1.6 orthologues in vertebrate species shows invariant amino acid sequence, except for zebrafish that has two substitutions C-terminal to the Pro-Gly-Ser-Pro motif. Accession number for vertebrate Nav1.6 orthologues are as follows: Green Spotted Pufferfish (CAG07463.1); Japanese Pufferfish (BAA07195.1); zebrafish (NP_001038648.1); chicken (XP_424477.2); mouse (NM_011323); rat (EDL86918.1); domestic dog (XP_534797.2); domestic cow (XP_873278.1); wild boar (XP_001927150.1); horse (XP_001916036.1); giant panda (FEB24314.1); rhesus monkey (XP_001090295.1); chimpanzee (XP_01141985.1); human (EAW58198.1).
FIGURE 3.
Activation of p38 reduces Nav1.6 peak current density in hippocampal neurons. A, representative traces of sodium currents from WT hippocampal neurons. Cells were held at −80 mV, and sodium currents were elicited by a series of step depolarizations from −80 to +40 mV in 5-mV increments. A 500-ms prepulse at −120 mV was applied before depolarization steps to allow channels to recover from inactivation. Neurons were incubated before recording for 30 min with the p38 inhibitor SB203580 or the control compound SB202474, followed by addition of anisomycin for 30 min. B, activated p38 reduced the peak sodium currents in WT hippocampal neurons but had no effect on peak currents in Scn8amedtg hippocampal neurons lacking Nav1.6. Numbers of recorded cells are given in parentheses. Means ± S.E.; *, p < 0.05. C, activation of p38 did not change voltage dependence of activation and steady-state fast-inactivation of VGSCs in WT hippocampal neurons. Means ± S.E. (n = 10–19). D, activation of p38 did not change voltage dependence of activation and steady-state fast-inactivation of VGSCs in Scn8amedtg hippocampal neurons. Means ± S.E.
We also examined the effect of p38 activation on gating properties of sodium channels in hippocampal neurons. Activation of p38 did not change voltage dependence of activation or fast-inactivation of sodium currents in hippocampal neurons from WT mice (Fig. 3C and Table 1) or Scn8amedtg mice (Fig. 3D and Table 1). These results are in agreement with our previous study, which showed that p38 activation reduces peak current density but does not alter biophysical properties of Nav1.6 in the DRG-derived cell line ND7/23 (13). Furthermore, these data are consistent with our hypothesis of p38-mediated internalization of the channel (13).
TABLE 1.
Activation of p38 has no effect on the biophysical properties of voltage-gated sodium currents in WT and Scn8amedtg mouse hippocampal neurons
Data are shown as means ± S.E. Hippocampal neurons were isolated from WT (C57/BL6) or Scn8amedtg mice and cultured for 24 h at 37 °C. Neurons were treated with the p38 inhibitor SB203580 or the inactive control compound SB202474 for 30 min, followed by incubation with anisomycin (10 μg/ml, 37 °C for 30 min) to activate p38 in the continued presence of SB203580 or SB202474.
| Activation | Steady-state fast-inactivation | |||||
|---|---|---|---|---|---|---|
| _V_½,act | k | n | _V_½,inact | k | n | |
| mV | mV | |||||
| C57/BL6 mice | ||||||
| SB203580 | −32.8 ± 1.3 | 6.00 ± 0.24 | 15 | −72.2 ± 0.7 | 5.86 ± 0.20 | 10 |
| SB202474 | −31.0 ± 0.9 | 6.34 ± 0.23 | 19 | −71.8 ± 0.9 | 5.59 ± 0.18 | 16 |
| Scn8amedtg | ||||||
| SB203580 | −27.4 ± 0.9 | 5.79 ± 0.24 | 13 | −64.1 ± 1.1 | 5.19 ± 0.11 | 13 |
| SB202474 | −29.1 ± 1.3 | 5.93 ± 0.50 | 10 | −66.2 ± 1.5 | 5.15 ± 0.14 | 9 |
Pharmacological Block of Endocytosis Abolishes pp38-mediated Current Reduction
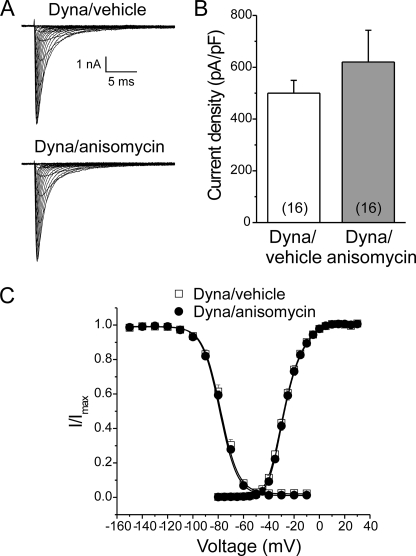
To investigate whether channel internalization underlies pp38-mediated Nav1.6 current reduction, we pharmacologically blocked endocytosis with Dynasore, which specifically inhibits GTPase activities of dynamin 1 and dynamin 2 (29, 30). WT hippocampal neurons were pretreated with Dynasore for 15 min, and p38 was then activated by anisomycin in the continued presence of Dynasore for an additional 30 min. Sodium currents were then measured by voltage clamp recordings (representative tracings shown in Fig. 4A). The pp38-mediated reduction of peak current density (see Fig. 3B, left) was completely blocked by treatment with Dynasore (Fig. 4B; vehicle control, 500 ± 49 pA/pF, n = 16, versus anisomycin, 620 ± 122 pA/pF, n = 16; p > 0.05). Similar to the results shown in Fig. 3, C and D, activation of p38 had no effect on voltage dependence of activation and steady-state fast-inactivation in hippocampal neurons upon block of endocytosis with Dynasore (Fig. 4C and Table 2). Taken together, these findings support the hypothesis that endocytosis of Nav1.6 channels underlies the pp38-mediated reduction in current density.
FIGURE 4.
Pharmacological block of endocytosis by Dynasore abolishes pp38-induced Nav1.6 current reduction. A, representative traces of sodium currents from WT hippocampal neurons. Cells were held at −100 mV, and sodium currents were elicited by a series of step depolarizations from −80 to +40 mV in 5-mV increments. A 500-ms prepulse at −120 mV was applied before depolarization steps to allow channels to recover from inactivation. Neurons were incubated before recording for 15 min with the dynamin inhibitor Dynasore (Dyna), followed by treatment with anisomycin or a vehicle control for 30 min in the continued presence of Dynasore. B, activation of p38 had no effect on peak sodium currents in WT hippocampal neurons upon block of endocytosis with Dynasore. Numbers of recorded cells are given in parentheses. Data are presented as means ± S.E. C, activation of p38 did not change voltage dependence of activation and steady-state fast-inactivation of VGSCs in WT hippocampal neurons in the presence of Dynasore. Means ± S.E.
TABLE 2.
Activation of p38 has no effect on the biophysical properties of voltage-gated sodium currents in WT mouse hippocampal neurons after block of endocytosis with Dynasore
Data are shown as means ± S.E. Hippocampal neurons were isolated from WT mice and cultured for 24 h at 37 °C. Neurons were treated with the dynamin inhibitor Dynasore (80 μm, 37 °C for 15 min), followed by incubation with anisomycin (10 μg/ml, 37 °C for 30 min) to activate p38 or DMSO as vehicle control in the continued presence of Dynasore.
| Activation | Steady-state fast inactivation | |||||
|---|---|---|---|---|---|---|
| _V_½,act | k | n | _V_½,inact | k | n | |
| mV | mV | |||||
| Dynasore/DMSO | −27.3 ± 0.8 | 6.75 ± 0.27 | 16 | −77.2 ± 1.3 | 7.25 ± 0.14 | 12 |
| Dynasore/anisomycin | −26.7 ± 0.7 | 6.57 ± 0.28 | 15 | −77.7 ± 0.8 | 7.53 ± 0.15 | 13 |
Internalization of Reporter Proteins by pp38 Requires Both Nav1.6/L1 and Nav1.6/C
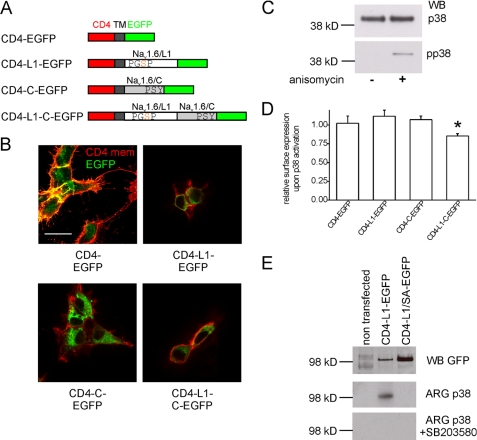
Because phosphorylation of Ser553 in Nav1.6/L1 is necessary for pp38-mediated current reduction (13), we have hypothesized that the phosphorylated Pro-Gly-Ser-Pro motif might recruit Nedd4 family proteins carrying a type IV WW domain, which then ubiquitinate the channel and induce its internalization (13). However, because Nedd4 family proteins also bind to P_X_Y motifs in the C termini of different VGSCs (17–20), including Nav1.6 (17), this second binding site might be necessary for the pp38-mediated regulation of Nav1.6. We generated a series of reporter proteins (Fig. 5A) to test whether one or both putative binding motifs are necessary for pp38-mediated internalization of Nav1.6. The reporter proteins consisted of the extracellular CD4 domain fused to the Nav1.6/L1 loop, the Nav1.6/C terminus, or both and an enhanced GFP tag, all in-frame. Previously, CD4 chimeras containing L2 loops or C termini of VGSCs were used in heterologous expression systems and neurons to investigate the role of specific sequence motifs in subcellular targeting (40–42). Thus, we reasoned that CD4-Nav1.6-EGFP chimeras (Fig. 5A) would be useful to assay pp38-mediated internalization in HEK293 cells.
FIGURE 5.
L1 and the C terminus of Nav1.6 are both necessary for pp38-mediated internalization of a reporter protein. A, scheme of the reporter proteins used for internalization assays (not drawn to scale). Chimeric proteins consisted of extracellular (red) and transmembrane (dark gray) domains of human CD4, Nav1.6/L1 (white), Nav1.6/C (light gray), and C-terminal enhanced GFP (green). Note that the Nav1.6/L1 fragments contain the sole phosphoacceptor site (Ser553) for p38 MAPK within the indicated Pro-Gly-Ser-Pro motif (13), whereas the C-terminal Nav1.6 domain contains a Pro-Ser-Tyr motif, which serves as a binding site for type I WW domains. B, HEK293 cells were transfected with the reporter proteins depicted in A. After 24 h, reporter proteins in the cell membrane were detected using a CD4 antibody without permeabilizing the cells. Red color (extracellular CD4 epitope) reflects reporter protein at the cell membrane, and green color (EGFP fluorescence) reflects the total level of reporter protein. Scale bar, 20 μm. C, HEK293 cells in culture produce p38 MAPK (upper panel, total p38). Stimulation with anisomycin for 30 min activates endogenous p38 in HEK293 cells, but p38 is not constitutively activated in HEK293 cells under standard culture conditions (lower panel, pp38). Representative blots of three independent experiments are shown. WB, Western blot. D, activated p38-mediated internalization of a reporter protein requires both L1 and the C terminus of Nav1.6. HEK293 cells transfected with different reporter proteins were treated for 30 min with anisomycin to activate p38. Reporter proteins at the cell membrane were probed with an antibody against CD4 after fixing but without permeabilizing the cells. After incubation with an HRP-conjugated secondary antibody and an HRP substrate, membrane expression of the reporter proteins was quantified by luminometry and normalized to cells treated with a vehicle control. Means ± S.E. (n = 4–8). *, p < 0.05 compared with all other conditions. E, activated p38 phosphorylates the reporter protein CD4-L1-EGFP at Ser553 in vitro. The reporter proteins CD4-L1-EGFP or CD4-L1/SA-EGFP, which carries the mutation S553A, were expressed in HEK293 cells, immunoprecipitated with an antibody against CD4, and phosphorylated by pp38. Protein loading of beads was analyzed by Western blotting against GFP (top panel). Kinase reactions were performed in the absence (middle panel) or presence (bottom panel) of the p38 inhibitor SB203580. Representative data are shown from three independent experiments.
To verify expression and trafficking of the reporter proteins, we stained extracellular CD4 in transfected cells without permeabilizing the membrane (Fig. 5B). Using a CD4-specific antibody, we detected the chimeras at the cell membrane (red color, Fig. 5B), as expected, and the enhanced GFP fluorescence provided an estimate of the total level of the fusion proteins (green color, Fig. 5B). Interestingly, we found that the control protein CD4-EGFP was detectable even in very fine filopodia of HEK293 cells, whereas the chimeric proteins containing Nav1.6/L1, Nav1.6/C, or both Nav1.6 domains were localized at the membrane but were excluded from the fine filopodia. The significance of this finding is not clear at this time. Next, we confirmed that anisomycin treatment of HEK293 cells activated p38. Western blots demonstrated that pp38 was not detectable in vehicle-treated cells, whereas treatment with anisomycin for 30 min activated p38 (Fig. 5C). To quantify changes in localization of the reporter proteins at the plasma membrane, we developed a quantitative luminometry-based assay in which we expressed the different reporter proteins in HEK293 cells, treated the cells with anisomycin to activate p38 or a vehicle control, and quantified extracellular CD4 levels by CD4-specific extracellular antibody-based luminometry without permeabilizing the cell membrane (Fig. 5D). In these experiments, we did not detect internalization of the reporter protein CD4-L1-EGFP, suggesting that the Pro-Gly-Ser-Pro motif in Nav1.6/L1 alone is not sufficient to induce internalization. To verify that the p38 phosphorylation site Ser553 in the chimeric protein is indeed phosphorylated by pp38, we immunoprecipitated the reporter protein from transiently transfected HEK293 cells using anti-CD4 antibody, and we tested the immunoprecipitated proteins as substrates for pp38 phosphorylation in an in vitro kinase assay (Fig. 5E). Equal loading of samples was verified by Western blot analysis using an antibody against GFP (Fig. 5D, top panel). The kinase assay shows that the CD4-L1-EGFP reporter protein was phosphorylated by pp38 in vitro. Importantly, the CD4-L1S553A-EGFP protein (Fig. 5D, middle panel) was not phosphorylated by pp38. To ensure specificity of the kinase reaction, the p38-specific inhibitor SB203580 was added to duplicate reactions, which completely abolished 32P incorporation, indicating that phosphorylation of CD4-L1-EGFP proteins was not the result of another kinase that may have co-immunoprecipitated with the CD4 fusion proteins. Because no internalization of the CD4-L1-EGFP reporter protein was detectable with our luminometric assay (Fig. 5D), we next tested whether the C-terminal domain of Nav1.6 is necessary for p38-mediated internalization of proteins carrying the Nav1.6/L1 sequence. The luminometric assay shows that activation of p38 does not induce internalization of the fusion protein CD4-C-EGFP, which carries the Nav1.6/C channel fragment, whereas the chimeric protein CD4-L1-C-EGFP, which carries both the Nav1.6/L1 and Nav1.6/C channel fragments, shows a significant reduction of surface expression by 15%. This indicates that both the L1 region and the C terminus of Nav1.6 are necessary to mediate p38-induced channel internalization.
C Terminus of Nav1.6 Recruits Nedd4-2 via a Pro-Ser-Tyr Motif
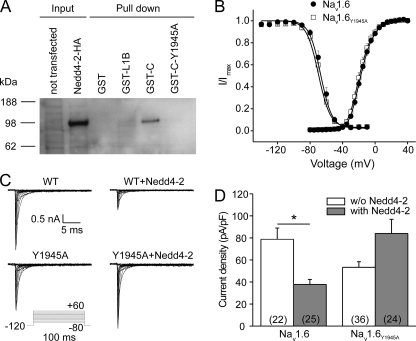
In addition to the hypothesized recruitment of Nedd4-2 after pp38-mediated phosphorylation of Ser553 in Nav1.6/L1 (13), the Pro-Ser-Tyr1945 motif in Nav1.6/C might be necessary for pp38-mediated regulation of Nav1.6, because it binds Nedd4 family proteins (17). This hypothesis is supported by the data shown in Fig. 5, indicating that both Nav1.6/L1 and Nav1.6/C are necessary for pp38-mediated internalization of a reporter protein. To determine whether both motifs can bind to Nedd4-2 and are necessary for modulation of the channel by pp38, we first analyzed interactions between Nav1.6/L1 and Nav1.6/C and Nedd4-2 in pulldown experiments. HA-tagged Nedd4-2 was expressed in HEK293 cells, and the transfected cell lysate was used in pulldown assays with purified GST-Nav1.6/L1B or GST-Nav1.6/C fusion proteins as baits. These experiments showed that Nav1.6/C, but not Nav1.6/L1B, a fragment of L1 that is a substrate for pp38 phosphorylation (13), was able to bind to Nedd4-2 (Fig. 6A). The mutation Y1945A in the Pro-Ser-Tyr motif of Nav1.6/C completely abolished binding of the fusion protein to Nedd4-2 (Fig. 6A), confirming the importance of the Pro-Ser-Tyr motif for interaction between Nav1.6/C and Nedd4-2 (17). Thus, it appears that the affinity of GST-Nav1.6/L1B to Nedd4-2 is not strong enough to form a binding complex in this assay.
FIGURE 6.
Nav1.6 mutation Y1945A abolishes both binding of Nedd4-2 to Nav1.6 and Nedd4-2-mediated reduction of Nav1.6 current density. A, Nedd4-2 binds to the C terminus of wild-type Nav1.6 but not to the C terminus of the mutant Nav1.6Y1945A or to the Nav1.6/L1 loop. GST-Nav1.6 fusion proteins (GST alone, GST fused to a truncated version of the Nav1.6/L1 region designated L1B, GST fused to the C terminus of Nav1.6, and GST fused to the C terminus of Nav1.6 carrying the mutation Y1945A) were used to pull down HA-tagged Nedd4-2. Bound Nedd4-2 was detected by Western blots against the HA tag. Left lanes, input cell lysates (5% of the amount used for pulldown). Right lanes, pulldown of Nedd4-2 by Nav1.6 fragments; representative blots of three independent experiments are shown. B, mutation Y1945A did not change activation or steady-state fast inactivation of Nav1.6. Voltage dependence of activation and steady-state fast-inactivation of wild-type Nav1.6 and the mutant Nav1.6Y1945A transiently expressed in ND7/23 cells are shown. Means ± S.E. (n = 7). C, representative sodium currents recorded from ND7/23 cells transiently co-transfected with Nedd4-2 and either wild-type Nav1.6 or the mutant Nav1.6Y1945A. Cells were held at −120 mV, and sodium currents were elicited by a series of step depolarizations from −80 to +60 mV in 10-mV increments. D, co-expression of Nedd4-2 reduced current density of wild-type Nav1.6 but not of Nav1.6Y1945A in ND7/23 cells. Numbers of recorded cells are given in parentheses. Means ± S.E., *, p < 0.05.
Nedd4-2-mediated Reduction of Nav1.6 Currents Depends upon the Pro-Ser-Tyr Motif in Nav1.6/C
Although Nedd4-2 has been shown to interact with the C terminus of Nav1.6 (Fig. 6A) (17), the functional effect of Nedd4-2 on Nav1.6 currents has not been previously tested. To investigate the effect of Nedd4-2 on Nav1.6 and the role of the C-terminal Pro-Ser-Tyr1945 motif in this interaction, we generated a mutant Nav1.6 channel (Nav1.6Y1945A), in which Tyr1945 was replaced by alanine. This mutant channel is predicted to lose interaction of its C terminus with Nedd4 proteins (Fig. 6A) (17). The current density of Nav1.6Y1945A in ND7/23 cells is not significantly different from that of WT channels (Nav1.6, 151.8 ± 32.5 pA/pF, n = 9, versus Nav1.6Y1945A, 138.5 ± 37.6 pA/pF, n = 10; p > 0.05), suggesting no constitutive interaction of channels and endogenous Nedd4-like proteins. Furthermore, there were no significant differences in voltage dependence of activation or steady-state fast-inactivation between WT and Nav1.6Y1945A channels (Fig. 6B and Table 3).
TABLE 3.
The mutation Y1945A does not alter the biophysical properties of Nav1.6 channels in ND7/23 cells
Data are shown as means ± S.E. ND7/23 cells were transiently transfected with Nav1.6-WT or Nav1.6Y1945A. Voltage-dependent sodium currents were measured in the presence of TTX (300 nm) 24 h after transfection.
| Activation | Steady-state fast inactivation | |||||
|---|---|---|---|---|---|---|
| _V_½,act | k | n | _V_½,inact | k | n | |
| mV | mV | |||||
| Nav1.6-WT | −17.0 ± 1.1 | 7.34 ± 0.31 | 7 | −65.4 ± 1.7 | 6.31 ± 0.41 | 7 |
| Nav1.6Y1945A | −18.5 ± 1.6 | 7.41 ± 1.56 | 7 | −68.0 ± 1.7 | 6.42 ± 0.17 | 7 |
To test the regulatory effect of Nedd4-2 on Nav1.6, we co-expressed Nav1.6 and Nedd4-2 in ND7/23 cells and recorded sodium currents by voltage clamp protocols. In these experiments, we did not detect transfection-mediated activation of p38 using Western blot analysis (data not shown). Fig. 6C shows representative traces of Nav1.6 currents in the presence or absence of Nedd4-2. Co-expression of Nav1.6 with Nedd4-2 robustly reduced Nav1.6 current density (Fig. 6, C and D) by more than 50% (Nav1.6 alone, 78.7 ± 10.3 pA/pF, n = 22, versus Nav1.6 + Nedd4-2, 37.8 ± 4.5 pA/pF, n = 25; p < 0.05). This indicates that Nedd4-2 can interact with full-length Nav1.6 and reduce its current density. To test whether the regulatory effect of Nedd4-2 on Nav1.6 depends upon the Pro-Ser-Tyr1945 motif within the C terminus of the channel, we examined the effect of Y1954A substitution on Nedd4-2-mediated regulation of Nav1.6 currents. In contrast to its effect on WT, co-expression with Nedd4-2 tended to increase the current density of Nav1.6Y1945A, although the difference was not statistically significant (Fig. 6, _C_ and _D_; Nav1.6Y1495A alone, 53.3 ± 5.1 pA/pF, _n_ = 36, _versus_ Nav1.6Y1495A + Nedd4-2, 84.0 ± 12.9 pA/pF, _n_ = 24, _p_ > 0.05). Taken together, these results demonstrate that Nedd4-2 reduces peak Nav1.6 current and that the C-terminal Pro-Ser-Tyr1945 motif of Nav1.6 is necessary for this effect.
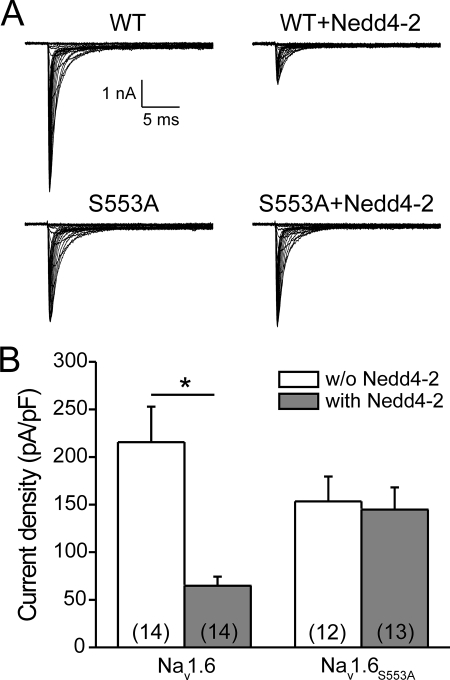
Nedd4-2-mediated Reduction of Nav1.6 Currents Depends upon the Pro-Gly-Ser-Pro Motif in Nav1.6/L1
To investigate if the putative Nedd4-binding motif Pro-Gly-Ser-Pro in Nav1.6/L1 is necessary for reduction of Nav1.6 currents by Nedd4-2, we used the mutant channel Nav1.6S553A. We co-expressed this mutant channel with Nedd4-2 in ND7/23 cells and recorded sodium currents by voltage clamp protocols. Again, we found a robust reduction of Nav1.6-WT currents by co-expression with Nedd4-2 (Fig. 7, A and B; Nav1.6 alone, 215.7 ± 37.2 pA/pF, n = 14, versus Nav1.6 + Nedd4-2, 64.9 ± 9.4 pA/pF, n = 14; p < 0.05). In contrast, currents of the mutant channel Nav1.6S553A were not reduced by co-expression of Nedd4-2 (Fig. 7, _A_ and _B_; Nav1.6S553A alone, 153.4 ± 26 pA/pF, _n_ = 12, _versus_ Nav1.6S553A + Nedd4-2, 144.7 ± 23.4 pA/pF, _n_ = 13, _p_ > 0.05). These results show that a functional Pro-Gly-Ser-Pro motif in Nav1.6/L1, similar to the Pro-Ser-Tyr motif in Nav1.6/C, is necessary for Nedd4-2-mediated reduction of Nav1.6 currents.
FIGURE 7.
Reduction of Nav1.6 currents by Nedd4-2 depends on the presence of a functional Pro-Gly-Ser-Pro motif in L1 of the channel. A, representative sodium currents recorded from ND7/23 cells transiently co-transfected with Nedd4-2 and either wild-type Nav1.6 or the mutant Nav1.6S553A. Cells were held at −120 mV, and sodium currents were elicited by a series of step depolarizations from −80 to +60 mV in 5-mV increments. B, co-expression of Nedd4-2 reduced current density of wild-type Nav1.6 but not of Nav1.6S553A in ND7/23 cells. Number of recorded cells are given in parentheses. Means± S.E., *, p < 0.05.
We also made the phosphomimetic S553D substitution to study the effect of a permanent negative charge at this site on the effect of Nedd4-2 on current density. However, Nav1.6S553D channels did not produce a sodium current when expressed in ND7/23 cells, and the mutant channel protein was not detectable in transiently transfected HEK293 cells by Western blot (data not shown). HEK293 cells were used for the biochemical assay because these cells do not produce significant levels of endogenous sodium channels, making it possible to use the pan-sodium channel antibody for Western blot analysis. These data suggest that the S553D substitution prevents channel maturation or stability, leading to its degradation, and support the hypothesis that the nonphosphorylated Pro-Gly-Ser-Pro motif may bind a channel partner that is necessary for channel biogenesis.
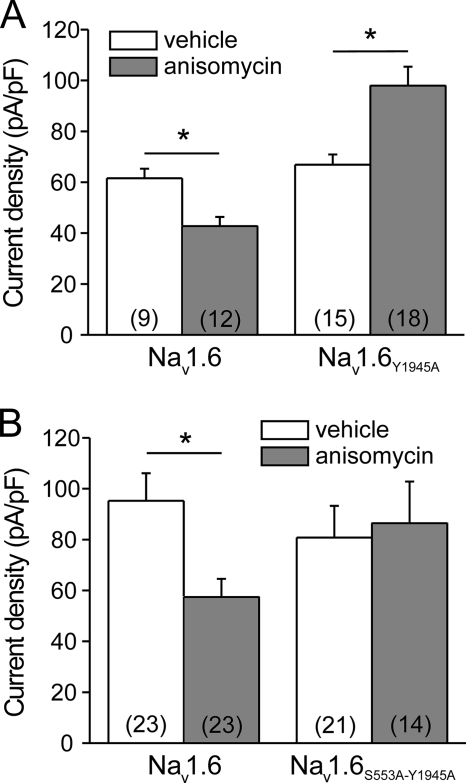
Anisomycin-induced Increase in Current Amplitude of the Mutant Nav1.6Y1945A
We hypothesized that the presence of the Pro-Ser-Tyr1945 motif in Nav1.6/C is required for the pp38-mediated reduction of current density; thus we investigated the effect of anisomycin-mediated p38 activation on the Nav1.6Y1945A mutant channel. Peak current densities of WT and Nav1.6Y1945A were comparable under control conditions (Fig. 8A; WT, vehicle control, 61.6 ± 3.7 pA/pF, n = 9; Nav1.6Y1945A, vehicle control, 66.9 ± 4.0 pA/pF, n = 15). Activation of p38 by anisomycin treatment significantly decreased the peak current density of Nav1.6-WT (42.7 ± 3.7 pA/pF, n = 12). Surprisingly, p38 activation significantly increased the peak current density of Nav1.6Y1945A (98.0 ± 7.4 pA/pF, n = 18). These results show that the C-terminal Pro-Ser-Tyr1945 motif of Nav1.6 is necessary for the pp38-mediated reduction of its peak currents and that loss of the Nedd4-binding site in the C terminus of Nav1.6 results in an effect of p38 activation in the opposite direction.
FIGURE 8.
Nav1.6 mutation Y1945A reverses the p38-mediated reduction of Nav1.6 peak current, and this effect requires Ser553 in Nav1.6/L1. A, ND7/23 cells were transiently transfected with wild-type Nav1.6 or the mutant Nav1.6Y1945A. Activation of p38 by anisomycin decreased the peak current density of Nav1.6-WT channels (left) but increased the peak current density of Nav1.6Y1945A mutant channels (right). B, ND7/23 cells were transiently transfected with wild-type Nav1.6 or the double mutant Nav1.6S553A/Y1945A. Disruption of the p38 phosphorylation site (Ser553 in Nav1.6/L1) abolished the anisomycin-induced increase of peak current density of the Nav1.6Y1945A mutant. Numbers of recorded cells are given in parentheses. Means ± S.E., *, p < 0.05.
Double Mutation Nav1.6S553A/Y1945A Abolishes the pp38-induced Current Increase of Nav1.6Y1945A
The unexpected increase in the current density of Nav1.6Y1945A upon treatment with anisomycin is analogous to the effect of pp38 on the Nav1.8 sodium channel (43), but it may not depend on pp38-mediated phosphorylation of the Pro-Gly-Ser-Pro motif in Nav1.6/L1. Previously, we have shown that S553A substitution completely blocks pp38-mediated reduction of Nav1.6 peak currents (13). To investigate whether phosphorylation of Ser553 is also necessary for the pp38-mediated increase in peak currents of Nav1.6Y1945A, we generated the double mutant Nav1.6S553A/Y1945A. As expected, anisomycin treatment reduced peak current density of Nav1.6-WT (Fig. 8B, left; vehicle control, 95.2 ± 10.9 pA/pF, n = 23, versus anisomycin, 57.4 ± 7.2 pA/pF, n = 23; p < 0.05). However, the pp38-mediated increase in Nav1.6Y1495A current density was abolished in the double mutant Nav1.6S553A/Y1945A (Fig. 8_B_, _right_; vehicle control/ 80.8 ± 12.5 pA/pF, _n_ = 21, _versus_ anisomycin/ 86.4 ± 16.4 pA/pF, _n_ = 14; _p_ > 0.05). Taken together, these results demonstrate that phosphorylation of Ser553 is necessary for both pp38-mediated increase in peak currents of Nav1.6Y1945A channels as well as pp38-mediated decrease in peak currents of WT Nav1.6 channels (13).
DISCUSSION
We have previously shown that activated p38 phosphorylates Nav1.6 exclusively at Ser553 within the Pro-Gly-Ser-Pro motif in Nav1.6/L1 (13), which is conserved in L1 of all vertebrate orthologues identified to date (Fig. 2). Although the L1 region of rat Nav1.8 contains two similar motifs (PRSP and PQSP), but at different positions within this loop, all other VGSC family members lack P_X_SP motifs in their L1 sequences (see supplemental Fig. 1). In agreement with our results, the lack of P_X_SP motifs in other TTX-S channels expressed in the central nervous system suggests that stress-induced pp38 modulation of hippocampal neuronal sodium current is unique to Nav1.6 channels. Phosphorylation of Ser553 by pp38 reduces Nav1.6 currents in the DRG-derived cell line ND7/23 (13), suggesting an adaptive mechanism that might limit sodium influx after injury (44, 45). Here, we provide evidence that in hippocampal neurons (i) p38 and Nav1.6 are co-expressed (Fig. 1), (ii) p38 activation reduces native Nav1.6 currents without affecting other endogenous TTX-S sodium currents (Fig. 3), and (iii) pharmacological block of endocytosis abrogates p38-mediated current reduction (Fig. 4). Taken together, these data are consistent with a model of Nav1.6 internalization induced by p38-mediated phosphorylation of Ser553 within L1 of the channel.
Although chimeric reporter proteins containing only Nav1.6/L1 or Nav1.6/C are not internalized upon p38 activation, the fusion of L1 and C terminus in one polypeptide restores, at least partially, the pp38-mediated internalization of the reporter protein from the plasma membrane (Fig. 5D). Furthermore, Nedd4-2 mediated reduction of Nav1.6 currents (Fig. 6F) depends upon both the Pro-Ser-Tyr motif in Nav1.6/C (Fig. 6F) and the Pro-Gly-Ser-Pro motif in Nav1.6/L1 (Fig. 7B). Mutating the Pro-Ser-Tyr motif in Nav1.6/C, which abolishes binding of Nedd4-2 to Nav1.6/C (Fig. 6A), surprisingly, reverses the pp38 effect on Nav1.6 by increasing the current density (Fig. 8A). Similar to the pp38-mediated current reduction of Nav1.6-WT, this effect depends upon phosphorylation of Ser553 in Nav1.6/L1 (Fig. 8B). Taken together, these data show that regulation of Nav1.6 by pp38 is a complex process that requires phosphorylation of Ser553 in Nav1.6/L1 but also depends on the binding of Nav1.6/C to Nedd4 ubiquitin ligases, and perhaps other proteins that remain to be identified.
Nav1.6 is the predominant VGSC at AIS and nodes of Ranvier in myelinated neurons and is also present in unmyelinated parallel fibers (1–4). Thus, acute regulation of Nav1.6 currents might modulate nerve impulse transmission and neuronal excitability and may allow neurons to adapt rapidly to stressful conditions. The presence of multiple VGSCs within the central nervous system neurons, which share similar biophysical and pharmacological properties, makes the analysis of Nav1.6 regulation challenging. To determine the contribution of effects on Nav1.6 to the neuronal stress response, we compared the impact of p38 activation on peak currents of endogenous TTX-S channels in hippocampal neurons from WT and Scn8amedtg mice, which lack Nav1.6 (6). We show here that p38 activation reduces endogenous TTX-S peak currents in hippocampal neurons from WT mice but not from Scn8amedtg mice, suggesting that other endogenous sodium channels present in these neurons are not regulated by pp38. Because activation of p38 does not alter biophysical properties of Nav1.6 currents in hippocampal neurons and in ND7/23 cells, we suggest that peak currents may be reduced by channel internalization. This hypothesis is supported by our finding that pp38-mediated current reduction in hippocampal neurons is abolished by the dynamin inhibitor Dynasore, which has been previously shown to block endocytosis in different cell types, including neurons (29, 30).
Sodium channel density at the plasma membrane is known to be regulated by cytosolic binding partners, which link channels to various internalization pathways. For example, SCLT1 (sodium channel-CLaThrin-linker 1; previously known as CAP1A), an isoform-specific adaptor protein, binds to both clathrin and Nav1.8 and facilitates internalization of the channel (46). In contrast, Nedd4 family ubiquitin ligases are ubiquitously expressed and can induce internalization of several sodium channels, including ENaC (47, 48) and some VGSCs (17–20). Previously, Nedd4 and Nedd4-2 were shown to bind to the Nav1.6/C region, but the functional impact of this interaction, as well as possible interactions with other intracellular regions of the channel, were not reported (17). Here, we present data that clearly demonstrate a functional effect of the interaction of Nedd4-2 and full-length Nav1.6, resulting in a reduction of peak sodium current.
Ubiquitin ligases induce internalization of target proteins with the outcome dependent on whether mono- or poly-ubiquitination takes place (reviewed in Ref. 14). Nedd4 family ubiquitin ligases (31) contain different numbers of type I WW domains (16) that bind to P_X_Y sequence motifs, which are present in the C termini of all VGSC family members except Nav1.4 and Nav1.9 channels (see supplemental Fig. 2). Here, we confirm that the C terminus of Nav1.6 binds to Nedd4-2 (Fig. 6A), and we demonstrate that co-expression of Nav1.6 with Nedd4-2 results in a robust reduction of Nav1.6 peak currents (Figs. 6D and 7B). This functional effect depends on the Pro-Ser-Tyr motif in Nav1.6/C (Fig. 6D), similar to Nav1.5 (19, 20) and Nav1.8 (17). Interestingly, we found that Nedd4-2-mediated current reduction of Nav1.6 also depends on a functional Pro-Gly-Ser-Pro motif in Nav1.6/L1, because regulation by Nedd4-2 is abolished in the mutant channel Nav1.6S553A. Although we did not activate p38 by anisomycin treatment in these experiments, these data suggest that phosphorylation of Ser553 in WT Nav1.6 takes place, which may be due to basal pp38 activity or activities of other MAPK that are able to phosphorylate SP motifs (e.g. ERK1/2 or JNK). Consistent with this hypothesis is our finding that purified recombinant Nav1.6/L1B produced in bacteria, which is not expected to be phosphorylated at Ser553, does not bind to Nedd4-2 (Fig. 6A), either because of too low affinity or because of phosphorylation-dependent folding of L1 that becomes permissive to Nedd4 binding. Unmasking of weak interactions by overexpression of Nedd4-2 in transfected cells or the marked strengthening of PGpSP affinity to Nedd4 may explain our data.
Previously, we hypothesized (13) that proteins containing type IV WW domains bind to the Pro-Gly-Ser-Pro motif in Nav1.6/L1 after phosphorylation of Ser553 by pp38. Thus, Nedd4-2 might be recruited to the channel after stress-induced p38 activation, causing ubiquitination and internalization of Nav1.6. However, a CD4-L1-EGFP reporter protein carrying the Pro-Gly-Ser-Pro motif was not internalized after p38 activation (Fig. 5), which indicates that Ser553 within the Pro-Gly-Ser-Pro motif is necessary but not sufficient to transduce the effects of pp38. Similarly, p38 activation had no effects on a reporter protein containing only the Nav1.6/C region. In contrast, a reporter protein carrying both Nav1.6/L1 and Nav1.6/C showed a 15% reduction in cell surface localization upon p38 activation (Fig. 5). This suggests that the interaction between the phosphorylated Nav1.6/L1 region and Nedd4 family proteins alone may not be robust enough to facilitate ubiquitination and subsequent internalization of the reporter protein. The additional Pro-Ser-Tyr motif of Nav1.6/C in the CD4-L1-C-EGFP protein could allow a co-operative binding of Nedd4 proteins to the phosphorylated Pro-Gly-Ser-Pro motif in L1, thus inducing ubiquitination and internalization of the protein. However, the reduction of Nav1.6 peak currents in ND7/23 cells upon p38 activation, which was in the range of 40–50% (Fig. 8) (13), was markedly higher than the 15% reduction of surface expression of the reporter protein. A possible explanation for this disparity could be that additional target sites for ubiquitination may be located outside of the Nav1.6/L1 and Nav1.6/C regions of the channel, resulting in an impaired ubiquitination of the chimeric protein compared with the full-length channel. Consistent with this hypothesis, ENaC sodium channel subunits, which are regulated by Nedd4 family proteins upon binding to C-terminal P_X_Y motifs, are ubiquitinated at sites within their N termini (48). Alternatively, the folded structure of the Nav1.6 fragments in the CD4-L1-C-EGFP reporter protein may not be as well suited for interaction with Nedd4 proteins as a result of their topology within the full-length channel protein.
Our data show that both the Pro-Ser-Tyr motif in Nav1.6/C and the Pro-Gly-Ser-Pro motif in Nav1.6/L1 play important roles in the regulation of this channel. In agreement with published data on VGSCs Nav1.5 (19, 20) and Nav1.8 (17), the mutant channel Nav1.6Y1945A is no longer regulated by Nedd4-2 (Fig. 6B). Surprisingly, activation of p38 increased the peak current of Nav1.6Y1945A (Fig. 8A), in contrast to the pp38-induced reduction of Nav1.6-WT peak current. Although the mechanistic details are not well understood, the increase in the peak current of Nav1.6Y1945A by pp38 is reminiscent of the pp38-induced increase in Nav1.8 current density in DRG neurons (43). However, the increase of the peak current of Nav1.6Y1945A by pp38 was abolished by the substitution S553A in the Pro-Gly-Ser-Pro motif in Nav1.6/L1 (Fig. 8B). This indicates that both possible outcomes of the dynamic regulation of Nav1.6 by pp38, a decrease in current density of Nav1.6-WT and an increase in current density of Nav1.6Y1945A, depends upon phosphorylation of Ser553 within Nav1.6/L1.
Taken together, the data presented here reveal a complex regulation of Nav1.6 by pp38. Stress-activated p38 induces phosphorylation of Nav1.6, which causes a reduction in Nav1.6 peak currents in native neurons and neuronal cell lines (this study and Ref. 13). Block of endocytosis abolishes this current reduction, suggesting that phosphorylation of Nav1.6 reduces the number of available channels by internalization. Thus neurons may modulate their excitability as an adaptive response toward cellular stress. However, our data suggest a dual function of the Pro-Gly-Ser-Pro motif within Nav1.6/L1, which can act as a switch that transduces pp38 effects in a positive or negative direction, depending on the ability of Nedd4-2 to bind the Pro-Ser-Tyr motif in the C terminus of the channel.
Supplementary Material
Supplemental Data
Acknowledgments
We thank Dr. Peter Snyder, University of Iowa, for the plasmid pSwick-Nedd4-2-HA and Bart Toftness for technical assistance.
*
This work was supported in part by grants from the National Multiple Sclerosis Society and the Rehabilitation Research Service and Medical Research Service, Department of Veterans Affairs. The Center for Neuroscience and Regeneration Research is a Collaboration of the Paralyzed Veterans of America and the United Spinal Association with Yale University.
3
The abbreviations used are:
VGSC
voltage-gated sodium channel
AIS
axon initial segment
DRG
dorsal root ganglia
MAPK
mitogen-activated protein kinase
Nav1.6/L1
intracellular loop 1 of Nav1.6
Nav1.6/C
intracellular C-terminal domain of Nav1.6
TTX
tetrodotoxin
BisTris
2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
WT
wild type
PBS
phosphate-buffered saline
HRP
horseradish peroxidase
GFP
green fluorescent protein
MOPS
4-morpholinepropanesulfonic acid
pF
picofarad
HA
hemagglutinin.
REFERENCES
- 1.Caldwell J. H., Schaller K. L., Lasher R. S., Peles E., Levinson S. R. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5616–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boiko T., Rasband M. N., Levinson S. R., Caldwell J. H., Mandel G., Trimmer J. S., Matthews G. (2001) Neuron 30, 91–104 [DOI] [PubMed] [Google Scholar]
- 3.Boiko T., Van Wart A., Caldwell J. H., Levinson S. R., Trimmer J. S., Matthews G. (2003) J. Neurosci. 23, 2306–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black J. A., Renganathan M., Waxman S. G. (2002) Mol. Brain Res. 105, 19–28 [DOI] [PubMed] [Google Scholar]
- 5.Meisler M. H., Kearney J., Escayg A., MacDonald B. T., Sprunger L. K. (2001) Neuroscientist 7, 136–145 [DOI] [PubMed] [Google Scholar]
- 6.Meisler M. H., Plummer N. W., Burgess D. L., Buchner D. A., Sprunger L. K. (2004) Genetica 122, 37–45 [DOI] [PubMed] [Google Scholar]
- 7.Craner M. J., Newcombe J., Black J. A., Hartle C., Cuzner M. L., Waxman S. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8168–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craner M. J., Hains B. C., Lo A. C., Black J. A., Waxman S. G. (2004) Brain 127, 294–303 [DOI] [PubMed] [Google Scholar]
- 9.Wolf J. A., Stys P. K., Lusardi T., Meaney D., Smith D. H. (2001) J. Neurosci. 21, 1923–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black J. A., Liu S., Waxman S. G. (2008) Glia 57, 1072–1081 [DOI] [PubMed] [Google Scholar]
- 11.Carrithers M. D., Chatterjee G., Carrithers L. M., Offoha R., Iheagwara U., Rahner C., Graham M., Waxman S. G. (2009) J. Biol. Chem. 284, 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craner M. J., Damarjian T. G., Liu S., Hains B. C., Lo A. C., Black J. A., Newcombe J., Cuzner M. L., Waxman S. G. (2005) Glia 49, 220–229 [DOI] [PubMed] [Google Scholar]
- 13.Wittmack E. K., Rush A. M., Hudmon A., Waxman S. G., Dib-Hajj S. D. (2005) J. Neurosci. 25, 6621–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abriel H., Staub O. (2005) Physiology 20, 398–407 [DOI] [PubMed] [Google Scholar]
- 15.Ingham R. J., Gish G., Pawson T. (2004) Oncogene 23, 1972–1984 [DOI] [PubMed] [Google Scholar]
- 16.Sudol M., Hunter T. (2000) Cell 103, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 17.Fotia A. B., Ekberg J., Adams D. J., Cook D. I., Poronnik P., Kumar S. (2004) J. Biol. Chem. 279, 28930–28935 [DOI] [PubMed] [Google Scholar]
- 18.Rougier J. S., van Bemmelen M. X., Bruce M. C., Jespersen T., Gavillet B., Apothéloz F., Cordonier S., Staub O., Rotin D., Abriel H. (2005) Am. J. Physiol. Cell Physiol. 288, C692–C701 [DOI] [PubMed] [Google Scholar]
- 19.van Bemmelen M. X., Rougier J. S., Gavillet B., Apothéloz F., Daidié D., Tateyama M., Rivolta I., Thomas M. A., Kass R. S., Staub O., Abriel H. (2004) Circ. Res. 95, 284–291 [DOI] [PubMed] [Google Scholar]
- 20.Abriel H., Kamynina E., Horisberger J. D., Staub O. (2000) FEBS Lett. 466, 377–380 [DOI] [PubMed] [Google Scholar]
- 21.Herzog R. I., Cummins T. R., Ghassemi F., Dib-Hajj S. D., Waxman S. G. (2003) J. Physiol. 551, 741–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rush A. M., Wittmack E. K., Tyrrell L., Black J. A., Dib-Hajj S. D., Waxman S. G. (2006) Eur. J. Neurosci. 23, 2551–2562 [DOI] [PubMed] [Google Scholar]
- 23.Wittmack E. K., Rush A. M., Craner M. J., Goldfarb M., Waxman S. G., Dib-Hajj S. D. (2004) J. Neurosci. 24, 6765–6775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herzog R. I., Liu C., Waxman S. G., Cummins T. R. (2003) J. Neurosci. 23, 8261–8270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cummins T. R., Dib-Hajj S. D., Herzog R. I., Waxman S. G. (2005) FEBS Lett. 579, 2166–2170 [DOI] [PubMed] [Google Scholar]
- 26.Rush A. M., Dib-Hajj S. D., Waxman S. G. (2005) J. Physiol. 564, 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood J. N., Bevan S. J., Coote P. R., Dunn P. M., Harmar A., Hogan P., Latchman D. S., Morrison C., Rougon G., Theveniau M., Wheatly S. (1990) Proc. R. Soc. Lond. B Biol. Sci. 241, 187–194 [DOI] [PubMed] [Google Scholar]
- 28.Brewer G. J., Torricelli J. R. (2007) Nat. Protoc. 2, 1490–1498 [DOI] [PubMed] [Google Scholar]
- 29.Kirchhausen T., Macia E., Pelish H. E. (2008) Methods Enzymol. 438, 77–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Macia E., Ehrlich M., Massol R., Boucrot E., Brunner C., Kirchhausen T. (2006) Dev. Cell 10, 839–850 [DOI] [PubMed] [Google Scholar]
- 31.Chen C., Matesic L. E. (2007) Cancer Metastasis Rev. 26, 587–604 [DOI] [PubMed] [Google Scholar]
- 32.Catterall W. A., Goldin A. L., Waxman S. G. (2005) Pharmacol. Rev. 57, 397–409 [DOI] [PubMed] [Google Scholar]
- 33.Schaller K. L., Caldwell J. H. (2000) J. Comp. Neurol. 420, 84–97 [PubMed] [Google Scholar]
- 34.Cummins T. R., Rush A. M., Estacion M., Dib-Hajj S. D., Waxman S. G. (2009) Nat. Protoc. 4, 1103–1112 [DOI] [PubMed] [Google Scholar]
- 35.Hedstrom K. L., Rasband M. N. (2006) J. Neurochem. 98, 1345–1352 [DOI] [PubMed] [Google Scholar]
- 36.Cano E., Mahadevan L. C. (1995) Trends Biochem. Sci. 20, 117–122 [DOI] [PubMed] [Google Scholar]
- 37.Raman I. M., Bean B. P. (1997) J. Neurosci. 17, 4517–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.García K. D., Sprunger L. K., Meisler M. H., Beam K. G. (1998) J. Neurosci. 18, 5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raman I. M., Sprunger L. K., Meisler M. H., Bean B. P. (1997) Neuron 19, 881–891 [DOI] [PubMed] [Google Scholar]
- 40.Garrido J. J., Fernandes F., Giraud P., Mouret I., Pasqualini E., Fache M. P., Jullien F., Dargent B. (2001) EMBO J. 20, 5950–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrido J. J., Giraud P., Carlier E., Fernandes F., Moussif A., Fache M. P., Debanne D., Dargent B. (2003) Science 300, 2091–2094 [DOI] [PubMed] [Google Scholar]
- 42.Garrido J. J., Fernandes F., Moussif A., Fache M. P., Giraud P., Dargent B. (2003) Biol. Cell 95, 437–445 [DOI] [PubMed] [Google Scholar]
- 43.Hudmon A., Choi J. S., Tyrrell L., Black J. A., Rush A. M., Waxman S. G., Dib-Hajj S. D. (2008) J. Neurosci. 28, 3190–3201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dargent B., Arsac C., Tricaud N., Couraud F. (1996) Neuroscience 73, 209–216 [DOI] [PubMed] [Google Scholar]
- 45.Paillart C., Boudier J. L., Boudier J. A., Rochat H., Couraud F., Dargent B. (1996) J. Cell Biol. 134, 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu C., Cummins T. R., Tyrrell L., Black J. A., Waxman S. G., Dib-Hajj S. D. (2005) Mol. Cell. Neurosci. 28, 636–649 [DOI] [PubMed] [Google Scholar]
- 47.Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 48.Staub O., Gautschi I., Ishikawa T., Breitschopf K., Ciechanover A., Schild L., Rotin D. (1997) EMBO J. 16, 6325–6336 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data