Maternal Rnf12/RLIM is required for imprinted X chromosome inactivation in mice (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 21.
Published in final edited form as: Nature. 2010 Oct 21;467(7318):977–981. doi: 10.1038/nature09457
Abstract
Two forms of XCI ensure the selective silencing of female sex chromosomes during mouse embryogenesis. Imprinted XCI begins with the detection of Xist RNA expression on the paternal X chromosome (Xp) around the four cell stage of embryonic development. In the embryonic tissues of the inner cell mass (ICM), a random form of XCI occurs in blastocysts which inactivates either the Xp or the maternal X chromosome (Xm) 1,2. Both forms of XCI require the non-coding Xist RNA which coats the inactive X chromosome (Xi) from which it is expressed. Xist plays crucial functions for the silencing of X-linked genes including Rnf12 3,4 encoding the ubiquitin ligase RLIM. Targeting a conditional knockout (KO) of Rnf12 to oocytes where RLIM accumulates to high levels, we find that the maternal transmission of the mutant X chromosome (Δm) leads to embryonic lethality due to defective imprinted XCI. We show that in Δm female embryos the initial formation of Xist clouds and Xp silencing is inhibited. In contrast, ES cells lacking RLIM are able to form Xist clouds and silence at least some X-linked genes during random XCI. These results assign crucial roles to the maternal deposit of Rnf12/RLIM for the initiation of imprinted XCI.
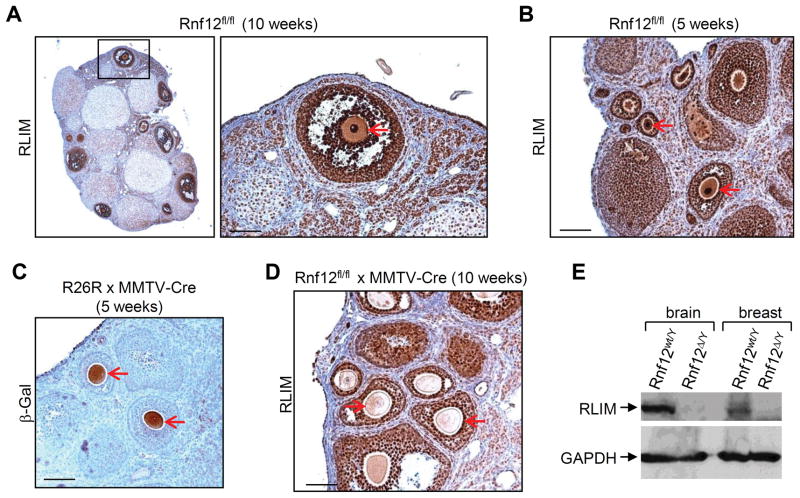
RING finger LIM domain-interacting protein (RLIM) is a ubiquitin ligase that regulates the activity of various transcription factors and cofactors 5–8. It is encoded by the Rnf12 gene 9 which is located around 500kb telomeric to the Xist gene on the X chromosome. During mouse embryogenesis RLIM mRNA and protein is ubiquitously expressed at E7.5/E8.0 5,10, in pre-implantation embryos at E3.5 (Fig. S1), and in mouse ES cells (Fig. S2). In ovaries, we detected particularly high levels of RLIM in oocytes, oocyte-supporting granulosa cells and follicle-surrounding theca cells (Fig. 1A). RLIM levels in pronuclei were high at all stages of oocyte differentiation in 10 and 5 week old mice (Fig. 1A, B). As oocytes in 5 week old females are immature, these results indicate that RLIM accumulates during oocyte maturation.
Fig. 1.
RLIM accumulates during oocyte maturation. Oocytes are indicated by red arrows in A–D. A) Immunohistochemical staining of ovary sections from a fl/fl female 10 weeks of age using RLIM antibodies. Right panel: Higher magnification of boxed region. B) RLIM staining of ovaries from 5 week old fl/fl animals. C) MMTV-LTR-induced Cre expression in oocytes. The Rosa26 loxP-Stop-LoxP –lacZ reporter strain mouse (R26R) was crossed to MMTV-Cre (line F) mice. Paraffin-embedded sections of ovaries of 5 weeks old females were stained with β-Gal antibodies. D) Ovarian section of a 10 weeks old Rnf12fl/fl x MMTV-Cre female stained with RLIM antibodies. Note the loss of RLIM detection in oocytes but not surrounding granulosa cells. E) Knockout of the Rnf12 gene leads to a loss of RLIM protein in mice. Upper panel: Western blot analysis of protein extracts prepared from brain and breast tissues of wt and Rnf12 KO male mice (wt/Y and Δ/Y, respectively) stained with RLIM antibodies. Lower panel: As control, the same blot was probed with GAPDH antibodies. Scale bars = 80 μm.
To generate a mouse model carrying a conditional Rnf12 allele we flanked the coding region of exon 5 with LoxP sites (Fig. S2). Exon 5 encodes 517 out of RLIM’s total of 600 amino acids 9 including the RING finger. We targeted the Rnf12 KO (Δ) to oocytes using transgenic mice that express Cre recombinase (Cre) under the control of the mouse mammary tumor virus long terminal repeat (MMTV-LTR). Several MMTV-Cre mouse lines exist some of which target the female germline 11. Cre expression in oocytes was verified by crossing MMTV-Cre (line F) mice to Rosa26-loxP-stop-loxP-lacZ (R26R) animals 12 (Fig. 1C). Ovaries from Rnf12fl/fl × MMTV-Cre (line F) females (fl/fl-Cre) showed lack of RLIM in oocytes but not in the surrounding granulosa cells or in stromal cells (Fig. 1D), confirming a Rnf12 KO in oocytes. The correct targeting was corroborated by the fact that we obtained KO males with germline deletion of the Rnf12 gene (Δ/Y), lacking RLIM in all somatic tissues examined (Fig. 1E; not shown). Because Δ/Y males are viable and fertile, these results demonstrate that Rnf12/RLIM is not required for basic cellular or developmental functions, for the maturation of oocytes or for meiotic sex chromosome inactivation.
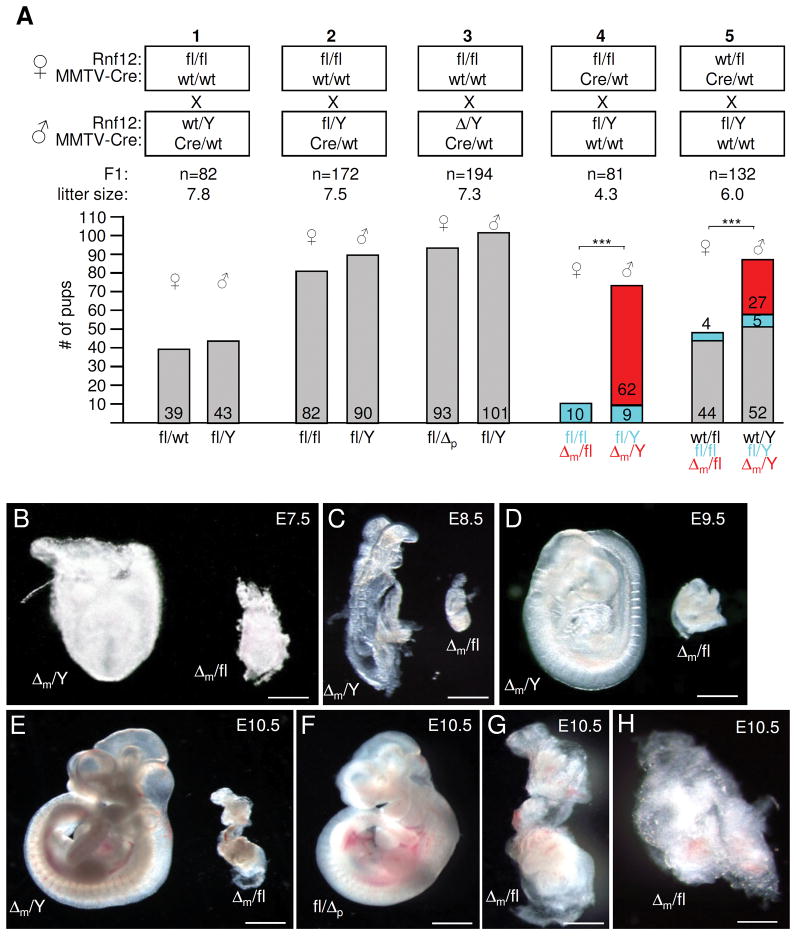
Pups born by fl/fl females displayed normal gender ratios and a normal transmission of the paternal X chromosomes (p) was observed in matings using wt, fl or Δp males (Fig. 2A, mating schemes 1–3). However, no female offspring carrying a maternally transmitted KO (Δm) allele was born by fl/fl-Cre or wt/fl-Cre females crossed with wt, fl or Δp males (Fig. 2A, schemes 4, 5; not shown) whereas the Δm allele transmitted efficiently to male pups. The probability of obtaining male versus female pups from fl/fl-Cre or wt/fl-Cre females was highly significant (P<2 × 10−8). In contrast, wt/fl-Cre females transmitted the wt allele normally and the probability of producing male wt/Y versus male fl/Y + Δm/Y offsprings was similar (P>0.13) (scheme 5). We also performed matings with the MMTV-Cre line D that does not target to the female germline 11 and observed normal Mendelian distributions for male and females pups (n=177; not shown) sired by fl/fl-Cre (line D) females. These findings combined with a decreased mean litter size for matings 4 and 5 indicated that the deletion of Rnf12 in the maternal germ line leads to embryonic lethality. As mice were bred in a congenic C57BL/6 background to eliminate strain-specific influences, our results reveal a sex-specific parent-of-origin effect.
Fig. 2.
A maternally transmitted Rnf12 deletion allele leads to early embryonic lethality specifically in females. Embryos were first photographed and then processed for genotyping in B–H. A) MMTV-Cre mediated loss of Δm females. Schematic diagram of born pups of indicated mating schemes (1–5). Parental genotypes of female (upper) and male (lower) mice with respect to Rnf12 and MMTV-Cre is shown and the total number (n) of F1 offsprings and the mean litter size is indicated. Number of offsprings (grouped in female and male) and their genotypes with respect to Rnf12 are indicated in the abscissa and ordinate, respectively. m (maternal) and p (paternal) indicate the origin of the KO (Δ) allele. In mating scheme 4 and 5, maternally transmitted wt, floxed and Δ alleles are indicated in grey, blue and red, respectively. Three asterisks indicate P values <1×10−7. B–E) Heterozygote Δm/fl female and homozygote Δm/Y littermates from a fl/Δp × fl/Y cross at E7.5 (A); E8.5 (B); E9.5 (C) and E10.5 (D). F) Heterozygote fl/Δp female at E10.5 from a fl/fl × Δ/Y cross. G) Best developed Δm/fl embryo (n=28) detected at E10.5 (magnification of the Δm/fl shown in F). H) Representative Δm female embryo at E10.5. Scale bars = 0.15 mm (B); 0.4 mm (C); 0.6 mm (D); 1 mm (E, F); 0.5 (G); 0,25 mm (H).
Next, we examined embryos that received either a Δm or Δp allele. While male Δm/Y embryos appeared normal, the maternal transmission of the KO allele to female conceptuses in the same litters resulted in severe growth defects that were apparent as early as E7.5 (Fig. 2B–E). However, generating this heterozygous genotype with paternal transmission of the KO allele resulted in female fl/Δp embryos that appeared normal (Fig. 2F). Embryonic components of Δm conceptuses differed in size and showed various degrees of disorganization (Figs 2G; H). The embryo shown in Fig. 2G was the largest observed female Δm embryo. No obvious differences in severity of growth defects between heterozygous Δm/fl and Δ/Δ female embryos were detected at E7.5, E8.5 or E9.5 (Fig. 2; Figs. S3A–C). We were unable to recover Δm female conceptuses at stages later than E11.5 presumably due to reabsorption. Quantifications of phenotypes showed that all recovered Δm female embryos displayed growth defects (Fig. S3D). In 25% of deciduas we were unable to recover a conceptus. As this corresponds to the number of missing female embryos expected for a Mendelian ratio it is highly likely that these correspond to Δm females.
Examining Δm blastocyst outgrowths at pre-implantation stages, trophoblast migration, cell number and expression of the early trophoblast marker Troma-1 13 appeared comparable to controls (Fig. S4A, B). For analyses at early post-implantation stages we sectioned through entire E5.5 and E6.5 deciduas sired by a fl/Δ-Cre × Δ/Y cross. From this mating around 50% of the embryos should correspond to Δm females. Indeed, Hematoxylin/Eosin (HE) stainings revealed around 50% of mildly or severely disorganized embryos (Fig. S4C, D). These results suggest that growth defects in Δm embryos first occur around implantation or very shortly thereafter. Next, we examined the effects of the Rnf12 KO on the development of extraembryonic tissues and analyzed placentae of Δm embryos in the cases when a conceptus was found. HE stainings revealed that most if not all tissues derived from the extraembryonic trophoblast were missing in Δm/fl placentae at E10.5 (n=4) and E9.5 (n=5), whereas the maternal deciduas appeared normal (Figs. S5A, B). This was accompanied by a lack of trophoblast markers PAI-1 and Cdx2 14,15 as early as E8.5 during placental development (n=4) (Fig. S5C; not shown). Because these phenotypes are reminiscent of those described for female Xist KO embryos caused by a paternally inherited _Xist_-KO allele 16, we hypothesized that Rnf12 may regulate XCI.
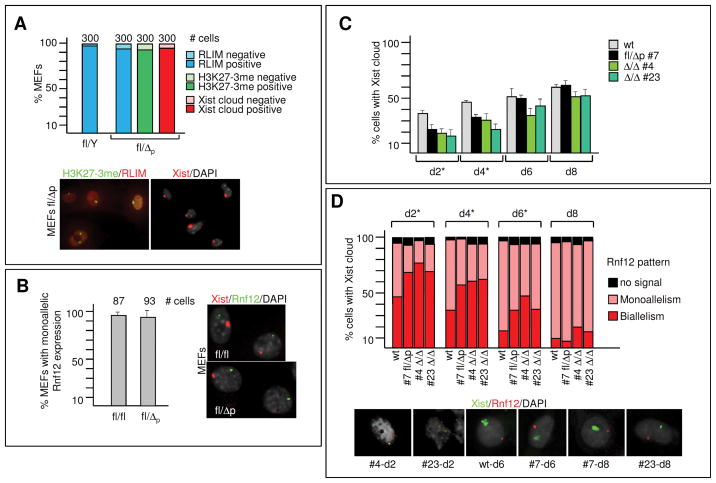
We first examined random XCI in wt/Δp adult females and found that virtually all somatic cells in ovaries of wt/Δp adult females (n=3) stained RLIM-positive (Fig. S6A), and general RLIM levels in somatic tissues were similar in wt/Δp and wt adult females (Fig. S6B). Because the ratio of fl/Δp female to male Δ/Y pups (Fig. 2A, scheme 3) was normal these results suggest that in wt/Δp adult females random XCI is skewed towards the mutated allele. Staining mouse embryonic fibroblasts (MEFs) of fl/Δp embryos at E12.5 with RLIM and H3K27me3 17 antibodies revealed that 94% (n=300 of 3 embryos) of MEFs stained positive for RLIM and H3K27me3 (Fig. 3A). In addition, in RNA fluorescent in-situ hybridization (RNA FISH) experiments using a double strand Xist probe that recognizes Xist and Tsix showed that 97% (n=300 of 3 embryos) of the stained MEFs showed specific Xist paints on one X chromosome.
Fig. 3.
Rnf12 is not required for initiation of random XCI. A) MEFs of E12.5 of fl/Δp and fl/Y males were co-stained with RLIM and H3K27me3 antibodies or with a Xist probe using RNA FISH. Upper panel: Summary graph representing 3 independent experiments. Error bars represent SD. Cells were scored for RLIM expression, the presence of H3K27me3 staining or Xist clouds. Lower left panel: Representative image with RLIM (red) and H3K27me3 (green) staining. Lower right panel: Representative image of Xist staining (red). B) Left panel: Summary graph of RNA FISH experiments on E12.5 fl/Δp MEFs co-stained with probes against Xist and Rnf12. Left panel: Representative images showing monoallelic expression. C, D) Time course of initiation of XCI (C) and silencing of Rnf12 (D) in ES cell lines. ES cells were EB differentiated for the indicated time before co-staining with RNA FISH using Xist and Rnf12 probes. Percentage of _Xist_-positive ES cells (C). Error bars represent SD. Asterisks indicate significant differences between wt and each of the three Δ cell lines (P < 0.05). Xist positive cells were scored for monoallelic or biallelic Rnf12 expression, or no signal (D). Upper panel: Summary graph; lower panel: representative images of various ES cells with biallelic and monoallelic Rnf12 expression. Asterisks indicate significant differences between wt and each of the three Δ cell lines (P < 0.05).
To test the status of the Rnf12 gene on the Xi, we performed RNA FISH with probes against Xist and Rnf12. For Rnf12 we used a 12 kb genomic probe located 5′ of the deletion site that recognizes wt and Δ transcripts of the Rnf12 gene equally well (Fig. S7B). Results revealed that 92% of MEFs that expressed Xist also expressed Rnf12 in a monoallelic fashion similar to fl/fl control MEFs (Fig. 3B). Next, we generated ES cells lacking Rnf12 and isolated the heterozygous fl/Δp line 7 and Δ/Δ ES lines 4, 6, 11 and 23 (Fig. S7A; not shown). Consistent with skewed random XCI we found that 98% of H3K27me3-positive EB-differentiated fl/Δp #7 ES cells expressed RLIM protein, whereas Δ/Δ ES cells did not (Fig. S7C, not shown). All Δ/Δ ES lines stained positive for H3K27me3 and developed Xist clouds upon differentiation (Fig. 3C, D; data not shown). A time course comparing wt female ES cells with lines fl/Δp #7, Δ/Δ #4 and Δ/Δ #23 revealed a slower initiation of XCI at days 2 and 4 during embryoid body (EB)-differentiation (Fig. 3C). However, at days 6 and 8 these differences were no longer significant. The rates of Xist cloud formation of Δ/Δ and fl/Δp ES cells were similar (P>0.05). To assess X silencing in mutant ES cells we co-hybridized cells with Xist and Rnf12 probes in RNA FISH at 2, 4, 6 and 8 days of EB-differentiation. Focusing on cells with Xist clouds (=100%) we compared distribution of Rnf12 signals in nuclei displaying either monoallelism, biallelism or no signal. Silencing of the Rnf12 gene was slower in all Rnf12 mutant ES cells at 2, 4 and 6 days of differentiation when compared to wt (P<0.05). Again, mutated ES cells did not differ significantly among themselves (Fig. 3D). Silencing of Pgk1, another X-linked gene, was also observed in Rnf12 mutant ES cells after 6 days of EB-differentiation (Fig. S7D). These results combined with our finding that in teratoma assays, injecting ES cells in kidney capsules of immunodeprived NOD-SKID mice, Δ/Δ cells participate in the formation of ectodermal, endodermal and mesodermal germ layers and derived cell types (Fig. S7E), indicate that ES cell lacking RLIM/Rnf12 initiate XCI. Our data also suggest that random XCI is skewed toward the mutated Rnf12 allele in fl/Δp females.
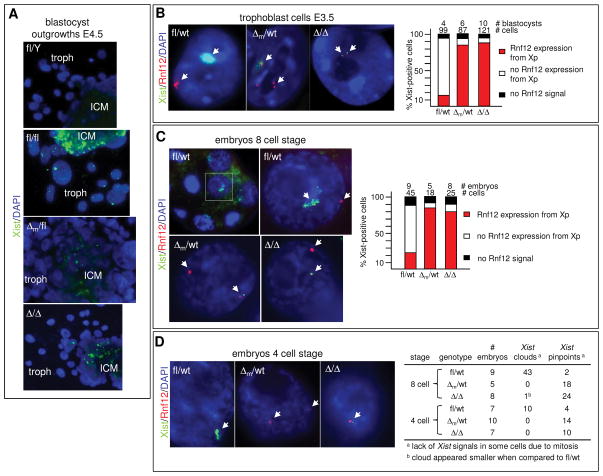
Because these results did not explain the observed embryonic lethalities (Fig. 2), we next investigated imprinted XCI in E3.5 and E4.5 blastocyst outgrowths via RNA FISH. When required, embryo gender was determined via isolation of the inner cell mass (ICM) after image recording and genotyping for the presence of the Zfy gene and, as control, β-actin using PCR. A high percentage of central cells in the ICM of control and Δm E4.5 blastocyst outgrowths developed Xist clouds or single pinpoints, indicating transcription foci (Fig. 4A; Movie S1 and S2; S8A). This suggests that in contrast to ES cells in culture, random XCI occurs with similar kinetics in blastocysts. However, although quantification of ICM stainings was not possible due to high cell density, the decreased number of E4.5 ICM cells staining positive for H3K27me3 17 in Δm when compared to wt blastocysts (Fig. S8B) suggested that imprinted XCI is inhibited in primitive endoderm cells. Importantly, focusing on trophoblasts that also undergo imprinted XCI, only around 10% of Δ/Δ and Δm cells displayed Xist clouds compared to more than 90% of fl/fl and fl/Δp cells (S9A–D). Higher magnification revealed the presence of pinpoints in around 20% of Δm trophoblasts at E4.5 and 30% at E3.5. Generally 1 pinpoint per trophoblast was detected although some cells developed 2 pinpoints likely due to the development of polyploidism 18. The parental origin of these Xist signals was not examined and because we used a double-stranded Xist probe we cannot distinguish between Xist and Tsix pinpoints. Co-stainings with antibodies directed against RLIM and H3K27me3 revealed that only around 10% of Δ/Δ trophoblasts displayed H3K27me3 signals compared to 94% in fl/fl trophoblasts (Fig. S10A–C). To quantitatively compare Xist expression in blastocysts and to monitor expression of _Tsix_2,19,20, we performed RT-qPCR on E3.5 and E4.5 blastocysts comparing Δ/Δ with fl/fl embryos (Fig. S11A). In agreement with results obtained in RNA FISH, Xist expression was reduced in Δ/Δ females. Tsix levels were only mildly affected in E3.5 embryos and decreased at E4.5, suggesting that RLIM does not induce Xist by repression of Tsix RNA transcription. Increased Pgk1 levels suggested defects at the level of Xp silencing.
Fig. 4.
Regulation of Xist cloud formation and X silencing by Rnf12 during imprinted XCI. Probes for RNA FISH were Xist (green) and Rnf12 (red). A) Representative E4.5 blastocyst outgrowths stained with Xist. ICM = inner cell mass; troph = trophoblasts. B) Representative trophoblasts of fl/wt, Δm/wt, and Δ/Δ stained with Xist and Rnf12 are shown. Right panel: For quantification, _Xist_-positive trophoblasts (clouds or pinpoints) were scored for co-localization with Rnf12, no co-localization with Rnf12, and no Rnf12 signal. Numbers of blastocysts and trophoblasts examined are indicated. C) Representative 8-cell stage embryos. Boxed area is magnified on the right. Right panel: Quantifications. D) Representative 4-cell stage embryos hybridized with Xist and Rnf12 probes. Right table: Summary of Xist signals detected in embryos at the 4 and 8 cell stages.
To investigate Xp silencing we co-hybridized E3.5 blastocyst outgrowths with the Xist probe and with probes recognizing X-linked genes Pgk1 or Rnf12. While both the Pgk1 or Rnf12 genes were efficiently silenced on the Xp in control trophoblasts, we observed at least two spots of Pgk1 or Rnf12 in a high percentage of Δm trophoblasts with at least one signal in proximity of a Xist pinpoint (Fig. 4B; S11B,C). Furthermore, the formation of nuclear compartments around the Xp that exclude Cot-1 RNA or general transcription factors such as TATA-Box binding protein (TBP) 21,22 was inhibited in Δm trophoblasts (Fig. S12; not shown). Defects in Rnf12 silencing were observed as early as the 8 cell stage (Fig. 4C). Because this is the earliest stage when X-linked genes are silenced 3,4, these results indicate that defects do not occur at the maintenance level. This is also in agreement with the finding that Δm cells did not form initial Xist clouds in 8 or 4 cell-staged embryos (Figs. 4C–D). However, because we detected one Xist transcription foci in a significant number of cells and Xist accumulation is regulated at the transcriptional level 23, our data are consistent with a crucial role for RLIM/Rnf12 for the transcriptional upregulation of Xist. The fact that a small but significant number of Δm trophoblasts developed Xist clouds (Fig. S9) indicates that RLIM is not absolutely required for Xist upregulation but rather for it to occur reliably at the appropriate time. Indeed, RLIM is able to modulate the transcriptional activity of various classes of transcription factors 5,8.
While our manuscript was in preparation a paper was published which showed that overexpression of RLIM/Rnf12 initiates random XCI in ES cells 24. While the reported data are in general agreement with our results, our finding that _Rnf12_−/− ES cells initiate random XCI was surprising, suggesting the existence of several competence factors that may compensate for the lack of RLIM/Rnf12. Our in vivo data show that RLIM/Rnf12 is required for imprinted XCI suggesting that it may be the only competence factor present in high concentrations during imprinted XCI. Indeed, the observed female parent-of origin effect in XCI in Δm females (Fig. 2) suggests that the maternal RLIM/Rnf12 protein/mRNA drives imprinted XCI and several observations support this view: 1. RLIM protein/mRNA accumulates to high levels in oocytes (Fig. 1); 2. RLIM is undetectable in two cell stage embryos carrying a Δm allele (Fig. S13); 3. Overexpression of RLIM protein can initiate random XCI 24; 4. The paternal contribution of RLIM/Rnf12 appears irrelevant for imprinted XCI (Fig. 2); and 5. It explains why imprinted XCI occurs in fl/Δp females (Figs. 2; S9). Indeed, the maternal deposit of mRNA/protein to control early embryogenesis represents a mechanism that is widespread among many species 25 including mice 26. Thus, we propose that maternal Rnf12/RLIM acts as a crucial regulator for the initiation of imprinted XCI in mice (Fig. S14).
Methods Summary
Mice were generated in which the coding region of exon 5 of the Rnf12 gene was flanked by loxP sites. Exon 5 was deleted in oocytes via MMTV-Cre. All embryos and blastocyst outgrowths examined were derived from natural matings. E3.5/E4.5 blastocysts were cultured for 2–3 days on gelatin-coated coverslips as described 27. Immunocytochemistry and Western blots were performed as reported 28. Paraffin-embedded sections of mouse tissues were stained with Hematoxylin/Eosin (Histoserv. Inc, Germantown, MD). Immunohistochemical stainings were performed in the DERC Morphology core at UMMS. Procedures used for establishing mouse ES cells have been described 29. RNA FISH experiments were performed as reported 21. Full Methods and associated references are available in the Supplementary Methods.
Supplementary Material
1
2
3
Acknowledgments
We thank V. Boyartchuk, T. Fazzio, E. Heard, P. Kaufman, O. Rando, D. Riethmacher and J. Sharp for advice and reagents and J. Zhu for statistics. I.B. is a member of the UMass DERC (DK32520). This work was supported from NIH grants R01CA131158 (NCI) and 5 P30 DK32520 (NIDDK) to I.B. and GM053234 to J.B.L.
Footnotes
Author contribution
J.S. and I.B. conceived and designed the experiments. M.B., I.H.-B., and I.B. generated the floxed Rnf12 mice. J.S. and Y.C. established and analyzed ES cell lines. J.S., M.B., H.M., M.B., N.T.-I., X.Z., and B.J. performed experiments. All authors analysed the data. I.B. wrote the manuscript.
Reprints and permissions information is available at npg.nature.com/reprintsandpermission. The authors declare no financial competing interests.
Reference List
- 1.Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- 2.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 3.Patrat C, et al. Dynamic changes in paternal X-chromosome activity during imprinted X-chromosome inactivation in mice. Proc Natl Acad Sci U SA. 2009;106:5198–5203. doi: 10.1073/pnas.0810683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bach I, et al. RLIM inhibits functional activity of LIM homeodomain transcription factors via recruitment of the histone deacetylase complex. Nat Genet. 1999;22:394–399. doi: 10.1038/11970. [DOI] [PubMed] [Google Scholar]
- 6.Ostendorff HP, et al. Ubiquitination-dependent cofactor exchange on LIM homeodomain transcription factors. Nature. 2002;416:99–103. doi: 10.1038/416099a. [DOI] [PubMed] [Google Scholar]
- 7.Gungor C, et al. Proteasomal selection of multiprotein complexes recruited by LIM homeodomain transcription factors. Proc Natl Acad Sci U SA. 2007;104:15000–15005. doi: 10.1073/pnas.0703738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnsen SA, et al. Regulation of estrogen-dependent transcription by the LIM cofactors CLIM and RLIM in breast cancer. Cancer Res. 2009;69:128–136. doi: 10.1158/0008-5472.CAN-08-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostendorff HP, et al. Functional characterization of the gene encoding RLIM, the corepressor of LIM homeodomain factors. Genomics. 2000;69:120–130. doi: 10.1006/geno.2000.6311. [DOI] [PubMed] [Google Scholar]
- 10.Ostendorff HP, et al. Dynamic expression of LIM cofactors in the developing mouse neural tube. Dev Dyn. 2006;235:786–791. doi: 10.1002/dvdy.20669. [DOI] [PubMed] [Google Scholar]
- 11.Wagner KU, et al. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 12.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 13.Oshima RG, Howe WE, Klier FG, Adamson ED, Shevinsky LH. Intermediate filament protein synthesis in preimplantation murine embryos. Dev Biol. 1983;99:447–455. doi: 10.1016/0012-1606(83)90294-4. [DOI] [PubMed] [Google Scholar]
- 14.Feinberg RF, et al. Plasminogen activator inhibitor types 1 and 2 in human trophoblasts. PAI-1 is an immunocytochemical marker of invading trophoblasts. Lab Invest. 1989;61:20–26. [PubMed] [Google Scholar]
- 15.Beck F, Erler T, Russell A, James R. Expression of Cdx-2 in the mouse embryo and placenta: possible role in patterning of the extra-embryonic membranes. Dev Dyn. 1995;204:219–227. doi: 10.1002/aja.1002040302. [DOI] [PubMed] [Google Scholar]
- 16.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 17.Plath K, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 18.Barlow PW, Sherman MI. The biochemistry of differentiation of mouse trophoblast: studies on polyploidy. J Embryol Exp Morphol. 1972;27:447–465. [PubMed] [Google Scholar]
- 19.Stavropoulos N, Lu N, Lee JT. A functional role for Tsix transcription in blocking Xist RNA accumulation but not in X-chromosome choice. Proc Natl Acad Sci U SA. 2001;98:10232–10237. doi: 10.1073/pnas.171243598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JT. Disruption of imprinted X inactivation by parent-of-origin effects at Tsix. Cell. 2000;103:17–27. doi: 10.1016/s0092-8674(00)00101-x. [DOI] [PubMed] [Google Scholar]
- 21.Hall LL, et al. An ectopic human XIST gene can induce chromosome inactivation in postdifferentiation human HT-1080 cells. Proc Natl Acad Sci U SA. 2002;99:8677–8682. doi: 10.1073/pnas.132468999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- 23.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 24.Jonkers I, et al. RNF12 Is an X-Encoded Dose-Dependent Activator of X Chromosome Inactivation. Cell. 2009;139:999–1011. doi: 10.1016/j.cell.2009.10.034. [DOI] [PubMed] [Google Scholar]
- 25.Nusslein-Volhard C, Frohnhofer HG, Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 26.Letterio JJ, et al. Maternal rescue of transforming growth factor-beta 1 null mice. Science. 1994;264:1936–1938. doi: 10.1126/science.8009224. [DOI] [PubMed] [Google Scholar]
- 27.Guidi CJ, et al. Disruption of Ini1 leads to peri-implantation lethality and tumorigenesis in mice. Mol Cell Biol. 2001;21:3598–3603. doi: 10.1128/MCB.21.10.3598-3603.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tursun B, et al. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 2005;19:2307–2319. doi: 10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung Y, et al. Embryonic and extraembryonic stem cell lines derived from single mouse blastomeres. Nature. 2006;439:216–219. doi: 10.1038/nature04277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3