The TRIM-NHL Protein TRIM32 Activates MicroRNAs and Prevents Self-Renewal in Mouse Neural Progenitors (original) (raw)
. Author manuscript; available in PMC: 2010 Nov 19.
SUMMARY
In the mouse neocortex, neural progenitor cells generate both differentiating neurons and daughter cells that maintain progenitor fate. Here, we show that the TRIM-NHL protein TRIM32 regulates protein degradation and microRNA activity to control the balance between those two daughter cell types. In both horizontally and vertically dividing progenitors, TRIM32 becomes polarized in mitosis and is concentrated in one of the two daughter cells. TRIM32 overexpression induces neuronal differentiation while inhibition of TRIM32 causes both daughter cells to retain progenitor cell fate. TRIM32 ubiquitinates and degrades the transcription factor c-Myc but also binds Argonaute-1 and thereby increases the activity of specific microRNAs. We show that Let-7 is one of the TRIM32 targets and is required and sufficient for neuronal differentiation. TRIM32 is the mouse ortholog of Drosophila Brat and Mei-P26 and might be part of a protein family that regulates the balance between differentiation and proliferation in stem cell lineages.
INTRODUCTION
The mammalian neocortex develops from a pseudostratified epithelium (Gotz and Huttner, 2005). Initially, neuroepithelial cells divide symmetrically into two daughter cells that maintain progenitor cell fate. Starting with embryonic day E10, however, neurogenesis begins and an increasing number of divisions become asymmetric, giving rise to one progenitor and one cell that differentiates into a neuron (Wodarz and Huttner, 2003). How distinct fates are generated in the two daughter cells of these asymmetric divisions is one of the key unresolved questions in developmental neurobiology (Knoblich, 2008).
Neural progenitor cells occupy the apical-most part of the neuroepithelium, which lines the ventricular cavity and is called the ventricular zone. They are called radial glia cells because they express glial markers and extend long radial fibers that extend apically to the ventricular surface and basally all the way to the pial surface (Fishell and Kriegstein, 2003; Gotz and Huttner, 2005). Radial glia cells undergo a cell-cycle-dependent movement called interkinetic nuclear migration: after completing S phase in the more basal areas of the ventricular zone, their nuclei move apically and mitotic divisions happen close to the ventricular lumen. After division, cells that retain progenitor fate remain in the ventricular zone while cells that exit the cell cycle migrate basally along the pial fiber to form the cortical plate. Later during neurogenesis, daughter cells can also become intermediate progenitors that migrate between the ventricular zone and the cortical plate and undergo one or more terminal divisions to form two or four neurons (Noctor et al., 2004; Haubensak et al., 2004).
In Drosophila, the asymmetric inheritance of cell fate determinants establishes distinct fates in the daughter cells of neural progenitors (Doe, 2008; Knoblich, 2008). In Drosophila neuroblasts, the proteins Numb, Prospero, and Brat are inherited by one of the two daughter cells. In this cell, Numb regulates endocytosis (Berdnik et al., 2002) and inhibits Notch signaling (Guo et al., 1996), while Prospero controls the transcription of cell-cycle and differentiation genes (Li and Vaessin, 2000; Choksi et al., 2006). Brat can act as a posttranscriptional regulator and controls the transcription factor dMyc, but how it acts on a molecular level is currently unclear (Lee et al., 2006b; Bello et al., 2006; Betschinger et al., 2006).
Prospero, Brat, and Numb as well as the proteins governing their asymmetric segregation have homologs in vertebrates. Numb is required for mouse neurogenesis (Zhong et al., 1996, 2000; Petersen et al., 2002; Li et al., 2003), but unlike in Drosophila, it is involved in recycling E-Cadherin in adherens junctions (Rasin et al., 2007). Mouse Prospero controls proliferation in the retina (Dyer et al., 2003) but has not been described to segregate asymmetrically. Finally, the stereotypic apical-basal orientation of the mitotic spindle does not seem to be conserved, and most neural progenitor divisions occur with a planar orientation (Kosodo et al., 2004; Noctor et al., 2008; Konno et al., 2008). Thus, it is currently not clear whether the asymmetric segregation of protein determinants is important for vertebrate neurogenesis at all.
Here, we analyze the functional conservation of Brat. Brat is an inhibitor of cell proliferation (Frank et al., 2002) whose asymmetric segregation restricts self-renewal to only one of the two daughter cells in Drosophila larval neuroblasts (Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006b). Brat can bind to the RNase Argonaute 1 (Ago1), but the functional significance of this interaction has not been determined (Neumuller et al., 2008). The Brat paralog Mei-P26, however, can inhibit microRNA activity by binding to Ago1 and thereby regulates proliferation in the Drosophila ovarian stem cell lineage (Neumuller et al., 2008). It reduces cell growth in cells that have lost contact with the ovarian stem cell niche. Since microRNAs are essential for self-renewal in ovarian stem cells (Hatfield et al., 2005; Park et al., 2007; Jin and Xie, 2007), this mechanism prevents uncontrolled proliferation in the Drosophila ovary. Our data show that the inhibitory effect on stem and progenitor cell proliferation is conserved in the Brat/Mei-P26 homolog TRIM32. In dividing cortical progenitor cells, TRIM32 is enriched in one of the two daughter cells and becomes upregulated during neuronal differentiation. TRIM32 is required and sufficient for suppressing self-renewal and inducing neuronal differentiation and acts both by degrading the transcription factor c-Myc and by activating certain microRNAs, among them the well-characterized stem cell regulator Let-7a (Bussing et al., 2008; Rybak et al., 2008; Wulczyn et al., 2007). Since Let-7a is required and sufficient for neuronal differentiation it might be a key target of TRIM32. Thus, we provide a potential explanation for the asymmetric outcome of mouse brain progenitor divisions and show that regulation of microRNA activity is important for the control of self-renewal.
RESULTS
TRIM32 Can Inhibit Proliferation
In Drosophila the TRIM-NHL proteins Brat and Mei-P26 control stem cell proliferation in neuroblasts and ovaries, respectively. TRIM-NHL proteins contain a C-terminal NHL domain, a coiled-coil region, one or more B boxes, and sometimes an N-terminal RING finger (Reymond et al., 2001; Figure S1A available online). In vertebrates, the TRIM-NHL proteins TRIM2, TRIM3, and TRIM32 are equally distant from Brat and Mei-P26. Since TRIM32 has previously been implicated in several inherited diseases (Frosk et al., 2002; Chiang et al., 2006), we decided to analyze its function in controlling mouse stem cell division.
To test whether TRIM32—like Brat—can inhibit proliferation, we overexpressed the gene in mouse NIH 3T3 fibroblasts (Figures S1B–S1G). In control fibroblasts overexpressing EGFP, 81% (n > 300) of all cells are positive for the proliferation marker anti-Ki67. 10.5% (n > 300) of these control cells are in mitosis and positive for anti-phospho-Histone H3. Forty-eight hours after transfection with an N-terminal EGFP-TRIM32 fusion, however, the number of Ki67-positive cells is reduced to 22% (n = 150) and only 2.5% (n > 300) of the cells are in mitosis. Anti-Fibrillarin staining reveals that the size of the nucleolus is reduced by 89% upon EGFP-TRIM32 transfection (Figures S1F and S1G). Since the nucleolus is the site of ribosomal RNA synthesis and its size often correlates with ribosome biogenesis (Frank et al., 2002), this might indicate that TRIM32—like Brat—can inhibit protein biosynthesis. However, TRIM32 expression does not induce apoptosis as the number of cells expressing activated Caspase-3 is not increased (Figure S1H). We conclude that TRIM32 can act as an inhibitor of cellular proliferation.
TRIM32 Distribution Is Polarized in Mitotic Mouse Neural Progenitors
To test the TRIM32 expression and localization, we raised an N-terminal peptide antibody. The antibody is specific since it detects a single band in immunoblots that disappears after TRIM32 RNAi (Figure S4, see below) and since anti-TRIM32 immunofluorescence disappears after preincubation of the antibody with the antigenic peptide (Figure S2B). At E14.5, the peak of mouse cortical neurogenesis (Gotz and Huttner, 2005), TRIM32 is strongly expressed in the cortical plate where it colocalizes with the neuronal marker TuJ1 (neuronal class III β-Tubulin). TRIM32 is also found in the ventricular zone (Figure 1A) starting at E12.5 (Figure S2A), while at later stages of development (E18.5, Figure S2A), expression becomes progressively stronger in the developing cortical layers and disappears from the ventricular zone. Thus, TRIM32 is highly expressed in differentiating neurons and at a lower levels in dividing progenitors.
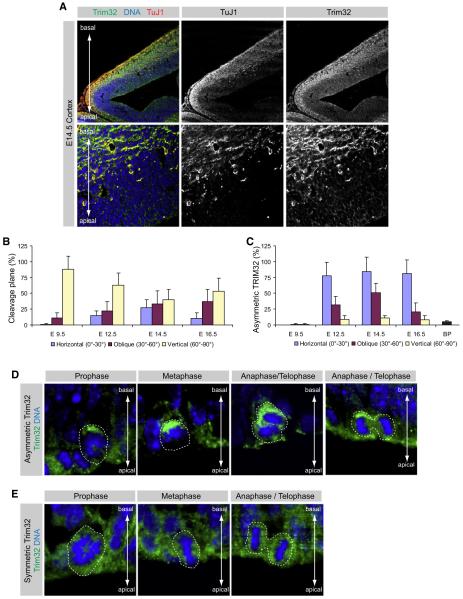
Figure 1. Trim32 Is Asymmetrically Distributed during Neurogenic Cell Divisions.
(A) Immunostainings of cryosections from the E14.5 mouse cortex labeled with the indicated markers (upper gray boxes). The lower panels show a high magnification of the ventricular zone (most apical), subventricular zone, and the apical part of the cortical plate (most basal).
(B) The fraction of dividing neural progenitors in anaphase or telophase with the indicated orientation of the cleavage plane is shown (mean ± standard error of the mean [SEM]). The angle of the cleavage plane was determined by calculating the angle between two virtual lines drawn at the ventricular surface and half way between the two sister chromatids.
(C) Diagram shows the fraction of those neural progenitor cells that divide at the ventricular surface or in the subventricular zone (basal progenitors [BP]) at the indicated stages, which shows a significant enrichment of TRIM32 at the basal pole (mean ± SEM).
(D and E) High-magnification immunostainings of dividing neural progenitor cells in different phases of the cell cycle (upper gray boxes) labeled with the indicated markers (left gray boxes). The dashed line highlights the cells that are in the indicated phase of cell cycle. The cell-cycle phases were identified by Hoechst staining for DNA.
Since Drosophila Brat segregates asymmetrically (Betschinger et al., 2006; Lee et al., 2006b; Bello et al., 2006), we followed the subcellular distribution of TRIM32 in dividing neural progenitors. In pro- and metaphase, TRIM32 is concentrated on the more basal side of the cell (Figures 1D and S2E–S2H). At E14.5, TRIM32 is preferentially inherited by one of the two daughter cells in 69% of the dividing progenitor cells during ana- and telophase (Figure S2C), while in other cells, it is equally inherited by both daughter cells (Figure 1E). The fraction of cells where TRIM32 is asymmetric is lower during earlier (E12.5) and later (E16.5) stages of neurogenesis and at E9.5 when most neuroepithelial cells divide symmetrically (Yingling et al., 2008) TRIM32 asymmetry is essentially never observed (Figure 1C). Quantification of imunofluorescence intensities in the two emerging daughter cells reveals that TRIM32 fluorescence is 4.7 times stronger in the basal half of the cell (Figure S2D, see Experimental Procedures for details) while intensities are not significantly different for Hsp70 (ratio 1.25 ± 0.42), a uniformly cytoplasmic protein (Butcher et al., 2000). Costaining of TRIM32 and Actin (outlining the whole cell cortex) reveals that this is not due to cell damage on the apical side (Figure S2G). A similar asymmetric localization is observed with a second, unrelated TRIM32 antibody (Figure S2E). Although TRIM32 is also highly expressed in basal progenitors, less than 5% (n > 100) of these cells show a polarized Trim32 staining (Figures 1C and S2J). TRIM32 asymmetry can also be seen in cultured neural stem cells (NSCs) undergoing asymmetric cell division (Qian et al., 1998, 2000). When stem cells are grown under conditions supporting progressive neuronal differentiation, 14.2% (n > 100) of the mitotic cells show an asymmetric TRIM32 distribution (Figures 2A and 2B). TRIM32 asymmetry is never observed when cells are grown under proliferative conditions where they divide symmetrically and do not give rise to differentiating neurons (data not shown). Thus, TRIM32 segregates asymmetrically in dividing mouse neural progenitor cells, although increased transcription or translation could also contribute to its higher expression level in differentiating neurons.
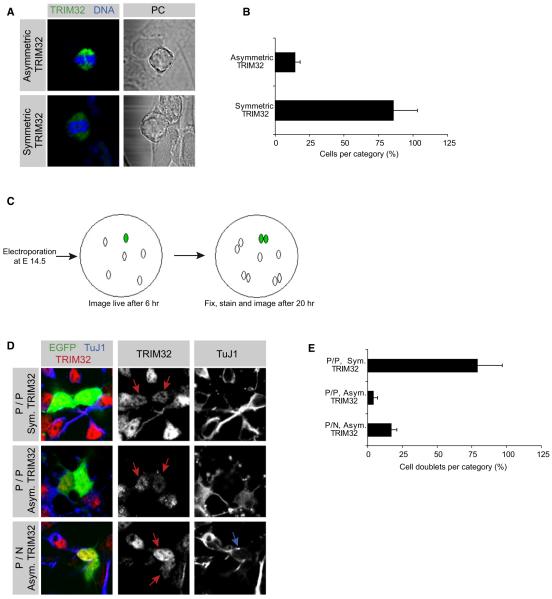
Figure 2. Unequal Distribution of Trim32 during Neuronal Differentiation.
(A) Immunostainings of NSCs in culture labeled with the indicated markers (upper gray boxes). The upper panels show NSCs dividing with asymmetrically localized TRIM32, while the lower panels show cells where TRIM32 is distributed symmetrically during the different phases of mitosis.
(B) The fraction of mitotic NSCs in culture that show an asymmetric or symmetric TRIM32 distribution is shown (mean ± SEM).
(C) The principle of the paired cell analysis is shown. Neural progenitors were electroporated at E14.5 via ex vivo electroporation with a plasmid coding for EGFP. The electroporation was followed by dissociation of the tissue and cultivation for 6 hr. Single cells were imaged live after 6 hr and fixed as doublets after 20 hr.
(D) Cell doublets from the paired cell analysis were immunostained as indicated (upper gray boxes) and grouped according to their TRIM32 and TuJ1 staining (left gray boxes). TuJ1-positive cells were counted as neurons (N) and TuJ1-negative cells were counted as progenitors (P). Red arrows point to transfected cells; the blue arrow points to a transfected TuJ1-positive neuron.
(E) The fraction of cell doublets showing the indicated division pattern is shown (mean ± SEM).
To test whether TRIM32 asymmetry correlates with spindle orientation, we costained brain sections for TRIM32 and the centrosomal marker γ-Tubulin (Figure S2F). In metaphase, TRIM32 localization is not correlated with the metaphase plate (Figure S2F), and in ana- and telophase TRIM32 asymmetry can be seen in divisions of any orientation (Figure 1C). Costaining for Nestin shows that TRIM32 is concentrated in the wider, apical-most part of the basal (pial) fiber and is nearly always inherited by the cell that inherits this fiber (Figure S2I). Thus, the asymmetric inheritance of TRIM32 is not instructed by epithelial apical-basal polarity but may be directed by the morphology of neural precursor cells.
TRIM32 Induces Differentiation in Cultured Neural Stem Cells
To address the effect of TRIM32 on neuronal differentiation and self-renewal, we electroporated TRIM32 overexpression or RNAi constructs in utero at E14.5 (Tabata and Nakajima, 2002). Electroporated brains were dissected and cells were grown in dissociated culture for 4 days. We used anti-Nestin to identify progenitor cells, anti-TuJ1 for early differentiating neurons, anti-MAP2 for late differentiating neurons, and anti-phospho-Histone H3 (p-H3) to determine mitotic activity. Electroporated cells were identified by cotransfection of EGFP or expression of TRIM32-EGFP.
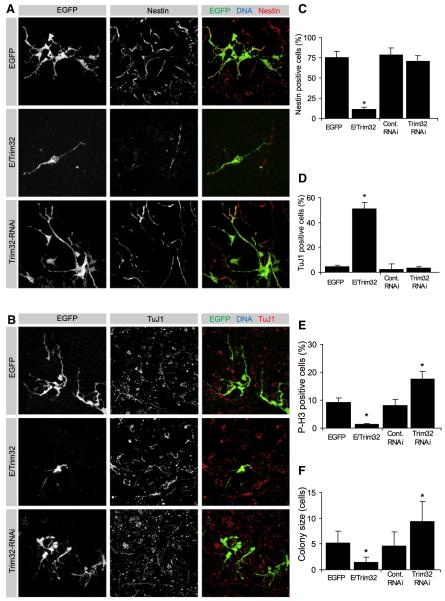
In control experiments (EGFP transfection), colonies consist of four to six cells, 9% (n > 100) of which are typically in mitosis (Figures 3E and 3F). Seventy-five percent of the transfected cells are Nestin positive (n > 100) while only 4% are TuJ1 positive and only 3% are MAP2 positive (Figures 3A–3D and S3). Upon TRIM32-EGFP transfection, however, GFP-positive cells remain individual and do not form colonies and only 1.5% of the transfected cells are p-H3 positive (n > 100). Only 12% of the transfected cells are Nestin positive (n > 100), while 51% of the cells become TuJ1 positive and 41% are positive for MAP2 (n > 100) (Figures 3A–3F). Thus, TRIM32 inhibits proliferation and induces differentiation in mouse neural progenitors.
Figure 3. Trim32 Is Sufficient to Induce Neuronal Cell Fate In Vitro.
(A and B) Neural progenitors were transfected at E14.5 via in utero electroporation with the indicated constructs (left gray boxes). The electroporation was followed by dissociation of the tissue and cultivation for 4 days. Immunostainings of the dissociated cells labeled as indicated (upper gray boxes) are shown.
(C–F) Diagrams show the fraction of transfected cells that are positive for the progenitor marker Nestin (C), the neuronal maker TuJ1 (D), the mitotic marker Phospho-Histone H3(P-H3) (E), and the colony size of transfected cells (F) (mean ± SEM; *p < 0.001 compared to EGFP).
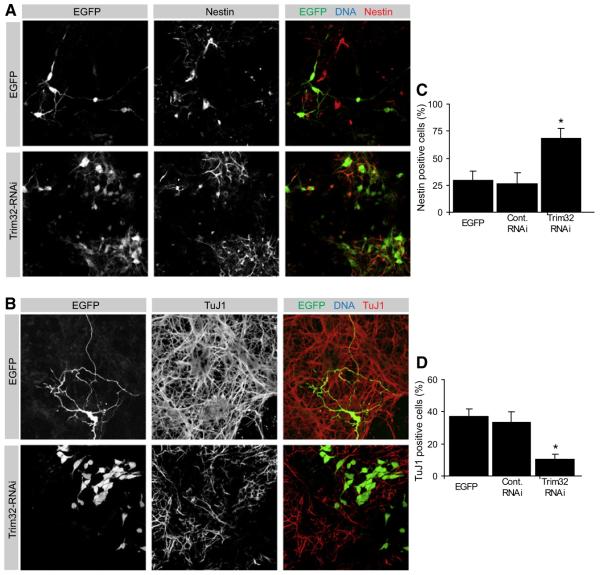
To ask whether TRIM32 is also required for neuronal differentiation, we used a hairpin RNAi construct that strongly reduces TRIM32 expression (Figures S4A and S4B). After 4 days, the differentiation pattern of progenitor cells lacking TRIM32 (71% Nestin-positive cells, 3% TuJ1-positive cells, n > 100) is very similar to that of controls (expressing EGFP or EGFP with a nonfunctional TRIM32 shRNA construct). However, colonies are larger (9 cells on average, n > 100) and have a higher mitotic index (18% p-H3-positive cells). After 6 days, however, control cells start to differentiate (30% Nestin-positive, 37% TuJ1-positive, n > 50) while cells lacking TRIM32 still retain progenitor status (68% Nestin-positive, 10.5% TuJ1-positive) (Figures 4A–4D). When cultured for longer times, EGFP transfected cells typically appear as small clusters of one to six cells with neuronal morphology while cells lacking TRIM32 grow into huge colonies of actively proliferating cells (Figure S4C). A second functional TRIM32 RNAi construct yields similar results (data not shown) while nonfunctional constructs and constructs expressing an unrelated shRNA have no effect. Thus, TRIM32 inhibits proliferation and is required for neuronal differentiation in cultured cortical progenitors.
Figure 4. Knockdown of Trim32 Is Sufficient for the Maintenance of Progenitor Status.
(A and B) Neural progenitors were transfected at E14.5 via in utero electroporation with the indicated constructs (left gray boxes). The electroporation was followed by dissociation of the tissue and cultivation for 6 days. Immunostainings of the dissociated cells labeled as indicated (upper gray boxes) are shown.
(C and D) Diagrams show the fraction of transfected cells that are positive for the progenitor marker Nestin (C) and the neuronal maker TuJ1 (D) (mean ± SEM; *p < 0.001 compared to EGFP).
To ask whether TRIM32 levels are indeed higher in the differentiating neuron, we used a culture system where neural progenitors can divide symmetrically into two progenitors (P/P divisions) or two neurons (N/N divisions) or undergo an asymmetric division to create one neuron and one progenitor cell (P/N divisions) (Qian et al., 1998, 2000) (Figures 2C–2E). Neural progenitors were electroporated at E14.5 with an EGFP expression construct. Single individual cells were identified by live imaging of EGFP expression after 6 hr and investigated by immunofluorescence after 20 hr when most of them had divided and appeared as doublets. Under our culture conditions (see Experimental Procedures), N/N divisions are not observed as TuJ1 levels are never high in both cells. In 79% of the analyzed cell pairs (n > 100), TuJ1 and TRIM32 expression are low in both daughter cells, indicating a P/P division (P/P Sym. TRIM32,Figures 2D and 2E). In 4% of the divisions, both daughter cells are negative for TuJ1 although TRIM32 levels are higher in one (P/P Asym. TRIM32, Figures 2D and 2E). In these cells, insufficient time might have passed after mitosis to allow significant upregulation of TuJ1. In 17% of the divisions, TuJ1 is upregulated in one of the two daughter cells (Figures 2D and 2E) indicating a P/N division. In all these cell pairs, TRIM32 levels are higher in the TuJ1-positive cell. Thus, TRIM32 levels are higher in the daughter cell that undergoes neuronal differentiation.
TRIM32 Regulates Neurogenesis In Vivo
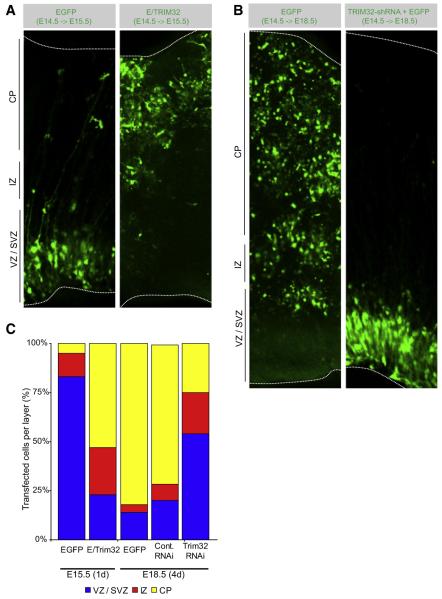
To ask whether TRIM32 also controls neurogenesis in vivo, we analyzed the consequences of TRIM32 overexpression or depletion for mouse brain development. EGFP or a TRIM32-EGFP fusion was introduced into cortical progenitors by in utero electroporation at E14.5. After 1 day, 83% (n > 100) of the EGFP transfected cells are still located in the ventricular or subventricular zone (Figures 5A and 5C) and express Nestin (Figure S5). In contrast, more than 53% (n > 50) of the TRIM32-EGFP transfected cells have entered the cortical plate (Figures 5A and 5C) and have become TuJ1 positive (Figure S5). Thus, TRIM32 overexpression induces neuronal differentiation in vivo.
Figure 5. Trim32 Regulates Neurogenesis In Vivo.
(A and B) Coronal cryosections of the cortex from embryonic brains at E15.5 (A) or E18.5 (B) that have been transfected as indicated (upper gray boxes) at E14.5. The indicated constructs were transfected into the neural progenitors by in utero electroporation. Transfected cells were identified by expression of EGFP. The cortical plate (CP) is the most basal layer followed by the intermediate zone (IZ) and the subventricular zone/ventricular zone, which is the most apical layer. Dotted lines indicate the lateral surface of the ventricular zone and the surface of the cortical plate.
(C) Diagram showing the proportion of transfected cells in VZ/SVZ, IZ, or CP at the indicated time points.
To test whether TRIM32 is also required for differentiation, we analyzed mice 4 days after in utero electroporation (Figures 5B and 5C). At this stage (E18.5), most control cells have migrated into the cortical plate (Figures 5B and 5C) and are TuJ1 positive (Figure S5). Upon TRIM32 inhibition by RNAi, however, only 25% (n > 100) of the cells have reached the cortical plate while 53% are still located in the ventricular or subventricular zone (Figures 5B and 5C) and are Nestin positive (Figure S5). Similar observations are made with a second functional TRIM32-RNAi construct while nonfunctional constructs and constructs expressing an unrelated shRNA are without effect (Figure 5C). Taken together, our data suggest that the higher levels of TRIM32 in one of the two daughter cells of dividing neural progenitors contribute to the differentiation of this cell into a cortical neuron while lower TRIM32 levels cause the other daughter cell to retain progenitor status.
TRIM32 Acts as an Ubiquitin Ligase to Mark c-Myc for Degradation
Since Brat can inhibit expression of the transcription factor dMyc (Betschinger et al., 2006), we tested whether TRIM32 has a similar activity. TRIM32 and c-Myc can be coimmunoprecipitated from mouse brain lysates indicating that the two proteins are present in a robust complex (Figure 6A). TRIM32 contains a RING finger that has ubiquitin ligase activity (Kudryashova et al., 2005) and might induce Myc degradation. Indeed, c-Myc levels are significantly reduced upon TRIM32 coexpression in 293T cells (Figure 6B). Upon proteasome inhibition by clasto-lactacystin-beta-lactone (Schwamborn et al., 2007) (Figure 6B) or MG-132 (data not shown), c-Myc degradation is prevented. Instead, polyubiquitinated forms of c-Myc appear that can be visualized by anti-HA staining upon cotransfection of HA-tagged ubiquitin (Treier et al., 1994). Thus, TRIM32 is a ubiquitin ligase for c-Myc that controls c-Myc protein stability.
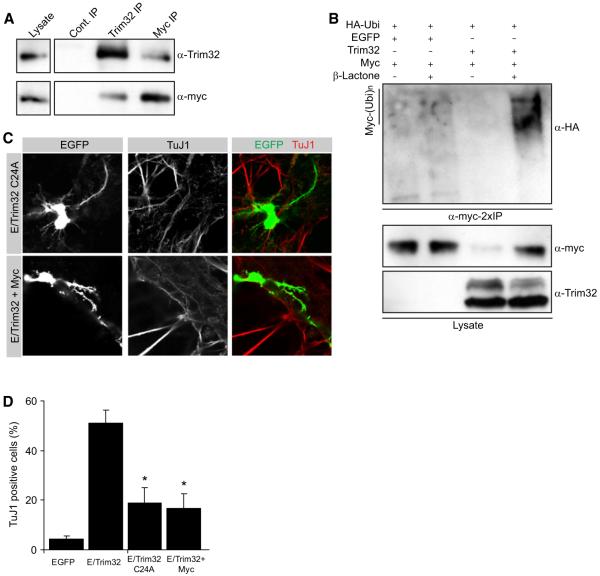
Figure 6. Trim32 Ubiquitinates Myc.
(A) Immunoblots of immunoprecipitations (IP) from embryonic brain lysates with antibodies against TRIM32 and Myc. As control-IP the anti-Trim32 antibody was preincubated with a blocking peptide.
(B) HEK293T cells were transfected as indicated and incubated overnight with clasto-Lactacystin-β-lactone (β-Lactone), and Myc was immunoprecipitated twice (2×IP) with an anti-Myc antibody. Ubiquitin-conjugated Myc (Myc-(Ubi)n) was detected with an anti-HA antibody labeling cotransfected HA-tagged ubiquitin. Myc and Trim32 in the lysate were detected with specific antibodies as indicated.
(C) Immunostainings of dissociated cells that have been transfected as indicated (left gray boxes) via ex vivo electroporation as neural progenitors at E14.5. After 4 days in culture the cells were fixed and labeled as indicated (upper gray boxes).
(D) Diagram showing the quantification for the relative amount of transfected cells that are positive for the neuronal maker TuJ1 (mean ± SEM; *p < 0.001 compared to EGFP).
To test the functional significance of c-Myc degradation, we mutated a catalytically important cysteine in the RING finger. The resulting mutant TRIM32-C24A no longer ubiquitinylates c-Myc and no longer targets c-Myc for degradation (Figure S6A). While TRIM32 transfection reduces the number of Ki67-positive fibroblasts to 22.1% (n > 100) TRIM32-C24A is less potent (40.2% Ki67-positive cells, n > 100) (Figures S6C and S6E). However, TRIM32 and TRIM32-C24A are equally potent in reducing the number of p-H3-positive mitotic cells (5.2% versus 7.3% p-H3-positive cells, respectively, Figures S6B and S6D). Thus, c-Myc degradation is essential for TRIM32 to induce cell-cycle exit, but other targets prevent mitosis even when c-Myc is not degraded.
To test the relevance of c-Myc degradation for neuronal differentiation, we coelectroporated TRIM32 and c-Myc or electroporated the mutant construct TRIM32-C24A. In both cases, electroporated cells still do not form colonies and do not divide 4 days after in utero electroporation (Figure 6C). Whereas TRIM32 increases the number of TuJ1-positive cells from 4.3% (control, EGFP transfection) to 51%, TRIM32-C24A or TRIM32 plus c-Myc cause only a mild increase to 18.9% or 16.7%, respectively (Figures 6C and 6D). Thus, c-Myc degradation is essential for TRIM32 to induce neuronal differentiation whereas mitotic proliferation is inhibited via other downstream pathways as well.
TRIM32 Binds Ago1 and Activates the MicroRNA Let-7a
Brat and Mei-P26 interact with Ago1 (Neumuller et al., 2008). Using coimmunoprecipitation from mouse brain lysates, we find that this protein interaction is conserved in TRIM32 (Figure 7A). A deletion analysis reveals that the C-terminal NHL domain is responsible for Ago1 binding (Figure 7B). Unlike c-Myc, however, Ago1 is not ubiquitinated by TRIM32 (data not shown). By radioactive RNA end-labeling experiments, we detect some large RNA fragments as well as RNA of approximately 21 nt, the size typical for processed microRNAs in both the TRIM32 and Ago1 immunoprecipitations (IPs) but not control IPs in which anti-TRIM32 is blocked by an antigenic peptide (Cont. IP) (Figure 7C). Thus, TRIM32 exists in a complex with Ago1 containing processed microRNAs.
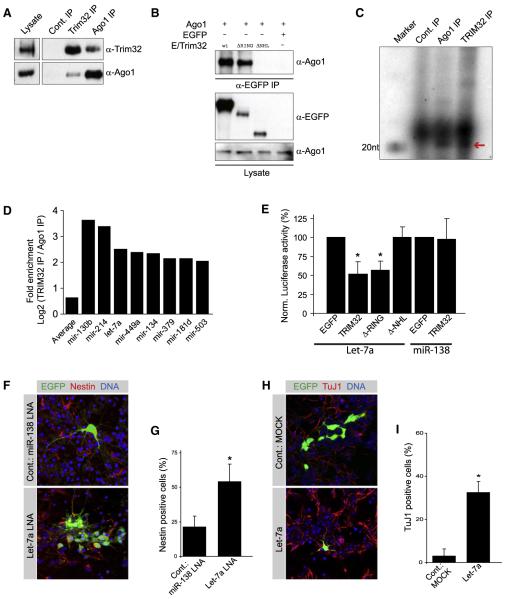
Figure 7. Neuronal Differentiation Is Regulated by TRIM32-Associated miRNAs.
(A) Immunoblots of immunoprecipitations (IP) from embryonic brain lysates with antibodies against TRIM32 and Ago1. As control-IP the anti-Trim32 antibody was preincubated with a blocking peptide.
(B) HEK293T cells were transfected as indicated and EGFP-tagged TRIM32 was immunoprecipitated with an anti-EGFP antibody. TRIM32-associated Ago1 was detected with an anti-Ago1 antibody. Ago1 and EGFP-tagged Trim32 in the lysate were detected with specific antibodies as indicated.
(C) Radioactive RNA end-labeling of immunoprecipitations (IP) from embryonic brain lysates with antibodies against TRIM32 and Ago1. As control-IP the anti-Trim32 antibody was preincubated with a blocking peptide. The Red arrow points to RNA with the size of approximately 21 nt.
(D) Diagram showing the fold enrichment of microRNAs that are more than 2-fold enriched in the TRIM32-IP compared to the Ago1-IP.
(E) Diagram showing the normalized activity of luciferase sensors for the microRNAs Let-7a and miR-138 when they are coexpressed with the indicated constructs.
(F and H) Neural progenitors were transfected at E14.5 via in utero electroporation with the indicated constructs (left gray boxes). The electroporation was followed by dissociation of the tissue and cultivation for 6 days (F) or 4 days (H). Immunostainings of the dissociated cells labeled as indicated (upper gray boxes) are shown.
(G and I) Diagrams show the fraction of transfected cells that are positive for the progenitor marker Nestin (G) or the neuronal maker TuJ1 (I) (mean ± SEM; *p < 0.001 compared to EGFP).
To determine the identity and the abundance of individual TRIM32-bound microRNAs, we used massive parallel sequencing (see Experimental Procedures). Bar-coded cDNA libraries were generated from 19–30 nucleotide long RNAs isolated from E14.5 mouse brain lysates or from Ago1, TRIM32, or control immunoprecipitates. While microRNAs are detected in all libraries, a set of 34 microRNAs is more than 4-fold enriched in the TRIM32 immunoprecipitate over the Ago1 immunoprecipitate (see Supplemental Data for all microRNA frequencies and complete sequencing data). Filtering for absence in the control immunoprecipitate and for significant expression in total brain lysate identifies a set of eight microRNAs that are strongly enriched in the TRIM32/Ago1 complex (Figure 7D). The effect of TRIM32 on the activity of associated and nonassociated microRNAs was determined in a luciferase assay (Schmitter et al., 2006). TRIM32 expression has no effect on the efficiency of miR-138, a microRNA that is not found in the TRIM32/Ago1 complex, but potentiates the efficiency of the associated microRNA Let-7a (Figure 7E) and all the other microRNAs significantly enriched in the TRIM32 complex (Figure S7). A TRIM32 deletion lacking the NHL domain has no effect, suggesting that the interaction with Ago1 is responsible for enhancing microRNA efficiency. Thus, TRIM32 enhances the activity of certain microRNAs.
One of the most prominent hits is the microRNA Let-7a. Because Let-7a controls proliferation in normal and malignant cells (Yu et al., 2007; Peng et al., 2008; Johnson et al., 2007) and is upregulated during neuronal differentiation (Sempere et al., 2004), we asked whether it can induce neuronal differentiation. Inhibition of miR-138 (which is expressed in brain but not regulated by TRIM32) by electroporation of LNAs (locked nucleic acids) does not prevent neuronal differentiation. Six days after electroporation only 21% of the transfected cells (n > 100) retain Nestin expression (Figures 7F and 7G). Transfection of LNAs targeting Let-7a, however, results in the formation of large colonies containing many Nestin-positive cells (54% of n > 100 cells), indicating that neuronal differentiation is inhibited. Conversely, cells expressing a Let-7a overexpression construct fail to form colonies 4 days after transfection (Figure 7H). 32.4% of these cells express the neuronal differentiation marker TuJ1, while in a control experiment (empty vector) only 2.9% of the cells are TuJ1 positive (Figures 7H and 7I). This effect is significant but not as strong as observed for TRIM32 overexpression (51% differentiating cells, see above). Together, these experiments indicate that Let-7a is an important—but not the only—downstream target of TRIM32.
DISCUSSION
Polarized Distribution of TRIM32 in Mitotic Neural Progenitor Cells
Our data suggest that the increased levels of TRIM32 in one of the two daughter cells contribute to the decision of this cell to undergo neuronal differentiation. Like Brat, TRIM32 localizes asymmetrically in mitosis. Brat is localized by binding to Miranda, which, in turn, is recruited to the basal side by the protein Lgl and excluded from the apical side by aPKC (Knoblich, 2008; Rolls et al., 2003). In fly neuroblasts, aPKC promotes self-renewal whereas Lgl inhibits proliferation (Lee et al., 2006a). Although Miranda is not conserved, mouse Lgl and aPKC have similar effects on neural progenitor proliferation. In Lgl knockout mice, neural precursors overproliferate and eventually die by apoptosis (Klezovitch et al., 2004). Removing one of the two aPKC mouse homologs does not affect the rate of neurogenesis (Imai et al., 2006), but depletion of its binding partner Par-3 results in premature cell-cycle exit of cortical progenitors (Costa et al., 2008). Despite these similarities, the precise mechanism by which TRIM32 localizes may be quite distinct. In Drosophila, the apical Par-3/6/aPKC complex directs the basal localization of Brat and Miranda (Wirtz-Peitz et al., 2008) but also orients the mitotic spindle along the apical-basal axis (Knoblich, 2008). In mice, however, the vast majority of progenitor divisions do not occur along the apical-basal axis (Konno et al., 2008; Noctor et al., 2008). TRIM32 is asymmetric even in those planar divisions and provides a suitable explanation for how unequal fates can be generated independently of cleavage plane orientation. Therefore, the relevance of TRIM32 segregation is independent of the somewhat conflicting results that have been reported for the fraction of horizontal versus vertical divisions (Haydar et al., 2003; Kosodo et al., 2004; Konno et al., 2008; Noctor et al., 2008). Since TRIM32 asymmetry does not follow the polarity set up by Par-3/6/aPKC, however, it is likely that it is established by mechanisms distinct from Drosophila.
What could those mechanisms be? TRIM32 often concentrates in the retracting basal fiber (Figure S2), a structure that is not present in Drosophila neuroblasts. TRIM32 might be present in the cytoplasm of the fiber and could be retained in the basal part of the cell during mitosis, when the fiber becomes extremely thin (Noctor et al., 2001) and its cytoplasm flows into the dividing progenitor. This would explain why TRIM32 is asymmetric even when the spindle is not oriented along the apical-basal axis. Since TRIM32-GFP expression prevents mitosis even at low levels, we cannot verify this observation by live imaging. The model would predict that the cell inheriting the basal fiber preferentially undergoes neuronal differentiation. This is in good agreement with some previous live-imaging studies (Miyata et al., 2001), but other studies have actually proposed that the fiber is maintained in mitosis and serves as a guide for migration of the newly formed neuron (Noctor et al., 2001). At the moment, we can therefore not exclude that other mechanisms contribute to the asymmetric localization of TRIM32.
How Does TRIM32 Affect Proliferation and Differentiation?
Our data suggest that TRIM32 acts through two distinct pathways. Through its N-terminal RING finger, TRIM32 ubiquitinylates c-Myc and targets it for proteasome-mediated degradation. High levels of c-Myc are important for the ability of NSCs to self-renew and make NSCs relatively easy targets for reprogramming into ES cells (Kim et al., 2008). Furthermore, the bFGF–SHP2–ERK–c-Myc–Bmi-1 pathway is critical for the self-renewal capacity of neural progenitor cells (Ke et al., 2007; Coskun et al., 2007), and Myc overexpression is known to promote neural progenitor proliferation in the mouse CNS (Fults et al., 2002). Therefore, a TRIM32-mediated reduction in the levels of c-Myc may well serve as a first step to induce neuronal differentiation. In agreement with this, overexpression of c-Myc in GFAP-positive astrocytes promotes formation of less differentiated Nestin-positive progenitor-like cells (Lassman et al., 2004) while a conditional ablation of the c-Myc ortholog N-Myc in mouse neuronal progenitor cells dramatically increases neuronal differentiation (Knoepfler et al., 2002).
Through its C-terminal NHL domain, TRIM32 acts as a potent activator of certain microRNAs. Although Drosophila Mei-P26 also binds Ago1 (Neumuller et al., 2008), it inhibits rather than enhances microRNAs, and the mechanisms by which TRIM32 and its invertebrate homologs regulate microRNAs may actually be quite distinct. This is consistent with the observation that microRNAs support self-renewal in Drosophila stem cells (Hatfield et al., 2005; Jin and Xie, 2007; Park et al., 2007) while they potentiate differentiation in mammalian stem cells (Alvarez-Garcia and Miska, 2005; Kanellopoulou et al., 2005; Yi et al., 2008; Esau et al., 2004; Tay et al., 2008). In particular, Let-7a has an antiproliferative effect (Peng et al., 2008), and its expression reduces tumor growth (Esquela-Kerscher et al., 2008) and can prevent self-renewal in breast cancer cells (Yu et al., 2007). In NSCs, Let-7a is expressed and upregulated during differentiation (Rybak et al., 2008). It is interesting to note that one of the targets for Let-7a is Myc (Sampson et al., 2007). Protein degradation and concomitant translational inhibition through microRNAs might be the key strategy through which TRIM32 induces differentiation in NSCs.
TRIM32 and Tumor Formation
Although brat and mei-P26 mutant flies develop tumors, TRIM32 has not been described as a tumor suppressor. In fact, several reports have even suggested that TRIM32 might induce rather than prevent tumor formation (Albor and Kulesz-Martin, 2007; Kano et al., 2008). TRIM32 is mutated in patients carrying limb girdle muscular dystrophy type 2H (Frosk et al., 2002). Since TRIM32 expression is upregulated during myogenic differentiation (Kudryashova et al., 2005), the muscular dystrophy in these patients could be explained by a differentiation defect in the satellite cell lineage analogous to the one we find in NSC lineages. TRIM32 has also been described as a gene potentially responsible for Bardet-Biedl syndrome and therefore has also been named BBS11 (Bardet-Biedl syndrome gene 11) (Chiang et al., 2006). Distinct TRIM32 mutations are responsible for the two diseases, but none of them seems to cause cancer since an increase in tumor formation is not described for any of the two diseases. Since TRIM32 is a bifunctional molecule, mutating only the RING or the NHL domain might not be sufficient to prevent the antiproliferative function of TRIM32. In Drosophila, tumors only form in a small subset of brat mutant neuroblasts (Bowman et al., 2008). In other neuroblasts, redundancy with other tumor suppressors prevents overproliferation. Should a similar degree of redundancy exist in vertebrates, this might explain why TRIM32 is not a common target for oncogenic mutations. A similar lack of a human tumor phenotype has been shown for the Drosophila tumor suppressor Lgl. In Drosophila, lgl mutant neuroblasts overproliferate and form brain tumors (Lee et al., 2006a). In mice, however, lgl mutant neural progenitors overproliferate initially but then die by apoptosis (Klezovitch et al., 2004). A vertebrate-specific mechanism that prevents tumorigenesis in response to stem cell overproliferation could provide an alternative explanation for the lack of tumor formation when TRIM32 function is compromised. Although such a mechanism has been suggested previously (Pan et al., 2007) the underlying mechanism remains unclear.
Our data establish TRIM-NHL proteins as a family of conserved stem cell regulators. The fact that Mei-P26 regulates stem cell proliferation in Drosophila ovaries (Neumuller et al., 2008) suggests that the function of this protein family might extend way beyond the brain. If this is the case, the presence of a catalytically active RING finger domain that could be inhibited by pharmaceutical compounds might make these proteins attractive targets for the manipulation of stem cell proliferation and the stimulation of regeneration in vivo.
EXPERIMENTAL PROCEDURES
Materials and Plasmids
The used plasmids and antibodies are listed in the Supplemental Experimental Procedures.
The rabbit anti-Trim32 antibody was raised against the N-terminal amino acids MESFTEEQLRPKLLH (Gramsch Laboratories). The final bleed was purified by affinity chromatography with the corresponding peptide. Except for Figure S2E (where the commercially available anti-TRIM32 antibody from Abnova was used), whenever a TRIM32 staining is shown, this antibody was used.
In Utero Electroporation
NSCs in the intact brain were transfected via in utero electroporation as described (Tabata and Nakajima, 2002). To transfect NSCs for dissociated cultures the in utero electroporation was followed by a dissociation of the brain tissue as it has been described previously (Saito and Nakatsuji, 2001). Briefly, after electroporation the complete brain was disintegrated and the dissociated cells were cultured in Neurobasalmedium with 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, 10ng/ml bFGF-2, and 1% N2 supplement (all from Invitrogen). Four days, six days, or ten days after transfection the cells were fixed with 4% PFA and processed for immunohistochemistry. For the paired cell analysis electroporated cells were seed into Labtek chambers (Labtek, Campbell). Six hours after electroporation transfect cells were imaged live; 14 hr later the cells were fixed and stained as described above. Transfected cells were reidentified according to their positions in the Labtek chambers. All animals used in this study were maintained and treated according to approved protocols and in accordance with institutional and national guidelines and regulations. Immunhistochemistry of cryosections and cultured cells were performed according to standard protocols (details are described in the Supplemental Experimental Procedures).
RNA End-labeling, Massive Parallel Sequencing of microRNAs, and microRNA Activity Measurement
For the RNA end-labeling material from IP was incubated with pCP (Amersham, 3.3 μM, 3000 Ci/mmol, 10 mCi/ml), DMSO, PAN buffer (500 mM Tris pH 7.6, 100 mM MgCl2, 100 mM MercaptoEtOH, 2 mM ATP, 1 mg/ml BSA), and RNA ligase (Roche) for 1 hr at 16 °C. Afterwards an RNA loading dye was added and the RNAs were separated on a 15% acrylamide gel. For the massive parallel sequencing, RNA species in the IPs smaller than 500 bases were enriched with the mirVana miRNA isolation kit (Ambion). The RNA sample was then separated on a denaturing 15% polyacrylamide (PAA) gel and stained with SYBRgreenII. The population of small RNA with a length of 19–29 bases was obtained by passive elution of the RNA from the gel. The eluted RNA was first poly(A)-tailed using poly(A) polymerase followed by ligation of an RNA adaptor to the 5μ-phosphate of the RNA. First-strand cDNA synthesis was then performed using an oligo(dT)-linker primer and M-MLV-RNase H-reverse transcriptase. The resulting cDNAs were then PCR-amplified to about 30 ng/μl using a high-fidelity Taq DNA polymerase. The bar-coded primers used for PCR amplification were designed for amplicon sequencing according to the instructions of 454 Live Sciences. The resulting cDNAs were obtained by fractionation on preparative 6% PAA-gels. cDNAs of the correct size were eluted, finally extracted with phenol/chloroform, and precipitated with Ethanol. These gel-purified cDNAs were submitted to 454 sequencing (done by Vertis Biotechnology AG, Freising).
The activity of microRNAs Let-7a and miR-138 was determined as previously described (Schmitter et al., 2006). Briefly the luciferase vectors pRL-let-7a or pGL3-miR-138 were cotransfected with pEGFP, EGFP-tagged TRIM32, or EGFP-tagged TRIM32 deletion constructs. To normalize the luciferase signal pRL-let-7a was coexpressed with a vector coding for firefly luciferase and pGL3-miR-138 was coexpressed with a vector coding for renilla luciferase.
Supplementary Material
materials and methods, supplementary figures
ACKNOWLEDGMENTS
The authors would like to thank Andrea Ballabio (Milan), Stephen Hann (Nashville), Dirk Bohmann (Rochester), Judy Lieberman (Boston), and Witold Filipowicz (Basel) for plasmids; Gunter Meister (Munich) for the anti-Ago1 antibody; Xiao-Bing Yuan (Shanghai) for teaching the in utero electroporation,;Jörg Betschinger for initiating production of the anti-TRIM32 antibody; Maria Novatchkova for sequence analysis; Katrin Heindl for help with the RNA end-labeling; and Angela M. Peer for excellent technical assistance. J.C.S. was supported by EMBO and HFSP. Work in J.A.K.’s lab is supported by the Austrian Academy of Sciences, the Wiener Wissenschafts-, Forschungs und Technologiefonds (WWTF), the Austrian Science Fund (FWF), the EU FP6 network ONCASYM, and the EU FP7 network EUROSYSTEM.
Footnotes
REFERENCES
- Albor A, Kulesz-Martin M. Novel initiation genes in squamous cell carcinomagenesis: a role for substrate-specific ubiquitylation in the control of cell survival. Mol. Carcinog. 2007;46:585–590. doi: 10.1002/mc.20344. [DOI] [PubMed] [Google Scholar]
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Berdnik D, Torok T, Gonzalez-Gaitan M, Knoblich JA. The endocytic protein alpha-Adaptin is required for numb-mediated asymmetric cell division in Drosophila. Dev. Cell. 2002;3:221–231. doi: 10.1016/s1534-5807(02)00215-0. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA. The tumor suppressors Brat and Numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell. 2008;14:535–546. doi: 10.1016/j.devcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Butcher BA, Gagliardo LF, ManWarren T, Appleton JA. Larvae-induced plasma membrane wounds and glycoprotein deposition are insufficient for Trichinella spiralis invasion of epithelial cells. Mol. Biochem. Parasitol. 2000;107:207–218. doi: 10.1016/s0166-6851(00)00189-4. [DOI] [PubMed] [Google Scholar]
- Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA, Kim KY, Huang J, et al. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc. Natl. Acad. Sci. USA. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev. Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Coskun V, Zhao J, Sun YE. Neurons or glia? Can SHP2 know it all? Sci. STKE. 2007;2007:pe58. doi: 10.1126/stke.4102007pe58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MR, Wen G, Lepier A, Schroeder T, Gotz M. Par-complex proteins promote proliferative progenitor divisions in the developing mouse cerebral cortex. Development. 2008;135:11–22. doi: 10.1242/dev.009951. [DOI] [PubMed] [Google Scholar]
- Doe CQ. Neural stem cells: balancing self-renewal with differentiation. Development. 2008;135:1575–1587. doi: 10.1242/dev.014977. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet. 2003;34:53–58. doi: 10.1038/ng1144. [DOI] [PubMed] [Google Scholar]
- Esau C, Kang X, Peralta E, Hanson E, Marcusson EG, Ravichandran LV, Sun Y, Koo S, Perera RJ, Jain R, et al. MicroRNA-143 regulates adipocyte differentiation. J. Biol. Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- Fishell G, Kriegstein AR. Neurons from radial glia: the consequences of asymmetric inheritance. Curr. Opin. Neurobiol. 2003;13:34–41. doi: 10.1016/s0959-4388(03)00013-8. [DOI] [PubMed] [Google Scholar]
- Frank DJ, Edgar BA, Roth MB. The Drosophila melanogaster gene brain tumor negatively regulates cell growth and ribosomal RNA synthesis. Development. 2002;129:399–407. doi: 10.1242/dev.129.2.399. [DOI] [PubMed] [Google Scholar]
- Frosk P, Weiler T, Nylen E, Sudha T, Greenberg CR, Morgan K, Fujiwara TM, Wrogemann K. Limb-girdle muscular dystrophy type 2H associated with mutation in TRIM32, a putative E3-ubiquitin-ligase gene. Am. J. Hum. Genet. 2002;70:663–672. doi: 10.1086/339083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fults D, Pedone C, Dai C, Holland EC. MYC expression promotes the proliferation of neural progenitor cells in culture and in vivo. Neoplasia. 2002;4:32–39. doi: 10.1038/sj.neo.7900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- Hatfield SD, Shcherbata HR, Fischer KA, Nakahara K, Carthew RW, Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Ang E, Jr., Rakic P. Mitotic spindle rotation and mode of cell division in the developing telencephalon. Proc. Natl. Acad. Sci. USA. 2003;100:2890–2895. doi: 10.1073/pnas.0437969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai F, Hirai S, Akimoto K, Koyama H, Miyata T, Ogawa M, Noguchi S, Sasaoka T, Noda T, Ohno S. Inactivation of aPKClambda results in the loss of adherens junctions in neuroepithelial cells without affecting neurogenesis in mouse neocortex. Development. 2006;133:1735–1744. doi: 10.1242/dev.02330. [DOI] [PubMed] [Google Scholar]
- Jin Z, Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano S, Miyajima N, Fukuda S, Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68:5572–5580. doi: 10.1158/0008-5472.CAN-07-6231. [DOI] [PubMed] [Google Scholar]
- Ke Y, Zhang EE, Hagihara K, Wu D, Pang Y, Klein R, Curran T, Ranscht B, Feng GS. Deletion of Shp2 in the brain leads to defective proliferation and differentiation in neural stem cells and early post-natal lethality. Mol. Cell. Biol. 2007;27:6706–6717. doi: 10.1128/MCB.01225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M, Scholer HR. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454:646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Klezovitch O, Fernandez TE, Tapscott SJ, Vasioukhin V. Loss of cell polarity causes severe brain dysplasia in Lgl1 knockout mice. Genes Dev. 2004;18:559–571. doi: 10.1101/gad.1178004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno D, Shioi G, Shitamukai A, Mori A, Kiyonari H, Miyata T, Matsuzaki F. Neuroepithelial progenitors undergo LGN-dependent planar divisions to maintain self-renewability during mammalian neurogenesis. Nat. Cell Biol. 2008;10:93–101. doi: 10.1038/ncb1673. [DOI] [PubMed] [Google Scholar]
- Kosodo Y, Roper K, Haubensak W, Marzesco AM, Corbeil D, Huttner WB. Asymmetric distribution of the apical plasma membrane during neurogenic divisions of mammalian neuroepithelial cells. EMBO J. 2004;23:2314–2324. doi: 10.1038/sj.emboj.7600223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudryashova E, Kudryashov D, Kramerova I, Spencer MJ. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005;354:413–424. doi: 10.1016/j.jmb.2005.09.068. [DOI] [PubMed] [Google Scholar]
- Lassman AB, Dai C, Fuller GN, Vickers AJ, Holland EC. Overexpression of c-MYC promotes an undifferentiated phenotype in cultured astrocytes and allows elevated Ras and Akt signaling to induce gliomas from GFAP-expressing cells in mice. Neuron Glia Biol. 2004;1:157–163. doi: 10.1017/s1740925x04000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Robinson KJ, Doe CQ. Lgl, Pins and aPKC regulate neuroblast self-renewal versus differentiation. Nature. 2006a;439:594–598. doi: 10.1038/nature04299. [DOI] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 2006b;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Li HS, Wang D, Shen Q, Schonemann MD, Gorski JA, Jones KR, Temple S, Jan LY, Jan YN. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron. 2003;40:1105–1118. doi: 10.1016/s0896-6273(03)00755-4. [DOI] [PubMed] [Google Scholar]
- Li L, Vaessin H. Pan-neural Prospero terminates cell proliferation during Drosophila neurogenesis. Genes Dev. 2000;14:147–151. [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–741. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage. Nature. 2008;454:241–245. doi: 10.1038/nature07014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J. Comp. Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan L, Chen S, Weng C, Call G, Zhu D, Tang H, Zhang N, Xie T. Stem cell aging is controlled both intrinsically and extrinsically in the Drosophila ovary. Cell Stem Cell. 2007;1:458–469. doi: 10.1016/j.stem.2007.09.010. [DOI] [PubMed] [Google Scholar]
- Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr. Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- Peng Y, Laser J, Shi G, Mittal K, Melamed J, Lee P, Wei JJ. Antiproliferative effects by Let-7 repression of high-mobility group A2 in uterine leiomyoma. Mol. Cancer Res. 2008;6:663–673. doi: 10.1158/1541-7786.MCR-07-0370. [DOI] [PubMed] [Google Scholar]
- Petersen PH, Zou K, Hwang JK, Jan YN, Zhong W. Progenitor cell maintenance requires numb and numblike during mouse neurogenesis. Nature. 2002;419:929–934. doi: 10.1038/nature01124. [DOI] [PubMed] [Google Scholar]
- Qian X, Goderie SK, Shen Q, Stern JH, Temple S. Intrinsic programs of patterned cell lineages in isolated vertebrate CNS ventricular zone cells. Development. 1998;125:3143–3152. doi: 10.1242/dev.125.16.3143. [DOI] [PubMed] [Google Scholar]
- Qian X, Shen Q, Goderie SK, He W, Capela A, Davis AA, Temple S. Timing of CNS cell generation: a programmed sequence of neuron and glial cell production from isolated murine cortical stem cells. Neuron. 2000;28:69–80. doi: 10.1016/s0896-6273(00)00086-6. [DOI] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, Sestan N. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat. Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih HP, Lee CY, Doe CQ. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol. 2003;163:1089–1098. doi: 10.1083/jcb.200306079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev. Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- Sampson VB, Rong NH, Han J, Yang Q, Aris V, Soteropoulos P, Petrelli NJ, Dunn SP, Krueger LJ. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–9770. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwamborn JC, Muller M, Becker AH, Puschel AW. Ubiquitination of the GTPase Rap1B by the ubiquitin ligase Smurf2 is required for the establishment of neuronal polarity. EMBO J. 2007;26:1410–1422. doi: 10.1038/sj.emboj.7601580. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata H, Nakajima K. Neurons tend to stop migration and differentiate along the cortical internal plexiform zones in the Reelin signal-deficient mice. J. Neurosci. Res. 2002;69:723–730. doi: 10.1002/jnr.10345. [DOI] [PubMed] [Google Scholar]
- Tay YM, Tam WL, Ang YS, Gaughwin PM, Yang H, Wang W, Liu R, George J, Ng HH, Perera RJ, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells, where it causes post-transcriptional attenuation of Nanog and LRH1. Stem Cells. 2008;26:17–29. doi: 10.1634/stemcells.2007-0295. [DOI] [PubMed] [Google Scholar]
- Treier M, Staszewski LM, Bohmann D. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell. 1994;78:787–798. doi: 10.1016/s0092-8674(94)90502-9. [DOI] [PubMed] [Google Scholar]
- Wirtz-Peitz F, Nishimura T, Knoblich JA. Linking cell cycle to asymmetric division: Aurora A phosphorylates the par complex to regulate numb localization. Cell. 2008;135:161–173. doi: 10.1016/j.cell.2008.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Huttner WB. Asymmetric cell division during neurogenesis in Drosophila and vertebrates. Mech. Dev. 2003;120:1297–1309. doi: 10.1016/j.mod.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Wulczyn FG, Smirnova L, Rybak A, Brandt C, Kwidzinski E, Ninnemann O, Strehle M, Seiler A, Schumacher S, Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing ‘stemness’. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yingling J, Youn YH, Darling D, Toyo-Oka K, Pramparo T, Hirotsune S, Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self-renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- Zhong W, Feder JN, Jiang MM, Jan LY, Jan YN. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Zhong W, Jiang MM, Schonemann MD, Meneses JJ, Pedersen RA, Jan LY, Jan YN. Mouse numb is an essential gene involved in cortical neurogenesis. Proc. Natl. Acad. Sci. USA. 2000;97:6844–6849. doi: 10.1073/pnas.97.12.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
materials and methods, supplementary figures