MicroRNA Regulation of DNA Repair Gene Expression in Hypoxic Stress (original) (raw)
. Author manuscript; available in PMC: 2010 Dec 6.
Abstract
Genetic instability is a hallmark of cancer; the hypoxic tumor microenvironment has been implicated as a cause of this phenomenon. MicroRNAs (miRs) are small, non-protein coding RNAs that can regulate various cellular pathways. We report here that two miRs, miR-210 and miR-373, are up-regulated in a hypoxia-inducible factor-1 alpha (HIF-1α)-dependent manner in hypoxic cells. Bioinformatics analyses suggested that these miRs could regulate factors implicated in DNA repair pathways. Forced expression of miR-210 was found to suppress the levels of RAD52, which is a key factor in homology-dependent repair (HDR); the forced expression of miR-373 led to a reduction in the nucleotide excision repair (NER) protein, RAD23B, as well as in RAD52. Consistent with these results, both RAD52 and RAD23B were found to be down-regulated in hypoxia, but in both cases, the hypoxia-induced down-regulation could be partially reversed by anti-sense inhibition of miR-210 and miR-373. Importantly, luciferase reportor assays indicated that miR-210 is capable of interacting with the 3′ untranslated region (UTR) of RAD52 and that miR-373 can act on the 3′ UTR of RAD23B. These results indicate that hypoxia-inducible miR-210 and miR-373 play roles in modulating the expression levels of key proteins involved in the HDR and NER pathways, providing new mechanistic insight into the impact of hypoxia on DNA repair and genetic instability in cancer.
Keywords: Hypoxia, Genetic Instability, microRNA, Homology-Dependent Repair (HDR), Nucleotide Excision Repair (NER), DNA Damage Response, Tumor Microenvironment
Introduction
Tumor cells exhibit a higher mutational frequency than their normal counterparts (1), providing the potential to adapt to otherwise hostile microenvironments. The mechanisms underlying this capacity have not been fully elucidated. However, models of genomic instability have been well-studied with respect to inherited mutations that lead to cancer-prone phenotypes, as in the case of hereditary non-polyposis colon cancer, which is linked to deficient mismatch repair (MMR) activity (2). Additionally, data indicate that the relatively higher rates of mutational frequencies in tumors may be partially attributed to the effects of tumor microenvironmental stresses, themselves (3). In addition to low pH and nutrient deprivation, hypoxia is a key component of the tumor microenvironment (4). Hypoxia leads to increased angiogenesis, upregulates glycolytic enzymes, and provides selective pressure for cells that are capable of evading apoptosis (5, 6).
Recent work has shown that hypoxia can also promote genetic instability by impacting the DNA repair capacity of tumor cells (7, 8). Due to the transcriptional down-regulation of the MLH1 and MSH2 genes and the BRCA1 and RAD51 genes, the MMR and the homology-dependent repair (HDR) pathways, respectively, are suppressed in hypoxic cells (9–11). In the case of MMR, MSH2 and MLH1 are down-regulated in a c-MYC-dependent manner, due to c-MYC’s displacement by MAX binding to the MLH1 promoter (8). The mechanism of this down-regulation also depends on histone deacetylation at the respective promoters (9). With respect to HDR, the down-regulation of two key factors, RAD51 and BRCA1, contributes to a functional loss of HDR activity and is mediated by E2F4/p130 transcriptional repression via increased occupancy at the respective proximal promoter regions, which is in response to hypoxic stress (11, 12).
Functionally, nucleotide excision repair (NER) was also found to be suppressed in hypoxic cells, as measured by a reduction in the removal of UV damage from reporter gene substrates, but the mechanism for the reduced repair was not elucidated (13). No changes in the expression levels of two key mediators in the pathway, XPA and XPD, were observed in hypoxia in one initial study (13).
While classical transcriptional factors like E2F and c-MYC have important roles in regulating genes implicated in cancer, recent work has identified a new level of genetic control invoking small non-protein coding RNAs, or microRNAs (miRs), which target mRNA destabilization (14), suppress target mRNA translation (14), or act through novel interactions with promoter sequences (15). In general, miRNAs bind to the 3′ untranslated regions (UTRs) of target mRNAs via imperfect base-pairing complementarity leading to degradation (16) or translation inhibition (17). The regulation of mRNAs by miRs has been shown to influence multiple cellular processes, including apoptosis, differentiation, and cell survival (18).
Here, we have examined miR expression in response to hypoxic stress as another potential mechanism that might alter the factors involved in DNA repair. We report that two key miRs, miR-210 and miR-373, are up-regulated in hypoxia in a hypoxia-inducible factor-1 alpha (HIF-1α)-dependent manner. We find that miR-210 targets RAD52, a member of the HDR pathway, while miR-373 can target both RAD52 and RAD23B. RAD23B forms a heterodimer with XPC and serves as the key DNA damage recognition complex in the NER pathway. Mechanistically, levels of both RAD52 and RAD23B were found to be down-regulated in hypoxic cells. Bioinformatics revealed miR-210 and miR-373 binding sites in the 3′ UTRs of these genes. In normoxic cells, the forced expression of miR-210 decreased endogenous levels of RAD52; the forced expression of miR-373 suppressed both RAD52 and RAD23B levels. In hypoxic cells, the inhibition of miR-210 and miR-373 partially reversed the hypoxia-induced down-regulation of RAD52 and RAD23B, respectively. The suppression of RAD52 by miR-210 and miR-373 offers a new explanation for the reduced HDR activity in hypoxic cells, while the down-regulation of RAD23B by miR-373 provides a new mechanism for the previously unexplained reduction of NER in hypoxia. These findings also identify a new role for miRs in the regulation of DNA repair in response to hypoxia.
Materials and Methods
Bioinformatics
Sequence information was analyzed by utilizing the UCSC Genome Browser (19). MiR sequence data were taken from miRBase (20–22). Target predictions were performed using miRBase Targets for miR-210 and RAD52 interactions, which uses the miRanda algorithm (23). For the interactions between miR-373 and both RAD23B and RAD52, the TargetScan algorithm was used. TargetScan predicts the biological targets of miRNAs by searching for the presence of conserved 8-mer and 7-mer sites matching the seed region of a miR (24–26).
Cell culture and treatment
HeLa cervical carcinoma and MCF-7 breast cancer cells were grown in DMEM supplemented with 10% fetal bovine serum (FBS). The HIF-1α nullizygous and parental mouse embryonic fibroblast (MEF) cell lines (27) were maintained in DMEM containing 10% FBS. Cells were exposed to hypoxia in culture as described previously (3).
Microarray analysis of miRs
Total RNA was harvested from HeLa cells exposed to normoxia or hypoxia (0.01% O2) for 24 h and sent for microarray analysis (LC Sciences, LLC, Houston, TX, USA). Total RNA (2 to 5 µg) was size fractionated using a YM-100 Microcon centrifugal filter (Millipore, Billerica, MA, USA) and the small RNAs (< 300 nt) isolated were 3’-extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent Cy3 and Cy5 dye labeling. The probes were complementary to target microRNAs (from miRBase, (http://microrna.sanger.ac.uk/sequences/) that contained a spacer segment of polyethylene glycol to extend the coding segment away from the substrate, which also enabled for homogenous hybridization melting temperatures. Hybridization images were collected using a GenePix 4000B laser scanner, (Molecular Devices, Sunnyvale, CA, USA) and digitized using Array-Pro image analysis software (Media Cybernetics, Silver Spring, MD, USA). Data were normalized by subtracting the background and utilizing a LOWESS (Locally-weighted Regression) filter, which reduces system-related variation. For two-color experiments, the ratio of the two sets of detected signals (log2-transformed, balanced) and p-values of the t-test were calculated. Differentially expressed signals having p-values less than 0.01 were considered to be significant.
Plasmids
The plasmid expressing miR-210 was prepared by amplifying genomic fragment encompassing precursor miR-210 and ~200-bp upstream- and downstream-flanking sequences using the following primers: F-5'-ATTGAATTCTGAAGTTGGGCCGAGAGC-3' and F-5'-ATTGAATTCCGCACACTGGTACCCTGG-3'. The fragment was cloned into the pBABE-puro vector. The miR-373 overexpression plasmid was constructed by cloning the genomic sequence of miR-373, as well as a ~200-bp flanking sequence into the pBABE-puro vector. This 571-bp genomic region was PCR amplified (GoTaq Green Master Mix, Promega, Madison, WI, USA) using the following forward and reverse primers, which contain the respective cleavage sequences for BamHI and SalI,: MIR373-F-5′-CATTGGGATCCCTGGGCGACAGAGCAAGACT-3′ and MIR373-R-5′-GATACGTCGACCATGCATGCTGCCTACCAAA-3′. The shRNA against HIF-1α was targeted against the sequence: 5′-GAAGGAACCTGATGCTTTA-3′, which is located 146-bp downstream of the start codon. Duplexes created to express the shRNA hairpin included the following oligonucleotides: F-5'-GATCCCCGAAGGAACCTGATGCTTT ATTCAAGAGATAAAGCATCAGGTTCCTTCTTTTTGGAAAT-3' and R-5′-CTAGATTTCC AAAAAGAAGGAACCTGATGCTTTATCTCTTGAATAAAGCATCAGGTTCCTTCGGG-3'.
The 3′ UTRs of RAD52 and RAD23B were cloned into the pMIR-REPORT-FLUC miRNA Expression Reporter Vector (Applied Biosystems). PCR was used to amplify the 3′ UTR of RAD52 (2.4–kbp) using the following forward and reverse primers containing the respective cleavage sequences for SpeI and MluI: RAD52NUF1-5′-GCATTGACTAGTTGGACCCACGCTCTGAAATC-3′ and RAD52UTR-5′-CGATACACGCGTTGACGTGATGCCAGAAGTG-3′. Similarly, PCR was used to amplify a 2.4-kbp region of the RAD23B 3′ UTR using the following forward and reverse primers containing the respective cleavage sequences for SpeI and MluI: RAD23NUTF-5′-GCATTGACTAGTGGGCTCATATCCACAAT-3′ and RAD23NUTR-5′-CGATACACGCGTAGAATCCTAGCCATCCAG-3′.
Real-time quantitative PCR (RTQ-PCR)
Total RNA was extracted from cells with the mirVana miRNA Isolation Kit (Applied Biosystems, Foster City, CA, USA). Synthesis of cDNA from total RNA was performed using reverse transcription primers that were specific to miR-210, miR-373, or an endogenous control miR, RNU19, and the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems). As miR-373 is expressed at relatively low levels, the cDNA synthesized from the reverse transcription reaction was further amplified using the TaqMan PreAmp Mix (Applied Biosystems). To analyze the mRNA expression of DEC1, RAD52, RAD23B, and 18S rRNA, total RNA was isolated as above, but was reverse transcribed into cDNA using the High-Capacity cDNA Archive Kit (Applied Systems). To assess miR and mRNA expression, the cDNAs prepared from the specific reverse transcription reactions were used in PCR reactions containing TaqMan Universal PCR Master Mix with No AmpErase UNG and premixed TaqMan assays (Applied Biosystems). The TaqMan assays for miRs only detect the mature miR species.
Reactions were carried out in a 96-well optical reaction plate with optical caps (Agilent Technologies, Santa Clara, CA, USA) in a Mx3000p Real-Time PCR Detection System spectrofluorometric thermal cycler (Agilent). Reactions proceeded with an initial 10 min incubation at 95°C followed by 40 cycles of amplification: 95°C for 15 sec and 60°C for 1 min. Fluorescence was measured in real-time; the cycle threshold (Ct) values were calculated using the Mx3000p algorithm (Agilent). Comparative quantitation was performed by comparing the Ct value obtained from the amplification of a given target miRNA to that determined for the normalizer, RNU19. For mRNA, 18S was used as a normalizer. Relative miRNA and mRNA abundance was calculated using the −ΔΔCt method.
Validation of miRNA regulation determined by the LC Sciences platform was performed using a TaqMan low-density array (Applied Biosystems) as a method for detecting miR expression. Total RNA was isolated as previously mentioned. RNA was reverse transcribed to produce cDNA using pooled reverse transcription primers (Applied Biosystems). After pooled cDNA synthesis, the samples were loaded into 384-well microfluidics cards and run on an ABI Prism 7900HT sequence detection system (Applied Biosystems). Internal controls on the card were used to normalize the data on each card, as well as between cards.
MiRNA forced expression and inhibition
To force the expression of miRs, pre-miRs were purchased as partially duplexed RNA molecules (Applied Biosystems). To inhibit miRs, single-stranded RNA oligonucleotides were purchased from Applied Biosystems. Both pre-miRs and anti-miRs were resuspended in nuclease-free water (50 µM) upon arrival. Transfection of the respective miRs (pre-miRs) or miR inhibitors (anti-miRs) specific for miR-210 (pre-miR: Cat#: AM17100, Product ID: PM10516; anti-miR: Cat#: AM17000, Product ID: AM10516), miR-373 (premiR: Cat#: AM17100, Product ID: PM11024; anti-miR: AM17000, Product ID: AM11024), or a negative control (Negative Control #1, pre-miR: Cat#: 17110; anti-miR: Cat#: 17010), was performed using the reverse transfection technique recommended for use with the siPORT NeoFX transfection reagent (Applied Biosystems). Transfections were carried out according to the manufacturer’s protocol using 3 µl of transfection reagent and a final concentration of 50 nM of each oligonucleotide per well on a 6-well plate. Medium was replaced at 24 h after the transfection.
The FuGENE 6 Transfection Reagent (Roche Applied Sciences, Indianapolis, IN, USA) was used to transfect the miR-210 and miR-373 pBABE constructs into MCF-7 cells. For each well on a 6-well plate, 1 µg of plasmid DNA and 3 µl of FuGENE 6 reagent was transfected, as indicated in the manufacturer’s protocol.
Western blot
Western analyses of whole cell lysates were performed as previously described (28). Nitrocellulose membranes were probed for RAD52 [(5H9) 1:500; GeneTex, Inc., San Antonio, TX, USA], or RAD23B [(HHR23B) 1:250; Rockland Immunochemicals, Inc., Gilbertsville, PA, USA], or HIF-1α [(Clone 54) 1:250; BD Biosciences, San Jose, CA, USA]. β-actin [(sc-47778)1:1000; Santa Cruz, Hercules, CA, USA] was used as a loading control. An anti-mouse IgG secondary antibody conjugated with horseradish peroxidase [(sc-2005) 1:1000; Santa Cruz] was used to detect RAD52, HIF-1α, and β-actin. An anti-goat IgG secondary antibody conjugated with horseradish peroxidase was used to detect RAD23B (1:1000; Rockland). Immunodetection was performed using an ECL kit (G.E. HealthCare, Piscataway, NJ, USA), according to the protocol supplied by G.E. HealthCare.
Luciferase assays
As indicated above, reporter constructs containing the 3′ UTRs of both RAD52 and RAD23B were designed to encompass the respective miR-210 and miR-373 binding sites. For transfection, 105 MCF-7 cells were seeded in triplicate into 12-well culture plates and transfected with .5 µg of each reporter construct by using the FuGENE 6 reagent. At the same time, .5 µg of either pBABE-miR-210 or pBABE-373 was also transfected into the cells. Renilla luciferase activity from a cotransfected pRL-SV40 control vector (2.5 ng/well) was used for normalization. Firefly and Renilla luciferase activities were measured by using the Dual-Luciferase Reporter Assay System kit (Promega), according to the manufacturer’s instructions.
Results
Hypoxia up-regulates miR expression
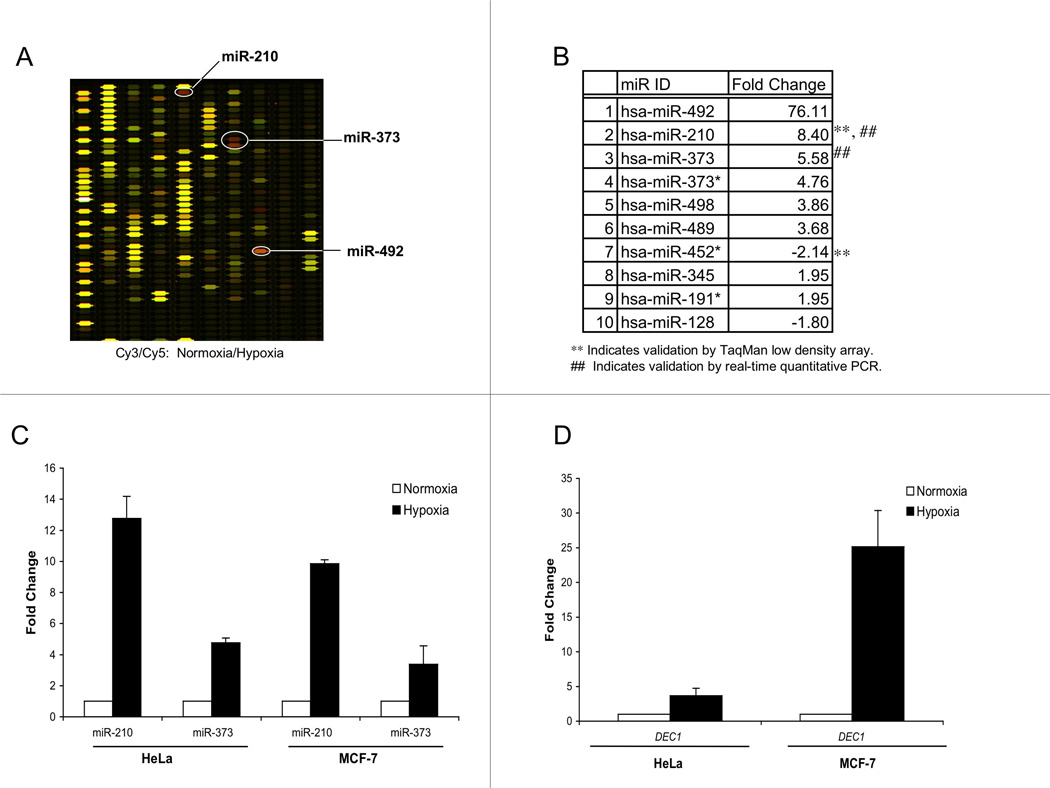
As miR regulation has been implicated in carcinogenesis and tumor growth, we hypothesized that hypoxia might regulate miR expression. After exposing HeLa cells to normoxia or hypoxia (0.01% O2) for 24 h, total RNA was harvested; the expression profiling experiments were performed utilizing RNA that was enriched for miRs. We compared the Cy3-labeled normoxia signal to the Cy5-labeled hypoxia signal (Fig. 1A). In addition to a previous report indicating that miR-210 is induced in hypoxia (29), we determined that miRs 492, 373, 373*, 498, 489, 345, and 191*, were up-regulated ≥ 1.95-fold (Fig. 1B). In contrast and among significant changes in expression, miRs 452* and 128 were down-regulated ≥ 1.80-fold. The findings with respect to both miRs 210 and 452* were also validated by performing TaqMan low-density array analyses in HeLa cells.
Fig. 1.
Microarray analyses indicate dynamic regulation of miRs in hypoxia. (A) Total RNA was extracted from HeLa cells that were exposed for 24 h to hypoxia (0.01% O2) or normoxia and processed by microarray analyses, where red (Cy 5) and green (Cy 3) indicate up- and down-regulation, respectively. The top 3 miRs regulated in hypoxia, miR-492, miR-373, and miR-492, are highlighted on the composite (Cy3 and Cy5) array diagram. (B) Hypoxia induces changes in miR expression, as indicated by the fold-changes in transcript levels detected in HeLa cells. (C) MiR-210 and miR-373 are up-regulated in HeLa and MCF-7 cells, as determined by RTQ-PCR. (D) The up-regulation of DEC1 in hypoxia HeLa and MCF-7 cells serves as a positive control for confirming physiologically relevant hypoxia. The error bars in C and D represent standard errors from triplicate experiments in all cases.
The most significantly up-regulated miRs, 492, 373, and 210, were further investigated using individual TaqMan MicroRNA Assays. In these experiments, we analyzed the expression levels of these miRs in HeLa, as well as in MCF-7 cells, after 48 h of hypoxia or normoxia, which is a timepoint that we have previously shown to be biologically relevant for changes in DNA repair activity (11). Both MCF-7 and HeLa cells had increased levels of miR-210; these were induced ~10–12 fold above basal levels in both cell lines (Fig. 1C). Additionally, the levels of miR-373 were increased ~3–4 fold in MCF-7 and HeLa cells in response to hypoxia. However, the expression level of miR-492 was below our detection threshold (data not shown); we were therefore not able to validate the initial results with respect to miR-492. We also measured the expression of DEC1; transcript levels were induced in hypoxia (Fig. 1D), serving as a positive control for the physiologic hypoxic response in both cell lines, which is in line with published reports (30, 31). In all cases, the levels of the respective miRs and mRNAs in normoxia and hypoxia were normalized to RNU19 and 18S expression, respectively.
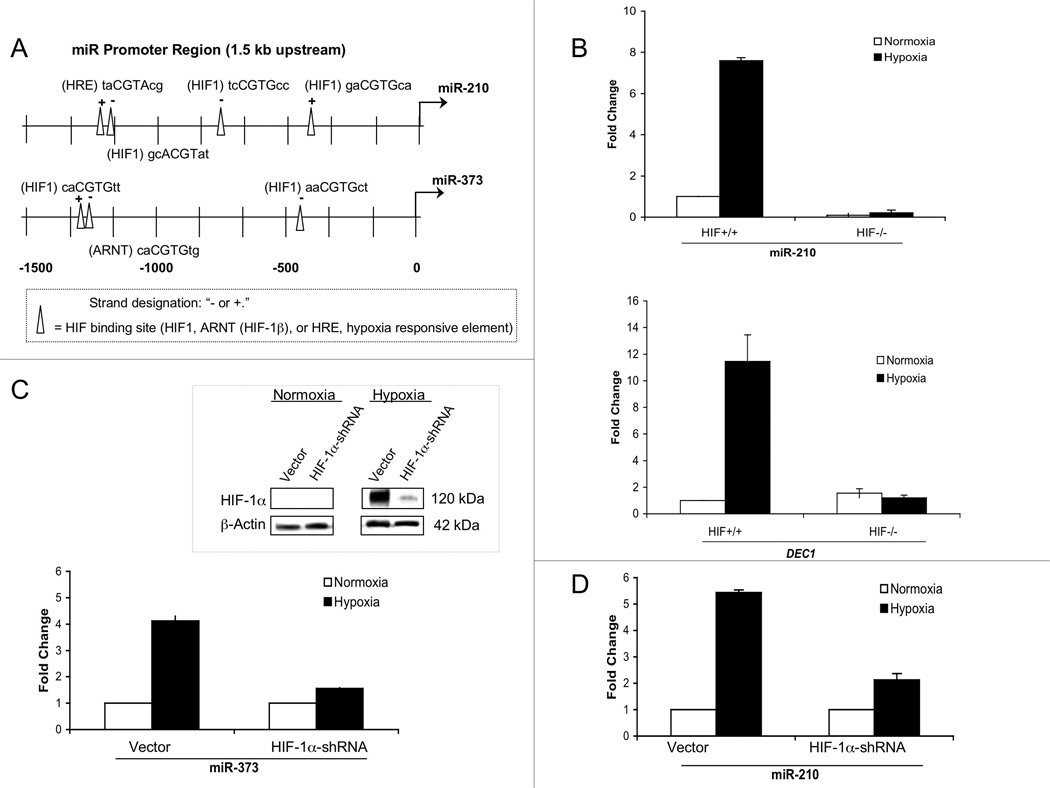
Based on the induction of miRs 210 and 373 in hypoxia, we sought to identify potential transcription factor binding sites in the promoters of these miRs, which could be responsible for mediating induction in hypoxia. As a starting point, we employed an in silico approach to search for binding sites for hypoxia inducible factor-1 (HIF-1), using consensus sequences of “cgtg” or “cgta” and the MatInspector tool from Genomatix Software GmbH (32). We found that the proximal promoter of miR-210 contained 4 such sites (Fig. 2A). Likewise, miR-373 contained 3 sites in the 1.5-kb region upstream of its transcriptional start site. To determine whether or not HIF-1α was involved in the regulation of miR-210 and miR-373, we utilized a matched set of MEFs that were either wild-type or knocked-out for HIF-1α (27). After hypoxia treatment for 48 h (0.01% O2), total RNA was harvested from the cells and analyzed by RTQ-PCR. The MEFs that were positive for HIF-1α were capable of inducing miR-210, which is in contrast to the MEFs that were deficient in HIF-1α (Fig. 2B, upper panel). In parallel, the hypoxia-inducible gene, DEC1, was seen to be up-regulated only in the HIF-1α-expressing MEFs (Fig. 2B, lower panel). Because there is no mouse homolog of human miR-373, we could not use the MEFs to study miR-373 regulation. Therefore, we expressed a HIF-1α-shRNA vector in human cells. Expression of this vector in MCF-7 cells blocked the accumulation of HIF-1α in hypoxia (Fig. 2C, inset). With regard to miR-373 expression, the HIF-1α-shRNA vector suppressed the induction of miR-373 in hypoxia cells, indicating a role for HIF-1α in the upregulation of miR-373 in response to hypoxia (Fig. 2C). HIF-1α-shRNA expression also reduced the induction of miR-210 in hypoxia (Fig. 2D), which is consistent with the findings based on the HIF-1α-proficient and -deficient MEFs.
Fig. 2.
HIF-1α is a key transcriptional activator of miR-210 and miR-373. (A) Bioinformatics analyses using Genomatix’s MatInspector identifies putative HIF-1 binding sites in the promoter regions of miR-210 and miR-373. (B) RTQ-PCR is used to analyze miR-210 regulation in MEFs with or without expression of HIF-1α. Hypoxia treatments were for 48 h at 0.01% O2. DEC1 induction in hypoxic MEFs serves as a control for confirming physiologically relevant hypoxia. (C) Induction of miR-373 in hypoxic cells is attenuated by the knock-down of HIF-1α via shRNA. MCF-7 cells infected with either an empty lentiviral vector or a lentiviral vector expressing HIF-1α-shRNA were exposed to normoxia or hypoxia and miR-373 levels were measured by RTQ-PCR. Knock-down of HIF-1α is achieved through HIF-1α-shRNA expression in MCF-7 cells under normoxia or hypoxia, as indicated (inset). (D) Induction of miR-210 in hypoxic cells is reduced by shRNA-mediated knock-down of HIF-1α, as in (C). Error bars in B, C, and D represent standard errors calculated from three replicates in all cases.
Hypoxia induces the down-regulation of DNA repair factors
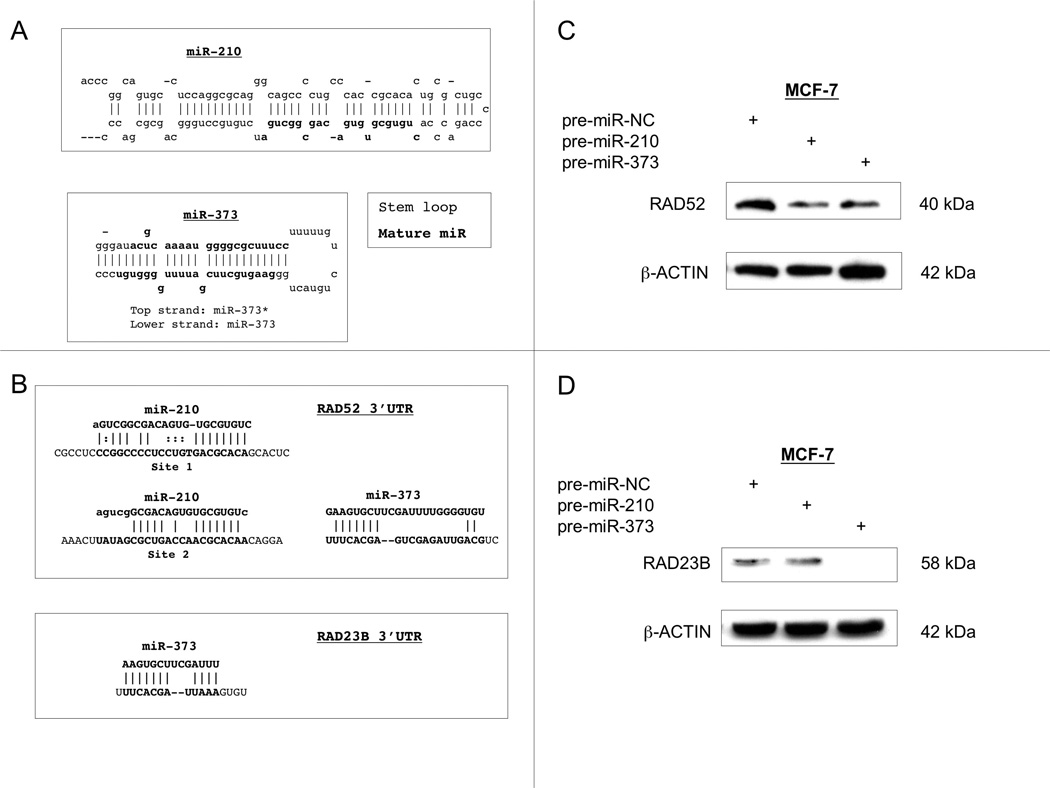
To determine the downstream impact of the miR upregulation, we performed in silico analyses to identify putative targets of miR-210 and miR-373. As the processed mature miRs are the functional molecules, we investigated the capacity of the miRs to bind possible target genes (Fig. 3A). Because of our interest in understanding the regulation of DNA repair genes in hypoxia, we employed miRBase (20–22) and TargetScan (24–26), which utilize different algorithms to examine the ability of miR-210 and miR-373 to interact with the 3′ UTRs of genes that are known to be involved in DNA repair activity. We determined that miR-210 was predicted to be capable of binding to the 3′ UTR of the HDR gene, RAD52, at 2 different sites, Site 1 and Site 2 (Fig. 3B). MiR-373 was predicted to bind to the 3′ UTRs of both RAD52 and the NER gene, RAD23B (Fig. 3B).
Fig. 3.
miR-210 and miR-373 regulates the DNA repair genes, RAD52 and RAD23B. (A) Bioinformatics analyses of miR-210 and miR-373 predict binding to the 3′ UTRs of genes implicated in DNA repair. The mature miRs (indicated in bold text) are processed from the stem-loop structures of miR-210 and miR-373. miR-373 is processed into two mature miRs, miR-373 and miR-373*, as indicated. (B) RAD52 contains two predicted binding sites for miR-210 in its 3′ UTR and one site for miR-373. RAD23B contains a single conserved site for miR-373 family binding, as indicated. (C) Forced expression of miR-210 and of miR-373 can down-regulate RAD52 protein levels. MCF-7 cells were transfected with the indicated pre-miRs (pre-miR sequences for miR-210, miR-373, and pre-miR-NC are 5′-CUGUGCGUGUGACAGCGGCUGA-3′ and 5′-GAAGUGCUUCGAUUUUGGGGUGU-3′, and 5′-AGUACUGCUUACGAUAC GGtt-3′ respectively, which are provided with an anti-sense passenger strand, creating partially double-stranded RNAs). Whole cell lysates were prepared from MCF-7 cells at 72 h post-transfection; RAD52 protein levels were assayed by Western blot. (D) Forced expression of miR-373, but not of miR-210, suppresses RAD23B protein levels. MCF-7 cells were transfected with pre-miRs as above; the total cell lysates were analyzed for RAD23B expression.
To examine the potential in vivo regulation of RAD52 and RAD23B by miR-210 and miR-373, we utilized transient forced expression of synthetic pre-miR molecules, which are partial duplexes comprised of the mature miR and a passenger strand. We detected the forced expression of the respective miRs by RTQ-PCR as early as 24 h and as late as 72 h after transfection (data not shown). After ensuring that the transfected miRs were expressed, we analyzed the effect on the protein expression of RAD52 and RAD23B by immunoblotting. We found that the overexpression of miR-210 suppressed the expression of RAD52, whereas miR-373 led to the decreased expression of both RAD52 and RAD23B (Figs. 3C and 3D). At the transcriptional level, we did not detect changes in the expression of either RAD52 or RAD23B (data not shown).
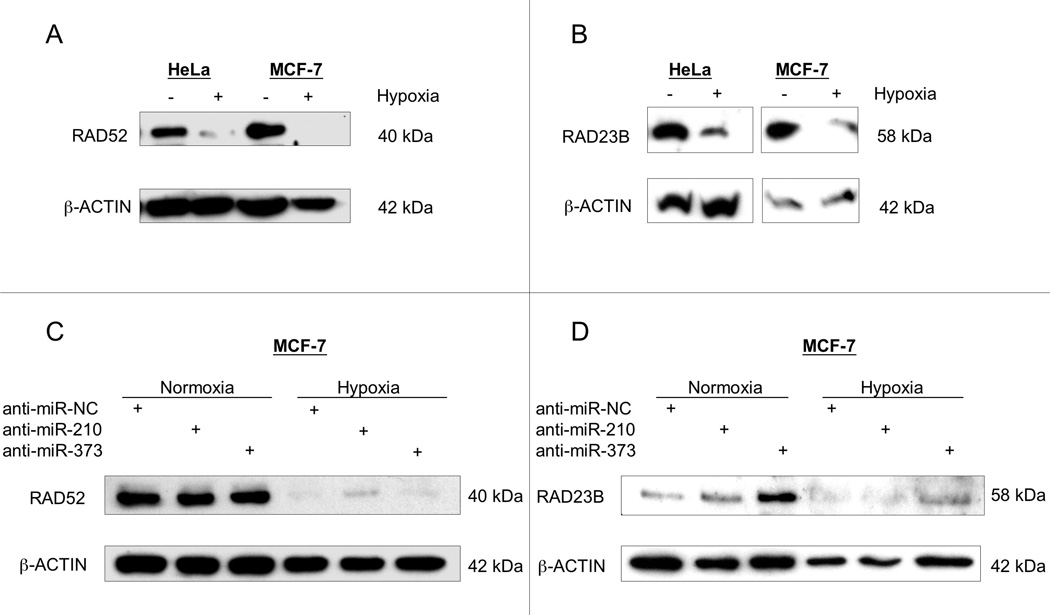
To further establish the physiological relevance of these observations, we next examined the expression of RAD52 and RAD23B in hypoxia. We performed immunoblotting experiments to analyze RAD52 and RAD23B levels in normoxic versus hypoxic (48 h, 0.01% O2) cells. In both HeLa and MCF-7 cells, RAD52 levels were decreased in response to hypoxia (Fig. 4A). Likewise, hypoxia also reduced the levels of RAD23B (Fig. 4B).
Fig. 4.
Hypoxia causes the down-regulation of RAD52 and RAD23B in pathways mediated by miR-210 and miR-373. (A) Western blotting indicates down-regulation of RAD52 expression in hypoxia (48 h, 0.01% O2) versus normoxia in HeLa and MCF-7 cells. (B) Western blot analysis shows RAD23B down-regulation in hypoxia versus normoxia in HeLa and MCF-7 cells. (C) Western blot analysis for RAD52 expression was performed on whole cell lysates in MCF-7 cells following pre-treatment (24 h) with anti-miR-210, anti-miR-373, or Negative Control #1 anti-miR (anti-miR-NC) and exposure to normoxia or hypoxia (48 h, 0.01% O2). The respective sequences for anti-miR-210, anti-miR-373, and anti-miR-NC are 5′-GACACGCACAC UGUCGCCGACU-3′ and 5′-CUUCACGAAGCUAAAACCCCACA-3′, and 5′-AAGUGGAUAUUGUUGCCAUC-3′. (D) RAD23B expression in MCF-7 cells after pre-treatment with anti-miR-210, anti-miR-373, or anti-miR-NC and exposure to normoxia or hypoxia is detected by Western blot.
As both RAD52 and RAD23B were down-regulated in hypoxia, we next asked whether miR-210 or miR-373 had roles in the suppression of either RAD52 or RAD23B. To probe the influence of these miRs on RAD52 and RAD23B levels in hypoxia, we used synthetic anti-sense oligonucleotides designed to target the mature forms of miR-210 or miR-373 and thereby inhibit their expression. The ability of such synthetic RNA-based anti-miRs to block miRNA activity has been established in prior studies (33). MCF-7 cells were transfected with the specific anti-sense molecules targeting miR-210, miR-373, or with scrambled anti-sense molecules 24 h prior to hypoxia or normoxia treatment. At 48 h post-transfection, the cells were harvested for the analysis of RAD52 and RAD23B expression by Western blot. In normoxic cells, the expression of RAD52 was unaffected by anti-miR-210 or anti-miR-373. However, in hypoxia cells, the suppression of RAD52 expression that occurs in hypoxia was somewhat alleviated by pre-treatment with anti-miR-210 (Fig. 4C). In the case of RAD23B expression, normoxic cells had increased RAD23B levels when treated with anti-miR-373 (Fig. 4D). In the hypoxic cells, pre-treatment with anti-miR-373 slightly rescued the levels of RAD23B relative to the otherwise hypoxia-suppressed levels in the other samples.
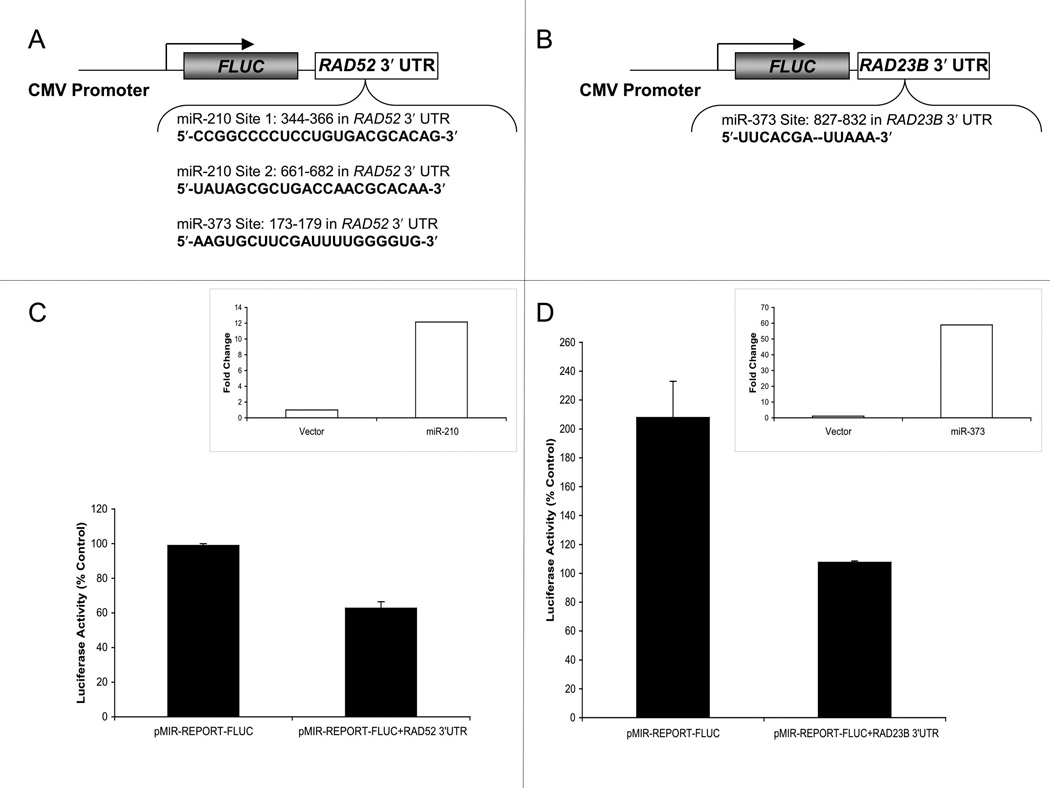
To test whether miR-210 and/or miR-373 may be implicated in the regulation of RAD52 or RAD23B via binding to the respective 3′ UTR targeting sites, we designed constructs utilizing the pMIR-REPORT-FLUC vector, which contains a multiple cloning site downstream of a luciferase reporter gene-coding region. The strategic cloning of the 3′ UTRs in these constructs allows the luciferase reporter to be subjected to regulation that mimics the putative miRNA target in the 3′ UTR of the respective gene. Thus, upon induction, miR-210 or 373 should be capable of binding to the respective target site(s) in the cloned 3′ UTR and thereby inhibit the protein expression of luciferase. For these experiments, ~2-kb genomic fragments, each comprising regions of the RAD52 and RAD23B 3′ UTRs, respectively, were inserted 3′ relative to the luciferase reporter gene (Figs. 5A and 5B). The resulting firefly luciferase reporter plasmids, pMIR-REPORT-FLUC+RAD52 3′ UTR or pMIR-REPORT-FLUC+RAD23B 3′ UTR were transiently co-transfected with constructs either expressing miR-210 or miR-373. Forced expression of miR-210 and miR-373 was validated by RTQ-PCR (Figs. 5C and 5D, insets). As controls, firefly luciferase constructs lacking the 3′ UTR targeting sequences were tested in separate samples. In each case, the transfection mixtures included a Renilla luciferase reporter vector, allowing a read-out for normalizing the transfection efficiency. At 24 h post-transfection in MCF-7 cells, lysates were harvested for the analysis of firefly luciferase activity, which was normalized to Renilla luciferase activity. In comparison to the vector control, the miR-210 overexpression vector reduced the luciferase expression from the vector containing the 3′ UTR of the RAD52 gene (Fig. 5C). Likewise, forced miR-373 expression vector led to reduced luciferase expression when the vector contained the 3′ UTR of the RAD23B gene (Fig. 5D). Taken together, these findings further suggest that miR-210 and miR-373 overexpression each can cause the down-regulation of RAD52 and RAD23B expression, respectively, via a mechanism that targets the 3′ UTR of these genes.
Fig. 5.
Luciferase reporter constructs indicate that miR-210 and miR-373 can act on the 3′ UTRs of the RAD52 or RAD23B genes, respectively. (A) Diagram of the pMIR-REPORT-FLUC+RAD52 3′ UTR vector indicates the sequences of the putative target sites for miR-210 and miR-373. (B) Diagram of the pMIR-REPORT-FLUC+RAD23B 3′ UTR vector shows the predicted binding site for miR-373. (C) MiR-210 suppresses the expression of the pMIR-REPORT+RAD52 3′ UTR vector. The pMIR-REPORT+RAD52 3′ UTR or the pMIR-REPORT vector containing only the luciferase coding region and no extra 3′ UTR were transfected into MCF-7 cells at the same time that a vector expressing miR-210 was introduced. The co-transfection of a vector expressing Renilla luciferase was used for normalization to control for the transfection efficiency. After 24 h, the cells were harvested and assayed for luciferase activity. Results are adjusted for transfection efficiency and normalized to the effect of an empty control pBABE vector. Error bars indicate stardard errors calculated from duplicate experiments, carried out in triplicate. (D) miR-373 suppresses the expression of pMIR-REPORT-FLUC+RAD23B 3′ UTR. Experiments were carried out as in (C), except that the forced expression of miR-373 was performed using pBABE-miR-373. Insets in (C) and (D) indicate the levels of miR-210 and miR-373 expression that were achieved via the pBABE-miR-210 and pBABE-miR-373 vectors, respectively, as quantitated by RTQ-PCR.
Discussion
Here, we have shown that hypoxic stress induces the expression of a series of miRs in human cells. One of these, miR-210, was predicted to target the mRNA for the DNA repair factor, RAD52; another, miR-373, was predicted to target both RAD52 and the NER damage recognition factor, RAD23B. Consistent with these in silico analyses, we found that the forced expression of either miR-210 or miR-373 reduced the levels of RAD52 and that miR-373 also suppressed RAD23B expression levels. In experiments comparing hypoxic to normoxic cells, we found that hypoxia induces the down-regulation of both RAD52 and RAD23B. The pre-treatment of cells with synthetic anti-sense oligonucleotides directed against miR-210 or miR-373 was found to reverse the hypoxia-mediated down-regulation of RAD52 or RAD23B, respectively. Interestingly, even in normoxic cells, anti-miR-373 treatment resulted in an up-regulation of RAD23B, indicating a key role for miR-373 in modulating the basal expression of RAD23B. On the other hand, the anti-miR-373 had an undetectable effect on the rescue of RAD52 supression in hypoxia cells, indicating that although the forced miR-373 expression can inhibit RAD52, miR-373 may have a lesser role in the suppression of RAD52 in hypoxic cells. Finally, experiments in which the 3′ UTRs of the RAD52 and RAD23B genes were inserted downstream of the firefly luciferase coding region in reporter vectors further demonstrated the roles of miR-210 and miR-373 in the regulation of these genes.
Our results indicated that both miR-210 and miR-373 are induced in hypoxia in a HIF-1α-dependent manner. This was previously observed for miR-210 (29) and is newly reported here for miR-373. These observations add to the complex network of HIF-1α-responsive pathways and provide a novel link between HIF-1α and DNA repair activity via miR pathways. Downstream of this regulation, our data indicate that synthetic anti-sense oligonucleotides specifically designed to target miR-210 or miR-373 can rescue RAD52 and RAD23B regulation in hypoxia. However, we note that this reversal is not complete. Indeed, several other groups have found that combinations of miRs can act together on an individual gene’s 3′ UTR (34, 35); moreover, no single miR may be sufficient to result in the complete inhibition of a protein (36). Hence, there may also be other miRs, as well as other factors, which may participate in the complex regulation of RAD52 and RAD23B. Nonetheless, miR-210 and miR-373 clearly have key modulatory roles.
RAD52 is an important factor in HDR. As a recombinational repair mediator, it is thought to assist in loading RAD51 onto DNA to form nucleoprotein filaments (37, 38). Functionally, our prior work has already established that HDR activity is reduced in hypoxic cells, using both episomal and chromosomal assays for HDR of double-strand breaks (10, 11). Initially, we attributed the observed reduction in HDR activity in hypoxic cells specifically to the down-regulation of BRCA1 and RAD51 due to transcriptional repression involving E2F4/p130 (11, 12). We can now include the miR-mediated suppression of RAD52 levels as another mechanism by which HDR is suppressed in hypoxia.
RAD23B is a key component of the XPC/RAD23B complex that mediates damage recognition in the NER pathway (39). Interestingly, we had previously reported that NER activity is functionally reduced in hypoxic cells using two different assays for UV damage repair (13), but we were unable to determine the mechanism at that time. Our finding here that RAD23B is suppressed by miR-373 in hypoxic cells provides new mechanistic insight into this phenomenon.
While our data clearly show that DNA repair factors are suppressed via a miR-dependent pathway in hypoxia, why would a cell down-regulate DNA repair under such conditions? It is conceivable that by shutting down these pathways, ATP pools may be preserved under metabolic stress. Conceptually, the shut down of certain DNA metabolic pathways may resemble features of the autophagy pathway, in which cellular components are sacrificed in the effort of self-preservation (40). Regardless of the reason, the consequences of decreased DNA repair include genomic instability, which confers a mutator phenotype on cancer cells in the adverse environment of a hypoxic tumor. This could be advantageous for the individual cancer cells, but would be deleterious for the host. Importantly, however, altered repair capacity many render hypoxic cells vulnerable to targeted strategies that exploit specific repair deficiencies, potentially providing an opportunity for designing new cancer therapies.
Acknowledgements
We thank Yuri Kim, Qun Lin, Yongming Ren, and Zhong Yun for their helpful advice, thoughful discussions, and generous sharing of reagents. We also thank the Glazer Lab members and Lisa Cabral for their assistance. This work was supported by grants from the NIH and the American Cancer Society to MEC (T32CA9159 and PF-07-263-01, respectively) and from the NIH to PMG (RO1ES005775 and PO1CA129186).
References
- 1.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci U S A. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buermeyer AB, Deschenes SM, Baker SM, Liskay RM. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds TY, Rockwell S, Glazer PM. Genetic instability induced by the tumor microenvironment. Cancer Res. 1996;56:5754–5757. [PubMed] [Google Scholar]
- 4.Hill RP. Tumor progression: potential role of unstable genomic changes. Cancer Metastasis Rev. 1990;9:137–147. doi: 10.1007/BF00046340. [DOI] [PubMed] [Google Scholar]
- 5.Shaw RJ. Glucose metabolism and cancer. Curr Opin Cell Biol. 2006;18:598–608. doi: 10.1016/j.ceb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Graeber TG, Osmanian C, Jacks T, Housman DE, Koch CJ, Lowe SW, Giaccia AJ. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 7.Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- 8.Bindra RS, Glazer PM. Co-repression of mismatch repair gene expression by hypoxia in cancer cells: role of the Myc/Max network. Cancer Lett. 2007;252:93–103. doi: 10.1016/j.canlet.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 9.Mihaylova VT, Bindra RS, Yuan J, Campisi D, Narayanan L, Jensen R, Giordano F, Johnson RS, Rockwell S, Glazer PM. Decreased expression of the DNA mismatch repair gene Mlh1 under hypoxic stress in mammalian cells. Mol Cell Biol. 2003;23:3265–3273. doi: 10.1128/MCB.23.9.3265-3273.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bindra RS, Gibson SL, Meng A, Westermark U, Jasin M, Pierce AJ, Bristow RG, Classon MK, Glazer PM. Hypoxia-induced down-regulation of BRCA1 expression by E2Fs. Cancer Res. 2005;65:11597–11604. doi: 10.1158/0008-5472.CAN-05-2119. [DOI] [PubMed] [Google Scholar]
- 12.Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- 13.Yuan J, Glazer PM. Mutagenesis induced by the tumor microenvironment. Mutat Res. 1998;400:439–446. doi: 10.1016/s0027-5107(98)00042-6. [DOI] [PubMed] [Google Scholar]
- 14.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 15.Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105:1608–1613. doi: 10.1073/pnas.0707594105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 18.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 26.Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson SL, Bindra RS, Glazer PM. Hypoxia-induced phosphorylation of Chk2 in an ataxia telangiectasia mutated-dependent manner. Cancer Res. 2005;65:10734–10741. doi: 10.1158/0008-5472.CAN-05-1160. [DOI] [PubMed] [Google Scholar]
- 29.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chakrabarti J, Turley H, Campo L, Han C, Harris AL, Gatter KC, Fox SB. The transcription factor DEC1 (stra13, SHARP2) is associated with the hypoxic response and high tumour grade in human breast cancers. Br J Cancer. 2004;91:954–958. doi: 10.1038/sj.bjc.6602059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koukourakis MI, Giatromanolaki A, Polychronidis A, Simopoulos C, Gatter KC, Harris AL, Sivridis E. Endogenous markers of hypoxia/anaerobic metabolism and anemia in primary colorectal cancer. Cancer Sci. 2006;97:582–588. doi: 10.1111/j.1349-7006.2006.00220.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 33.Fasanaro P, D'Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine-kinase ligand Ephrin-A3. J Biol Chem. 2008 doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai EC, Tam B, Rubin GM. Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev. 2005;19:1067–1080. doi: 10.1101/gad.1291905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuzin A, Kundu M, Brody T, Odenwald WF. The Drosophila nerfin-1 mRNA requires multiple microRNAs to regulate its spatial and temporal translation dynamics in the developing nervous system. Dev Biol. 2007;310:35–43. doi: 10.1016/j.ydbio.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benson FE, Baumann P, West SC. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 38.Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 39.Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 40.Bialik S, Kimchi A. Autophagy and tumor suppression: recent advances in understanding the link between autophagic cell death pathways and tumor development. Adv Exp Med Biol. 2008;615:177–200. doi: 10.1007/978-1-4020-6554-5_9. [DOI] [PubMed] [Google Scholar]