Association analyses of 249,796 individuals reveal eighteen new loci associated with body mass index (original) (raw)
. Author manuscript; available in PMC: 2011 May 1.
Published in final edited form as: Nat Genet. 2010 Oct 10;42(11):937–948. doi: 10.1038/ng.686
Abstract
Obesity is globally prevalent and highly heritable, but the underlying genetic factors remain largely elusive. To identify genetic loci for obesity-susceptibility, we examined associations between body mass index (BMI) and ~2.8 million SNPs in up to 123,865 individuals, with targeted follow-up of 42 SNPs in up to 125,931 additional individuals. We confirmed 14 known obesity-susceptibility loci and identified 18 new loci associated with BMI (P<5×10−8), one of which includes a copy number variant near GPRC5B. Some loci (MC4R, POMC, SH2B1, BDNF) map near key hypothalamic regulators of energy balance, and one is near GIPR, an incretin receptor. Furthermore, genes in other newly-associated loci may provide novel insights into human body weight regulation.
Obesity is a major and increasingly prevalent risk factor for multiple disorders, including type 2 diabetes and cardiovascular disease1,2. While lifestyle changes have driven its prevalence to epidemic proportions, heritability studies provide evidence for a substantial genetic contribution (h2~40–70%) to obesity risk3,4. BMI is an inexpensive, non-invasive measure of obesity that predicts the risk of related complications5. Identifying genetic determinants of BMI could lead to a better understanding of the biological basis of obesity.
Genome-wide association (GWA) studies of BMI have previously identified ten loci with genome-wide significant (P < 5×10−8) associations in or near FTO, MC4R, TMEM18, GNPDA2, BDNF, NEGR1, SH2B1, ETV5, MTCH2, and KCTD156–10. Many of these genes are expressed or known to act in the central nervous system, highlighting a likely neuronal component to the predisposition to obesity9. This pattern is consistent with results in animal models and studies of monogenic human obesity, where neuronal genes, particularly those expressed in the hypothalamus and involved in regulation of appetite or energy balance, are known to play a major role in susceptibility to obesity11–13.
The ten previously identified loci account for only a small fraction of the variation in BMI. Furthermore, power calculations based on the effect sizes of established variants have suggested that increasing the sample size would likely lead to the discovery of additional variants9. To identify more loci associated with BMI, we expanded the GIANT (Genetic Investigation of ANtropometric Traits) consortium GWA meta-analysis to include a total of 249,769 individuals of European ancestry.
Results
Stage 1 GWA studies identify novel loci associated with BMI
We first conducted a meta-analysis of GWA studies of BMI and ~2.8 million imputed or genotyped SNPs using data from 46 studies including up to 123,865 individuals (Online Methods, Supplementary Fig. 1 and Supplementary Note). This stage 1 analysis revealed 19 loci associated with BMI at P < 5×10−8 (Table 1, Fig. 1a and Supplementary Table 1). These 19 loci included all ten loci from previous GWA studies of BMI6–10, two loci previously associated with body weight10 (FAIM2 and SEC16B) and one locus previously associated with waist circumference14(near TFAP2B). The remaining six loci, near GPRC5B, MAP2K5/LBXCOR1, TNNI3K, LRRN6C, FLJ35779/HMGCR, and PRKD1, have not previously been associated with BMI or other obesity-related traits.
Table 1.
Stage 1 and stage 2 results of the 32 SNPs that were associated with BMI at genome-wide significance (P < 5.10−8) levels.
| SNP | Nearest gene | Other nearby genes* | Chr | Position** (bp) | Alleles** | Frequency effect allele (%) | Per allele change in BMI beta (se)*** | Explained variance (%) | Stage 1_P_-value | Stage 2_P_-value | Stage 1 + 2 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Effect | Other | n | _P_-value | ||||||||||
| Previous BMI loci | |||||||||||||
| rs1558902 | FTO | 16 | 52,361,075 | a | t | 42% | 0.39 (0.02) | 0.34% | 2.05E-62 | 1.007E-60 | 192,344 | 4.8E-120 | |
| rs2867125 | TMEM18 | 2 | 612,827 | c | t | 83% | 0.31 (0.03) | 0.15% | 2.42E-22 | 4.42E-30 | 197,806 | 2.77E-49 | |
| rs571312 | MC4R B | 18 | 55,990,749 | a | c | 24% | 0.23 (0.03) | 0.10% | 1.82E-22 | 3.19E-21 | 203,600 | 6.43E-42 | |
| rs10938397 | GNPDA2 | 4 | 44,877,284 | g | a | 43% | 0.18 (0.02) | 0.08% | 4.35E-17 | 1.45E-15 | 197,008 | 3.78E-31 | |
| rs10767664 | BDNFB,M | 11 | 27,682,562 | a | t | 78% | 0.19 (0.03) | 0.07% | 5.53E-13 | 1.17E-14 | 204,158 | 4.69E-26 | |
| rs2815752 | NEGR1C,Q | 1 | 72,585,028 | a | g | 61% | 0.13 (0.02) | 0.04% | 1.17E-14 | 2.29E-09 | 198,380 | 1.61E-22 | |
| rs7359397 | SH2B1Q,B,M | APOB48RQ,M, SULT1A2Q,M, AC138894.2M, ATXN2LM, TUFMQ | 16 | 28,793,160 | t | c | 40% | 0.15 (0.02) | 0.05% | 1.75E-10 | 7.89E-12 | 204,309 | 1.88E-20 |
| rs9816226 | ETV5 | 3 | 187,317,193 | t | a | 82% | 0.14 (0.03) | 0.03% | 7.61E-14 | 1.15E-06 | 196,221 | 1.69E-18 | |
| rs3817334 | MTCH2Q,M | NDUFS3 Q , CUGBP1 Q | 11 | 47,607,569 | t | c | 41% | 0.06 (0.02) | 0.01% | 4.79E-11 | 1.10E-03 | 191,943 | 1.59E-12 |
| rs29941 | KCTD15 | 19 | 39,001,372 | g | a | 67% | 0.06 (0.02) | 0.00% | 1.31E-09 | 2.40E-02 | 192,872 | 3.01E-09 | |
| Previous waist & weight loci | |||||||||||||
| rs543874 | SEC16B | 1 | 176,156,103 | g | a | 19% | 0.22 (0.03) | 0.07% | 1.66E-13 | 2.41E-11 | 179,414 | 3.56E-23 | |
| rs987237 | TFAP2B | 6 | 50,911,009 | g | a | 18% | 0.13 (0.03) | 0.03% | 5.97E-16 | 2.40E-06 | 195,776 | 2.90E-20 | |
| rs7138803 | FAIM2 | 12 | 48,533,735 | a | g | 38% | 0.12 (0.02) | 0.04% | 3.96E-11 | 7.82E-08 | 200,064 | 1.82E-17 | |
| rs10150332 | NRXN3 | 14 | 79,006,717 | c | t | 21% | 0.13 (0.03) | 0.02% | 2.03E-07 | 2.86E-05 | 183,022 | 2.75E-11 | |
| Newly identified BMI loci | |||||||||||||
| rs713586 | RBJ | ADCY3Q, M, POMCQ,B | 2 | 25,011,512 | c | t | 47% | 0.14 (0.02) | 0.06% | 1.80E-07 | 1.44E-16 | 230,748 | 6.17E-22 |
| rs12444979 | GPRC5BC,Q | IQCK Q | 16 | 19,841,101 | c | t | 87% | 0.17 (0.03) | 0.04% | 4.20E-11 | 8.13E-12 | 239,715 | 2.91E-21 |
| rs2241423 | MAP2K5 | LBXCOR1 M | 15 | 65,873,892 | g | a | 78% | 0.13 (0.02) | 0.03% | 1.15E-10 | 1.59E-09 | 227,950 | 1.19E-18 |
| rs2287019 | QPCTL | GIPRB,M | 19 | 50,894,012 | c | t | 80% | 0.15 (0.03) | 0.04% | 3.18E-07 | 1.40E-10 | 194,564 | 1.88E-16 |
| rs1514175 | TNNI3K | 1 | 74,764,232 | a | g | 43% | 0.07 (0.02) | 0.02% | 1.36E-09 | 7.04E-06 | 227,900 | 8.16E-14 | |
| rs13107325 | SLC39A8Q,M | 4 | 103,407,732 | t | c | 7% | 0.19 (0.04) | 0.03% | 1.37E-07 | 1.93E-07 | 245,378 | 1.50E-13 | |
| rs2112347 | FLJ35779 M | HMGCR B | 5 | 75,050,998 | t | g | 63% | 0.10 (0.02) | 0.02% | 4.76E-08 | 8.29E-07 | 231,729 | 2.17E-13 |
| rs10968576 | LRRN6C | 9 | 28,404,339 | g | a | 31% | 0.11 (0.02) | 0.02% | 1.88E-08 | 3.19E-06 | 216,916 | 2.65E-13 | |
| rs3810291 | TMEM160 Q | ZC3H4 Q | 19 | 52,260,843 | a | g | 67% | 0.09 (0.02) | 0.02% | 1.04E-07 | 1.59E-06 | 233,512 | 1.64E-12 |
| rs887912 | FANCL | 2 | 59,156,381 | t | c | 29% | 0.10 (0.02) | 0.03% | 2.69E-06 | 1.72E-07 | 242,807 | 1.79E-12 | |
| rs13078807 | CADM2 | 3 | 85,966,840 | g | a | 20% | 0.10 (0.02) | 0.02% | 9.81E-08 | 5.32E-05 | 237,404 | 3.94E-11 | |
| rs11847697 | PRKD1 | 14 | 29,584,863 | t | c | 4% | 0.17 (0.05) | 0.01% | 1.11E-08 | 2.25E-04 | 241,667 | 5.76E-11 | |
| rs2890652 | LRP1B | 2 | 142,676,401 | c | t | 18% | 0.09 (0.03) | 0.02% | 2.38E-07 | 9.47E-05 | 209,068 | 1.35E-10 | |
| rs1555543 | PTBP2 | 1 | 96,717,385 | c | a | 59% | 0.06 (0.02) | 0.01% | 7.65E-07 | 4.48E-05 | 243,013 | 3.68E-10 | |
| rs4771122 | MTIF3 | GTF3A Q | 13 | 26,918,180 | g | a | 24% | 0.09 (0.03) | 0.02% | 1.20E-07 | 8.24E-04 | 198,577 | 9.48E-10 |
| rs4836133 | ZNF608 | 5 | 124,360,002 | a | c | 48% | 0.07 (0.02) | 0.01% | 7.04E-07 | 1.88E-04 | 241,999 | 1.97E-09 | |
| rs4929949 | RPL27A | TUB B | 11 | 8,561,169 | c | t | 52% | 0.06 (0.02) | 0.01% | 7.57E-08 | 1.00E-03 | 249,791 | 2.80E-09 |
| rs206936 | NUDT3 | HMGA1 B | 6 | 34,410,847 | g | a | 21% | 0.06 (0.02) | 0.01% | 2.81E-06 | 7.39E-04 | 249,777 | 3.02E-08 |
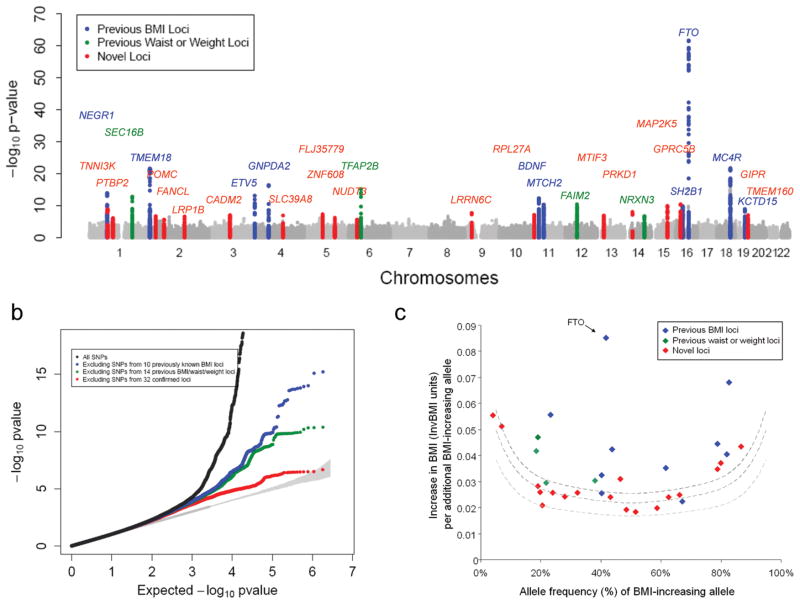
Figure 1. Genome-wide association results for the BMI meta-analysis.
(a) Manhattan plot showing the significance of association between all SNPs and BMI in the stage 1 meta-analysis, highlighting SNPs previously reported to show genome-wide significant association with BMI (blue), weight or waist circumference (green), and the 18 new regions described here (red). The 19 SNPs that reached genome-wide significance at Stage 1 (13 previously reported and 6 new) are listed in Table 1). (b) Quantile-quantile (Q-Q) plot of SNPs in stage 1 meta-analysis (black) and after removing any SNPs within 1 Mb of the 10 previously reported genome-wide significant hits for BMI (blue), after additionally excluding SNPs from the four loci for waist/weight (green) and after excluding SNPs from all 32 confirmed loci (red). The plot was abridged at the Y-axis (at P < 10−20) to better visualise the excess of small_P_-values after excluding the 32 confirmed loci (Supplementary Fig. 3shows full-scale Q-Q plot). The shaded region is the 95% concentration band. (c) Plot of effect size (in inverse normally transformed units (invBMI)) versus effect allele frequency of newly identified and previously identified BMI variants after stage 1 + stage 2 analysis; including the 10 previously identified BMI loci (blue), the four previously identified waist and weight loci (green) and the 18 newly identified BMI loci (blue). The dotted lines represent the minimum effect sizes that could be identified for a given effect-allele frequency with 80% (upper line), 50% (middle line), and 10% (lower line) power, assuming a sample size of 123,000 individuals and a α-level of 5×10−8.
Stage 2 follow-up leads to additional novel loci for BMI
To identify additional BMI-associated loci and to validate the loci that reached genome-wide significance in stage 1 analyses, we examined SNPs representing 42 independent loci (including the 19 genome-wide significant loci) with stage 1 P < 5×10−6. Variants were considered to be independent if the pair-wise linkage disequilibrium (LD;r_2) was less than 0.1 and if they were separated by at least 1 Mb. In stage 2, we examined these 42 SNPs in up to 125,931 additional individuals (79,561 newly genotyped individuals from 16 different studies and 46,370 individuals from 18 additional studies for which GWA data were available; Table 1, Supplementary Note, and Online Methods). In a joint analysis of stage 1 and stage 2 results, 32 of the 42 SNPs reached_P < 5×10−8. Even after excluding SNPs within these 32 confirmed BMI loci, we still observed an excess of small _P_-values compared to the distribution expected under the null hypothesis (Fig. 1b), suggesting that more BMI loci remain to be uncovered.
The 32 confirmed associations included all 19 loci with_P_ < 5×10−8 at stage 1, 12 additional novel loci near RBJ/ADCY3/POMC, QPCTL/GIPR, SLC39A8, TMEM160, FANCL, CADM2, LRP1B, PTBP2, MTIF3/GTF3A, ZNF608, RPL27A/TUB, NUDT3/HMGA1, and one locus (NRXN3) previously associated with waist circumference15 (Table 1, Supplementary Table 1, Supplementary Fig. 1 and 2). In all, our study increased the number of loci robustly associated with BMI from 10 to 32. Four of the 22 new loci were previously associated with body weight10 or waist circumference14,15, whereas 18 loci had not previously associated with any obesity-related trait in the general population. Whilst we confirmed all loci previously established by large-scale GWA studies for BMI6–10and waist circumference14,15, four loci identified by GWA studies for early-onset or adult morbid obesity16,17 [at_NPC1_ (rs1805081; P = 0.0025),MAF (rs1424233; P = 0.25),PTER (rs10508503; P = 0.64), and_TNKS/MSRA_ (rs473034; P = 0.23)] showed limited or no evidence of association with BMI in our study.
As expected, the effect sizes of the 18 newly discovered loci are slightly smaller, for a given minor allele frequency, than those of the previously identified variants (Table 1and Fig. 1c). The increased sample size also brought out more signals with low minor allele frequency. The BMI-increasing allele frequencies for the 18 newly identified variants ranged from 4% to 87%, covering more of the allele frequency spectrum than previous, smaller GWA studies of BMI (24%–83%)9,10(Table 1 and Fig. 1c).
We tested for evidence of non-additive (dominant or recessive) effects, SNP×SNP interaction effects and heterogeneity by sex or study among the 32 BMI-associated SNPs (Online Methods). We found no evidence for any such effects (P > 0.001, no significant results after correcting for multiple testing) (Supplementary Tables 1 and Supplementary Note).
Impact of 32 confirmed loci on BMI, obesity, body size, and other metabolic traits
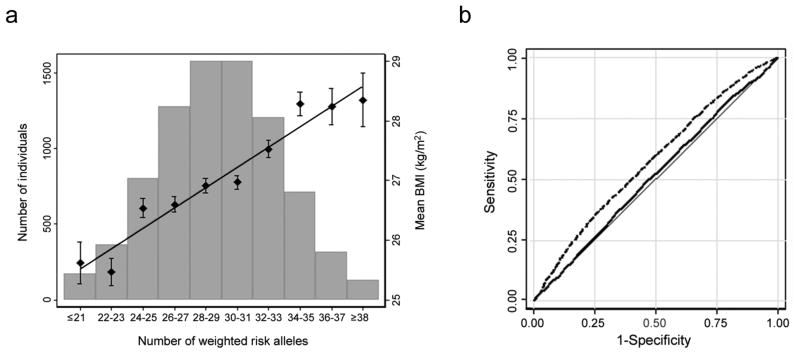
Together, the 32 confirmed BMI loci explained 1.45% of the inter-individual variation in BMI of the stage 2 samples, with the_FTO_ SNP accounting for the largest proportion of the variance (0.34%) (Table 1). To estimate the cumulative effect of the 32 variants on BMI, we constructed a genetic-susceptibility score that sums the number of BMI-increasing alleles weighted by the overall stage 2 effect sizes in the ARIC study (N = 8,120), one of our largest population-based studies (Online Methods). For each unit increase in the genetic-susceptibility score, approximately equivalent to one additional risk allele, BMI increased by 0.17 kg/m2, equivalent to a 435–551 g gain in body weight in adults of 160–180 cm in height. The difference in average BMI between individuals with a high genetic-susceptibility score (≥38 BMI-increasing alleles, 1.5% (n=124) of the ARIC sample) and those with a low genetic-susceptibility score (≤21 BMI-increasing alleles, 2.2% (n=175) of the ARIC sample) was 2.73 kg/m2, equivalent to a 6.99 to 8.85 kg body weight difference in adults 160–180 cm in height (Fig. 2a). Still, we note that the predictive value for obesity risk and BMI of the 32 variants combined was modest, although statistically significant (Fig. 2b, Supplementary Fig. 4). The area under the receiver operating characteristic (ROC) curve for prediction of risk of obesity (BMI ≥ 30 kg/m2) using age, age2 and sex only was 0.515 (P = 0.023 compared to AUC of 0.50), which increased to 0.575 (P < 10−5) when also the 32 confirmed SNPs were included in the model (Fig. 2b). The area under the ROC for the 32 SNPs only was 0.574 (P < 10−5).
Figure 2. Combined impact of risk alleles on BMI/obesity.
(a) Combined effect of risk alleles on average BMI in the population-based Atherosclerosis Risk in Communities (ARIC) study (n = 8,120 individuals of European descent). For each individual, the number of “best guess” replicated (n = 32) risk alleles from imputed data (0,1,2) per SNP was weighted for their relative effect sizes estimated from the stage 2 data. Weighted risk alleles were summed for each individual and the overall individual sum was rounded to the nearest integer to represent the individual’s risk allele score (range 16–44). Along the x-axis, individuals in each risk allele category are shown (grouped ≤21 and ≥38 at the extremes), and the mean BMI (+/− SEM) is plotted (y axis on right), with the line representing the regression of the mean BMI values across the risk-allele scores. The histogram (y-axis on left) represents the number of individuals in each risk-score category. (b) The area under the ROC curve (AUC) of two different models predicting the risk of obesity (BMI = ≥30 kg/m2) in the n = 8,120 genotyped individuals of European descent in the ARIC Study. Model 1, represented by the solid line, includes age, age2, and sex (AUC = 0.515,P = 0.023 for difference from AUCnull = 0.50). Model 2, represented by the dashed line, includes age, age2, sex, and the n = 32 confirmed BMI SNPs (AUC = 0.0575, P < 10−5 for difference from AUCnull = 0.50). The difference between both AUCs is significant (P< 10−4).
All 32 confirmed BMI-increasing alleles showed directionally consistent effects on risk of being overweight (BMI ≥25 kg/m2) or obese (≥30 kg/m2) in stage 2 samples, with 30 of 32 variants achieving at least nominally significant associations. The BMI-increasing alleles increased the odds of overweight by 1.013 to 1.138-fold, and the odds for being obese by 1.016- to 1.203-fold (Supplementary Table 2). In addition, 30 of the 32 loci also showed directionally consistent effects on the risk of extreme and early-onset obesity in a meta-analysis of seven case-control studies of adults and children (binomial sign test P = 1.3×10−7) (Supplementary Table 3). The BMI-increasing allele observed in adults also increased the BMI in children and adolescents with directionally consistent effects observed for 23 of the 32 SNPs (binomial sign test P = 0.01). Furthermore, in family-based studies, the BMI-increasing allele was over-transmitted to the obese offspring for 24 of the 32 SNPs (binomial sign test _P_= 0.004) (Supplementary Table 3). As these studies in extreme obesity cases, children and families were relatively small (Nrange = 354 − 15,251) compared to the overall meta-analyses, their power was likely insufficient to confirm association for all 32 loci. Nevertheless, these results show that the effects are unlikely to reflect population stratification and that they extend to BMI differences throughout the life course.
All BMI-increasing alleles were associated with increased body weight, as expected from the correlation between BMI and body weight (Supplementary Table 2). To confirm an effect of the loci on adiposity rather than general body size, we tested association with body fat percentage, which was available in a subset of the stage 2 replication samples (n = 5,359–28,425) (Supplementary Table 2). The BMI-increasing allele showed directionally consistent effects on body fat percentage at 31 of the 32 confirmed loci (binomial sign test _P_= 1.54×10−8) (Supplementary Table 2).
We also examined the association of the BMI loci with metabolic traits (type 2 diabetes18, fasting glucose, fasting insulin, indices of beta-cell function (HOMA-B) and insulin resistance (HOMA-IR)19, and blood lipid levels20) and with height (Supplementary Tables 2 and 4). Although many nominal associations are expected because of known correlations between BMI and most of these traits and because of overlap in samples, several associations stand out as possible examples of pleiotropic effects of the BMI-associated variants. Particularly interesting is the variant in the GIPR locus where the BMI-increasing allele is also associated with increased fasting glucose levels and lower 2-hour glucose levels (Supplementary Table 4)19,21. The direction of the effect is opposite to what would be expected due to the correlation between obesity and glucose intolerance, but is consistent with the suggested roles of _GIPR_in glucose and energy metabolism (see below)22. Three loci show strong associations (P < 10−4) with height (MC4R,RBJ/ADCY3/POMC and MTCH2/NDUFS3). Because BMI is weakly correlated with height (and indeed, the BMI-associated variants as a group show no consistent effect on height), these associations are also suggestive of pleiotropy. Interestingly, analogous to the effects of severe mutations in POMC and MC4R on height and weight23,24, the BMI-increasing alleles of the variants near these genes were associated with decreased (POMC) and increased (MC4R) height, respectively (Supplementary Table 2).
Potential functional roles and pathways analyses
Although associated variants typically implicate genomic regions rather than individual genes, we note that some of the 32 loci include candidate genes with established connections to obesity. Several of the 10 previously identified loci are located in or near genes that encode neuronal regulators of appetite or energy balance, including MC4R12,25,BDNF26, and_SH2B1_11,27. Each of these genes has been tied to obesity, not only in animal models, but also by rare human variants that disrupt each of these genes and lead to severe obesity24,28,29. Using the automated literature search programme, Snipper (Online Methods), we identified various genes within the novel loci with potential biological links to obesity-susceptibility (Supplementary Note). Among the novel loci, the location of rs713586 near POMC provides further support for a role of neuroendocrine circuits that regulate energy balance in susceptibility to obesity. POMC encodes several polypeptides including α-MSH, a ligand of the MC4R gene product30, and rare mutations in_POMC_ also cause human obesity23,29,31.
In contrast, the locus near GIPR, which encodes a receptor of gastric inhibitory polypeptide (GIP), suggests a role for peripheral biology in obesity. GIP, which is expressed in the K cell of the duodenum and intestine, is an incretin hormone that mediates incremental insulin secretion in response to oral intake of glucose. The variant associated with BMI is in strong LD (r_2 = 0.83) with a missense SNP in_GIPR (rs1800437, Glu354Gln) that has recently been shown to influence the glucose and insulin response to an oral glucose challenge 21. Although no human phenotype is known to be caused by mutations in GIPR, mice with disruption of_Gipr_ are resistant to diet-induced obesity32. The association of a variant in_GIPR_ with BMI suggests that there may be a link between incretins/insulin secretion and body weight regulation in humans as well.
To systematically identify biological connections among the genes located near the 32 confirmed SNPs, and to potentially identify new pathways associated with BMI, we performed pathway-based analyses using MAGENTA33. Specifically, we tested for enrichment of BMI genetic associations in biological processes or molecular functions that contain at least one gene from the 32 confirmed BMI loci (Online Methods). Using annotations from the KEGG, Ingenuity, PANTHER, and Gene Ontology databases, we found evidence of enrichment for pathways involved in the platelet-derived growth factor (PDGF) signaling (PANTHER, P = 0.0008, FDR = 0.0061), translation elongation (PANTHER, P = 0.0008, FDR = 0.0066), hormone or nuclear hormone receptor binding (Gene Ontology, P < 0.0005, FDR < 0.0085), homeobox transcription (PANTHER, P = 0.0001, FDR = 0.011), regulation of cellular metabolism (Gene Ontology, P = 0.0002, FDR = 0.031), neurogenesis and neuron differentiation (Gene Ontology, P < 0.0002, FDR < 0.034), protein phosphorylation (PANTHER, P = 0.0001, FDR = 0.045) and numerous other pathways related to growth, metabolism, immune and neuronal processes (Gene Ontology, P < 0.002, FDR < 0.046) (Supplementary Table 5).
Identifying possible functional variants
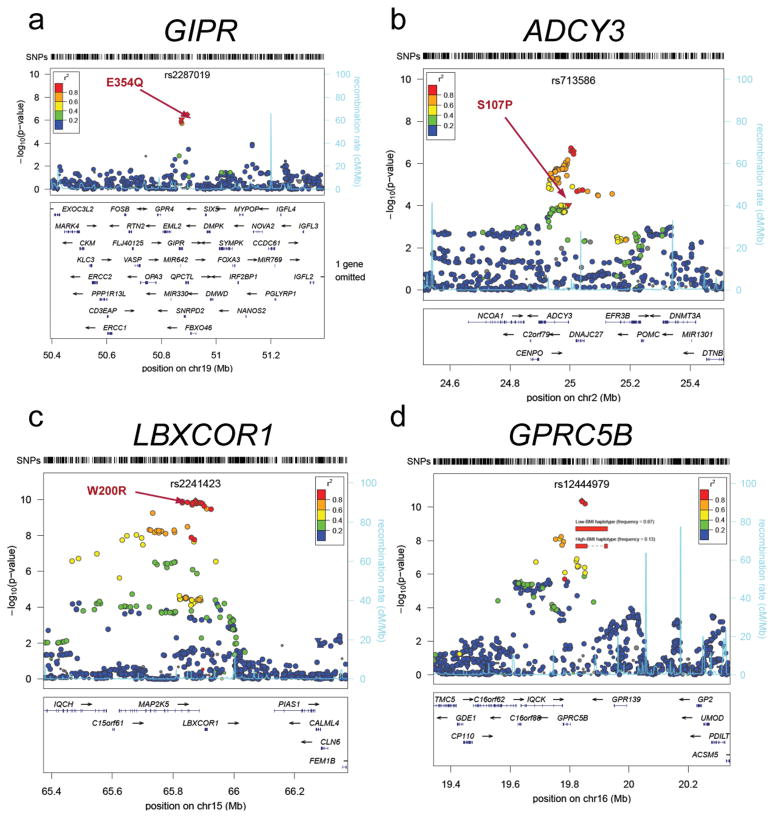
We used data from the 1000 Genomes Project and the HapMap Consortium to explore whether the 32 confirmed BMI SNPs were in LD (r_2 ≥ 0.75) with common missense SNPs or copy number variants (CNVs) (Online Methods). Non-synonymous variants in LD with our signals were present in the BDNF, SLC39A8, FLJ35779/HMGCR, QPCTL/GIPR, MTCH2, ADCY3, and LBXCOR1 genes. In addition, the rs7359397 signal was in LD with coding variants in several genes including_SH2B1, ATNX2L, APOB48R, SULT1A2, and_AC138894.2_ (Table 1,Fig. 3, Supplementary Table 6 and Supplementary Fig. 2). Furthermore, two SNPs tagged common CNVs. The first CNV was previously identified and is a 45-kb deletion near NEGR19. The second CNV is a 21-kb deletion that lies 50kb upstream of GPRC5B; the deletion allele is tagged by the T-allele of rs12444979 (_r_2 = 1) (Fig. 3). Although the correlations with potentially functional variants does not prove that these variants are indeed causal, these provide first clues as to which genes and variants at these loci might be prioritized for fine-mapping and functional follow-up.
Figure 3. Regional plots of selected replicating BMI loci with missense and CNV variants.
SNPs are plotted by position on chromosome against association with BMI (−log10 _P-_value). The SNP name shown on the plot was the most significant SNP after stage 1 meta-analysis. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP (taken from pairwise r2 values from the HapMap CEU database, www.hapmap.org). Genes, position of exons, and direction of transcription from UCSC genome browser (http://genome.ucsc.edu) are noted. Hashmarks represent SNP positions available in the meta-analysis. (a, b, c) Missense variants noted with their amino acid change for the gene noted above the plot. (d) Structural haplotypes and BMI association signal in the GPRC5B region. A 21 kb deletion polymorphism is associated with 4 SNPs (r2=1.0) that comprise the best haplogroup associating with BMI. Plots were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom).
As many of the 32 BMI loci harbor multiple genes, we examined whether gene expression (eQTL) analyses could also direct us to positional candidates. Gene expression data were available for human brain, lymphocytes, blood, subcutaneous and visceral adipose tissue, and liver34–36(Online Methods,Table 1 and Supplementary Table 7). Significant_cis_-associations, defined at the tissue-specific level, were observed between 14 BMI-associated alleles and expression levels (Table 1 and Supplementary Table 7). In several cases, the BMI-associated SNP was the most significant SNP or explained a substantial proportion of the association with the most significant SNP for the gene transcript in conditional analyses (P_adj>0.05). These significant associations included NEGR1, ZC3H4, TMEM160,MTCH2, NDUFS3, GTF3A, ADCY3, APOB48R, SH2B1,TUFM, GPRC5B, IQCK,SLC39A8, SULT1A1, and_SULT1A2 (Table 1 andSupplementary Table 7), making these genes higher priority candidates within the associated loci. However, we note that some BMI-associated variants were correlated with the expression of multiple nearby genes, making it difficult to determine the most relevant gene.
Evidence for the existence of additional associated variants
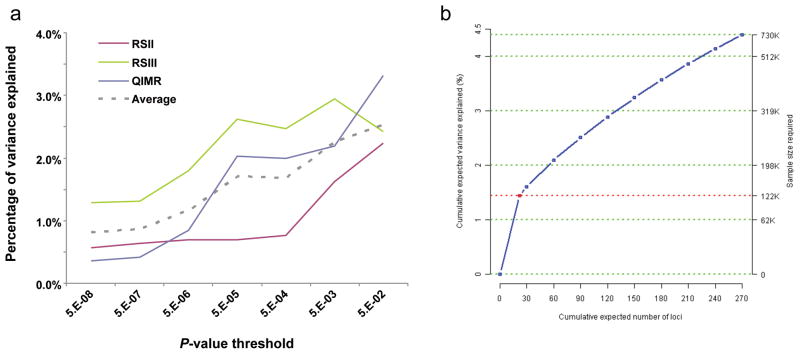
Because the variants identified by this large study explain only 1.45% of the variance in BMI (2–4% of genetic variance based on an estimated heritability of 40–70%), we considered how much the explained phenotypic variance could be increased by including more SNPs at various degrees of significance in a polygene model using an independent validation set (Online Methods)37. We found that including SNPs associated with BMI at lower significance levels (up to_P_ > 0.05) increased the explained phenotypic variance in BMI to 2.5%, or 4% to 6% of genetic variance (Fig. 4a). In a separate analysis, we estimated the total number of independent BMI-associated variants that are likely to exist with similar effect sizes to the 32 confirmed here (Online Methods)38. Based on the effect size and allele frequencies of the 32 replicated loci observed in stage 2 and the power to detect association in the combined stage 1 and stage 2, we estimated that there are 284 (95% CI: 132–510) loci with similar effect sizes as the currently observed ones, which together would account for 4.5% (95% CI: 3.1–6.8%) of the variation in BMI or 6–11% of the genetic variation (based on an estimated heritability of 40–70%) (Supplementary Table 8). In order to detect 95% of these loci, a sample size of approximately 730,000 subjects would be needed (Fig. 4b). This method does not account for the potential of loci of smaller effect than those identified here to explain even more of the variance and thus provides an estimated lower bound of explained variance. These two analyses strongly suggest that larger GWA studies will continue to identify additional novel associated loci, but also indicate that even extremely large studies focusing on variants with allele frequencies above 5% will not account for a large fraction of the genetic contribution to BMI.
Figure 4. Phenotypic variance explained by common variants.
(a) Variance explained is higher when SNPs not reaching genome-wide significance are included in the prediction model. The y-axis represents the proportion of variance explained at different_P-_value thresholds from stage 1 meta-analysis. Results are given for three studies (RSII, RSIII, QIMR), which were not included in the meta-analysis, after exclusion of all samples from The Netherlands (for RSII and RSIII) and the United Kingdom (for QIMR) from the discovery analysis for this sub-analysis. The dotted line represents the weighted average of the explained variance of three validation sets. (b) Cumulative number of susceptibility loci expected to be discovered, including those we have already identified and others that have yet to be detected, by the expected percentage of phenotypic variation explained and sample size required for a one-stage GWA study assuming a GC correction is utilized. The projections are based on loci that achieved a significance level of P < 5×10−8 in the joint analysis of stage 1 and stage 2 and the distribution of their effect sizes in stage 2. The dotted red line corresponds to the expected phenotypic variance explained by the 22 loci that are expected to be discovered in a one-stage GWAS with the sample size of stage 1 of this study.
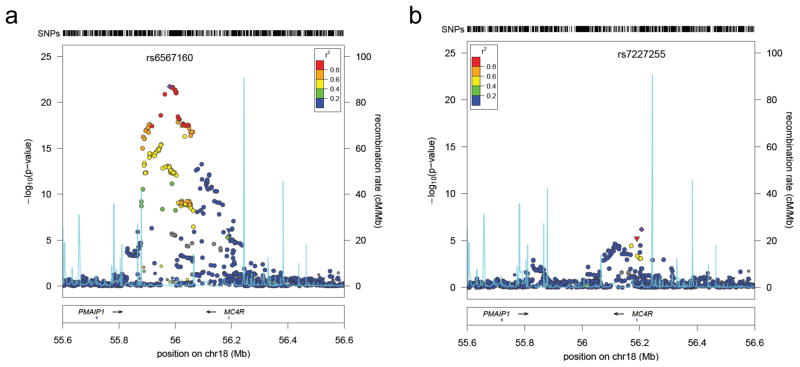
We examined whether selecting only a single variant from each locus for follow-up led us to underestimate the fraction of phenotypic variation explained by the associated loci. To search for additional independent loci at each of the 32 associated BMI loci, we repeated our GWA meta-analysis, conditioning on the 32 confirmed SNPs. Using a significance threshold of 5 × 10−6 for SNPs at known loci, we identified one apparently independent signal at the MC4R locus; rs7227255 was associated with BMI (P = 6.56 × 10−7) even after conditioning for the most strongly associated variant near_MC4R_ (rs571312) (Fig. 5). Interestingly, rs7227255 is in perfect LD (r2= 1) with a relatively rare MC4R missense variant (rs2229616, V103I, minor allele frequency = 1.7%) that has been associated with BMI in two independent meta-analyses39,40. Furthermore, mutations at the MC4R locus are known to influence early-onset obesity24,41, supporting the notion that allelic heterogeneity may be a frequent phenomenon in the genetic architecture of obesity.
Figure 5. Second signal at the MC4R locus contributing to BMI.
SNPs are plotted by position in a 1 Mb window of chromosome 18 against association with BMI ( log10 _P-_value). Panel (a) highlights the most significant SNP in stage 1 meta-analysis, panel (b) the most significant SNP after conditional analysis where the model included the most strongly associated SNP from panel A as a covariate. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP (taken from pairwise r2 values from the HapMap CEU database, www.hapmap.org). Genes, exons, and direction of transcription from UCSC genome browser (genome.ucsc.edu) are noted. Hashmarks at the top of the figure represent positions of SNPs in the meta-analysis. Regional plots were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom).
Discussion
Using a two-stage genome-wide association meta-analysis of up to 249,796 individuals of European descent, we have identified 18 additional loci that are associated with BMI at genome-wide significance, bringing the total number of such loci to 32. We estimate that more than 250 (i.e. 284 predicted loci – 32 confirmed loci) common variant loci with effects on BMI similar to those described here remain to be discovered, and even larger numbers of loci with smaller effects. A substantial proportion of these loci should be identifiable through larger GWA studies and/or by targeted follow-up of top signals selected from our stage 1 analysis. The latter approach is already being implemented through large-scale genotyping of samples informative for BMI using a custom array (the Metabochip) designed to support follow-up of thousands of promising variants in hundreds of thousands of individuals.
The combined effect on BMI of the associated variants at the 32 loci is modest, and even when we try to account for as-yet-undiscovered variants with similar properties, we estimate that these common variant signals account for only 6–11% of the genetic variation in BMI. There is a strong expectation that additional variance and biology will be explained using complementary approaches that capture variants not examined in the current study, such as lower frequency variants and short insertion-deletion polymorphisms. There is good reason to believe (based on our findings at MC4R and other loci_– POMC, BDNF, SH2B1 –_ which feature both common and rare variant associations) that a proportion of such low-frequency and rare causal variation will map to the loci already identified by GWA studies.
A primary goal of human genetic discovery is to improve understanding of the biology of conditions such as obesity42. One particularly interesting finding in this regard is the association between BMI and common variants near GIPR, which may indicate a causal contribution of variation in postprandial insulin secretion to the development of obesity. In most cases, the loci identified by the present study harbor few, if any, annotated genes with clear connections to the biology of weight regulation. This reflects our still limited understanding of the biology of BMI and obesity-related traits and is in striking contrast with the results from equivalent studies of certain other traits (such as autoimmune diseases or lipid levels). Thus, these results suggest that much novel biology remains to be uncovered, and that GWA studies may provide an important entry point. In particular, further examination of the associated loci through a combination of resequencing and fine-mapping to find causal variants, and genomic and experimental studies designed to assign function, could uncover novel insights into the biology of obesity.
In conclusion, we have performed GWA studies in large samples to identify numerous genetic loci associated with variation in BMI, a common measure of obesity. Because current lifestyle interventions are largely ineffective in addressing the challenges of growing obesity43,44, new insights into biology are critically needed to guide the development and application of future therapies and interventions.
Supplementary Material
1
Supplementary Figure 1 Study design. Stage 1 - Meta analysis of genome-wide association data was performed in stage 1 across 46 studies of white European Ancestry. A total of 42 SNPs representing the best associating (P < 10−6) loci (shown) were taken forward for replication. Nineteen of these SNPs (loci in bold) reached already genome-wide significance at stage 1. Stage 2 – The 42 SNPs were genotyped in 16 de novo replication studies and extracted from 18 in silico replication studies, all adults of European ancestry and were tested for association with BMI. In a joint analyses of stage 1 and stage 2 data, 32 SNPs (loci in bold) reached genome-wide significance (P < 5×10−8). Follow-up analyses – The 32 confirmed loci were taken forward for additional analyses.
Supplementary Figure 2 Regional plots of the 32 confirmed BMI loci with missense and CNV variants. SNPs are plotted by position on chromosome against association with BMI (−log10 _P_-value). The SNP name shown on the plot was the most significant SNP after stage 1 meta-analysis. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP (taken from pairwise r2 values from the HapMap CEU database, www.hapmap.org). Genes, position of exons, and direction of transcription from UCSC genome browser (genome.ucsc.edu) are noted. Hash-marks represent SNP positions available in the meta-analysis. Plots were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom).
Supplementary Figure 3 Quantile-quantile plot of SNPs at stage 1 GIANT meta-analysis (black) and after removing any SNPs within 1 Mb of the 10 previously reported genome-wide significant hits for BMI (blue), after additionally excluding the four loci for waist/weight (green) and after excluding all 32 confirmed loci (red).
Supplementary Figure 4 Relationship between the predicted BMI, based on the 32 confirmed BMI loci combined, and the actual BMI in the ARIC Study (N=8,120). Panel (a) shows the comparison of the predicted BMI (grey), with the actual BMI (green) and quartile ranges (orange) in sets of 500 individuals, suggesting good average predictions. Panel (b) shows the comparison of the individual predicted and observed BMI values, suggesting poor individual predictions.
Supplementary Table 1 The 42 SNPs, associated with BMI at P < 5.10−6 at stage 1, that were taken forward for replication in stage 2.
Supplementary Table 2 Association of 32 replicated SNPs with other anthropometric traits.
Supplementary Table 3 Association between 32 replicated SNPs with risk of extreme obesity in children and adults, and with BMI in population-based childhood studies.
Supplementary Table 4 Association of the 32 confirmed BMI SNPs with metabolic traits.
Supplementary Table 5 Gene set enrichment analysis (MAGENTA) of biological pathways with one or more genes from the 32 confirmed BMI loci, using the BMI meta-analysis.
Supplementary Table 6 Non-synonymous or splice-site variants in linkage disequilibrium (r2 > 0.75) with lead SNPs.
Supplementary Table 7 Significant associations between BMI SNPs and cis gene expression (_cis_-eQTLs) in lymphocyte, blood, adipose and brain tissues.
Supplementary Table 8 Estimated number of BMI loci for each of the effect sizes observed in Stage 2 for the SNPs that reached a genome-wide significance of 5×10−8 in the joint analysis of stage 1 and stage 2, given the power to detect the association in the joint analysis of stage 1 and stage 2.
Acknowledgments
A full list of acknowledgments appears in the Supplementary Note.
Academy of Finland (10404, 77299, 104781, 114382, 117797, 120315, 121584, 124243, 126775, 126925, 127437, 129255, 129269, 129306, 129494, 129680, 130326, 209072, 210595, 213225, 213506, 216374); ADA Mentor-Based Postdoctoral Fellowship; Amgen; Agency for Science, Technology and Research of Singapore (A*STAR); ALF/LUA research grant in Gothenburg; Althingi (the Icelandic Parliament); AstraZeneca; Augustinus Foundation; Australian National Health and Medical Research Council (241944, 389875, 389891, 389892, 389938, 442915, 442981, 496739, 496688, 552485 and 613672); Australian Research Council (ARC grant DP0770096); Becket Foundation; Biocenter (Finland); Biomedicum Helsinki Foundation, Boston Obesity Nutrition Research Center; British Diabetes Association (1192); British Heart Foundation (97020; PG/02/128); Busselton Population Medical Research Foundation; Cambridge Institute for Medical Research; Cambridge NIHR Comprehensive Biomedical Research Centre; CamStrad (UK); Cancer Research UK; Centre for Medical Systems Biology (The Netherlands); Centre for Neurogenomics and Cognitive Research (The Netherlands); Chief Scientist Office of the Scottish Government; Contrat Plan Etat Région (France); Danish Centre for Health Technology Assessment; Danish Diabetes Association; Danish Heart Foundation; Danish Pharmaceutical Association; Danish Research Council; Deutsche Forschungsgemeinschaft (DFG; HE 1446/4-1); Department of Health (UK); Diabetes UK; Diabetes & Inflammation Laboratory; Donald W. Reynolds Foundation; Dresden University of Technology Funding Grant; Emil and Vera Cornell Foundation; Erasmus Medical Center (Rotterdam); Erasmus University (Rotterdam); European Commission (DG XII; QLG1-CT-2000-01643, QLG2-CT-2002-01254, LSHC-CT-2005, LSHG-CT-2006-018947, LSHG-CT-2004-518153, LSH-2006-037593, LSHM-CT-2007-037273, HEALTH-F2-2008-ENGAGE, HEALTH-F4-2007-201413, HEALTH-F4-2007-201550, FP7/2007-2013, 205419, 212111, 245536, SOC 95201408 05F02, WLRT-2001-01254); Federal Ministry of Education and Research (Germany) (01AK803, 01EA9401, 01GI0823, 01GI0826, 01GP0209, 01GP0259, 01GS0820, 01GS0823, 01GS0824, 01GS0825, 01GS0830, 01GS0831, 01IG07015, 01KU0903, 01ZZ9603, 01ZZ0103, 01ZZ0403, 03ZIK012); Federal State of Mecklenburg-West Pomerania; European Social Fund; Eve Appeal; Finnish Diabetes Research Foundation; Finnish Foundation for Cardiovascular Research; Finnish Foundation for Pediatric Research, Finnish Medical Society; Finska Läkaresällskapet, Päivikki and Sakari Sohlberg Foundation, Folkhalsan Research Foundation; Fond Européen pour le Développement Régional (France); Fondation LeDucq (Paris, France); Foundation for Life and Health in Finland; Foundation for Strategic Research (Sweden); Genetic Association Information Network; German Research Council (KFO-152) German National Genome Research Net ‘NGFNplus’ (FKZ 01GS0823); German Research Center for Environmental Health; Giorgi-Cavaglieri Foundation; GlaxoSmithKline; Göteborg Medical Society; Great Wine Estates Auctions; Gyllenberg Foundation; Health Care Centers in Vasa, Närpes and Korsholm; Healthway, Western Australia; Helmholtz Center Munich; Helsinki University Central Hospital, Hjartavernd (the Icelandic Heart Association); INSERM (France); Ib Henriksen Foundation; IZKF (B27); Jalmari and Rauha Ahokas Foundation; Juho Vainio Foundation; Juvenile Diabetes Research Foundation International (JDRF); Karolinska Institute; Knut and Alice Wallenberg Foundation; Leenaards Foundation; Lundbeck Foundation Centre of Applied Medical Genomics for Personalized Disease Prediction, Prevention and Care (LUCAMP); Lundberg Foundation; Marie Curie Intra-European Fellowship; Medical Research Council (UK) (G0000649, G0000934, G9521010D, G0500539, G0600331, G0601261, PrevMetSyn); Ministry of Cultural Affairs and Social Ministry of the Federal State of Mecklenburg-West Pomerania; Ministry for Health, Welfare and Sports (Netherlands); Ministry of Education (Finland); Ministry of Education, Culture and Science (Netherlands); Ministry of Internal Affairs and Health (Denmark); Ministry of Science, Education and Sport of the Republic of Croatia (216-1080315-0302); Ministry of Science, Research and the Arts Baden-Württemberg; Montreal Heart Institute Foundation; Municipal Health Care Center and Hospital in Jakobstad; Municipality of Rotterdam; Närpes Health Care Foundation; National Cancer Institute; National Health and Medical Research Council of Australia; National Institute for Health Research Cambridge Biomedical Research Centre; National Institute for Health Research Oxford Biomedical Research Centre; National Institute for Health Research comprehensive Biomedical Research Centre; National Institutes of Health (263-MA-410953, AA07535, AA10248, AA014041, AA13320, AA13321, AA13326, CA047988, CA65725, CA87969, CA49449, CA67262, CA50385, DA12854, DK58845, DK46200, DK062370, DK063491, DK072193, HG002651, HL084729, HHSN268200625226C, HL71981, K23-DK080145, K99-HL094535, M01-RR00425, MH084698, N01-AG12100, NO1-AG12109, N01-HC15103, N01-HC25195, N01-HC35129, N01-HC45133, N01-HC55015, N01-HC55016, N01-HC55018, N01-HC55019, N01-HC55020, N01-N01HC-55021, N01-HC55022, N01-HC55222, N01-HC75150, N01-HC85079, N01-HC85080, N01-HG-65403, N01-HC85081, N01-HC85082; N01-HC85083; N01-HC85084; N01-HC85085; N01-HC85086, N02-HL64278, P30-DK072488, R01-AG031890, R01-DK073490, R01-DK075787, R01DK068336, R01DK075681, R01-HL59367, R01-HL086694, R01-HL087641, R01-HL087647, R01-HL087652, R01-HL087676, R01-HL087679, R01-HL087700, R01-HL088119, R01-MH59160, R01-MH59565, R01-MH59566, R01-MH59571, R01-MH59586, R01-MH59587, R01-MH59588, R01-MH60870, R01-MH60879, R01-MH61675, R01-MH63706, R01-MH67257, R01-MH79469, R01-MH79470, R01-MH81800, RL1-MH083268, UO1-CA098233, U01-DK062418, U01-GM074518, U01-HG004402, U01-HG004399, U01-HL72515, U01-HL080295, U01-HL084756, U54-RR020278, T32-HG00040, UL1-RR025005, Z01-HG000024); National Alliance for Research on Schizophrenia and Depression (NARSAD); Netherlands Genomics Initiative/Netherlands Consortium for Healthy Aging (050-060-810); Netherlands Organisation for Scientific Research (NWO) (904-61-090, 904-61-193, 480-04-004, 400-05-717, SPI 56-464-1419, 175.010.2005.011, 911-03-012); Nord-Trøndelag County Council; Nordic Center of Excellence in Disease Genetics; Novo Nordisk Foundation; Norwegian Institute of Public Health; Ollqvist Foundation; Oxford NIHR Biomedical Research Centre; Organization for the Health Research and Development (10-000-1002); Paavo Nurmi Foundation; Paul Michael Donovan Charitable Foundation; Perklén Foundation; Petrus and Augusta Hedlunds Foundation; Pew Scholar for the Biomedical Sciences; Public Health and Risk Assessment, Health & Consumer Protection (2004310); Research Foundation of Copenhagen County; Research Institute for Diseases in the Elderly (014-93-015; RIDE2); Robert Dawson Evans Endowment; Royal Society (UK); Royal Swedish Academy of Science; Sahlgrenska Center for Cardiovascular and Metabolic Research (CMR, no. A305: 188); Siemens Healthcare, Erlangen, Germany; Sigrid Juselius Foundation; Signe and Ane Gyllenberg Foundation; Science Funding programme (UK); Social Insurance Institution of Finland; Söderberg’s Foundation; South Tyrol Ministry of Health; South Tyrolean Sparkasse Foundation; State of Bavaria; Stockholm County Council (560183); Susan G. Komen Breast Cancer Foundation; Swedish Cancer Society; Swedish Cultural Foundation in Finland; Swedish Foundation for Strategic Research; Swedish Heart-Lung Foundation; Swedish Medical Research Council (8691, K2007-66X-20270-01-3, K2010-54X-09894-19-3, K2010-54X-09894-19-3, 2006-3832); Swedish Research Council; Swedish Society of Medicine; Swiss National Science Foundation (33CSCO-122661, 310000-112552, 3100AO-116323/1); Torsten and Ragnar Söderberg’s Foundation; Université Henri Poincaré-Nancy 1, Région Lorraine, Communauté Urbaine du Grand Nancy; University Hospital Medical funds to Tampere; University Hospital Oulu; University of Oulu, Finland (75617); Västra Götaland Foundation; Walter E. Nichols, M.D., and Eleanor Nichols endowments; Wellcome Trust (068545, 072960, 075491, 076113, 077016, 079557, 079895, 081682, 083270, 085301, 086596); Western Australian DNA Bank; Western Australian Genetic Epidemiology Resource; Yrjö Jahnsson Foundation.
Footnotes
Author contributions
A full list of author contributions appears in the Supplementary Note.
Competing interests statement
The authors declare competing financial interests. A full list of competing interests appears in the Supplementary Note.
References
- 1.Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults--The Evidence Report. National Institutes of Health. Obes Res. 1998;6 (Suppl 2):51S–209S. [PubMed] [Google Scholar]
- 2.Lewis CE, et al. Mortality, health outcomes, and body mass index in the overweight range: a science advisory from the American Heart Association. Circulation. 2009;119:3263–71. doi: 10.1161/CIRCULATIONAHA.109.192574. [DOI] [PubMed] [Google Scholar]
- 3.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. Jama. 1986;256:51–4. [PubMed] [Google Scholar]
- 4.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–51. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 5.Taylor AE, et al. Comparison of the associations of body mass index and measures of central adiposity and fat mass with coronary heart disease, diabetes, and all-cause mortality: a study using data from 4 UK cohorts. Am J Clin Nutr. 2010;91:547–56. doi: 10.3945/ajcn.2009.28757. [DOI] [PubMed] [Google Scholar]
- 6.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–94. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 11.Ren D, et al. Neuronal SH2B1 is essential for controlling energy and glucose homeostasis. J Clin Invest. 2007;117:397–406. doi: 10.1172/JCI29417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huszar D, et al. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell. 1997;88:131–41. doi: 10.1016/s0092-8674(00)81865-6. [DOI] [PubMed] [Google Scholar]
- 13.O’Rahilly S, Farooqi IS. Human obesity as a heritable disorder of the central control of energy balance. Int J Obes (Lond) 2008;32 (Suppl 7):S55–61. doi: 10.1038/ijo.2008.239. [DOI] [PubMed] [Google Scholar]
- 14.Lindgren CM, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heard-Costa NL, et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41:157–9. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 17.Scherag A, et al. Two new Loci for body-weight regulation identified in a joint analysis of genome-wide association studies for early-onset extreme obesity in French and german study groups. PLoS Genet. 2010;6:e1000916. doi: 10.1371/journal.pgen.1000916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–45. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–8. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIntosh CH, Widenmaier S, Kim SJ. Glucose-dependent insulinotropic polypeptide (Gastric Inhibitory Polypeptide; GIP) Vitam Horm. 2009;80:409–71. doi: 10.1016/S0083-6729(08)00615-8. [DOI] [PubMed] [Google Scholar]
- 23.Farooqi IS, et al. Heterozygosity for a POMC-null mutation and increased obesity risk in humans. Diabetes. 2006;55:2549–53. doi: 10.2337/db06-0214. [DOI] [PubMed] [Google Scholar]
- 24.Farooqi IS, et al. Clinical spectrum of obesity and mutations in the melanocortin 4 receptor gene. N Engl J Med. 2003;348:1085–95. doi: 10.1056/NEJMoa022050. [DOI] [PubMed] [Google Scholar]
- 25.Marsh DJ, et al. Response of melanocortin-4 receptor-deficient mice to anorectic and orexigenic peptides. Nat Genet. 1999;21:119–22. doi: 10.1038/5070. [DOI] [PubMed] [Google Scholar]
- 26.Unger TJ, Calderon GA, Bradley LC, Sena-Esteves M, Rios M. Selective deletion of Bdnf in the ventromedial and dorsomedial hypothalamus of adult mice results in hyperphagic behavior and obesity. J Neurosci. 2007;27:14265–74. doi: 10.1523/JNEUROSCI.3308-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Z, Zhou Y, Carter-Su C, Myers MG, Jr, Rui L. SH2B1 enhances leptin signaling by both Janus kinase 2 Tyr813 phosphorylation-dependent and -independent mechanisms. Mol Endocrinol. 2007;21:2270–81. doi: 10.1210/me.2007-0111. [DOI] [PubMed] [Google Scholar]
- 28.Gray J, et al. Hyperphagia, severe obesity, impaired cognitive function, and hyperactivity associated with functional loss of one copy of the brain-derived neurotrophic factor (BDNF) gene. Diabetes. 2006;55:3366–71. doi: 10.2337/db06-0550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochukova EG, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–70. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coll AP, Loraine Tung YC. Pro-opiomelanocortin (POMC)-derived peptides and the regulation of energy homeostasis. Mol Cell Endocrinol. 2009;300:147–51. doi: 10.1016/j.mce.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Krude H, et al. Obesity due to proopiomelanocortin deficiency: three new cases and treatment trials with thyroid hormone and ACTH4–10. J Clin Endocrinol Metab. 2003;88:4633–40. doi: 10.1210/jc.2003-030502. [DOI] [PubMed] [Google Scholar]
- 32.Miyawaki K, et al. Inhibition of gastric inhibitory polypeptide signaling prevents obesity. Nat Med. 2002;8:738–42. doi: 10.1038/nm727. [DOI] [PubMed] [Google Scholar]
- 33.Segrè AV, et al. Common Inherited Variation in Mitochondrial Genes is not Enriched for Associations with Type 2 Diabetes or Related Glycemic Traits. PLoS Genetics. 2010 doi: 10.1371/journal.pgen.1001058. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 35.Myers AJ, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–9. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 36.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 37.Purcell SM, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–52. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J-H, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nat Genet. 2010 doi: 10.1038/ng.610. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young EH, et al. The V103I polymorphism of the MC4R gene and obesity: population based studies and meta-analysis of 29 563 individuals. Int J Obes (Lond) 2007;31:1437–41. doi: 10.1038/sj.ijo.0803609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stutzmann F, et al. Non-synonymous polymorphisms in melanocortin-4 receptor protect against obesity: the two facets of a Janus obesity gene. Hum Mol Genet. 2007;16:1837–44. doi: 10.1093/hmg/ddm132. [DOI] [PubMed] [Google Scholar]
- 41.Yeo GS, et al. Mutations in the human melanocortin-4 receptor gene associated with severe familial obesity disrupts receptor function through multiple molecular mechanisms. Hum Mol Genet. 2003;12:561–74. doi: 10.1093/hmg/ddg057. [DOI] [PubMed] [Google Scholar]
- 42.Hirschhorn JN. Genomewide association studies--illuminating biologic pathways. N Engl J Med. 2009;360:1699–701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 43.Lemmens VE, Oenema A, Klepp KI, Henriksen HB, Brug J. A systematic review of the evidence regarding efficacy of obesity prevention interventions among adults. Obes Rev. 2008;9:446–55. doi: 10.1111/j.1467-789X.2008.00468.x. [DOI] [PubMed] [Google Scholar]
- 44.Anderson JW, Konz EC, Frederich RC, Wood CL. Long-term weight-loss maintenance: a meta-analysis of US studies. Am J Clin Nutr. 2001;74:579–84. doi: 10.1093/ajcn/74.5.579. [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 47.Guan Y, Stephens M. Practical issues in imputation-based association mapping. PLoS Genet. 2008;4:e1000279. doi: 10.1371/journal.pgen.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abecasis GR, Wigginton JE. Handling marker-marker linkage disequilibrium: pedigree analysis with clustered markers. Am J Hum Genet. 2005;77:754–67. doi: 10.1086/497345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 51.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet. 2010;86:581–91. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
Supplementary Figure 1 Study design. Stage 1 - Meta analysis of genome-wide association data was performed in stage 1 across 46 studies of white European Ancestry. A total of 42 SNPs representing the best associating (P < 10−6) loci (shown) were taken forward for replication. Nineteen of these SNPs (loci in bold) reached already genome-wide significance at stage 1. Stage 2 – The 42 SNPs were genotyped in 16 de novo replication studies and extracted from 18 in silico replication studies, all adults of European ancestry and were tested for association with BMI. In a joint analyses of stage 1 and stage 2 data, 32 SNPs (loci in bold) reached genome-wide significance (P < 5×10−8). Follow-up analyses – The 32 confirmed loci were taken forward for additional analyses.
Supplementary Figure 2 Regional plots of the 32 confirmed BMI loci with missense and CNV variants. SNPs are plotted by position on chromosome against association with BMI (−log10 _P_-value). The SNP name shown on the plot was the most significant SNP after stage 1 meta-analysis. Estimated recombination rates (from HapMap) are plotted in cyan to reflect the local LD structure. The SNPs surrounding the most significant SNP are color-coded to reflect their LD with this SNP (taken from pairwise r2 values from the HapMap CEU database, www.hapmap.org). Genes, position of exons, and direction of transcription from UCSC genome browser (genome.ucsc.edu) are noted. Hash-marks represent SNP positions available in the meta-analysis. Plots were generated using LocusZoom (http://csg.sph.umich.edu/locuszoom).
Supplementary Figure 3 Quantile-quantile plot of SNPs at stage 1 GIANT meta-analysis (black) and after removing any SNPs within 1 Mb of the 10 previously reported genome-wide significant hits for BMI (blue), after additionally excluding the four loci for waist/weight (green) and after excluding all 32 confirmed loci (red).
Supplementary Figure 4 Relationship between the predicted BMI, based on the 32 confirmed BMI loci combined, and the actual BMI in the ARIC Study (N=8,120). Panel (a) shows the comparison of the predicted BMI (grey), with the actual BMI (green) and quartile ranges (orange) in sets of 500 individuals, suggesting good average predictions. Panel (b) shows the comparison of the individual predicted and observed BMI values, suggesting poor individual predictions.
Supplementary Table 1 The 42 SNPs, associated with BMI at P < 5.10−6 at stage 1, that were taken forward for replication in stage 2.
Supplementary Table 2 Association of 32 replicated SNPs with other anthropometric traits.
Supplementary Table 3 Association between 32 replicated SNPs with risk of extreme obesity in children and adults, and with BMI in population-based childhood studies.
Supplementary Table 4 Association of the 32 confirmed BMI SNPs with metabolic traits.
Supplementary Table 5 Gene set enrichment analysis (MAGENTA) of biological pathways with one or more genes from the 32 confirmed BMI loci, using the BMI meta-analysis.
Supplementary Table 6 Non-synonymous or splice-site variants in linkage disequilibrium (r2 > 0.75) with lead SNPs.
Supplementary Table 7 Significant associations between BMI SNPs and cis gene expression (_cis_-eQTLs) in lymphocyte, blood, adipose and brain tissues.
Supplementary Table 8 Estimated number of BMI loci for each of the effect sizes observed in Stage 2 for the SNPs that reached a genome-wide significance of 5×10−8 in the joint analysis of stage 1 and stage 2, given the power to detect the association in the joint analysis of stage 1 and stage 2.