Making sense of ubiquitin ligases that regulate p53 (original) (raw)
Abstract
The functions of p53 most highly associated with the well-studied tumor suppressor are its abilities to induce cell cycle arrest and apoptosis in response to cellular stresses. Recent progress underscores that p53 is a multi-functional protein with activities that range beyond tumor suppression to normal homeostasis, metabolism, fertility and differentiation. A unifying theme of these studies is that p53 is first and foremost a transcription factor; and control of p53 protein stability determines its ability to carry out this task. There are an expanding number of E3-ubiquitin ligase proteins that target p53 for ubiquitin tagging and protein degradation. This review discusses these many effectors of p53 protein degradation, and our task is to provide some level of understanding as to their differences and their similarities. Further, we propose how some degree of specialization may be assigned to the E3-ligases, in their navigation toward a common goal of regulating p53 protein levels, and emphasize that better understanding of the mechanisms involved in E3-ligase functions is needed to further their potential as therapeutic targets.
Key words: tumor suppressor, proteasome, protein degradation, tissue-specificity, developmental expression, cancer, E3-ligase
The tumor suppressor p53 receives considerable attention within and without the cell, as a key player in tumor prevention and the most frequently mutated gene in human cancers.1,2 A majority of human tumors harbor mutations in the TP53 gene and, in other cases, p53 is inactivated by other mechanisms.1,2 The functions of p53 in preventing propagation of DNA damage and genome instability are of major import and multiple signaling pathways converge on p53 to control its protein stability and activities. On and off signals in the form of post-translational modifications (PTMs), when enzymes covalently link chemical groups to the amino acid structure of p53, are read into actions that dictate stability, protein partner and chromatin interactions, as well as subcellular localization, of p53. Recent studies offer an expanding list of p53 regulatory functions that additionally impact development, differentiation and metabolism.3 The power of p53, to shut down the cell cycle or induce cellular death, is held at bay during normal cellular homeostasis, but is activated when trouble threatens. Single-cell analyses reveal that p53 is post-translationally regulated not only in response to stress that threatens genomic stability, but also during normal DNA replication.4 Although the majority of control is exercised at a post-translational level, p53 mRNA levels are also controlled at post-transcriptional levels by micro-RNAs (miRNAs), e.g., miRNA-504, which targets p53 message for degradation.5
The major outcome of p53 regulation is alteration of its nuclear concentration and, thus, its ability to interact with chromatin and function as a transcription factor (Fig. 1).6–9 This is primarily achieved by a highly regulated balancing act between p53 protein degradation and protein synthesis. At the fulcrum of this balance is a collection of proteins that ligate ubiquitin to lysine (K) residues, as a PTM of p53 and regulate protein stability. Ubiquitin is added at a specific amino acid residue as a growing chain or monomeric unit, which flags p53 for elimination by the proteasome complex or modifies activity of p53, respectively.10,11 The E3-ubiquitin ligase determines target protein selection and interaction, bringing a complex of E1/E2/ubiquitin together in association with the target protein to complete transfer and linkage of a ubiquitin moiety.12 Ubiquitin polymerization versus monomer addition is determined by the lysine amino acid (K48 versus K63) of ubiquitin that is covalently linked to p53.13,14 Conjugation of a single ubiquitin molecule to one or more lysines of a target protein is involved in a variety of cellular processes, including protein trafficking, DNA repair and transcriptional regulation.15 Recent, comprehensive reviews that discuss the collection of proteins, which regulate p53 protein levels by ubiquitination and are themselves regulated by p53, are available.11,16 Here, we update this growing number of E3-ligases and further discuss a potential biological rationale for the large group of proteins intent on negative regulation of p53 via ubiquitin addition.
Figure 1.
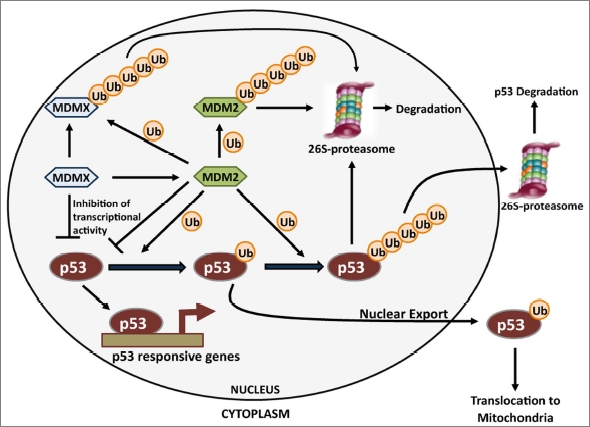
Complex control of p53 protein levels. Mdm2 as an example: An intricate regulatory mechanism controls ubiquitination of p53 and dictates its activity. MDM2 in partnership with MDMX poly-ubiquitinates p53, which is targeted for proteasomal degradation. MDM2 also directs self-destruction and MDMX degradation, mediated by its RING-domain. Mono-ubiquitination of p53 by MDM2 leads to its nuclear export. Direct binding with MDMX may also render p53 transcriptionally inactive. p53 upregulates transcriptional activation of MDM2 to complete a negative feedback loop.
The Principle Player: Mdm2
The paradigm of p53 modification by the addition of ubiquitin and subsequent regulation of protein stability seemed straight-forward with the discovery of Mdm2 and its RING-domain-dependent ubiquitination of p53.14,17–19 In fact, deletion of Mdm2 from the mouse genome illustrates the consequences of unleashing p53 from regulatory control by Mdm2. Knockout of Mdm2 is early embryonic lethal, a phenotype that is rescued by loss of Trp53 expression.20,21 Mdm2 has its homologue in humans, where the protein is referred to as HDM2 while the gene remains MDM2, but no Mdm2 homologue exists in invertebrates even where p53 is conserved.22 Efforts of a number of labs define an autoregulatory circuit between the E3-ligase Mdm2 and p53; where p53 activates expression of MDM2, Mdm2/HDM2 in turn associates with p53, p53 is ubiquitinated and degraded by the proteasome complex and MDM2 expression declines; the loop continues to maintain both Mdm2/HDM2 and p53 at low levels in normal cells (Fig. 1).23 Further studies revealed how Mdm2-directed stability of p53 is regulated in response to stress signaling, which temporarily breaks the Mdm2-p53 circuitry and allows p53 protein levels to increase.24 Mdm2 levels are themselves highly regulated and finely tuned;24 aberrant Mdm2 may either contribute to oncogenesis or tumor suppression, as recently reviewed in ref. 25.
Repression of p53 activity by Mdm2 may be achieved by two major mechanisms: (1) by promoting p53 degradation and (2) by disrupting the ability of p53 to function as a transcription factor.17,19,26 Mdm2 binds p53 at its amino (N)-terminus to block the transactivation domain of p53;27 additionally a second binding site for Mdm2 within the DNA-binding domain (DBD) of p53 is known.28,29 Interestingly, some mutant forms of p53, which contain single point mutations within the DBD site of Mdm2 interaction, have increased levels of Mdm2-dependent ubiquitination but are not degraded.29 Mdm2 has a highly similar partner called MdmX or Mdm4, which is a RING-domain protein defective in ubiquitin ligase activity. MdmX has considerable influence on Mdm2 and p53:30 MdmX inhibits p53 transcriptional functions by direct binding and interacts with Mdm2 via their respective RING domains (Fig. 1).31,32 Multiple upstream modifying enzymes regulate both p53-Mdm2 interactions and protein levels during the activation of p53.33 During this process, Mdm2 may ubiquitinate itself and its partner, MdmX.24,34
Further investigations show that Mdm2-mediated regulation of p53 is more complex than initially thought, as Mdm2 also influences subcellular localization and transcriptional potential of p53 (Fig. 1). By conjugating a single ubiquitin onto one or more lysines within the carboyx (C)-terminus and DBD of p53, Mdm2 enhances the nuclear export of p53.35,36 How Mdm2 activity is regulated to effect mono- versus poly-ubiquitination of p53 is not well understood. Several reports suggest that other proteins, including p300,37 YY1,38 Gankyrin,39 KAP140 and Siva1,41 may modulate Mdm2-mediated poly-ubiquitination of p53, while others support a model where the levels of Mdm2 determine the type of ubiquitin conjugated to p53.35 Mdm2 also inhibits p53 by promoting its conjugation to the ubiquitin-like molecule NEDD8.42 Modifications of C-terminal lysines of p53 by ubiquitination, neddylation, sumoylation or acetylation affect interactions between p53 and Mdm2 to compete for Mdm2-mediated ubiquitination.43,44
Nothing is Simple about Regulation of p53
With further studies of p53 regulation, the model of Mdm2 as sole arbiter of p53 protein levels began to erode. Genetic knock-in methodology was used to create mice that express E3-ligase deficient Mdm2 (a C462A mutant), which retains the ability to bind to p53 and theoretically inhibit p53 transcriptional activity without ubiquitination.45 Surprisingly, these mice exhibit p53-dependent embryonic lethality, similar to Mdm2-null mice, and also retain ubiquitination and degradation of Mdm2. These results suggest that protein-protein interactions between p53 and Mdm2 are insufficient to block p53-mediated apoptosis during embryonic development and, more importantly, Mdm2-independent pathways must exist that regulate protein stability of both p53 and Mdm2 by ubiquitin modification. Using a switchable, endogenous p53 mouse model that allows rapid and reversible toggling of endogenous p53 between nucleus and cytoplasm, Evan and colleagues showed that, although p53 is simultaneously active in all tested tissue of Mdm2-deficient mice, its levels fall rapidly after functional restoration post DNA-damage,46 suggesting that a negative feedback loop remains operable even in absence of Mdm2. Other reports of in vivo, mouse knock-in models that express mutant forms of p53 (p53-6KR or p53-7KR), lacking the majority of ubiquitination sites for Mdm2, show that mutant p53 has a normal half-life and is stabilized and activated by stress.47,48 Thus, alternative pathways for p53-degradation likely exist independent of Mdm2.
Benchimol and colleagues were the first to show that Mdm2 is not alone as an E3-ligase capable of controlling p53 protein levels by ubitiquin-tagging p53 for degradation.49 The identification of Pirh2, another RING-domain E3-ligase that mediates degradation of p53 by direct interaction,49 was soon followed by discoveries of several other proteins that regulate p53 at the level of protein stability. Table 1 presents a summary of numerous E3-ligases that target p53 and underscores the major premise of this review: Why are there so many E3-ligases that negatively regulate p53? We will address this question in a teleological manner by assuming there is specificity among these negative regulators of p53. We offer the multi-component What, When and Where Hypothesis of specificity: (1) E3-ligase proteins are expressed in distinct tissues or cell types at specific times of development. A corollary to this is that tissue- or stage-specific expression of the required E2 or co-regulators dictates function of an E3-ligase. (2) Interactions between p53 and an E3-ligase are determined by readout of a p53 “PTM code”, e.g., situational signaling to p53 leads to modification of specific residues, which likely determine protein-protein interactions, including those with E3-ligases. (3) An E3-ligase mediates ubiquitination of a defined residue of p53 that determines a specific outcome or timing of delayed versus immediate degradation. (4) An E3-ligase itself acquires specific PTMs under a specific condition that determines its interaction with p53 and ultimately the status of p53 activity. (5) Targeting p53 for ubiquitination by an E3-ligase is specific to the isoform of p53 under different situations.
Table 1.
Ubiquitin e3-ligases that regulate p53 activity
| Ligase | Type | p53-responsive | E2 | Ub | p53-Lysine residue | p53 status | Mode |
|---|---|---|---|---|---|---|---|
| Mdm2 | RING | Yes | UbcH5b | Mono/Poly | K370, K372, K373, K381, K382, K386 | Degradation | Direct |
| Pirh2 | RING | Yes | UbcH5b | Poly | - | Degradation | Direct |
| Cop1 | RING | Yes | UbcH5b | Poly | - | Degradation | Direct |
| TRIM24 | RING | - | UbcH8 | Poly | - | Degradation | Direct |
| ARF-BP1 | HECT | No | UbcH5c | Poly | - | Degradation | Direct |
| CARP1/2 | RING | - | - | Poly | - | Degradation | Direct |
| TOPORS | RING | - | UbcH5a/c, UbcH6 | Poly | - | Degradation | Direct |
| Synoviolin | RING | - | UbcH5c | Poly | - | Degradation | Direct |
| CHIP | - | - | UbcH5b | Poly | - | Degradation | Direct |
| JFK | RING | - | - | Poly | - | Degradation | Skp1-Cul1-Rbx1-F-box |
| MKRN1 | RING | No | - | - | K291, K292 | Degradation | Direct |
| E4orf6 and E1B55K | RING | - | UbcH5a | Poly | - | Degradation | ElonginB-Cul5-Rbx1 |
| ICP0 | RING | - | UbcH5a, UbcH6 | Poly | - | Translocation to Nuclear Foci | Direct |
| MSL2 | RING | - | - | Poly | K351, K357 | Nuclear export | Direct |
| CUL7 | - | - | - | Mono/Di | - | Transcriptional inactivation | Direct |
| WWP1 | HECT | Yes | UbcH5c | Mono/Poly | - | Nuclear Export | Direct |
| Ubc13 | - | Repressed | Ubc13 | Poly | - | Prevent Tetramerization | Direct |
| E4F1 | - | - | - | Poly | K320 | Translocation to Chromatin | Direct |
| BZLF1 | - | - | UbcH5a/c | Poly | - | Degradation | ElonginB/c-Cul2/5-SOCS |
Embryonic Expression of E3-Ligases that Target p53
None of the regulators, listed in Table 1, has been scrutinized at the level of Mdm2, especially in development of mouse models of function. Lack of Mdm2 causes embryonic lethality and, in the category of embryonic regulation of p53, Mdm2 is the principle player.20,21 Its functional partner, MdmX (Mdm4), which lacks the enzymatic capacity to ubiquitinate p53, also induces embryonic lethality when deleted but at a later stage of development.50 In consideration of the other E3-ligases of p53, important questions are whether these other E3-ligases of p53 are expressed during mouse embryonic development, as well as when and where expression occurs. To address these questions, databases such as EMAGE (www.emouseatlas.org/emage/home.php) offer collected information regarding expression of specific genes during mouse embryonic development. Comparison of whole-mount in situ expression analysis allows some assessment of specific E3-ligases that target p53. Here, we present interpretation of more robust, available data and not all members listed in Table 1 are analyzed.
The primary literature reveals that Mdm2 expression during mouse embryogenesis is detectable, by in situ analysis, in neural folds and in migratory neural crest cells, 7.5–9 dpc. After these stages of critical neural crest development, Mdm2 is expressed more ubiquitously in the embryo.51 Interestingly, embryonic lethality in Mdm2-null mouse models occurs during peri-implantation; thus, curbing p53 from massive cell death occurs much earlier than stages of detectable Mdm2 expression.52 This underscores the possibility that E3-ligases of p53 may acquire specific functions at different stages of development and/or in specific cells.
In contrast to detectable Mdm2 expression, Trim24 at 10.5 dpc is visibly expressed at the tip of the hindlimb bud and in the facial primordia, primarily the nasal eminence, with more mesenchymal appearance. Cop1, at the same stage of development, is also expressed in the facial primordia but with additional expression in the forelimb bud and ear. CARP1 at 10.5 dpc is expressed in the outflow tract of the developing heart, the distal forelimb bud, as well as the facial primordia. Makorin Ring Finger Protein-1 (MKRN1) displays expression in developing neural tissues, including the forebrain and the ear at 10.5 dpc. ARF-BP1 appears to be ubiquitously expressed but available data are compiled at a later stage of 14.5 dpc. At this same stage, Synoviolin1 is primarily expressed in selected bone and cartilaginous sites of the embryo. These examples of embryonic expression support the hypothesis that the many E3-ligases of p53 display distinct expression patterns during embryonic development.
Further analyses are needed to encompass adult tissues and expression levels, in order to address a major question of the roles that E3-ligases may play in specific tumor development and potential treatments based on these functions. An intriguing aspect of p53 regulation, which is less studied than most, is that the p53 gene encodes other expressed p53 protein isoforms, which have been associated with specific cancers. These isoforms are expressed as a result of secondary promoter use and alternative splicing, which create a form of p53 truncated at the N-terminus.53 As the N-terminus is the site of interaction with Mdm2, additional E3-ligases might provide a “failsafe” mechanism to keep these truncated p53 proteins in check and regulate their protein stability. Additionally, work with mouse models suggests that specificity in regulation of full-length p53 occurs at a tissue-specific level. When normal mice are exposed to damage by 5 Gy of IR, p53 accumulates in the nuclei of cells and induces apoptosis in spleen, thymus, bone marrow, intestine and ependyma. p53 levels also increase in kidney, osteocytes, myocardium and salivary glands but no cell death occurs.54 In liver, skeletal muscle and brain tissue, there is no response to IR at the level of p53 stability or apoptosis.54 In an intriguing study, Mdm2 was shown to regulate mutant p53 protein stability as well as normal p53 protein levels in a mouse model, but this regulatory control was lacking in liver tissue.55
Partners of E3-Ligase Function
Tissue-specificity may lie at the level of E3-ligase expression or whether the specific E2 needed for function is expressed (see Table 1). Although UbcH5 is a common E2-partner of many E3-ligases, there are a notable few that are more active or exclusively active with other E2 proteins. Even UbcH5 has its a, b and c subtypes and their expression and interactions with E3-ligases offer another level of regulation for specificity. Additionally one E2 in particular, Ubc13, can directly ubiquitinate p53 without intervention of a partner E3.56
E3-ligase functions are influenced by proteins that interact with or regulate the E3-ligase, in addition to an E2 protein. MdmX is a well-known example of a regulatory partner that alters Mdm2 activity and even its stability.30,31 Whether MdmX activity is more or less influential in specific cell types or tissues is unknown and other previously noted modifiers of Mdm2 functions, p300,37 YY1,38 Gankyrin,39 KAP140 and Siva1,41 offer other regulatory nodes. As an example of potential tissue-specificity and its effects, deletion of Trim24 in mice led to tumor development in the liver.57 These tumors are responsive to retinoic acid treatment, suggesting that Trim24 primarily functions as a co-regulator of the retinoic acid receptor rather than an E3-ligase of p53 in the liver. Whether E3-ligases interact with p53 in a direct or indirect manner (Table 1), protein partners and upstream regulation likely influence their activities, stability or expression.
Evolution of E3-Ligases that Target p53
Evolutionary conservation may offer some sense of the most basic of E3-ligase functions and whether a single ancestral E3 was duplicated or multiple E3-ligases arose separately over time. A review by Abrams and colleagues suggests that control of p53 protein stability may be restricted to vertebrate development although orthologs and structural conservation of p53 exist across invertebrate phyla.22 Mdm2 and MdmX are not conserved in flies and worms, but recent work from our laboratory suggest that other regulators of p53, which serve as E3-ligases in mammalian cells, are active in invertebrates. We used mosaic deletion analysis in Drosophila to assess the outcome of bonus loss of function.58 Bonus is the single Drosophila representative of the TIF1 sub-family of TRIM (RING/B-box/Coiled-coil or Tripartite Motif) proteins and closest in homology to TRIM24 among the TRIM24/28/33 members. Loss of bonus induces apoptosis and cleavage of Caspase 3. Cell death is apparently due to unrestrained D-p53 activity, as RNAi-mediated depletion of D-p53 rescues the phenotype. Drosophila bonus may be an ancestral negative regulator of p53; molecular studies are needed to show that ubiquitination and E3-ligase activity are the basis of this regulation. Evolutionary comparisons between all E3-ligase regulators of p53 and conserved proteins are needed to reveal comparable pathways and conserved functions.
Upstream Regulation and Downstream Response
The most striking aspect of p53-signaling, in response to stress or inductive signaling, is the tightly regulated process of post-transcriptional modification of p53, which guide p53 activities primarily by dictating p53-interactions with proteins that control: (1) the levels of p53, (2) the ability of p53 to bind to DNA, (3) the subcellular localization of p53, (4) interaction with regulatory proteins, and thus determine the physiological outcome of p53 activation. A comparison can be made to chromatin, where PTMs of specific histone residues are interpreted by histone “reader” proteins via specific, specialized domains, while other specialized enzymes “write” the PTM code. A few readers and writers identified for histones are known to interact and/or modify p53 as well.59–62
Covalent modifications of p53 occur on more than 40 different amino acid residues and likely provide fine-tuning and specificity of p53 activation. These PTMs range from phosphorylation, acetylation, methylation, ubiquitination, sumoylation, neddylation, glycosylation, ribosylation and more recently _O_-GlcNAcylation.10,63–67 Depending on the type of stress, specific signaling events are activated that target precise residues on p53 protein resulting in a stress-to-enzyme-to-residue specific modification on p53 protein.68 Discussing all these modifications and the enzymes responsible (writers) in detail is beyond the scope of this review, however several comprehensive reviews cover this topic in some depth.7,10,11,14,63–66,68,69 Table 2 summarizes and updates some of the significant modifications of p53 and the enzymes that create or abolish them.68 Though these modifications may be crucial for p53 functions in cultured cells, in vivo models suggest that PTMs play subtle, modulatory roles in regulating p53 functions.66,70 Therefore, the key regulatory switch in turning on and off p53 response may be direct or indirect regulation of p53 stability through ubiquitin modification.
Table 2.
p53 post-translational modifications and modifying enzymes
| Residue of p53 | Modification | Modifiers | De-modifiers |
|---|---|---|---|
| S6 | P | JNK2, CKIδ/ε | - |
| S9 | P | CK1δ/ε | - |
| S15 | P | ATM, ATR, DNA-PK, AMPK, mTOR, RSK2, CDK5 | PP1, PPM1D |
| T18 | P | TTK, CHK2, VRK1 | - |
| S20 | P | CHK2, PIK3 | - |
| S33 | P | CDK9, CDK5, CAK/CDK7, GSK3β, P38K | - |
| S37 | P | ATR, PRAK | PP1, PP2A |
| S46 | P | CDK5, AMPKα, HIPK2, P38K, PKCδ, DYRK2 | PP2A |
| T55 | P | ERK2, TAF1 | PP2A, B56Y |
| T81 | P | JNK2 | - |
| K120 | Ac | TIP60 | - |
| T155 | P | CSN | - |
| K164 | Ac | P300 (KAT3B) | - |
| S215 | P | STKI5 | - |
| E255, E258, E259 | R | PARP-1 (poly ADP-ribosylase) | - |
| K305 | Ac | P300 (KAT3B) | - |
| S313, S314 | P | CHK1, CHK2 | - |
| S315 | P | STKI5, CDK9, CDK2 | CDC14A |
| L320 | Ac | PCAF (KAT2B) | HDAC1 |
| S366 | P | CHK2, IκBK2 | - |
| K370 | Me1, Me2 | SMYD2 (KMT3C) | LSD1 |
| K372 | Me1 | SET7/9 (KMT3) | - |
| K373 | Ac | P300 (KAT3B) | HDAC1 |
| S376 | P | PKC, GSK3β | - |
| T377 | P | CHK1, CHK2 | - |
| S378 | P | PKC, CHK1, CHK2 | - |
| K382 | Ac | P300 (KAT3B) | HDAC1, SIRT1 |
| K382 | Me2 | SeT8 (KMT5A) | - |
| K386 | S | SUMO-1 | - |
| T387 | P | CHK1 | - |
| S392 | P | CDK9, PKR, FACT (CK2) | PP1 |
| K320, K321, K370, K372, K373 | N8 | FBXO11, MDM2 | - |
| K120, K132, K139, K164 | Ub | - | - |
| K291, K292 | Ub | MKRN1 | - |
| K320 | Ub | E4F1 | - |
| K351, K357 | Ub | MSL2 | - |
| K101, K370, K372, K373, K381, K382, K386 | Ub | MDM2 | - |
One possibility is that among the collection of E3-ligases are specific readers of p53 PTMs, which evolved in order to respond to particular inductive signals that write a pattern of PTMs. The type of structural domains known to interact with modified lysine residues, such as Bromo with acetylated lysines as well as PHD and chromodomain with specific patterns of methylated lysines, are present in a subset of E3-ligases that target p53. For example, TRIM24 possesses a tandem PHD-Bromo domain unit, but the ability of TRIM24 to recognize p53 that is acetylated and/or methylated is untested. Cop1 has a WD40-domain, which can mediate protein-protein interactions, not notably dependent on PTMs. ARF-BP1 has a WWE domain that acts similarly but preferentially in the ubiquitin pathway. Other specific proteins that target cytoplasmic p53 for ubiquitin-mediated degradation, proteins p300 and CREB-binding protein (CBP), have Bromo domains as well as poly-ubiquitin ligase (E4) activities that tag p53 for proteolysis.71 These proteins are histone acetyl transferases that mediate acetylation of target proteins, e.g., p53 and histones H3 and H4, in addition to recognizing the acetylated motif. Acetylation of p53 occurs during activation of p53, in response to a variety of stresses;44,72 thus, these E4 proteins may also be involved in the termination of a stress-induced p53-response.
Protein-protein interactions between E3-ligases and p53 do not necessarily rely on independent, structural domain recognition, but occur as a result of allosteric recognition of surfaces or interfaces created by p53 protein folding or charge. Interestingly, viral protein BZLF1 directly functions as an adaptor component of the ECS (ElonginB/C-Cul2/5-SOCS-box protein) ubiquitin ligase complex and targets phosphorylated p53, induced by viral lytic replication.73 Signaling to E3-ligases, as well as p53, alters interactions between them, e.g., phosphorylation of S166, S188, S395, Y276 or Y394 of Mdm2 disrupts the p53-Mdm2 association.33 An E3-ligase may be regulated by signaling, as a result of stress or other regulatory pathways, which induce PTMs or the association of protein partners that modify the feedback loops of regulation that exist between p53 and its E3-ligase partners.23
E3-Ligases Targeted to Specific Outcomes
Mouse knock-in models have been created that express mutant forms of p53 (p53-6KR or p53-7KR), where C-terminal lysines are mutated and cannot be ubiquitinated, acetylated or methylated. These mice are viable and their mutant p53 has a normal half-life and response to stress.47,48 Elucidation of alternative pathways, which act in addition to Mdm2, began with Pirh2,49 and offered some idea of whether specific E3-ligases are involved in response to certain stimuli or to different aspects of p53 regulation. The primary function of Pirh2 may be terminating the p53 response to stimuli, because Pirh2 interacts preferentially with formed p53-tetramers, an association stimulated during stress activation of p53.74 COP1, another E3-ligase with a RING domain, negatively regulates p53 levels by ubiquitination but its targeted residues are unknown.75 Similar to Mdm2, the genes encoding Pirh2 and COP1 are regulated by p53 and, like other E3-ligases listed in Table 1, are thought to participate in autoregulatory feedback loops. Mdm2, Pirh2, COP1 and additional E3-ligases, including TRIM24,58 TOPORS,76 ARF-BP1,77 WWP1,78 ICP0,79 Ubc13,56 CUL7,80 CARP1 and CARP2,81 Synoviolin,82 CHIP,83 E4F1,84 E4orf6 and E1B55K,85 MSL2,86 the human kelch domain containing F-box protein, JFK,87 and Makorin Ring Finger Protein-1, MKRN1,88 mediate K48-linked polyubiquitination of p53 and degradation. However, mono- or K63-linked ubiquitination is linked to other functions, e.g., nuclear export and cytosolic localization of p53, as well as alteration of p53 transcriptional activities. Limited conjugation of ubiquitin by Mdm2, WWP1 and MSL2 induces nuclear export of p53; ICP0 promotes accumulation of ubiquitinated p53 at nuclear foci; E4F1, an atypical E3-ligase, forms K48-linked mono-, di- or tri-ubiquitinated p53 that activates cell cycle arrest at G0/G1; whereas, CUL7 generates mono- or di-ubiquitinated p53 to repress p53 transcriptional activities by unknown mechanisms.78–80,84,86 Ubc-13 is an E2-enzyme, capable of independently ubiquitinating p53, prevent p53 tetramerization.56 Thus, p53-ubiquitination can come in several flavors and the specific E3-ligase involved in modification of p53 can greatly alter the outcome.
So Many E3-ligases that Target p53
In this discussion, we offer multiple ways to parse the E3-ligases of p53 and suggest specialization among them in their roles as controllers of p53. Further development of animal models, analysis in multiple model organisms and testing of numerous hypotheses are needed to sort out the roles of the many E3-ligases active in regulation of p53. Recent evidence that p53 exhibits cycles of induction and decay, even in normal, unstressed cells,4 offers further frontiers to determinations of relevant PTMs of p53, the signaling pathways involved and whether any known E3-ligases, or those yet to be discovered, are involved in this regulatory program. Together these p53 regulators may alter p53 functions, terminate p53-signaling, and/or maintain p53 at low levels bearable by normal, unstressed cells. Targeting E3-ligases to inhibit ubiquitin-mediated degradation and restore p53 functions holds considerable therapeutic promise. The multiple layers of negative and positive regulation involved in controlling p53 offer challenges to understanding these critical pathways that regulate p53 stability.
Acknowledgements
We apologize to colleagues for our failure to cite their work due to space limitations. We thank Y. Furuta for his assistance in interpretation of expression analyses. Research in the Barton laboratory is supported by the Mitchell Foundation, CellCentric, Ltd., and grants from the National Institutes of Health (GM081627 and DK070824). A.K.J. is an Odyssey Fellow supported by the Odyssey Program and The Laura and John Arnold Foundation at The University of Texas MD Anderson Cancer Center.
Footnotes
References
- 1.Soussi T, Beroud C. Assessing TP53 status in human tumours to evaluate clinical outcome. Nat Rev Cancer. 2001;1:233–240. doi: 10.1038/35106009. [DOI] [PubMed] [Google Scholar]
- 2.Hainaut P, Hollstein M. p53 and human cancer: The first ten thousand mutations. Adv Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 4.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu W, Chan CS, Wu R, Zhang C, Sun Y, Song JS, et al. Negative Regulation of Tumor Suppressor p53 by MicroRNA miR-504. Mol Cell. 38:689–699. doi: 10.1016/j.molcel.2010.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 7.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 8.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 9.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951–961. doi: 10.1038/sj.cdd.4401916. [DOI] [PubMed] [Google Scholar]
- 10.Carter S, Vousden KH. Modifications of p53: competing for the lysines. Curr Opin Genet Dev. 2009;19:18–24. doi: 10.1016/j.gde.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 11.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 13.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 14.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307–315. doi: 10.1016/j.molcel.2006.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 16.Jain AK, Barton MC. Regulation of p53: TRIM24 enters the RING. Cell Cycle. 2009;8:3668–3674. doi: 10.4161/cc.8.22.9979. [DOI] [PubMed] [Google Scholar]
- 17.Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 18.Honda R, Tanaka H, Yasuda H. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 1997;420:25–27. doi: 10.1016/s0014-5793(97)01480-4. [DOI] [PubMed] [Google Scholar]
- 19.Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 20.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 21.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 22.Lu WJ, Abrams JM. Lessons from p53 in non-mammalian models. Cell Death Differ. 2006;13:909–912. doi: 10.1038/sj.cdd.4401922. [DOI] [PubMed] [Google Scholar]
- 23.Wu X, Bayle JH, Olson D, Levine AJ. The p53-mdm-2 autoregulatory feedback loop. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 24.Stommel JM, Wahl GM. Accelerated MDM2 auto-degradation induced by DNA-damage kinases is required for p53 activation. EMBO J. 2004;23:1547–1556. doi: 10.1038/sj.emboj.7600145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 24:1580–1589. doi: 10.1101/gad.1941710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 27.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu GW, Rudiger S, Veprintsev D, Freund S, Fernandez-Fernandez MR, Fersht AR. The central region of HDM2 provides a second binding site for p53. Proc Natl Acad Sci USA. 2006;103:1227–1232. doi: 10.1073/pnas.0510343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shimizu H, Burch LR, Smith AJ, Dornan D, Wallace M, Ball KL, Hupp TR. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J Biol Chem. 2002;277:28446–28458. doi: 10.1074/jbc.M202296200. [DOI] [PubMed] [Google Scholar]
- 30.Marine JC, Francoz S, Maetens M, Wahl G, Toledo F, Lozano G. Keeping p53 in check: essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13:927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 31.Uldrijan S, Pannekoek WJ, Vousden KH. An essential function of the extreme C-terminus of MDM2 can be provided by MDMX. EMBO J. 2007;26:102–112. doi: 10.1038/sj.emboj.7601469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh RK, Iyappan S, Scheffner M. Hetero-oligomerization with MdmX rescues the ubiquitin/Nedd8 ligase activity of RING finger mutants of Mdm2. J Biol Chem. 2007;282:10901–10907. doi: 10.1074/jbc.M610879200. [DOI] [PubMed] [Google Scholar]
- 33.Meek DW, Knippschild U. Posttranslational modification of MDM2. Mol Cancer Res. 2003;1:1017–1026. [PubMed] [Google Scholar]
- 34.Kawai H, Wiederschain D, Kitao H, Stuart J, Tsai KK, Yuan ZM. DNA damage-induced MDMX degradation is mediated by MDM2. J Biol Chem. 2003;278:45946–45953. doi: 10.1074/jbc.M308295200. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Brooks CL, Wu-Baer F, Chen D, Baer R, Gu W. Mono- versus poly-ubiquitination: differential control of p53 fate by Mdm2. Science. 2003;302:1972–1975. doi: 10.1126/science.1091362. [DOI] [PubMed] [Google Scholar]
- 36.Nie L, Sasaki M, Maki CG. Regulation of p53 nuclear export through sequential changes in conformation and ubiquitination. J Biol Chem. 2007;282:14616–14625. doi: 10.1074/jbc.M610515200. [DOI] [PubMed] [Google Scholar]
- 37.Grossman SR, Deato ME, Brignone C, Chan HM, Kung AL, Tagami H, et al. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 2003;300:342–344. doi: 10.1126/science.1080386. [DOI] [PubMed] [Google Scholar]
- 38.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, et al. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Higashitsuji H, Itoh K, Sakurai T, Nagao T, Sumitomo Y, Masuda T, et al. The oncoprotein gankyrin binds to MDM2/HDM2, enhancing ubiquitylation and degradation of p53. Cancer Cell. 2005;8:75–87. doi: 10.1016/j.ccr.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 40.Wang C, Ivanov A, Chen L, Fredericks WJ, Seto E, Rauscher FJ, 3rd, Chen J. MDM2 interaction with nuclear corepressor KAP1 contributes to p53 inactivation. EMBO J. 2005;24:3279–3290. doi: 10.1038/sj.emboj.7600791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Du W, Jiang P, Li N, Mei Y, Wang X, Wen L, et al. Suppression of p53 activity by Siva1. Cell Death Differ. 2009;16:1493–1504. doi: 10.1038/cdd.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118:83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 43.Carter S, Bischof O, Dejean A, Vousden KH. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat Cell Biol. 2007;9:428–435. doi: 10.1038/ncb1562. [DOI] [PubMed] [Google Scholar]
- 44.Tang Y, Zhao W, Chen Y, Zhao Y, Gu W. Acetylation is indispensable for p53 activation. Cell. 2008;133:612–626. doi: 10.1016/j.cell.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Itahana K, Mao H, Jin A, Itahana Y, Clegg HV, Lindstrom MS, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 46.Ringshausen I, O'Shea CC, Finch AJ, Swigart LB, Evan GI. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell. 2006;10:501–514. doi: 10.1016/j.ccr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 47.Feng L, Lin T, Uranishi H, Gu W, Xu Y. Functional analysis of the roles of posttranslational modifications at the p53 C terminus in regulating p53 stability and activity. Mol Cell Biol. 2005;25:5389–5395. doi: 10.1128/MCB.25.13.5389-5395.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krummel KA, Lee CJ, Toledo F, Wahl GM. The C-terminal lysines fine-tune p53 stress responses in a mouse model but are not required for stability control or transactivation. Proc Natl Acad Sci USA. 2005;102:10188–10193. doi: 10.1073/pnas.0503068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- 50.Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29:92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 51.Daujat S, Neel H, Piette J. Preferential expression of Mdm2 oncogene during the development of neural crest and its derivatives in mouse early embryogenesis. Mech Dev. 2001;103:163–165. doi: 10.1016/s0925-4773(01)00339-2. [DOI] [PubMed] [Google Scholar]
- 52.Chavez-Reyes A, Parant JM, Amelse LL, de Oca Luna RM, Korsmeyer SJ, Lozano G. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 2003;63:8664–8669. [PubMed] [Google Scholar]
- 53.Bourdon JC. p53 and its isoforms in cancer. Br J Cancer. 2007;97:277–282. doi: 10.1038/sj.bjc.6603886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Midgley CA, Owens B, Briscoe CV, Thomas DB, Lane DP, Hall PA. Coupling between gamma irradiation, p53 induction and the apoptotic response depends upon cell type in vivo. J Cell Sci. 1995;108:1843–1848. doi: 10.1242/jcs.108.5.1843. [DOI] [PubMed] [Google Scholar]
- 55.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laine A, Topisirovic I, Zhai D, Reed JC, Borden KL, Ronai Z. Regulation of p53 localization and activity by Ubc13. Mol Cell Biol. 2006;26:8901–8913. doi: 10.1128/MCB.01156-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Khetchoumian K, Teletin M, Tisserand J, Mark M, Herquel B, Ignat M, et al. Loss of Trim24 (Tif1alpha) gene function confers oncogenic activity to retinoic acid receptor alpha. Nat Genet. 2007;39:1500–1506. doi: 10.1038/ng.2007.15. [DOI] [PubMed] [Google Scholar]
- 58.Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, et al. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci USA. 2009;106:11612–11616. doi: 10.1073/pnas.0813177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu L, Scolnick DM, Trievel RC, Zhang HB, Marmorstein R, Halazonetis TD, Berger SL. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang J, Perez-Burgos L, Placek BJ, Sengupta R, Richter M, Dorsey JA, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 61.Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 62.Huang J, Dorsey J, Chuikov S, Perez-Burgos L, Zhang X, Jenuwein T, et al. G9a and Glp methylate lysine 373 in the tumor suppressor p53. J Biol Chem. 285 doi: 10.1074/jbc.M109.062588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murray-Zmijewski F, Slee EA, Lu X. A complex barcode underlies the heterogeneous response of p53 to stress. Nat Rev Mol Cell Biol. 2008;9:702–712. doi: 10.1038/nrm2451. [DOI] [PubMed] [Google Scholar]
- 64.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 65.Olsson A, Manzl C, Strasser A, Villunger A. How important are post-translational modifications in p53 for selectivity in target-gene transcription and tumour suppression? Cell Death Differ. 2007;14:1561–1575. doi: 10.1038/sj.cdd.4402196. [DOI] [PubMed] [Google Scholar]
- 66.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 67.Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- 68.Meek DW, Anderson CW. Posttranslational Modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 70.Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9:831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 71.Shi D, Pop MS, Kulikov R, Love IM, Kung AL, Grossman SR. CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53. Proc Natl Acad Sci USA. 2009;106:16275–16280. doi: 10.1073/pnas.0904305106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 73.Sato Y, Kamura T, Shirata N, Murata T, Kudoh A, Iwahori S, et al. Degradation of phosphorylated p53 by viral protein-ECS E3 ligase complex. PLoS Pathog. 2009;5:1000530. doi: 10.1371/journal.ppat.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sheng Y, Laister RC, Lemak A, Wu B, Tai E, Duan S. Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat Struct Mol Biol. 2008;15:1334–1342. doi: 10.1038/nsmb.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 76.Rajendra R, Malegaonkar D, Pungaliya P, Marshall H, Rasheed Z, Brownell J, et al. Topors functions as an E3 ubiquitin ligase with specific E2 enzymes and ubiquitinates p53. J Biol Chem. 2004;279:36440–36444. doi: 10.1074/jbc.C400300200. [DOI] [PubMed] [Google Scholar]
- 77.Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 78.Laine A, Ronai Z. Regulation of p53 localization and transcription by the HECT domain E3 ligase WWP1. Oncogene. 2007;26:1477–1483. doi: 10.1038/sj.onc.1209924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boutell C, Everett RD. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and Ubiquitinates p53. J Biol Chem. 2003;278:36596–36602. doi: 10.1074/jbc.M300776200. [DOI] [PubMed] [Google Scholar]
- 80.Andrews P, He YJ, Xiong Y. Cytoplasmic localized ubiquitin ligase cullin 7 binds to p53 and promotes cell growth by antagonizing p53 function. Oncogene. 2006;25:4534–4548. doi: 10.1038/sj.onc.1209490. [DOI] [PubMed] [Google Scholar]
- 81.Yang W, Rozan LM, McDonald ER, 3rd, Navaraj A, Liu JJ, Matthew EM, et al. CARPs are ubiquitin ligases that promote MDM2-independent p53 and phospho-p53ser20 degradation. J Biol Chem. 2007;282:3273–3281. doi: 10.1074/jbc.M610793200. [DOI] [PubMed] [Google Scholar]
- 82.Yamasaki S, Yagishita N, Sasaki T, Nakazawa M, Kato Y, Yamadera T, et al. Cytoplasmic destruction of p53 by the endoplasmic reticulum-resident ubiquitin ligase ‘Synoviolin’. EMBO J. 2007;26:113–122. doi: 10.1038/sj.emboj.7601490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Esser C, Scheffner M, Hohfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 84.Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 85.Querido E, Blanchette P, Yan Q, Kamura T, Morrison M, Boivin D, et al. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001;15:3104–3117. doi: 10.1101/gad.926401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kruse JP, Gu W. MSL2 promotes Mdm2-independent cytoplasmic localization of p53. J Biol Chem. 2009;284:3250–3263. doi: 10.1074/jbc.M805658200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun L, Shi L, Li W, Yu W, Liang J, Zhang H, et al. JFK, a Kelch domain-containing F-box protein, links the SCF complex to p53 regulation. Proc Natl Acad Sci USA. 2009;106:10195–10200. doi: 10.1073/pnas.0901864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lee EW, Lee MS, Camus S, Ghim J, Yang MR, Oh W, et al. Differential regulation of p53 and p21 by MKRN1 E3 ligase controls cell cycle arrest and apoptosis. EMBO J. 2009;28:2100–2113. doi: 10.1038/emboj.2009.164. [DOI] [PMC free article] [PubMed] [Google Scholar]