Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases (original) (raw)
Abstract
Summary: The mitogen-activated protein kinases (MAPKs) regulate diverse cellular programs by relaying extracellular signals to intracellular responses. In mammals, there are more than a dozen MAPK enzymes that coordinately regulate cell proliferation, differentiation, motility, and survival. The best known are the conventional MAPKs, which include the extracellular signal-regulated kinases 1 and 2 (ERK1/2), c-Jun amino-terminal kinases 1 to 3 (JNK1 to -3), p38 (α, β, γ, and δ), and ERK5 families. There are additional, atypical MAPK enzymes, including ERK3/4, ERK7/8, and Nemo-like kinase (NLK), which have distinct regulation and functions. Together, the MAPKs regulate a large number of substrates, including members of a family of protein Ser/Thr kinases termed MAPK-activated protein kinases (MAPKAPKs). The MAPKAPKs are related enzymes that respond to extracellular stimulation through direct MAPK-dependent activation loop phosphorylation and kinase activation. There are five MAPKAPK subfamilies: the p90 ribosomal S6 kinase (RSK), the mitogen- and stress-activated kinase (MSK), the MAPK-interacting kinase (MNK), the MAPK-activated protein kinase 2/3 (MK2/3), and MK5 (also known as p38-regulated/activated protein kinase [PRAK]). These enzymes have diverse biological functions, including regulation of nucleosome and gene expression, mRNA stability and translation, and cell proliferation and survival. Here we review the mechanisms of MAPKAPK activation by the different MAPKs and discuss their physiological roles based on established substrates and recent discoveries.
INTRODUCTION
Mitogen-activated protein kinases (MAPKs) are protein Ser/Thr kinases that convert extracellular stimuli into a wide range of cellular responses. MAPKs are among the most ancient signal transduction pathways and are widely used throughout evolution in many physiological processes (396). All eukaryotic cells possess multiple MAPK pathways, which coordinately regulate gene expression, mitosis, metabolism, motility, survival, apoptosis, and differentiation. In mammals, 14 MAPKs have been characterized into seven groups (Fig. 1). Conventional MAPKs comprise the extracellular signal-regulated kinases 1/2 (ERK1/2), c-Jun amino (N)-terminal kinases 1/2/3 (JNK1/2/3), p38 isoforms (α, β, γ, and δ), and ERK5 (reviewed in references 54, 198, and 264). Atypical MAPKs have nonconforming particularities and comprise ERK3/4, ERK7, and Nemo-like kinase (NLK) (reviewed in reference 71). By far the most extensively studied groups of mammalian MAPKs are the ERK1/2, JNKs, and p38 isoforms, but recent studies have shed some light on the regulation and function of other groups of MAPKs.
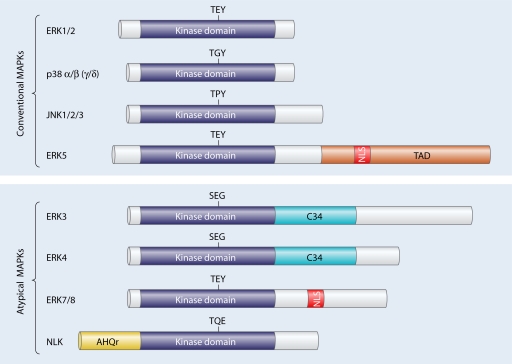
FIG. 1.
Schematic representation of the overall structures of conventional and atypical MAPKs. All MAPKs contain a Ser/Thr kinase domain flanked by N- and C-terminal regions of different lengths. Different additional domains are also present in some MAPKs, including a transactivation domain (TAD), a nuclear localization sequence (NLS), a region conserved in ERK3 and ERK4 (C34), and a domain rich in Ala, His, and Glu (AHQr). The respective MAPKAPKs activated by the MAPKs are show in light gray next to the cognate MAPK. The γ and δ isoforms of p38 are in parentheses to indicate that they have not been shown to promote MAPKAPK activation.
Each group of conventional MAPKs is composed of a set of three evolutionarily conserved, sequentially acting kinases: a MAPK, a MAPK kinase (MAPKK), and a MAPKK kinase (MAPKKK) (Fig. 2). The MAPKKKs, which are protein Ser/Thr kinases, are often activated through phosphorylation and/or as a result of their interaction with a small GTP-binding protein of the Ras/Rho family in response to extracellular stimuli. MAPKKK activation leads to the phosphorylation and activation of a MAPKK, which then stimulates MAPK activity through dual phosphorylation on Thr and Tyr residues within a conserved Thr-X-Tyr motif located in the activation loop of the kinase domain subdomain VIII (Fig. 1). Phosphorylation of these residues is essential for enzymatic activities, as was originally demonstrated for ERK2 (288).
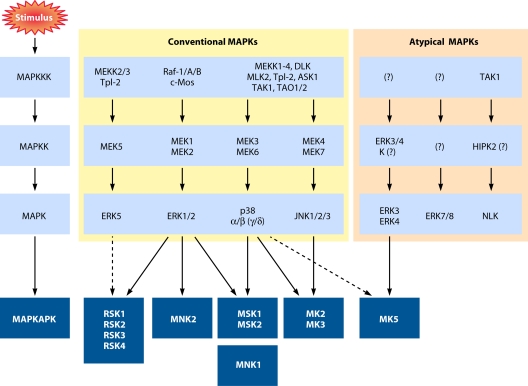
FIG. 2.
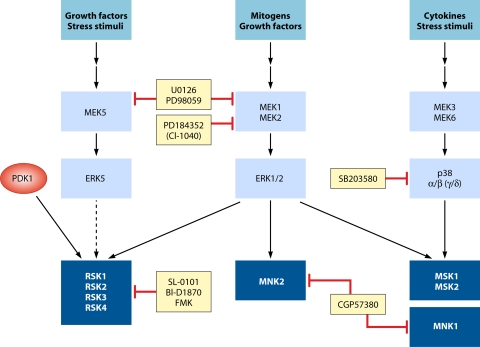
MAPK signaling cascades leading to activation of the MAPKAPKs. Mitogens, cytokines, and cellular stresses promote the activation of different MAPK pathways, which in turn phosphorylate and activate the five subgroups of MAPKAPKs, including RSK, MSK, MNK, MK2/3, and MK5. Dotted lines indicate that, although reported, substrate regulation by the respective kinase remains to be thoroughly demonstrated. The γ and δ isoforms of p38 are in parentheses to indicate that they have not been shown to promote MAPKAPK activation.
Less is known about the exact molecular mechanisms involved in activation of atypical MAPKs, as they do not share many characteristics of conventional MAPKs (71). One determining feature of atypical MAPKs is that these proteins are not organized into classical three-tiered kinase cascades. In addition, the Thr-X-Tyr motif is absent in ERK3/4 and NLK, where a Gly or Glu residue replaces the Tyr. ERK7 contains the motif Thr-Glu-Tyr in its activation loop, but phosphorylation of these residues appears to be catalyzed by ERK7 itself, rather than by an upstream MAPKK. Nevertheless, once activated, conventional and atypical MAPKs phosphorylate target substrates on Ser or Thr followed by a Pro residue, making them Pro-directed kinases, and therefore have limited specificity in their consensus phosphorylation motifs. In addition to the transient kinase-substrate interaction, MAPK substrate selectivity is also conferred by specific interaction domains termed docking sites. Scaffolding proteins also mediate MAPK cascade specificity by simultaneously binding several components and organizing pathways in specific modules.
The wide range of functions regulated by the MAPKs is mediated through phosphorylation of several substrates, including members of a family of protein kinases termed MAPK-activated protein kinases (MAPKAPKs) (Fig. 2) (123, 301). This family comprises the p90 ribosomal S6 kinases (RSKs) (48), mitogen- and stress-activated kinases (MSKs) (14), MAPK-interacting kinases (MNKs) (44), MAPK-activated protein kinase 2/3 (MK2/3) (293), and MK5 (267). MAPKAPK family members represent an additional enzymatic and amplification step in the MAPK catalytic cascades. In addition, they control a wide range of biological functions and thereby increase the range of action regulated by activated MAPK modules. This article gives an overview of the different groups of mammalian MAPKs, as well as describing our current understanding of the properties, regulation, and function of the MAPK-activated protein kinases.
THE CONVENTIONAL MAPKs
The ERK1/2 Module
Identification.
ERK1 was the first mammalian MAPK to be cloned and characterized. It was originally found to be phosphorylated on Tyr and Thr residues in response to growth factors (70, 184, 281), and both ERK1 and ERK2 cDNAs were cloned in the early 1990s (35, 36). ERK1 and ERK2 share 83% amino acid identity (Fig. 1) and are expressed to various extents in all tissues, with particularly high levels in the brain, skeletal muscle, thymus, and heart (36). Alternatively spliced isoforms have been described for ERK1 (ERK1b and ERK1c) (325, 418) and ERK2 (ERK2b) (135), and these appear to be activated by specific agonists and may have a different subcellular localization and tissue distribution than full-length proteins (278, 326).
Activation mechanisms and inhibitors.
ERK1 and ERK2 are activated by growth factors, including platelet-derived growth factor (PDGF), epidermal growth factor (EGF), and nerve growth factor (NGF), and in response to insulin (36). They are also activated by ligands for heterotrimeric G protein-coupled receptors (GPCRs), cytokines, osmotic stress, and microtubule disorganization (278). The mammalian ERK1/2 module consists of the MAPKKKs A-Raf, B-Raf, and Raf-1, the MAPKKs MEK1 and MEK2, and the MAPKs ERK1 and ERK2 (Fig. 2). While Raf isoforms are the primary MAPKKKs in the ERK1/2 module, the protein kinases MEKK1, Mos, and Tpl2 (also known as Cot) are additional MAPKKKs utilized in a more restricted cell type- and stimulus-specific manner (reviewed in references 278 and 326).
The ERK1/2 module is activated principally by cell surface receptors, such as receptor tyrosine kinases (RTKs). Ligand-induced receptor dimerization promotes receptor activation and autophosphorylation of Tyr residues in the intracellular domain. These phosphorylated residues serve as specific binding sites for proteins that contain Src homology 2 (SH2) or phosphotyrosine-binding (PTB) domains, such as Grb2 (growth factor receptor-bound protein 2). The best-characterized route of Ras activation occurs at the plasma membrane and is mediated by SOS (son of sevenless), a guanine nucleotide exchange factor (GEF). SOS is recruited from the cytosol to the plasma membrane as a result of its interaction with Grb2, and it stimulates the exchange of GDP bound to Ras by GTP that is required for a positive regulation of Ras activity. This nucleotide exchange allows Ras to interact directly with its target effectors, one of which is Raf, the initiating kinase of the ERK1/2 module. Regulation of both Ras and Raf is crucial for the proper maintenance of cell proliferation, as activating mutations in these genes lead to oncogenesis (181). Activated Raf binds to and phosphorylates the dual-specificity kinases MEK1/2, which in turn, phosphorylate ERK1/2 within a conserved Thr-Glu-Tyr (TEY) motif in their activation loop (Fig. 1).
MEK1/2 inhibitors have been used extensively to implicate ERK1/2 in a wide array of biological events. Two companies developed MEK1/2 inhibitors in the mid-1990s. One class is typified by PD98059 (8, 98) and the other by U0126 (112). These inhibitors are not competitive with respect to ATP, and they appear to interact with the inactive unphosphorylated kinase more strongly than the active phosphorylated species. This interaction is thought to prevent the phosphorylation of MEK1/2 and/or the conformational transition that generates the activated enzyme (8). More recently, additional noncompetitive inhibitors of MEK1/2 with greater bioavailability (PD184352 and PD0325901) have been developed and entered clinical trials as potential anticancer agents (116). These compounds were tested in a recent study against a panel of recombinant protein kinases (18), which recommended that PD184352 or PD0325901 be used to inhibit MEK1/2 in cells and that the structurally unrelated compound U0126 be used to verify the results.
Substrates and biological functions.
In quiescent cells, all components of the ERK1/2 module have a cytoplasmic localization, but upon extracellular stimulation, a significant proportion of ERK1/2 accumulates in the nucleus (53, 213). While the mechanisms involved in nuclear accumulation of ERK1/2 remain incompletely understood, nuclear retention, dimerization, phosphorylation, and release from cytoplasmic anchors have been shown to play a role (reviewed in reference 272). More recently, a new nuclear translocation mechanism for ERK1/2 was identified and is based on a novel nuclear translocation sequence (NTS) located within the kinase insert domain (420). Phosphorylation of this domain upon stimulation allows ERK1/2 to interact with nuclear importing proteins, which mediates the translocation of ERK1/2 into the nucleus via nuclear pores. Upon stimulation, ERK1/2 phosphorylate a large number of substrates (reviewed in reference 416). Some of these are found localized in the cytoplasm (death-associated protein kinase [DAPK], tuberous sclerosis complex 2 [TSC2], RSK, and MNK) and the nucleus (NF-AT, Elk-1, myocyte enhancer factor 2 [MEF2], c-Fos, c-Myc, and STAT3), whereas others have been found associated with membranes (CD120a, Syk, and calnexin) or the cytoskeleton (neurofilaments and paxillin).
The ERK1/2 module plays a central role in the control of cell proliferation. ERK1/2 activity is rapidly stimulated by mitogenic agents, and in normal cells, sustained activation of these kinases is required for efficient G1- to S-phase progression. ERK1/2 control cell proliferation via several mechanisms, including the induction of positive regulators of the cell cycle (reviewed in reference 235). ERK1/2 phosphorylate and activate the transcription factor Elk-1, which is involved in expression of immediate-early (IE) genes, such as that for c-Fos (132). ERK1/2 stabilize c-Fos protein through direct phosphorylation (245), thereby allowing c-Fos to associate with c-Jun and form transcriptionally active AP-1 complexes (395). AP-1 activity is required for expression of cyclin D1 (327), a protein that interacts with cyclin-dependent kinases (CDKs) and permits G1/S transition and cell cycle progression. In addition, ERK1/2 extend the MAPK cascade by phosphorylating and activating MAPKAPK family members, including RSKs, MSKs, and MNKs (Fig. 1 and 2). These protein kinases are important regulators of ERK1/2-dependent biological processes and are discussed below in further detail.
The p38 MAPK Module
Identification.
Identified simultaneously by three groups in 1994, p38α (also known as CSBP, mHOG1, RK, and SAPK2) is the archetypal member of a second MAPK module that is generally more responsive to stress stimuli (142, 208, 295). p38α is 50% identical to ERK2 and bears significant homology to the product of the budding yeast hog1 gene, which is activated in response to hyperosmolarity (142, 208, 295). Since identification of p38α, three additional isoforms have been found, i.e., p38β, p38γ, and p38δ (reviewed in reference 76). Whereas p38α and p38β are ubiquitously expressed in cell lines and tissues, p38γ and p38δ have more restricted expression patterns and may have specialized functions (168). Because p38α is generally more highly expressed than p38β, most of the published literature on p38 MAPKs refers to the former.
Activation mechanisms and inhibitors.
In mammalian cells, the four p38 isoforms are strongly activated by various environmental stresses and inflammatory cytokines, including oxidative stress, UV irradiation, hypoxia, ischemia, interleukin-1 (IL-1), and tumor necrosis factor alpha (TNF-α) (reviewed in reference 76). TNF-α and IL-1 activate p38 isoforms by promoting the recruitment of TRAF adaptor proteins to the intracellular domains of their cognate receptors (37). TRAF recruitment promotes activation of various MAPKKKs involved in the activation of the p38 isoforms. The p38 isoforms are also activated by GPCRs (134), as well as by the Rho family GTPases Rac and Cdc42 (17).
MKK3 and MKK6 are thought to be the major protein kinases responsible for p38 activation (89, 143, 344), but MKK4 has also been shown to possess some activity toward p38 (233). While MKK6 activates all p38 isoforms, MKK3 is somewhat more selective, as it preferentially phosphorylates the α, δ, and γ isoforms. The specificity in p38 activation is thought to result from the formation of functional complexes between MKK3/6 and different p38 isoforms and from the selective recognition of the activation loop of p38 isoforms by MKK3/6. Activation of the p38 isoforms results from the MKK3/6-catalyzed phosphorylation of a conserved Thr-Gly-Tyr (TGY) motif in their activation loops (Fig. 1). MKK3/6 are activated by a plethora of MAPKKKs, including MEKK1 to -3, MLK2/3, ASK1, Tpl2, TAK1, and TAO1/2 (76).
Most stimuli that activate p38 MAPKs also stimulate JNK isoforms, and many MAPKKKs in the p38 module are shared by the JNK module (Fig. 2). Identification of the anti-inflammatory drug SB203580 and its close relative SB202190 has been exploited in thousands of studies to delineate the functions of p38. These drugs specifically target and inhibit the p38α and p38β isoforms by acting as competitive inhibitors of ATP binding (208). BIRB0796 is a more potent inhibitor of p38α and p38β that inhibits kinase activity by a novel mechanism that indirectly competes with the binding of ATP (283). A recent study in which these inhibitors were tested against a panel of purified protein kinases concluded that all three compounds have suitable potency and selectivity for their use as p38 MAPK inhibitors in cell-based assays (18).
Substrates and biological functions.
p38 isoforms are present in the nuclei and cytoplasm of quiescent cells (20) and have been shown to accumulate in the nuclei of cells subjected to certain stresses (277). While the mechanisms involved in the nucleocytoplasmic shuttling of p38 isoforms remain elusive, the MAPK-activated protein kinases MK2, MK3, and MK5 have been shown to play roles as cytoplasmic anchors for these kinases (122). Upon stimulation, p38 isoforms phosphorylate a large number of substrates in many cellular compartments, including the cytoplasm (cPLA2, MNK1/2, MK2/3, HuR, Bax, and Tau) and the nucleus (ATF1/2/6, MEF2, Elk-1, GADD153, Ets1, p53, and MSK1/2) (76).
The p38 module plays a critical role in normal immune and inflammatory responses (reviewed in reference 76). p38 is activated by numerous extracellular mediators of inflammation, including chemoattractants, cytokines, chemokines, and bacterial lipopolysaccharide (LPS). A major function of p38 isoforms is the production of proinflammatory cytokines. p38 can regulate cytokine expression by modulating transcription factors, such as NF-κB (180), or at the mRNA level, by modulating their stability and translation through the regulation of MNK1 (44) and MK2/3 (293). p38α appears to be the main p38 isoform involved in the inflammatory response, as its deletion in epithelial cells was found to reduce proinflammatory gene expression (186).
The p38 MAPKs have also been shown to play roles in cell proliferation and survival. p38α negatively regulates cell cycle progression at both the G1/S and G2/M transitions by a number of mechanisms, including downregulation of cyclins and upregulation of CDK inhibitors (365). Although some studies have reported prosurvival functions for p38α, many more have associated p38α activity with the induction of apoptosis by cellular stresses. These effects can be mediated by transcriptional and posttranscriptional mechanisms, which affect death receptors, survival pathways or pro- and antiapoptotic Bcl-2 family proteins (77). In addition, the p38 isoforms can extend the MAPK kinase cascade by phosphorylating and activating several MAPKAPK family members, including MSKs, MNK1, MK2/3, and MK5 (Fig. 1 and 2). As briefly suggested above, these kinases are important regulators of p38-dependent processes and will be discussed in greater detail.
The JNK Module
Identification.
The first JNK (also known as stress-activated protein kinase [SAPK]) family member was originally identified as a cycloheximide-activated MAP-2 kinase and as a kinase activity that could be affinity purified using c-Jun protein bound to beads (153, 199). It was subsequently found that stress stimuli promote JNK phosphorylation on Thr and Tyr residues, much like what had been observed for the ERKs (200, 202). There are three known JNK isoforms, JNK1 to -3 (also known as SAPKγ, SAPKα, and SAPKβ, respectively), which were independently cloned by two groups in the mid-1990s (88, 201). The JNKs are greater than 85% identical to each other and are encoded by three distinct genes giving rise to 10 or more spliced forms ranging in molecular mass from 46 to 55 kDa (88, 140, 201). JNK1 and JNK2 have a broad tissue distribution, whereas JNK3 seems to be localized primarily to neuronal tissues, testis, and cardiac myocytes (30).
Activation mechanisms and inhibitors.
Much like the p38 MAPKs, the JNK isoforms are strongly activated in response to various cellular stresses, including heat shock, ionizing radiation, oxidative stress, DNA-damaging agents, cytokines, UV irradiation, DNA and protein synthesis inhibitors, and growth factor deprivation, and to a lesser extent by growth factors, some GPCR ligands, and serum (reviewed in reference 31). Activation of JNK isoforms requires dual phosphorylation on Thr and Tyr residues within a conserved Thr-Pro-Tyr (TPY) motif in their activation loops (Fig. 1). The MAPKKs that catalyze this reaction are known as MKK4 (also known as SEK1) and MKK7, which appear to cooperate in the phosphorylation and activation of the JNKs (206). MKK4/7 are phosphorylated and activated by several MAPKKKs, including MEKK1 to -4, MLK1 to -3, Tpl-2, DLK, TAO1/2, TAK1, and ASK1/2 (Fig. 2) (198, 382).
With regard to JNK inhibitors, two reversible ATP-competitive inhibitors have been widely used in the last decade, namely, SP600125 (also known as JNK inhibitor II) (22) and AS601245 (JNK inhibitor V) (46). However, a recent study which tested the specificity of these compounds concluded that they had poor selectivity when tested against a panel of purified protein kinases (18). A recent wave of new molecules have been described in the literature over the last 2 to 3 years (reviewed in reference 31), most of which appear to specifically target JNK3.
Substrates and biological functions.
Like ERK1/2 and p38 MAPKs, a proportion of activated JNKs have been shown to relocalize from the cytoplasm to the nucleus following stimulation (238). The transcription factor c-Jun is a well-described substrate for JNKs, as its phosphorylation on Ser63/73 was found to increase c-Jun-dependent transcription (reviewed in reference 393). Certain studies have shown that there are functional differences between JNK isoforms with regard to the regulation of c-Jun (306), but more recent evidence using chemical genetics indicated that both JNK1 and JNK2 are positive regulators of c-Jun expression as well as cell proliferation (164). Additional transcription factors have been shown to be phosphorylated by JNK, including p53, ATF-2, NF-ATc1, Elk-1, HSF-1, STAT3, c-Myc, and JunB (31, 278), but very little is known about the contribution of each JNK family member to their regulation. While some cytoplasmic targets of JNK are known, the fact that stimulated JNK does not exhibit an exclusive nuclear localization suggests that many other cytoplasmic substrates remain to be identified.
JNK1 and JNK2 have been shown to play important roles in the control of cell proliferation. Through c-Jun, JNK activity promotes AP-1 complex formation and transcription of genes containing AP-1-binding sites, including genes that control the cell cycle, such as cyclin D1 (306). In addition to having been implicated in the differentiation of hematopoietic populations, JNK isoforms play an important role in the apoptotic response to cellular stresses (90). Mouse embryonic fibroblasts (MEFs) isolated from _Jnk1_−/−/_Jnk2_−/− knockout mice are resistant to apoptosis induced by DNA-damaging agents and UV irradiation (368). The inactivation of JNK1 and JNK2 was found to inhibit cytochrome c release, indicating a role for the JNK module in the intrinsic apoptotic pathway. Unlike ERK1/2 and p38 MAPKs, there are currently no known MAPKAPK family members directly activated by the JNK isoforms.
The ERK5 Module
Identification.
The ERK5 (also known as BMK1, for big MAP kinase 1) pathway was identified independently by three groups using different approaches (106, 209, 434). ERK5 is twice the size of other MAPKs (∼100 kDa), and its N-terminal half contains a kinase domain similar to that of ERK1/2, with 51% amino acid identity to ERK2. ERK5 has a unique C-terminal half that contains a nuclear localization signal (NLS) and a proline-rich region (Fig. 1). Three spliced forms of ERK5 have been reported, i.e., ERK5a, ERK5b, and ERK5c (411). ERK5 is expressed to various extents in all tissues, with particularly high levels in the brain, thymus, and spleen (412). ERK5 is essential for early embryonic development and is required for normal development of the vascular system as well as cell survival (282, 340, 412).
Activation mechanisms and inhibitors.
ERK5 activity is increased in response to growth factors, serum, oxidative stress, and hyperosmolarity (reviewed in reference 383). This is correlated with the dual phosphorylation of Thr and Tyr residues within a conserved Thr-Glu-Tyr (TEY) motif in the activation loop of the kinase domain. MEK5 was identified as the MAPKK that phosphorylates ERK5 on these residues (106, 434), and other known MEKs do not appear to influence ERK5 activity (Fig. 2). MEK5 is phosphorylated and activated by both MEKK2 and MEKK3 in a stimulus- and cell type-dependent manner (383), but the signaling cascade at this level is not entirely specific to ERK5, since MEKK2/3 also stimulate the JNK and p38 MAPK modules. Finally, WNK1 was recently identified as a potential kinase upstream of MEKK2/3 (408). While PD98059 and U0126 were identified as MEK1/2 inhibitors, these molecules also efficiently inhibit MEK5 (175, 239). Interestingly, MEK5 is less sensitive to PD184352, which can be used as a tool to delineate MEK1/2- and MEK5-dependent functions.
Substrates and biological functions.
ERK5 localizes to the cytoplasm in resting cells and translocates to the nucleus when coexpressed with activated MEK5 or upon stimulation (383). A number of molecules have been identified as ERK5 substrates, including the myocyte enhancer factor 2 (MEF2) family of transcriptional factors, Sap1a (ETS domain transcription factor), c-Myc, SGK, connexin 43 (Cx43), and Bad (reviewed in references 147 and 383).
At the cellular level, ERK5 was found to regulate cell survival and proliferation via several mechanisms. Similarly to ERK1/2, activated ERK5 promotes G1/S transition by driving cyclin D1 expression (244). In addition, the regulation of SGK by ERK5 was found to be essential for S-phase entry in response to growth factors (147). Alternatively, ERK5 has been shown to phosphorylate and activate the RSK family of protein kinases (280), thereby increasing this MAPK kinase cascade by one step (Fig. 2).
THE ATYPICAL MAPKs
ERK3/4
Identification.
ERK3 was cloned in 1991 by homology screening of a rat cDNA library using a probe derived from ERK1 sequences (35). Subsequent cloning of the human (234, 435) and mouse (372) orthologs helped established that ERK3 possesses a C-terminal extension of 178 amino acids (aa), yielding a protein with a molecular mass of ∼100 kDa. The cDNAs of human and rat ERK4 were isolated using a method similar to that used for ERK3 (126, 135). Whereas it was originally described as a 557-aa protein, resequencing of the human cDNA revealed that ERK4 is a 578-aa protein with a molecular mass of ∼70 kDa (71). ERK3 and ERK4 have very similar protein structures, and their kinase domains display 73% amino acid identity. ERK3/4 are considered atypical because their activation loop lacks a phosphoacceptor Tyr residue and contains the Ser-Glu-Gly motif (Fig. 1). The precise function of the C-terminal extension found in ERK3/4 remains elusive, but characterization of this region suggests that it plays a role in subcellular targeting (173). The N-terminal region of ERK3, but not ERK4, is involved in the degradation of ERK3 by the ubiquitin-proteasome pathway (72).
Activation mechanisms and inhibitors.
The ERK3/4 module remains poorly characterized (Fig. 2). Although the Ser residue in the activation loop of ERK3 is phosphorylated in vivo (55, 56, 73, 86), no stimuli have been found to promote ERK3/4 phosphorylation or activity. Although ERK3 was shown to autophosphorylate in vitro (55), a kinase activity toward ERK3 has also been partially purified (55, 56), suggesting that a MAPKK for ERK3 and/or ERK4 may exist. At present, there are no known specific inhibitors of ERK3 and ERK4.
Substrates and biological functions.
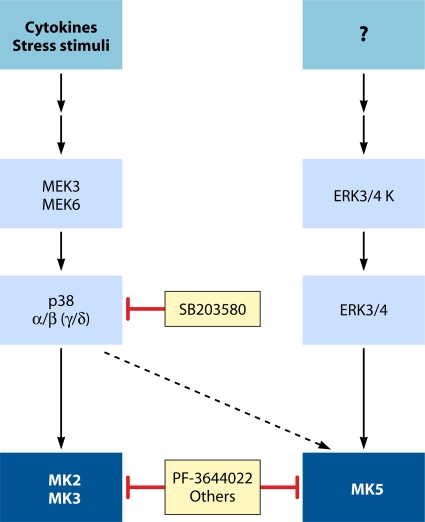
The only known substrate of ERK3/4 is the MAPK-activated protein kinase MK5, which was identified by several groups as a bona fide ERK3/4 phosphorylation target (4, 179, 318, 322). While MK5 is also very poorly understood, the activation mechanisms of MK5 and potential biological functions are described below.
Whereas the biological role of ERK4 is presently unknown, ERK3 has been shown to participate in a number of biological functions, including cell proliferation, cell cycle progression, and cell differentiation. Targeted disruption of the Mapk6 gene (encoding ERK3) leads to intrauterine growth restriction and early neonatal death, suggesting that ERK3 plays an important role during embryogenesis (189). ERK3 has been suggested to be a negative regulator of cell proliferation (73, 173). ERK3 phosphorylation and protein stability were shown to be regulated by Cdk1 and the Cdc14 phosphatase, suggesting that ERK3 function is regulated throughout mitosis (357). ERK3-mediated proliferation arrest may be linked with its putative role in cellular differentiation. Indeed, it was demonstrated that ERK3 expression increases with the differentiation state of neuronal cells as well as myoblasts (35, 73).
ERK7
Identification.
ERK7 was cloned in 1999 by PCR amplification from a rat cDNA library using degenerate primers derived from the kinase domain sequence of ERK1 (2). Thereafter, a probe derived from the sequence of ERK7 was used to identify its human ortholog, which was termed ERK8 (3). ERK7 and ERK8 display 69% amino acid identity, which is lower than what is typical for rodent and human orthologs (71). While ERK7 displays approximately 45% amino acid identity with the kinase domain of ERK1, it is considered atypical partly because it contains a C-terminal extension that is not present in conventional MAPKs (Fig. 1). The precise role of this C-terminal extension is unclear, but it has been shown to play roles in the subcellular localization and autoactivation of ERK7 (1, 2). As is the case for ERK3, the N-terminal region of ERK7 promotes its degradation by the ubiquitin-proteasome pathway (197).
Activation mechanisms and inhibitors.
The ERK7 module is poorly characterized, as there are currently no known MAPKKs involved in its activation (Fig. 2). Despite this, the activation loop of ERK7/8 is composed of a Thr-Glu-Tyr motif, suggesting that a classical MAPKK may exist. In the case of ERK7, these residues appear to be constitutively phosphorylated, as they are not modulated by classical stimuli of conventional MAPKs (1, 2). Evidence suggests that they are regulated by autophosphorylation or, at the very least, that ERK7 activity is required for their regulation (1, 2). Consistent with this, ERK8 has also been shown to autophosphorylate in vitro and in vivo on activation loop residues (3, 188). Conversely to the case for ERK7, certain stimuli of conventional MAPKs have been shown to regulate ERK8 phosphorylation, including serum and H2O2 (3, 188). In addition, expression of an oncogenic allele of Src promotes kinase-inactive ERK8 phosphorylation at the Thr-Glu-Tyr motif (3), suggesting that an unidentified MAPKK phosphorylates ERK8 in trans. At present, there are no known catalytic inhibitors of ERK7/8, complicating the study of these enigmatic kinases.
Substrates and biological functions.
While no in vivo ERK7 substrates have been identified thus far, a number of proteins have been shown to be phosphorylated by ERK7 in vitro, including classical substrates of conventional MAPKs, such as myelin basic protein (MBP), c-Fos, and c-Myc (2). In the case of ERK8, only MBP has been shown to be a productive substrate for this kinase in vitro (188). Despite the lack of bona fide ERK7/8 substrates, both protein kinases play important biological functions, notably in the regulation of cell proliferation (2) and in the response to estrogens (152) and glucocorticoids (307). With regard to MAPKAPKs, there is currently no evidence that ERK7 plays a role in their activation.
NLK
Identification.
Nemo-like kinase (NLK) was identified in 1994 by PCR using degenerate primers derived from conventional MAPK sequences (39). NLK is the ortholog of Nemo, a protein kinase previously identified in Drosophila (60). NLK displays 45% amino acid identity to the kinase domain of ERK2. It is considered atypical because it possesses N- and C-terminal extensions not present in conventional MAPKs and has a single phosphorylatable residue in its activation loop (Fig. 1). The N-terminal extension is not well conserved between NLK orthologs, and its function remains obscure. The C-terminal extension is conserved from worm to human and may contribute to the interaction of NLK with specific substrates (161).
Activation mechanisms and inhibitors.
NLK is activated by stimuli of the Wnt pathway, including Wnt-1 and Wnt-5a (176). Several cytokines have also been reported to activate NLK, including IL-6, granulocyte colony-stimulating factor (G-CSF), and transforming growth factor β (TGF-β) (192, 256). The NLK module is incompletely understood, but the MAPKKK TAK-1 has been reported by several groups to promote NLK activation (161, 256, 336). There are currently no known MAPKKs that directly regulate NLK, but its activation loop is composed of a Thr-Gln-Glu motif (Fig. 1). This motif is reminiscent of the Thr-X-Glu sequence also found in the activation loop of Cdk1, and consistent with this, the kinase domain of NLK is 38% identical to that of Cdk1. Potential MAPKKs that could regulate NLK activation include the kinase homeodomain-interacting protein kinase 2 (HIPK2) (176). This protein was shown to phosphorylate NLK in vitro and in vivo, but at present it is unclear whether HIPK2 phosphorylates the activation loop of NLK or promotes its autophosphorylation. Consistent with the latter, NLK was found by several groups to autophosphorylate in vitro (39, 176).
Substrates and biological functions.
Several NLK substrates have been identified to date, including transcription factors of the T-cell factor/lymphoid enhancer factor (TCF/LEF) family (161), as well as STAT3 (192). In response to Wnt stimulation, NLK has been shown to regulate the β-catenin pathway positively or negatively (161, 336). In Caenorhabditis elegans, NLK activation by TAK-1 promotes phosphorylation of the TCF/LEF transcription factor POP1, a suppressor of β-catenin-dependent transcription (289). Phosphorylation of POP1 by NLK was found to inhibit its activity, thereby promoting Wnt-dependent establishment of the embryo anteroposterior axis. NLK was also shown to directly phosphorylate the proto-oncogene c-Myb and thereby promote its degradation in response to Wnt signaling (176). With regard to MAPKAPKs, there is currently no evidence that NLK plays a role in their activation.
DOCKING INTERACTIONS
MAPK Docking Domains
D domains.
MAPK signaling efficiency and specificity can be achieved in part through specialized docking motifs present in components of the cascade. At least two types of docking interactions between MAPKs and their substrates have been identified, activators and inactivating phosphatases, and both require interaction of short linear sequence motifs present within substrates with a complementary pocket or groove on the kinase. The first docking motif involved in MAPK interaction is the D domain (also referred to as the D site, δ domain, or DEJL domain), which consists of a core of basic residues followed by a hydrophobic patch (Lys/Arg-Lys/Arg-Xaa2-6-Φ-X-Φ, where Φ is a hydrophobic residue, such as Leu, Iso or Val) (reviewed in reference 360). MAPK interactions with D domains have been mapped by mutagenesis, hydrogen exchange-mass spectrometry, and X-ray crystallography (324, 358). Although D domains can sometimes be recognized by more than one group of MAPKs, they are thought to increase signaling specificity and efficacy. D domains lie either upstream or downstream of the phosphoacceptor site and are present on many MAPK regulatory proteins and substrates, including MAPKAPKs (reviewed in references 107 and 123).
DEF domains.
The second major MAPK docking site, known as the DEF domain (Docking site for ERK, FXFP; also called the F site or DEF site), has been identified in a number of ERK1/2 substrates. DEF domains are generally characterized by a Phe-Xaa-Phe-Pro sequence, where one of the Phe residues can also be a Tyr (111, 163, 245). This domain is typically located between 6 and 20 amino acids C terminal to the phosphoacceptor site. DEF domains are required for efficient binding to ERK1/2 (210) and have been shown to be required for ERK1/2-mediated substrate phosphorylation (329). Although generally described as a docking site found in ERK1/2 substrates, the DEF domain in the transcription factor SAP-1 contributes to efficient phosphorylation by p38α (125). Currently, no DEF domains have been identified in MAPKAPKs.
CD domain.
Two groups independently identified a conserved C-terminal common docking (CD) domain outside the catalytic region of ERK, p38, and JNK involved in D domain interactions (304, 358). The CD domain contains acidic and hydrophobic residues, which are necessary for establishing electrostatic and hydrophobic interactions with the positively charged and hydrophobic residues of D domains, respectively (107, 358). The CD domain is prolonged by a specific 2-aa patch which is neutral in ERK1/2 (TT motif) and acidic in p38 isoforms (ED motif), forming a docking groove for their interacting partners. The importance of these docking interactions was nicely demonstrated by ED/TT motif swapping, which rendered ERK2 capable of binding MK3, a normally exclusive p38 substrate (359). It is important to note that the conserved CD domain is dispensable for the interaction of ERK3 and ERK4 with MK5. A recent study demonstrated, using peptide overlay assays, a novel MK5 interaction motif within ERK3/4 that is essential for binding to the C-terminal region of MK5 (5). While MK5 represents the first described ERK3/4 substrate utilizing such an interaction motif, it will be interesting to determine how many more use similar docking mechanisms.
MAPKAPK Docking Motifs
Properties.
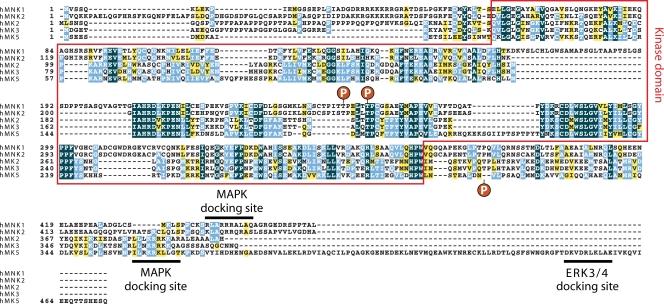
Except for MK5 binding to ERK3/4, all MAPKAPK family members interact with their cognate MAPKs through their C-terminally located D domains, which do not match the best-characterized consensus sequence for such motifs (360). Instead, D domains found in MAPKAPKs appear to fit the kinase interaction motif (KIM) consensus sequence (224). This D domain-related motif corresponds to a hydrophobic residue closely followed by two positively charged Lys or Arg residues (Leu-Xaa2-Lys/Arg-Lys/Arg-Xaa5-Leu) (Fig. 3). A similar motif has also been reported in several other MAPK substrates, including the tyrosine phosphatase PTP-SL (273) and the cyclic AMP (cAMP)-specific phosphodiesterase PDE4D (224).
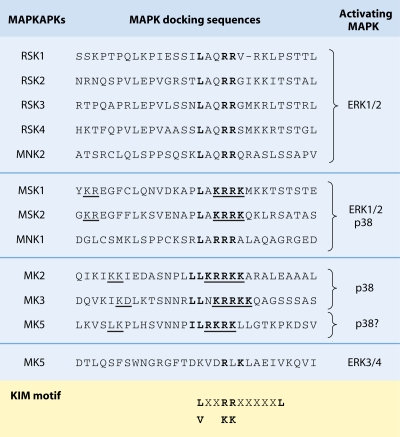
FIG. 3.
Alignment of MAPK-binding sequences from MAPKAPKs. MAPKs bind regions termed D domains that are found in all MAPKAPKs and required for efficient activation. The D domains, shown in boldface, are characterized by a stretch of positively charged residues surrounded by hydrophobic residues. Some D domains mediate the specific interaction with ERK1/2 or p38, while others are necessary for the interaction with both upstream activators. In some MAPKAPKs, the D domain region overlaps with an NLS (underlined). ERK3/4 binding to MK5 requires a degenerate D domain in the C-terminal region of the protein. MAPK-binding sequences in MAPKAPKs do not fit the optimal consensus sequence for D domains but rather fit a D domain-like sequence termed the kinase-interacting motif (KIM). Similar alignments were previously demonstrated in recent review articles (adapted from references 123 and 301 with permission).
MAPK docking specificity arises from variations in D domain sequences, whereby the number of contiguous basic residues appears to correlate with MAPK specificity (338). Generally, ERK1/2-specific MAPKAPKs have two contiguous basic residues, ERK1/2/p38-specific MAPKAPKs have three and four, and p38-specific MAPKAPKs have four and five (Fig. 3). The amount and location of hydrophobic residues within D domains may also regulate specificity, as p38-activated MAPKAPKs have at least two hydrophobic residues before the basic stretch (Fig. 3). Consistent with this, the CD motif in p38 kinases contains more contiguous acidic residues than ERK1/2, suggesting that specific electrostatic interactions between charged residues in CD and D domains provide specificity. Elucidation of the three-dimensional structure of the p38α/MK2 complex (361, 394) demonstrated that a specific Lys residue, present only in MK2 and MK3, specifically interacts with the acidic ED domain of p38α.
As mentioned above, a completely different type of interaction is responsible for MK5 binding to ERK3/4. While the D domain of MK5 has been shown to mediate its interaction with p38, this domain is not required for docking to ERK3/4 (318, 322). A region near the C terminus of MK5 has been shown to mediate binding to ERK3/4, which appears to contain a degenerate D domain sequence (Fig. 3) (4, 179). Additional studies will be required to understand the exact requirements of this atypical interaction region.
Regulation of docking interaction.
D domain-mediated interactions are very stable and are often required for preexisting complexes between inactive MAPKs and their substrates. In some cases, MAPK activation regulates docking interactions with MAPKAPKs, a phenomenon that was first described for the interaction between ERK1/2 and RSK family members (302). Indeed, RSK isoforms bind to ERK1/2 in quiescent cells, but following mitogenic stimulation, the complex transiently dissociates for the duration of ERK1/2 activation. Autophosphorylation of a Ser residue near the D domain of RSK was found to promote ERK1/2 dissociation (302), suggesting that a Ser/Thr phosphatase regulates formation of the corresponding inactive complex. Interestingly, MSK isoforms also have autophosphorylation sites near their D domains (14), but whether ERK1/2 or p38 docking is regulated by phosphorylation remains unknown. MNK1 has been shown to interact more strongly with dephosphorylated ERK2 (389), suggesting that MNK1 phosphorylation may also regulate ERK1/2 or p38 docking. Conversely, phosphorylation of ERK3 and ERK4 in their activation loop sites was found to stabilize their interaction with MK5 (86, 266), indicating that MAPKAPK docking interactions are not always weakened upon MAPK activation.
Docking and subcellular localization.
Despite having similar general structures, a major difference between RSKs and MSKs is their subcellular localization. While MSK1/2 are constitutively found in the nucleus due to the presence of a bipartite NLS within their MAPK docking sequence, RSK1 to -3 are cytoplasmic enzymes in quiescent cells that translocate to the nucleus upon ERK1/2 stimulation (53). RSK4 does not abide by this rule, as it remains cytoplasmic following most types of stimulation (100). The molecular mechanism involved in RSK translocation remains elusive but likely involves regulated docking to ERK1/2 (302). All MNK isoforms contain a polybasic sequence in their N termini that functions as a potent NLS. MNK1 also contains a functional CRM1-type nuclear export signal (NES) and has been shown to shuttle between the cytoplasm and the nucleus (231, 263). The localization of MNK2 is dictated by alternative splicing, as the long form of MNK2 (MNK2A) was also found to shuttle between the cytoplasm and the nucleus, whereas the short form (MNK2B) localizes primarily in the nuclear compartment (312). MK2, MK3, and MK5 are much more dependent on their upstream MAPKs for their localization. All three MAPKAPKs display a functional NLS that overlaps with their D domains (Fig. 3), and as a result, expression of p38α was shown to promote the nuclear export of MK3 and MK5 (251, 359). p38-mediated phosphorylation of MK2/3 was shown to regulate their nuclear export through a mechanism that involves unmasking of the C-terminal NES found within these kinases (236, 249). In the case of ERK3/4-dependent regulation of MK5, this interaction was found to promote cytoplasmic accumulation of MK5 in a D domain-independent manner (4, 179, 318, 322).
MAPK-ACTIVATED PROTEIN KINASES
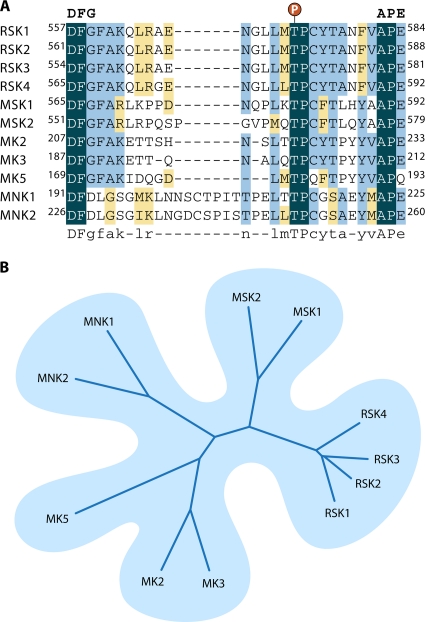
The MAPK-activated protein kinase (MAPKAPK) family contains 11 members (Fig. 4) that are activated by various stimuli depending on their upstream activating kinases (Fig. 1 and 2). Based on homologies within their kinase domains, the MAPKAPKs belong to the calcium/calmodulin-dependent protein kinase (CAMK) family. Of these, the RSK and MSK isoforms contain an additional kinase domain within the same polypeptide, belonging to the AGC (containing PKA, PKG, and PKC families) family of protein kinases (Fig. 4). All MAPKAPK family members share similar activation loop sequences that are targeted for phosphorylation by their cognate upstream MAPKs (Fig. 5 A). Based on overall sequence and activation segment homologies, the MAPKAPKs can be classified into five subgroups, the RSKs, MSKs, MNKs, MK2/3, and MK5 (Fig. 5B), which are discussed in greater detail in the following sections.
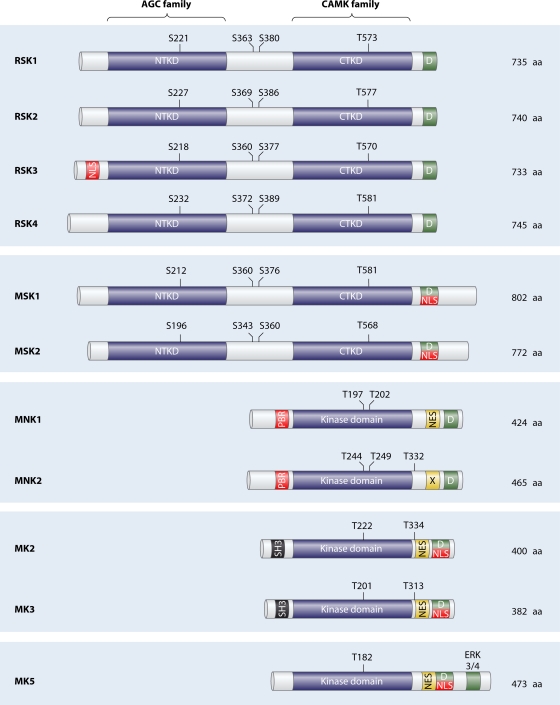
FIG. 4.
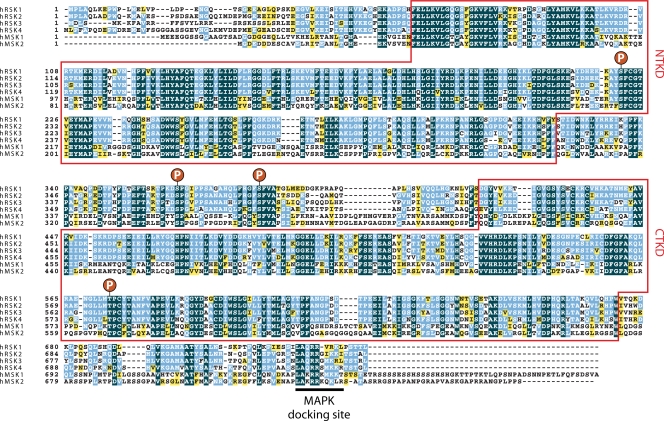
Schematic representation of the overall structure of the MAPKAPKs. While RSK1/2/3/4 and MSK1/2 are composed of two nonidentical kinase domains, the MNKs and MK2/3/5 are single-headed kinases that display homology to the CTKD of RSKs and MSKs. The NTKDs of the RSKs and MSKs are members of the AGC family of kinases, which also includes Akt, PKA, and PKC. The CTKDs of RSKs and MSKs and the kinase domains of MK2/3/5 belong to the CAMK family of kinases, which also include AMPK, DAPK, and CAMK1/2. The amino acid composition of each MAPKAPK along with phosphorylation site numbering refers to the human nomenclature. Only the full-length form of each MAPKAPK protein is included in this diagram. NES, nuclear export signal; NLS, nuclear localization signal; PBR, polybasic region; SH3, Src homology 3 domain; X, nonfunctional NES; D, D domain or MAPK docking site; ERK3/4, docking region for ERK3/4; NTKD, N-terminal kinase domain; CTKD, C-terminal kinase domain.
FIG. 5.
Sequence analysis of the different MAPKAPKs. (A) Alignment of sequences around activation loops of MAPKAPKs reveal conservation of the MAPK phosphoacceptor residue followed by a Pro. In the case of RSK and MSK, activation loop sequences are from their respective C-terminal kinase domains. (B) Phylogenetic tree of MAPKAPK family members. All sequences used for the construction of this tree were human. The relative similarities between all MAPKAPKs reflected by this tree suggest that the MAPKAPKs are comprised within five groups, the RSK, MSK, MNK, MK2/3, and MK5 subfamilies. The CLUSTAL X program was used to generate the multiple alignments on which the tree was based.
RSK
Identification and protein structure.
RSK was first identified in Xenopus laevis extracts (108), and orthologs have since been found throughout metazoans. The human RSK family contains four isoforms (RSK1 [298], RSK2 and RSK3 [174], and RSK4 [299]) that are 73 to 80% identical to each other (Fig. 6). A notable feature of the RSK subfamily of MAPKAPKs is that during evolution, the genes for two distinct protein kinases have fused, generating a single kinase capable of receiving an upstream activating signal from ERK1/2 to the RSK carboxyl-terminal kinase domain (CTKD) and transmitting, with high efficiency and fidelity, an activating input to the RSK amino-terminal kinase domain (NTKD) (Fig. 6). The two kinase domains are distinct and functional (114, 172) and are connected by a regulatory linker region of about 100 aa (Fig. 6). The NTKD belongs to the AGC family of kinases, which also includes protein kinase A (PKA), PKC, Akt, and the S6Ks. The CTKD belongs to the CAMK family, which also includes AMP-activated protein kinase (AMPK), DAPK, and CAMK1/2, and is homologous to the kinase domains found in other MAPKAPKs (Fig. 4). The only known purpose of the CTKD is to activate the NTKD via autophosphorylation (29, 114, 380). In addition, RSK isoforms contain a MAPK docking site that is required for activation by ERK1/2 (127, 338). The D domain in RSK1 to -4 consists of Leu-Ala-Gln-Arg-Arg, where only the Leu and Arg residues are essential for ERK1/2 docking (Fig. 3) (302). Two additional basic residues located C terminal to the D domain also contribute to ERK1/2 docking, but their presence is not essential for RSK1 activation (302).
FIG. 6.
Alignment of the amino acid sequence of the MAPKAPKs containing two kinase domains. Sequences comprising the kinase domains and its subregions are boxed in red and reveal regions of highest homology. The conserved activation loop threonine residue is shown, as well as other conserved phosphorylation sites. The MAPK-binding domain is identified by a line.
Tissue expression and subcellular localization.
Expression of the RSK1 to -3 mRNAs (but not that of RSK4 mRNA) has been shown to be ubiquitous in every human tissue tested (422). While these results support functional redundancy, some tissue variations have been reported and suggest RSK isoform-specific functions. RSK1 is expressed predominantly in the kidney, lung, and pancreas, whereas both RSK2 and RSK3 are highly expressed in skeletal muscle, heart, and pancreas (7, 241, 422). In the brain, RSK expression is as follows: RSK1 is expressed predominantly in the granular cell layer of the cerebellum; RSK2 in the neocortex, hippocampus, and cerebellum; and RSK3 in the cerebral cortex, the dentate gyrus, and the amygdala (149, 422). RSK4 expression is much lower than those of other RSKs. RSK4 was found to be expressed in the brain, heart, cerebellum, kidney, and skeletal muscle, whereas other tissues, such as lung, liver, pancreas, and adipose tissue, showed no detectable RSK4 expression (100).
During mouse development, RSK2 was found to be expressed at a very low level compared to RSK3, whose mRNA is very abundant in fetal tissues (191). RSK3 expression can be detected in the ventricular zone, a site of high proliferative activity. Conversely, RSK1 expression is strongest in the neuroepithelium of the forming neural tube, whereas at later stages it decreases dramatically and becomes undetectable in the nervous system. These results are consistent with a temporal regulation of RSK1 and RSK3 expression and support the requirement of RSK1 in early and RSK3 in later development of the nervous system. At late stages of development, RSK1 is highly expressed in regions harboring highly proliferating cells. These include liver, lung, thymus, olfactory, and gut epithelia. RSK4 is ubiquitously expressed throughout development but at a very low level in all tissues examined (191). While both the Rsk1 and Rsk3 genes give rise to only one transcript, Northern analysis of Rsk2 expression revealed the alternative use of two different polyadenylation sites giving rise to two transcripts of 3.5 and 8.5 kb (422). Similarly, two secondary Rsk4 transcripts (5 and 9 kb) also exist, but whether they result from alternative splicing or alternative polyadenylation remains unknown (415).
At the subcellular level, RSK1 to -3 are usually present in the cytoplasm of quiescent cells, but upon stimulation, a significant proportion of these proteins translocates to the nucleus (53, 213, 284, 375, 429). Within minutes of stimulation, RSK1 was shown to accumulate transiently at the plasma membrane, where it presumably receives additional inputs necessary for activation before nuclear translocation (284). RSK4 appears to be predominantly cytoplasmic (100), but in contrast to the case for other RSK isoforms, RSK4 does not significantly accumulate in the nucleus following mitogenic stimulation. The mechanisms involved in RSK translocation to the nucleus remain elusive, but the small death effector domain protein PEA-15 (phosphoprotein enriched in astrocytes 15 kDa) has been shown to interact with RSK2 and inhibit its nuclear translocation (375). Interestingly, RSK2 was recently found to localize to stress granules upon oxidative stress (102), suggesting the possibility that PEA-15 may also localize to these structures. While the mechanisms responsible for the nuclear translocation of the RSK isoforms remain unknown, RSK3 is the only human isoform to possess an optimal NLS, consisting of Lys-Lys-Xaa10-Leu-Arg-Arg-Lys-Ser-Arg, but the functionality of this domain has never been tested. Compared to other RSK isoforms, RSK3 is often found associated with insoluble cellular fractions, indicating that RSK3 may localize to specific cellular compartments (P. P. Roux, unpublished data).
Activation mechanisms and inhibitors.
RSK activation requires ordered phosphorylation events mediated by ERK1/2, phosphoinositide-dependent protein kinase 1 (PDK1), and RSK autophosphorylation. All RSK isoforms, including C. elegans and Drosophila melanogaster RSK orthologs, contain the four essential phosphorylation sites (Ser221, Ser363, Ser380, and Thr573 in human RSK1) responsive to mitogenic stimulation (Fig. 4) (78). The current model of RSK activation is that RSK and ERK1/2 form an inactive complex in quiescent cells (156, 428). Upon mitogenic stimulation, ERK1/2 (and probably ERK5) phosphorylate Thr573 located in the activation loop of the CTKD (280, 338, 352) and Thr359/Ser363 in the linker region (78). Activation of the CTKD leads to autophosphorylation at Ser380 located within a hydrophobic motif (380), which creates a docking site for PDK1 (119). For RSK2, this interaction has been shown to increase the catalytic activity of PDK1 by severalfold, indicating that this motif functions to both recruit and activate PDK1. PDK1 is required for mitogenic stimulation of RSK1 to -3, but surprisingly, RSK4 does not appear to require PDK1 to maintain its high basal activity (100). PDK1 association with RSK1 to -3 leads to phosphorylation of Ser221 in the activation loop of the NTKD (167, 285), resulting in full RSK activation (Fig. 7). Recent evidence indicates that RSK2 is also phosphorylated on Tyr residues in response to fibroblast growth factor receptor (FGFR) (177) and Src activation (178). These phosphorylation events were found to stabilize ERK1/2 binding to RSK2 and to promote subsequent activation of RSK2, suggesting an alternative mechanism for RSK activation in human tumors with activated FGFR3 signaling and in response to normal EGF receptor activation.
FIG. 7.
Signaling cascades leading to activation of the RSKs, MSKs, and MNKs. RSK1/2/3/4 are activated by two inputs originating from ERK1/2 or ERK5, as well as PDK1 enzymes. Similarly, MSK1/2 are activated by ERK1/2 but also by stimuli that activate the p38 module. Depending on the isoforms, MNKs can be stimulated by either ERK1/2 or both ERK1/2 and p38. Different inhibitors of components within these cascades are also shown. Dotted lines indicate that, although reported, substrate regulation by the respective kinase remains to be thoroughly demonstrated.
Mutational inactivation of the CTKD was shown to only partially inhibit activation of the NTKD of RSK1 (62, 302), suggesting that Ser380 phosphorylation may also occur in a CTKD-independent manner (67, 284). Interestingly, the related MK2/3 enzymes were found to phosphorylate Ser380 in certain cell types, which may explain how various stresses that stimulate p38 lead to RSK activation (419). Aside from being involved in RSK phosphorylation, ERK1/2 may also promote RSK1 activation by facilitating its recruitment to the plasma membrane, as suggested by the constitutive activation of a RSK mutant with a myristoylation sequence (284). The process of RSK activation is closely linked to ERK1/2 activity, and MEK1/2 inhibitors (U0126, PD98059, and PD184352) have been used extensively to study RSK function. Recently, three different classes of RSK inhibitors targeting the NTKD (SL-0101 and BI-D1870) or the CTKD (fluoromethyl ketone [FMK]) have been identified (68, 308, 339). While BI-D1870 and SL-0101 are competitive inhibitors with respect to ATP, FMK is an irreversible inhibitor that covalently modifies the CTKDs of RSK1, RSK2, and RSK4. These compounds have been tested against a panel of protein kinases and found to be relatively specific for the RSK isoforms (Fig. 7) (18).
A recent study identified a new point of cross talk between the PKA and ERK1/2 signaling pathways (49). Inactive RSK1 was found to interact with the PKA regulatory I subunit and thereby sensitize PKA to cAMP. However, activation of RSK promotes its interaction with the PKA catalytic subunit, which was found to decrease the ability of cAMP to stimulate PKA. RSK inactivation may require the phosphatase PP2Cδ, which was found to associate with RSK1 to -4 (92). Inactivation of RSK1 may also involve its autophosphorylation at Ser732, which was found to promote ERK/RSK dissociation and correlate with reduced RSK kinase activity (302).
Substrates and biological functions.
An important clue about the physiological roles of RSK came from the finding that mutations in the Rps6ka3 gene (coding for RSK2) were the cause of Coffin-Lowry syndrome (CLS) (Table 1) (370). CLS is an X-linked mental retardation syndrome characterized by psychomotor retardation and facial, hand, and skeletal malformations (145). Numerous mutations have been identified in the Rps6ka3 gene, most of which result in truncated or inactive RSK2 proteins (85). RSK2 knockout mice have impaired learning and cognitive functions, as well as poor coordination compared to wild-type littermates (99, 271). In addition, they develop a progressive skeletal disease, osteopenia, due to cell-autonomous defects in osteoblast activity (80). Both c-Fos and ATF4 transcription factors have been proposed to be critical targets of RSK2 mediating its effects in osteoblasts (80, 413). RSK2 knockout mice display additional phenotypes. They are approximately 15% smaller than their wild-type littermates, with a specific loss of white adipose tissue that is accompanied by reduced serum levels of the adipocyte-derived peptide leptin (103). RSK1/RSK2/RSK3 triple knockout mice are viable, but no other information regarding their phenotype has yet been reported (101). The Rps6ka6 gene (which codes for RSK4) is located on chromosome X and was suggested to be involved in nonspecific X-linked mental retardation, but definitive evidence remains to be provided (415). Interestingly, deletion of Drosophila RSK was found to result in defects in learning and conditioning (274), but whether these deficits result from the specific loss of RSK activity or deregulated ERK activation or function will require further investigation. Recent evidence has shown that deletion of the Rps6ka6 gene in the mouse results in early developmental defects, but limited information which would explain how RSK4 regulates embryogenesis was provided (74).
TABLE 1.
Physiological roles of the MAPKAPKs
| MAPKAPK | Gene symbol | Alternate name(s) | Human gene location | Associated disease(s) (reference[s]) | Phenotype(s) of knockout mouse (reference[s]) |
|---|---|---|---|---|---|
| RSK1 | Rps6ka1 | HU-1, MAPKAP-K1A, S6K-alpha 1, p90RSK1 | 1p36-p35 | Expressed at high level in breast and prostate tumors (65, 339) | Viable (101) |
| RSK2 | Rps6ka3 | HU-3, MAPKAP-K1B, MRX19, S6K-alpha 3, ISPK-1, p90RSK2 | Xp22.13 | Coffin-Lowry syndrome (370), expressed at a higher level in breast and prostate tumors (65, 339) | Viable and fertile, but knockout animals weigh 10% less than wild-type counterparts (99); reduction in adipose mass, as well as reduced serum leptin levels (103); impaired cognition and coordination skills (271); impaired T-lymphocyte activation (216) |
| RSK3 | Rps6ka2 | HU-2, MAPKAP-K1C, S6K-alpha 2, p90RSK3 | 6q27 | Potential tumor suppressor in ovarian cancer (28), RSK3 mutations are common in cancer (136) | Viable (101) |
| RSK4 | Rps6ka6 | S6K-alpha 6, p90RSK4 | Xq21 | Possible role in nonspecific X-linked mental retardation (415), aberrant expression in breast cancer (362) | Developmental defects (74) |
| MSK1 | Rps6ka5 | MSPK1, RLPK | 14q31-q32.1 | None reported | No obvious phenotype (15) |
| MSK2 | Rps6ka4 | RSK-B | 11q11-q13 | Located in the locus of Bardet-Biedl syndrome (437) | MSK2 and MSK1/2 double knockouts have no obvious phenotype (398); MSK1/2 play a negative role in innate immune system signaling (10) |
| MNK1 | Mknk1 | 1p34.1 | None reported | No obvious phenotype (373) | |
| MNK2 | Mknk2 | 19p13.3 | None reported | No obvious phenotype (373) | |
| MK2 | Mapkapk2 | MAPKAP-K2 | 1q32.1 | None reported | Knockout mice show increased resistance to endotoxic shock and increased susceptibility to infections (195, 211) |
| MK3 | Mapkapk3 | MAPKAP-K3, 3pK | 3p21.2 | Commonly deleted in small cell lung cancer (334) | No obvious phenotype; deletion of MK3 worsens the phenotype of the MK2 knockout mouse (292) |
| MK5 | Mapkapk5 | MAPKAP-K5, PRAK | 12q24.12 | None reported | No obvious phenotype (331); MK5-deficient mice are more susceptible to skin cancer (351) |
The substrate specificity of RSK1 for target phosphorylation has been determined using synthetic peptide libraries and was found to require the minimum sequences Arg/Lys-Xaa-Arg-Xaa-Xaa-pSer/Thr or Arg-Arg-Xaa-pSer/Thr (212). These data were confirmed using an arrayed positional scanning peptide library, except that results indicated a stronger requirement for Arg residues at positions −3 and −5 than previously thought (P. P. Roux, unpublished data). These analyses also revealed that RSK1 prefers to phosphorylate Ser rather than Thr residues by a factor of at least 5-fold, and consistent with this, the majority of RSK substrates found to date are phosphorylated on Ser residues (Fig. 8).
FIG. 8.
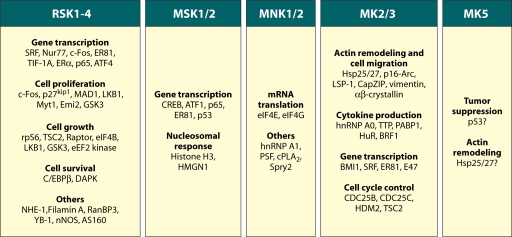
Biological functions and substrates of the MAPKAPKs. Upon activation, the RSKs, MSKs, MNKs, and MK2/3/5 phosphorylate several substrates and regulate many biological responses. The list of substrates indicated in this figure is not exhaustive but emphasizes the many important substrates identified to date for the different MAPKAPKs.
Although a number of RSK functions can be deduced from the nature of its substrates, data from many groups point toward roles for the RSKs in nuclear signaling, cell cycle progression and cell proliferation, cell growth and protein synthesis, and cell survival. Whereas more substrates have been identified for RSK2 than for any other RSK isoforms, most studies have not determined isoform selectivity. Therefore, many known substrates of RSK2 may be shared by different RSK family members. In addition, many AGC family members share the same consensus phosphorylation motif as RSKs and MSKs, suggesting that substantial overlap in function may exist between RSK and other AGC-related kinases. Indeed, several RSK substrates have been shown to be targeted by Akt, including TSC2, glycogen synthase kinase 3 (GSK3), and p27kip1. This also needs to be kept in mind when analyzing phenotypes of knockout mice, as compensation between MK2 and MK3, and probably also other MAPKAPK family members, has been shown to occur.
(i) Nuclear signaling.
Activated RSK phosphorylates several transcription factors, some of which contribute to the IE gene response or are IE gene products themselves. Two studies using human cells from CLS patients and primary fibroblasts isolated from _Rsk2_−/− mice showed that RSK2 mediates mitogen-induced c-Fos transcription (40, 84). Two distinct mechanisms were proposed, including activation of the Elk1/serum response factor (SRF) complex as well as phosphorylation of the cAMP response element-binding protein (CREB) by RSK2. While CREB was also shown by others to be phosphorylated by RSK2 at Ser133 (133, 407), more recent evidence indicated that the related MSK protein kinases are the predominant CREB kinases operating in somatic cells (398). While the MSKs were shown to be required following mitogenic stimulation and cellular stresses, some residual CREB phosphorylation was still present in cells derived from _Msk1_−/−_Msk2_−/− mice, suggesting that RSK and/or PKA cooperates with MSK1/2 in the phosphorylation of CREB. Related to this, histone H3 was also suggested to be regulated by RSK, but conclusive evidence from the same group demonstrated that MSK1/2 fulfill this function in response to both stress and mitogenic stimulations (341). At the posttranslational level, RSK1 phosphorylates the IE gene products SRF (287), c-Fos (51), and Nur77 (82, 114, 400).
RSK also interacts with the ETS transcription factor ER81 and enhances ER81-dependent transcription by phosphorylating Ser191 and Ser216 (404). ER81 performs many essential functions in homeostasis, signaling response, and development, potentially implicating RSK1 in these processes. The transcription initiator factor TIF-1A also becomes phosphorylated by ERK1/2 and RSK2 following serum stimulation on two Ser residues important for TIF-1A function (427). TIF-1A is required for RNA polymerase I transcription and rRNA synthesis, suggesting that RSK signaling regulates growth-related transcription initiation. The estrogen receptor α (ERα) is an ERK1/2 substrate that becomes activated following stimulation with growth factors. RSK1 associates with and phosphorylates ERα on Ser167, which increases ERα-mediated transcription (170, 410). Another transcription factor, microphthalmia (mi), has been shown to be phosphorylated by the RSKs (406) and has been linked to malignant melanoma (261).
Additional nuclear factors are regulated by RSK, including the NF-κB transcription factor. RSK1 phosphorylates the inhibitory factors IκBα and IκBβ on sites that promote their degradation, thereby stimulating NF-κB activity (131, 317, 409). RSK was also suggested to promote p65 phosphorylation at Ser536 (32, 424), but the role of RSK in phosphorylation of this site remains a subject of debate (94). RSK2 phosphorylates the CREB family member activating factor 4 (ATF4) (413), a transcription factor required for the timely onset of osteoblast differentiation during development. RSK2 was found to be required for osteoblast differentiation and function, suggesting a mechanism by which loss of RSK2 leads to CLS-associated skeletal abnormalities. The transcriptional coactivator CREB-binding protein (CBP) has been identified as a binding target of RSK1 (247). Stimulation of the Ras/MAPK pathway promotes the interaction between RSK1 and CBP (247, 388), but the exact outcome of this association remains to be determined. Interestingly, CBP also interacts with transcription factors that are phosphorylated by RSK1 and RSK2, such as CREB, c-Fos, ER81, ERα, and NF-κB, suggesting that RSK may help in the recruitment of CBP and p300 cofactors to promoters regulated by these transcription factors.
(ii) Cell cycle progression and cell proliferation.
Based on the nature of its substrates, RSK is thought to play roles in cell cycle progression and cell proliferation. A recently identified inhibitor of RSK, termed SL-0101 (339), has been tested for its antiproliferative potency. Treatment of cells with SL0101 or RNA interference against RSK1 and RSK2 was found to inhibit proliferation of human prostate and breast cancer cell lines (P. P. Roux, unpublished data) (65, 339). Accordingly, RSK1 and RSK2 were shown to be overexpressed in tumors of the breast and prostate (65, 339), indicating that these two isoforms positively regulate cancer cell proliferation.
The best-characterized molecular mechanism implicating RSK in cell cycle progression involves the regulation of c-Fos, which promotes expression of cyclin D1 during G1/S transition. RSK2-mediated phosphorylation of c-Fos at Ser362 promotes its stability and oncogenic properties and was found to be essential for osteosarcoma formation in mice (52, 80, 245). RSK1 and RSK2 may also promote G1-phase progression by phosphorylating the CDK2 inhibitor p27kip1 on Thr198 (120, 205). RSK-mediated phosphorylation of p27kip1 promotes its association with 14-3-3, which prevents its translocation to the nucleus (120). MAX dimerization protein 1 (MAD1) is a suppressor of Myc-mediated cell proliferation and transformation and was recently identified as an RSK1/2 substrate (436). Phosphorylation by RSK on Ser145 accelerated the degradation of MAD1 by the ubiquitin-proteasome pathway, resulting in increased Myc-dependent transcription. RSK2 phosphorylates the tumor suppressor LKB1, a kinase found mutated in the cancer-prone Peutz-Jeghers syndrome (309). Phosphorylation of LKB1 on Ser431 was not found to affect its activity or membrane association (309), but its phosphorylation was shown to be necessary to prevent LKB1-mediated growth suppression in melanomas through an unknown mechanism (430).
RSK2 appears to be a critical regulator of cell transformation, as ectopic expression of RSK2 was found to increase proliferation as well as anchorage-independent transformation (59). Interestingly, RSK was recently identified as a key effector of Ras/ERK-mediated epithelial-mesenchymal transition (EMT) (93). The transcriptional program initiated by RSK was found to coordinately modulate the extracellular environment, the intracellular motility apparatus, and receptors mediating communication between these compartments to stimulate motility and invasion. In accordance with a role for RSK1 and RSK2 in tumorigenesis, FGFR3 has been recently shown to promote hematopoietic transformation by activating RSK2 in a two-step fashion, by facilitating both ERK-RSK2 interaction and subsequent phosphorylation of RSK2 by ERK (177).
Although RSK3 was initially suggested to play positive roles in cell proliferation (429), this isoform was recently shown to act as a potential tumor suppressor in ovarian cancer (28). RSK3 mutations have been found in a cancer genome sequencing study which evaluated their likelihood of being driver mutations to be very high, providing strong evidence that RSK3 is a tumor suppressor (136). Overexpression of RSK3 was found to reduce proliferation by causing G1 arrest and increasing apoptosis, but the mechanisms by which RSK3 negatively regulates cell proliferation are currently unknown. One hypothesis is that overexpression of RSK3 may promote a negative feedback loop that suppresses the Ras/ERK cascade (95), but a thorough investigation will be required to fully validate this. The role of RSK4 in proliferation is even more enigmatic. Recent evidence suggests that RSK4 plays an inhibitory role during embryogenesis by negatively regulating RTK signaling (246). RSK4 was also found to participate in p53-dependent cell growth arrest (23) and in oncogene-induced cellular senescence (220), indicating that this isoform behaves somewhat like a tumor suppressor. Consistent with this, exogenous expression of RSK4 resulted in decreased breast cancer cell proliferation and increased accumulation of cells in the G0/G1 phase of the cell cycle (363).
The role of RSK in G2/M progression has been demonstrated by many groups using the preferred model of Xenopus oocyte maturation. Immature oocytes are arrested in the G2 phase of the first meiotic cell division. Addition of progesterone induces the synthesis of the MAPKKK c-Mos, which in turn activates the MEK1-ERK-RSK cascade, leading to M-phase entry and subsequent maturation to an unfertilized egg. M-phase entry is controlled in part by Cdc2, which is a CDK normally kept in check by dual phosphorylation on both Thr14 and Tyr15 by the inhibitory kinase Myt1. RSK2 is the prominent RSK isoform in Xenopus oocytes (24), and using this model system, RSK2 was shown to contribute to the control of the meiotic cell cycle at several critical points (316). One mechanism by which RSK2 participates in the progression of oocytes through the G2/M phase of meiosis I is through phosphorylation and inhibition of the Myt1 kinase (137, 260, 305). It remains unknown whether this mechanism is conserved in other species, but recent efforts demonstrated that Akt can also act as a Myt1 kinase in starfish oocytes (257).
Another way by which RSK can modulate the meiotic cell cycle in Xenopus is through ERK-mediated metaphase II arrest, an activity known as cytostatic factor (CSF) (25, 138). RSK1 phosphorylates and activates in vitro the kinase Bub1, a mediator of anaphase-promoting complex (APC) inhibition (319), suggesting that RSK1-mediated Bub1 activation contributes, at least in part, to metaphase II arrest (371). Emi2 (also called Erp1) is another APC inhibitor that was initially thought to function independently from the ERK pathway. Three independent studies recently demonstrated that Emi2 is in fact a substrate for RSK. Phosphorylation of Emi2 by RSK promotes Emi2-PP2A association, facilitating Emi2 dephosphorylation at specific Cdc2 phosphorylation sites, which in turn enhances Emi2 stability and function (160, 255, 405).
(iii) Cell growth and protein synthesis.
RSK1 was originally identified as an in vitro ribosomal protein S6 (rpS6) kinase (108, 109), but S6K1 and S6K2 were later shown to be the predominant rpS6 kinases operating in somatic cells (64). Recent evidence with _S6k1_−/−_S6k2_−/− cells (265) and rapamycin-treated cells (303) suggests that the RSK isoforms also contribute to rpS6 phosphorylation in vivo. Whereas S6K1/2 phosphorylate all sites on rpS6, RSK1/2 specifically phosphorylate Ser235 and Ser236 in response to Ras/MAPK pathway activation (303). These findings indicated that rpS6 phosphorylation also occurs in an mTOR-independent manner. RSK-mediated rpS6 phosphorylation was found to facilitate assembly of the translation preinitiation complex and to correlate with increased cap-dependent translation (303), providing an additional oncogene- and mitogen-regulated input linking the Ras/MAPK signaling pathway to the regulation of translation initiation.
Ras/MAPK signaling was also found to stimulate mTOR activity through the regulation of the tuberous sclerosis complex (TSC). mTOR is a master regulator of protein synthesis and cell growth, and its activity is controlled by several growth-related pathways. RSK and ERK phosphorylate TSC2 at Ser1798 and Ser664, respectively, which negatively regulates the guanine nucleotide-activating protein (GAP) activity of TSC2 toward the small GTPase Rheb (222, 291, 300). More recently, RSK was shown to phosphorylate Raptor, an important interacting partner of mTOR, providing another link between the Ras/MAPK and mTOR signaling pathways (47). RSK may also regulate mRNA translation through the phosphorylation of GSK3β (11, 353). RSK1-mediated phosphorylation of GSK3β on Ser9 inhibits its kinase activity and thereby releases inhibition on the translation initiation factor eukaryotic initiation factor 2B (eIF2B) (385). Interestingly, activated GSK3β and the LKB1-activated kinase AMPK were both shown to phosphorylate and activate TSC2 (158, 159), suggesting that RSK may inhibit TSC2 activity using direct and indirect mechanisms. Finally, RSK was shown to phosphorylate the eEF2 kinase (386) and the translation initiation factor eIF4B (323), underscoring the involvement of RSK at multiple levels of the pathway leading to protein synthesis.
(iv) Cell survival.
RSK1 and RSK2 have been shown to positively regulate cell survival in different cell types. Both RSK isoforms phosphorylate the proapoptotic protein Bad on Ser112, thereby enhancing its ability to bind, and be inactivated by, cytosolic 14-3-3 proteins (34, 332). RSK1 also promotes survival of hepatic stellate cells by phosphorylating C/EBPβ Thr217 in response to the hepatotoxin CCI4 (41). Phosphorylation of Thr217 was suggested to create a functional XEVD caspase inhibitory box that binds and inhibits caspases 1 and 8. More recently, RSK1 and RSK2 were shown to phosphorylate and inactivate death-associated protein kinase (DAPK). Phosphorylation of DAPK at Ser289 inhibits its proapoptotic activity and results in increased cell survival in response to mitogenic stimulation (12). DAPK behaves as a tumor suppressor, and its expression is commonly silenced in tumors through DNA methylation (26). RSK also promotes cell survival through transcription-dependent mechanisms. Indeed, RSK2-mediated phosphorylation of the transcription factor CREB was shown to promote survival of primary neurons through increased transcription of survival-promoting genes, including those for Bcl-2, Bcl-XL, and Mcl-1 (34, 407). More recently, RSK1 was found to promote survival through the activation of the transcription factor NF-κB (131, 317, 423).
(v) Other substrates.
In addition to regulating transcriptional and translational programs related to cell growth and proliferation, the RSK isoforms have been shown to phosphorylate many more substrates involved in diverse cellular processes. RSK2 phosphorylates the Na+/H+ exchanger isoform 1 (NHE-1) (356), a key member of a family of exchangers that regulate intracellular pH and cell volume (355). RSK2-mediated phosphorylation of NHE-1 on Ser703 was found to regulate mitogen-dependent Na+/H+ exchange and intracellular pH (356). RSK2 may also phosphorylate on Ser1152 the cell adhesion molecule L1 (402), a protein that becomes hyperphosphorylated during periods of high neuronal activity, suggesting the involvement of RSK2 in neurite outgrowth. Recently, RSK1 was suggested to play a role in membrane ruffling by phosphorylating the cytoskeleton-associated protein filamin A on Ser2152 (403). Ser2152 was previously shown to be phosphorylated by PAK1 and to be necessary for membrane ruffling in response to PAK1 activation (374), suggesting that RSK1 may play a similar role in actin reorganization.
Ran-binding protein 3 (RanBP3) phosphorylation by RSK modulates nucleocytoplasmic protein transport. Both RSK and Akt phosphorylate Ser58 of RanBP3, and that phosphorylation event contributes to the formation of a Ran gradient essential for nucleocytoplasmic transport, kinetochore function, spindle assembly, microtubule dynamics, and other mitotic events (417). RSK phosphorylates the transcription/translation factor Y box-binding protein-1 (YB-1) at Ser102 and thereby promotes its nuclear accumulation (349). YB-1 is overexpressed in a number of cancer types, suggesting that RSK signaling may contribute to mediating the oncogenic functions of YB-1. RSK1 phosphorylates neuronal nitric oxide synthase (nNOS) on Ser847 in cells treated with mitogens, leading to the inhibition of NOS activity (342). Following EGF treatment, nNOS was phosphorylated by RSK1 in hippocampal and cerebellar neurons, suggesting a novel role for RSK in the regulation of nitric oxide function in the brain. RSK has been shown to phosphorylate the Akt substrate of 160 kDa (AS160), a protein implicated in the translocation of the glucose transporter 4 (GLUT4) to the plasma membrane in response to insulin (128). Deregulation of GLUT4 translocation occurs early in the pathophysiology of insulin resistance and type 2 diabetes.
MSK
Identification and protein structure.
The mitogen- and stress-activated kinases 1 and 2 (MSK1 and MSK2) were discovered by two groups through genome-wide homology searches (83, 253). Simultaneously, MSK2 was detected in a yeast two-hybrid screen using p38α as bait (268). Human MSK1 (also known as RSK-like protein kinase or RLPK) and MSK2 (RSK-B) are 63% identical to each other and display significant homology to the RSKs (about 40% identity) (Fig. 5B). Similar to the RSKs, MSK1/2 possess two autonomous kinase domains within the same polypeptide (Fig. 6), a feature that is conserved in MSK orthologs from different species, such as C. elegans (C54G4) and Drosophila (JIL-1) (169). Although the role of C54G4 is unknown, JIL-1 is essential for viability in Drosophila and was found to mediate histone H3 phosphorylation (169). Similar to the RSKs, the NTKD of MSK1/2 belongs to the AGC family of kinases. The CTKD of MSK1/2 has a CAMK-like sequence and is mostly homologous to the kinase domain of MK2/3 (about 40% amino acid identity). Both MSK isoforms interact with ERK1/2 and p38 isoforms through their MAPK-binding domains (Leu-Ala-Lys-Arg-Arg-Lys), located near the C terminus of the protein (Fig. 3) (366).
Tissue expression and subcellular localization.
Analysis of MSK1 and MSK2 expression in different tissues revealed that they are ubiquitously expressed, with predominant expression of MSK1 and MSK2 in the brain, heart, placenta, and skeletal muscle (83). Interestingly, the Rps6ka4 gene (coding for MSK2) maps to the BBS1 locus on chromosome 11 (437). Bardet-Biedl syndrome (BBS) is a genetically heterogeneous disorder characterized primarily by retinal dystrophy, obesity, polydactyly, renal malformations, and learning disabilities (183). Although MSK2 inactivation has not yet been shown to contribute to BBS, it is worth noting that clinical symptoms of BBS are somewhat similar to those of CLS, a syndrome caused by RSK2 inactivation (Table 1).
The C terminus of MSK1/2 contains a functional bipartite NLS (Lys-Arg-Xaa14-Lys-Arg-Arg-Lys-Gln-Lys in MSK2) (Fig. 4), conferring an almost exclusively nuclear localization in both serum-starved and stimulated cells (83, 268). Consistent with such localization, MSK1/2 have been shown to regulate mainly nuclear events (14, 378). Although MSKs do not appear to translocate following activation, expression of MSK2 was found to regulate the localization of ectopically expressed p38α and ERK1 (268). These results indicate that MSK1/2 may control the cellular localization of their upstream activators, ERK1/2 and p38, a finding that was also observed with the closely related MK2/3/5 (20).
Activation mechanisms and inhibitors.
In cells, MSK1 and MSK2 are potently activated by mitogens and stress stimuli that promote ERK1/2 and p38 activation (83, 268). In vitro, both ERK1/2 and p38 directly phosphorylate the same sites on MSK1/2, resulting in their activation (83, 230). Consistent with these original observations, specific inhibitors of p38α and p38β (SB203580) and MEK1/2 (U0126, PD98059, and PD184352) block MSK1/2 activation in a stimulus-dependent manner (Fig. 7) (83, 366, 398). Contribution of the ERK5 module in MSK activation has not been studied in detail, but certain evidence suggests that ERK5 does not regulate MSK1/2 phosphorylation and activation (229).
MSK and RSK share many characteristics that first suggested they would have similar activation mechanisms (118). First, as stated above, they both contain two distinct and functional kinase domains, a rare characteristic for protein kinases (Fig. 4). Inactivation of either kinase domain through mutation of conserved residues completely blocks NTKD activity of MSK1/2 (83, 268, 367), indicating that unlike the case for the RSKs (29, 62, 302), MSK1/2 activation critically requires CTKD activation (62). Second, RSKs and MSKs share the four key phosphorylation sites essential for activation (Fig. 4), but regulation of these sites was found to be very different in MSKs. While PDK1-mediated phosphorylation of the NTKD was shown to be essential for RSK activation, analysis of _Pdk1_-null cells revealed that this kinase is not required for MSK1 activation (69, 399). Consistent with this, phosphorylation of the NTKD activation loop is mediated by autophosphorylation in MSK isoforms (229, 230). Whereas PDK1 is essential for activation of RSK1 to -3 (119), RSK4 appears to use autophosphorylation mechanisms similar to those of MSKs for activation (100), suggesting that they may share other similarities.
The current model of MSK activation suggests that MSK1/2 interact with ERK1/2 and p38 isoforms via the C-terminal MAPK docking site (Fig. 3). These upstream MAPKs then phosphorylate three Pro-directed residues in MSK1: a site in the linker between the two kinase domains (Ser360), the activation loop of the CTKD (Thr581), and a site in the C terminus (Thr700). Phosphorylation of Thr700 acts to reduce CTKD inhibition caused by an autoinhibitory C-terminal sequence (230). In combination with Thr581, phosphorylation of Thr700 activates the CTKD, which can then autophosphorylate two sites in the linker region (Ser376 and Ser381) as well as the activation loop of the NTKD (Ser212). This results in NTKD activation, which can then phosphorylate exogenous substrates (14, 378).
Based on their homology to the RSKs, the MSKs were predicted to contain additional phosphorylation sites in their C-terminal regions. Consistent with this, three residues (Ser750, Ser752, and Ser758) were found to be autophosphorylated by the NTKD upon MSK activation (229). While similar sites in RSK isoforms regulate ERK1/2 docking (302), these sites do not appear to play a similar function in MSK1 (229). Although the MAPK-binding site of MSK1/2 is located near their C termini, MSK1 and MSK2 have an additional 20 to 50 aa C terminal to the docking site, suggesting that this region contains additional elements required for ERK1/2 and p38 association. Finally, several additional phosphorylation sites were recently identified in MSK1, but these are not required for MSK1 activation (230, 330). While the kinase(s) responsible for the phosphorylation of these sites remains to be identified, recent evidence suggest that casein kinase 2 (CK2) regulates MSK1 phosphorylation at multiple C-terminal residues following UV irradiation (162, 330).
Several compounds have been reported to inhibit MSK1/2 in cells (378), but none of them are selective, as they inhibit several kinases related to MSKs. MEK1/2 and p38 inhibitors can be used to prevent MSK activation, but both MAPK modules may need to be inhibited simultaneously to completely block MSK activation.
Substrates and biological functions.
Although mice deficient in MSK1 and MSK2 do not display obvious phenotypes (15, 398), a relatively recent study indicated that MSK1/2-deficient animals are hypersensitive to lipopolysaccharide-induced endotoxic shock and showed prolonged inflammation in a model of toxic contact eczema (10) (Table 1). The authors of that study suggested that a lack of MSK1/2 signaling results in decreased expression of the phosphatase DUSP1 and the cytokine IL-10, which are normally involved in a negative feedback mechanism responsible for dampening the innate immune response.
The substrate specificity of the MSKs resembles that of other AGC family members, particularly RSK and PKA, with the minimal consensus substrate sequence Arg-Xaa-Xaa-pSer/Thr (83). In cells, both MSK and RSK can be activated by similar signals, suggesting possible redundancy in substrate phosphorylation. This may not be the case in vivo, however, as MSK1/2 localize primarily in the nuclei of quiescent and activated cells (83, 268), suggesting that they predominantly regulate nuclear substrates (Fig. 8). In addition, the MSKs do not require PDK1 in order to become activated, indicating that they may not depend on the same upstream pathways for activation. Studies from several groups indicated that MSK1/2 regulate gene expression at multiple levels, by playing active roles in transcriptional regulation and chromatin remodeling in response to stress and mitogens (14, 378). While deletion of both MSK isoforms in the mouse did not yield obvious developmental defects (398), this lack of a specific phenotype may be due to overlapping functions between AGC and MAPKAPK family members. This may not be the case in Drosophila, as deletion of the MSK ortholog JIL-1 is lethal (387), perhaps due to lower genetic redundancy (169). In Drosophila, JIL-1 is a key regulator of chromatin structure that functions to maintain euchromatic domains while counteracting heterochromatization and gene silencing (387). JIL-1 regulates histone H3 phosphorylation on Ser10, which was shown to inhibit the spread of heterochromatin markers to ectopic locations on the chromosome arms (19). These studies suggest that MSK1/2 play similar roles in gene regulation, which may affect several physiological functions, including both immunity and neuronal functions.
(i) Transcriptional regulation.
MSKs phosphorylate many transcription factors and thereby increase their stability and/or activity (378). As mentioned for the RSKs, the transcription factor CREB was found to be an MSK1/2 substrate. Upon their discovery, MSK1/2 were shown to phosphorylate CREB on Ser133 in vitro with a Km value much lower than those of PKA, RSK2, and MK2 (83, 268), raising the possibility that MSK1 rather than RSK2 mediates the mitogen-stimulated phosphorylation of CREB. Since then, work from several groups using different pathway inhibitors has supported the involvement of MSK1/2 in CREB phosphorylation (141, 207). The most convincing evidence for MSK-dependent phosphorylation of CREB comes from knockout mice. MEFs isolated from _Msk1_−/−_Msk2_−/− mice were found to have greatly reduced CREB phosphorylation in response to stress and mitogens (398). The transcriptional activity of CREB requires its phosphorylation at Ser133, which recruits the CBP and p300 cofactors. Activated CREB participates in the transcriptional activation of several IE genes, such as those for c-Fos, JunB, and Egr1 (219). The knockout of both MSK1 and MSK2 resulted in a 50% reduction in c-Fos and JunB gene transcription in MEFs subjected to stress but only a minimal reduction in response to mitogenic stimulation (398), suggesting again that RSKs and MSKs may collaborate in the regulation of IE genes in response to mitogens. In addition to CREB, MSKs were also shown to regulate another CRE-binding factor, the activating factor 1 (ATF1). Phosphorylation of ATF1 at Ser63 appears to follow the same rule as CREB. Indeed, MSKs have been convincingly shown to be the primary ATF1 kinases in cells (398), but residual ATF1 phosphorylation in _Msk1_−/−_Msk2_−/− cells suggests the involvement of a collaborating kinase.
The activity of the NF-κB transcription factor is highly regulated by posttranslational modification, and phosphorylation of the p65 subunit at Ser276 was attributed to MSKs (379). As with CREB, phosphorylation of p65 was first reported to be regulated by PKA, which associates with the IκB kinase complex (431). In this context, p65 phosphorylation was found to promote p65 interaction with the cofactors CBP and p300, which in turn acetylate histones and NF-κB bound to promoters (432). Loss of MSK1/2 in mouse fibroblasts resulted in reduced TNF-α-mediated activation of NF-κB (50, 379), suggesting that activation of MSK1/2 is important for NF-κB-dependent transcription.
Other transcription factors that may be regulated by MSKs include STAT3 (signal transducer and activator of transcription 3) and ER81. Phosphorylation of STAT3 at Ser727 in response to UV irradiation and erythropoietin (EPO) was shown to require MSK1 activity (397, 426). In the case of ER81, MSK1 was found to mediate its phosphorylation at Ser191 and Ser216, two residues that control its transcriptional activity (166). While STAT3 and ER81 may be bona fide substrates of MSKs, validation experiments with cells derived from _Msk1_−/−_Msk2_−/− animals are still lacking. These will be necessary, as other MAPKAPKs also phosphorylate these transcription factors. MSK2 was also found to inhibit the transcription factor p53 in the absence of stress stimuli (218). This basal inhibition of p53 by MSK2 occurred independently of its kinase activity and of upstream MAPK signaling, suggesting that MSK2 acts to suppress p53 transcriptional activity under basal conditions.
(ii) Regulation of the chromatin environment.
The primary roles of MSK1 and MSK2 may be as mediators of the nucleosomal response, which has the main goal of promoting gene relaxation and activation. The nucleosomal response refers to the rapid phosphorylation of histone H3 on Ser10 and of HMGN1 (also known as HMG-14) on Ser6 that occurs concomitantly with IE gene induction in response to a wide variety of stimuli (364). Histone H3 is a component of the nucleosome, and its C-terminal tail is the site of many posttranslational modifications, including phosphorylation, acetylation, methylation, and ubiquitination. Phosphorylation of histone H3 on Ser10 in response to mitogens and stress is associated with gene activation, suggesting that the ERK1/2 and p38 modules may regulate common nuclear effectors. Although RSK1 and RSK2 had initially been shown to phosphorylate histone H3 in vitro and in vivo (310, 315), the MSKs were later convincingly shown to be the primary histone H3 kinases (341). Indeed, stress- and mitogen-induced phosphorylation of histone H3 was found to be completely inhibited in primary embryonic fibroblasts from _Msk1_−/−_Msk2_−/− animals (341), ruling out the possible involvement of other kinases such as the RSKs (81). Recently, MSK1-mediated phosphorylation of histone H3 at Ser10 was found to be required for tumor promoter-induced cell transformation (187).
In addition to Ser10, MSKs have also been shown to phosphorylate histone H3 on Ser28, a site also associated with IE gene induction (341). Although conserved from yeast to human, the molecular mechanisms by which phosphorylation of histone H3 regulates transcription remain unclear. Ser10 and Ser28 lie in putative 14-3-3-binding sequences, which may dislodge potential transcriptional repressors from activated promoter regions (223). Phosphorylation of histone H3 was found to inhibit binding of the transcriptional repressor HP1 (421), suggesting a possible mechanism by which MSK may regulate transcription (14). Alternatively, 14-3-3 proteins may act as scaffolds for the recruitment of SWI/SNF complexes at IE gene promoters and thereby promote chromatin remodeling upon activation of MSK1/2 (97).
As mentioned above, MSKs also regulate HMGN1, another chromatin-associated protein (341). While it is not a core component of nucleosomes, HMGN1 was shown to regulate chromatin compaction in vitro (155). Studies with HMGN1-null cells revealed that it suppresses histone H3 phosphorylation and IE gene expression (215), suggesting that MSK-mediated phosphorylation of HMGN1 may reduce its interaction with nucleosomes and thereby enable MSK1/2 to phosphorylate core histones (378).
(iii) Other substrates.
Although this was not validated using cells derived from _Msk1_−/−_Msk2_−/− animals, the MSKs have been suggested to phosphorylate the proapoptotic protein Bad (328) and the translational inhibitor 4E-BP1 (217).
MNK
Identification and protein structure.
The MAPK-interacting kinases 1 and 2 (MNK1 and MNK2) were discovered simultaneously by two teams in 1997. The first group used a two-hybrid screen designed to identify ERK2-binding proteins (389). The second approach consisted of a phosphorylation screen looking for ERK1 substrates (121). The catalytic domain of human MNK1 displays approximately 70% amino acid identity with MNK2 (Fig. 9), and MNK orthologs have been identified in other species. MNK1 displays relatively high homology (51% amino acid identity) to a large protein kinase in Drosophila termed LK6, which was found to localize to centrosomes and to regulate microtubule organization (185). MNK1 also displays sequence homology (46% amino acid identity) with the C. elegans protein kinase mnk-1 (R166.5), and knockdown of this protein is embryonic lethal in the worm (343). The catalytic domain of MNK1/2 belongs to the CAMK family of kinases and is most similar to the CTKD of the RSKs (33% amino acid identity) and MK2/3 (389) (Fig. 4). Compared to those of other MAPKAPKs, the kinase domains of MNK1 and MNK2 have several particularities. First, the MNKs have a DFD (Asp-Phe-Asp) motif in subdomain VII, whereas most protein kinases, including MAPKAPK family members, have DFG (Fig. 5A). Second, the kinase domain of MNK1/2 has two unusual short inserts, one in the activation loop (Fig. 5A) and the other after the APE motif of subdomain VIII (44). Interestingly, these features are also present in Drosophila LK6, suggesting that they may serve a function in MNK activation and/or substrate recognition.
FIG. 9.
Alignment of the amino acid sequence of the MAPKAPKs containing a single kinase domain. Sequences comprising the kinase domains and its subregions are boxed in red and reveal regions of highest homology. The conserved activation loop threonine residue is shown, as well as other conserved phosphorylation sites. The MAPK-binding domain is identified by a line.
Tissue expression and subcellular localization.
Both the Mnk1 and Mnk2 genes generate two spliced isoforms, a long form (MNK1A and MNK2A) and a shorter version (MNK1B and MNK2B) that lacks the C-terminal MAPK-binding motif (259, 335). The MAPK-binding domain of MNK1A consists of Leu-Ala-Arg-Arg-Arg (Fig. 3) and mediates the interaction of MNK1 with both ERK1/2 and p38 (121, 389). The D domain present in MNK2A contains only two contiguous basic residues (Leu-Ala-Gln-Arg-Arg) and is very similar to the motif found in RSKs (Fig. 3). Consistent with this, MNK2A preferentially interacts with ERK1/2 (389).
Both MNK isoforms contain a polybasic sequence lying N terminal to the kinase domain (Fig. 4) (312). This sequence is involved in the recognition of eIF4G, as is discussed below, but also serves as an NLS that promotes MNK nuclear import (263, 275). Indeed, using a two-hybrid strategy, the NLS of MNK1 has been shown to mediate binding to importin α, which is an intracellular receptor protein involved in nuclear import (263, 390). Although all MNKs contain this sequence, not all isoforms are nuclear, and this is partly due to the presence of a CRM1-type NES in their C termini (263). The NES of MNK1 was shown to be functional through the use of leptomycin B (which blocks CRM1-dependent nuclear export), as treatment with the drug traps MNK1 in the nucleus and revealed that MNK1 is actively shuttling between the cytoplasm and the nucleus (231, 263). The C termini of MNK1B and MNK2B do not contain such a motif, which correlates with their mostly nuclear localization (259, 312). Because MNK2A lacks critical residues important for NES function (Fig. 4), its cytoplasmic localization is more enigmatic. Data suggest that its extreme C-terminal domain interferes with binding of its polybasic region to importin α, which may affect its entry into the nucleus (312).
The expression of MNK1 and MNK2 has not been studied extensively, but both proteins are expressed in all adult tissues, with the exception of the brain, where levels are greatly reduced compared to those in other tissues (389). The expression of both proteins was shown to be especially abundant in skeletal muscle, suggesting a higher requirement for MNK activity in this tissue.
Activation mechanisms and inhibitors.
The basal activities and regulation of MNKs by MAPK agonists vary depending on the isoform. MNK1A has low basal activity in cells and is responsive to agonists of both ERK1/2 and p38 isoforms (Fig. 7) (121, 384, 389, 390). In contrast, MNK1B has high basal activity in quiescent cells, and this activity is not significantly affected by inhibitors of the ERK1/2 and p38 modules (258), consistent with the fact that MNK1B does not contain a MAPK-binding domain. With respect to MNK2, the long isoform (MNK2A) displays high basal activity in quiescent cells, and this activity can be slightly enhanced by agonists of ERK1/2 but not p38 (311). MNK2B has very low activity under most types of stimulation, and it is unclear which circumstances will promote its activity (312). There are several potential reasons for differences in the levels of activity between MNK1A and MNK2A, some of which involves features of both the C-terminal region and the catalytic domain. This was nicely illustrated through the generation of MNK1/2 chimeras with inverted C-terminal regions (262).
Phosphopeptide analysis of MNK1 and MNK2 revealed the presence of several MAPK-stimulated phosphorylation sites (311, 390). Phosphorylation of two proline-directed sites within the activation loop of MNKs (Thr209 and Thr214 in MNK1A) was found to be essential for activation (Fig. 4), as replacement of both sites by Ala residues in MNK1/2 results in inactive kinases (311). Another important residue was found to be phosphorylated in MNK1/2 (Thr344 in MNK1A), but mutation of this phosphorylation site yielded different outcomes in MNK1 and MNK2. While replacement by an Ala residue did not affect MNK1A but completely disrupted MNK2A activity, replacement of Thr344 by an Asp residue resulted in a constitutively activated MNK1A enzyme while not affecting MNK2A activity (121, 311, 389). Deletion of the C-terminal 91 aa containing the MAPK-binding motif was also shown to render MNK1A inactive (121), suggesting that ERK/p38 docking and phosphorylation of several important regulatory sites are required for efficient MNK1 and MNK2 activation.
The compound CGP57380 has been described as an MNK inhibitor (190) and used in cell-based assays for this purpose in several studies. However, a recent study indicated that this compound was a relatively weak inhibitor of MNKs, with 50% inhibitory concentrations (IC50s) in the low-micromolar range (18). In addition, CGP57380 was tested against an extended panel, and many protein kinases were inhibited with similar potencies, including MKK1, CK1, and BRSK2. These studies indicated that CGP57380 is not a specific inhibitor of MNK isoforms, and results obtained from its use in cell-based assays should be interpreted with caution. The use of cells derived from _Mnk1_−/−_Mnk2_−/− knockout animals (373) should provide valuable information and should be favored over the use of CGP57380 to validate substrates and biological functions.
Substrates and biological functions.
MNKs have been reported to phosphorylate a number of different substrates, including components of the translational machinery and some mRNA-binding proteins and splicing factors (Fig. 8). The exact consensus phosphorylation motif of MNKs has not been determined, and even based on known phosphorylated sites in MNK substrates, no clear consensus that would help for prediction of sites emerges.
(i) eIF4E and eIF4G.
Regulation of protein synthesis by the eukaryotic initiation factor 4F (eIF4F) complex plays an important role in controlling cell growth and proliferation (reviewed in reference 286). In higher eukaryotes, the eIF4F complex consists of three subunits: eIF4A, eIF4E, and eIF4G. eIF4E recruits mRNAs to the complex through binding of their 5′-cap structure, the _N_7-methylguanosine cap. eIF4E also interacts with the scaffold protein eIF4G, which recruits the RNA helicase eIF4A to the complex, unwinds the secondary structure in the mRNA, and facilitates ribosome scanning to the initiating codon. eIF4E is phosphorylated on Ser209 in response to stress and mitogen stimulation, and this phosphorylation event was shown to be dependent on both the ERK1/2 and p38 modules (115). MNK1 was found to be recruited to the eIF4F complex through its association with the C terminus of eIF4G (276), making it a likely candidate kinase for eIF4E. Consistent with this idea, MNK1 and MNK2 were shown to phosphorylate eIF4E at Ser209 in response to stress and mitogen stimulation (190, 311, 389). More recently, this phosphorylation event was shown to be completely absent in cells derived from _Mnk1_−/−_Mnk2_−/− animals, indicating that the MNKs are the predominant eIF4E kinases operating in cells (373). eIF4G was also suggested to be phosphorylated by MNK1/2 (275), but more experimentation will be required to confirm this regulation and its biological significance.
Although eIF4E phosphorylation has been studied for over 2 decades, no clear consensus has emerged regarding its biological significance, and this topic has been highly debated (313). Phosphorylation of eIF4E has been shown to decrease its affinity for the cap structure of mRNA (314), but the exact role of eIF4E phosphorylation in the regulation of translation remains elusive. Several lines of evidence suggest that MNKs are negative regulators of protein synthesis. Overexpression of MNK was found to decrease cap-dependent translation in Aplysia neurons (294), suggesting that hyperphosphorylation of eIF4E decreases protein synthesis rates. Treatment of cells with the MNK1/2 inhibitor CGP57380 was found to slightly enhance cap-dependent translation (190), consistent with the idea that MNKs may play a negative role in eIF4E-dependent translation. Conversely, recent work has suggested that MNK1/2 positively regulate mRNA translation and polysome assembly (27, 425), but these studies generally relied on CGP57380 to inhibit MNK1/2 activity and eIF4E phosphorylation, and as indicated above, this inhibitor is not very selective and may have pleiotropic effects. Despite having undetectable eIF4E phosphorylation at Ser209, mice deficient in MNK1 and MNK2 develop normally, and cells derived from these animals have normal global protein synthesis and cap-dependent translation (373). These findings suggest that the role of eIF4E phosphorylation may not be evident under normal growth conditions and that MNK1/2-dependent eIF4E phosphorylation may be involved in more specialized functions, such as in response to oncogene activation and cellular stress. Consistent with this idea, expression of a constitutively activated form of MNK1 was shown to promote tumorigenesis in the mouse, through a mechanism that may involve expression of the antiapoptotic protein Mcl-1 (392).
(ii) Other substrates.
Two RNA-binding proteins were shown to be regulated by MNKs, the heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and the polypyrimidine tract-binding protein (PTB)-associated splicing factor (PSF). hnRNP A1 is a very abundant nuclear protein that plays an important role in mRNA metabolism. The MNKs were shown to phosphorylate hnRNP A1 at two potential sites, Ser192 and Ser310, in response to T-cell activation (45). Phosphorylation of hnRNP A1 was found to decrease its ability to interact with the TNF-α mRNA, suggesting that MNKs may play key roles in the regulation of specific messages. hnRNP A1 also becomes phosphorylated in response to osmotic stress in a p38-dependent manner (377), suggesting that MNKs may also regulate translation of specific mRNAs under stress conditions. PSF is a nuclear protein involved in transcription and RNA processing. Together with p54nrb, PSF forms a transcription-splicing factor implicated in diverse biological functions. MNKs were shown to phosphorylate PSF at two sites in vitro, Ser8 and Ser283, which increase its affinity in vivo for the TNF-α mRNA (43), suggesting again that MNKs may regulate the fate of specific mRNAs.
Cytoplasmic phospholipase A2 (cPLA2) plays a key role in the production of eicosanoids, which are second messengers with important functions in immunity and inflammation. MNK1 was shown to phosphorylate cPLA2 on Ser727 in vitro (150), a phosphorylation event that depends on the p38 module in cells. Phosphorylation of cPLA2 on Ser727 was shown to correlate with enzyme activation, suggesting that MNK1 may regulate cPLA2-mediated arachidonate release. It is important to note that the p38-activated protein kinases MSK1 and MK5 were also shown to phosphorylate cPLA2 in vitro, suggesting that other MAPKAPK family members may participate in the phosphorylation of Ser727 and regulation of cPLA2. Finally, MNK1 has been shown to regulate the phosphorylation of Sprouty 2 (Spry2), a membrane-associated protein that suppresses ERK activation and/or signaling (42). Recent work shows that MNK1 phosphorylates Spry2 on two sites, Ser112 and Ser121, and replacement of these sites by Ala residues caused destabilization of Spry2 (79). The MNK inhibitor also led to degradation of Spry2, suggesting that MNK1-mediated phosphorylation of Spry2 may prolong its half-life and thus its capacity to inhibit ERK signaling.
MK2/3
Identification and protein structure.
The MAPK-activated protein kinase 2 (MK2) was discovered as an ERK1/2-activated protein kinase that could phosphorylate heat shock protein 25 (Hsp25) and Hsp27 (346). It was 2 years later that two groups determined that MK2 was in fact stimulated by the p38 module in response to stress stimuli (117, 295). MAPK-activated protein kinase 3 (MK3) was discovered a few years later by two groups, using a two-hybrid screen for p38-interacting proteins (232) and by analyzing genes commonly deleted in small-cell lung cancer (334). A single homolog kinase of MK2/3 has been identified in Drosophila and C. elegans, which share about 60% amino acid identity with human MK2 (204). Yeasts lack obvious structural homologues of MK2, but functionally homologous kinases that also respond to p38 (Hog1 in yeast) do exist; these are Rck1 and Rck2 in budding yeast (279) and Srk1 (also known as Mkp1) and Mkp2 in fission yeast (16).
MK2 is highly homologous to MK3 (75% amino acid identity), and their kinase domains are most similar (35 to 40% identity) to CAMK, and the CTKD of RSK (347, 438) (Fig. 9). Vertebrate MK2/3 contain an N-terminal proline-rich region that interacts with the Src homology 3 (SH3) of c-Abl in vitro (270). The C terminus of MK2 contains a functional bipartite NLS (Lys-Lys-Xaa10-Lys-Arg-Arg-Lys-Lys) that is also present in MK3 (Fig. 3). The NLS found in both MK2 and -3 encompasses a D domain (Leu-Leu-Lys-Arg-Arg-Lys-Lys in MK2) (Fig. 4) that mediates specific interaction with p38α and p38β (337). MK2 and MK3 also possess a functional NES (Met-Thr-Ser-Ala-Leu-Ala-Thr-Met-Arg-Val) located N terminal to the NLS and D domain (Fig. 4). The NES in MK2 triggers its nuclear export following stimulation, which can be inhibited by leptomycin B (20, 104).
Tissue expression and subcellular localization.
MK2 and MK3 mRNAs are expressed at detectable levels in most tissues analyzed, with a predominant expression in the heart, skeletal muscle, and kidney (105, 334, 346). The Mk2 gene was shown to give rise to two alternatively spliced transcripts (58, 346), and more recently, a similar observation was made with the mouse Mk3 gene (240). The shorter MK2 and MK3 isoforms (MK2S and MK3S) lack part of the C-terminal region and thus the nuclear import/export sequences and the MAPK-binding domain. MK3S also lacks catalytic subdomains IX, X, and XI but, surprisingly, displayed detectable in vitro kinase activity toward Hsp27 (240). MK2S is not well represented in expressed sequence tag (EST) databases, suggesting that this isoform may be the result of a rare alternative splicing event (122).
While MK2S and MK3S are mostly cytoplasmic enzymes (240, 438), MK2 and MK3 are located predominantly in the nuclei of quiescent cells (104, 250). Upon stress stimulation, both enzymes are rapidly exported to the cytoplasm using a CRM1-dependent mechanism (20, 104, 321). While the long form of MK3 translocates upon stimulation, MK3S is degraded in a stress- and p38-dependent manner (240). Intramolecular unmasking of the NES found in MK2/3 was shown to be regulated by phosphorylation. Indeed, nuclear export of MK2 appears to be mediated by kinase activation, as phosphomimetic mutation of Thr334 enhances the cytoplasmic localization of MK2 (104). For this reason, it is thought that MK2 and MK3 contain a constitutively active NLS and a phosphorylation-regulated NES.
Activation mechanisms and inhibitors.
MK2 and MK3 are activated under various stress conditions that stimulate the p38 isoforms (117, 139, 232, 295), such as UV irradiation, heat shock, oxidative stress, hyperosmolarity, and different cytokines. Through the use of a p38-specific inhibitor (SB203580) (66) and p38α-deficient cells (6), MK2 activation was found to be completely dependent on p38α (Fig. 10). Activated p38α promotes MK2 phosphorylation at Thr222 within its activation loop (Fig. 5A), Ser272 within subdomain X of the kinase domain, and Thr334 located in a hinge region that controls MK2 autoinhibition (21). The crystal structure of MK2 has been resolved (394) and suggests that Thr334 phosphorylation may serve as a switch for MK2 nuclear import and export (236). Upon activation, phosphorylation of Thr334 is thought to release the autoinhibitory helix from the core of the kinase domain, thereby exposing the NES and promoting nuclear export (104). This MK2 conformational change was demonstrated by fluorescence resonance energy transfer (FRET) analysis of a green fluorescent protein (GFP)-MK2 fusion protein, which revealed that MK2 activation correlates with an open conformation that is detectable only in the cytoplasm of activated cells (249). In contrast to the regulated NES, the C-terminal NLS is active independently of MK2 phosphorylation state, allowing this kinase to shuttle between the nucleus and the cytoplasm. As part of the NLS overlaps with the MAPK docking domain, it is possible that p38 docking regulates NLS function, but this has not been addressed.
FIG. 10.
Signaling cascades leading to activation of MK2/3 and MK5. MK2/3 have been shown to be activated by both ERK1/2 and p38 kinases. Conversely, MK5 was initially shown to be regulated by p38, but recent data suggest a stronger link with ERK3 and ERK4. Different inhibitors of components within these cascades are also shown. Dotted lines indicate that, although reported, substrate regulation by the respective kinase remains to be thoroughly demonstrated.
MK2 forms a stable complex with p38α, and each protein mutually stabilizes its partner (122). Indeed, knockout studies revealed decreased levels of p38α in MK2-deficient cells (196), and MK2 expression was shown to be reduced in p38α-deficient cells (350). The reason for this is unclear, but it was suggested that MK2 may compete with other partners of p38α involved in its inactivation. Early studies have suggested that p38α is exported from the nucleus in a complex with MK2 (20), but more recent findings indicate that this is unlikely (293). A large number of studies have used p38 inhibitors to prevent MK2/3 activation and study their biological functions. However, several pharmaceutical companies have recently reported new small-molecule inhibitors of MK2 (124), some of which are reversible ATP-competitive compounds (e.g., PF-3644022) that suppress MK2-dependent functions in cells (243).
Substrates and biological functions.
Experimental evidence supports a role for the p38 module in cytokine production (142, 208), cell migration (148, 269), actin remodeling (296), cell cycle control (9), and gene expression (87). Interestingly, targeted deletion of the mouse Mk2 gene provided convincing evidence that although p38 regulates a large set of substrates, MK2 appears to be a key player in these p38-dependent biological processes (122, 293).
MK2 and MK3 have comparable substrate preferences and phosphorylate the same residues in Hsp25 and Hsp27 with similar kinetic constants (66). The optimal sequence required for efficient phosphorylation by MK2/3 is Φ-Xaa-Arg-Xaa-(Leu/Asn)-pSer/Thr-(Iso/Val/Phe/Leu)-Xaa, where Φ is a bulky hydrophobic residue (66, 227, 334). While MK2 and MK3 appear to be equals in vitro, the in vivo situation is quite different, as MK2 activity was shown to be higher than that of MK3 (292). Consistent with this, MK3 was shown to partly compensate for the loss of MK2 in mice, whereas deletion of MK3 alone had very little effect (Table 1). MK2 and MK3 regulate a number of substrates, some of which are described in detail below (Fig. 8).
(i) Actin remodeling and cell migration.
The first MK2/3 substrates to be identified were Hsp25 and Hsp27 (221, 232, 348), which are ATP-independent chaperones that keep unfolded proteins in a folding-competent state before they can be refolded by Hsp70 (194). Hsp27 is of particular interest because it forms large oligomers which can act as molecular chaperones and protect cells from heat shock and oxidative stress (203). Once phosphorylated by MK2/3 at several residues, Hsp27 loses its ability to form large oligomers and is unable to block actin polymerization (203, 290). These findings suggest that MK2 serves a homeostatic function aimed at regulating actin dynamics that would otherwise be destabilized during stress (113, 139). Other MK2/3 substrates that are involved in actin remodeling include the p16 subunit (p16-Arc) (333) of the seven-member actin-related protein 2/3 complex (Arp2/3), lymphocyte-specific protein 1 (LSP-1) (157), and F-actin-capping protein Z-interacting protein (CapZIP) (110). Others, such as vimentin (57) and αβ-crystallin (182), may interact with intermediate filaments and microtubules. Remodeling of the actin cytoskeleton is a prerequisite for cell migration, and phosphorylation of Hsp27 by MK2/3 was shown to contribute to cell motility (237). Consistent with this, MK2-deficient neutrophils have defects in chemotaxis and altered chemokinesis (146). The exact contribution of other MK2/3 substrates involved in actin remodeling remains elusive.
(ii) Cytokine production.
Following stimulation of the p38 module with LPS, MK2 regulates the inflammatory response through posttranscriptional mechanisms. MK2 was found to increase production of TNF-α and IL-6 by promoting translation and/or stability of their mRNAs (195, 248). MK2-deficient mice are less sensitive to LPS-induced endotoxic shock but more susceptible to bacterial infection (211), consistent with an impaired inflammatory response. While deletion of MK3 alone does not significantly alter cytokine production, absence of both MK2 and MK3 further impairs the inflammatory response, suggesting significant overlap in function (292). The stability of many mRNAs, including those of IL-6 and TNF-α, depends on AU-rich elements (AREs) located in their 3′ untranslated regions (3′-UTRs). Several proteins bind ARE-containing mRNAs, many of which are specifically regulated by MK2/3 (248, 401). Consistent with this, MK2 has been shown to bind and/or phosphorylate hnRNP A0 (297), tristetraprolin (TTP) (225), poly(A)-binding protein 1 (PABP1) (33), human R-antigen (HuR) (144, 369), and butyrate response factor 1 (BRF1) (226). MK2-dependent phosphorylation of TTP creates functional 14-3-3-binding sites (61) that inhibit TTP-dependent degradation of ARE-containing transcripts and thereby contributes to LPS-induced TNF-α expression (38, 154, 345). Alternatively, MK2-mediated phosphorylation of TTP may reduce its ability to promote deadenylation by inhibiting the recruitment of the CAF1 deadenylase, as suggested in a recent study (228).
(iii) Transcriptional regulation.
The polycomb group family, originally identified in Drosophila as a repressor of homeotic genes, represents epigenetic chromatin modifiers with a transcriptional silencing function (376). Recent evidence indicates that polycomb group proteins may be targets for MK2 (414) and MK3 (381). Indeed, MK2/3 bind the human polyhomeotic protein 2 (HPH2), which is a component of the large and dynamic polycomb repressive complex 1 (PRC1). Following stress stimulation, MK2/3 phosphorylate components of the complex, such as BMI1, a protein involved in the self-renewal of hematopoietic stem cells. Interestingly, a recent study demonstrated that hematopoietic stem cells in MK2-deficient mice are reduced in number and show an impaired ability for competitive repopulation in vivo (320).
In addition, MK2 was shown to contribute to the phosphorylation of SRF (151) and ER81 (165). Comparison of wild-type and MK2-deficient cells revealed that MK2 is the major SRF kinase induced by stress (151), suggesting a role for MK2 in stress-mediated IE gene response. Both MK2 and -3 interact with the basic helix-loop-helix (bHLH) transcription factor E47 in vivo and phosphorylate E47 in vitro (250). MK2-mediated phosphorylation of E47 was found to repress E47 transcriptional activity, suggesting that MK2/3 may regulate tissue-specific gene expression and cell differentiation (250).
(iv) Cell cycle control.
MK2 has recently been shown to phosphorylate CDC25B and CDC25C in response to UV irradiation (227). CDC25C is an evolutionarily conserved dual-specificity phosphatase that directs dephosphorylation of cyclin B-bound Cdc2 and triggers entry into mitosis (13). Upon DNA damage, CDC25 is inhibited by the Chk1 and Chk2 protein kinases, and MK2 appears to work in parallel to these enzymes to promote the G2/M checkpoint in response to stress (227). Intriguingly, RSK was also found to phosphorylate CDC25C in Xenopus oocytes (63), suggesting that RSK may also contribute to a G2/M cell cycle arrest under certain circumstances.
p53 stability is regulated by HDM2, a RING domain protein that acts as an E3 ligase to ubiquitinate p53 and target it for degradation. Phosphorylation of HDM2 on Ser166 by Akt was shown to enhance HDM2 activity and promote the degradation of p53 (433). Interestingly, MK2 was also found to phosphorylate HDM2 on Ser157 and Ser166, resulting in HDM2 activation and degradation of p53 in response to stimuli of the p38 module (391). These results suggest that MK2 may act to dampen the extent and duration of the p53 response to stress and DNA damage, but more experimentation will be required to test this model. Consistent with this scenario, a recent study showed decreased HDM2 phosphorylation and increased p53 levels in MK2-deficient mice subjected to a skin carcinogenesis model (171).
(v) Other substrates.
14-3-3 proteins normally interact with a number of signaling pathway components, including protein kinases, phosphatases, and transcription factors (reviewed in reference 242). The generation of 14-3-3-binding sites appears to be important for MK2-dependent functions, as 14-3-3 interacts with several MK2 substrates in a stress-inducible manner. In addition to TTP, which was described above, MK2 promotes 14-3-3 binding to Hsp27 (96), CDC25B and CDC25C (227), and TSC2 (214) to modulate their function. TSC2 normally functions as a negative regulator of mTOR activation, and MK2-mediated phosphorylation of TSC2 at Ser1210 may regulate mTOR-dependent protein synthesis in response to certain stresses.
MK5
Identification and protein structure.
MK5 (also known as p38-regulated/activated protein kinase [PRAK]) was discovered simultaneously by two groups searching EST databases for proteins with sequence homology to MK2 (252, 254). While MK2 and MK3 display 75% amino acid identity, MK5 is more distantly related to MK2/3, with 38% homology to these enzymes (Fig. 9). MK5 and MK2/3 may have originated from a common ancestral protein, because all three proteins display similar amino acid identities to Drosophila and C. elegans homologs of MK2. Like that of MK2/3, the kinase domain of MK5 is most similar to CAMK-related kinases (Fig. 4).
MK5 has a structure similar to that of MK2/3, consisting of a kinase domain flanked by a short N-terminal region lacking the SH3 domain-binding motif and a 100-aa C-terminal extension that contains a functional NLS, NES, and MAPK-binding motif (Fig. 4). The NLS (Arg-Lys-Arg-Lys) of MK5 is functional, as Ala substitutions disrupt its nuclear localization (251, 321). The NLS also overlaps with a MAPK docking site (Ile-Leu-Arg-Lys-Arg-Lys-Leu-Leu) that mediates interaction with p38α and p38β (Fig. 3) (321). Interaction of p38 with MK5 was found to be weaker than that with MK2, and consistent with this, MK5 does not appear to stabilize p38 in vivo (331). The NES of MK5 (Leu-Arg-Val-Ser-Leu-Arg-Pro-Leu-His-Ser) is also functional and is sufficient to trigger CRM1-dependent nuclear export (321). Finally, the C-terminal region of MK5 contains a new domain necessary for interaction with ERK3 and ERK4 (5).
Tissue expression and subcellular localization.
MK5 expression appears to be ubiquitous, with predominant expression in the heart, skeletal muscle, pancreas, and lung (130, 252, 254, 267). The human Mk5 gene encodes two alternatively spliced transcripts of 471 and 473 aa, but the relevance of having two nearly identical isoforms is currently unknown. In mice, four MK5 isoforms have been identified, in addition to the full-length protein, resulting from both exon skipping and alternative splice site activation during pre-mRNA processing (91). The biological significance of these different isoforms is still unknown, but these proteins appear to have distinct subcellular localizations, catalytic activities, and tissue expression. When overexpressed, full-length MK5 localizes to the nuclei of quiescent cells, but upon cellular stress, MK5 translocates to the cytoplasm (251, 321). Interestingly, expression of exogenous p38α with MK5 has been shown to relocalize MK5 to the cytoplasm, suggesting that p38 docking interferes with the function of its NLS (321).
Activation mechanisms and inhibitors.
Despite MK5 and MK2/3 being structurally related, the activation mechanisms of MK5 are quite different from those of MK2/3 (Fig. 10). Several groups have shown that under conditions of overexpression, p38 phosphorylates MK5 in response to cellular stress (251, 252, 254, 321). Analysis of _in vitro_-phosphorylated MK5 revealed that p38 phosphorylates the activation loop residue of MK5 (Thr182) (Fig. 5A), which is essential for kinase activation (252). However, certain experimental evidence suggests that MK5 is not a physiological p38 substrate. First, endogenous MK5 is not significantly activated by classical p38 stimuli, such as arsenite and sorbitol (331). Second, MK5 is not involved in the inflammatory response induced by LPS, a strong agonist of the p38 module (331). Indeed, MK5-deficient mice are not resistant to endotoxic shock caused by LPS and do not have impaired cytokine production. Third, no interaction between endogenous MK5 and p38 has ever been reported (322, 331, 351).
More recently, MK5 was also shown to be a substrate of ERK3 and ERK4. These atypical MAPKs regulate MK5 activity when overexpressed, but unlike for the p38 isoforms, this was also demonstrated under physiological conditions (4, 179, 318, 322). Indeed, endogenous MK5 kinase activity was shown to be reduced in _Erk3_−/− MEFs (322), and the interaction between ERK3/4 and MK5 was demonstrated at the endogenous level (179, 322). While ERK3/4 phosphorylate MK5 at its activation loop residue Thr182 (Fig. 5A), ERK3 was also shown to act as a scaffolding protein by promoting the autophosphorylation and activation of MK5 (318). ERK3 and ERK4 can also regulate the subcellular localization of MK5 (4, 179, 318, 322). When expressed independently, MK5 and ERK3 localize mainly in the nucleus, whereas ERK4 displays a cytoplasmic localization. Coexpression of MK5 with ERK3 or ERK4 promotes MK5 relocalization to the cytoplasm, and this event was shown to be facilitated by activation loop phosphorylation of ERK3 and ERK4 (86, 266).
Substrates and biological functions.
The exact biological function of MK5 is unknown, but recent evidence suggests that MK5 may play a role in oncogene-induced senescence (351) and actin remodeling (129) (Fig. 8). MK5 was originally thought to share many substrates with MK2, such as Hsp27 and glycogen synthase (252), but this was questioned by the characterization of the MK5-deficient mouse (331). Despite the relatively high similarity between MK2/3 and MK5 (Fig. 9), MK5-deficient mice do not display any of the phenotypic changes seen in MK2-deficient animals (195). Indeed, disruption of the Mk5 gene in mice of mixed genetic background did not give rise to detectable phenotypic changes (331) (Table 1). However, inactivation of MK5 into a C57BL/6 genetic background resulted in lethality at embryonic day 11.5 (E11.5) with incomplete penetrance (318). ERK3 and MK5 mRNAs were shown to be coexpressed in space and time during mouse embryogenesis, suggesting an important role for this signaling module during development.
(i) Tumor suppression.
Certain observations suggest that ERK3 may play some roles in tumor suppression, including its apparent negative regulatory effect on cell cycle progression, cell proliferation, and migration (73, 75, 173). MK5 may be an important ERK3 effector protein mediating these effects, as it was recently shown to promote tumor suppression and oncogene-induced senescence (351). Indeed, MK5-deficient mice from a mixed genetic background were found to be more susceptible to skin carcinogenesis induced by the mutagen dimethylbenzanthracene (DMBA), an agent that causes skin tumors that are 90% positive for Ras mutations. Consistent with these findings, MK5 was shown to be essential for Ras-induced senescence in primary mouse and human fibroblasts. Reintroduction of a kinase-defective MK5 mutant could not restore Ras-induced senescence in MK5-deficient cells, suggesting that MK5 may regulate an essential substrate involved in cellular senescence. The same group demonstrated that MK5 phosphorylates p53 at Ser37, a residue that promotes its transcriptional activity (351). Interestingly, this residue is not located within a consensus for MAPKAPKs but rather is followed by a Pro residue, suggesting that this site is a more likely candidate for phosphorylation by MAPKs. Although the role of MK5 in oncogene-induced senescence appeared to require activation of the p38 module, the potential roles of ERK3 and ERK4 were not determined in that study, and there may be important roles for these atypical MAPKs.
(ii) Actin remodeling.
A potential role for MK5 in cytoskeletal organization was also suggested. Overexpression of MK5 was reported to increase HeLa cell migration and F-actin production (354). The same study also showed that 14-3-3ɛ interacts with and inhibits MK5, resulting in reduced phosphorylation of Hsp27, cell migration, and actin filament dynamics. Similar findings were observed in PC12 cells, where knockdown of MK5 reduced forskolin-induced F-actin levels (129). Recent evidence suggests that F-actin rearrangement requires MK5-mediated phosphorylation of Hsp27 (193, 194), but more experimentation with genetic models lacking MK2/3 or MK5 will be required to confirm the role of MK5 in actin remodeling.
CONCLUSIONS AND PERSPECTIVES
MAPKAPK family members display relatively high homology and are activated by similar mechanisms involving conserved sequences in their kinase domains. Despite these facts, activation of each MAPKAPK family member results in regulation of specific substrates and cellular functions. While all MAPKAPKs appear to have independent functions, these kinases may also act in concert to mediate global biological responses. For example, MAPKAPK activity may generally be needed under mitogenic or stressful conditions, where, for example, stress-induced p38 activation results in increased cytokine production. In this instance, cytokine gene transcription may require chromatin remodeling by MSKs, increased mRNA stability by MK2/3, and increased translation mediated by MNKs.
Many reports have established the role of MAPKAPKs in various biological processes, including the response to mitogens, oncogenes, stress, and inflammation, as well as the regulation of proliferation, differentiation, and survival in particular cell types. There is also ample information on pathway components and their regulatory mechanisms. However, very little is known about in vivo functions, and detailed molecular information on how these signaling molecules regulate particular cellular processes is still scarce. Outstanding questions that should be addressed in the future include (i) the extent of functional redundancy and interplay between MAPKAPK family members, (ii) how cross talk with other signaling pathways contributes to context-specific responses, (iii) the identification of bona fide substrates that are responsible for specific function, and (iv) the physiological and pathological roles of MAPKAPK family members.
The application of systems biology approaches and high-throughput genomic and proteomic techniques may provide valuable insights into these important questions. Moreover, the use of genetically modified mice to modulate expression of the MAPKAPKs in a time- and tissue-specific manner will be very useful to elucidate in vivo functions. The identification of MK5 as a potential mediator of oncogene-induced senescence or of MK2 as an essential mediator of the inflammatory response underscores the fact that the generated knowledge may be translatable into new therapeutic opportunities. It is likely that we are just beginning to understand the cellular processes regulated by the MAPKAPKs, and future studies should be most enlightening.
Acknowledgments
We thank Sylvain Meloche, Audrey Carrière, and Yves Romeo for critical reading of the manuscript.
Research in the Roux laboratory is supported by the Terry Fox Foundation through the Cancer Research Society Research Institute (CCSRI) and by a Career Development Award from the Human Frontier Science Program Organization (HFSPO). Philippe Roux holds a Canada Research Chair in Signal Transduction and Proteomics. Marie Cargnello is recipient of a doctoral award from the Cole Foundation. IRIC core facilities are supported by the Fonds de la Recherche en Santé du Québec (FRSQ).
Biography
 Marie Cargnello received her B.Sc. and M.Sc. degrees in molecular and cellular biology in 2007 from the University of Cergy-Pontoise, France. In 2008, she joined the system biology Ph.D. program at the Institute for Research in Immunology and Cancer, University of Montreal, Canada, where she is a graduate student in the group of Dr. Philippe Roux. Her research interests focus on the signaling events that control cell growth and proliferation and especially how they regulate translation and protein synthesis.
Marie Cargnello received her B.Sc. and M.Sc. degrees in molecular and cellular biology in 2007 from the University of Cergy-Pontoise, France. In 2008, she joined the system biology Ph.D. program at the Institute for Research in Immunology and Cancer, University of Montreal, Canada, where she is a graduate student in the group of Dr. Philippe Roux. Her research interests focus on the signaling events that control cell growth and proliferation and especially how they regulate translation and protein synthesis.
 Philippe P. Roux is an Assistant Professor of Pathology and Cell Biology at the University of Montreal, Canada. He is Principal Investigator at the Institute for Research in Immunology and Cancer (IRIC), where he leads the Laboratory of Cell Signaling and Proteomics. He has held the Canada Research Chair in Signal Transduction and Proteomics since 2006. He received his B.Sc. in biological sciences (1995) and M.Sc. in microbiology and immunology (1997) from the University of Montreal and then received his Ph.D. in molecular neuroscience (2002) from McGill University, Canada. After postdoctoral studies on cancer cell signaling in the laboratory of Professor John Blenis at Harvard Medical School, Boston, he joined the University of Montreal as a faculty member. His group is interested mainly in understanding how mitogens and oncogenes regulate cell growth and proliferation. His research focuses on signal transduction pathways that are often deregulated in metabolic diseases and cancer.
Philippe P. Roux is an Assistant Professor of Pathology and Cell Biology at the University of Montreal, Canada. He is Principal Investigator at the Institute for Research in Immunology and Cancer (IRIC), where he leads the Laboratory of Cell Signaling and Proteomics. He has held the Canada Research Chair in Signal Transduction and Proteomics since 2006. He received his B.Sc. in biological sciences (1995) and M.Sc. in microbiology and immunology (1997) from the University of Montreal and then received his Ph.D. in molecular neuroscience (2002) from McGill University, Canada. After postdoctoral studies on cancer cell signaling in the laboratory of Professor John Blenis at Harvard Medical School, Boston, he joined the University of Montreal as a faculty member. His group is interested mainly in understanding how mitogens and oncogenes regulate cell growth and proliferation. His research focuses on signal transduction pathways that are often deregulated in metabolic diseases and cancer.
REFERENCES
- 1.Abe, M. K., et al. 2001. ERK7 is an autoactivated member of the MAPK family. J. Biol. Chem. 276**:**21272-21279. [DOI] [PubMed] [Google Scholar]
- 2.Abe, M. K., W. L. Kuo, M. B. Hershenson, and M. R. Rosner. 1999. Extracellular signal-regulated kinase 7 (ERK7), a novel ERK with a C-terminal domain that regulates its activity, its cellular localization, and cell growth. Mol. Cell. Biol. 19**:**1301-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abe, M. K., et al. 2002. ERK8, a new member of the mitogen-activated protein kinase family. J. Biol. Chem. 277**:**16733-16743. [DOI] [PubMed] [Google Scholar]
- 4.Aberg, E., et al. 2006. Regulation of MAPK-activated protein kinase 5 activity and subcellular localization by the atypical MAPK ERK4/MAPK4. J. Biol. Chem. 281**:**35499-35510. [DOI] [PubMed] [Google Scholar]
- 5.Aberg, E., et al. 2009. Docking of PRAK/MK5 to the atypical MAPKs ERK3 and ERK4 defines a novel MAPK interaction motif. J. Biol. Chem. 284**:**19392-19401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams, R. H., et al. 2000. Essential role of p38alpha MAP kinase in placental but not embryonic cardiovascular development. Mol. Cell 6**:**109-116. [PubMed] [Google Scholar]
- 7.Alcorta, D. A., et al. 1989. Sequence and expression of chicken and mouse rsk: homologs of Xenopus laevis ribosomal S6 kinase. Mol. Cell. Biol. 9**:**3850-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alessi, D. R., A. Cuenda, P. Cohen, D. T. Dudley, and A. R. Saltiel. 1995. PD 098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. J. Biol. Chem. 270**:**27489-27494. [DOI] [PubMed] [Google Scholar]
- 9.Ambrosino, C., and A. R. Nebreda. 2001. Cell cycle regulation by p38 MAP kinases. Biol. Cell 93**:**47-51. [DOI] [PubMed] [Google Scholar]
- 10.Ananieva, O., et al. 2008. The kinases MSK1 and MSK2 act as negative regulators of Toll-like receptor signaling. Nat. Immunol. 9**:**1028-1036. [DOI] [PubMed] [Google Scholar]
- 11.Angenstein, F., W. T. Greenough, and I. J. Weiler. 1998. Metabotropic glutamate receptor-initiated translocation of protein kinase p90rsk to polyribosomes: a possible factor regulating synaptic protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 95**:**15078-15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anjum, R., P. P. Roux, B. A. Ballif, S. P. Gygi, and J. Blenis. 2005. The tumor suppressor DAP kinase is a target of RSK-mediated survival signaling. Curr. Biol. 15**:**1762-1767. [DOI] [PubMed] [Google Scholar]
- 13.Aressy, B., and B. Ducommun. 2008. Cell cycle control by the CDC25 phosphatases. Anticancer Agents Med. Chem. 8**:**818-824. [DOI] [PubMed] [Google Scholar]
- 14.Arthur, J. S. 2008. MSK activation and physiological roles. Front. Biosci. 13**:**5866-5879. [DOI] [PubMed] [Google Scholar]
- 15.Arthur, J. S., and P. Cohen. 2000. MSK1 is required for CREB phosphorylation in response to mitogens in mouse embryonic stem cells. FEBS Lett. 482**:**44-48. [DOI] [PubMed] [Google Scholar]
- 16.Asp, E., and P. Sunnerhagen. 2003. Mkp1 and Mkp2, two MAPKAP-kinase homologues in Schizosaccharomyces pombe, interact with the MAP kinase StyI. Mol. Genet. Genomics 268**:**585-597. [DOI] [PubMed] [Google Scholar]
- 17.Bagrodia, S., B. Derijard, R. J. Davis, and R. A. Cerione. 1995. Cdc42 and PAK-mediated signaling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J. Biol. Chem. 270**:**27995-27998. [DOI] [PubMed] [Google Scholar]
- 18.Bain, J., et al. 2007. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408**:**297-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao, X., H. Deng, J. Johansen, J. Girton, and K. M. Johansen. 2007. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect variegation at pericentric sites in Drosophila heterochromatin. Genetics 176**:**1355-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ben-Levy, R., S. Hooper, R. Wilson, H. F. Paterson, and C. J. Marshall. 1998. Nuclear export of the stress-activated protein kinase p38 mediated by its substrate MAPKAP kinase-2. Curr. Biol. 8**:**1049-1057. [DOI] [PubMed] [Google Scholar]
- 21.Ben-Levy, R., et al. 1995. Identification of novel phosphorylation sites required for activation of MAPKAP kinase-2. EMBO J. 14**:**5920-5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett, B. L., et al. 2001. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. U. S. A. 98**:**13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berns, K., et al. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428**:**431-437. [DOI] [PubMed] [Google Scholar]
- 24.Bhatt, R. R., and J. E. Ferrell, Jr. 2000. Cloning and characterization of Xenopus Rsk2, the predominant p90 Rsk isozyme in oocytes and eggs. J. Biol. Chem. 275**:**32983-32990. [DOI] [PubMed] [Google Scholar]
- 25.Bhatt, R. R., and J. E. Ferrell, Jr. 1999. The protein kinase p90 rsk as an essential mediator of cytostatic factor activity. Science 286**:**1362-1365. [DOI] [PubMed] [Google Scholar]
- 26.Bialik, S., and A. Kimchi. 2004. DAP-kinase as a target for drug design in cancer and diseases associated with accelerated cell death. Semin. Cancer Biol. 14**:**283-294. [DOI] [PubMed] [Google Scholar]
- 27.Bianchini, A., et al. 2008. Phosphorylation of eIF4E by MNKs supports protein synthesis, cell cycle progression and proliferation in prostate cancer cells. Carcinogenesis 29**:**2279-2288. [DOI] [PubMed] [Google Scholar]
- 28.Bignone, P. A., et al. 2007. RPS6KA2, a putative tumour suppressor gene at 6q27 in sporadic epithelial ovarian cancer. Oncogene 26**:**683-700. [DOI] [PubMed] [Google Scholar]
- 29.Bjorbaek, C., Y. Zhao, and D. E. Moller. 1995. Divergent functional roles for p90rsk kinase domains. J. Biol. Chem. 270**:**18848-18852. [DOI] [PubMed] [Google Scholar]
- 30.Bode, A. M., and Z. Dong. 2007. The functional contrariety of JNK. Mol. Carcinog. 46**:**591-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogoyevitch, M. A., K. R. Ngoei, T. T. Zhao, Y. Y. Yeap, and D. C. Ng. 2010. c-Jun N-terminal kinase (JNK) signaling: recent advances and challenges. Biochim. Biophys. Acta 1804**:**463-475. [DOI] [PubMed] [Google Scholar]
- 32.Bohuslav, J., L. F. Chen, H. Kwon, Y. Mu, and W. C. Greene. 2004. p53 induces NF-kappaB activation by an IkappaB kinase-independent mechanism involving phosphorylation of p65 by ribosomal S6 kinase 1. J. Biol. Chem. 279**:**26115-26125. [DOI] [PubMed] [Google Scholar]
- 33.Bollig, F., et al. 2003. Affinity purification of ARE-binding proteins identifies polyA-binding protein 1 as a potential substrate in MK2-induced mRNA stabilization. Biochem. Biophys. Res. Commun. 301**:**665-670. [DOI] [PubMed] [Google Scholar]
- 34.Bonni, A., et al. 1999. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 286**:**1358-1362. [DOI] [PubMed] [Google Scholar]
- 35.Boulton, T. G., et al. 1991. ERKs: a family of protein-serine/threonine kinases that are activated and tyrosine phosphorylated in response to insulin and NGF. Cell 65**:**663-675. [DOI] [PubMed] [Google Scholar]
- 36.Boulton, T. G., et al. 1990. An insulin-stimulated protein kinase similar to yeast kinases involved in cell cycle control. Science 249**:**64-67. [DOI] [PubMed] [Google Scholar]
- 37.Bradley, J. R., and J. S. Pober. 2001. Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20**:**6482-6491. [DOI] [PubMed] [Google Scholar]
- 38.Brook, M., et al. 2006. Posttranslational regulation of tristetraprolin subcellular localization and protein stability by p38 mitogen-activated protein kinase and extracellular signal-regulated kinase pathways. Mol. Cell. Biol. 26**:**2408-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brott, B. K., B. A. Pinsky, and R. L. Erikson. 1998. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc. Natl. Acad. Sci. U. S. A. 95**:**963-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruning, J. C., et al. 2000. Ribosomal subunit kinase-2 is required for growth factor-stimulated transcription of the c-Fos gene. Proc. Natl. Acad. Sci. U. S. A. 97**:**2462-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Buck, M., V. Poli, T. Hunter, and M. Chojkier. 2001. C/EBPbeta phosphorylation by RSK creates a functional XEXD caspase inhibitory box critical for cell survival. Mol. Cell 8**:**807-816. [DOI] [PubMed] [Google Scholar]
- 42.Bundschu, K., U. Walter, and K. Schuh. 2006. The VASP-Spred-Sprouty domain puzzle. J. Biol. Chem. 281**:**36477-36481. [DOI] [PubMed] [Google Scholar]
- 43.Buxade, M., N. Morrice, D. L. Krebs, and C. G. Proud. 2008. The PSF.p54nrb complex is a novel Mnk substrate that binds the mRNA for tumor necrosis factor alpha. J. Biol. Chem. 283**:**57-65. [DOI] [PubMed] [Google Scholar]
- 44.Buxade, M., J. L. Parra-Palau, and C. G. Proud. 2008. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front. Biosci. 13**:**5359-5373. [DOI] [PubMed] [Google Scholar]
- 45.Buxade, M., et al. 2005. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23**:**177-189. [DOI] [PubMed] [Google Scholar]
- 46.Carboni, S., et al. 2004. AS601245 (1,3-benzothiazol-2-yl (2-[[2-(3-pyridinyl) ethyl] amino]-4 pyrimidinyl) acetonitrile): a c-Jun NH2-terminal protein kinase inhibitor with neuroprotective properties. J. Pharmacol. Exp. Ther. 310**:**25-32. [DOI] [PubMed] [Google Scholar]
- 47.Carriere, A., et al. 2008. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr. Biol. 18**:**1269-1277. [DOI] [PubMed] [Google Scholar]
- 48.Carriere, A., H. Ray, J. Blenis, and P. P. Roux. 2008. The RSK factors of activating the Ras/MAPK signaling cascade. Front. Biosci. 13**:**4258-4275. [DOI] [PubMed] [Google Scholar]
- 49.Chaturvedi, D., H. M. Poppleton, T. Stringfield, A. Barbier, and T. B. Patel. 2006. Subcellular localization and biological actions of activated RSK1 are determined by its interactions with subunits of cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 26**:**4586-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen, L. F., et al. 2005. NF-κB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 25**:**7966-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen, R. H., C. Abate, and J. Blenis. 1993. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc. Natl. Acad. Sci. U. S. A. 90**:**10952-10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen, R. H., P. C. Juo, T. Curran, and J. Blenis. 1996. Phosphorylation of c-Fos at the C-terminus enhances its transforming activity. Oncogene 12**:**1493-1502. [PubMed] [Google Scholar]
- 53.Chen, R. H., C. Sarnecki, and J. Blenis. 1992. Nuclear localization and regulation of erk- and rsk-encoded protein kinases. Mol. Cell. Biol. 12**:**915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen, Z., et al. 2001. MAP kinases. Chem. Rev. 101**:**2449-2476. [DOI] [PubMed] [Google Scholar]
- 55.Cheng, M., T. G. Boulton, and M. H. Cobb. 1996. ERK3 is a constitutively nuclear protein kinase. J. Biol. Chem. 271**:**8951-8958. [DOI] [PubMed] [Google Scholar]
- 56.Cheng, M., et al. 1996. Characterization of a protein kinase that phosphorylates serine 189 of the mitogen-activated protein kinase homolog ERK3. J. Biol. Chem. 271**:**12057-12062. [DOI] [PubMed] [Google Scholar]
- 57.Cheng, T. J., and Y. K. Lai. 1998. Identification of mitogen-activated protein kinase-activated protein kinase-2 as a vimentin kinase activated by okadaic acid in 9L rat brain tumor cells. J. Cell Biochem. 71**:**169-181. [PubMed] [Google Scholar]
- 58.Chevalier, D., and B. G. Allen. 2000. Two distinct forms of MAPKAP kinase-2 in adult cardiac ventricular myocytes. Biochemistry 39**:**6145-6156. [DOI] [PubMed] [Google Scholar]
- 59.Cho, Y. Y., et al. 2007. Ribosomal s6 kinase 2 is a key regulator in tumor promoter induced cell transformation. Cancer Res. 67**:**8104-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi, K. W., and S. Benzer. 1994. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell 78**:**125-136. [DOI] [PubMed] [Google Scholar]
- 61.Chrestensen, C. A., et al. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J. Biol. Chem. 279**:**10176-10184. [DOI] [PubMed] [Google Scholar]
- 62.Chrestensen, C. A., and T. W. Sturgill. 2002. Characterization of the p90 ribosomal S6 kinase 2 carboxyl-terminal domain as a protein kinase. J. Biol. Chem. 277**:**27733-27741. [DOI] [PubMed] [Google Scholar]
- 63.Chun, J., et al. 2005. Phosphorylation of Cdc25C by pp90Rsk contributes to a G(2) cell cycle arrest in Xenopus cycling egg extracts. Cell Cycle 4. [DOI] [PubMed]
- 64.Chung, J., C. J. Kuo, G. R. Crabtree, and J. Blenis. 1992. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell 69**:**1227-1236. [DOI] [PubMed] [Google Scholar]
- 65.Clark, D. E., et al. 2005. The serine/threonine protein kinase, p90 ribosomal S6 kinase, is an important regulator of prostate cancer cell proliferation. Cancer Res. 65**:**3108-3116. [DOI] [PubMed] [Google Scholar]
- 66.Clifton, A. D., P. R. Young, and P. Cohen. 1996. A comparison of the substrate specificity of MAPKAP kinase-2 and MAPKAP kinase-3 and their activation by cytokines and cellular stress. FEBS Lett. 392**:**209-214. [DOI] [PubMed] [Google Scholar]
- 67.Cohen, M. S., H. Hadjivassiliou, and J. Taunton. 2007. A clickable inhibitor reveals context-dependent autoactivation of p90 RSK. Nat. Chem. Biol. 3**:**156-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cohen, M. S., C. Zhang, K. M. Shokat, and J. Taunton. 2005. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science 308**:**1318-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Collins, B. J., M. Deak, J. S. Arthur, L. J. Armit, and D. R. Alessi. 2003. In vivo role of the PIF-binding docking site of PDK1 defined by knock-in mutation. EMBO J. 22**:**4202-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cooper, J. A., D. F. Bowen-Pope, E. Raines, R. Ross, and T. Hunter. 1982. Similar effects of platelet-derived growth factor and epidermal growth factor on the phosphorylation of tyrosine in cellular proteins. Cell 31**:**263-273. [DOI] [PubMed] [Google Scholar]
- 71.Coulombe, P., and S. Meloche. 2007. Atypical mitogen-activated protein kinases: structure, regulation and functions. Biochim. Biophys. Acta 1773**:**1376-1387. [DOI] [PubMed] [Google Scholar]
- 72.Coulombe, P., G. Rodier, E. Bonneil, P. Thibault, and S. Meloche. 2004. N-terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol. Cell. Biol. 24**:**6140-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coulombe, P., G. Rodier, S. Pelletier, J. Pellerin, and S. Meloche. 2003. Rapid turnover of extracellular signal-regulated kinase 3 by the ubiquitin-proteasome pathway defines a novel paradigm of mitogen-activated protein kinase regulation during cellular differentiation. Mol. Cell. Biol. 23**:**4542-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cox, B. J., et al. 2010. Phenotypic annotation of the mouse X chromosome. Genome Res. 20**:**1154-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Crowe, D. L. 2004. Induction of p97MAPK expression regulates collagen mediated inhibition of proliferation and migration in human squamous cell carcinoma lines. Int. J. Oncol. 24**:**1159-1163. [PubMed] [Google Scholar]
- 76.Cuadrado, A., and A. R. Nebreda. 2010. Mechanisms and functions of p38 MAPK signalling. Biochem. J. 429**:**403-417. [DOI] [PubMed] [Google Scholar]
- 77.Cuenda, A., and S. Rousseau. 2007. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim. Biophys. Acta 1773**:**1358-1375. [DOI] [PubMed] [Google Scholar]
- 78.Dalby, K. N., N. Morrice, F. B. Caudwell, J. Avruch, and P. Cohen. 1998. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90rsk that are inducible by MAPK. J. Biol. Chem. 273**:**1496-1505. [DOI] [PubMed] [Google Scholar]
- 79.DaSilva, J., L. Xu, H. J. Kim, W. T. Miller, and D. Bar-Sagi. 2006. Regulation of sprouty stability by Mnk1-dependent phosphorylation. Mol. Cell. Biol. 26**:**1898-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.David, J. P., et al. 2005. Essential role of RSK2 in c-Fos-dependent osteosarcoma development. J. Clin. Invest. 115**:**664-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davie, J. R. 2003. MSK1 and MSK2 mediate mitogen- and stress-induced phosphorylation of histone H3: a controversy resolved. Sci. STKE 2003**:**PE33. [DOI] [PubMed] [Google Scholar]
- 82.Davis, I. J., T. G. Hazel, R. H. Chen, J. Blenis, and L. F. Lau. 1993. Functional domains and phosphorylation of the orphan receptor Nur77. Mol. Endocrinol. 7**:**953-964. [DOI] [PubMed] [Google Scholar]
- 83.Deak, M., A. D. Clifton, L. M. Lucocq, and D. R. Alessi. 1998. Mitogen- and stress-activated protein kinase-1 (MSK1) is directly activated by MAPK and SAPK2/p38, and may mediate activation of CREB. EMBO J. 17**:**4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Cesare, D., S. Jacquot, A. Hanauer, and P. Sassone-Corsi. 1998. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc. Natl. Acad. Sci. U. S. A. 95**:**12202-12207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Delaunoy, J., et al. 2001. Mutations in the X-linked RSK2 gene (RPS6KA3) in patients with Coffin-Lowry syndrome. Hum. Mutat. 17**:**103-116. [DOI] [PubMed] [Google Scholar]
- 86.Deleris, P., et al. 2008. Activation loop phosphorylation of the atypical MAP kinases ERK3 and ERK4 is required for binding, activation and cytoplasmic relocalization of MK5. J. Cell Physiol. 217**:**778-788. [DOI] [PubMed] [Google Scholar]
- 87.de Nadal, E., and F. Posas. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 29**:**4-13. [DOI] [PMC free article] [PubMed]
- 88.Derijard, B., et al. 1994. JNK1: A protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76**:**1025-1037. [DOI] [PubMed] [Google Scholar]
- 89.Derijard, B., et al. 1995. Independent human MAP kinase signal transduction pathways defined by MEK and MKK isoforms. Science 267**:**682-685. [DOI] [PubMed] [Google Scholar]
- 90.Dhanasekaran, D. N., and E. P. Reddy. 2008. JNK signaling in apoptosis. Oncogene 27**:**6245-6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dingar, D., et al. 2010. Characterization of the expression and regulation of MK5 in the murine ventricular myocardium. Cell Signal. 22**:**1063-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Doehn, U., S. Gammeltoft, S. H. Shen, and C. J. Jensen. 2004. p90 ribosomal S6 kinase 2 is associated with and dephosphorylated by protein phosphatase 2Cdelta. Biochem. J. 382**:**425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Doehn, U., et al. 2009. RSK is a principal effector of the RAS-ERK pathway for eliciting a coordinate promotile/invasive gene program and phenotype in epithelial cells. Mol. Cell 35**:**511-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Douillette, A., et al. 2006. The proinflammatory actions of angiotensin II are dependent on p65 phosphorylation by the IkappaB kinase complex. J. Biol. Chem. 281**:**13275-13284. [DOI] [PubMed] [Google Scholar]
- 95.Douville, E., and J. Downward. 1997. EGF induced SOS phosphorylation in PC12 cells involves P90 RSK-2. Oncogene 15**:**373-383. [DOI] [PubMed] [Google Scholar]
- 96.Dreiza, C. M., et al. 2005. Transducible heat shock protein 20 (HSP20) phosphopeptide alters cytoskeletal dynamics. FASEB J. 19**:**261-263. [DOI] [PubMed] [Google Scholar]
- 97.Drobic, B., B. Perez-Cadahia, J. Yu, S. K. Kung, and J. R. Davie. 2010. Promoter chromatin remodeling of immediate-early genes is mediated through H3 phosphorylation at either serine 28 or 10 by the MSK1 multi-protein complex. Nucleic Acids Res. 38**:**3196-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dudley, D. T., L. Pang, S. J. Decker, A. J. Bridges, and A. R. Saltiel. 1995. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc. Natl. Acad. Sci. U. S. A. 92**:**7686-7689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dufresne, S. D., et al. 2001. Altered extracellular signal-regulated kinase signaling and glycogen metabolism in skeletal muscle from p90 ribosomal S6 kinase 2 knockout mice. Mol. Cell. Biol. 21**:**81-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dummler, B. A., et al. 2005. Functional characterization of human RSK4, a new 90-kDa ribosomal S6 kinase, reveals constitutive activation in most cell types. J. Biol. Chem. 280**:**13304-13314. [DOI] [PubMed] [Google Scholar]
- 101.Dumont, J., M. Umbhauer, P. Rassinier, A. Hanauer, and M. H. Verlhac. 2005. p90Rsk is not involved in cytostatic factor arrest in mouse oocytes. J. Cell Biol. 169**:**227-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eisinger-Mathason, T. S., et al. 2008. Codependent functions of RSK2 and the apoptosis-promoting factor TIA-1 in stress granule assembly and cell survival. Mol. Cell 31**:**722-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.El-Haschimi, K., et al. 2003. Insulin resistance and lipodystrophy in mice lacking ribosomal S6 kinase 2. Diabetes 52**:**1340-1346. [DOI] [PubMed] [Google Scholar]
- 104.Engel, K., A. Kotlyarov, and M. Gaestel. 1998. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 17**:**3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Engel, K., K. Plath, and M. Gaestel. 1993. The MAP kinase-activated protein kinase 2 contains a proline-rich SH3-binding domain. FEBS Lett. 336**:**143-147. [DOI] [PubMed] [Google Scholar]
- 106.English, J. M., C. A. Vanderbilt, S. Xu, S. Marcus, and M. H. Cobb. 1995. Isolation of MEK5 and differential expression of alternatively spliced forms. J. Biol. Chem. 270**:**28897-28902. [DOI] [PubMed] [Google Scholar]
- 107.Enslen, H., and R. J. Davis. 2001. Regulation of MAP kinases by docking domains. Biol. Cell 93**:**5-14. [DOI] [PubMed] [Google Scholar]
- 108.Erikson, E., and J. L. Maller. 1985. A protein kinase from Xenopus eggs specific for ribosomal protein S6. Proc. Natl. Acad. Sci. U. S. A. 82**:**742-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Erikson, E., and J. L. Maller. 1986. Purification and characterization of a protein kinase from Xenopus eggs highly specific for ribosomal protein S6. J. Biol. Chem. 261**:**350-355. [PubMed] [Google Scholar]
- 110.Eyers, C. E., et al. 2005. The phosphorylation of CapZ-interacting protein (CapZIP) by stress-activated protein kinases triggers its dissociation from CapZ. Biochem. J. 389**:**127-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fantz, D. A., D. Jacobs, D. Glossip, and K. Kornfeld. 2001. Docking sites on substrate proteins direct extracellular signal-regulated kinase to phosphorylate specific residues. J. Biol. Chem. 276**:**27256-27265. [DOI] [PubMed] [Google Scholar]
- 112.Favata, M. F., et al. 1998. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 273**:**18623-18632. [DOI] [PubMed] [Google Scholar]
- 113.Fernando, P., L. A. Megeney, and J. J. Heikkila. 2003. Phosphorylation-dependent structural alterations in the small hsp30 chaperone are associated with cellular recovery. Exp. Cell Res. 286**:**175-185. [DOI] [PubMed] [Google Scholar]
- 114.Fisher, T. L., and J. Blenis. 1996. Evidence for two catalytically active kinase domains in pp90rsk. Mol. Cell. Biol. 16**:**1212-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Flynn, A., R. G. Vries, and C. G. Proud. 1997. Signalling pathways which regulate eIF4E. Biochem. Soc Trans 25**:**192S. [DOI] [PubMed] [Google Scholar]
- 116.Fremin, C., and S. Meloche. 2010. From basic research to clinical development of MEK1/2 inhibitors for cancer therapy. J. Hematol. Oncol. 3**:**8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Freshney, N. W., et al. 1994. Interleukin-1 activates a novel protein kinase cascade that results in the phosphorylation of Hsp27. Cell 78**:**1039-1049. [DOI] [PubMed] [Google Scholar]
- 118.Frodin, M., and S. Gammeltoft. 1999. Role and regulation of 90 kDa ribosomal S6 kinase (RSK) in signal transduction. Mol. Cell Endocrinol. 151**:**65-77. [DOI] [PubMed] [Google Scholar]
- 119.Frodin, M., C. J. Jensen, K. Merienne, and S. Gammeltoft. 2000. A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1. EMBO J. 19**:**2924-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fujita, N., S. Sato, and T. Tsuruo. 2003. Phosphorylation of p27Kip1 at threonine 198 by p90 ribosomal protein S6 kinases promotes its binding to 14-3-3 and cytoplasmic localization. J. Biol. Chem. 278**:**49254-49260. [DOI] [PubMed] [Google Scholar]
- 121.Fukunaga, R., and T. Hunter. 1997. MNK1, a new MAP kinase-activate protein kinase, isolated by a novel expression screening method for identifying protein kinase substrates. EMBO J. 16**:**1921-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gaestel, M. 2006. MAPKAP kinases—MKs—two's company, three's a crowd. Nat. Rev. Mol. Cell. Biol. 7**:**120-130. [DOI] [PubMed] [Google Scholar]
- 123.Gaestel, M. 2008. Specificity of signaling from MAPKs to MAPKAPKs: kinases' tango nuevo. Front. Biosci. 13**:**6050-6059. doi:http://dx.doi.org/10.2741/3136. [DOI] [PubMed] [Google Scholar]
- 124.Gaestel, M., A. Mengel, U. Bothe, and K. Asadullah. 2007. Protein kinases as small molecule inhibitor targets in inflammation. Curr. Med. Chem. 14**:**2214-2234. [DOI] [PubMed] [Google Scholar]
- 125.Galanis, A., S. H. Yang, and A. D. Sharrocks. 2001. Selective targeting of MAPKs to the ETS domain transcription factor SAP-1. J. Biol. Chem. 276**:**965-973. [DOI] [PubMed] [Google Scholar]
- 126.Garcia, J. I., G. Zalba, S. D. Detera-Wadleigh, and C. de Miguel. 1996. Isolation of a cDNA encoding the rat MAP-kinase homolog of human p63mapk. Mamm. Genome 7**:**810-814. [DOI] [PubMed] [Google Scholar]
- 127.Gavin, A. C., and A. R. Nebreda. 1999. A MAP kinase docking site is required for phosphorylation and activation of p90(rsk)/MAPKAP kinase-1. Curr. Biol. 9**:**281-284. [DOI] [PubMed] [Google Scholar]
- 128.Geraghty, K. M., et al. 2007. Regulation of multisite phosphorylation and 14-3-3 binding of AS160 in response to IGF-1, EGF, PMA and AICAR. Biochem. J. 407**:**231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gerits, N., et al. 2007. Modulation of F-actin rearrangement by the cyclic AMP/cAMP-dependent protein kinase (PKA) pathway is mediated by MAPK-activated protein kinase 5 and requires PKA-induced nuclear export of MK5. J. Biol. Chem. 282**:**37232-37243. [DOI] [PubMed] [Google Scholar]
- 130.Gerits, N., et al. 2009. The transcriptional regulation and cell-specific expression of the MAPK-activated protein kinase MK5. Cell Mol. Biol. Lett. 14**:**548-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ghoda, L., X. Lin, and W. C. Greene. 1997. The 90-kDa ribosomal S6 kinase (pp90rsk) phosphorylates the N-terminal regulatory domain of IkappaBalpha and stimulates its degradation in vitro. J. Biol. Chem. 272**:**21281-21288. [DOI] [PubMed] [Google Scholar]
- 132.Gille, H., et al. 1995. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 14**:**951-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ginty, D. D., A. Bonni, and M. E. Greenberg. 1994. Nerve growth factor activates a Ras-dependent protein kinase that stimulates c-fos transcription via phosphorylation of CREB. Cell 77**:**713-725. [DOI] [PubMed] [Google Scholar]
- 134.Goldsmith, Z. G., and D. N. Dhanasekaran. 2007. G protein regulation of MAPK networks. Oncogene 26**:**3122-3142. [DOI] [PubMed] [Google Scholar]
- 135.Gonzalez, F. A., D. L. Raden, M. R. Rigby, and R. J. Davis. 1992. Heterogeneous expression of four MAP kinase isoforms in human tissues. FEBS Lett. 304**:**170-178. [DOI] [PubMed] [Google Scholar]
- 136.Greenman, C., et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446**:**153-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gross, S. D., A. L. Lewellyn, and J. L. Maller. 2001. A constitutively active form of the protein kinase p90Rsk1 is sufficient to trigger the G2/M transition in Xenopus oocytes. J. Biol. Chem. 276**:**46099-46103. [DOI] [PubMed] [Google Scholar]
- 138.Gross, S. D., M. S. Schwab, A. L. Lewellyn, and J. L. Maller. 1999. Induction of metaphase arrest in cleaving Xenopus embryos by the protein kinase p90Rsk. Science 286**:**1365-1367. [DOI] [PubMed] [Google Scholar]
- 139.Guay, J., et al. 1997. Regulation of actin filament dynamics by p38 MAP kinase-mediated phosphorylation of heat shock protein 27. J. Cell Sci. 110**:**357-368. [DOI] [PubMed] [Google Scholar]
- 140.Gupta, S., et al. 1996. Selective interaction of JNK protein kinase isoforms with transcription factors. EMBO J. 15**:**2760-2770. [PMC free article] [PubMed] [Google Scholar]
- 141.Hamm, J., D. R. Alessi, and R. M. Biondi. 2002. Bi-functional, substrate mimicking RNA inhibits MSK1-mediated cAMP-response element-binding protein phosphorylation and reveals magnesium ion-dependent conformational changes of the kinase. J. Biol. Chem. 277**:**45793-45802. [DOI] [PubMed] [Google Scholar]
- 142.Han, J., J. D. Lee, L. Bibbs, and R. J. Ulevitch. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 265**:**808-811. [DOI] [PubMed] [Google Scholar]
- 143.Han, J., et al. 1996. Characterization of the structure and function of a novel MAP kinase kinase (MKK6). J. Biol. Chem. 271**:**2886-2891. [DOI] [PubMed] [Google Scholar]
- 144.Han, Q., et al. 2002. Rac1-MKK3-p38-MAPKAPK2 pathway promotes urokinase plasminogen activator mRNA stability in invasive breast cancer cells. J. Biol. Chem. 277**:**48379-48385. [DOI] [PubMed] [Google Scholar]
- 145.Hanauer, A., and I. D. Young. 2002. Coffin-Lowry syndrome: clinical and molecular features. J. Med. Genet. 39**:**705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hannigan, M. O., et al. 2001. Abnormal migration phenotype of mitogen-activated protein kinase-activated protein kinase 2−/− neutrophils in Zigmond chambers containing formyl-methionyl-leucyl-phenylalanine gradients. J. Immunol. 167**:**3953-3961. [DOI] [PubMed] [Google Scholar]
- 147.Hayashi, M., and J. D. Lee. 2004. Role of the BMK1/ERK5 signaling pathway: lessons from knockout mice. J. Mol. Med. 82**:**800-808. [DOI] [PubMed] [Google Scholar]
- 148.Hedges, J. C., et al. 1999. A role for p38(MAPK)/HSP27 pathway in smooth muscle cell migration. J. Biol. Chem. 274**:**24211-24219. [DOI] [PubMed] [Google Scholar]
- 149.Heffron, D., and J. W. Mandell. 2005. Differential localization of MAPK-activated protein kinases RSK1 and MSK1 in mouse brain. Brain Res. Mol. Brain Res. 136**:**134-141. [DOI] [PubMed] [Google Scholar]
- 150.Hefner, Y., et al. 2000. Serine 727 phosphorylation and activation of cytosolic phospholipase A2 by MNK1-related protein kinases. J. Biol. Chem. 275**:**37542-37551. [DOI] [PubMed] [Google Scholar]
- 151.Heidenreich, O., et al. 1999. MAPKAP kinase 2 phosphorylates serum response factor in vitro and in vivo. J. Biol. Chem. 274**:**14434-14443. [DOI] [PubMed] [Google Scholar]
- 152.Henrich, L. M., et al. 2003. Extracellular signal-regulated kinase 7, a regulator of hormone-dependent estrogen receptor destruction. Mol. Cell. Biol. 23**:**5979-5988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hibi, M., A. Lin, T. Smeal, A. Minden, and M. Karin. 1993. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 7**:**2135-2148. [DOI] [PubMed] [Google Scholar]
- 154.Hitti, E., et al. 2006. Mitogen-activated protein kinase-activated protein kinase 2 regulates tumor necrosis factor mRNA stability and translation mainly by altering tristetraprolin expression, stability, and binding to adenine/uridine-rich element. Mol. Cell. Biol. 26**:**2399-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hock, R., T. Furusawa, T. Ueda, and M. Bustin. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 17**:**72-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hsiao, K.-M., S.-Y. Chou, S.-J. Shih, and J. E. Ferrell, Jr. 1994. Evidence that inactive p42 mitogen-activated protein kinase and inactive Rsk exist as a heterodimer in vivo. Proc. Natl. Acad. Sci. U. S. A. 91**:**5480-5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Huang, C. K., L. Zhan, Y. Ai, and J. Jongstra. 1997. LSP1 is the major substrate for mitogen-activated protein kinase-activated protein kinase 2 in human neutrophils. J. Biol. Chem. 272**:**17-19. [DOI] [PubMed] [Google Scholar]
- 158.Inoki, K., et al. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126**:**955-968. [DOI] [PubMed] [Google Scholar]
- 159.Inoki, K., T. Zhu, and K. L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115**:**577-590. [DOI] [PubMed] [Google Scholar]
- 160.Inoue, D., M. Ohe, Y. Kanemori, T. Nobui, and N. Sagata. 2007. A direct link of the Mos-MAPK pathway to Erp1/Emi2 in meiotic arrest of Xenopus laevis eggs. Nature 446**:**1100-1104. [DOI] [PubMed] [Google Scholar]
- 161.Ishitani, T., et al. 1999. The TAK1-NLK-MAPK-related pathway antagonizes signalling between beta-catenin and transcription factor TCF. Nature 399**:**798-802. [DOI] [PubMed] [Google Scholar]
- 162.Jacks, K. A., and C. A. Koch. 2010. Differential regulation of mitogen- and stress-activated protein kinase-1 and -2 (MSK1 and MSK2) by CK2 following UV radiation. J. Biol. Chem. 285**:**1661-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Jacobs, D., G. J. Beitel, S. G. Clark, H. R. Horvitz, and K. Kornfeld. 1998. Gain-of-function mutations in the Caenorhabditis elegans lin-1 ETS gene identify a C-terminal regulatory domain phosphorylated by ERK MAP kinase. Genetics 149**:**1809-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Jaeschke, A., et al. 2006. JNK2 is a positive regulator of the cJun transcription factor. Mol. Cell 23**:**899-911. [DOI] [PubMed] [Google Scholar]
- 165.Janknecht, R. 2001. Cell type-specific inhibition of the ETS transcription factor ER81 by mitogen-activated protein kinase-activated protein kinase 2. J. Biol. Chem. 276**:**41856-41861. [DOI] [PubMed] [Google Scholar]
- 166.Janknecht, R. 2003. Regulation of the ER81 transcription factor and its coactivators by mitogen- and stress-activated protein kinase 1 (MSK1). Oncogene 22**:**746-755. [DOI] [PubMed] [Google Scholar]
- 167.Jensen, C. J., et al. 1999. 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1. J. Biol. Chem. 274**:**27168-27176. [DOI] [PubMed] [Google Scholar]
- 168.Jiang, Y., et al. 1996. Characterization of the structure and function of a new mitogen-activated protein kinase (p38beta). J. Biol. Chem. 271**:**17920-17926. [DOI] [PubMed] [Google Scholar]
- 169.Jin, Y., et al. 1999. JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol. Cell 4**:**129-135. [DOI] [PubMed] [Google Scholar]
- 170.Joel, P. B., et al. 1998. pp90rsk1 regulates estrogen receptor-mediated transcription through phosphorylation of Ser-167. Mol. Cell. Biol. 18**:**1978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Johansen, C., et al. 2009. MK2 regulates the early stages of skin tumor promotion. Carcinogenesis 30**:**2100-2108. [DOI] [PubMed] [Google Scholar]
- 172.Jones, S. W., E. Erikson, J. Blenis, J. L. Maller, and R. L. Erikson. 1988. A Xenopus ribosomal protein S6 kinase has two apparent kinase domains that are each similar to distinct protein kinases. Proc. Natl. Acad. Sci. U. S. A. 85**:**3377-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Julien, C., P. Coulombe, and S. Meloche. 2003. Nuclear export of ERK3 by a CRM1-dependent mechanism regulates its inhibitory action on cell cycle progression. J. Biol. Chem. 278**:**42615-42624. [DOI] [PubMed] [Google Scholar]
- 174.Julien, L.-A., and P. P. Roux. 2007. Rsk3. UCSD-Nat. Mol. Pages, doi: 10.1038/mp.a002101.01. [DOI]
- 175.Kamakura, S., T. Moriguchi, and E. Nishida. 1999. Activation of the protein kinase ERK5/BMK1 by receptor tyrosine kinases. Identification and characterization of a signaling pathway to the nucleus. J. Biol. Chem. 274**:**26563-26571. [DOI] [PubMed] [Google Scholar]
- 176.Kanei-Ishii, C., et al. 2004. Wnt-1 signal induces phosphorylation and degradation of c-Myb protein via TAK1, HIPK2, and NLK. Genes Dev. 18**:**816-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kang, S., et al. 2007. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell 12**:**201-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kang, S., et al. 2008. Epidermal growth factor stimulates RSK2 activation through activation of the MEK/ERK pathway and src-dependent tyrosine phosphorylation of RSK2 at Tyr-529. J. Biol. Chem. 283**:**4652-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kant, S., et al. 2006. Characterization of the atypical MAPK ERK4 and its activation of the MAPK-activated protein kinase MK5. J. Biol. Chem. 281**:**35511-35519. [DOI] [PubMed] [Google Scholar]
- 180.Karin, M. 2006. Nuclear factor-kappaB in cancer development and progression. Nature 441**:**431-436. [DOI] [PubMed] [Google Scholar]
- 181.Karnoub, A. E., and R. A. Weinberg. 2008. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell. Biol. 9**:**517-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kato, K., et al. 1998. Phosphorylation of alphaB-crystallin in mitotic cells and identification of enzymatic activities responsible for phosphorylation. J. Biol. Chem. 273**:**28346-28354. [DOI] [PubMed] [Google Scholar]
- 183.Katsanis, N., J. R. Lupski, and P. L. Beales. 2001. Exploring the molecular basis of Bardet-Biedl syndrome. Hum. Mol. Genet. 10**:**2293-2299. [DOI] [PubMed] [Google Scholar]
- 184.Kazlauskas, A., and J. A. Cooper. 1988. Protein kinase C mediates platelet-derived growth factor-induced tyrosine phosphorylation of p42. J. Cell Biol. 106**:**1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kidd, D., and J. W. Raff. 1997. LK6, a short lived protein kinase in Drosophila that can associate with microtubules and centrosomes. J. Cell Sci. 110**:**209-219. [DOI] [PubMed] [Google Scholar]
- 186.Kim, C., et al. 2008. The kinase p38 alpha serves cell type-specific inflammatory functions in skin injury and coordinates pro- and anti-inflammatory gene expression. Nat. Immunol. 9**:**1019-1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kim, H. G., et al. 2008. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res. 68**:**2538-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Klevernic, I. V., et al. 2006. Characterization of the reversible phosphorylation and activation of ERK8. Biochem. J. 394**:**365-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Klinger, S., et al. 2009. Loss of Erk3 function in mice leads to intrauterine growth restriction, pulmonary immaturity, and neonatal lethality. Proc. Natl. Acad. Sci. U. S. A. 106**:**16710-16715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Knauf, U., C. Tschopp, and H. Gram. 2001. Negative regulation of protein translation by mitogen-activated protein kinase-interacting kinases 1 and 2. Mol. Cell. Biol. 21**:**5500-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Kohn, M., H. Hameister, M. Vogel, and H. Kehrer-Sawatzki. 2003. Expression pattern of the Rsk2, Rsk4 and Pdk1 genes during murine embryogenesis. Gene Expr. Patterns 3**:**173-177. [DOI] [PubMed] [Google Scholar]
- 192.Kojima, H., et al. 2005. STAT3 regulates Nemo-like kinase by mediating its interaction with IL-6-stimulated TGFbeta-activated kinase 1 for STAT3 Ser-727 phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 102**:**4524-4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kostenko, S., M. Johannessen, and U. Moens. 2009. PKA-induced F-actin rearrangement requires phosphorylation of Hsp27 by the MAPKAP kinase MK5. Cell Signal. 21**:**712-718. [DOI] [PubMed] [Google Scholar]
- 194.Kostenko, S., and U. Moens. 2009. Heat shock protein 27 phosphorylation: kinases, phosphatases, functions and pathology. Cell Mol. Life Sci. 66**:**3289-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kotlyarov, A., et al. 1999. MAPKAP kinase 2 is essential for LPS-induced TNF-alpha biosynthesis. Nat. Cell Biol. 1**:**94-97. [DOI] [PubMed] [Google Scholar]
- 196.Kotlyarov, A., et al. 2002. Distinct cellular functions of MK2. Mol. Cell. Biol. 22**:**4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kuo, W. L., et al. 2004. ERK7 expression and kinase activity is regulated by the ubiquitin-proteosome pathway. J. Biol. Chem. 279**:**23073-23081. [DOI] [PubMed] [Google Scholar]
- 198.Kyriakis, J. M., and J. Avruch. 2001. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 81**:**807-869. [DOI] [PubMed] [Google Scholar]
- 199.Kyriakis, J. M., and J. Avruch. 1990. pp54 microtubule-associated protein 2 kinase. A novel serine/threonine protein kinase regulated by phosphorylation and stimulated by poly-l-lysine. J. Biol. Chem. 265**:**17355-17363. [PubMed] [Google Scholar]
- 200.Kyriakis, J. M., and J. Avruch. 1996. Sounding the alarm: protein kinase cascades activated by stress and inflammation. J. Biol. Chem. 271**:**24313-24316. [DOI] [PubMed] [Google Scholar]
- 201.Kyriakis, J. M., et al. 1994. The stress-activated protein kinase subfamily of c-Jun kinases. Nature 369**:**156-160. [DOI] [PubMed] [Google Scholar]
- 202.Kyriakis, J. M., D. L. Brautigan, T. S. Ingebritsen, and J. Avruch. 1991. pp54 microtubule-associated protein-2 kinase requires both tyrosine and serine/threonine phosphorylation for activity. J. Biol. Chem. 266**:**10043-10046. [PubMed] [Google Scholar]
- 203.Lambert, H., S. J. Charette, A. F. Bernier, A. Guimond, and J. Landry. 1999. HSP27 multimerization mediated by phosphorylation-sensitive intermolecular interactions at the amino terminus. J. Biol. Chem. 274**:**9378-9385. [DOI] [PubMed] [Google Scholar]
- 204.Larochelle, S., and B. Suter. 1995. The Drosophila melanogaster homolog of the mammalian MAPK-activated protein kinase-2 (MAPKAPK-2) lacks a proline-rich N-terminus. Gene 163**:**209-214. [DOI] [PubMed] [Google Scholar]
- 205.Larrea, M. D., et al. 2009. RSK1 drives p27Kip1 phosphorylation at T198 to promote RhoA inhibition and increase cell motility. Proc. Natl. Acad. Sci. U. S. A. 106**:**9268-9273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Lawler, S., Y. Fleming, M. Goedert, and P. Cohen. 1998. Synergistic activation of SAPK1/JNK1 by two MAP kinase kinases in vitro. Curr. Biol. 8**:**1387-1390. [DOI] [PubMed] [Google Scholar]
- 207.Lee, C. W., et al. 2003. Lysophosphatidic acid stimulates CREB through mitogen- and stress-activated protein kinase-1. Biochem. Biophys. Res. Commun. 305**:**455-461. [DOI] [PubMed] [Google Scholar]
- 208.Lee, J. C., et al. 1994. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature 372**:**739-746. [DOI] [PubMed] [Google Scholar]
- 209.Lee, J. D., R. J. Ulevitch, and J. Han. 1995. Primary structure of BMK1: a new mammalian map kinase. Biochem. Biophys. Res. Commun. 213**:**715-724. [DOI] [PubMed] [Google Scholar]
- 210.Lee, T., et al. 2004. Docking motif interactions in MAP kinases revealed by hydrogen exchange mass spectrometry. Mol. Cell 14**:**43-55. [DOI] [PubMed] [Google Scholar]
- 211.Lehner, M. D., et al. 2002. Mitogen-activated protein kinase-activated protein kinase 2-deficient mice show increased susceptibility to Listeria monocytogenes infection. J. Immunol. 168**:**4667-4673. [DOI] [PubMed] [Google Scholar]
- 212.Leighton, I. A., K. N. Dalby, F. B. Caudwell, P. T. Cohen, and P. Cohen. 1995. Comparison of the specificities of p70 S6 kinase and MAPKAP kinase-1 identifies a relatively specific substrate for p70 S6 kinase: the N-terminal kinase domain of MAPKAP kinase-1 is essential for peptide phosphorylation. FEBS Lett. 375**:**289-293. [DOI] [PubMed] [Google Scholar]
- 213.Lenormand, P., et al. 1993. Growth factors induce nuclear translocation of MAP kinases (p42mapk and p44mapk) but not of their activator MAP kinase kinase (p45mapkk) in fibroblasts. J. Cell Biol. 122**:**1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Li, Y., K. Inoki, P. Vacratsis, and K. L. Guan. 2003. The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J. Biol. Chem. 278**:**13663-13671. [DOI] [PubMed] [Google Scholar]
- 215.Lim, J. H., et al. 2004. Chromosomal protein HMGN1 modulates histone H3 phosphorylation. Mol. Cell 15**:**573-584. [DOI] [PubMed] [Google Scholar]
- 216.Lin, J. X., R. Spolski, and W. J. Leonard. 2008. Critical role for Rsk2 in T-lymphocyte activation. Blood 111**:**525-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu, G., Y. Zhang, A. M. Bode, W. Y. Ma, and Z. Dong. 2002. Phosphorylation of 4E-BP1 is mediated by the p38/MSK1 pathway in response to UVB irradiation. J. Biol. Chem. 277**:**8810-8816. [DOI] [PubMed] [Google Scholar]
- 218.Llanos, S., A. Cuadrado, and M. Serrano. 2009. MSK2 inhibits p53 activity in the absence of stress. Sci. Signal 2**:**ra57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Lonze, B. E., and D. D. Ginty. 2002. Function and regulation of CREB family transcription factors in the nervous system. Neuron 35**:**605-623. [DOI] [PubMed] [Google Scholar]
- 220.Lopez-Vicente, L., et al. 2009. Regulation of replicative and stress-induced senescence by RSK4, which is down-regulated in human tumors. Clin. Cancer Res. 15**:**4546-4553. [DOI] [PubMed] [Google Scholar]
- 221.Ludwig, S., et al. 1996. 3pK, a novel mitogen-activated protein (MAP) kinase-activated protein kinase, is targeted by three MAP kinase pathways. Mol. Cell. Biol. 16**:**6687-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ma, L., Z. Chen, H. Erdjument-Bromage, P. Tempst, and P. P. Pandolfi. 2005. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121**:**179-193. [DOI] [PubMed] [Google Scholar]
- 223.Macdonald, N., et al. 2005. Molecular basis for the recognition of phosphorylated and phosphoacetylated histone h3 by 14-3-3. Mol. Cell 20**:**199-211. [DOI] [PubMed] [Google Scholar]
- 224.MacKenzie, S. J., G. S. Baillie, I. McPhee, G. B. Bolger, and M. D. Houslay. 2000. ERK2 mitogen-activated protein kinase binding, phosphorylation, and regulation of the PDE4D cAMP-specific phosphodiesterases. The involvement of COOH-terminal docking sites and NH2-terminal UCR regions. J. Biol. Chem. 275**:**16609-16617. [DOI] [PubMed] [Google Scholar]
- 225.Mahtani, K. R., et al. 2001. Mitogen-activated protein kinase p38 controls the expression and posttranslational modification of tristetraprolin, a regulator of tumor necrosis factor alpha mRNA stability. Mol. Cell. Biol. 21**:**6461-6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Maitra, S., et al. 2008. The AU-rich element mRNA decay-promoting activity of BRF1 is regulated by mitogen-activated protein kinase-activated protein kinase 2. RNA 14**:**950-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Manke, I. A., et al. 2005. MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell 17**:**37-48. [DOI] [PubMed] [Google Scholar]
- 228.Marchese, F. P., et al. 2010. MAPKAP kinase 2 blocks tristetraprolin-directed mRNA decay by inhibiting CAF1 deadenylase recruitment. J. Biol. Chem. 285**:**27590-275600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.McCoy, C. E., D. G. Campbell, M. Deak, G. B. Bloomberg, and J. S. Arthur. 2005. MSK1 activity is controlled by multiple phosphorylation sites. Biochem. J. 387**:**507-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.McCoy, C. E., et al. 2007. Identification of novel phosphorylation sites in MSK1 by precursor ion scanning MS. Biochem. J. 402**:**491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.McKendrick, L., E. Thompson, J. Ferreira, S. J. Morley, and J. D. Lewis. 2001. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol. Cell. Biol. 21**:**3632-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.McLaughlin, M. M., et al. 1996. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J. Biol. Chem. 271**:**8488-8492. [DOI] [PubMed] [Google Scholar]
- 233.Meier, R., J. Rouse, A. Cuenda, A. R. Nebreda, and P. Cohen. 1996. Cellular stresses and cytokines activate multiple mitogen-activated-protein kinase kinase homologues in PC12 and KB cells. Eur. J. Biochem. 236**:**796-805. [DOI] [PubMed] [Google Scholar]
- 234.Meloche, S., B. G. Beatty, and J. Pellerin. 1996. Primary structure, expression and chromosomal locus of a human homolog of rat ERK3. Oncogene 13**:**1575-1579. [PubMed] [Google Scholar]
- 235.Meloche, S., and J. Pouyssegur. 2007. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene 26**:**3227-3239. [DOI] [PubMed] [Google Scholar]
- 236.Meng, W., et al. 2002. Structure of mitogen-activated protein kinase-activated protein (MAPKAP) kinase 2 suggests a bifunctional switch that couples kinase activation with nuclear export. J. Biol. Chem. 277**:**37401-37405. [DOI] [PubMed] [Google Scholar]
- 237.Menon, M. B., N. Ronkina, J. Schwermann, A. Kotlyarov, and M. Gaestel. 2009. Fluorescence-based quantitative scratch wound healing assay demonstrating the role of MAPKAPK-2/3 in fibroblast migration. Cell Motil. Cytoskeleton 66**:**1041-1047. [DOI] [PubMed] [Google Scholar]
- 238.Mizukami, Y., K. Yoshioka, S. Morimoto, and K. Yoshida. 1997. A novel mechanism of JNK1 activation. Nuclear translocation and activation of JNK1 during ischemia and reperfusion. J. Biol. Chem. 272**:**16657-16662. [DOI] [PubMed] [Google Scholar]
- 239.Mody, N., J. Leitch, C. Armstrong, J. Dixon, and P. Cohen. 2001. Effects of MAP kinase cascade inhibitors on the MKK5/ERK5 pathway. FEBS Lett. 502**:**21-24. [DOI] [PubMed] [Google Scholar]
- 240.Moise, N., et al. 2010. Characterization of a novel MK3 splice variant from murine ventricular myocardium. Cell Signal. 22**:**1502-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Moller, D. E., C. H. Xia, W. Tang, A. X. Zhu, and M. Jakubowski. 1994. Human rsk isoforms: cloning and characterization of tissue-specific expression. Am. J. Physiol. 266**:**351-359. [DOI] [PubMed] [Google Scholar]
- 242.Morrison, D. K. 2009. The 14-3-3 proteins: integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19**:**16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Mourey, R. J., et al. 2010. A benzothiophene inhibitor of mitogen-activated protein kinase-activated protein kinase 2 inhibits tumor necrosis factor alpha production and has oral anti-inflammatory efficacy in acute and chronic models of inflammation. J. Pharmacol. Exp. Ther. 333**:**797-807. [DOI] [PubMed] [Google Scholar]
- 244.Mulloy, R., S. Salinas, A. Philips, and R. A. Hipskind. 2003. Activation of cyclin D1 expression by the ERK5 cascade. Oncogene 22**:**5387-5398. [DOI] [PubMed] [Google Scholar]
- 245.Murphy, L. O., S. Smith, R. H. Chen, D. C. Fingar, and J. Blenis. 2002. Molecular interpretation of ERK signal duration by immediate early gene products. Nat. Cell Biol. 4**:**556-564. [DOI] [PubMed] [Google Scholar]
- 246.Myers, A. P., L. B. Corson, J. Rossant, and J. C. Baker. 2004. Characterization of mouse Rsk4 as an inhibitor of fibroblast growth factor-RAS-extracellular signal-regulated kinase signaling. Mol. Cell. Biol. 24**:**4255-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Nakajima, T., et al. 1996. The signal-dependent coactivator CBP is a nuclear target for pp90RSK. Cell 86**:**465-474. [DOI] [PubMed] [Google Scholar]
- 248.Neininger, A., et al. 2002. MK2 targets AU-rich elements and regulates biosynthesis of tumor necrosis factor and interleukin-6 independently at different post-transcriptional levels. J. Biol. Chem. 277**:**3065-3068. [DOI] [PubMed] [Google Scholar]
- 249.Neininger, A., H. Thielemann, and M. Gaestel. 2001. FRET-based detection of different conformations of MK2. EMBO Rep. 2**:**703-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Neufeld, B., et al. 2000. Serine/Threonine kinases 3pK and MAPK-activated protein kinase 2 interact with the basic helix-loop-helix transcription factor E47 and repress its transcriptional activity. J. Biol. Chem. 275**:**20239-20242. [DOI] [PubMed] [Google Scholar]
- 251.New, L., Y. Jiang, and J. Han. 2003. Regulation of PRAK subcellular location by p38 MAP kinases. Mol. Biol. Cell 14**:**2603-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.New, L., et al. 1998. PRAK, a novel protein kinase regulated by the p38 MAP kinase. EMBO J. 17**:**3372-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.New, L., et al. 1999. Cloning and characterization of RLPK, a novel RSK-related protein kinase. J. Biol. Chem. 274**:**1026-1032. [DOI] [PubMed] [Google Scholar]
- 254.Ni, H., X. S. Wang, K. Diener, and Z. Yao. 1998. MAPKAPK5, a novel mitogen-activated protein kinase (MAPK)-activated protein kinase, is a substrate of the extracellular-regulated kinase (ERK) and p38 kinase. Biochem. Biophys. Res. Commun. 243**:**492-496. [DOI] [PubMed] [Google Scholar]
- 255.Nishiyama, T., K. Ohsumi, and T. Kishimoto. 2007. Phosphorylation of Erp1 by p90rsk is required for cytostatic factor arrest in Xenopus laevis eggs. Nature 446**:**1096-1099. [DOI] [PubMed] [Google Scholar]
- 256.Ohkawara, B., et al. 2004. Role of the TAK1-NLK-STAT3 pathway in TGF-beta-mediated mesoderm induction. Genes Dev. 18**:**381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Okumura, E., et al. 2002. Akt inhibits Myt1 in the signalling pathway that leads to meiotic G2/M-phase transition. Nat. Cell Biol. 4**:**111-116. [DOI] [PubMed] [Google Scholar]
- 258.O'Loghlen, A., V. M. Gonzalez, T. Jurado, M. Salinas, and M. E. Martin. 2007. Characterization of the activity of human MAP kinase-interacting kinase Mnk1b. Biochim. Biophys. Acta 1773**:**1416-1427. [DOI] [PubMed] [Google Scholar]
- 259.O'Loghlen, A., et al. 2004. Identification and molecular characterization of Mnk1b, a splice variant of human MAP kinase-interacting kinase Mnk1. Exp. Cell Res. 299**:**343-355. [DOI] [PubMed] [Google Scholar]
- 260.Palmer, A., A. C. Gavin, and A. R. Nebreda. 1998. A link between MAP kinase and p34(cdc2)/cyclin B during oocyte maturation: p90(rsk) phosphorylates and inactivates the p34(cdc2) inhibitory kinase Myt1. EMBO J. 17**:**5037-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Park, H. Y., and B. A. Gilchrest. 2002. More on MITF. J. Invest. Dermatol. 119**:**1218-1219. [DOI] [PubMed] [Google Scholar]
- 262.Parra, J. L., M. Buxade, and C. G. Proud. 2005. Features of the catalytic domains and C termini of the MAPK signal-integrating kinases Mnk1 and Mnk2 determine their differing activities and regulatory properties. J. Biol. Chem. 280**:**37623-37633. [DOI] [PubMed] [Google Scholar]
- 263.Parra-Palau, J. L., G. C. Scheper, M. L. Wilson, and C. G. Proud. 2003. Features in the N and C termini of the MAPK-interacting kinase Mnk1 mediate its nucleocytoplasmic shuttling. J. Biol. Chem. 278**:**44197-44204. [DOI] [PubMed] [Google Scholar]
- 264.Pearson, G., et al. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocrinol. Rev. 22**:**153-183. [DOI] [PubMed] [Google Scholar]
- 265.Pende, M., et al. 2004. S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 24**:**3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Perander, M., et al. 2008. The Ser(186) phospho-acceptor site within ERK4 is essential for its ability to interact with and activate PRAK/MK5. Biochem. J. 411**:**613-622. [DOI] [PubMed] [Google Scholar]
- 267.Perander, M., S. M. Keyse, and O. M. Seternes. 2008. Does MK5 reconcile classical and atypical MAP kinases? Front. Biosci. 13**:**4617-4624. [DOI] [PubMed] [Google Scholar]
- 268.Pierrat, B., J. S. Correia, J. L. Mary, M. Tomas-Zuber, and W. Lesslauer. 1998. RSK-B, a novel ribosomal S6 kinase family member, is a CREB kinase under dominant control of p38alpha mitogen-activated protein kinase (p38alphaMAPK). J. Biol. Chem. 273**:**29661-29671. [DOI] [PubMed] [Google Scholar]
- 269.Piotrowicz, R. S., E. Hickey, and E. G. Levin. 1998. Heat shock protein 27 kDa expression and phosphorylation regulates endothelial cell migration. FASEB J. 12**:**1481-1490. [DOI] [PubMed] [Google Scholar]
- 270.Plath, K., K. Engel, G. Schwedersky, and M. Gaestel. 1994. Characterization of the proline-rich region of mouse MAPKAP kinase 2: influence on catalytic properties and binding to the c-abl SH3 domain in vitro. Biochem. Biophys. Res. Commun. 203**:**1188-1194. [DOI] [PubMed] [Google Scholar]
- 271.Poirier, R., et al. 2007. Deletion of the Coffin-Lowry syndrome gene Rsk2 in mice is associated with impaired spatial learning and reduced control of exploratory behavior. Behav. Genet. 37**:**31-50. [DOI] [PubMed] [Google Scholar]
- 272.Pouyssegur, J., V. Volmat, and P. Lenormand. 2002. Fidelity and spatio-temporal control in MAP kinase (ERKs) signalling. Biochem. Pharmacol. 64**:**755-763. [DOI] [PubMed] [Google Scholar]
- 273.Pulido, R., A. Zuniga, and A. Ullrich. 1998. PTP-SL and STEP protein tyrosine phosphatases regulate the activation of the extracellular signal-regulated kinases ERK1 and ERK2 by association through a kinase interaction motif. EMBO J. 17**:**7337-7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Putz, G., F. Bertolucci, T. Raabe, T. Zars, and M. Heisenberg. 2004. The S6KII (rsk) gene of Drosophila melanogaster differentially affects an operant and a classical learning task. J. Neurosci. 24**:**9745-9751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Pyronnet, S., et al. 1999. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18**:**270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Quan, L. T., A. Caputo, R. C. Bleackley, D. J. Pickup, and G. S. Salvesen. 1995. Granzyme B is inhibited by the cowpox virus serpin cytokine response modifier A. J. Biol. Chem. 270**:**10377-10379. [DOI] [PubMed] [Google Scholar]
- 277.Raingeaud, J., et al. 1995. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J. Biol. Chem. 270**:**7420-7426. [DOI] [PubMed] [Google Scholar]
- 278.Raman, M., W. Chen, and M. H. Cobb. 2007. Differential regulation and properties of MAPKs. Oncogene 26**:**3100-3112. [DOI] [PubMed] [Google Scholar]
- 279.Ramne, A., E. Bilsland-Marchesan, S. Erickson, and P. Sunnerhagen. 2000. The protein kinases Rck1 and Rck2 inhibit meiosis in budding yeast. Mol. Gen. Genet. 263**:**253-261. [DOI] [PubMed] [Google Scholar]
- 280.Ranganathan, A., G. W. Pearson, C. A. Chrestensen, T. W. Sturgill, and M. H. Cobb. 2006. The MAP kinase ERK5 binds to and phosphorylates p90 RSK. Arch. Biochem. Biophys. 449**:**8-16. [DOI] [PubMed] [Google Scholar]
- 281.Ray, L. B., and T. W. Sturgill. 1988. Insulin-stimulated microtubule-associated protein kinase is phosphorylated on tyrosine and threonine in vivo. Proc. Natl. Acad. Sci. U. S. A. 85**:**3753-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Regan, C. P., et al. 2002. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc. Natl. Acad. Sci. U. S. A. 99**:**9248-9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Regan, J., et al. 2002. Pyrazole urea-based inhibitors of p38 MAP kinase: from lead compound to clinical candidate. J. Med. Chem. 45**:**2994-3008. [DOI] [PubMed] [Google Scholar]
- 284.Richards, S. A., V. C. Dreisbach, L. O. Murphy, and J. Blenis. 2001. Characterization of regulatory events associated with membrane targeting of p90 ribosomal S6 kinase 1. Mol. Cell. Biol. 21**:**7470-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Richards, S. A., J. Fu, A. Romanelli, A. Shimamura, and J. Blenis. 1999. Ribosomal S6 kinase 1 (RSK1) activation requires signals dependent on and independent of the MAP kinase ERK. Curr. Biol. 9**:**810-820. [DOI] [PubMed] [Google Scholar]
- 286.Richter, J. D., and N. Sonenberg. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433**:**477-480. [DOI] [PubMed] [Google Scholar]
- 287.Rivera, V. M., et al. 1993. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol. Cell. Biol. 13**:**6260-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Robbins, D. J., et al. 1993. Regulation and properties of extracellular signal-regulated protein kinases 1 and 2 in vitro. J. Biol. Chem. 268**:**5097-5106. [PubMed] [Google Scholar]
- 289.Rocheleau, C. E., et al. 1997. Wnt signaling and an APC-related gene specify endoderm in early C. elegans embryos. Cell 90**:**707-716. [DOI] [PubMed] [Google Scholar]
- 290.Rogalla, T., et al. 1999. Regulation of Hsp27 oligomerization, chaperone function, and protective activity against oxidative stress/tumor necrosis factor alpha by phosphorylation. J. Biol. Chem. 274**:**18947-18956. [DOI] [PubMed] [Google Scholar]
- 291.Rolfe, M., L. E. McLeod, P. F. Pratt, and C. G. Proud. 2005. Activation of protein synthesis in cardiomyocytes by the hypertrophic agent phenylephrine requires the activation of ERK and involves phosphorylation of tuberous sclerosis complex 2 (TSC2). Biochem. J. 388**:**973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ronkina, N., et al. 2007. The mitogen-activated protein kinase (MAPK)-activated protein kinases MK2 and MK3 cooperate in stimulation of tumor necrosis factor biosynthesis and stabilization of p38 MAPK. Mol. Cell. Biol. 27**:**170-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Ronkina, N., A. Kotlyarov, and M. Gaestel. 2008. MK2 and MK3—a pair of isoenzymes? Front. Biosci. 13**:**5511-5521. [DOI] [PubMed] [Google Scholar]
- 294.Ross, G., J. R. Dyer, V. F. Castellucci, and W. S. Sossin. 2006. Mnk is a negative regulator of cap-dependent translation in Aplysia neurons. J. Neurochem. 97**:**79-91. [DOI] [PubMed] [Google Scholar]
- 295.Rouse, J., et al. 1994. A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins. Cell 78**:**1027-1037. [DOI] [PubMed] [Google Scholar]
- 296.Rousseau, S., F. Houle, J. Landry, and J. Huot. 1997. p38 MAP kinase activation by vascular endothelial growth factor mediates actin reorganization and cell migration in human endothelial cells. Oncogene 15**:**2169-2177. [DOI] [PubMed] [Google Scholar]
- 297.Rousseau, S., et al. 2002. Inhibition of SAPK2a/p38 prevents hnRNP A0 phosphorylation by MAPKAP-K2 and its interaction with cytokine mRNAs. EMBO J. 21**:**6505-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Roux, P. P. 2007. Rsk1. UCSD-Nat. Mol. Pages, doi: 10.1038/mp.a002099.01. [DOI]
- 299.Roux, P. P. 2007. Rsk4. UCSD-Nat. Mol. Pages, doi: 10.1038/mp.a002102.01. [DOI]
- 300.Roux, P. P., B. A. Ballif, R. Anjum, S. P. Gygi, and J. Blenis. 2004. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc. Natl. Acad. Sci. U. S. A. 101**:**13489-13494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68**:**320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Roux, P. P., S. A. Richards, and J. Blenis. 2003. Phosphorylation of p90 ribosomal S6 kinase (RSK) regulates extracellular signal-regulated kinase docking and RSK activity. Mol. Cell. Biol. 23**:**4796-4804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Roux, P. P., et al. 2007. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J. Biol. Chem. 282**:**14056-14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Rubinfeld, H., T. Hanoch, and R. Seger. 1999. Identification of a cytoplasmic-retention sequence in ERK2. J. Biol. Chem. 274**:**30349-30352. [DOI] [PubMed] [Google Scholar]
- 305.Ruiz, E. J., M. Vilar, and A. R. Nebreda. 2010. A two-step inactivation mechanism of Myt1 ensures CDK1/cyclin B activation and meiosis I entry. Curr. Biol. 20**:**717-723. [DOI] [PubMed] [Google Scholar]
- 306.Sabapathy, K., et al. 2004. Distinct roles for JNK1 and JNK2 in regulating JNK activity and c-Jun-dependent cell proliferation. Mol. Cell 15**:**713-725. [DOI] [PubMed] [Google Scholar]
- 307.Saelzler, M. P., et al. 2006. ERK8 down-regulates transactivation of the glucocorticoid receptor through Hic-5. J. Biol. Chem. 281**:**16821-16832. [DOI] [PubMed] [Google Scholar]
- 308.Sapkota, G. P., et al. 2007. BI-D1870 is a specific inhibitor of the p90 RSK (ribosomal S6 kinase) isoforms in vitro and in vivo. Biochem. J. 401**:**29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Sapkota, G. P., et al. 2001. Phosphorylation of the protein kinase mutated in Peutz-Jeghers cancer syndrome, LKB1/STK11, at Ser431 by p90(RSK) and cAMP-dependent protein kinase, but not its farnesylation at Cys(433), is essential for LKB1 to suppress cell vrowth. J. Biol. Chem. 276**:**19469-19482. [DOI] [PubMed] [Google Scholar]
- 310.Sassone-Corsi, P., et al. 1999. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science 285**:**886-891. [DOI] [PubMed] [Google Scholar]
- 311.Scheper, G. C., N. A. Morrice, M. Kleijn, and C. G. Proud. 2001. The mitogen-activated protein kinase signal-integrating kinase Mnk2 is a eukaryotic initiation factor 4E kinase with high levels of basal activity in mammalian cells. Mol. Cell. Biol. 21**:**743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Scheper, G. C., et al. 2003. The N and C termini of the splice variants of the human mitogen-activated protein kinase-interacting kinase Mnk2 determine activity and localization. Mol. Cell. Biol. 23**:**5692-5705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Scheper, G. C., and C. G. Proud. 2002. Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation? Eur. J. Biochem. 269**:**5350-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Scheper, G. C., et al. 2002. Phosphorylation of eukaryotic initiation factor 4E markedly reduces its affinity for capped mRNA. J. Biol. Chem. 277**:**3303-3309. [DOI] [PubMed] [Google Scholar]
- 315.Schmitt, A., G. J. Gutierrez, P. Lenart, J. Ellenberg, and A. R. Nebreda. 2002. Histone H3 phosphorylation during Xenopus oocyte maturation: regulation by the MAP kinase/p90Rsk pathway and uncoupling from DNA condensation. FEBS Lett. 518**:**23-28. [DOI] [PubMed] [Google Scholar]
- 316.Schmitt, A., and A. R. Nebreda. 2002. Signalling pathways in oocyte meiotic maturation. J. Cell Sci. 115**:**2457-2459. [DOI] [PubMed] [Google Scholar]
- 317.Schouten, G. J., et al. 1997. IκBα is a target for the mitogen-activated 90 kDa ribosomal S6 kinase. EMBO J. 16**:**3133-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Schumacher, S., et al. 2004. Scaffolding by ERK3 regulates MK5 in development. EMBO J. 23**:**4770-4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Schwab, M. S., et al. 2001. Bub1 is activated by the protein kinase p90(Rsk) during Xenopus oocyte maturation. Curr. Biol. 11**:**141-150. [DOI] [PubMed] [Google Scholar]
- 320.Schwermann, J., et al. 2009. MAPKAP kinase MK2 maintains self-renewal capacity of haematopoietic stem cells. EMBO J. 28**:**1392-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Seternes, O. M., et al. 2002. Both binding and activation of p38 mitogen-activated protein kinase (MAPK) play essential roles in regulation of the nucleocytoplasmic distribution of MAPK-activated protein kinase 5 by cellular stress. Mol. Cell. Biol. 22**:**6931-6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Seternes, O. M., et al. 2004. Activation of MK5/PRAK by the atypical MAP kinase ERK3 defines a novel signal transduction pathway. EMBO J. 23**:**4780-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Shahbazian, D., et al. 2006. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. EMBO J. 25**:**2781-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Sharrocks, A. D., S. H. Yang, and A. Galanis. 2000. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem. Sci. 25**:**448-453. [DOI] [PubMed] [Google Scholar]
- 325.Shaul, Y. D., and R. Seger. 2006. ERK1c regulates Golgi fragmentation during mitosis. J. Cell Biol. 172**:**885-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Shaul, Y. D., and R. Seger. 2007. The MEK/ERK cascade: from signaling specificity to diverse functions. Biochim. Biophys. Acta 1773**:**1213-1226. [DOI] [PubMed] [Google Scholar]
- 327.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 20**:**2390-2400. [DOI] [PubMed] [Google Scholar]
- 328.She, Q. B., W. Y. Ma, S. Zhong, and Z. Dong. 2002. Activation of JNK1, RSK2, and MSK1 is involved in serine 112 phosphorylation of Bad by ultraviolet B radiation. J. Biol. Chem. 277**:**24039-24048. [DOI] [PubMed] [Google Scholar]
- 329.Sheridan, D. L., Y. Kong, S. A. Parker, K. N. Dalby, and B. E. Turk. 2008. Substrate discrimination among mitogen-activated protein kinases through distinct docking sequence motifs. J. Biol. Chem. 283**:**19511-19520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Shi, Y., et al. 2009. Casein kinase 2 interacts with human mitogen- and stress-activated protein kinase MSK1 and phosphorylates it at multiple sites. BMB Rep. 42**:**840-845. [DOI] [PubMed] [Google Scholar]
- 331.Shi, Y., et al. 2003. Elimination of protein kinase MK5/PRAK activity by targeted homologous recombination. Mol. Cell. Biol. 23**:**7732-7741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Shimamura, A., B. A. Ballif, S. A. Richards, and J. Blenis. 2000. Rsk1 mediates a MEK-MAP kinase cell survival signal. Curr. Biol. 10**:**127-135. [DOI] [PubMed] [Google Scholar]
- 333.Singh, S., et al. 2003. Identification of the p16-Arc subunit of the Arp 2/3 complex as a substrate of MAPK-activated protein kinase 2 by proteomic analysis. J. Biol. Chem. 278**:**36410-36417. [DOI] [PubMed] [Google Scholar]
- 334.Sithanandam, G., et al. 1996. 3pK, a new mitogen-activated protein kinase-activated protein kinase located in the small cell lung cancer tumor suppressor gene region. Mol. Cell. Biol. 16**:**868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Slentz-Kesler, K., et al. 2000. Identification of the human Mnk2 gene (MKNK2) through protein interaction with estrogen receptor beta. Genomics 69**:**63-71. [DOI] [PubMed] [Google Scholar]
- 336.Smit, L., et al. 2004. Wnt activates the Tak1/Nemo-like kinase pathway. J. Biol. Chem. 279**:**17232-17240. [DOI] [PubMed] [Google Scholar]
- 337.Smith, J. A., et al. 2000. Creation of a stress-activated p90 ribosomal S6 kinase. The carboxyl-terminal tail of the MAPK-activated protein kinases dictates the signal transduction pathway in which they function. J. Biol. Chem. 275**:**31588-31593. [DOI] [PubMed] [Google Scholar]
- 338.Smith, J. A., C. E. Poteet-Smith, K. Malarkey, and T. W. Sturgill. 1999. Identification of an extracellular signal-regulated kinase (ERK) docking site in ribosomal S6 kinase, a sequence critical for activation by ERK in vivo. J. Biol. Chem. 274**:**2893-2898. [DOI] [PubMed] [Google Scholar]
- 339.Smith, J. A., et al. 2005. Identification of the first specific inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected role for RSK in cancer cell proliferation. Cancer Res. 65**:**1027-1034. [PubMed] [Google Scholar]
- 340.Sohn, S. J., B. K. Sarvis, D. Cado, and A. Winoto. 2002. ERK5 MAPK regulates embryonic angiogenesis and acts as a hypoxia-sensitive repressor of vascular endothelial growth factor expression. J. Biol. Chem. 277**:**43344-43351. [DOI] [PubMed] [Google Scholar]
- 341.Soloaga, A., et al. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22**:**2788-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Song, T., et al. 2007. p90 RSK-1 associates with and inhibits neuronal nitric oxide synthase. Biochem. J. 401**:**391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Sonnichsen, B., et al. 2005. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans. Nature 434**:**462-469. [DOI] [PubMed] [Google Scholar]
- 344.Stein, B., H. Brady, M. X. Yang, D. B. Young, and M. S. Barbosa. 1996. Cloning and characterization of MEK6, a novel member of the mitogen-activated protein kinase kinase cascade. J. Biol. Chem. 271**:**11427-11433. [DOI] [PubMed] [Google Scholar]
- 345.Stoecklin, G., et al. 2004. MK2-induced tristetraprolin: 14-13-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J. 23**:**1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Stokoe, D., et al. 1992. MAPKAP kinase-2; a novel protein kinase activated by mitogen-activated protein kinase. EMBO J. 11**:**3985-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Stokoe, D., B. Caudwell, P. T. Cohen, and P. Cohen. 1993. The substrate specificity and structure of mitogen-activated protein (MAP) kinase-activated protein kinase-2. Biochem. J. 296**:**843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Stokoe, D., K. Engel, D. G. Campbell, P. Cohen, and M. Gaestel. 1992. Identification of MAPKAP kinase 2 as a major enzyme responsible for the phosphorylation of the small mammalian heat shock proteins. FEBS Lett. 313**:**307-313. [DOI] [PubMed] [Google Scholar]
- 349.Stratford, A. L., et al. 2008. Y-box binding protein-1 serine 102 is a downstream target of p90 ribosomal S6 kinase in basal-like breast cancer cells. Breast Cancer Res. 10**:**R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Sudo, T., K. Kawai, H. Matsuzaki, and H. Osada. 2005. p38 mitogen-activated protein kinase plays a key role in regulating MAPKAPK2 expression. Biochem. Biophys. Res. Commun. 337**:**415-421. [DOI] [PubMed] [Google Scholar]
- 351.Sun, P., et al. 2007. PRAK is essential for ras-induced senescence and tumor suppression. Cell 128**:**295-308. [DOI] [PubMed] [Google Scholar]
- 352.Sutherland, C., D. G. Campbell, and P. Cohen. 1993. Identification of insulin-stimulated protein kinase-1 as the rabbit equivalent of rskmo-2: identification of two threonines phosphorylated during activation by mitogen-activated protein kinase. Eur. J. Biochem. 212**:**581-588. [DOI] [PubMed] [Google Scholar]
- 353.Sutherland, C., I. A. Leighton, and P. Cohen. 1993. Inactivation of glycogen synthase kinase-3 beta by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem. J. 296**:**15-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Tak, H., et al. 2007. 14-3-3epsilon inhibits MK5-mediated cell migration by disrupting F-actin polymerization. Cell Signal. 19**:**2379-2387. [DOI] [PubMed] [Google Scholar]
- 355.Takahashi, E., J. Abe, and B. C. Berk. 1997. Angiotensin II stimulates p90rsk in vascular smooth muscle cells. A potential Na(+)-H+ exchanger kinase. Circ. Res. 81**:**268-273. [DOI] [PubMed] [Google Scholar]
- 356.Takahashi, E., et al. 1999. p90(RSK) is a serum-stimulated Na+/H+ exchanger isoform-1 kinase. Regulatory phosphorylation of serine 703 of Na+/H+ exchanger isoform-1. J. Biol. Chem. 274**:**20206-20214. [DOI] [PubMed] [Google Scholar]
- 357.Tanguay, P. L., G. Rodier, and S. Meloche. 2010. C-terminal domain phosphorylation of ERK3 controlled by Cdk1 and Cdc14 regulates its stability in mitosis. Biochem. J. 428**:**103-111. [DOI] [PubMed] [Google Scholar]
- 358.Tanoue, T., M. Adachi, T. Moriguchi, and E. Nishida. 2000. A conserved docking motif in MAP kinases common to substrates, activators and regulators. Nat. Cell Biol. 2**:**110-116. [DOI] [PubMed] [Google Scholar]
- 359.Tanoue, T., R. Maeda, M. Adachi, and E. Nishida. 2001. Identification of a docking groove on ERK and p38 MAP kinases that regulates the specificity of docking interactions. EMBO J. 20**:**466-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Tanoue, T., and E. Nishida. 2003. Molecular recognitions in the MAP kinase cascades. Cell Signal. 15**:**455-462. [DOI] [PubMed] [Google Scholar]
- 361.ter Haar, E., P. Prabhakar, X. Liu, and C. Lepre. 2007. Crystal structure of the p38 alpha-MAPKAP kinase 2 heterodimer. J. Biol. Chem. 282**:**9733-9739. [DOI] [PubMed] [Google Scholar]
- 362.Thakur, A., et al. 2007. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol. Cancer Res. 5**:**171-181. [DOI] [PubMed] [Google Scholar]
- 363.Thakur, A., et al. 2008. Anti-invasive and antimetastatic activities of ribosomal protein S6 kinase 4 in breast cancer cells. Clin. Cancer Res. 14**:**4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Thomson, S., et al. 1999. The nucleosomal response associated with immediate-early gene induction is mediated via alternative MAP kinase cascades: MSK1 as a potential histone H3/HMG-14 kinase. EMBO J. 18**:**4779-4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Thornton, T. M., and M. Rincon. 2009. Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int. J. Biol. Sci. 5**:**44-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Tomas-Zuber, M., J. L. Mary, F. Lamour, D. Bur, and W. Lesslauer. 2001. C-terminal elements control location, activation threshold, and p38 docking of ribosomal S6 kinase B (RSKB). J. Biol. Chem. 276**:**5892-5899. [DOI] [PubMed] [Google Scholar]
- 367.Tomas-Zuber, M., J. L. Mary, and W. Lesslauer. 2000. Control sites of ribosomal S6 kinase B and persistent activation through tumor necrosis factor. J. Biol. Chem. 275**:**23549-23558. [DOI] [PubMed] [Google Scholar]
- 368.Tournier, C., et al. 2000. Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288**:**870-874. [DOI] [PubMed] [Google Scholar]
- 369.Tran, H., F. Maurer, and Y. Nagamine. 2003. Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2. Mol. Cell. Biol. 23**:**7177-7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Trivier, E., et al. 1996. Mutations in the kinase Rsk-2 associated with Coffin-Lowry syndrome. Nature 384**:**567-570. [DOI] [PubMed] [Google Scholar]
- 371.Tunquist, B. J., M. S. Schwab, L. G. Chen, and J. L. Maller. 2002. The spindle checkpoint kinase bub1 and cyclin e/cdk2 both contribute to the establishment of meiotic metaphase arrest by cytostatic factor. Curr. Biol. 12**:**1027-1033. [DOI] [PubMed] [Google Scholar]
- 372.Turgeon, B., M. K. Saba-El-Leil, and S. Meloche. 2000. Cloning and characterization of mouse extracellular-signal-regulated protein kinase 3 as a unique gene product of 100 kDa. Biochem. J. 346**:**169-175. [PMC free article] [PubMed] [Google Scholar]
- 373.Ueda, T., R. Watanabe-Fukunaga, H. Fukuyama, S. Nagata, and R. Fukunaga. 2004. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol. Cell. Biol. 24**:**6539-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Vadlamudi, R. K., et al. 2002. Filamin is essential in actin cytoskeletal assembly mediated by p21-activated kinase 1. Nat. Cell Biol. 4**:**681-690. [DOI] [PubMed] [Google Scholar]
- 375.Vaidyanathan, H., and J. W. Ramos. 2003. RSK2 activity is regulated by its interaction with PEA-15. J. Biol. Chem. 278**:**32367-32372. [DOI] [PubMed] [Google Scholar]
- 376.Valk-Lingbeek, M. E., S. W. Bruggeman, and M. van Lohuizen. 2004. Stem cells and cancer; the polycomb connection. Cell 118**:**409-418. [DOI] [PubMed] [Google Scholar]
- 377.van der Houven van Oordt, W., et al. 2000. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J. Cell Biol. 149**:**307-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Vermeulen, L., W. V. Berghe, I. M. Beck, K. De Bosscher, and G. Haegeman. 2009. The versatile role of MSKs in transcriptional regulation. Trends Biochem. Sci. 34**:**311-318. [DOI] [PubMed] [Google Scholar]
- 379.Vermeulen, L., G. De Wilde, P. Van Damme, W. Vanden Berghe, and G. Haegeman. 2003. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1). EMBO J. 22**:**1313-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Vik, T. A., and J. W. Ryder. 1997. Identification of serine 380 as the major site of autophosphorylation of Xenopus pp90rsk. Biochem. Biophys. Res. Commun. 235**:**398-402. [DOI] [PubMed] [Google Scholar]
- 381.Voncken, J. W., et al. 2005. MAPKAP kinase 3pK phosphorylates and regulates chromatin association of the polycomb group protein Bmi1. J. Biol. Chem. 280**:**5178-5187. [DOI] [PubMed] [Google Scholar]
- 382.Wagner, E. F., and A. R. Nebreda. 2009. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer. 9**:**537-549. [DOI] [PubMed] [Google Scholar]
- 383.Wang, X., et al. 2006. Activation of extracellular signal-regulated protein kinase 5 downregulates FasL upon osmotic stress. Cell Death Differ. 13**:**2099-2108. [DOI] [PubMed] [Google Scholar]
- 384.Wang, X., et al. 1998. The phosphorylation of eukaryotic initiation factor eIF4E in response to phorbol esters, cell stresses, and cytokines is mediated by distinct MAP kinase pathways. J. Biol. Chem. 273**:**9373-9377. [DOI] [PubMed] [Google Scholar]
- 385.Wang, X., M. Janmaat, A. Beugnet, F. E. Paulin, and C. G. Proud. 2002. Evidence that the dephosphorylation of Ser(535) in the epsilon-subunit of eukaryotic initiation factor (eIF) 2B is insufficient for the activation of eIF2B by insulin. Biochem. J. 367**:**475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Wang, X., et al. 2001. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. EMBO J. 20**:**4370-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Wang, Y., W. Zhang, Y. Jin, J. Johansen, and K. M. Johansen. 2001. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105**:**433-443. [DOI] [PubMed] [Google Scholar]
- 388.Wang, Z., B. Zhang, M. Wang, and B. I. Carr. 2003. Persistent ERK phosphorylation negatively regulates cAMP response element-binding protein (CREB) activity via recruitment of CREB-binding protein to pp90RSK. J. Biol. Chem. 278**:**11138-11144. [DOI] [PubMed] [Google Scholar]
- 389.Waskiewicz, A. J., A. Flynn, C. G. Proud, and J. A. Cooper. 1997. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. EMBO J. 16**:**1909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Waskiewicz, A. J., et al. 1999. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol. Cell. Biol. 19**:**1871-1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Weber, H. O., et al. 2005. HDM2 phosphorylation by MAPKAP kinase 2. Oncogene 24**:**1965-1972. [DOI] [PubMed] [Google Scholar]
- 392.Wendel, H. G., et al. 2007. Dissecting eIF4E action in tumorigenesis. Genes Dev. 21**:**3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Weston, C. R., and R. J. Davis. 2002. The JNK signal transduction pathway. Curr. Opin. Genet. Dev. 12**:**14-21. [DOI] [PubMed] [Google Scholar]
- 394.White, A., C. A. Pargellis, J. M. Studts, B. G. Werneburg, and B. T. Farmer, 2nd. 2007. Molecular basis of MAPK-activated protein kinase 2:p38 assembly. Proc. Natl. Acad. Sci. U. S. A. 104**:**6353-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Whitmarsh, A. J., and R. J. Davis. 1996. Transcription factor AP-1 regulation by mitogen-activated protein kinase signal transduction pathways. J. Mol. Med. 74**:**589-607. [DOI] [PubMed] [Google Scholar]
- 396.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79**:**143-180. [DOI] [PubMed] [Google Scholar]
- 397.Wierenga, A. T., I. Vogelzang, B. J. Eggen, and E. Vellenga. 2003. Erythropoietin-induced serine 727 phosphorylation of STAT3 in erythroid cells is mediated by a MEK-, ERK-, and MSK1-dependent pathway. Exp. Hematol. 31**:**398-405. [DOI] [PubMed] [Google Scholar]
- 398.Wiggin, G. R., et al. 2002. MSK1 and MSK2 are required for the mitogen- and stress-induced phosphorylation of CREB and ATF1 in fibroblasts. Mol. Cell. Biol. 22**:**2871-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Williams, M. R., et al. 2000. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr. Biol. 10**:**439-448. [DOI] [PubMed] [Google Scholar]
- 400.Wingate, A. D., D. G. Campbell, M. Peggie, and J. S. Arthur. 2006. Nur77 is phosphorylated in cells by RSK in response to mitogenic stimulation. Biochem. J. 393**:**715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Winzen, R., et al. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18**:**4969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wong, E. V., A. W. Schaefer, G. Landreth, and V. Lemmon. 1996. Involvement of p90rsk in neurite outgrowth mediated by the cell adhesion molecule L1. J. Biol. Chem. 271**:**18217-18223. [DOI] [PubMed] [Google Scholar]
- 403.Woo, M. S., Y. Ohta, I. Rabinovitz, T. P. Stossel, and J. Blenis. 2004. Ribosomal S6 kinase (RSK) regulates phosphorylation of filamin A on an important regulatory site. Mol. Cell. Biol. 24**:**3025-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Wu, J., and R. Janknecht. 2002. Regulation of the ETS transcription factor ER81 by the 90-kDa ribosomal S6 kinase 1 and protein kinase A. J. Biol. Chem. 277**:**42669-42679. [DOI] [PubMed] [Google Scholar]
- 405.Wu, J. Q., et al. 2007. Control of Emi2 activity and stability through Mos-mediated recruitment of PP2A. Proc. Natl. Acad. Sci. U. S. A. 104**:**16564-16569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Wu, M., et al. 2000. c-Kit triggers dual phosphorylations, which couple activation and degradation of the essential melanocyte factor Mi. Genes Dev. 14**:**301-312. [PMC free article] [PubMed] [Google Scholar]
- 407.Xing, J., D. D. Ginty, and M. E. Greenberg. 1996. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 273**:**959-963. [DOI] [PubMed] [Google Scholar]
- 408.Xu, B. E., et al. 2004. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J. Biol. Chem. 279**:**7826-7831. [DOI] [PubMed] [Google Scholar]
- 409.Xu, S., H. Bayat, X. Hou, and B. Jiang. 2006. Ribosomal S6 kinase-1 modulates interleukin-1beta-induced persistent activation of NF-kappaB through phosphorylation of IkappaBbeta. Am. J. Physiol. Cell Physiol. 291**:**C1336-C1345. [DOI] [PubMed] [Google Scholar]
- 410.Yamnik, R. L., and M. K. Holz. 2010. mTOR/S6K1 and MAPK/RSK signaling pathways coordinately regulate estrogen receptor alpha serine 167 phosphorylation. FEBS Lett. 584**:**124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Yan, C., H. Luo, J. D. Lee, J. Abe, and B. C. Berk. 2001. Molecular cloning of mouse ERK5/BMK1 splice variants and characterization of ERK5 functional domains. J. Biol. Chem. 276**:**10870-10878. [DOI] [PubMed] [Google Scholar]
- 412.Yan, L., et al. 2003. Knockout of ERK5 causes multiple defects in placental and embryonic development. BMC Dev. Biol. 3**:**11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Yang, X., et al. 2004. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117**:**387-398. [DOI] [PubMed] [Google Scholar]
- 414.Yannoni, Y. M., M. Gaestel, and L. L. Lin. 2004. P66(ShcA) interacts with MAPKAP kinase 2 and regulates its activity. FEBS Lett. 564**:**205-211. [DOI] [PubMed] [Google Scholar]
- 415.Yntema, H. G., et al. 1999. A novel ribosomal S6-kinase (RSK4; RPS6KA6) is commonly deleted in patients with complex X-linked mental retardation. Genomics 62**:**332-343. [DOI] [PubMed] [Google Scholar]
- 416.Yoon, S., and R. Seger. 2006. The extracellular signal-regulated kinase: multiple substrates regulate diverse cellular functions. Growth Factors. 24**:**21-44. [DOI] [PubMed] [Google Scholar]
- 417.Yoon, S. O., et al. 2008. Ran-binding protein 3 phosphorylation links the Ras and PI3-kinase pathways to nucleocytoplasmic transport. Mol. Cell 29**:**362-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Yung, Y., Z. Yao, T. Hanoch, and R. Seger. 2000. ERK1b, a 46-kDa ERK isoform that is differentially regulated by MEK. J. Biol. Chem. 275**:**15799-15808. [DOI] [PubMed] [Google Scholar]
- 419.Zaru, R., N. Ronkina, M. Gaestel, J. S. Arthur, and C. Watts. 2007. The MAPK-activated kinase Rsk controls an acute Toll-like receptor signaling response in dendritic cells and is activated through two distinct pathways. Nat. Immunol. 8**:**1227-1235. [DOI] [PubMed] [Google Scholar]
- 420.Zehorai, E., Z. Yao, A. Plotnikov, and R. Seger. 2010. The subcellular localization of MEK and ERK—a novel nuclear translocation signal (NTS) paves a way to the nucleus. Mol. Cell Endocrinol. 314**:**213-220. [DOI] [PubMed] [Google Scholar]
- 421.Zeng, W., A. R. Ball, Jr., and K. Yokomori. 2010. HP1: Heterochromatin binding proteins working the genome. Epigenetics 5**:**287-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Zeniou, M., T. Ding, E. Trivier, and A. Hanauer. 2002. Expression analysis of RSK gene family members: the RSK2 gene, mutated in Coffin-Lowry syndrome, is prominently expressed in brain structures essential for cognitive function and learning. Hum. Mol. Genet. 11**:**2929-2940. [DOI] [PubMed] [Google Scholar]
- 423.Zhang, L., et al. 2005. Dual pathways for nuclear factor kappaB activation by angiotensin II in vascular smooth muscle: phosphorylation of p65 by IkappaB kinase and ribosomal kinase. Circ. Res. 97**:**975-982. [DOI] [PubMed] [Google Scholar]
- 424.Zhang, L., Y. Ma, J. Zhang, J. Cheng, and J. Du. 2005. A new cellular signaling mechanism for angiotensin II activation of NF-kappaB: an IkappaB-independent, RSK-mediated phosphorylation of p65. Arterioscler. Thromb. Vasc. Biol. 25**:**1148-1153. [DOI] [PubMed] [Google Scholar]
- 425.Zhang, M., et al. 2008. Inhibition of polysome assembly enhances imatinib activity against chronic myelogenous leukemia and overcomes imatinib resistance. Mol. Cell. Biol. 28**:**6496-6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Zhang, Y., G. Liu, and Z. Dong. 2001. MSK1 and JNKs mediate phosphorylation of STAT3 in UVA-irradiated mouse epidermal JB6 cells. J. Biol. Chem. 276**:**42534-42542. [DOI] [PubMed] [Google Scholar]
- 427.Zhao, J., X. Yuan, M. Frodin, and I. Grummt. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11**:**405-413. [DOI] [PubMed] [Google Scholar]
- 428.Zhao, Y., C. Bjorbaek, and D. E. Moller. 1996. Regulation and interaction of pp90(rsk) isoforms with mitogen-activated protein kinases. J. Biol. Chem. 271**:**29773-29779. [DOI] [PubMed] [Google Scholar]
- 429.Zhao, Y., C. Bjorbaek, S. Weremowicz, C. C. Morton, and D. E. Moller. 1995. RSK3 encodes a novel pp90rsk isoform with a unique N-terminal sequence: growth factor-stimulated kinase function and nuclear translocation. Mol. Cell. Biol. 15**:**4353-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Zheng, B., et al. 2009. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol. Cell 33**:**237-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Zhong, H., H. SuYang, H. Erdjument-Bromage, P. Tempst, and S. Ghosh. 1997. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell 89**:**413-424. [DOI] [PubMed] [Google Scholar]
- 432.Zhong, H., R. E. Voll, and S. Ghosh. 1998. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol. Cell 1**:**661-671. [DOI] [PubMed] [Google Scholar]
- 433.Zhou, B. P., et al. 2001. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat. Cell Biol. 3**:**973-982. [DOI] [PubMed] [Google Scholar]
- 434.Zhou, G., Z. Q. Bao, and J. E. Dixon. 1995. Components of a new human protein kinase signal transduction pathway. J. Biol. Chem. 270**:**12665-12669. [DOI] [PubMed] [Google Scholar]
- 435.Zhu, A. X., Y. Zhao, D. E. Moller, and J. S. Flier. 1994. Cloning and characterization of p97MAPK, a novel human homolog of rat ERK-3. Mol. Cell. Biol. 14**:**8202-8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Zhu, J., J. Blenis, and J. Yuan. 2008. Activation of PI3K/Akt and MAPK pathways regulates Myc-mediated transcription by phosphorylating and promoting the degradation of Mad1. Proc. Natl. Acad. Sci. U. S. A. 105**:**6584-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Zhu, S., and D. S. Gerhard. 1998. A transcript map of an 800-kb region on human chromosome 11q13, part of the candidate region for SCA5 and BBS1. Hum. Genet. 103**:**674-680. [DOI] [PubMed] [Google Scholar]
- 438.Zu, Y. L., et al. 1994. The primary structure of a human MAP kinase activated protein kinase 2. Biochem. Biophys. Res. Commun. 200**:**1118-1124. [DOI] [PubMed] [Google Scholar]