PolyQ Repeat Expansions in ATXN2 Associated with ALS Are CAA Interrupted Repeats (original) (raw)
Abstract
Amyotrophic lateral sclerosis (ALS) is a devastating, rapidly progressive disease leading to paralysis and death. Recently, intermediate length polyglutamine (polyQ) repeats of 27–33 in ATAXIN-2 (ATXN2), encoding the ATXN2 protein, were found to increase risk for ALS. In ATXN2, polyQ expansions of ≥34, which are pure CAG repeat expansions, cause spinocerebellar ataxia type 2. However, similar length expansions that are interrupted with other codons, can present atypically with parkinsonism, suggesting that configuration of the repeat sequence plays an important role in disease manifestation in ATXN2 polyQ expansion diseases. Here we determined whether the expansions in ATXN2 associated with ALS were pure or interrupted CAG repeats, and defined single nucleotide polymorphisms (SNPs) rs695871 and rs695872 in exon 1 of the gene, to assess haplotype association. We found that the expanded repeat alleles of 40 ALS patients and 9 long-repeat length controls were all interrupted, bearing 1–3 CAA codons within the CAG repeat. 21/21 expanded ALS chromosomes with 3CAA interruptions arose from one haplotype (GT), while 18/19 expanded ALS chromosomes with <3CAA interruptions arose from a different haplotype (CC). Moreover, age of disease onset was significantly earlier in patients bearing 3 interruptions vs fewer, and was distinct between haplotypes. These results indicate that CAG repeat expansions in ATXN2 associated with ALS are uniformly interrupted repeats and that the nature of the repeat sequence and haplotype, as well as length of polyQ repeat, may play a role in the neurological effect conferred by expansions in ATXN2.
Introduction
Amyotrophic lateral sclerosis (ALS, also referred to as Lou Gehrig's disease) is a progressive, fatal neurodegenerative disease caused by the degeneration of motor neurons [1]–[3]. Approximately 10% of ALS cases are familial, with the remainder of cases being sporadic. To date, 12 genetic loci have been identified associated with familial ALS. Among these, mutations in superoxide dismutase 1 (SOD1) account for 15–20% of cases of familial ALS [4]. Trans- activate reponse DNA-binding protein (TARDBP, or TDP-43) is a major disease protein of the ubiquitin-positive cytoplasmic inclusions in ALS without SOD1 mutations [5]. Mutations in the TDP-43 coding gene TARDBP were later found in multiple cases of familial and sporadic ALS [6]–[9], indicating that TDP-43 plays a critical role in disease pathogenesis.
Spinocerebellar ataxia 2 (SCA2) is an autosomal dominant disease caused by an expanded CAG trinucleotide repeat encoding glutamine within the open reading frame of the gene encoding the ataxin 2 protein, ATAXIN2 (ATXN2) [10]–[12]. As with other polyglutamine (polyQ) diseases, the length of the CAG repeat expansion is inversely correlated with disease onset and severity [13]. CAG repeats in normal alleles of ATXN2 are variable in length, although by far the most common allele carries 22 repeats. In the normal ATXN2 allele, the CAG repeat region encoding the glutamine domain is typically interrupted by one or more CAA repeats (also encoding glutamine). The CAG repeat region within expanded alleles of ATXN2 associated with SCA2 disease are 34–59 in length [10]–[12], [14]–[19]. Similar to other polyQ diseases, SCA2 is for the most part a “pure” repeat disease. That is, the expanded allele is comprised of an uninterrupted sequence of CAGs encoding glutamine. However, four years after the 1996 identification of expansions in ATXN2 as the molecular basis of SCA2, a pathogenic length expansion bearing an “interrupted” repeat sequence was first reported [20]. It is now recognized that lower-range polyQ repeat expansions in ATXN2, of 33–49 in length, can be associated with levo-dopa responsive parkinsonism [21]–[30]. Intriguingly, in the situations of parkinsonism sequenced to date, the CAG repeat region has been found to not be a pure CAG repeat run, but rather to be interrupted with one or more CAA (or other) codons [21]–[30]. Additional data suggests that two SNPs (rs695871 and rs695872) in exon 1 of the ATXN2 gene, where the CAG repeat region occurs, predominant in distinct patterns in normal versus SCA2 individuals: the GT haplotype predominates in controls, whereas the CC haplotype is associated with disease [19].
Recently, intermediate-length polyQ repeats in ATXN2 were found to be a significant risk factor for ALS [31]. Consistent with the association between ATXN2 and ALS, it was previously known that SCA2 can present with motor neuron features that mimic ALS [32], [33]. Thus, it appears that individuals with repeat expansions in ATXN2 can present with SCA2, parkinsonism, or ALS depending upon the length of the repeat. Moreover, the finding that _ATXN2_-linked parkinsonism may be associated with an interrupted CAG repeat region, versus the pure CAG repeats typical of SCA2, raises the possibility that the CAG purity of the repeat expansion may contribute to disease manifestation.
Given these data, we sequenced the polyQ repeat region within ATXN2 in the cases of ALS and controls harboring ATXN2 polyQ repeats of 27 and higher [31]. Our results show that the repeats associated with increased risk for ALS are uniformly comprised of interrupted CAG repeats. All of the sequences are interrupted by 1–3 CAA codons; none of the repeats harbors a pure CAG repeat. Further, SNP analysis using the derived cleaved amplified polymorphic sequences (dCAPS) technique [34] revealed that the alleles with 3 CAA codons are the GT haplotype, whereas alleles with fewer CAA codons are predominantly the CC haplotype. Age of disease onset in subjects with ALS and 3 CAA repeat interruptions, compared to those with fewer, revealed that the age of onset was significantly earlier; age of disease onset also differed by haplotype. This is despite the fact that the average repeat length is shorter in subjects with 3 CAA interruptions. These results highlight that an interplay of the CAG repeat sequence configuration, haplotype association, as well as repeat length, may play a prominent role in disease manifestation in _ATXN2_-associated neuropathologies.
Results
CAA interruptions in the CAG repeat region of ATXN2 in ALS
We characterized the repeat sequence in 40 of the 45 ALS cases with intermediate length polyQ repeats in the ATXN2 gene of 27–33 in length and nine of the control cases [31]; these constituted a subset of 40 ALS cases we previously reported with expanded ATXN2 alleles for which motor neuron disease was the initial presentation and from which we were able to obtain amplifiable DNA. All 40 cases met El Escorial criteria for ALS; in addition, for 19/40 cases (those from the University of Pennsylvania Center for Neurodegenerative Disease Research (CNDR)) clinical charts were available and reviewed by a neurologist to confirm the diagnosis of ALS. With the exception of one previously described individual with ataxic features late in the course of disease (case A14, Table 1; [31]), these 19 well-characterized cases did not demonstrate features atypical of ALS such as parkinsonism or ataxia.
Table 1. Sequence of the polyQ repeat region of expanded allele of ALS patients.
| Case ID | Expanded allele | Age at Onset | CAG Pattern | SNP expanded allele** | CAA# | Pattern |
|---|---|---|---|---|---|---|
| A32 | 29 | 49 | 20-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A48 | 29 | 72 | 20-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A35 | 30 | 62 | 21-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A33 | 30 | 72 | 21-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A42 | 31 | 74 | 21-9 | CC | 1 | OOOOOOOOOOOOOOOOOOOOO•OOOOOOOOO |
| A37 | 31 | 61 | 21-9 | CC | 1 | OOOOOOOOOOOOOOOOOOOOO•OOOOOOOOO |
| A43 | 31 | 58 | 22-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A50 | 32 | 77 | 22-9 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOO•OOOOOOOOO |
| A47 | 32 | 79 | 23-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A51 | 32 | 55 | 23-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A49 | 32 | 53 | 23-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| A53 | 33 | 52 | 23-9 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOOO•OOOOOOOOO |
| A52 | 33 | 70 | 23-9 | CC | 1 | OOOOOOOOOOOOOOOOOOOOOOO•OOOOOOOOO |
| A30 | 28 | 55 | 13-5-8 | CC | 2 | OOOOOOOOOOOOO•OOOOO•OOOOOOOO |
| A41 * | 29 | 35 | 13-6-8 | CC | 2 | OOOOOOOOOOOOO•OOOOOO•OOOOOOOO |
| A34 | 30 | 58 | 13-7-8 | CC | 2 | OOOOOOOOOOOOO•OOOOOOO•OOOOOOOO |
| A38 | 31 | 56 | 8-13-8 | GT | 2 | OOOOOOOO•OOOOOOOOOOOOO•OOOOOOOO |
| A40 | 31 | 65 | 13-7-9 | CC | 2 | OOOOOOOOOOOOO•OOOOOOO•OOOOOOOOO |
| A45 | 32 | 54 | 13-8-9 | CC | 2 | OOOOOOOOOOOOO•OOOOOOOO•OOOOOOOOO |
| A14 | 27 | 30 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A17 * | 27 | 65 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A16 | 27 | not known | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A18 | 27 | 53 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A19 | 27 | 59 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A20 | 27 | 57 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A21 | 27 | 60 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A22 | 27 | 46 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A23 | 27 | 49 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A24 | 27 | 53 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A12 | 27 | 66 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A25 | 27 | 48 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A27 | 27 | 51 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A26 | 27 | 47 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A28 | 27 | 62 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A10 | 27 | 51 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A11 | 27 | 61 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A13 | 27 | 59 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| A31 | 29 | 64 | 8-4-4-10 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOOOO |
| A39 | 31 | 63 | 8-4-8-8 | GT | 3 | OOOOOOOO•OOOO•OOOOOOOO•OOOOOOOO |
| A46 | 32 | 42 | 8-4-9-8 | GT | 3 | OOOOOOOO•OOOO•OOOOOOOOO•OOOOOOOO |
The ATXN2 repeat region was amplified by polymerase chain reaction from genomic DNA samples. Because the repeats were intermediate in length compared to the normal allele (22 or 23), it was difficult to cleanly separate the allele bearing the longer repeat from the normal allele. Thus, we were unable to sequence the DNA directly from the PCR product and determine the sequence with high integrity. Instead, the amplification products were separated by size on a 4% agarose gel, then extracted, and expanded alleles subcloned and sequenced. Multiple clones for each allele were sequenced to assure against PCR-introduced changes.
The length of the normal allele of the ALS patients was typically 22; this is consistent with previous findings that a repeat of 22 is the most common normal allele [16], [17], [19], [35]. For the longer alleles, in ALS patients (n = 40 with ATXN2 expansion), the repeats ranged from 27 to 33, and in controls (n = 9 with ATXN2 expansions) from 27 to 32. Sequencing of the repeat region revealed that none of the polyQ repeats in patients with ALS were comprised of pure CAG repeat expansions. Rather, all were interrupted by at least 1 CAA codon, in stark contrast to the situation observed in SCA2. Interruptions of the repeat sequence are typical of the intermediate expansion repeats observed in control individuals [19]. Note that such interruptions do not affect the protein sequence, as CAA also encodes glutamine. Analysis of the repeat length patterns indicated that the repeat region was interrupted by 1, 2 or 3 CAA codons. Aside from the striking finding that none of the repeats were pure CAG repeats, we also observed among the Q27 repeats a monomorphic sequence pattern of 3 CAA interruptions in an 8-4-4-8 configuration.
In order to determine whether or not the polyQ repeat sequence was distinct among patients with repeat lengths within the ALS-susceptibility range and controls, we also sequenced 9 neurologically normal individuals with repeat lengths of 27 and longer (Table 2). This analysis confirmed that the polyQ-encoding DNA sequence was also interrupted in controls, and, moreover, bore interruption patterns found among the ALS patients. Thus, the controls of 27 had the same repeat configuration as the ALS patients; there was also no distinguishing feature among the controls with repeat lengths of 29, 30 and 32—these DNAs showed interrupted repeat patterns also found among ALS patients. Taken together, these data indicate that intermediate-length polyQ repeats in the ATXN2 gene associated with ALS are CAA interrupted repeats bearing interruptions of 1, 2 or 3 CAA codons.
Table 2. Sequence of the polyQ repeat region of expanded alleles of controls.
| Case ID | Atx2 length | Age at sampling | CAG Pattern | SNP expanded allele | CAA # | Pattern |
|---|---|---|---|---|---|---|
| N11 | 27 | 80 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| N12 | 27 | 65 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| N13 | 27 | 62<1/emph> | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| N14 | 27 | 61 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| N15 | 27 | 74 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| N16 | 27 | 73 | 8-4-4-8 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOO |
| N22 | 29 | 59 | 8-4-4-10 | GT | 3 | OOOOOOOO•OOOO•OOOO•OOOOOOOOOO |
| N23 | 30 | 57 | 21-8 | CC | 1 | OOOOOOOOOOOOOOOOOOOOO•OOOOOOOO |
| N24 | 32 | 59 | 13-8-9 | CC | 2 | OOOOOOOOOOOOO•OOOOOOOO•OOOOOOOOO |
Association of the haplotype of rs695871 and rs695872 with CAA interruptions in ATXN2
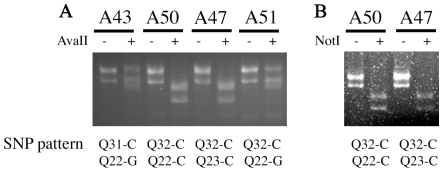
Two SNPs are found in exon 1 which bears the polyQ domain of ATXN2. rs695871 is 177 bp upstream of the repeat, and bears either a G or C base; the polymorphism affects the amino acid sequence, changing a Val to a Leu. The second SNP rs695872 is a T or C, and is silent, with no effect on the protein sequence. Others have previously reported that these two SNPs define two distinct haplotypes, a CC haplotype that appears to be evolutionarily ancestral and a GT haplotype of more recent origin; the CC haplotype predominates among SCA2 expanded alleles [19], [36]. Using dCAPS, to introduce a restriction enzyme site depending upon the polymophism to allow the SNPs to be defined by polymerase chain amplification followed by restriction enzyme analysis [34], we defined these polymorphisms in the ALS patients and control cases (Figure 1; Tables 1 and 2). This analysis showed that the 1 and 2 CAA interruption situations were predominantly associated with the CC haplotype (18/19 expanded chromosomes), whereas the 3 CAA interrupted alleles were uniformly the GT haplotype (21/21 chromosomes). This distinction was also the case for the intermediate length alleles among the controls.
Figure 1. SNP analysis by dCAPS method.
Analysis of restriction fragments for SNP (A) rs695871 and (B) rs695872. For SNP rs695871 (C or G, AvaII digestion) analysis of a Q32/Q22 patient, the amplified products were Q32:359bp and Q22:329bp. If Q32 is linked to a C, AvaII will digest the product into two fragments of 313 bp and 46 bp. If Q32 is linked to a G, AvaII will not cut. If Q22 is linked to a C, AvaII will digest it into fragments of 46 bp and 283 bp. If Q22 is linked to a G, AvaII will not cut. In the case of C/G heterozygotes, some uncut hybrid product remained. For SNP rs695872 (C or T, NotI digestion) analysis of a Q32/Q22 patient, the amplified products were Q32:308bp and Q22:278bp. If Q32 is linked to a C, NotI will digest the product into two fragments of 254 bp and 64 bp. If Q32 is linked to a T, NotI will not cut. If Q22 is linked to a C, NotI will digest it into fragments of 64 bp and 214 bp. If Q22 is linked to a T, NotI will not cut. In the case of C/T heterozygotes, some uncut hybrid product remained. In the gels, the low molecular weight bands (46, 64 bp) are largely undetectable; the pattern is scored by whether the higher bands are cut or not.
Association of the number of CAA interruptions and haplotype in ATXN2 with age of disease onset among the ALS patients
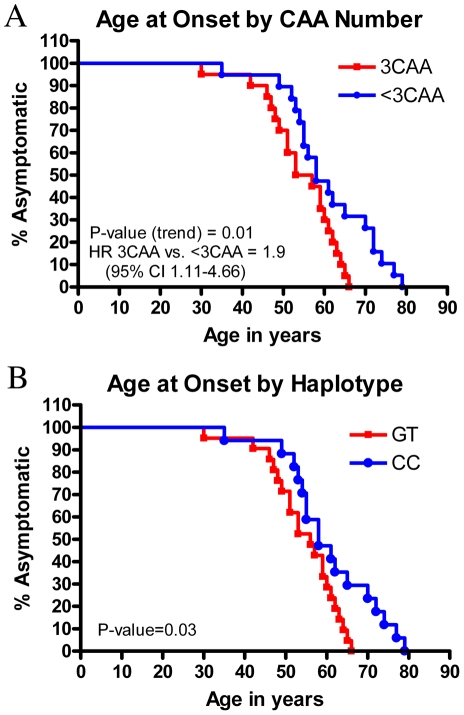
We considered whether the number of CAA interruptions within the repeat region and/or the genetic background in the form of SNP haplotype might result in differences in disease features among the 40 ALS patients. We evaluated whether age at onset, disease duration (for postmortem cases, n = 15), gender and presence/absence of atypical features differed by number of CAA interruptions or SNP haplotype. We found that age at onset among the ALS patients differed significantly depending on the number of CAA interruptions and the haplotype in the expanded allele, with individuals with more CAA interruptions and of the GT haplotype developing disease earlier (Figure 2, number of interruptions p-value for trend = 0.01, 3CAA vs <3CAA HR 1.9; haplotype, p = 0.03; log rank test). This is despite the fact that ALS patients with 3CAA interruptions have a shorter average repeat length (27.5) versus patients with fewer interruptions (30.8). Normally, for polyQ repeat expansion diseases, the relationship between repeat length and disease onset is such that longer repeats are associated with an earlier age of disease onset [13], [37]; thus, this trend for an earlier age of disease onset does not correlate with average repeat length. Other clinical features, such as gender, disease duration, presence/absence of atypical features, did not differ among the groups.
Figure 2. Age of disease onset by repeat interruptions and haplotype.
Age of disease onset by (A) repeat interruptions or by (B) haplotype for ALS patients bearing either 3CAA interruptions within the polyQ repeat region, versus patients with fewer than 3 CAA interruptions. The patients with 3 interruptions show an earlier disease onset, with a hazard ratio (HR) of 1.9, compared to patients with fewer interruptions. This is despite that patients with 3 CAA interruptions have an average repeat length that is lower (27.5) compared to patients with fewer interruptions (30.8). The age of disease onset is also significantly different when broken down by haplotype.
Discussion
Multiple faces of ATXN2 in neurological disease
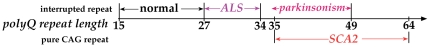
PolyQ expansions in ATXN2 are now associated with three clinically distinct neurological diseases; these different situations are associated with different repeat lengths and sequence configuration of the DNA repeat region (Figure 3). If the polyQ domain within ATXN2 is composed of a pure CAG repeat with a length ≥34, this presents with SCA2 [10]–[12], [14]–[19]. However, repeat expansions in ATXN2 can also present with levo-dopa responsive parkinsonism; about half of these situations have been sequenced. The repeat lengths range from 33 to 49, and in the situations sequenced, the polyQ domain is not a pure CAG repeat run, but rather is comprised of an interrupted CAG repeat region [22]–[30]. Further, recent studies indicate that intermediate-length polyQ repeats in ATXN2 of 27 to 33 are associated with increased risk for ALS [31], although the precise repeat length cut-off is likely to vary depending upon the population [38]. Here we show that ALS-associated polyQ repeats in ATXN2 are uniformly interrupted CAG repeats, comprised of interruptions of 1–3 CAA codons. We also observed a haplotype distinction between individuals bearing 3CAA interruptions and fewer. Note that CAA also encodes glutamine, thus the repeat region within the protein will remain a pure polyQ domain. These data suggest that the length of the polyQ repeat, as well as the CAG-repeat purity of the repeat region and the haplotype, influence disease presentation.
Figure 3. The spectrum of diseases associated with ATXN2 polyQ repeat expansions.
Although the repeat in ATXN2 is polymorphic normally, it is typically 22. In this cohort of controls, we found that only 1.4% (12/980 total) had repeats greater than 27 (range 27–31), whereas 4.7% of this cohort of ALS patients [31] had repeats of 27–33 (43/915 sporadic, 3/65 familial); the exact cut-off associated with risk for ALS may vary in different populations [38]. The repeats of ALS and high range controls from this study are interrupted by 1–3 CAA codons. ATXN2 repeats associated with parkinsonism range from 34 to 49, and studies to date indicate that the repeats are interrupted [21], [27]. Repeat expansions associated with SCA2 are greater than 34, and are typically pure CAG repeats [12], [14], [37].
RNA toxicity in _ATXN2_-related neuropathology
That interrupted and uninterrupted polyQ repeat expansions with similar length in ATXN2 lead to different neurological presentations (SCA2 or parkinsonism) highlights the possibility that repeat sequence configuration, as well as repeat length, is an important arbiter in clinical manifestation and phenotypic variability in _ATXN2_-associated pathologies. SCA2 is known to show nigral atrophy and motor neuron loss; that is, the clinical manifestations of parkinsonism and motor neuron disease are within the spectrum of SCA2 pathology [39]–[41]. Intriguingly, select situations appear biased strongly toward one neurological outcome or another; interestingly, this may be influenced by not only the length of the repeat, but also the purity of the CAG repeat region encoding the polyQ expansion.
PolyQ disease is classically viewed as a purely protein-based disease, with dominant toxicity conferred by the expanded polyQ domain [13], [37]. However, some data suggest that the situation may be more complicated, and that pathogenic mechanisms beyond toxicity or abnormal interactions solely due to the polyQ-containing protein may contribute to polyQ expansion diseases. For example, toxicity at the level of the expanded repeat RNA is conferred by CAG expansions in Drosophila [42], C elegans and mammals [43], [44]. Notably, CAA interruptions within the repeat sequence for SCA3 shift the toxicity curve, such that expression of a polyQ protein of identical amino acid sequence, but encoded by a CAA/G interrupted repeat region, is less toxic [42]. In the context of human disease, interruptions may present with different manifestations, or different clinical features of disease being dominant. In potential support of this idea, interruption of the CAG repeat expansion by a CAT codon within the ATXN1 polyQ-encoding domain was reported to cause later onset and milder disease in a rare case of SCA1 compared to pure, uninterrupted repeats of similar length [45]. An intriguing situation also occurs in SCA17, which is due to polyQ repeat expansions in TATA-box binding protein (TBP) [46]–[48]. Normally, the polyQ repeat region within TBP is interrupted by multiple CAA codons, and remains so in the expanded disease situation. Intriguingly, the threshold for a pathogenic repeat length is higher for repeat expansions for SCA17 (∼44) compared to that of most of the dominantly inherited spinocerebellar ataxias (∼37) [13], [49]. It is tempting to speculate that this shifted toxicity curve for SCA17 may be due to the interrupted repeat sequence of the polyQ domain within TBP.
Our findings are consistent with the idea that both the presence of any CAA interruptions and the presence of increasing numbers of CAA interruptions may influence phenotypic presentation. Specifically, while the overwhelming majority of SCA patients have pure CAG repeat expansions [10]–[12], [37], [41], 40/40 ALS patients in this study had at least one CAA interruption. Among these 40 patients with CAA interruptions, the number of interruptions as well as the haplotype may matter. Here we found an earlier age at onset for those ALS patients with more interruptions and who also share the GT haplotype.
The 3CAA-interrupted chromosomes arose on one genetic background (the GT haplotype), and most <3CAA-interrupted chromosomes arose on a different genetic background (the CC haplotype). We found both an association between earlier age at onset and number of CAA interruptions as well as SNP haplotype. Moreover, the fact that the ALS patients are not associated with a single haplotype may explain why ATXN2 has not emerged as a significant contributor to ALS from genome wide association (GWA) studies [50]–[53]. Intriguingly, however, a SNP linked to a gene that interacts with ATXN2, Ataxin-2 binding protein, has been identified as significant [50], [54]. Our previous study found an earlier age of disease onset in a small cohort of ALS patients (n = 65), when comparing those bearing a longer ATXN2 repeat to those with a normal length repeat [31]; it is possible that those data were driven by patients with 3CAA interruptions in the repeat sequence, of the GT haplotype. As with all such findings, such trends require confirmation among larger and additional sets of patients.
How might the sequence within the repeat region influence disease presentation? Whether the repeat sequence is a pure CAG or an interrupted CAG is predicted to influence somatic instability of the repeat, with a pure repeat being far more unstable and tending to expand [19], [55]–[57]. CAA interruptions have been proposed to play a critical role in conferring stability to the CAG repeat in ATXN2, with their absence predisposing ATXN2 alleles towards instability and pathogenic expansion to SCA2 disease [19]. Interruptions in a CAG repeat RNA by CAA codons are predicted to affect the secondary structure [58]. Even the interruption of a single CAA codon can have a notable effect on structure and free energy of folding. Moreover, if, as with other expanded repeat RNAs such as CGG expanded RNA of FXTAS [59], [60] and expanded CTG repeats of myotonic dystrophy [61], the CAG repeat RNA interacts with select proteins, the altered structure due to CAA interruptions may result in differential interactions. Such differential protein or other interactions may in part contribute to the distinct disease manifestations. The purity of the repeat may also influence the level of the mRNA and/or level of the translated protein. Such an influence of the RNA may be notable for ALS, as defects in RNA processing and metabolism may particularly relevant. The association of mutations in TDP43 and fused in sarcoma (FUS)/translocated in liposarcoma (TLS), two RNA-binding proteins with striking structural and functional similarities [62], as well as ATXN2, which itself is an RNA binding protein, in ALS [31], suggests a prominent role for RNA binding proteins and RNA-dependent processes in the disease.
Methods
Ethics statement
Samples from the CNDR were obtained with written informed consent under Institutional Board Approval of the University of Pennsylvania. Samples from Coriell were obtained with permission from the Coriell Institute for Medical Research Institutional Review Board. The individual submitters who contributed DNA samples to Coriell received written informed consent from all patients (or guardians of patients) participating in the study (consent for research).
PolyQ repeat amplification and sequencing
The polyglutamine repeat region of the ATXN2 gene was amplified from genomic DNA isolated from B-lymphocytes of controls and ALS patients. Patient samples were from the CNDR (19/40 ALS samples with 27 or more repeats) and Coriell (21/40 ALS samples with 27 or more repeats), controls were from Coriell; for additional details see [31]. Amplification was carried out with the primers SCA2-S2 (5′-CGCCGCGTTCCGGCGTCTCC-3′) and SCA2-B (5′-CGGGCTTGCGGACATTGG-3′), using Takara LA taq with GC buffer (Takara RR02AG, Shiga, Japan) plus 5% DMSO (Sigma, Cat# D8779). The amplification products were separated on a 4% agarose gel, purified, then sub-cloned into the pGEMT Easy vector (Promega Cat.# A1360, Madison, Wisconsin) and transformed into DH5alpha competent cells (Invitrogen Cat. #18265017, Carlsbad, CA). Colonies were screened by amplification to identify those bearing the expanded allele. For each expanded allele, 2–6 clones from at least two independent PCR reactions were prepared by Qiagen mini-prep and sequenced using the T7 primer from the pGEMT Easy vector.
SNP genotyping
The dCAPS technique [34] was used to analyze SNP rs695871 and SNP rs695872 (www.ncbi.nlm.nih.gov/SNP/) within exon1 of the human ATXN2 gene. For SNP695871, Primer SCA2-AVAII (5′-ctcccggcggctccttggtctcggcggg CCTCCCCGCCCCTTCGTGGTC-3′, mismatch underlined) was used such that one mismatch was introduced to generate a recognition site for the restriction endonuclease AvaII (recognition site 5′-G′G(A/T)CC-3′). Products covering the SNP and polyQ repeat region were amplified with primers SCA2-AVAII and SCA2-B (5′-CGGGCTTGCGGACATTGG-3′) using LA taq DNA polymerase as above. Undigested and AvaII-digested products were separated on a 4% agarose gel in parallel. AvaII could only cut the amplified product if the SNP were a C and not a G. Thus, the SNP (C or G) and linkage to the normal or expanded allele were determined by the digestion pattern. For SNP695872, Primer SCA2-NOTI (5′-ccttctccccctcgccagcccgggcgcccctccggccgcgccaacccgcg CCTCCCCGCTCGGCGGCCG-3′, mismatch underlined) was used such that one mismatch was introduced to generate a recognition site for the restriction endonuclease NotI (recognition site 5′-GC′GGCCGC-3′). Products spanning the SNP and polyQ region were amplified with primers SCA2-NOTI and SCA2-B. Undigested and NotI-digested products were separated on a 4% agarose gel in parallel. NotI could only cut the amplified product if the SNP were a C and not T. Thus, the SNP (C or T) and the linkage to the normal or expanded allele were determined by the digestion pattern.
Statistics
Survival curve analyses using log-rank tests were used to compare age at onset, disease duration, gender, and presence/absence of atypical features between subgroups of ALS patients with 1,2 and 3CAA interruptions, and between subgroups of ALS patients with different genetic backgrounds.
Acknowledgments
We are truly grateful for the dedication of the patients and their families and for their invaluable contributions to this research.
Footnotes
Competing Interests: N.B. and A.D.G. are inventors on patent applications. A.D.G. is an inventor on patents and patent applications which have been licensed to FoldRx. This does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: This work was supported in part by a Pilot grant from the University of Pennsylvania Institute on Aging (V.M.V.D., A.D.G.), a National Institutes of Health (NIH) Director's New Innovator Award 1DP2OD004417-01 (A.D.G.), 1R01NS065317-01 (A.D.G.), P01 AG-09215 (N.B.), AG-10124 (J.Q.T., V.M.V.D.) and AG-17586, (V.M.-Y.L., V.M.V.D.). A.D.G. is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts. A.C.P. is supported by a Burroughs Wellcome Fund Career Award, NIH K08 AG-033101-01 and the Benaroya Fund. N.M.B. is an Investigator of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nat Rev Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 2.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 4.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 5.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 6.Gitcho MA, Baloh RH, Chakraverty S, Mayo K, Norton JB, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kabashi E, Valdmanis PN, Dion P, Spiegelman D, McConkey BJ, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 8.Kuhnlein P, Sperfeld AD, Vanmassenhove B, Van Deerlin V, Lee VM, et al. Two German kindreds with familial amyotrophic lateral sclerosis due to TARDBP mutations. Arch Neurol. 2008;65:1185–1189. doi: 10.1001/archneur.65.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- 11.Sanpei K, Takano H, Igarashi S, Sato T, Oyake M, et al. Identification of the spinocerebellar ataxia type 2 gene using a direct identification of repeat expansion and cloning technique, DIRECT. Nat Genet. 1996;14:277–284. doi: 10.1038/ng1196-277. [DOI] [PubMed] [Google Scholar]
- 12.Imbert G, Saudou F, Yvert G, Devys D, Trottier Y, et al. Cloning of the gene for spinocerebellar ataxia 2 reveals a locus with high sensitivity to expanded CAG/glutamine repeats. Nat Genet. 1996;14:285–291. doi: 10.1038/ng1196-285. [DOI] [PubMed] [Google Scholar]
- 13.Gusella JF, MacDonald ME. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 14.Cancel G, Durr A, Didierjean O, Imbert G, Burk K, et al. Molecular and clinical correlations in spinocerebellar ataxia 2: a study of 32 families. Hum Mol Genet. 1997;6:709–715. doi: 10.1093/hmg/6.5.709. [DOI] [PubMed] [Google Scholar]
- 15.Geschwind DH, Perlman S, Figueroa CP, Treiman LJ, Pulst SM. The prevalence and wide clinical spectrum of the spinocerebellar ataxia type 2 trinucleotide repeat in patients with autosomal dominant cerebellar ataxia. Am J Hum Genet. 1997;60:842–850. [Google Scholar]
- 16.Lorenzetti D, Bohlega S, Zoghbi HY. The expansion of the CAG repeat in ataxin-2 is a frequent cause of autosomal dominant spinocerebellar ataxia. Neurology. 1997;49:1009–1013. doi: 10.1212/wnl.49.4.1009. [DOI] [PubMed] [Google Scholar]
- 17.Giunti P, Sabbadini G, Sweeney MG, Davis MB, Veneziano L, et al. The role of the SCA2 trinucleotide repeat expansion in 89 autosomal dominant cerebellar ataxia families. Frequency, clinical and genetic correlates. Brain. 1998;121(Pt 3):459–467. doi: 10.1093/brain/121.3.459. [DOI] [PubMed] [Google Scholar]
- 18.Saleem Q, Choudhry S, Mukerji M, Bashyam L, Padma MV, et al. Molecular analysis of autosomal dominant hereditary ataxias in the Indian population: high frequency of SCA2 and evidence for a common founder mutation. Hum Genet. 2000;106:179–187. doi: 10.1007/s004390051026. [DOI] [PubMed] [Google Scholar]
- 19.Choudhry S, Mukerji M, Srivastava AK, Jain S, Brahmachari SK. CAG repeat instability at SCA2 locus: anchoring CAA interruptions and linked single nucleotide polymorphisms. Hum Mol Genet. 2001;10:2437–2446. doi: 10.1093/hmg/10.21.2437. [DOI] [PubMed] [Google Scholar]
- 20.Costanzi-Porrini S, Tessarolo D, Abbruzzese C, Liguori M, Ashizawa T, et al. An interrupted 34-CAG repeat SCA-2 allele in patients with sporadic spinocerebellar ataxia. Neurology. 2000;54:491–493. doi: 10.1212/wnl.54.2.491. [DOI] [PubMed] [Google Scholar]
- 21.Kim JM, Hong S, Kim GP, Choi YJ, Kim YK, et al. Importance of low-range CAG expansion and CAA interruption in SCA2 Parkinsonism. Arch Neurol. 2007;64:1510–1518. doi: 10.1001/archneur.64.10.1510. [DOI] [PubMed] [Google Scholar]
- 22.Gwinn-Hardy K, Chen JY, Liu HC, Liu TY, Boss M, et al. Spinocerebellar ataxia type 2 with parkinsonism in ethnic Chinese. Neurology. 2000;55:800–805. doi: 10.1212/wnl.55.6.800. [DOI] [PubMed] [Google Scholar]
- 23.Shan DE, Soong BW, Sun CM, Lee SJ, Liao KK, et al. Spinocerebellar ataxia type 2 presenting as familial levodopa-responsive parkinsonism. Ann Neurol. 2001;50:812–815. doi: 10.1002/ana.10055. [DOI] [PubMed] [Google Scholar]
- 24.Furtado S, Farrer M, Tsuboi Y, Klimek ML, de la Fuente-Fernandez R, et al. SCA-2 presenting as parkinsonism in an Alberta family: clinical, genetic, and PET findings. Neurology. 2002;59:1625–1627. doi: 10.1212/01.wnl.0000035625.19871.dc. [DOI] [PubMed] [Google Scholar]
- 25.Lu CS, Wu Chou YH, Kuo PC, Chang HC, Weng YH. The parkinsonian phenotype of spinocerebellar ataxia type 2. Arch Neurol. 2004;61:35–38. doi: 10.1001/archneur.61.1.35. [DOI] [PubMed] [Google Scholar]
- 26.Ragothaman M, Sarangmath N, Chaudhary S, Khare V, Mittal U, et al. Complex phenotypes in an Indian family with homozygous SCA2 mutations. Ann Neurol. 2004;55:130–133. doi: 10.1002/ana.10815. [DOI] [PubMed] [Google Scholar]
- 27.Charles P, Camuzat A, Benammar N, Sellal F, Destee A, et al. Are interrupted SCA2 CAG repeat expansions responsible for parkinsonism? Neurology. 2007;69:1970–1975. doi: 10.1212/01.wnl.0000269323.21969.db. [DOI] [PubMed] [Google Scholar]
- 28.Modoni A, Contarino MF, Bentivoglio AR, Tabolacci E, Santoro M, et al. Prevalence of spinocerebellar ataxia type 2 mutation among Italian Parkinsonian patients. Mov Disord. 2007;22:324–327. doi: 10.1002/mds.21228. [DOI] [PubMed] [Google Scholar]
- 29.Socal MP, Emmel VE, Rieder CR, Hilbig A, Saraiva-Pereira ML, et al. Intrafamilial variability of Parkinson phenotype in SCAs: novel cases due to SCA2 and SCA3 expansions. Parkinsonism Relat Disord. 2009;15:374–378. doi: 10.1016/j.parkreldis.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Wang JL, Xiao B, Cui XX, Guo JF, Lei LF, et al. Analysis of SCA2 and SCA3/MJD repeats in Parkinson's disease in mainland China: genetic, clinical, and positron emission tomography findings. Mov Disord. 2009;24:2007–2011. doi: 10.1002/mds.22727. [DOI] [PubMed] [Google Scholar]
- 31.Elden A, Kim H-J, Hart M, Chen-Plotkin A, Johnson B, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased susceptibility to amyotrophic lateral sclerosis. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Infante J, Berciano J, Volpini V, Corral J, Polo JM, et al. Spinocerebellar ataxia type 2 with Levodopa-responsive parkinsonism culminating in motor neuron disease. Mov Disord. 2004;19:848–852. doi: 10.1002/mds.20090. [DOI] [PubMed] [Google Scholar]
- 33.Nanetti L, Fancellu R, Tomasello C, Gellera C, Pareyson D, et al. Rare association of motor neuron disease and spinocerebellar ataxia type 2 (SCA2): a new case and review of the literature. J Neurol. 2009;256:1926–1928. doi: 10.1007/s00415-009-5237-9. [DOI] [PubMed] [Google Scholar]
- 34.Neff MM, Neff JD, Chory J, Pepper AE. dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: experimental applications in Arabidopsis thaliana genetics. Plant J. 1998;14:387–392. doi: 10.1046/j.1365-313x.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- 35.Riess O, Laccone FA, Gispert S, Schols L, Zuhlke C, et al. SCA2 trinucleotide expansion in German SCA patients. Neurogenetics. 1997;1:59–64. doi: 10.1007/s100480050009. [DOI] [PubMed] [Google Scholar]
- 36.Ramos EM, Martins S, Alonso I, Emmel VE, Saraiva-Pereira ML, et al. Common origin of pure and interrupted repeat expansions in spinocerebellar ataxia type 2 (SCA2). Am J Med Genet B Neuropsychiatr Genet. 2009;153B:524–531. doi: 10.1002/ajmg.b.31013. [DOI] [PubMed] [Google Scholar]
- 37.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 38.Lee T, Li YR, Ingre C, Weber M, Grehl T, et al. Ataxin-2 intermediate-length polyglutamine expansions in European ALS patients. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoche F, Baliko L, den Dunnen W, Steinecker K, Bartos L, et al. Spinocerebellar Ataxia Type 2 (SCA2): Identification of Early Brain Degeneration in One Monozygous Twin in the Initial Disease Stage. Cerebellum. 2010 doi: 10.1007/s12311-010-0239-9. [DOI] [PubMed] [Google Scholar]
- 40.Ishida C, Komai K, Yonezawa K, Sakajiri KI, Nitta E, et al. An autopsy case of an aged patient with spinocerebellar ataxia type 2. Neuropathology. 2010 doi: 10.1111/j.1440-1789.2010.01176.x. [DOI] [PubMed] [Google Scholar]
- 41.Lastres-Becker I, Rub U, Auburger G. Spinocerebellar ataxia 2 (SCA2). Cerebellum. 2008;7:115–124. doi: 10.1007/s12311-008-0019-y. [DOI] [PubMed] [Google Scholar]
- 42.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hsu RJ, Hsiao KM, Lin MJ, Li CY, Wang LC, et al. Long Tract of Untranslated CAG Repeats Is Deleterious in Transgenic Mice. PLoS One. 2011;6:e16417. doi: 10.1371/journal.pone.0016417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang LC, Chen KY, Pan H, Wu CC, Chen PH, et al. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0522-4. DOI: 10.1007/s00018-010-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsuyama Z, Izumi Y, Kameyama M, Kawakami H, Nakamura S. The effect of CAT trinucleotide interruptions on the age at onset of spinocerebellar ataxia type 1 (SCA1). J Med Genet. 1999;36:546–548. [PMC free article] [PubMed] [Google Scholar]
- 46.Nakamura K, Jeong SY, Uchihara T, Anno M, Nagashima K, et al. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum Mol Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- 47.Zuhlke C, Hellenbroich Y, Dalski A, Kononowa N, Hagenah J, et al. Different types of repeat expansion in the TATA-binding protein gene are associated with a new form of inherited ataxia. Eur J Hum Genet. 2001;9:160–164. doi: 10.1038/sj.ejhg.5200617. [DOI] [PubMed] [Google Scholar]
- 48.Koide R, Kobayashi S, Shimohata T, Ikeuchi T, Maruyama M, et al. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum Mol Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 49.van Roon-Mom WM, Reid SJ, Faull RL, Snell RG. TATA-binding protein in neurodegenerative disease. Neuroscience. 2005;133:863–872. doi: 10.1016/j.neuroscience.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 50.Cronin S, Berger S, Ding J, Schymick JC, Washecka N, et al. A genome-wide association study of sporadic ALS in a homogenous Irish population. Hum Mol Genet. 2008;17:768–774. doi: 10.1093/hmg/ddm361. [DOI] [PubMed] [Google Scholar]
- 51.Cronin S, Tomik B, Bradley DG, Slowik A, Hardiman O. Screening for replication of genome-wide SNP associations in sporadic ALS. Eur J Hum Genet. 2009;17:213–218. doi: 10.1038/ejhg.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dunckley T, Huentelman MJ, Craig DW, Pearson JV, Szelinger S, et al. Whole-genome analysis of sporadic amyotrophic lateral sclerosis. N Engl J Med. 2007;357:775–788. doi: 10.1056/NEJMoa070174. [DOI] [PubMed] [Google Scholar]
- 53.van Es MA, Veldink JH, Saris CG, Blauw HM, van Vught PW, et al. Genome-wide association study identifies 19p13.3 (UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41:1083–1087. doi: 10.1038/ng.442. [DOI] [PubMed] [Google Scholar]
- 54.Cronin S, Blauw HM, Veldink JH, van Es MA, Ophoff RA, et al. Analysis of genome-wide copy number variation in Irish and Dutch ALS populations. Hum Mol Genet. 2008;17:3392–3398. doi: 10.1093/hmg/ddn233. [DOI] [PubMed] [Google Scholar]
- 55.Pearson CE, Eichler EE, Lorenzetti D, Kramer SF, Zoghbi HY, et al. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 56.Gao R, Matsuura T, Coolbaugh M, Zuhlke C, Nakamura K, et al. Instability of expanded CAG/CAA repeats in spinocerebellar ataxia type 17. Eur J Hum Genet. 2008;16:215–222. doi: 10.1038/sj.ejhg.5201954. [DOI] [PubMed] [Google Scholar]
- 57.Pearson CE, Nichol Edamura K, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat Rev Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 58.Sobczak K, Krzyzosiak WJ. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J Biol Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 59.Sofola OA, Jin P, Qin Y, Duan R, Liu H, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jin P, Duan R, Qurashi A, Qin Y, Tian D, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 62.Lagier-Tourenne C, Cleveland DW. Rethinking ALS: the FUS about TDP-43. Cell. 2009;136:1001–1004. doi: 10.1016/j.cell.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]