ARNO and ARF6 Regulate Axonal Elongation and Branching through Downstream Activation of Phosphatidylinositol 4-Phosphate 5-Kinase α (original) (raw)
Abstract
In the developing nervous system, controlled neurite extension and branching are critical for the establishment of connections between neurons and their targets. Although much is known about the regulation of axonal development, many of the molecular events that regulate axonal extension remain unknown. ADP-ribosylation factor nucleotide-binding site opener (ARNO) and ADP-ribosylation factor (ARF)6 have important roles in the regulation of the cytoskeleton as well as membrane trafficking. To investigate the role of these molecules in axonogenesis, we expressed ARNO and ARF6 in cultured rat hippocampal neurons. Expression of catalytically inactive ARNO or dominant negative ARF6 resulted in enhanced axonal extension and branching and this effect was abrogated by coexpression of constitutively active ARF6. We sought to identify the downstream effectors of ARF6 during neurite extension by coexpressing phosphatidyl-inositol-4-phosphate 5-Kinase α [PI(4)P 5-Kinase α] with catalytically inactive ARNO and dominant negative ARF6. We found that PI(4)P 5-Kinase α plays a role in neurite extension and branching downstream of ARF6. Also, expression of inactive ARNO/ARF6 depleted the actin binding protein mammalian ena (Mena) from the growth cone leading edge, indicating that these effects on axonogenesis may be mediated by changes in cytoskeletal dynamics. These results suggest that ARNO and ARF6, through PI(4)P 5-Kinase α, regulate axonal elongation and branching during neuronal development.
INTRODUCTION
During the development of the nervous system, neurite outgrowth is necessary for the formation of the highly specific pattern of connections between nerve cells. Importantly, neurons are polarized cells with distinct domains, axons, and dendrites, which serve very different functions in impulse transmission and integration (Craig and Banker, 1994). The differentiation of axons and dendrites has been the subject of intensive investigation (Luo et al., 1996a; Li et al., 2000; Scott and Luo, 2001; Luo, 2002; Ng et al., 2002; Whitford et al., 2002); however, the molecular mechanism of differential growth of these processes remains incompletely understood. Some aspects of these processes seem similar. For example, during navigation, the growth cones of axons and dendrites extend filopodia and lamellipodia by changing the dynamics of the plasma membrane and the organization of the actin cytoskeleton (Craig et al., 1995; Gallo and Letourneau, 2000; Kamiguchi and Lemmon, 2000). These events result in neurite outgrowth, retraction, or branching (Tessier-Lavigne and Goodman, 1996; Song and Poo, 1999). The small GTPases of the Rho subfamily are important regulators of the actin cytoskeleton (for review, see Hall, 1998) and have been shown to be critical in the regulation of both axonal and dendritic extension (for review, see Luo, 2000). However, in addition to the Rho-family of GTPases, members of the ADP-ribosylation factor (ARF) family of small GTPases play an essential role in membrane trafficking and cytoskeletal rearrangement. In nonneuronal cells, ARF6 regulates endocytic membrane traffic, alters cortical actin (Radhakrishna et al., 1996, 1999; D'Souza-Schorey et al., 1997; Radhakrishna and Donaldson, 1997; Frank et al., 1998b; Boshans et al., 2000), and modulates cell adhesion events (Palacios et al., 2001; Santy and Casanova, 2001). ARF proteins activate the lipid-modifying enzymes phospholipase D (PLD) and phosphatidyl-inositol-4-phosphate 5-Kinase [PI(4)P 5-Kinase] (Exton, 1997; Honda et al., 1999) and indirectly, Rac1 (Santy and Casanova, 2001). Also, the ARF-GTP exchange factor (GEF) ARF nucleotide-binding site opener (ARNO) and ARF6 modulate epithelial cell migration through downstream activation of both Rac1 and PLD (Santy and Casanova, 2001). These data indicate that ARNO and ARF6 are key players in regulating actin dynamics and membrane trafficking in various cell types.
In neurons, ARF proteins may regulate axonal elongation because treatment with brefeldin A inhibited axonal growth (Jareb and Banker, 1997; Hess et al., 1999). ARNO is a brefeldin A-insensitive GEF that catalyzes exchange on ARF1 and ARF6 (Chardin et al., 1996; Frank et al., 1998a). We have shown that ARNO is present during neuritogenesis (Hernandez-Deviez et al., 2002), and morphological and biochemical studies revealed high expression of both ARF6 and ARNO mRNA and protein in the developing hippocampus (Suzuki et al., 2001, 2002; Hernandez-Deviez et al., 2002). In addition, overexpression of inactive ARNO and ARF6 caused increased branching of dendritic processes, and this effect was reversed by expression of Rac1 (Hernandez-Deviez et al., 2002). However, the function of ARNO and ARF6 and their effectors during axonal extension remains unclear. Recent studies in chick retinal neurons (Albertinazzi et al., 2003) and Aplysia motor neurons (Huh et al., 2003) have suggested that ARF6 regulates neurite extension. Here, we find that expression of inactive forms of ARNO and ARF6 result in enhanced axonal elongation and branching in developing hippocampal neurons. Strikingly, these effects were reversed by coexpression of type I PI(4)P 5-Kinase α with catalytically inactive ARNO or dominant negative ARF6. The mechanism of ARNO action may be mediated through the actin binding protein mammalian ena (Mena), because cells expressing mutant ARNO or ARF6 showed specific depletion of Mena from the growth cone leading edge. This is consistent with the proposed negative regulatory role for Mena in cell migration (Bear et al., 2000; Krause et al., 2002) and may indicate a negative regulatory role in axonal extension. We propose a model in which ARNO/ARF6, through stimulation of PI(4)P 5-K, regulates the activity of actin binding proteins at the growth cone and subsequently regulates axonal extension.
MATERIALS AND METHODS
Reagents, Antibodies, and DNA Constructs
Cell culture media were obtained from Invitrogen (Carlsbad, CA). The following antibodies were used: mouse anti-myc 9E10, mouse anti-MAP2, mouse anti-FLAG M2, mouse anti-vinculin, mouse anti-β tubulin, mouse anti-gelsolin (Sigma-Aldrich, St. Louis, MO), rabbit anti-profilin (Cytoskeleton, Denver, CO), mouse anti-tau (Zymed Laboratories, South San Francisco, CA), mouse anti-HA (Roche Diagnostics, Indianapolis, IN), mouse anti-Mena (BD Biosciences, San Diego, CA), rabbit anti-myc (Upstate Biotechnology, Lake Placid, NY), mouse anti-ERM 13H9 (gift from Dr. F. Solomon, Massachusetts Institute of Technology, Cambridge, MA). Secondary antibodies were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). Rhodamine-conjugated phalloidin was from Molecular Probes (Eugene, OR). C-myc-tagged ARNO and hemagglutinin (HA)-tagged ARF6 constructs were generated as described previously (Frank et al., 1998b; Altschuler et al., 1999). FLAG-tagged Rac1 constructs were provided by Dr. J. Settleman (Harvard Medical School, Boston, MA).
GFP-ARNO-E156K was generated with the QuikChange TM XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) by using the following primers: 5′-3′ GCTTTCGCCTACCCGGAAAGGCCCAGAAAATTGACC, and 5′-3′ CGAAAGCGGATGGGCCTTTCCGGGTCTTTTAACTTG.
Cell Culture and Transfection
Fetuses were obtained at E17 or E18 from timed pregnant Sprague Dawley rats (Harlan, Indianapolis, IN). Primary neuronal cultures were prepared based on published protocols (Banker and Goslin, 1991). Hippocampi were dissociated by treatment with trypsin (0.05% for 15 min at 37°C), followed by trituration. Dissociated cells were plated at low density (350,000 cells/60-mm dish) on poly-l-lysine–treated coverslips in glia-conditioned Neurobasal medium in the presence of supplement B27 and kept at 37°C under 5% CO2. All animal protocols were reviewed and approved by the Institutional Animal Care Use and Committee of the University of Arizona.
cDNAs were expressed in the hippocampal neurons by transient transfection by using Effectene transfection reagent according to manufacturer's directions (QIAGEN, Valencia, CA). Single transfection with cDNAs encoding ARNO, ARNO-E156K, PI(4)P 5-K α, and PI(4)P 5-K α-D266A and coexpression of ARNO-E156K and PI(4)P 5-K α was performed after 1 d in vitro, whereas transfection with ARF6, ARF6-T27N, and ARF6-Q67L and coexpression of ARNO-E156K and ARF6-Q67L or PI(4)P 5-K α, and coexpression of ARF6-T27N and PI(4)P 5-K α plasmids were performed after 5 d in vitro. Viability after transfection was >85%.
Immunofluorescence Microscopy
For quantitative analysis, neurons were fixed after 9 d in vitro in 4% paraformaldehyde. After fixation, cells were permeabilized and blocked with 10% goat serum, 0.05% saponin, and then incubated with specific antibodies followed by incubation with fluorophore-conjugated secondary antibodies. After washing, coverslips were mounted in Aqua Polymount (Polyscience, Warrington, PA). Fluorescent images of single optical sections were obtained using a Leica TCS 4D laser scanning confocal microscope (Arizona Research Laboratory, Division of Biotechnology, University of Arizona) by using a 20× (numerical aperture [NA] 0.6) objective. Simultaneous two-channel recording was performed with pinhole sizes of 40–90 μm by using excitation wavelengths of 488/588 nm, a 510/580 double diachronic mirror, and a 515–545 band-pass fluorescein isothiocyanate filter together with 590-nm long-pass filter. Images were processed using Adobe Photoshop software (Adobe Systems, Mountain View, CA).
For cytoskeletal protein studies, fluorescent images were acquired using the DeltaVision restoration microscopy system (Applied Precision, Issaquah, WA) by using an 100 × (NA 1.35) objective. The z-stacks were software deconvolved on a Silicon Graphics Workstation (SGI, Mountain View, CA) by using measured point-spread functions to create the final images. Images were processed and merged using Adobe Photoshop software (Adobe Systems). To facilitate comparison, identical imaging and processing parameters were used for all figures.
Quantification of Axonal Length and Branching
Morphometric analyses of axonal length and branching were performed by analyzing single optical sections of confocal images with a SimplePCI Image Analysis System (Compix, Pittsburgh, PA). Confocal images were acquired with the 20× (NA 0.6) objective. The software was calibrated to the microscope magnification. Axons were identified based on their characteristic morphology or as microtubule associate protein 2 negative processes, and transfected cells were identified using antibodies against the epitope tag (Table 1). Axonal length was determined by tracing the entire length of the process, and total length was calculated. Axonal branching was determined by counting the number of branch points along the axon. For each construct, 10 transfected cells were analyzed.
Table 1.
List of plasmids used for transfection of rat hippocampal neurons
| Plasmid | Mutation |
|---|---|
| ARNO-myc | Wild type |
| ARNO-E156K-myc | Catalytically inactive |
| ARNO-GFP | Wild type |
| ARNO-E156K-GFP | Catalytically inactive |
| ARF6-HA | Wild type |
| ARF6-T27N-HA | Dominant negative |
| ARF6-Q67L-HA | Constitutively active |
| Rac1-FLAG | Wild type |
| Rac1-N17-FLAG | Dominant negative |
| PI(4)P 5K α-HA | Wild type |
One-way analysis of variance was used to compare mean changes in the relative length of axons. Results were expressed as the mean of axonal length in microns. Data are presented as means ± SD (n = 10). The p value of <0.0001 was considered highly significant.
Quantification of Mena Distribution in the Axonal Growth Cone
Hippocampal neurons were transfected with _myc_-tagged ARNO wild-type and ARNO-E156K, labeled with anti-Mena antibody, and visualized by deconvolution microscopy. Sixteen axonal growth cones were imaged per treatment and the distribution of Mena was scored as present at the leading lamellae and filopodia, at the growth cone base, or depleted from the growth cone. Identical imaging and processing parameters were used for each plasmid.
RESULTS
Expression of Catalytically Inactive ARNO Increases Axonal Length and Branching
ARNO has been shown to act as a nucleotide exchange factor for both ARF1 and ARF6 in vitro (Chardin et al., 1996; Frank et al., 1998a). To study the role that ARF family members play during axonogenesis, we first expressed wild-type and catalytically inactive forms of the ARF-GEF ARNO in embryonic day 17 (E17) rat hippocampal neurons in culture. ARNO is recruited to membrane phosphoinositides via its pleckstrin homology domain (Chardin et al., 1996) and, when the inactive form is overexpressed, it may displace endogenous ARNO and inhibit nucleotide exchange on ARF molecules. (Beraud-Dufour et al., 1998). This catalytically inactive form of ARNO has been used previously to study inhibition of ARF molecules. (Frank et al., 1998b; Caumont et al., 2000; Santy and Casanova, 2001; Hernandez-Deviez et al., 2002; Huh et al., 2003)
Expression of catalytically inactive ARNO resulted in a dramatic effect on axonal elongation and branching. The majority of untransfected cells exhibited a single axonal extension that emerged from the cell body and formed approximately four to five branches (Figure 1, a–c); neurons expressing ARNO wild type showed no statistically significant effects upon axonal length and complexity (Figure 1, a, b, and d), suggesting downstream effectors are limiting in the effect of ARNO upon axonogenesis at this stage of axonal development. In contrast, cells expressing catalytically inactive ARNO had a long axon that emitted numerous collateral branches that were also long, resulting in a nearly sixfold increase in total axonal length and a fourfold increase in the average number of axonal branches (Figure 1, a, b, and e). Because ARNO has been shown to be present during early events in neuronal development, these results suggest that ARNO could regulate axonal growth and branching.
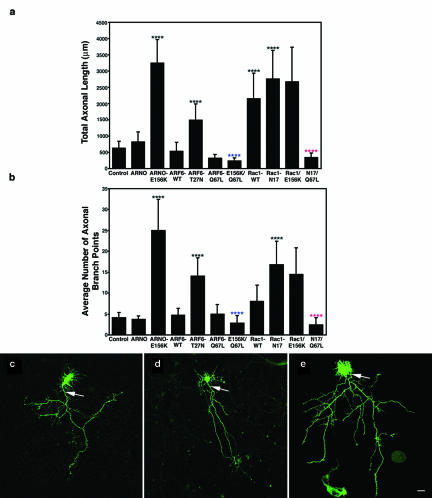
Figure 1.
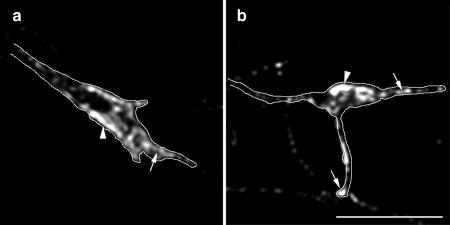
ARNO mediates axonal length and branching through an ARF6-dependent pathway. Morphometric analysis of the effects of expression of wild-type or dominant negative mutant forms of ARNO and ARF6 on axonal outgrowth and branching. (a) Effects on total axonal length. Expression of ARNO-E156K and ARF6-T27N, Rac1 or Rac1-N17 resulted in increased axonal length, whereas expression of wild-type ARNO, wild-type ARF6 or ARF6-Q67L resulted in no quantifiable effect upon axonal length. Coexpression of ARF6-Q67L reversed the effect of expression of ARNO-E156K or Rac1N17 upon axonal length. (b) Effects on the number of axonal branch points. Expression of ARNO-E156K, ARF6-T27N, and Rac1-N17 increased the number of axonal branch points, whereas expression of wild-type ARNO, wild-type ARF6 or ARF6-Q67L did not affect the number of axonal branch points. Coexpression of ARF6-Q67L reversed the effect of expression of ARNO-E156K and Rac1-N17 upon axonal branching. Data are presented as the means ± SD (n = 10). p < 0.0001 (****) with respect to control (****, blue), with respect to ARNO-E156K (****, red), and with respect to Rac1-N17. (c–e) Hippocampal neurons transfected with green fluorescent protein (GFP) vector alone (c), GFP-ARNO wild-type (d), and GFP-ARNO-E156K (e) were fixed after 6 d in culture and visualized by confocal microscopy. (c and d) In cells expressing GFP alone (c) or GFP-ARNO wild type (d), the axon (arrow) extends some distance away from cell body emitting some collateral branches. (e) Cell expressing GFP-ARNO-E156K. The axonal process (arrow) is very long with numerous collateral branches. The elaboration of the dendritic tree (Hernandez-Deviez et al., 2002) cannot be visualized at this magnification. Bars, 25 μm (e–g).
Effects of ARNO upon Axonogenesis Are Mediated by Downstream Activation of ARF6
ARNO has been shown to have GEF activity on both ARF1 and ARF6 in vitro (Chardin et al., 1996; Frank et al., 1998a; Monier et al., 1998), and both ARF proteins are present in developing hippocampus (Hernandez-Deviez et al., 2002). To determine whether ARNO effects could be mediated through ARF6, we next examined the consequences of expression of wild-type and mutant forms of ARF6 upon axonogenesis. If ARNO acts through ARF6, we would expect that neurons expressing dominant negative ARF6 would exhibit longer and more branched neurons.
Cells expressing dominant negative ARF6 (ARF6-T27N) exhibited an axonal phenotype similar to that seen upon expression of inactive ARNO (Figure 1, a and b), resulting in a twofold increase in axonal length (Figure 1a), and nearly twofold increase in axonal complexity (Figure 1b). However, cells expressing wild-type ARF6 or constitutively active ARF6 (ARF6-Q67L), had no statistically significant effects upon axonal length and branching compared with untransfected neurons (Figure 1, a and b). This is again evidence that there is a limiting amount of ARNO/ARF6 downstream substrates at this stage of hippocampal development. However, under conditions when ARF6 function is limited, we would expect ARF6 transgenes to restore normal function. We tested whether expression of constitutively active ARF6 would rescue the observed effects of catalytically inactive ARNO on axonogenesis. In fact, coexpression of ARNO-E156K and ARF6-Q67L resulted in a dramatic reduction in the total length and in the average number of collateral branches of the axon compared with cells expressing inactive ARNO alone (Figure 1, a and b). These results indicate that ARNO functions upstream of ARF6 to regulate axonal elongation and branching.
Because ARNO has been shown to act as a GEF for ARF1 as well as ARF6, we attempted to study directly the effects of ARF1 expression upon axonogenesis. Unfortunately, expression of either wild-type or mutant forms of ARF1 resulted in cell death, undoubtedly due to inhibition of the secretory pathway (Lippincott-Schwartz et al., 1998). However, previous work is inconsistent with ARNO's function in axonal growth being mediated by ARF1. Incubation of neurons with brefeldin A, a drug that is known to inhibit ARF1 GEFs, inhibits axonal extension (Jareb and Banker, 1997; Hess et al., 1999), in contrast with our finding that ARNO inhibition increases axonal extension.
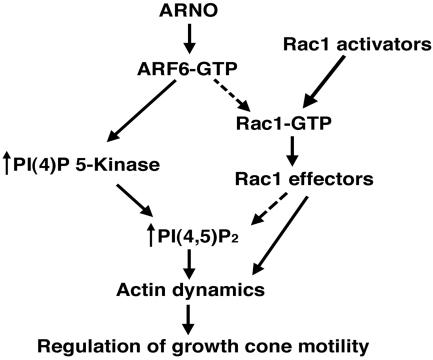
ARF6 has been suggested to reside near the top of a diverging signaling cascade (Figure 2), which ultimately results in reorganization of the actin cytoskeleton. In one pathway, ARF6 activates Rac1, with subsequent rearrangement of the actin cytoskeleton and lamellipodial extension. In the other pathway, ARF6 activates PI(4)P 5-Kinase, resulting in increased production of phophatidylinositol-4,5-biphosphate (PIP2) and subsequent rearrangement of actin through effects upon actin binding proteins. If the effects of ARNO/ARF6 upon axonogenesis are mediated through Rac1, coexpression of Rac1 with inactive ARNO should reverse the enhanced axonal phenotype caused by mutant ARNO/ARF6 alone. Alternatively, if these effects are mediated by downstream activation of PI(4)P 5-Kinase, coexpression of PI(4)P 5-Kinase with inactive ARNO or ARF6 should also reverse the enhanced axonal growth phenotype caused by expression of mutant ARNO/ARF6 alone.
Figure 2.
ARNO/ARF6 signaling pathways that could modulate actin reorganization in the axonal growth cone. ARNO stimulates GTP exchange on ARF6, thereby increasing the amount of active ARF6. In one pathway, activated ARF6 stimulates the lipid-modifying enzyme PI(4)P 5-Kinase, leading to local increases in plasma membrane PIP2 and changes in actin dynamics. Alternatively, activation of Rac1 by upstream Rac1 activators or indirectly by ARF6-GTP results in stimulation of actin polymerization. Both of these pathways could ultimately regulate axonal growth cone dynamics.
Rac1 has been shown to regulate axonal outgrowth (Luo et al., 1994, 1996b; Jin and Strittmatter, 1997; Albertinazzi et al., 1998; Kaufmann et al., 1998; Kuhn et al., 1999), and is activated downstream of ARF6 during Madin-Darby canine kidney cell migration (Santy and Casanova, 2001). Recently, we found that ARF6 can regulate dendritic growth via downstream activation of Rac1 (Hernandez-Deviez et al., 2002). To determine whether Rac1 is a downstream effector of ARF6 during axonogenesis, we first investigated the effects of overexpression of wild-type or dominant-negative mutant Rac1 (Rac1-N17) on axonal length and branching. As shown by others (Luo et al., 1994, 1996b; Jin and Strittmatter, 1997; Albertinazzi et al., 1998; Kaufmann et al., 1998; Kuhn et al., 1999), overexpression of wild-type Rac1 resulted in a threefold increase in axonal length (Figure 1a), but no quantifiable effects upon axonal branching (Figure 1b). Interestingly, overexpression of Rac1-N17 resulted in a 4.5-fold increase in axonal length and a twofold increase in axonal branching (Figure 1, a and b), a phenotype consistent with the results seen with ARNO-E156K and ARF6-T27N.
If the effects of Rac1-N17 reflect an upstream regulation by ARF6, coexpression of Rac1-N17 with activated ARF6 should result in increased branching and axonal length. In fact, coexpression of constitutively active ARF6 with dominant-negative Rac1 resulted in reduction in both axonal length and branching compared with cells expressing dominant-negative Rac1 alone (Figure 1, a and b), suggesting that ARF6 and Rac1 are in parallel pathways. Also, if Rac1 is being activated downstream of ARF6 during axonogenesis, we would predict that coexpression of Rac1 with ARNO-E156K would reduce the enhanced axonal branching seen with ARNO-E156K. However, coexpression of Rac1 and ARNO-E156K did not cause an additive effect upon axonal length or decrease axonal branching (Figure 1, a and b), again suggesting that ARNO/ARF6 effects upon axonal extension and branching are not mediated by ultimate downstream activation of Rac1. These results suggest that regulation of axonal length and branching is through two independent pathways using both Rac1 and other ARF6 downstream effectors (see DISCUSSION) (Exton, 1997; Honda et al., 1999).
Effects of ARF6 upon Axonogenesis Are Mediated by Phosphatidyl-inositol-4-phosphate 5-Kinase α
Because ARF6 is known to activate the lipid-modifying enzyme PI(4)P-5K (Exton, 1997; Honda et al., 1999), we next tested whether this enzyme could regulate axonal extension downstream of ARF6 but independently of Rac1. Recent studies have shown that type I PI(4)P 5-Kinase α modulates neurite outgrowth, implicating this enzyme in neurite remodeling (van Horck et al., 2002; Yamazaki et al., 2002), but the upstream components of the pathway remain unknown. We first investigated the effects of expression of mouse type I PI(4)P 5-K α during axonal development. Consistent with previous findings in neuroblastoma cells (van Horck et al., 2002), hippocampal neurons expressing PI(4)P 5-Kinase α showed no statistically significant effects upon either axonal length and branching (Figure 3, a–c). However, coexpression of PI(4)P 5-Kinase α and ARNO-E156K resulted in a fivefold reduction in the total axonal length and fourfold decrease in the average number of axonal collateral branches compared with cells expressing catalytically inactive ARNO alone (Figure 3, a, b, and d). If these effects are being mediated through ARF6, we expect that coexpression of ARF6T27N and PI(4)P 5-Kinase α would result in shorter axons compared with cells expressing ARF6-T27N alone. In fact, coexpression of these molecules resulted in a threefold reduction in the total axonal length and an 11-fold decrease in the average number of collateral branches, indicating that ARF6-T27N is upstream of PI(4)P 5-Kinase α (Figure 3, a and b). Together, these results suggest that the ARNO/ARF6 pathway can regulate axonal length through downstream activation of PI(4)P 5-Kinase α.
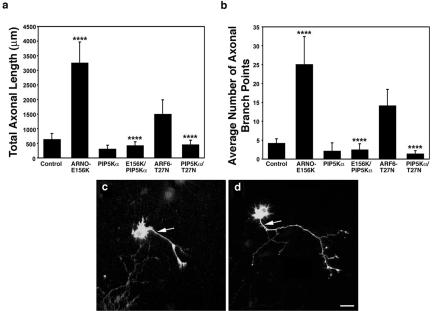
Figure 3.
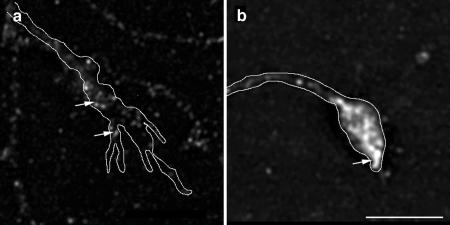
PI(4)P 5-Kinase α is downstream of ARNO/ARF6 during axonal elongation and branching. Morphometric analysis of the effect of expression of PI(4)P 5-kinase α on axonal outgrowth and branching. (a) Effect on total axonal length. Expression of PI(4)P 5-Kinase α alone resulted in no quantifiable effects upon axonal length compared with control, whereas coexpression of PI(4)P 5-Kinase α reversed the effect of expression of ARNO-E156K or ARF6-T27N upon axonal length. (b) Effect on the number of axonal branch points. Expression of PI(4)P 5-Kinase α resulted in no quantifiable effects upon the number of axonal branch points, and coexpression of PI(4)P 5-Kinase α reversed the effect of expression of ARNO-E156K or ARF6-T27N upon axonal branching. Data are presented as the means ± SD (n = 10). p < 0.0001 (****). (c and d) Hippocampal neurons transfected with PI(4)P 5-Kinase α (c) and ARNO-E156K and PI(4)P 5-Kinase α (d) were visualized by confocal microscopy. (c) Cell expressing PI(4)P 5-Kinase α. The axon (arrow) is short with very few collateral branches. (d) Cell expressing ARNO-E156K and PI(4)P 5-Kinase α. The axonal process (arrow) is short with fewer collateral branches compared with cells expressing ARNO-E156K (compare with Figure 1e). Bars, 25 μm (e–g).
ARNO/ARF6 Regulate Neurite Elongation through Changes in the Distribution of Cytoskeletal Proteins
ARNO and ARF6 modulate the reorganization of the cortical actin cytoskeleton in fibroblast cell lines (Radhakrishna et al., 1996; D'Souza-Schorey et al., 1997; Frank et al., 1998b) and changes in the activity of ARF6, through their effects upon Rho family GTPases and PI(4)P 5-Kinase, may modulate the proteins associated with actin dynamics. Several studies have shown the importance of cytoskeletal proteins in controlling actin dynamics, growth cone motility, axonal guidance, and fibroblast motility (Renaudin et al., 1999; Furnish et al., 2001; Bear et al., 2002; Bamburg, 2003). This prompted us to examine the effects of ARF6 inhibition on the distribution of actin-modulating proteins in the growth cone. To study this issue, we chose cells at 5–6 d in vitro (developmental stage 3–4) (Dotti et al., 1988) because they frequently have expanded, flattened growth cones at the tips of their extending processes. At these developmental stages, cells expressing catalytically inactive ARNO exhibited similar effects upon axon length and branching compared with cells expressing this plasmid at later developmental stages (our unpublished data). Because recent studies have shown that members of the Ena/VASP family regulate cell motility, axonal extension, and navigation (Lanier et al., 1999; Wills et al., 1999; Bear et al., 2000, 2002), we first examined the distribution of Mena in neurons expressing wild-type and inactive ARNO. As shown in Figure 4, Mena is depleted from the growth cone leading edge in cells expressing catalytically inactive ARNO. This depletion is not due to competition with expressed ARNO for membrane binding sites, because in cells expressing wild-type ARNO Mena is localized in a punctate distribution at the tips of growth cones identical to the pattern seen in untransfected cells (Figure 4, a–c). Quantification of Mena distribution showed that, in untransfected cells and cells expressing wild-type ARNO, Mena was always at the leading edge of lamellae and filopodia. However, in cells expressing ARNO-E156K, Mena was localized either at the growth cone base or was completely depleted from the growth cone (Table 2). We were not able to determine whether this depletion was due to decreased synthesis, increased degradation, or redistribution of Mena, because low transfection efficiency in neurons makes it impossible to quantify the effects of expression of catalytically inactive ARNO upon Mena protein levels.
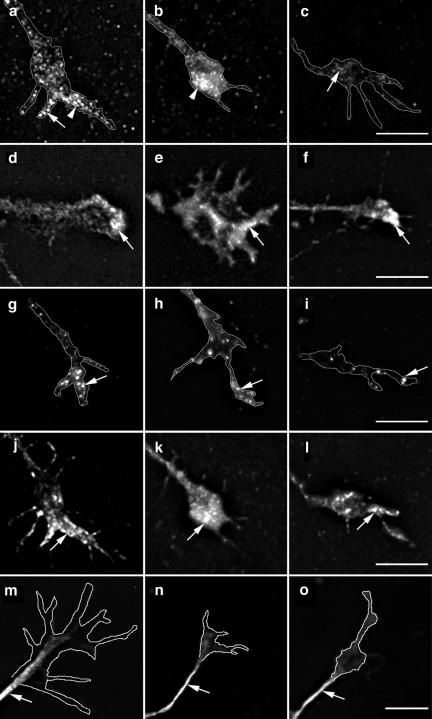
Figure 4.
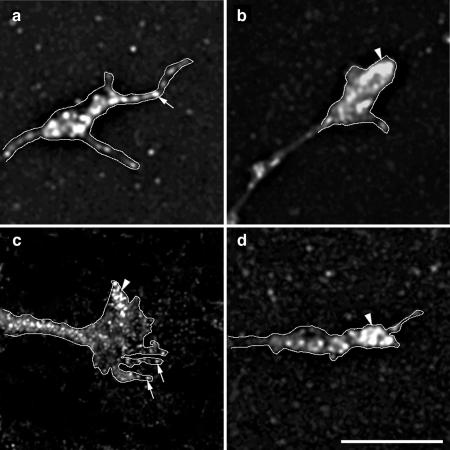
Depletion of Mena from the growth cone plasma membrane in cells expressing inactive ARNO. Hippocampal neurons were transfected with _myc_-tagged ARNO wild-type and ARNO-E156K and labeled with anti-Mena antibody (a–c), anti-vinculin antibody (d–f), anti-ERM antibody (g–i), rhodamine-conjugated phalloidin (j–l), or anti-β-tubulin antibody (m–o), and visualized by deconvolution microscopy. The left column shows growth cones of untransfected cells, the middle column shows growth cones of cells expressing ARNO wild-type, and the right column shows growth cones of cells expressing ARNO-E156K. All images were taken and processed using identical parameters. (a and b) In untransfected cells or cells expressing ARNO wild type, Mena is localized at the growth cone filopodia (arrow) and lamellipodia (arrowhead). (c) In the growth cone of a cell expressing ARNO-E156K, Mena is depleted from the growth cone (arrow). (d–f) Vinculin is localized at the distal region of the growth cone in all cells (arrow). (g–i) ERM proteins are localized at the growth cone filopodia (arrow). (j–l) F-actin is localized at the filopodial/lamellipodial extensions of growth cones (arrow). (m–o) β-Tubulin labeling is present in axonal shafts but not in the central domain of the growth cone. Growth cones have been outlined to facilitate the visualization of the distribution of labeling. Bars, 5 μm.
Table 2.
Quantification of Mena distribution in the axonal growth cone
The redistribution of Mena could be caused by changes in the number of actin barbed ends available for Mena binding, or secondarily by influencing the activity of other cytoskeletal proteins. Because cytoskeletal proteins such as vinculin and Ezrin-Radixin-Moesin (ERM) are concentrated at the growth cone and bind to PIP2 (Varnum-Finney and Reichardt, 1994; Renaudin et al., 1999), we next tested their distribution in cells expressing wild-type or inactive ARNO. However, there was no change in the distribution of vinculin (Figure 4, d–f) or ERM (Figure 4, g–i) in the growth cone filopodia of cells expressing either wild-type or catalytically inactive ARNO. We also found the distribution of filamentous actin and microtubules to be unaffected by wild-type ARNO and ARNO-E156K expression (Figure 4, j–o).
Gelsolin is an actin filament severing and capping protein that produces numerous short and capped actin filaments (Cunningham et al., 1991; Westberg et al., 1999; Furnish et al., 2001) and codistribution of ARF6 and gelsolin has been demonstrated in fibroblasts (Radhakrishna et al., 1996). Gelsolin and profilin are actin-binding proteins whose activities are negatively regulated by PIP2. Because ARF6, through activation of PI(4)P 5-Kinase α, would increase PIP2 levels, we next tested the distribution of these molecules in cells expressing ARNO-E156K. However, in cells expressing inactive ARNO, we did not detect any changes in the distribution of either gelsolin (Figure 5, a and b) or the PIP2 binding protein profilin (our unpublished data) in the growth cone.
Figure 5.
Gelsolin distribution in the growth cone of cells expressing ARNO-E156K. Hippocampal neurons were transfected with _myc_-tagged ARNO-E156K labeled with anti-gelsolin antibody and visualized by deconvolution microscopy. All images were taken and processed using identical parameters. (a) Growth cone of untransfected cell. (b) Growth cone of cell expressing ARNO-E156K. In untransfected cells or cells expressing ARNO-E156K, gelsolin is localized at the growth cone filopodia (arrows) and lamellipodia (arrowheads). Growth cones have been outlined to facilitate the visualization of the distribution of labeling. Bars, 5 μm.
Because we have found that dominant negative ARF6 also caused an increase in axonal growth and branching, we also examined the distribution of Mena in the growth cones of cells expressing ARF6-T27N. As shown in Figure 6, Mena is depleted from the growth cone of cells expressing ARF6-T27N (Figure 6a), but not from the growth cone of cells expressing Rac-N17, which also promotes axonal growth (Figure 6b and Table 2). This result underscores the idea that multiple pathways are active in the regulation of axonal extension.
Figure 6.
ARF6-T27N but not Rac1-N17 depletes Mena from the growth cone plasma membrane. Hippocampal neurons were transfected with either HA-tagged ARF6-T27N or FLAG-tagged Rac1-N17 labeled with anti-Mena antibody and visualized by deconvolution microscopy. All images were taken and processed using identical parameters. (a) In the growth cone of a cell expressing ARF6-T27N, Mena is depleted from the growth cone (arrow). (b) Growth cone of cell expressing Rac1-N17, Mena is localized at the leading lamellae of the growth cone (arrow). Growth cones have been outlined to facilitate the visualization of the distribution of labeling. Bars, 5 μm.
The specificity of the depletion of Mena from the growth cone by ARNO-E156K is illustrated by the fact that, under conditions where we have demonstrated normal axonal growth, as during expression of ARF6-Q67L and PI(4)P 5-Kinase, Mena is found concentrated in the growth cone (Figure 7, a and b). In addition, reversal of increased axonal growth by coexpression of ARNO-E156K and either ARF6-Q67L or PI(4)P 5-Kinase was correlated with the return of a normal distribution of Mena to the growth cone leading edge (Figure 7, c and d). These results suggest that changes in actin dynamics induced by ARF6 could modulate Mena localization, resulting in regulation of axonal extension and pathfinding.
Figure 7.
Coexpression of either constitutively active ARF6 or PI(4)P 5-Kinase α restores Mena distribution to the plasma membrane in ARNO-E156K growth cones. Hippocampal neurons were transfected with HA-tagged ARF6-Q67L, PI(4)P 5-Kinase α, and myc-tagged ARNO-E156K, labeled with anti-Mena antibody, and visualized by deconvolution microscopy. (a-b) In cells expressing either ARF6-Q67L or PI(4)P 5-Kinase α, Mena is localized along filopodial (arrows) and lamellipodial (arrowhead) extensions of the growth cone. (c and d) Coexpression of ARNO-E156K and either ARF6-Q67L (c) or PI(4)P 5-Kinase α (d), returns Mena to growth cone filopodia (arrows) and lamellipodia (arrowhead) (compare with Figure 4c). All images were taken and processed using identical parameters. Growth cones have been outlined to facilitate the visualization of the distribution of labeling. Bars, 5 μm.
DISCUSSION
The results reported here support a role for ARF6 and its downstream lipid-modifying effectors in the controlled extension and branching of axonal processes. These effects of ARNO and ARF6 upon axonal growth are distinct from our previous findings in dendrites (Hernandez-Deviez et al., 2002), in that the enhancement of axonal length and branching caused by expression of mutant ARF6 and ARNO are mediated by the PI(4)P 5-Kinase α pathway rather than through the small GTPase Rac1. These effects may be through changes in the dynamics of the actin cytoskeleton, because expression of inactive ARNO or ARF6 resulted in a specific depletion of the actin-binding protein Mena from the growth cone. These data suggest that, during development, ARNO and ARF6 play an important role in the regulation of axonal elongation and sprouting through effects upon membrane structure and cytoskeleton dynamics.
During axonal development three distinct events occur: elongation, pathfinding, and branching. Rho GTPases have been implicated in the regulation of many aspects of axonal development (Luo, 2000) because progressive deletion of the three Drosophila Rac GTPases result in differential defects in axon outgrowth, guidance, and branching (Ng et al., 2002). We found that catalytically inactive ARNO and ARF6 caused both an increase in axonal length and in axonal collateral branches. Although ARF6 and Rac1 seem to act in concert to modulate dendritic development in hippocampal neurons (Hernandez-Deviez et al., 2002), our results suggest that Rac1 is not downstream of ARF6 in axonal extension and branching and must reflect an additional regulatory pathway. This contrasts with recent reports showing that neurite extension in chick retinal cells and Aplysia motor neurons is activated by ARF6 (Huh et al., 2003) and ARF6/PIX/p95-APP1 (Albertinazzi et al., 2003). Whether these discrepancies reflect differences in cell type or in the regulation of axonal elongation remains unclear.
In hippocampal neurons, it seems that ARF6 modulates axonal extension and branching through effects upon membrane structure. ARF6 stimulates PI(4)P 5-Kinase (Honda et al., 1999) and PIP2 has been shown to be enriched at sites of ARF6 activation (Brown et al., 2001). It has recently been shown that dominant negative type I PI(4)P 5-Kinase α promotes neurite elongation in neuroblastoma cells, suggesting that PI(4,5)P2 negatively regulates neuritogenesis (van Horck et al., 2002; Yamazaki et al., 2002). We found that coexpression of type I PI(4)P 5-Kinase α and catalytically inactive ARNO or dominant negative ARF6 dramatically reversed the increased axonal length and branching of cells expressing inactive ARNO alone, indicating that PI(4)P 5-Kinase α is downstream of ARF6 in regulating axonal extension.
In neurons expressing catalytically inactive ARNO, Mena, but not other cytoskeletal-associated proteins, was depleted from the growth cone and expression of constitutively active ARF6 or PI(4)P 5-Kinase resulted in reassociation of Mena with the growth cone filopodia and lamellipodia. Although Ena/VASP proteins mediate actin polymerization (Laurent et al., 1999) and have been shown to be localized to the leading edge of migrating cells and at growth cone tips (Gertler et al., 1996), recent studies have shown that depletion of Mena from the plasma membrane promotes fibroblast motility (Bear et al., 2000). This apparent paradox is incompletely understood, but recent studies suggest that Mena may act to control actin filament length by acting as an “anticapping” molecule at the barbed ends, resulting in longer, more flexible filaments (Bear et al., 2002; Krause et al., 2002). Because extension of lamellipodia seems to require short filaments in highly branched networks (Bear et al., 2002), antagonizing Mena should result in increased extension of the leading edge. In fact, incubation of neurons with low concentrations of the barbed-end capping drug cytochalasin B or D causes depletion of Mena from the growth cone and increased neurite growth (Marsh and Letourneau, 1984; Bear et al., 2002). Because ARF6 activation of PI(4)P 5-Kinase increases PIP2 levels, this would inhibit gelsolin, inducing uncapping of actin filaments, resulting in free barbed ends and subsequently Mena localization at the plasma membrane. The ultimate consequence of this increased PIP2 would be to restrict growth cone motility and axonal extension. Further ultrastructural analysis of the actin filaments awaits additional investigation.
ARNO and ARF6 play an essential role in both actin dynamics and membrane traffic in various cell types (Radhakrishna et al., 1996; D'Souza-Schorey et al., 1997; Radhakrishna and Donaldson, 1997; Frank et al., 1998b; Radhakrishna et al., 1999; Boshans et al., 2000; Palacios et al., 2001) and ARF6, through phosphatidylinositol kinases, has been implicated in the regulation of synaptic vesicle recycling (Krauss et al., 2003) and regulated exocytosis (Aikawa and Martin, 2003). Here, we have presented data that suggest that ARF6 activation regulates axonal elongation and branching through modulation of the growth cone cytoskeleton. However, we have also found that expression of inactive ARNO causes a redistribution of specialized endosomes to the axonal but not dendritic plasma membrane (Hernández-Deviez and Wilson, unpublished data), suggesting that ARNO regulates membrane cycling in neurons and may also control axonal elongation and branching by increasing membrane available for growth.
Axon regeneration after injury is blocked by various growth-inhibitory proteins, and efforts have been made to develop strategies to promote axonal regrowth in injured adult neurons. Small GTPases are common targets of many of these inhibitors (Fitch and Silver, 1997; Fournier and Strittmatter, 2001). Recently, it has been found that injured axons regrow on inhibitory substrates when Rho GTPase is inactivated (Lehmann et al., 1999). In addition, pharmacological inhibition of its major downstream effector, Rhokinase, promotes axonal outgrowth (Borisoff et al., 2003; Fournier et al., 2003). These findings suggest that small GTPases as well as their effectors constitute potential targets to disrupt inhibitory signaling. Our study suggests that manipulation of the active state of ARNO, ARF6, and PI(4)P 5-Kinase are potential targets to promote axonal regeneration after injury.
Acknowledgments
We thank Drs. J. Settleman for providing the Rac plasmids, F. Solomon for anti-ERM antibody, and M. Durán for assistance with the statistical analysis. We also thank Drs. M. Ramaswami, C. Gregorio, R. Levine, and A. McElhinny for critical reading of the manuscript. This work was supported by National Institutes of Health grants DK43329 (to J.M.W.) and AI32991 (to J.E.C). D.H.D. was supported by Consejo Nacional de Investigaciones Científicas y Tecnológicas, Venezuela and University of Los Andes, Mérida-Venezuela.
References
- Aikawa, Y., and Martin, T.F. (2003). ARF6 regulates a plasma membrane pool of phosphatidylinositol(4,5)bisphosphate required for regulated exocytosis. J. Cell Biol. 162, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertinazzi, C., Gilardelli, D., Paris, S., Longhi, R., and de Curtis, I. (1998). Overexpression of a neural-specific rho family GTPase, cRac1B, selectively induces enhanced neuritogenesis and neurite branching in primary neurons. J. Cell Biol. 142, 815–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertinazzi, C., Za, L., Paris, S., and De Curtis, I. (2003). ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol. Biol. Cell 14, 1295–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschuler, Y., Liu, S., Katz, L., Tang, K., Hardy, S., Brodsky, F., Apodaca, G., and Mostov, K. (1999). ADP-ribosylation factor 6 and endocytosis at the apical surface of Madin-Darby canine kidney cells. J. Cell Biol. 147, 7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg, J.R. (2003). Introduction to cytoskeletal dynamics and pathfinding of neuronal growth cones. J. Histochem. Cytochem. 51, 407–409. [DOI] [PubMed] [Google Scholar]
- Banker, G., and Goslin, K. (1991). Culturing Nerve Cells. Cambridge, MA: The MIT Press.
- Bear, J.E., Loureiro, J.J., Libova, I., Fassler, R., Wehland, J., and Gertler, F.B. (2000). Negative regulation of fibroblast motility by Ena/VASP proteins. Cell 101, 717–728. [DOI] [PubMed] [Google Scholar]
- Bear, J.E., et al. (2002). Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell 109, 509–521. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour, S., Robineau, S., Chardin, P., Paris, S., Chabre, M., Cherfils, J., and Antonny, B. (1998). A glutamic finger in the guanine nucleotide exchange factor ARNO displaces Mg2+ and the beta-phosphate to destabilize GDP on ARF1. EMBO J. 17, 3651–3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisoff, J.F., Chan, C.C., Hiebert, G.W., Oschipok, L., Robertson, G.S., Zamboni, R., Steeves, J.D., and Tetzlaff, W. (2003). Suppression of Rho-kinase activity promotes axonal growth on inhibitory CNS substrates. Mol. Cell Neurosci. 22, 405–416. [DOI] [PubMed] [Google Scholar]
- Boshans, R.L., Szanto, S., van Aelst, L., and D'Souza-Schorey, C. (2000). ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, F.D., Rozelle, A.L., Yin, H.L., Balla, T., and Donaldson, J.G. (2001). Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 154, 1007–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caumont, A.S., Vitale, N., Gensse, M., Galas, M.C., Casanova, J.E., and Bader, M.F. (2000). Identification of a plasma membrane-associated guanine nucleotide exchange factor for ARF6 in chromaffin cells. Possible role in the regulated exocytotic pathway. J. Biol. Chem. 275, 15637–15644. [DOI] [PubMed] [Google Scholar]
- Chardin, P., Paris, S., Antonny, B., Robineau, S., Beraud-Dufour, S., Jackson, C.L., and Chabre, M. (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature 384, 481–484. [DOI] [PubMed] [Google Scholar]
- Craig, A.M., and Banker, G. (1994). Neuronal polarity. Annu. Rev. Neurosci. 17, 267–310. [DOI] [PubMed] [Google Scholar]
- Craig, A.M., Wyborski, R.J., and Banker, G. (1995). Preferential addition of newly synthesized membrane protein at axonal growth cones. Nature 375, 592–594. [DOI] [PubMed] [Google Scholar]
- Cunningham, C.C., Stossel, T.P., and Kwiatkowski, D.J. (1991). Enhanced motility in NIH 3T3 fibroblasts that overexpress gelsolin. Science 251, 1233–1236. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Boshans, R.L., McDonough, M., Stahl, P.D., and Van Aelst, L. (1997). A role for POR1, a Rac1-interacting protein, in ARF6-mediated cytoskeletal rearrangements. EMBO J. 16, 5445–5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti, C.G., Sullivan, J.M., and Banker, G. (1988). The establishment of polarity by hippocampal neurons in culture. J. Neurosci. 8, 1454–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exton, J.H. (1997). Regulation of phosphoinositide phospholipases by G-proteins. Adv. Exp. Med. Biol. 400A, 3–8. [DOI] [PubMed] [Google Scholar]
- Fitch, M.T., and Silver, J. (1997). Glial cell extracellular matrix: boundaries for axon growth in development and regeneration. Cell Tissue Res. 290, 379–384. [DOI] [PubMed] [Google Scholar]
- Fournier, A.E., and Strittmatter, S.M. (2001). Repulsive factors and axon regeneration in the CNS. Curr. Opin. Neurobiol. 11, 89–94. [DOI] [PubMed] [Google Scholar]
- Fournier, A.E., Takizawa, B.T., and Strittmatter, S.M. (2003). Rho kinase inhibition enhances axonal regeneration in the injured CNS. J. Neurosci. 23, 1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, S., Upender, S., Hansen, S.H., and Casanova, J.E. (1998a). ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J. Biol. Chem. 273, 23–27. [DOI] [PubMed] [Google Scholar]
- Frank, S.R., Hatfield, J.C., and Casanova, J.E. (1998b). Remodeling of the actin cytoskeleton is coordinately regulated by protein kinase C and the ADP-ribosylation factor nucleotide exchange factor ARNO. Mol. Biol. Cell 9, 3133–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnish, E.J., Zhou, W., Cunningham, C.C., Kas, J.A., and Schmidt, C.E. (2001). Gelsolin overexpression enhances neurite outgrowth in PC12 cells. FEBS Lett. 508, 282–286. [DOI] [PubMed] [Google Scholar]
- Gallo, G., and Letourneau, P.C. (2000). Neurotrophins and the dynamic regulation of the neuronal cytoskeleton. J. Neurobiol. 44, 159–173. [DOI] [PubMed] [Google Scholar]
- Gertler, F.B., Niebuhr, K., Reinhard, M., Wehland, J., and Soriano, P. (1996). Mena, a relative of VASP and Drosophila Enabled, is implicated in the control of microfilament dynamics. Cell 87, 227–239. [DOI] [PubMed] [Google Scholar]
- Hall, A. (1998). Rho GTPases and the actin cytoskeleton. Science 279, 509–514. [DOI] [PubMed] [Google Scholar]
- Hernandez-Deviez, D.J., Casanova, J.E., and Wilson, J.M. (2002). Regulation of dendritic development by the ARF exchange factor ARNO. Nat. Neurosci. 5, 623–624. [DOI] [PubMed] [Google Scholar]
- Hess, D.T., Smith, D.S., Patterson, S.I., Kahn, R.A., Skene, J.H., and Norden, J.J. (1999). Rapid arrest of axon elongation by brefeldin A: a role for the small GTP-binding protein ARF in neuronal growth cones. J. Neurobiol. 38, 105–115. [DOI] [PubMed] [Google Scholar]
- Honda, A., et al. (1999). Phosphatidylinositol 4-phosphate 5-kinase alpha is a downstream effector of the small G protein ARF6 in membrane ruffle formation. Cell 99, 521–532. [DOI] [PubMed] [Google Scholar]
- Huh, M., Han, J.H., Lim, C.S., Lee, S.H., Kim, S., Kim, E., and Kaang, B.K. (2003). Regulation of neuritogenesis and synaptic transmission by msec7–1, a guanine nucleotide exchange factor, in cultured Aplysia neurons. J. Neurochem. 85, 282–285. [DOI] [PubMed] [Google Scholar]
- Jareb, M., and Banker, G. (1997). Inhibition of axonal growth by brefeldin A in hippocampal neurons in culture. J. Neurosci. 17, 8955–8963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Z., and Strittmatter, S.M. (1997). Rac1 mediates collapsin-1-induced growth cone collapse. J. Neurosci. 17, 6256–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiguchi, H., and Lemmon, V. (2000). Recycling of the cell adhesion molecule L1 in axonal growth cones. J. Neurosci. 20, 3676–3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann, N., Wills, Z.P., and Van Vactor, D. (1998). Drosophila Rac1 controls motor axon guidance. Development 125, 453–461. [DOI] [PubMed] [Google Scholar]
- Krause, M., Bear, J.E., Loureiro, J.J., and Gertler, F.B. (2002). The Ena/VASP enigma. J. Cell Sci. 115, 4721–4726. [DOI] [PubMed] [Google Scholar]
- Krauss, M., Kinuta, M., Wenk, M.R., De Camilli, P., Takei, K., and Haucke, V. (2003). ARF6 stimulates clathrin/AP-2 recruitment to synaptic membranes by activating phosphatidylinositol phosphate kinase type Igamma. J. Cell Biol. 162, 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, T.B., Brown, M.D., Wilcox, C.L., Raper, J.A., and Bamburg, J.R. (1999). Myelin and collapsin-1 induce motor neuron growth cone collapse through different pathways: inhibition of collapse by opposing mutants of rac1. J. Neurosci. 19, 1965–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier, L.M., Gates, M.A., Witke, W., Menzies, A.S., Wehman, A.M., Macklis, J.D., Kwiatkowski, D., Soriano, P., and Gertler, F.B. (1999). Mena is required for neurulation and commissure formation. Neuron 22, 313–325. [DOI] [PubMed] [Google Scholar]
- Laurent, V., Loisel, T.P., Harbeck, B., Wehman, A., Grobe, L., Jockusch, B.M., Wehland, J., Gertler, F.B., and Carlier, M.F. (1999). Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J. Cell Biol. 144, 1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann, M., Fournier, A., Selles-Navarro, I., Dergham, P., Sebok, A., Leclerc, N., Tigyi, G., and McKerracher, L. (1999). Inactivation of Rho signaling pathway promotes CNS axon regeneration. J. Neurosci. 19, 7537–7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Van Aelst, L., and Cline, H.T. (2000). Rho GTPases regulate distinct aspects of dendritic arbor growth in Xenopus central neurons in vivo. Nat. Neurosci. 3, 217–225. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Cole, N., and Presley, J. (1998). Unravelling Golgi membrane traffic with green fluorescent protein chimeras. Trends Cell Biol. 8, 16–20. [DOI] [PubMed] [Google Scholar]
- Luo, L. (2000). Rho GTPases in neuronal morphogenesis. Nat. Rev. Neurosci. 1, 173–180. [DOI] [PubMed] [Google Scholar]
- Luo, L. (2002). Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol. 18, 601–635. [DOI] [PubMed] [Google Scholar]
- Luo, L., Hensch, T.K., Ackerman, L., Barbel, S., Jan, L.Y., and Jan, Y.N. (1996a). Differential effects of the Rac GTPase on Purkinje cell axons and dendritic trunks and spines. Nature 379, 837–840. [DOI] [PubMed] [Google Scholar]
- Luo, L., Jan, L., and Jan, Y.N. (1996b). Small GTPases in axon outgrowth. Perspect. Dev. Neurobiol. 4, 199–204. [PubMed] [Google Scholar]
- Luo, L., Liao, Y.J., Jan, L.Y., and Jan, Y.N. (1994). Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes Dev. 8, 1787–1802. [DOI] [PubMed] [Google Scholar]
- Marsh, L., and Letourneau, P.C. (1984). Growth of neurites without filopodial or lamellipodial activity in the presence of cytochalasin B. J. Cell Biol. 99, 2041–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monier, S., Chardin, P., Robineau, S., and Goud, B. (1998). Overexpression of the ARF1 exchange factor ARNO inhibits the early secretory pathway and causes the disassembly of the Golgi complex. J. Cell Sci. 111, 3427–3436. [DOI] [PubMed] [Google Scholar]
- Ng, J., Nardine, T., Harms, M., Tzu, J., Goldstein, A., Sun, Y., Dietzl, G., Dickson, B.J., and Luo, L. (2002). Rac GTPases control axon growth, guidance and branching. Nature 416, 442–447. [DOI] [PubMed] [Google Scholar]
- Palacios, F., Price, L., Schweitzer, J., Collard, J.G., and D'Souza-Schorey, C. (2001). An essential role for ARF6-regulated membrane traffic in adherens junction turnover and epithelial cell migration. EMBO J. 20, 4973–4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna, H., Al-Awar, O., Khachikian, Z., and Donaldson, J.G. (1999). ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112, 855–866. [DOI] [PubMed] [Google Scholar]
- Radhakrishna, H., and Donaldson, J.G. (1997). ADP-ribosylation factor 6 regulates a novel plasma membrane recycling pathway. J. Cell Biol. 139, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishna, H., Klausner, R.D., and Donaldson, J.G. (1996). Aluminum fluoride stimulates surface protrusions in cells overexpressing the ARF6 GTPase. J. Cell Biol. 134, 935–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin, A., Lehmann, M., Girault, J., and McKerracher, L. (1999). Organization of point contacts in neuronal growth cones. J. Neurosci. Res. 55, 458–471. [DOI] [PubMed] [Google Scholar]
- Santy, L.C., and Casanova, J.E. (2001). Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E.K., and Luo, L. (2001). How do dendrites take their shape? Nat. Neurosci. 4, 359–365. [DOI] [PubMed] [Google Scholar]
- Song, H.J., and Poo, M.M. (1999). Signal transduction underlying growth cone guidance by diffusible factors. Curr. Opin. Neurobiol. 9, 355–363. [DOI] [PubMed] [Google Scholar]
- Suzuki, I., Owada, Y., Suzuki, R., Yoshimoto, T., and Kondo, H. (2001). Localization of mRNAs for six ARFs (ADP-ribosylation factors) in the brain of developing and adult rats and changes in the expression in the hypoglossal nucleus after its axotomy. Brain Res. Mol. Brain Res. 88, 124–134. [DOI] [PubMed] [Google Scholar]
- Suzuki, I., Owada, Y., Suzuki, R., Yoshimoto, T., and Kondo, H. (2002). Localization of mRNAs for subfamily of guanine nucleotide-exchange proteins (GEP) for ARFs (ADP-ribosylation factors) in the brain of developing and mature rats under normal and postaxotomy conditions. Brain Res. Mol. Brain Res. 98, 41–50. [DOI] [PubMed] [Google Scholar]
- Tessier-Lavigne, M., and Goodman, C.S. (1996). The molecular biology of axon guidance. Science 274, 1123–1133. [DOI] [PubMed] [Google Scholar]
- van Horck, F.P., Lavazais, E., Eickholt, B.J., Moolenaar, W.H., and Divecha, N. (2002). Essential role of type I(alpha) phosphatidylinositol 4-phosphate 5-kinase in neurite remodeling. Curr. Biol. 12, 241–245. [DOI] [PubMed] [Google Scholar]
- Varnum-Finney, B., and Reichardt, L.F. (1994). Vinculin-deficient PC12 cell lines extend unstable lamellipodia and filopodia and have a reduced rate of neurite outgrowth. J. Cell Biol. 127, 1071–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westberg, J.A., Zhang, K.Z., and Andersson, L.C. (1999). Regulation of neural differentiation by normal and mutant (G654A, amyloidogenic) gelsolin. FASEB J. 13, 1621–1626. [DOI] [PubMed] [Google Scholar]
- Whitford, K.L., Dijkhuizen, P., Polleux, F., and Ghosh, A. (2002). Molecular control of cortical dendrite development. Annu. Rev. Neurosci. 25, 127–149. [DOI] [PubMed] [Google Scholar]
- Wills, Z., Bateman, J., Korey, C.A., Comer, A., and Van Vactor, D. (1999). The tyrosine kinase Abl and its substrate enabled collaborate with the receptor phosphatase Dlar to control motor axon guidance. Neuron 22, 301–312. [DOI] [PubMed] [Google Scholar]
- Yamazaki, M., Miyazaki, H., Watanabe, H., Sasaki, T., Maehama, T., Frohman, M.A., and Kanaho, Y. (2002). Phosphatidylinositol 4-phosphate 5-kinase is essential for ROCK-mediated neurite remodeling. J. Biol. Chem. 277, 17226–17230. [DOI] [PubMed] [Google Scholar]