DYRK1A protein kinase promotes quiescence and senescence through DREAM complex assembly (original) (raw)
Abstract
In the absence of growth signals, cells exit the cell cycle and enter into G0 or quiescence. Alternatively, cells enter senescence in response to inappropriate growth signals such as oncogene expression. The molecular mechanisms required for cell cycle exit into quiescence or senescence are poorly understood. The DREAM (DP, RB [retinoblastoma], E2F, and MuvB) complex represses cell cycle-dependent genes during quiescence. DREAM contains p130, E2F4, DP1, and a stable core complex of five MuvB-like proteins: LIN9, LIN37, LIN52, LIN54, and RBBP4. In mammalian cells, the MuvB core dissociates from p130 upon entry into the cell cycle and binds to BMYB during S phase to activate the transcription of genes expressed late in the cell cycle. We used mass spectroscopic analysis to identify phosphorylation sites that regulate the switch of the MuvB core from BMYB to DREAM. Here we report that DYRK1A can specifically phosphorylate LIN52 on serine residue 28, and that this phosphorylation is required for DREAM assembly. Inhibiting DYRK1A activity or point mutation of LIN52 disrupts DREAM assembly and reduces the ability of cells to enter quiescence or undergo Ras-induced senescence. These data reveal an important role for DYRK1A in the regulation of DREAM activity and entry into quiescence.
Keywords: retinoblastoma-like protein p130, growth arrest, cell cycle, Down syndrome, phosphorylation, cellular senescence
The ability of cells to exit from the cell cycle and enter into the G0 or quiescence state is important for cell differentiation, tissue development, and prevention of tumorigenesis. Inactivation of proteins that control the ability of cells to enter the G0 state results in increased proliferation and tumor formation as well as defects in differentiation (for review, see Malumbres and Barbacid 2001; Vidwans and Su 2001; Massague 2004; Koreth and van den Heuvel 2005; Miller et al. 2007). The E2F family of transcription factors includes both activators and repressors that control the expression of genes required for cell cycle progression (Blais and Dynlacht 2004; Dimova and Dyson 2005; McClellan and Slack 2007; van den Heuvel and Dyson 2008). Binding of repressor E2F4 to the promoters of cell cycle-dependent genes has been demonstrated by chromatin immunoprecipitation (ChIP) in many cell-based and animal models of quiescence (Takahashi et al. 2000; Rayman et al. 2002; Cam et al. 2004; Conboy et al. 2007). The activity of E2F transcription factors is regulated by the retinoblastoma (RB) family. In mammalian cells, the RB family proteins—including pRB (RB1), p107 (RBL1), and p130 (RBL2)—bind to and inhibit several forms of E2F (for review, see Cobrinik 2005). Binding and inhibition of E2Fs by RB family members are relieved by CDK (cyclin-dependent kinase) protein kinases that phosphorylate all RB proteins in a cell cycle-dependent manner (Cobrinik 2005; Malumbres and Barbacid 2009).
Mouse genetic studies revealed that RB family proteins function redundantly to control entry into the G0 state and specifically in development and tumor suppression (for review, see Cobrinik 2005; Dannenberg and te Riele 2006). Despite the apparent redundancy in cell cycle control, RB family members differ in their cell cycle-dependent expression and preferential binding of specific E2Fs. Notably, p130 accumulates in cells entering quiescence in response to various growth arrest signals such as serum deprivation, confluency, or increased p16INK4a expression (Smith et al. 1996; Cam et al. 2004). This accumulation is in part mediated by phosphorylation of p130 by GSK3 (glycogen synthase kinase 3), which promotes its stability (Litovchick et al. 2004). High levels of expression in G0 and a preferential interaction with E2F4/5 make p130 a predominant RB family member bound to a repressor E2F4 in quiescent cells (Cam et al. 2004).
Mass spectroscopy proteomic analysis in human cell lines revealed that p130 interacts with E2F4, DP1, RBBP4, LIN9, LIN37, LIN52, and LIN54 in G0/G1 but not in S phase (Litovchick et al. 2007; Schmit et al. 2007). Drosophila orthologs of LIN9, LIN37, and LIN54 were first identified as Myb-interacting proteins (MIPs) (Beall et al. 2002), and later were shown to be a part of nearly identical RBF/E2F2/dMyb complexes independently purified by two groups (Korenjak et al. 2004; Lewis et al. 2004). These complexes were named dREAM (RBF, E2F2 and MIPs) or MMB (Myb–MuvB) because all subunits of these complexes except for Myb have also been identified in Caenorhabditis elegans and belong to the SynMuv B class of genes (Harrison et al. 2006; Fay and Yochem 2007). Further proteomic analysis revealed that human RBBP4, LIN9, LIN37, LIN52, and LIN54 form a stable complex (referred to as the MuvB core) that binds to BMYB in S phase (Litovchick et al. 2007; Schmit et al. 2007). Since no interaction was detected between BMYB and p130/E2F4 in human cells, these studies show that the MuvB core alternatively binds to p130 in G0/G1 and to BMYB in S phase. These respective complexes are referred to as the DREAM complex (DP, RB-like, E2F4, and MuvB) and the MMB complex (MYB–MuvB). The MuvB core can bind to p107, especially in cells depleted of p130 with RNAi (Litovchick et al. 2007; Pilkinton et al. 2007a; Schmit et al. 2007). However, no interaction between the MuvB core and pRB was detected by mass spectroscopic analysis of LIN9-, LIN37-, and LIN54-interacting proteins (Litovchick et al. 2007).
Analysis of the target genes of the RB/E2F complexes in flies and humans revealed both overlapping and unique functions. Human DREAM complex binds to the promoters of >800 cell cycle-regulated genes during G0 and contributes to their repression, while the MMB complex is required for expression of a subset of these genes (Osterloh et al. 2007; Pilkinton et al. 2007b). Interestingly, the fly dREAM/MMB complex displays both transcriptional repressor and activator functions in the distinct classes of targets, including the developmentally and cell cycle-regulated genes (Georlette et al. 2007). ChIP and microarray analysis (ChIP–chip) of the fly dREAM/MMB target genes showed that both Myb and E2F components were present at the majority of targeted promoters, consistent with their presence in the same protein complex. However, the gene expression changes observed in the cells treated with E2F2- or dMyb-specific RNAi revealed subsets of predominantly E2F- or Myb-regulated genes. Interestingly, these genes had a higher enrichment of either E2F or Myb consensus binding sites in their promoters, correlated with a relatively stronger binding of the corresponding factors (Georlette et al. 2007). Therefore, it appears that, even within the context of a single protein complex, E2F2 and dMyb are responsible for the binding and regulation of the specific classes of the target genes.
These studies have revealed that, although the overall organization of the multisubunit RB/E2F repressor complexes is highly conserved in evolution, there are important differences (for review, see van den Heuvel and Dyson 2008). In mammalian cells, the switch of the MuvB core between DREAM and MMB could reflect a specialized function of these complexes in the precise timing of the cell cycle-regulated gene expression. The mechanism that triggers binding of the MuvB core to p130/E2F4/DP1, resulting in the DREAM complex assembly, could be critical for entry into quiescence in response to various growth arrest signals. To identify this mechanism, we used proteomic analysis to determine whether any of the shared subunits were differentially phosphorylated in the context of the DREAM or MMB complexes.
Results
DREAM is phosphorylated in vivo
The MuvB core—consisting of LIN9, LIN37, LIN52, LIN54, and RBBP4—binds to p130/E2F4/DP1 to form the DREAM complex in G0/G1 and to BMYB to form MMB during S phase. To determine whether there were any specific differences in the composition of the MuvB core when bound to p130 or BMYB, we performed 43 large-scale immunoprecipitations for endogenous and HA-tagged p130 and endogenous and V5-tagged LIN9, LIN37, and LIN54, as reported earlier (Litovchick et al. 2007), as well as endogenous and V5-tagged LIN52 and endogenous BMYB (Supplemental Table S1; this study). Each sample was analyzed independently by multidimensional protein identification technology (MudPIT) proteomic analysis (Swanson et al. 2009). We observed that each of the proteins included in this analysis coprecipitated all components of the MuvB core—including LIN9, LIN37, LIN52, LIN54, and RBBP4—while there was never any detectable interaction between BMYB and p130/E2F4/DP1 (Supplemental Fig. S1A,B). The RB-like p107 was detected in a subset of LIN37, LIN52, and LIN54 immunoprecipitates but not in BMYB samples prepared from S-phase-synchronized cells (Supplemental Table S2). In agreement with our earlier analysis, pRB was not detected in any of the samples (Litovchick et al. 2007). Therefore, we could not confirm previous reports of binding between p107 and BMYB in S phase or between LIN9 and pRB (Gagrica et al. 2004; Schmit et al. 2007). Our proteomic analysis revealed that human LIN9, LIN37, LIN52, LIN54, and RBBP4 interact with each other to form the MuvB core complex. The MuvB core can bind to either p130 or p107 and E2F4/DP1 to form the DREAM complex, or to BMYB without an RB-like protein to form the MMB complex. This switch represents a major difference from the Drosophila complex, where RB, E2F, DP and MYB orthologs all interact with the MuvB core to form a single dREAM/MMB complex (Korenjak et al. 2004; Lewis et al. 2004).
Although there were no differences in the components of the MuvB core when bound to p130 or BMYB, we asked whether there were any phosphorylated residues specific for each of the complexes. The complete proteomic data set was merged to search for serine, threonine, and tyrosine phosphorylation in the DREAM and MMB subunits as described previously (Xiang et al. 2007). Use of the multiple baits for immunoprecipitation of the DREAM and MMB complexes combined with the high sensitivity of MudPIT resulted in high sequence coverage and detection of phosphorylated residues in most of the proteins included in the analysis (Table 1; Supplemental Table S3).
Table 1.
Phosphorylation sites detected in the DREAM and BMYB–MuvB complex subunits by immunoprecipitation/MS/MS
We compared the results of our phosphorylation search with previously reported data and confirmed the presence of several known phosphorylation sites in p130 and BMYB, as well as sites in LIN52, LIN37, and DP1 detected in prior large-scale phosphoproteomic studies (Table 1; Supplemental Table S2 and references therein). In addition, we detected novel phosphorylation sites in p130, BMYB, LIN9, LIN37, LIN52, LIN54, DP1, and E2F4 (Table 1). No phosphorylated residues were detected in RBBP4, E2F5, and DP2 despite the significant sequence coverage obtained by tandem mass spectra (MS/MS) analysis for these proteins (75.8%, 46%, and 42.2%, respectively).
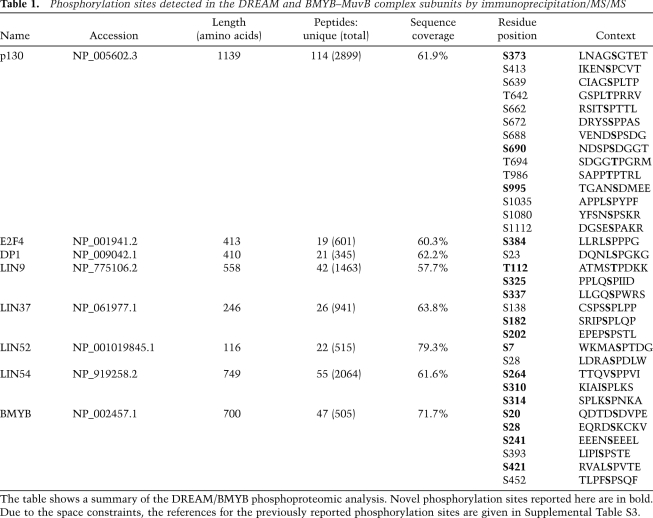
We examined this phosphoproteome data set to determine whether any of the identified phosphorylation events in the MuvB core were specific for a given immunoprecipitation condition. To approximate the extent of modification for a given residue, we compared the number of spectra corresponding to a phosphorylated residue with the total number of spectra—phosphorylated and nonphosphorylated—detected for the corresponding peptide (Fig. 1; Supplemental Fig. S2). This analysis revealed that Ser 28 (S28) in LIN52 was present exclusively in the phosphorylated form (33 phosphorylated out of 33 total spectra) when coimmunoprecipitated with p130 (Fig. 1A). In contrast, this residue was detected in both phosphorylated and unphosphorylated forms in the merged data set containing the peptide information for all immunoprecipitates (73 phosphorylated out of 136 total spectra) when LIN52 was directly immunoprecipitated (13 phosphorylated out of 38 total) or coprecipitated by MuvB core components or BMYB (Fig. 1B,C; Supplemental Fig. S2). In addition, Ser 182 (S182) in LIN37 was detected only in the phosphorylated form when coprecipitated with p130 or LIN52, while both phosphorylated and unphosphorylated forms were detected in LIN37, LIN9, LIN54, or BMYB immunoprecipitates (Fig. 1; Supplemental Fig. S2).
Figure 1.
Analysis of DREAM phosphorylation reveals a critical role of LIN52-S28. (A–C) Detection of phosphorylated amino acid residues in the LIN proteins by the immunoprecipitation/MS/MS of p130, BMYB, and merged data set of all samples. The graphs show spectral counts of peptides where phosphorylated amino acids were detected (gray bars), and the total spectral counts for the corresponding peptides (white bars). Boxes indicate sites that were always phosphorylated in p130 immunoprecipitation samples. (D) Mutational analysis of LIN52-S28 and LIN37-S182 residues. Ectopically expressed V5-tagged wild-type or the indicated mutants of LIN52 and LIN37 were immunoprecipitated using anti-V5 antibody, and the binding of p130 and BMYB was detected by immunoblot. (E) Immunoprecipitation/Western blot assay shows that endogenous p130 interacts with the wild type but not the S28A-LIN52 mutant. Expression of LIN52 constructs was detected by immunoprecipitation/Western blot with the V5 tag antibody. (F) Immunoprecipitation/Western blot reveals that the immunoprecipitates for p130 and BMYB contain different species of LIN52. (G) Western blot shows that the upper form of LIN52 coprecipitated with p130, LIN9, or LIN37 collapses to the lower form with phosphatase treatment. (H) T98G cells were synchronized at various stages of the cell cycle by serum starvation and readdition, and LIN52 was detected in the extracts by immunoblot.
LIN52-S28 is required for DREAM assembly
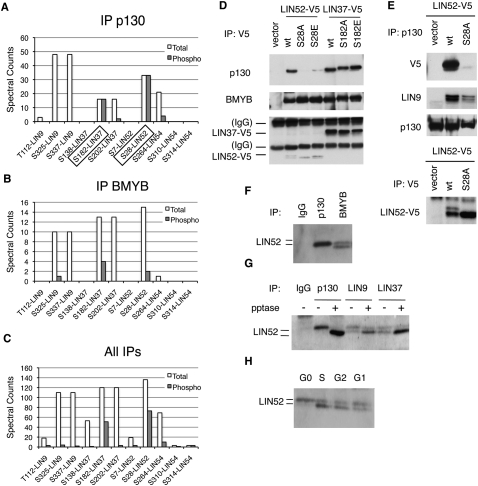
The specific enrichment of pS28-LIN52 and pS182-LIN37 in p130 immunoprecipitations suggested that these phosphorylation events could be required for the DREAM complex assembly. To test this, we generated T98G cell lines stably expressing epitope-tagged wild-type or mutant alleles of LIN52 and LIN37 with point substitutions at residues S28 and S182, respectively. When LIN52-S28 was substituted with alanine, the mutant LIN52-S28A was unable to interact with p130, while the substitution of S28 with the phosphomimetic glutamate residue (S28E) resulted in weak p130 binding (Fig. 1D,E). Point substitution of LIN52-S28 did not affect the interaction with BMYB, LIN9, and LIN37 (Fig. 1D; Supplemental Fig. S3A). Therefore, an intact LIN52-S28 residue was required for binding to p130, consistent with a predominance of the phosphorylated S28 LIN52 in the p130 immunoprecipitates detected by mass spectroscopy analysis. Conversely, substitution of residue S182 in LIN37 with alanine or glutamate did not affect LIN37 binding to p130, LIN9, or BMYB (Fig. 1D; Supplemental Fig. S3B), indicating that the phosphorylation of LIN37 at S182 was not required for these interactions.
We observed that the phosphorylation status of LIN52 affected its gel migration pattern. While the ectopically expressed wild-type LIN52 migrated as a doublet in the SDS–polyacrylamide gel, the S28A mutant appeared as a single faster-migrating band (Fig. 1D,E). Importantly, only a single slower-migrating band coimmunoprecipitated with p130, while both slower- and faster-migrating forms were observed when endogenous LIN52 was coprecipitated with BMYB or LIN9 or LIN37 (Fig. 1F,G). Phosphatase treatment of p130, LIN9, and LIN37 immunoprecipitations resulted in a relative decrease of the upper form of LIN52, indicating that these gel migration differences were due to phosphorylation (Fig. 1G). The slower-migrating form of the endogenous LIN52 was also enriched in cell extracts prepared from G0-arrested human T98G cells and cells entering G1 compared with the cells in S or G2 (Fig. 1H). Therefore, the mass spectroscopy proteomic analysis, point substitution mutant analysis, and gel migration pattern of LIN52 all confirmed independently that only phosphorylated S28-LIN52 was present in the p130/DREAM complex. These results demonstrate a requirement of LIN52-S28 phosphorylation for p130 binding, and indicate that phosphorylation of LIN52 can occur in the G0 quiescent state.
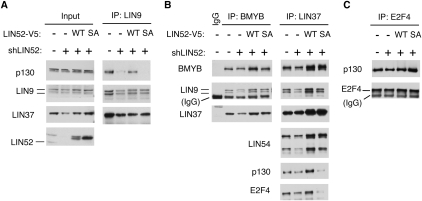
In cells stably expressing S28A-LIN52, we observed that p130 coprecipitated significantly less LIN9 compared with cells expressing wild-type LIN52 (Fig. 1E). This result indicated a dominant-negative effect of S28A-LIN52 on the interaction between p130 and LIN9. To test whether an intact LIN52 was required for the interaction between p130 and the MuvB core, we depleted LIN52 by RNAi in T98G cells. We observed that, in the cells where LIN52 was depleted, the binding between p130 and LIN9 was significantly reduced, accompanied by a decrease of LIN9 and LIN37 expression levels (Supplemental Fig. S4A,B). We then generated cell lines with stable knockdown of LIN52 with an shRNA targeting the 5′ untranslated region of the LIN52 mRNA. In these cells, the expression of LIN52 as well as LIN9 and LIN37 was also reduced, and the interaction between LIN9 and p130 was disrupted (Fig. 2A). Notably, when LIN52 expression was rescued with V5-tagged wild-type or S28A LIN52 resistant to the shLIN52, the protein levels of LIN9 and LIN37 were restored to wild-type levels (Fig. 2A,B). However, only expression of wild-type LIN52, but not the LIN52-S28A mutant, restored the interaction between LIN9 and p130 as well as between LIN37 and p130/E2F4 (Fig. 2A,B). We examined the contribution of LIN52 to the interaction between BMYB and the MuvB core subunits LIN9 and LIN37. Depletion of LIN52 by shRNA resulted in decreased expression of the components of the MuvB core in T98G cells (Fig. 2B) and HeLa cells (Supplemental Fig. S4C). However, both the wild-type and S28A mutant forms of LIN52 were capable of binding to BMYB, LIN9, and LIN37 and forming an intact MMB complex (Fig. 2B). Notably, although depletion of LIN52 or expression of LIN52-S28A disrupted the binding of the MuvB core to p130 and E2F4, it did not affect the interaction between p130 and E2F4 (Fig. 2B,C), consistent with our earlier finding that E2F4 and the MuvB core bind independently to p130 (Litovchick et al. 2007). These results support a specific role of pS28-LIN52 in the MuvB core for binding to p130/E2F4, resulting in the formation of the DREAM complex.
Figure 2.
Intact LIN52 is required for the DREAM complex assembly. (A) Immunoprecipitation/Western blot assays show that only the wild-type LIN52 allele can rescue the binding of p130 to LIN9 and LIN37 in cells with reduced expression of LIN52. LIN52 was depleted in T98G cells by stable expression of LIN52-shRNA and rescued by wild-type LIN52 or the S28A mutant. The DREAM complex was assayed by immunoprecipitation/Western blot. Input panels show the levels of the proteins in cell extracts. (B) The immunopreciptiation/Western blot assays show that both wild-type and the S28A mutant of LIN52 bind to BMYB and up-regulate the levels of LIN37 in LIN52-depleted cells. (C) The interaction between p130 and E2F4 is independent of LIN52, as shown by immunoprecipitation/Western blot.
DYRK1A binds and phosphorylates LIN52
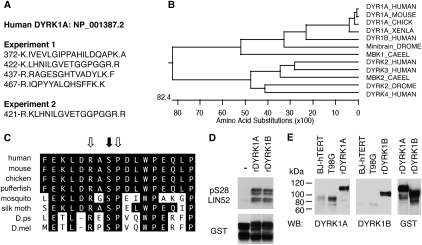
Given that LIN52 is phosphorylated in vivo, we searched the LIN52 MudPIT proteomic data set for the presence of a protein kinase (Supplemental Table S2). Indeed, in addition to all subunits of the DREAM complex and BMYB, the DYRK1A protein kinase was detected in at least two independent LIN52 immunoprecipitations but not in the control samples (Fig. 3A; Supplemental Table S2). Mammals have five DYRK genes, with DYRK1A and DYRK1B most highly related to each other and to the Drosophila Minibrain kinase and C. elegans MBK1 (Fig. 3B; Aranda et al. 2010). In addition, the sequence surrounding the S28 residue in LIN52 closely resembles the DYRK1A phosphorylation consensus motif RX(X)(S/T)P and is highly conserved in LIN52 orthologs from other species (Fig. 3C; Himpel et al. 2000; Woods et al. 2001).
Figure 3.
DYRK1A interacts with LIN52 in vivo and phosphorylates LIN52-S28. (A) Positions and amino acid sequences of DYRK1A peptides detected in two independent LIN52 immunoprecipitation/MS-MS experiments. Dots indicate tryptic cleavage sites. (B) Clustal W alignment of DYRK1A orthologs from different species reveals a protein kinase family that is highly conserved in evolution. (C) Amino acid sequence around LIN52-S28 (black arrow), including the DYRK1A consensus motif (white arrows), is evolutionarily conserved. (D) In vitro kinase assay shows that both recombinant DYRK1A and DYRK1B can phosphorylate LIN52. Phosphorylated or total GST-LIN52 was detected by immunoblots with phospho-S28-LIN52 antibody and anti-GST antibody, respectively. (E) DYRK1A and DYRK1B were detected in BJ-hTERT and T98G cell extracts by immunoblot using recombinant purified GST-tagged proteins (rDYRK1A or rDYRK1B, 30 ng per lane) as a reference. The rDYRK1A and rDYRK1B samples were also blotted for GST to ensure equal loading.
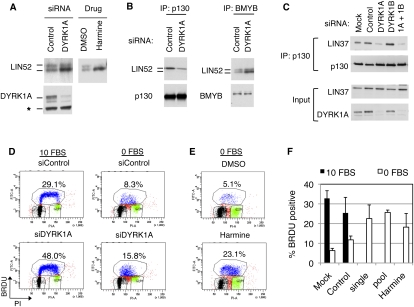
We tested purified recombinant DYRK1A and DYRK1B and found them both to be capable of phosphorylating S28 of LIN52 in an in vitro kinase assay (Fig. 3D). Although both DYRK1A and DYRK1B could specifically phosphorylate LIN52-S28 in vitro, we observed that human BJ-hTERT fibroblasts and T98G cells expressed DYRK1A, while DYRK1B was not readily detectable (Fig. 3E). Furthermore, when expression vectors for DYRK1A or DYRK1B were transfected into T98G cells, the endogenous LIN52 could coprecipitate overexpressed DYRK1A more efficiently than DYRK1B (Supplemental Fig. S5A). These results are consistent with the proteomic detection of DYRK1A but not DYRK1B in LIN52 immunoprecipitates. To determine whether DYRK1A phosphorylates LIN52 in vivo, we depleted DYRK1A in T98G cells by siRNA. As shown in Figure 4A, depletion of DYRK1A resulted in the accumulation of unphosphorylated faster-migrating LIN52. Similarly, when T98G cells were treated with harmine, a specific small molecule inhibitor of DYRK1A (Bain et al. 2007), the phosphorylation of LIN52 was significantly reduced (Fig. 4A), supporting the hypothesis that DYRK1A phosphorylates LIN52 in vivo.
Figure 4.
DYRK1A contributes to the DREAM complex assembly and entry into quiescence. (A) Immunoblot of LIN52 in the extracts from T98G cells incubated with DYRK1A-specific siRNA (48 h) or 10 μM harmine (20 h). (B) Depletion of DYRK1A in T98G cells by RNAi reduces the binding of LIN52 to p130 but not BMYB, as shown by immunoprecipitation/Western assay. (C) Depletion of DYRK1A but not DYRK1B in T98G cells by RNAi results in a decreased binding of p130 to LIN37, as shown by immunoprecipitation/Western blot. (D,E) Treatment of T98G cells by DYRK1A siRNA (a pool of four oligos) or treatment with 10 μM harmine interferes with G0/G1 growth arrest upon serum deprivation, as shown by FACS analysis of BRDU-labeled DNA. Cells were incubated with 10% FBS (10 FBS) or 0% FBS (0 FBS) for 24 h before the assay. (F) The graph shows the average percentage of BrDU-positive cells ± SD measured in three independent experiments shown in D and E. Two-tailed _t_-test: 0 FBS control versus siDYRK1A single and pool, P = 0.00008 and 0.014, respectively.
Given that phosphorylated S28 residue in LIN52 was required for the MuvB core binding to p130/E2F4 and DREAM assembly, we tested whether depletion of DYRK1A could affect this interaction. When T98G cells were transfected with DYRK1A-specific siRNA, the interaction between p130 and LIN52 was decreased (Fig. 4B). In contrast, binding between BMYB and LIN52 was increased upon the depletion of DYRK1A in these cells. Notably, upon DYRK1A depletion, BMYB coprecipitated relatively more unphosphorylated LIN52 compared with the control RNAi knockdown, consistent with the finding that BMYB could bind to both forms of LIN52 and that the phosphorylation state of S28 of LIN52 did not affect the MMB complex assembly. In addition, RNAi-mediated depletion of DYRK1A but not DYRK1B in T98G cells reduced the interaction between p130 and LIN37 (Fig. 4C). This finding is consistent with the role of LIN52 in promoting p130 binding to the MuvB core and strongly indicates that DYRK1A contributes to the assembly of the DREAM complex by phosphorylation of LIN52. Although we were unable to find a specific role for DYRK1B in LIN52 phosphorylation in the cell lines we tested, ectopic overexpression of DYRK1B in T98G cells was able to rescue the effect of the depletion of DYRK1A by siRNA (Supplemental Fig. S5B), indicating that DYRK1B could potentially phosphorylate LIN52 in cell types that express high levels of this kinase, such as myoblasts (Aranda et al. 2010).
DYRK1A is required for growth arrest
Upon removal of serum from the culture media, T98G cells enter a G0/quiescent state characterized by decreased levels of DNA synthesis. Under these conditions, the DREAM complex binds to the promoters of many E2F-dependent gene targets and contributes to their repression (Litovchick et al. 2007). Given the contribution of DYRK1A to LIN52 phosphorylation and DREAM assembly, we asked whether DYRK1A was required for entry into a G0 quiescent state. Cells in which DYRK1A levels were reduced by RNAi displayed increased DNA synthesis under serum-starved conditions when compared with the cells transfected with nontargeting siRNA (Fig. 4D,F). Similarly, T98G cells that were serum-starved in the presence of the DYRK1A inhibitor harmine had higher levels of DNA synthesis relative to control-treated cells (Fig. 4E,F). These results indicate that DYRK1A is required for DREAM complex assembly and entry into G0/quiescence.
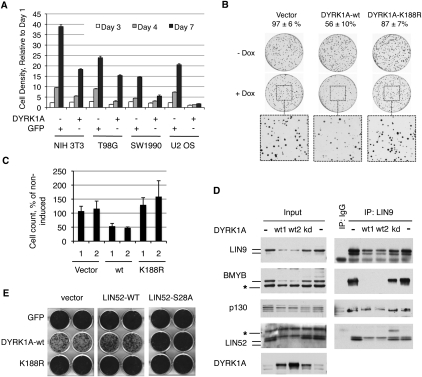
Given that DYRK1A was required for entry into quiescence, we tested whether overexpression of DYRK1A could inhibit cell proliferation. DYRK1A was stably overexpressed using retroviral infection in a panel of cultured cells, resulting in significantly slower growth rates compared with the control cell lines expressing GFP (Fig. 5A). U-2 OS osteosarcoma cells appeared particularly sensitive to growth arrest upon overexpression of DYRK1A (Fig. 5A; Supplemental Fig. S6A). We engineered tetracycline-inducible (tet-on) expression of wild-type or kinase-inactive K188R mutant alleles of DYRK1A (Kentrup et al. 1996) in U-2 OS cells and determined whether the catalytic activity of DYRK1A was required for growth suppression effect in a colony formation assay. As shown in Figure 5B, induced expression of wild-type DYRK1A but not kinase-inactive DYRK1A-K188R resulted in a significant suppression of colony formation of the U-2 OS cells. Consistent with this finding, induced expression in these cells of the wild-type DYRK1A but not the K188R mutant led to the appearance of a growth-arrested flattened cell phenotype (Supplemental Fig. S6B). We isolated two stable clones from each of the tet-inducible U2OS cell lines expressing vector, wild-type, or kinase-inactive DYRK1A and compared their growth rate in a cell proliferation assay. The proliferation rate was reduced by ∼50% when wild-type DYRK1A was induced with doxycycline compared with the uninduced cells, while the cells expressing the DYRK1A-K188R proliferated slightly faster when induced (Fig. 5C). The viability of U-2 OS cells was not affected by induction of the DYRK1A alleles (data not shown). Notably, we observed a dramatic increase in the phosphorylated form of LIN52 upon expression of wild-type DYRK1A but not DYRK1A-K188R (Fig. 5D). Furthermore, in wild-type DYRK1A-expressing cells, the binding of LIN9 to p130 was readily detectable, while LIN9 binding to BMYB was reduced (Fig. 5D). Therefore, expression of active DYRK1A in U-2 OS cells resulted in preferential recruitment of the MuvB core into the DREAM complex, down-regulation of the MMB complex, and growth suppression.
Figure 5.
Overexpression of DYRK1A suppresses cell proliferation. (A) Overexpression of DYRK1A causes growth suppression. The indicated cell lines were transduced with retroviruses to express DYRK1A or GFP and were used for cell proliferation assay after antibiotic selection. The graph shows the density of the cultures determined by crystal violet staining relative to day 1 (average of the triplicate samples ± SD). The normalized OD values at day 7 were significantly different for all pairs of DYRK1A- and GFP-expressing cell lines (two-tailed _t_-test, P < 0.005). (B) Active but not kinase-dead DYRK1A suppresses colony growth of U2OS cells. Tet-on U-2 OS cells were treated as described in the Materials and Methods. Colony counts of induced samples relative to uninduced (taken as 100%) are shown above the images. (C) Overexpression of active but not kinase-dead DYRK1A suppresses proliferation of U2OS cells. Two clones of each type of cell line were grown ±doxycycline and counted. Induced cell counts are shown as percentage of uninduced (average values ± SD of two experiments, each done in triplicate). (D) Overexpression of active but not kinase-dead DYRK1A in U-2 OS cells affects the DREAM and BMYB–MuvB complexes, as shown by immunoprecipitation/Western blots. DYRK1A alleles were expressed in U-2 OS cells by retroviral infection and by antibiotic selection. Two wild-type-expressing samples (wt1 and wt2) with different levels of DYRK1A are shown to emphasize a potent effect of DYRK1A on the DREAM and BMYB–MuvB complexes in U-2 OS cells. (E) Overexpression of LIN52-S28A but not the wild type can override the growth suppression effect of DYRK1A in NIH 3T3 cells. GFP, DYRK1A, or the DYRK1A-K188R mutant were introduced into NIH 3T3 cell lines expressing the LIN52 alleles or vector. After antibiotic selection, the cell lines were equally plated, grown for 7 d, and stained by crystal violet dye. Images of two representative wells for each condition are shown.
Since overexpressed DYRK1A likely phosphorylates other targets in addition to LIN52 that could contribute to the observed growth inhibition, we tested whether the expression of LIN52-S28A in cells could suppress this effect. We generated NIH 3T3 cell lines stably expressing vector, wild-type, or S28A mutant LIN52 and then introduced GFP (control), active DYRK1A, or the DYRK1A-K188R mutant. Immediately after antibiotic selection, the resulting set of nine cell lines was assayed for cell proliferation. As shown in Figure 5E and Supplemental Figure S6D, expression of the LIN52-S28A mutant could indeed rescue the growth suppression effect caused by overexpression of active DYRK1A. A similar result was also obtained in T98G cells (data not shown), indicating that phosphorylation of LIN52 and DREAM complex assembly significantly contribute to the growth suppression by DYRK1A.
DYRK1A and intact LIN52 are both required for oncogenic Ras-induced senescence
Oncogene-induced senescence is characterized by permanent cell cycle arrest in G0/G1 with induction of senescence-associated β-galactosidase (SA-β-gal) activity (Serrano et al. 1997). In a previously reported gene expression profiling study in human IMR90 fibroblasts, expression of the oncogenic HRAS-G12V allele resulted in a significant down-regulation of cell cycle-regulated genes (Mason et al. 2004). We previously identified >800 DREAM target genes using ChIP–chip analysis (Litovchick et al. 2007). A comparison of the DREAM target genes with the genes that were repressed in the HRAS-G12V-expressing fibroblasts revealed a highly significant overlap (P = 2.2e−16, exact Fisher test) (Supplemental Fig. S7A).
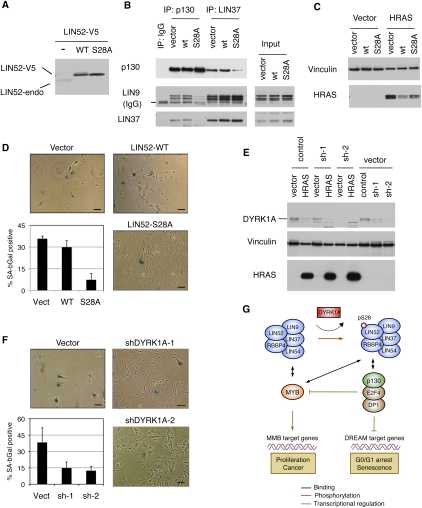
To determine whether DREAM contributes to the repression of genes down-regulated in cells undergoing cellular senescence, we tested whether disruption of the DREAM complex assembly could interfere with Ras-induced senescence in immortalized human BJ-hTERT fibroblasts (Hahn et al. 1999). We generated BJ-hTERT cell lines stably expressing wild-type LIN52 or the S28A mutant. We noted that expression of V5-epitope-tagged LIN52 led to a reduced expression of endogenous LIN52 (Fig. 6A). As expected, we observed that expression of LIN52-S28A led to disruption of the DREAM complex, as shown by significantly reduced binding of p130 to LIN9 and LIN37 compared with cells expressing wild-type LIN52 (Fig. 6B). These cell lines were infected by retroviruses carrying the oncogenic HRAS-G12V allele or empty vector and were assayed for cellular senescence on day 14 post-infection (Supplemental Fig. S7B). Expression of HRAS-G12V was comparable in wild-type LIN52- and LIN52-S28A-expressing cells and was slightly lower than in the empty vector-expressing cells (Fig. 6C). Remarkably, cells expressing the LIN52-S28A mutant resulted in significantly reduced SA-β-gal activity and increased proliferation in response to HRAS-G12V compared with vector control- or wild-type LIN52-expressing cells (Fig. 6D; Supplemental Fig. S7C). Since S28A-LIN52 specifically disrupts DREAM complex assembly, these results indicate that an intact DREAM complex is required for oncogenic RAS-induced senescence.
Figure 6.
DYRK1A and intact LIN52-S28A contribute to oncogenic Ras-induced senescence. (A) Western blot shows that overexpression of either the wild type or LIN52-S28A mutant in BJ-hTERT cells results in down-regulation of the endogenous LIN52 protein. (B) The LIN52-S28A mutant but not wild-type LIN52 disrupts the DREAM complex in BJ-hTERT fibroblasts, as shown by immunoprecipitation/Western blot assay. (C) The indicated BJ-hTERT cell lines were infected with retroviruses to express HRAS-G12V or empty vector, and the expression of HRAS-G12V was assayed by Western blot. Vinculin was used as a loading control. (D) BJ-hTERT cells expressing LIN52 alleles or vector were infected with HRAS-G12V and stained for SA-β-gal on day 14 post-infection. Representative images of the cells are shown as well as average values ± SD of three independent experiments, each counting >100 cells per condition. Two-tailed _t_-test: vector versus LIN52 wild type (LIN52-WT) and S28A mutant (LIN52-S28A), P = 0.24 and 0.01, respectively. Bar, 100 μM. (E) BJ-hTERT cells were transduced with either control or two different DYRK1A-specific shRNA lentiviruses, followed by infection with retrovirus encoding HRAS-G12V. Western blot shows the levels of DYRK1A and HRAS-G12V in these cells. Vinculin was used as a loading control. (F) Ras-induced senescence assay in BJ-hTERT cell lines treated with DYRK1A-shRNA. The cells were infected and processed as in D. Two-tailed _t_-test: vector versus shDYRK1A-1 and shDYRK1A-2, P = 0.09 and 0.05, respectively. Bar, 100 μM. (G) The model shows how DYRK1A promotes the DREAM complex assembly, G0/G1 arrest, and senescence. DYRK1A phosphorylation of the Ser 28 residue in the LIN52 subunit of the MuvB core promotes the DREAM complex assembly and repression of E2F target genes such as BMYB. MuvB core subunits are shaded in blue.
Since DYRK1A was required for DREAM assembly and entry into the G0-arrested state in RNAi experiments, we tested whether DYRK1A was also required for oncogenic HRAS-induced senescence. We generated BJ-hTERT cell lines stably expressing DYRK1A targeting shRNA and infected them with the HRAS-G12V-expressing retrovirus. The expression levels of HRAS-G12V in the control and DYRK1A-depleted cell lines were similar (Fig. 6E). Similar to BJ-hTERT cells expressing LIN52-S28A, the DYRK1A-depleted cell lines displayed significantly reduced levels of SA-β-gal activity in response to oncogenic HRAS when compared with the cells expressing the nontargeting shRNA (Fig. 6F). Together, these results indicate that DYRK1A and the DREAM assembly are required for oncogenic RAS-induced growth arrest and senescence.
Discussion
Evolutionarily conserved RB/E2F complexes control the expression of cell cycle-dependent genes in various organisms. The function of these complexes is regulated by a family of cell cycle-dependent protein kinases that relieves the RB-mediated inhibition of E2Fs, resulting in activation of E2F-dependent transcription and progression of the cell cycle. In addition to CDK regulation, the RB family member p130 protein stability is increased by GSK3 phosphorylation. Our results here indicate that DYRK1A serves as an additional protein kinase that activates the p130/E2F4-containing DREAM complex (Fig. 6). DYRK1A directly phosphorylates the DREAM subunit LIN52 on S28, and this phosphorylation is required for binding to p130. Moreover, this phosphorylation is also required for the interaction between p130/E2F4 and all other subunits of the MuvB core because either point substitution of LIN52-S28 or depletion of DYRK1A by RNAi can interfere with the DREAM complex assembly. Conversely, phosphorylation of S28 is not required for LIN52 binding to BMYB or the other MuvB core proteins. Inhibiting DYRK1A phosphorylation of LIN52-S28 results in loss of the ability of cells to enter the G0 growth-arrested state upon serum deprivation or undergo oncogenic RAS-induced senescence. On the other hand, overexpression of DYRK1A in cell lines leads to a marked inhibition of cell proliferation, increased phosphorylation of LIN52, down-regulation of the MMB complex, and the recruitment of the MuvB core into the DREAM complex. Importantly, expression of the LIN52-S28A mutant rescues the growth suppression effect of DYRK1A in NIH 3T3 and T98G cells, indicating that this growth inhibition requires the DREAM complex assembly.
This study reveals that DYRK1A plays an important role in suppression of mammalian cell proliferation. DYRK1A protein levels do not change throughout the cell cycle (L Litovchick and J DeCaprio, unpubl.); therefore, its activity could be regulated by other mechanisms, including subcellular localization, autophosphorylation, and interaction with regulatory proteins such as 14-3-3 and SPRED1 (for review, see Aranda et al. 2010). Specific up-regulation of DYRK1A activity toward LIN52 by the tumor suppressor protein kinase LATS2 is described in the coordinated study by Tschöp et al. (2011). Overexpression of LATS2 suppresses proliferation of cancer cell lines, while the loss of LATS2 is observed in many types of cancer and has been shown to promote proliferation and oncogenic transformation of the primary fibroblasts (for review, see Visser and Yang 2010). Future studies will hopefully provide a deeper insight into the relationship between LATS2, DYRK1A regulation, and DREAM complex assembly, and into the role of this emerging pathway in the entry into quiescence and tumor suppression.
Our findings presented here implicate DYRK1A in cell cycle control in mammalian cells. Curiously, based on structural similarity, DYRK1A belongs to the same CMGC group of protein kinases that includes well known regulators of the cell cycle processes, such as CDKs, mitogen-activated protein kinases (MAPKs), GSKs, and cdc2-like kinases (Kannan and Neuwald 2004; Park et al. 2009). DYRK proteins are well characterized in yeast, Dictyostelium discoideum, C. elegans, Drosophila, and mammals (for review, see Aranda et al. 2010). Many of the DYRK proteins function in various aspects of cell cycle regulation, differentiation, and stress response pathways. Yeast DYRK proteins Yak1p and Pom1p are implicated in the control of the cell cycle and nutrient-sensing pathways, and coupling of the cell size and cell cycle progression (Aranda et al. 2010). In Dictyostelium, one of the three known DYRK proteins, YakA, regulates exit from the cell cycle upon starvation and the transition from growing phase to the developmental phase (Souza et al. 1998; Taminato et al. 2002).
Drosophila Minibrain is closely related to mammalian DYRK1A and DYRK1B, and was named because of the marked reduction of the brain size in hypomorphic mutant flies (Tejedor et al. 1995). The region that harbors the serine residue homologous to human LIN52-S28 is conserved in the Drosophila protein, so it will be interesting to learn whether it could be phosphorylated by Minibrain, and whether this phosphorylation could affect the structural composition or the function of the dREAM/MMB complex. The function of the mnb homolog DYRK1A in neurogenesis is well supported by evidence from both transgenic mouse models and human disease (for review, see Park et al. 2009). Dyrk1A−/− mice die during mid-gestation, while the heterozygous animals show decreased viability and reduction of body and brain size, with regional-specific increases and decreases in neuronal density (Fotaki et al. 2002). In humans, the DYRK1A gene is located within the Down syndrome (DS)-critical region on chromosome 21, and is a strong candidate contributor to the neurological abnormalities associated with this syndrome (Park et al. 2009). The exact mechanisms linking the trisomy of the chromosome 21 region that includes DYRK1A as well as other genes with the pathological phenotypes observed in DS are not well understood. However, a detailed analysis of several available mouse models of DS indicates that increased dosage of the DYRK1A gene is sufficient to induce learning defects in transgenic animals (Altafaj et al. 2001; Bouchikhi et al. 2009).
More than 20 different protein substrates for DYRK1A have been reported to date, including some with well-documented functions important for neuronal differentiation, cell survival, and metabolism (for review, see Aranda et al. 2010). Most likely, the DS brain phenotype results from multilevel defects in neuronal genesis, differentiation, and function, mediated by increased dosage of DYRK1A and other genes in the DS-critical region (Gardiner et al. 2010). The role of cell proliferation and cell cycle exit in DS pathogenesis is not well understood. However, a decreased proliferation capacity, elongated cell cycle, and reduced expression of cell cycle-dependent genes have been shown in specific regions of the brain in DS patients as well as in a mouse model of DS (Contestabile et al. 2007, 2009; Hewitt et al. 2010). In addition, overexpression of DYRK1A in the embryonic mouse neocortex by in utero electroporation results in inhibition of neural cell proliferation without affecting cell fate and layer positioning (Yabut et al. 2010). Therefore, it is possible that at least some of the neurogenesis defects associated with DS could be attributed to a decreased neuronal proliferation due to overexpression of DYRK1A and increased DREAM formation during development.
In addition to the potential contribution of DYRK1A and DREAM to decreased neuronal proliferation and premature cell cycle exit, there is a strong correlation between increased DYRK1A dosage and the reduced incidence of solid tumors observed in DS (Baek et al. 2009). We observed that DYRK1A activity and intact LIN52-S28 are both required for oncogenic Ras-induced senescence in the immortalized human fibroblasts. Oncogene-induced senescence in premalignant lesions in mouse models as well as in some human tumors could serve as a protective tumor suppressor mechanism (for review, see Collado and Serrano 2010). Since DYRK1A was able to suppress the proliferation of several human cancer cell lines, it will be interesting to investigate whether this kinase could serve as a tumor suppressor in humans. Therefore, our finding of the key role of DYRK1A in promoting the DREAM complex function and the cell cycle exit could reveal a novel important pathway to understand cancer pathogenesis. Our data could also bring insight into the pathogenesis of DS, and potentially lead to strategies to improve the cognitive functions of affected individuals.
Materials and methods
Cell lines, plasmids, and reagents
Established cell lines were obtained from American Type Culture Collection. Primary BJ human skin fibroblasts were immortalized by infection with hTERT-pWZL retroviral vector (gift of R. DePinho) at passage 5 and selection on blasticidin (Invitrogen). pBabe-Hygro retroviral vectors encoding human LIN37 and LIN52 with a C-terminal V5-6His epitope tag (Litovchick et al. 2007) were used for site-directed mutagenesis (QuikChange; Agilent). HRAS-G12V-pBabe-Puro and HRAS-G12V-pWZL-Hygro were from Addgene (plasmids 9051 [W. Hahn] and 18749 [S. Lowe], respectively).
Tet-on U-2 OS cells were generated using pRev Tet-On system (Clontech Takara Bio). Human DYRK1A cDNA (gift of W. Hahn) was subcloned into tet-inducible pRevTre vector and used to generate the kinase-inactive DYRK1A-K188R by site-directed mutagenesis (QuikChange; Agilent). GFP-HA-Flag, DYRK1A-HA-Flag, and DYRK1B-HA-Flag constructs were prepared using Gateway cloning using entry clones from Harvard PlasmID repository and pMSCV-CTAP retroviral vector (gift of M. Sowa).
Antibodies, siRNA oligonucleotides, and shRNA constructs are described in the Supplemental Material.
Cell cycle analysis
Cell cycle synchronization using serum deprivation and readdition was performed as described (Litovchick et al. 2004). DNA synthesis was assayed by incubation of the cells with BrdU/FdU (GE Healthcare LifeSciences) for 1 h, followed by detection using FITC BrdU Flow kit (BD Biosciences) and FACS analysis.
Recombinant proteins and kinase assays
Purified GST-tagged DYRK1A (PV3785) and DYRK1B (PV4649) proteins were from Invitrogen. The bacterial expression construct for GST-LIN52 was generated by subcloning human LIN52 into a pGEX-2T vector. GST-tagged LIN52 was expressed in bacteria and purified using glutathione-Sepharose 4B (GE Healthcare). In vitro kinase assay was performed by incubating 20 ng of GST-LIN52 and 10 ng of GST-DYRK1A and GST-DYRK1B in a buffer containing 20 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM DTT, and 100 μM ATP for 30 min at room temperature. Phosphorylation at the S28 residue in LIN52 was detected by immunoblotting with rabbit phospho-S28-specific antibody (custom-made by New England Peptide, Inc.).
MudPIT analysis and PTM search
LIN52 was purified from the T98G cell line expressing LIN52-V5 using either anti-peptide antibody against the endogenous LIN52 or V5-epitope tag and was analyzed by MudPIT mass spectroscopy analysis (Litovchick et al. 2007; Swanson et al. 2009). BMYB was purified from S-phase T98G cells using anti-peptide antibody specific to BMYB (Bethyl Laboratories, Inc.). MS/MS were interpreted using SEQUEST against a database of 61,788 sequences, consisting of 30,709 human proteins (downloaded from NCBI on 2009-07-02) as described in Swanson et al. (2009). Peptide hits from multiple runs were compared using CONTRAST, and relative protein levels were estimated using distributed normalized spectral abundance factors (dNSAFs) (Florens and Washburn 2006). The MS/MS data sets were searched for serine, threonine, and tyrosine phosphorylation (Δ mass + 80) as described in Xiang et al. (2007). The raw mass spectrometric data are available at Proteome Commons Tranche (https://proteomecommons.org/tranche) using the identifiers provided in Supplemental Table S1.
RAS-induced senescence
RAS-induced senescence assay was performed as described (Serrano et al. 1997). HRAS-G12V-pBabe-Puro was used in BJ-hTERT cell lines expressing LIN52 alleles, while HRAS-G12V-pWZL-Hygro was used in BJ-hTERT cell lines transduced with pGIPZ-based lentiviruses. Infected cells were selected using 1 μg/mL puromycin and 200 μg/mL hygromycin, respectively. At day 7 post-infection, the cells were counted and replated. The cells were then grown for seven more days and used for SA-β-gal Senescence Colorimetric Assay (Sigma-Aldrich). Stained cells were overlaid with 70% glycerol and photographed at 10× magnification using a Nikon Eclipse E300 microscope and Spot digital camera (Diagnostic Instruments, Inc.). At least nine nonoverlapping fields were photographed for each condition, and digital images were used to determine the number of SA-β-gal-positive and SA-β-gal-negative cells (>100 cells for each condition).
Cell proliferation assays
NIH 3T3, T98G, U-2 OS, and SW1990 cells were infected with retroviruses to express GFP or human DYRK1A, selected on puromycin (1 μg/mL) for 5 d, and seeded in triplicate onto 12-well plates (2 × 104 cells per well). Cell density was measured using crystal violet (Dannenberg et al. 2004). All cell density values were normalized to day 1.
For cell proliferation assay, tet-on U2 OS cell lines were seeded onto six-well plates in triplicate at 104 cells per well and grown ±doxycycline (0.5 μg/mL) for 5 d. Cells were counted using ViaCount assay and the Guava flow cytometer (Millipore). Two different stable clones for each cell line were analyzed in two independent experiments.
For colony formation assays, tet-on U2 OS cells were infected with inducible DYRK1A alleles or vector retroviruses and selected on antibiotic for 5 d. Viable cells were then counted, and two sets of cell lines were seeded in triplicate at 104 cells per 10-cm plate. The cells were allowed to grow for 2 wk while the cell culture medium (±doxycycline) was replaced every 4 d. The plates were then stained using crystal violet (Sigma) and scanned to obtain TIFF images that were quantified using QuantityOne software (Bio-Rad).
Acknowledgments
We are grateful to M. Padi for help with bioinformatics analysis, and to R. Toddings and Y. Skversky for technical assistance. We thank Eric McIntush (Bethyl Laboratories) for antibodies, and N. Dyson and W.G. Kaelin Jr. for helpful suggestions. This work was supported in part by Public Health Service grants P01CA050661, RO1CA93804, and R01CA63113 to J.A.D., and by the DOD New Investigator Award W81XWH-08-1-0048 to L.L.
Footnotes
Supplemental material is available for this article.
References
- Altafaj X, Dierssen M, Baamonde C, Marti E, Visa J, Guimera J, Oset M, Gonzalez JR, Florez J, Fillat C, et al. 2001. Neurodevelopmental delay, motor abnormalities and cognitive deficits in transgenic mice overexpressing Dyrk1A (minibrain), a murine model of Down's syndrome. Hum Mol Genet 10: 1915–1923 [DOI] [PubMed] [Google Scholar]
- Aranda S, Laguna A, de la Luna S 2010. DYRK family of protein kinases: evolutionary relationships, biochemical properties, and functional roles. FASEB J 25: 449–462 [DOI] [PubMed] [Google Scholar]
- Baek KH, Zaslavsky A, Lynch RC, Britt C, Okada Y, Siarey RJ, Lensch MW, Park IH, Yoon SS, Minami T, et al. 2009. Down's syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 459: 1126–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P 2007. The selectivity of protein kinase inhibitors: a further update. Biochem J 408: 297–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall EL, Manak JR, Zhou S, Bell M, Lipsick JS, Botchan MR 2002. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature 420: 833–837 [DOI] [PubMed] [Google Scholar]
- Blais A, Dynlacht BD 2004. Hitting their targets: an emerging picture of E2F and cell cycle control. Curr Opin Genet Dev 14: 527–532 [DOI] [PubMed] [Google Scholar]
- Bouchikhi F, Anizon F, Moreau P 2009. Synthesis, kinase inhibitory potencies and in vitro antiproliferative activity of isoindigo and 7′-azaisoindigo derivatives substituted by Sonogashira cross-coupling. Eur J Med Chem 44: 2705–2710 [DOI] [PubMed] [Google Scholar]
- Cam H, Balciunaite E, Blais A, Spektor A, Scarpulla RC, Young R, Kluger Y, Dynlacht BD 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell 16: 399–411 [DOI] [PubMed] [Google Scholar]
- Cobrinik D 2005. Pocket proteins and cell cycle control. Oncogene 24: 2796–2809 [DOI] [PubMed] [Google Scholar]
- Collado M, Serrano M 2010. Senescence in tumours: evidence from mice and humans. Nat Rev Cancer 10: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conboy CM, Spyrou C, Thorne NP, Wade EJ, Barbosa-Morais NL, Wilson MD, Bhattacharjee A, Young RA, Tavare S, Lees JA, et al. 2007. Cell cycle genes are the evolutionarily conserved targets of the E2F4 transcription factor. PLoS ONE 2: e1061 doi: 10.1371/journal.pone.0001061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contestabile A, Fila T, Ceccarelli C, Bonasoni P, Bonapace L, Santini D, Bartesaghi R, Ciani E 2007. Cell cycle alteration and decreased cell proliferation in the hippocampal dentate gyrus and in the neocortical germinal matrix of fetuses with Down syndrome and in Ts65Dn mice. Hippocampus 17: 665–678 [DOI] [PubMed] [Google Scholar]
- Contestabile A, Fila T, Cappellini A, Bartesaghi R, Ciani E 2009. Widespread impairment of cell proliferation in the neonate Ts65Dn mouse, a model for Down syndrome. Cell Prolif 42: 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, te Riele HP 2006. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl Cell Differ 42: 183–225 [DOI] [PubMed] [Google Scholar]
- Dannenberg JH, Schuijff L, Dekker M, van der Valk M, te Riele H 2004. Tissue-specific tumor suppressor activity of retinoblastoma gene homologs p107 and p130. Genes Dev 18: 2952–2962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ 2005. The E2F transcriptional network: old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Fay DS, Yochem J 2007. The SynMuv genes of Caenorhabditis elegans in vulval development and beyond. Dev Biol 306: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florens L, Washburn MP 2006. Proteomic analysis by multidimensional protein identification technology. Methods Mol Biol 328: 159–175 [DOI] [PubMed] [Google Scholar]
- Fotaki V, Dierssen M, Alcantara S, Martinez S, Marti E, Casas C, Visa J, Soriano E, Estivill X, Arbones ML 2002. Dyrk1A haploinsufficiency affects viability and causes developmental delay and abnormal brain morphology in mice. Mol Cell Biol 22: 6636–6647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagrica S, Hauser S, Kolfschoten I, Osterloh L, Agami R, Gaubatz S 2004. Inhibition of oncogenic transformation by mammalian Lin-9, a pRB-associated protein. EMBO J 23: 4627–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner K, Herault Y, Lott IT, Antonarakis SE, Reeves RH, Dierssen M 2010. Down syndrome: from understanding the neurobiology to therapy. J Neurosci 30: 14943–14945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georlette D, Ahn S, MacAlpine DM, Cheung E, Lewis PW, Beall EL, Bell SP, Speed T, Manak JR, Botchan MR 2007. Genomic profiling and expression studies reveal both positive and negative activities for the Drosophila Myb MuvB/dREAM complex in proliferating cells. Genes Dev 21: 2880–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA 1999. Creation of human tumour cells with defined genetic elements. Nature 400: 464–468 [DOI] [PubMed] [Google Scholar]
- Harrison MM, Ceol CJ, Lu X, Horvitz HR 2006. Some C. elegans class B synthetic multivulva proteins encode a conserved LIN-35 Rb-containing complex distinct from a NuRD-like complex. Proc Natl Acad Sci 103: 16782–16787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt CA, Ling KH, Merson TD, Simpson KM, Ritchie ME, King SL, Pritchard MA, Smyth GK, Thomas T, Scott HS, et al. 2010. Gene network disruptions and neurogenesis defects in the adult Ts1Cje mouse model of Down syndrome. PLoS ONE 5: e11561 doi: 10.1371/journal.pone.0011561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himpel S, Tegge W, Frank R, Leder S, Joost HG, Becker W 2000. Specificity determinants of substrate recognition by the protein kinase DYRK1A. J Biol Chem 275: 2431–2438 [DOI] [PubMed] [Google Scholar]
- Kannan N, Neuwald AF 2004. Evolutionary constraints associated with functional specificity of the CMGC protein kinases MAPK, CDK, GSK, SRPK, DYRK, and CK2α. Protein Sci 13: 2059–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kentrup H, Becker W, Heukelbach J, Wilmes A, Schurmann A, Huppertz C, Kainulainen H, Joost HG 1996. Dyrk, a dual specificity protein kinase with unique structural features whose activity is dependent on tyrosine residues between subdomains VII and VIII. J Biol Chem 271: 3488–3495 [DOI] [PubMed] [Google Scholar]
- Korenjak M, Taylor-Harding B, Binne UK, Satterlee JS, Stevaux O, Aasland R, White-Cooper H, Dyson N, Brehm A 2004. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell 119: 181–193 [DOI] [PubMed] [Google Scholar]
- Koreth J, van den Heuvel S 2005. Cell-cycle control in Caenorhabditis elegans: how the worm moves from G1 to S. Oncogene 24: 2756–2764 [DOI] [PubMed] [Google Scholar]
- Lewis PW, Beall EL, Fleischer TC, Georlette D, Link AJ, Botchan MR 2004. Identification of a Drosophila Myb–E2F2/RBF transcriptional repressor complex. Genes Dev 18: 2929–2940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Chestukhin A, DeCaprio JA 2004. Glycogen synthase kinase 3 phosphorylates RBL2/p130 during quiescence. Mol Cell Biol 24: 8970–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litovchick L, Sadasivam S, Florens L, Zhu X, Swanson SK, Velmurugan S, Chen R, Washburn MP, Liu XS, DeCaprio JA 2007. Evolutionarily conserved multisubunit RBL2/p130 and E2F4 protein complex represses human cell cycle-dependent genes in quiescence. Mol Cell 26: 539–551 [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M 2001. To cycle or not to cycle: a critical decision in cancer. Nat Rev Cancer 1: 222–231 [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M 2009. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9: 153–166 [DOI] [PubMed] [Google Scholar]
- Mason DX, Jackson TJ, Lin AW 2004. Molecular signature of oncogenic ras-induced senescence. Oncogene 23: 9238–9246 [DOI] [PubMed] [Google Scholar]
- Massague J 2004. G1 cell-cycle control and cancer. Nature 432: 298–306 [DOI] [PubMed] [Google Scholar]
- McClellan KA, Slack RS 2007. Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle 6: 2917–2927 [DOI] [PubMed] [Google Scholar]
- Miller JP, Yeh N, Vidal A, Koff A 2007. Interweaving the cell cycle machinery with cell differentiation. Cell Cycle 6: 2932–2938 [DOI] [PubMed] [Google Scholar]
- Osterloh L, Von Eyss B, Schmit F, Rein L, Hubner D, Samans B, Hauser S, Gaubatz S 2007. The human synMuv-like protein LIN-9 is required for transcription of G2/M genes and for entry into mitosis. EMBO J 26: 144–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Song WJ, Chung KC 2009. Function and regulation of Dyrk1A: towards understanding Down syndrome. Cell Mol Life Sci 66: 3235–3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilkinton M, Sandoval R, Colamonici OR 2007a. Mammalian Mip/LIN-9 interacts with either the p107, p130/E2F4 repressor complex or B-Myb in a cell cycle-phase-dependent context distinct from the Drosophila dREAM complex. Oncogene 26: 7535–7543 [DOI] [PubMed] [Google Scholar]
- Pilkinton M, Sandoval R, Song J, Ness SA, Colamonici OR 2007b. Mip/LIN-9 regulates the expression of B-Myb and the induction of cyclin A, cyclin B, and CDK1. J Biol Chem 282: 168–175 [DOI] [PubMed] [Google Scholar]
- Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, Watson RJ, te Riele H, Dynlacht BD 2002. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev 16: 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmit F, Korenjak M, Mannefeld M, Schmitt K, Franke C, von Eyss B, Gagrica S, Hanel F, Brehm A, Gaubatz S 2007. LINC, a human complex that is related to pRB-containing complexes in invertebrates regulates the expression of G2/M genes. Cell Cycle 6: 1903–1913 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Smith EJ, Leone G, DeGregori J, Jakoi L, Nevins JR 1996. The accumulation of an E2F-p130 transcriptional repressor distinguishes a G0 cell state from a G1 cell state. Mol Cell Biol 16: 6965–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza GM, Lu S, Kuspa A 1998. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development 125: 2291–2302 [DOI] [PubMed] [Google Scholar]
- Swanson SK, Florens L, Washburn MP 2009. Generation and analysis of multidimensional protein identification technology datasets. Methods Mol Biol 492: 1–20 [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Rayman JB, Dynlacht BD 2000. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev 14: 804–816 [PMC free article] [PubMed] [Google Scholar]
- Taminato A, Bagattini R, Gorjao R, Chen G, Kuspa A, Souza GM 2002. Role for YakA, cAMP, and protein kinase A in regulation of stress responses of Dictyostelium discoideum cells. Mol Biol Cell 13: 2266–2275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach KF, Pongs O 1995. minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron 14: 287–301 [DOI] [PubMed] [Google Scholar]
- Tschöp K, Conery AR, Litovchick L, DeCaprio JA, Settleman J, Harlow E, Dyson N 2011. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev (this issue). doi: 10.1101/gad.2000211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel S, Dyson NJ 2008. Conserved functions of the pRB and E2F families. Nat Rev Mol Cell Biol 9: 713–724 [DOI] [PubMed] [Google Scholar]
- Vidwans SJ, Su TT 2001. Cycling through development in Drosophila and other metazoa. Nat Cell Biol 3: E35–E39 doi: 10.1038/35050681 [DOI] [PubMed] [Google Scholar]
- Visser S, Yang X 2010. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle 9: 3892–3903 [DOI] [PubMed] [Google Scholar]
- Woods YL, Cohen P, Becker W, Jakes R, Goedert M, Wang X, Proud CG 2001. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bɛ at Ser539 and the microtubule-associated protein tau at Thr212: potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem J 355: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Takeo S, Florens L, Hughes SE, Huo LJ, Gilliland WD, Swanson SK, Teeter K, Schwartz JW, Washburn MP, et al. 2007. The inhibition of polo kinase by matrimony maintains G2 arrest in the meiotic cell cycle. PLoS Biol 5: e323 doi: 10.1371/journal.pbio.0050323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabut O, Domogauer J, D'Arcangelo G 2010. Dyrk1A overexpression inhibits proliferation and induces premature neuronal differentiation of neural progenitor cells. J Neurosci 30: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]