Differential curvature sensing and generating activities of dynamin isoforms provide opportunities for tissue-specific regulation (original) (raw)
Abstract
Dynamin 1 (Dyn1) and Dyn2 are neuronal and ubiquitously expressed isoforms, respectively, of the multidomain GTPase required for clathrin-mediated endocytosis (CME). Although they are 79% identical, Dyn1 and Dyn2 are not fully functionally redundant. Through direct measurements of basal and assembly-stimulated GTPase activities, membrane binding, self-assembly, and membrane fission on planar and curved templates, we have shown that Dyn1 is an efficient curvature generator, whereas Dyn2 is primarily a curvature sensor. Using Dyn1/Dyn2 chimeras, we identified the lipid-binding pleckstrin homology domain as being responsible for the differential in vitro properties of these two isoforms. Remarkably, their in vitro activities were reversed by a single amino acid change in the membrane-binding variable loop 3. Reconstitution of KO mouse embryo fibroblasts showed that both the pleckstrin homology and the Pro/Arg-rich domains determine the differential abilities of these two isoforms to support CME. These domains are specific to classical dynamins and are involved in regulating their activity. Our findings reveal opportunities for fundamental differences in the regulation of Dyn1, which mediates rapid endocytosis at the synapse, vs. Dyn2, which regulates early and late events in CME in nonneuronal cells.
Keywords: synaptic vesicle recycling, membrane remodeling, curvature generation, protein–membrane interactions
The large atypical GTPase dynamin plays a dual role in clathrin-mediated endocytosis (CME) (1). In nonneuronal cells, dynamin is recruited to nascent clathrin-coated pits (CCPs) (2, 3), where it functions in early rate-limiting stages to monitor the maturation of productive CCPs and the turnover of abortive CCPs (4, 5). At late stages, a burst of dynamin recruitment (6) and self-assembly into collar-like structures at the necks of deeply invaginated CCPs positions dynamin to directly catalyze membrane fission and clathrin-coated vesicle (CCV) release (1, 7, 8).
Caenorhabditis elegans and Drosophila express only a single dynamin isoform, whereas mammals encode three isoforms, each of which is expressed as different splice variants (8). The first identified and most studied isoform, dynamin 1 (Dyn1), is primarily expressed in neurons and is specifically required for rapid endocytosis after synaptic vesicle release (9). Dyn2 is ubiquitously expressed and required for CME in nonneuronal cells (10). Previous overexpression studies showed that dominant negative mutants of either isoform inhibit CME in nonneuronal cells and led to the suggestion that they were functionally redundant (11). However, more recent reconstitution studies in neurons from Dyn1 KO mice (9) or conditional Dyn2 KO mouse fibroblasts (10) showed that Dyn1 and Dyn2 were not fully functionally redundant. Thus, despite sharing 79% sequence identity, Dyn2 could only weakly rescue the specific defect in rapid synaptic vesicle uptake in the neuron (9), whereas Dyn1 was less effective than Dyn2 at supporting CME in Dyn2 null mouse fibroblasts (10). These reciprocal findings suggest a more fundamental mechanistic difference between these two isoforms. The explanation for these differential activities and their significance remains unknown.
Dynamin consists of five functionally defined domains (1, 7, 12), three of which are conserved among all dynamin-related family members. These include the N-terminal G domain, which mediates GTP hydrolysis, and a middle domain and GTPase effector domain (GED) that together form an α-helical stalk involved in quaternary structure and self-assembly. Two domains that are specific to classical dynamins are the pleckstrin homology (PH) domain, which mediates lipid interactions, and the C-terminal Pro/Arg domain (PRD), which mediates interactions with dynamin's SH3 domain-containing partners. Each of these domains is conserved between classical dynamins, with the greatest variation occurring in the PRD.
A number of assays have been developed to individually follow the different biochemical activities of dynamin. These include assays for basal and assembly-stimulated GTPase activities on liposomes (13) and preformed lipid nanotubes (14), fluorescence-based assays for membrane binding and self-assembly on lipid templates (15, 16), assays for curvature generation and deformation of liposomes (17, 18), and assays for dynamin-catalyzed membrane fission, either from planar-supported bilayers with excess membrane reservoir (SUPER templates) or membrane tubules drawn from these templates (18, 19). To date, most of the biochemical characterization has been directed to Dyn1, and only a few studies have used the ubiquitously expressed isoform Dyn2 (20–22). Fewer still have directly compared Dyn1 and Dyn2 with respect to their biochemical properties (21, 22). In these reports, Dyn2 was found to have a higher propensity to self-assemble and therefore, showed higher basal and assembly-stimulated GTPase activities. However, both studies measured GTP hydrolysis under low-salt conditions and/or with microtubules as substrates. None of the many activities of Dyn1 and Dyn2 on membrane templates, including binding, self-assembly, and catalysis of membrane fission, have been compared under physiological conditions. Therefore, equipped with an established toolset to directly measure multiple aspects of dynamin's in vitro activities, we have revisited the comparison of Dyn1 and Dyn2 seeking new insight into their isoform-specific in vivo activities.
Results
Dyn1 but Not Dyn2 Mediates Fission from SUPER Templates in Vitro.
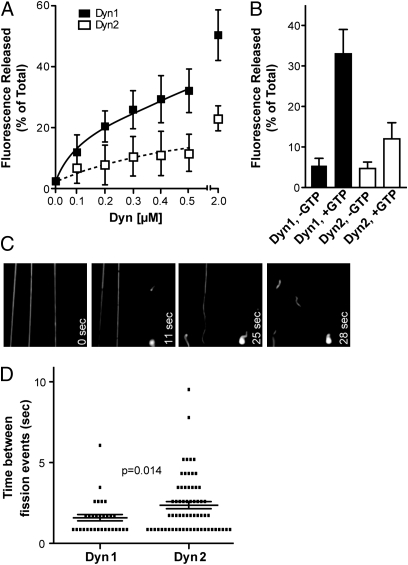
We recently showed that Dyn1 alone was sufficient to catalyze membrane fission and vesicle release from SUPER templates (18). SUPER templates are formed by depositing membrane bilayers onto 5-μm silica beads under conditions that produce an excess membrane reservoir available for budding events (18, 23). By incorporating trace amounts of Rhodamine phosphatidylethanolamine (RhPE) into the bilayer, membrane fission can be measured as the release of fluorescently labeled vesicles into the supernatant after sedimentation. As previously shown, when incubated with SUPER templates in the presence of GTP, Dyn1 was able to catalyze membrane fission and vesicle release (Fig. 1 A and B). However, Dyn2 exhibited approximately fourfold reduced ability under these conditions (Fig. 1 A and B). This finding was unexpected given that Dyn2 supports CME and presumably, membrane fission at CCPs in nonneuronal cells more effectively than Dyn1 (10). Therefore, we tested whether Dyn2 could execute the scission of a membrane tether, which more closely resembles the narrow neck of a CCP. In contrast with the inability to mediate fission from the planar SUPER templates, Dyn2 could catalyze fission on these curved membrane tethers (Fig. 1_C_ and Movie S1), albeit at a slightly but significantly slower rate than Dyn1 (Fig. 1_D_).
Fig. 1.
Differential ability of Dyn1 and Dyn2 to release vesicles from planar membranes. (A) The indicated concentrations of Dyn1 or Dyn2 were incubated with SUPER templates for 30 min at room temperature. Membrane fission was measured by the release of fluorescently labeled vesicles into the supernatant after sedimentation of the SUPER templates (shown are averages ± SD, n = 5). (B) Average fission activity of 0.5 μM Dyn1 or Dyn2 (n ≥ 21). (C) Time-lapse images showing fission activity of Dyn2 on membrane tethers drawn from SUPER templates and imaged in the presence of an oxygen scavenger system. Dyn2 was added at a final concentration of 0.5 μM, and images were taken in 1.4-s intervals (Movie S1). (D) Quantification of fission activity of Dyn1 and Dyn2 with membrane tethers as templates. Dyn1 or Dyn2 was added at a final concentration of 0.5 μM, and movies were taken with 0.866-s time-lapse intervals. In an attempt to quantify fission activity, the time between fission events on an individual tether was determined. Data are presented as a scattered plot and were analyzed for significance with a Mann–Whitney Test using PRISM (Graphpad) statistical software.
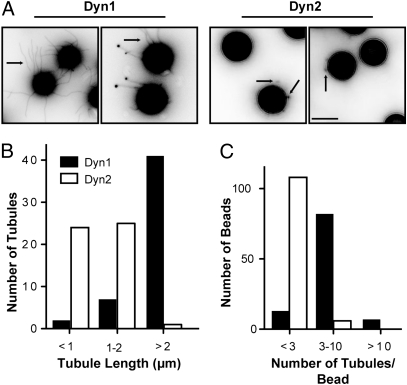
The ability of Dyn2 to mediate fission on membrane tethers but not from SUPER templates is reminiscent of the behavior of a Dyn1 mutant, I533A, that disrupts the hydrophobic character of the variable loop 1 (VL1) in the PH domain (19). VL1 makes a shallow insertion into the lipid bilayer (15) that is necessary for efficient curvature generation. We, therefore, compared the curvature-generating activities of Dyn1 and Dyn2 by incubating them with SUPER templates in the absence of GTP. As previously shown (18, 19), Dyn1 could generate long membrane tubules under these conditions. In contrast, incubation with Dyn2 produced much shorter and many fewer tubules (Fig. 2_A_); these differences were quantified in Fig. 2 B and C, respectively.
Fig. 2.
Differential ability of Dyn1 and Dyn2 to generate curvature from planar membranes. (A) Tubulation of SUPER templates. Dyn1 or Dyn2 (0.5 μM) were incubated with SUPER templates for 10 min at room temperature in the absence of GTP and imaged in the presence of an oxygen scavenger system. Images are inverted in contrast for clarity, and arrows indicate tubules. (Scale bar: 5 μm.) (B) Quantification of tubule length. Length of tubules was determined using ImageJ (National Institutes of Health; n > 50 tubules). (C) Quantification of number of tubules per bead (n > 100 beads).
Dyn2 Activity Is Highly Sensitive to Membrane Curvature.
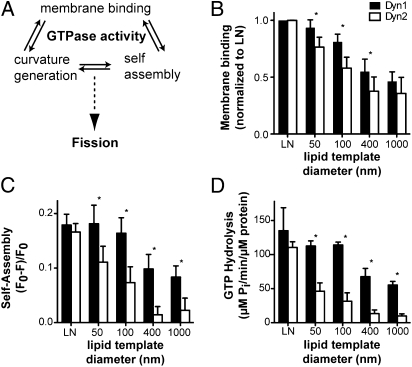
Structure function analyses of a series of Dyn1 mutants have revealed that its ability to catalyze membrane fission depends on its interdependent abilities to (i) bind and self-assemble onto membranes, (ii) generate high curvature on these membranes, and (iii) hydrolyze GTP (Fig. 3_A_) (18, 19). To gain further insights into their differential ability to catalyze membrane fission, we subjected Dyn1 and Dyn2 to a series of assays to directly measure these specific activities (Methods). In addition, given the ability of Dyn2 to catalyze fission on membrane tubules but not planar templates, we compared the curvature sensitivities of these two isoforms by using highly curved lipid nanotubes (∼30 nm) or liposomes of different diameters (50–1,000 nm) and hence, different membrane curvatures as templates.
Fig. 3.
Differential curvature sensitivity of Dyn1 and Dyn2 on lipid templates. (A) The assembly-stimulated GTPase activity of dynamin requires its ability to bind membranes, self-assemble, and generate curvature. Although these activities are interdependent, each can be directly measured. All four of these activities combine and are required for dynamin-catalyzed membrane fission. (B) Membrane binding activity of Dyn1 and Dyn2. Dyn1-G532C-NBD and Dyn2-S532C-NBD were mixed with WT Dyn1 and Dyn2 at a ratio of 1:10 and incubated at room temperature with lipid templates of different curvature (LN, lipid nanotubes, estimated at 30 nm diameter) for 10 min to achieve steady-state binding in the absence of nucleotide. The fold change of fluorescence at 530 nm was measured and normalized to the fold change observed with lipid nanotubes. Data are shown as averages ± SD (n = 9). (C) Self-assembly of Dyn1 and Dyn2 on lipid templates for 10 min to achieve steady-state assembly. Dyn1-BODIPY and Dyn2-S702C-BODIPY were incubated with lipid templates of different curvature in the absence of nucleotide at room temperature, and the assembly-dependent quenching of BODIPY fluorescence was monitored at 510 nm. Data are shown as averages ± SD (n = 5). (D) Lipid-stimulated GTPase activity of Dyn1 and Dyn2. Dynamin (0.5 μM) was incubated with lipid templates of different curvature in the presence of 1 mM GTP at 37 °C. Released Pi was determined using a colorimetric malachite green assay. Data are shown as averages ± SD (n = 4). *P ≤ 0.01
We first used a fluorescence-based assay that measures the binding of nitrobenz-2-oxa-1,3-diazol-4-yl (NBD) -labeled dynamin to lipid templates (15). Direct sedimentation analyses established that the degree of binding of Dyn1 and Dyn2 to nanotubes was indistinguishable (Fig. S1). However, the fold increase in NBD fluorescence after membrane binding differed between Dyn1 and Dyn2 (nine- vs. sixfold, respectively); therefore, the data in Fig. 3_B_ were normalized to the maximum fluorescence obtained on reaching steady-state binding to nanotubes. As previously reported for Dyn1 (15, 24), both isoforms exhibited curvature sensitivity and bound to a greater extent to nanotubes and small liposomes than larger-diameter liposomes (Fig. 3_B_). However, the extent of Dyn2 binding to liposomes of intermediate size (50–400 nm in diameter) was significantly less than Dyn1, suggesting greater curvature sensitivity. This isoform-specific difference was much more pronounced when we directly measured self-assembly by fluorescence self-quenching of BODIPY-labeled dynamin. Although the extent of Dyn2 self-assembly on nanotubes was equal to that of Dyn1 (Fig. 3_C_), its ability to self-assemble on liposomes was strongly reduced and indeed, was barely detectable above background for liposomes ≥400 nm in diameter.
Contrary to previous studies performed under low-salt conditions, the basal GTPase activities of Dyn1 and Dyn2 measured at physiological salt concentrations and in the absence of membranes were indistinguishable (1.03 ± 0.08 μM Pi released/min/μM protein for Dyn1 and 0.98 ± 0.09 μM Pi released/min/μM protein for Dyn2, average ± SD, n ≥ 4), confirming that the underlying mechanisms of GTP hydrolysis are conserved (25). Similarly, when measured on lipid nanotubes, the assembly-stimulated GTPase activities of Dyn1 and Dyn2 were not significantly different (Fig. 3_D_), consistent with their equivalent abilities to bind and assemble on these templates. However, when assayed in the presence of progressively larger diameter liposomes, Dyn2 exhibited much greater curvature dependence than Dyn1 (Fig. 3_D_). Dyn1 was stimulated equally by lipid templates ≤100 nm in diameter, whereas Dyn2 was >70% less active on 100-nm liposomes. Although Dyn1 exhibited ∼50% reduced activity when assayed in the presence of liposomes ≥400 nm in diameter, as previously reported (26), the assembly-stimulated GTPase activity of Dyn2 was reduced by ∼10-fold (Fig. 3_D_). The differential curvature sensitivity of Dyn2 vs. Dyn1 closely paralleled that observed for self-assembly (Fig. 3_C_), but it was sharper than that observed for membrane binding (Fig. 3_B_). This is consistent with evidence that dynamin–dynamin interactions and not simply membrane binding are essential for stimulated GTPase activity (14, 25).
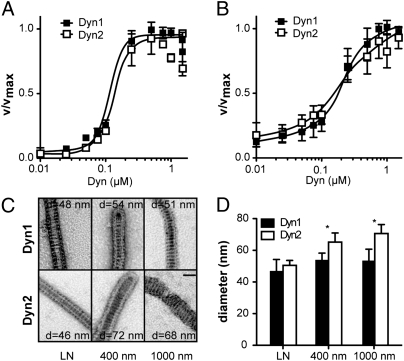
Recent structural studies have shown that dynamin's assembly-stimulated GTPase activity requires the dimerization of GTPase domains that are aligned across adjacent rungs of assembled dynamin spirals (25). This dimerization, in turn, requires membrane binding and curvature generation to correctly position adjacent dynamin molecules for efficient catalysis, and it is reflected in the high degree of cooperation in assembly-stimulated GTPase activity (14). The reduced activities of Dyn2 on large-diameter lipid templates could reflect either differences in the nature of dynamin–membrane interactions (hence, its ability to generate membrane curvature) or differences in dynamin–dynamin interactions required for self-assembly. To distinguish these possibilities, we compared the cooperative behavior of Dyn1 and Dyn2 after normalizing their GTPase activities to the maximal activity observed. When lipid nanotubes were used as membrane templates, both isoforms showed a similarly high cooperative behavior (Fig. 4_A_). In the presence of 100-nm liposomes, although Dyn2 exhibited significantly (P < 10−4) impaired activity relative to Dyn1 (Fig. 3_D_), the cooperative behavior of the two isoforms was statistically insignificant (Fig. 4_B_ and Methods). The very low activity of Dyn2 on larger-diameter liposomes precluded us from using these liposomes for comparison. Nevertheless, these data suggest that the two isoforms are not significantly different with respect to their propensity for intermolecular interactions. Instead, we speculate that the reduced assembly-stimulated GTPase activity of Dyn2 reflects its reduced ability to generate curvature and thus, create the optimized register of assembled molecules for efficient catalysis.
Fig. 4.
Differential abilities of Dyn1 and Dyn2 to self-assemble and generate curvature on lipid templates. Concentration dependence and cooperative behavior of Dyn1 and Dyn2 GTPase activity on (A) nanotubes or (B) 100-nm liposomes are shown. Data (average ± SD, n = 3) are normalized to maximum activity. (C) Electron micrographs of Dyn1 and Dyn2 assembled onto lipid nanotubes (LN) or 400- or 1,000-nm liposomes as indicated. Dynamin was incubated with lipid templates for 20 min at room temperature, and then, it was adsorbed to a grid and imaged by negative-stain EM. Diameter (d) is in nanometers. (Scale bar: 50 nm.) (D) Quantification of diameter of dynamin-decorated lipid templates (n ≥ 4).
To directly observe whether Dyn2 would assemble on liposomes, we performed EM using membrane templates of different curvature (Fig. 4_C_). As expected for the similar biochemical activities of Dyn1 and Dyn2 on nanotubes, both isoforms assembled efficiently onto these curved membrane templates, yielding decorated tubes of 47 ± 7 nm and 51 ± 3 nm, respectively. As previously shown (17), Dyn1 was also able to bind and assemble onto 400- or 1,000-nm liposomes to generate decorated membrane tubules with nearly identical diameters (Fig. 4 C and D). By contrast, Dyn2-coated tubules were less frequently observed after incubation with either 400- or 1,000-nm liposomes, and instead, we observed large clusters of decorated tubules on the grid. Moreover, when present, the diameters of the assembled tubules were larger than for Dyn1 and varied with the size of the original liposome (Fig. 4_D_). Together, these data establish that the ability of Dyn2 to self-assemble and generate curvature is reduced relative to Dyn1, and consequently, the ubiquitously expressed Dyn2 isoform exhibits significantly greater curvature sensitivity than the neuronal-specific Dyn1.
PH Domain of Dyn2 Confers Curvature Selectivity.
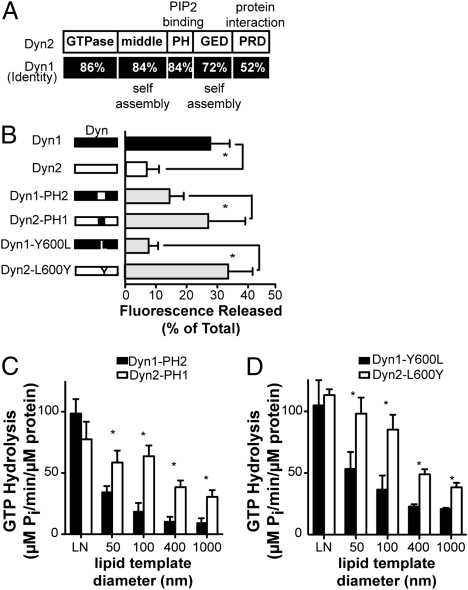
To identify which domain(s) might confer these isoform-specific activities, we generated a series of Dyn1/2 domain-swap chimeras (Fig. 5_A_ and Fig. S2_A_). The highest degree of sequence divergence between these two isoforms occurs within the PRD, but this domain is not required for assembly-stimulated GTPase activity (27) or dynamin-catalyzed membrane fission (Fig. S3_A_). Previous studies had shown that the GTPase domain confers isoform-specific signaling functions on Dyn2, but these functions were independent of its function in CME (28). Moreover, given that the basal and assembly-stimulated GTPase activities (at least on lipid nanotubes) of Dyn1 and Dyn2 were indistinguishable, we considered it unlikely that this domain accounted for their differential activities. The similar cooperative behavior of Dyn1 and Dyn2 also suggested that the middle domain and GED, which mediate intermolecular interactions, were also unlikely candidates. Rather, we were struck by the functional similarities between Dyn2 and the I533A mutant in the VL1 loop of Dyn1 (19). Therefore, we first tested the involvement of the PH domain by generating the domain-swap chimeras Dyn1-PH2 and Dyn2-PH1 and measuring their fission and GTPase activities. Strikingly, Dyn2-PH1 acquired the ability to release vesicles from SUPER templates, whereas Dyn1-PH2 showed a decrease of fission activity to levels nearer to Dyn2 (Fig. 5_B_). Similarly, the assembly-stimulated GTPase activities of Dyn1-PH2 and Dyn2-PH1 showed swapped curvature dependence compared with their respective WTs (Fig. 5_C_ compared with Fig. 3_D_). From these data, we conclude that the PH domain is primarily responsible for the differential selectivity of Dyn2 to highly curved membranes.
Fig. 5.
A tyrosine residue in the PH domain VL3 confers isoform-specific curvature sensitivity and membrane fission activity. (A) Domain structure of Dyn1 and Dyn2 and sequence identity between both isoforms. Dyn1-PH2 and Dyn2-PH1 chimeras, as illustrated in B, were generated by seamless cloning (Fig. S2_A_). (B) Fission activity of parent chimeric Dyn1 and Dyn2 or single amino acid-substituted Dyn1 and Dyn2 (0.5 μM), as indicated, was determined after incubation with SUPER templates by sedimentation and release of fluorescent vesicles into the supernatant. Data shown are averages ± SD (n ≥ 16, *P ≤ 0.001). Fluorescence released in the absence of GTP was subtracted as a background. (C and D) Stimulated GTPase activity of chimeric dynamins (C) or single amino acid substitution at position 600 in VL3 (D) is shown. Dynamins (0.5 μM) were incubated with lipid templates of different curvature, and released Pi was determined using a colorimetric malachite green assay. Data are shown as averages ± SD (n = 3, *P < 0.02).
Previously, it was shown that rapid, compensatory endocytosis could be potently inhibited by microinjection of the PH domain from Dyn1 but not Dyn2 (29). Mutation of only two nonconserved amino acids (S532 and L533) in the Dyn2 PH domain VL1 to match those in Dyn1 (G532 and I533) was sufficient to confer inhibitory properties on the Dyn2 PH domain. To determine whether these residues were sufficient to confer the isoform-specific behaviors on Dyn1 and Dyn2, we generated the corresponding mutants Dyn1-SL and Dyn2-GI and examined their in vitro properties. The activities of these proteins were indistinguishable from their WT parents (Fig. S3_B_). Therefore, despite its importance in curvature generation (19), differences in the VL1 between Dyn1 and Dyn2 were not sufficient to account for the differential in vitro activities of these two isoforms. Alignment of the PH domain from human, mouse, and rat Dyn1 and Dyn2 revealed a third conserved variation within VL3, Dyn1-Y600 and Dyn2-L600 (Fig. S2_B_). Changing this single amino acid was sufficient to reverse the fission activities of Dyn1 and Dyn2 (Fig. 5_B_) as well as the curvature sensitivity for their assembly-stimulated GTPase activities (Fig. 5_D_).
Differential Dyn1 and Dyn2 Activities in Cultured Fibroblasts.
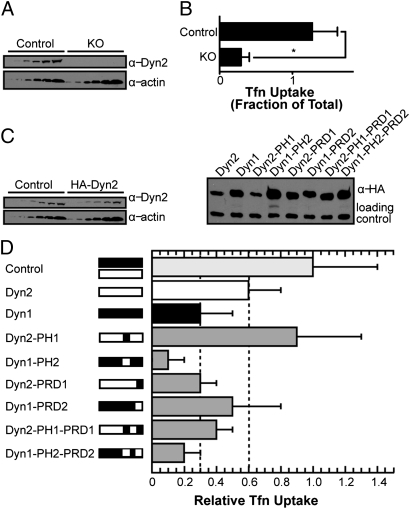
We have shown that Dyn2 has a reduced ability to assemble onto and catalyze vesicle formation from planar templates in vitro because of its reduced ability to generate membrane curvature. Paradoxically, we previously showed that Dyn2 was more effective at supporting CME than Dyn1 (10). To better understand which properties of Dyn2 might be responsible for the differential activities in vivo, we compared the abilities of Dyn1 and Dyn2 and their chimeras to support CME in fibroblasts. For these experiments, we used a recently generated mouse embryonic fibroblast (MEF) cell line encoding a conditional KO for both Dyn1 and Dyn2 (30). Induction of Cre recombinase in these cells leads to efficient excision and KO of both Dyn2 (Fig. 6_A_) and Dyn1 (30) and inhibits CME by >75% as measured by uptake of biotinylated transferrin (b-Tfn) (Fig. 6_B_). MEFs were reconstituted using bicistronic retroviral expression constructs encoding the various HA-tagged dynamin constructs and GFP driven by an internal ribosome entry site (IRES). FACS of GFP expression was used to select cells that express low levels of exogenous dynamin (Fig. 6_C_ Left). Control cells were infected with retroviruses expressing only GFP and also subjected to FACS sorting.
Fig. 6.
The PH domain and the PRD confer isoform-specific activities in vivo. (A) Conditional Dyn1/Dyn2 null mouse embryo fibroblasts were treated with 4-OHT for 4 d to induce Cre recombinase for KO of endogenous dynamins. Control cells are Dyn1/2-conditional null MEFs reconstituted with retroviruses expressing GFP only and not treated with 4-OHT. Western blot analysis of Dyn2 expression levels before (Control) and after (KO) 4-OHT treatment. Different amounts of cell lysates (1×, 2×, 5×, 10×, 20×, and 50× from left to right per cell line) were loaded and probed using an antibody against Dyn2. A representative blot is shown, and actin serves as a loading control. (B) CME activity was measured by internalization of b-Tfn for 5 min at 37 °C as determined by ELISA and is expressed relative to total surface bound. Data shown are average ± SD (n = 8, *P < 0.05). (C) Western blot analysis of the expression of HA-tagged dynamin proteins in conditional null MEFs after KO of endogenous dynamins. For quantification, see Table 1. Loading control is a background band stained by the HA antibody. (Left) Western blot of control cells and HA-Dyn2 reconstituted cells (different amounts of cell lysate are loaded in each lane). (Right) Western blot of cells reconstituted with indicated dynamins (same amounts of cell lysate are loaded in each lane). (D) Clathrin-mediated endocytosis in control or KO cells expressing indicated dynamin constructs. Uptake of b-Tfn was measured as described above and then normalized for different expression levels of the dynamin constructs to obtain a measure of relative Tfn uptake activity. Data are shown as averages ± SD of n ≥ 3 experiments (P < 0.05 compared with Dyn2).
Dyn1 can be highly overexpressed in cultured cells without affecting its viability; however, Dyn2 was shown to be cytotoxic when overexpressed (28, 31). Consequently, despite our best efforts to select cell lines with comparable levels of dynamin expression, we found that, after expansion for experiments (typically 2–3 wk), cells consistently expressed more HA-Dyn1 and its derivatives than HA-Dyn2 (Fig. 6_C_ Right). The differing expression levels in these selected populations likely reflect adaptation to the cytotoxic effects of Dyn2 as well as the need for higher levels of Dyn1 expression to support CME (10). Therefore, to more directly compare the abilities of these different dynamin variants to support CME, we normalized measured values for the rate of Tfn uptake to the expression level of the various constructs, as determined by quantitative Western blotting, relative to that of HA-Dyn2 (Table 1). We term this measurement relative Tfn uptake. By these criteria, cells that were reconstituted with Dyn2 exhibited ∼60% relative Tfn uptake compared with control (Fig. 6_D_ and Table 1). The inability to fully reconstitute CME in these double KO cells compared with previous studies with Dyn2 KO cells (10) likely reflects the complete KO of both dynamin isoforms, which might reduce the ability to reconstitute activity with a single splice variant of dynamin. Consistent with previous results (10), the relative Tfn uptake activity of KO cells reconstituted with Dyn1 was significantly lower than that of Dyn2-reconstituted cells (Fig. 6_D_), although at its higher level of expression (Fig. 6_C_ Right), HA-Dyn1 could support Tfn uptake (Table 1) as previously shown (10). Notably, normalization assumes that CME efficiency will be proportional to dynamin expression levels. Indeed, although this relationship is unlikely to be linear, we previously showed that, at low levels of expression, HA-Dyn1 only weakly supports CME in Dyn2 conditional null fibroblasts but that at higher levels, it was equally able to support CME (10). Importantly, the relative activities of the various chimeric molecules studied here were qualitatively similar in comparing either the normalized (Fig. 6_D_) or raw data (Table 1).
Table 1.
Quantification of dynamin expression and relative Tfn uptake
| Dynamin | Tfn uptake a (fraction of total) | Fold overexpression b (relative to HA-Dyn2) | Relative Tfn uptake a/b |
|---|---|---|---|
| Control | 1.3 ± 0.4 | 1.3 ± 0.1 | 1.0 ± 0.4 |
| KO | 0.3 ± 0.1 | n.a. | n.a. |
| Dyn2 | 0.6 ± 0.3 | 1.0 ± 0.0 | 0.6 ± 0.2 |
| Dyn1 | 0.6 ± 0.2 | 2.2 ± 0.9 | 0.3 ± 0.2 |
| Dyn2-PH1 | 0.8 ± 0.3 | 0.9 ± 0.1 | 0.9 ± 0.4 |
| Dyn1-PH2 | 0.2 ± 0.1 | 3.2 ± 1.9 | 0.1 ± 0.1 |
| Dyn2-PRD1 | 0.5 ± 0.1 | 1.7 ± 0.4 | 0.3 ± 0.1 |
| Dyn1-PRD2 | 1.0 ± 0.2 | 2.0 ± 0.9 | 0.5 ± 0.3 |
| Dyn2-PH1-PRD1 | 0.7 ± 0.1 | 1.9 ± 0.3 | 0.4 ± 0.1 |
| Dyn1-PH2-PRD2 | 0.4 ± 0.1 | 2.1 ± 0.7 | 0.2 ± 0.1 |
PH Domain Contributes to Dynamin Activities in MEFs.
We next tested whether the PH domain swaps between dynamin isoforms would be sufficient to reverse their abilities to reconstitute CME. Cells expressing the Dyn2-PH1 chimera exhibited higher, albeit more variable, relative Tfn uptake than the parent Dyn2. Conversely, cells expressing the Dyn1-PH2 chimera exhibited significantly lower relative Tfn uptake than the parent Dyn1 (Fig. 6_D_ and Table 1). These in vivo results are consistent with our in vitro finding that the PH domain of Dyn1 confers the ability to more efficiently mediate vesicle formation than the Dyn2 PH domain. These results, however, do not explain why Dyn2 more effectively supports CME than Dyn1.
Regulation of Dynamin Through the PRD Domain.
The greatest sequence divergence between Dyn1 and Dyn2 is in the PRD (Fig. 5_A_), which is required for targeting dynamin to CCPs (32). We have previously shown that Dyn1 was less effectively targeted to CCPs than Dyn2 (10). The differential extent of localization to CCPs was fully reversed by exchanging PRDs in that Dyn1-PRD2 chimera was localized more effectively to CCPs than Dyn2-PRD1 (Fig. S4). We next compared the abilities of these chimeras to support CME and found that replacing the Dyn2 PRD with that from Dyn1 significantly decreased its relative Tfn uptake activity (Fig. 6_D_ and Table 1). Conversely, Dyn1-PRD2 was significantly more effective at supporting Tfn uptake than Dyn1 (Fig. 6_D_). These data suggest that the differential in vivo activities of the two isoforms are, in part, because of differential targeting to CCPs.
Finally, to test if there were synergistic effects of these two domains, we generated chimeras swapping both the PH domain and the PRD. Once again, the chimera bearing the Dyn1 PH domain was more active than its counterpart bearing the Dyn2 PH domain (Fig. 6_D_) (compare Dyn2-PH1-PRD1 with Dyn2-PRD1 and Dyn1-PRD2 with Dyn1-PH2-PRD2). The chimera bearing the Dyn2 PRD was more active than its counterpart bearing the Dyn1 PRD (compare Dyn1-PH2-PRD2 with Dyn1-PH2 and Dyn2-PH1 with Dyn2-PH1-PRD1). Thus, these data confirm that the PH domain of Dyn2 negatively affects dynamin activity in MEFs, whereas the PRD of Dyn2 positively affects dynamin activity. Because neither of these double-swap chimeras were as active as Dyn2 in supporting CME, these data also suggest that other regions of dynamin are important for isoform-differential activities in vivo.
Discussion
Dyn1 Is a More Potent Curvature Generator, Whereas Dyn2 Is a More Effective Curvature Sensor.
Despite their high degree of conservation, we have identified unexpected quantitative differences in the biochemical activities of the neuronal-specific isoform Dyn1 and the ubiquitously expressed isoform Dyn2. Most notably, Dyn1 can mediate vesicle formation from SUPER templates (18), whereas Dyn2 has little activity. Through the use of multiple assays that directly and quantitatively measure distinct in vitro activities of dynamin, including basal and assembly-stimulated GTPase activities, membrane binding, self-assembly, curvature generation, and curvature-sensing abilities, we were able to identify the exact nature of the biochemical difference. Namely, Dyn1 can efficiently generate curvature on a variety of membrane templates, whereas the activity of Dyn2 shows much greater sensitivity to curvature. Analysis of chimeric Dyn1/2 proteins established that the PH domain was sufficient to confer the distinct fission and curvature-sensing activities of the two isoforms. Strikingly, these activities could be attributed to a single amino acid substitution (tyrosine to leucine) in the membrane-interacting VL3 of the PH domain.
PH Domain Negatively Affects Dyn2 Function in Vivo.
Paradoxically, although Dyn1 is much more effective than Dyn2 at catalyzing vesicle formation from SUPER templates, Dyn2 is more effective than Dyn1 in reconstituting CME in dynamin-deficient mouse fibroblasts (10). At the synapse, the opposite is true; Dyn1 (or Dyn3) can fully reconstitute rapid synaptic vesicle recycling in neurons isolated from Dyn1 KO mice, whereas Dyn2 is significantly less effective (9). The differential in vivo activity of Dyn2 in fibroblasts is not because of its PH domain. On the contrary, a Dyn2-PH1 chimera was more effective than WT Dyn2, whereas a Dyn1-PH2 chimera was less effective than WT Dyn1. Thus, both in vivo and in vitro, constructs bearing the Dyn1 PH domain are more effective at catalyzing vesicle formation. This suggests that the PH domain of Dyn2 negatively regulates its ability to catalyze membrane fission. A comparison of the sequences of VL3 in the PH domains of Dyn1, Dyn2, and Dyn3 from mouse, rat, and human shows that, in all cases, Dyn1 and Dyn3 encode a tyrosine (Y600 in Dyn1) in the otherwise conserved VL3, whereas Dyn2 isoforms all encode a leucine at this position. Interestingly, the single isoforms expressed in C. elegans and Drosophila also encode a tyrosine at this position. Thus, mammals seem to express an evolutionarily conserved, ubiquitously expressed isoform that exhibits greater curvature sensitivity and weakened ability to catalyze vesicle release from planar membranes than its evolutionary progenitor.
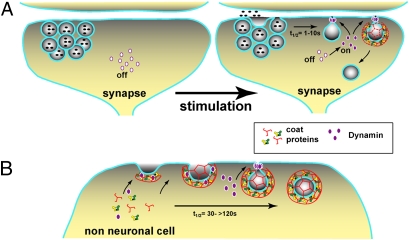
These results beg the questions of why Dyn2 evolved to be more sensitive to membrane curvature than Dyn1 and why the curvature-sensing PH domain regulates dynamin function in vivo. We propose that these results reflect the differential requirements for the regulation of CME in neuronal vs. nonneuronal cells. At synapses after synaptic vesicle release, the potent curvature-generating activity of Dyn1 may be required to quickly capture membrane and pinch off vesicles. The stimulus-induced retrieval of synaptic vesicle contents takes places within 1–10 s; however, the mechanisms and proteins required for this rapid uptake remain a matter of debate (33). Given its in vitro properties, we suggest that Dyn1 can support this fast type of endocytosis, because it can rapidly and efficiently bind, squeeze, and stabilize the necks of nascent pits, perhaps even without the help of accessory proteins or the clathrin coat (Fig. 7_A_). In contrast, in nonneuronal cells, as a curvature sensor, Dyn2 is well-suited for its dual role in CME (1). Dyn2 is efficiently recruited to nascent CCPs through its PRD (32). However, given its biochemical properties, other factors including coat assembly and recruitment of Bin/amphiphysin/Rvs domain-containing proteins would be needed to generate a narrow neck of sufficient curvature to trigger Dyn2 assembly. In this way, Dyn2 is positioned to sense when CCP maturation is complete and mediate membrane fission only after other factors have triggered its assembly switch (Fig. 7_B_).
Fig. 7.
Differential activities of dynamin isoforms at the synapse and in fibroblasts provide opportunities for tissue-specific regulation. (A) Dyn1 is the major isoform at the synapse, and it functions during rapid endocytosis after synaptic vesicle release. Phosphorylation of the PRD inhibits Dyn self-assembly and membrane binding (37). On stimulation by an action potential, Ca2+ influx triggers the release of synaptic vesicles docked at the membrane and activates the phosphatase calcineurin, which in turn, dephosphorylates and activates Dyn1 and other endocytic proteins (44). We propose that Dyn1 can support rapid (_t_1/2 = 1–10 s) membrane fission and retrieval of synaptic vesicle membrane components because of its powerful PH domain, potentially with or without the help of coat proteins. In contrast, Dyn2 in fibroblasts (B) supports a more classical (_t_1/2 = 30 to >120 s) form of endocytosis and is regulated by an assembly switch. Dyn2 is recruited to nascent CCPs through their PRDs, where it can regulate CCP maturation and the formation of a deeply invaginated pit by coat and accessory proteins as illustrated. After the neck is sufficiently narrow, additional Dyn2 is recruited to the membrane through its PH domain, where it can mediate fission. Thus, the differential properties of Dyn1 and Dyn2, as curvature generators or sensors, respectively, are critical for their tissue-specific activities.
Dyn2 PRD Is Required for Targeting Dyn2 to CCPs.
The greatest sequence divergence between Dyn1 and Dyn2 occurs in their PRDs, which are only ∼50% identical. However, they retain their proline-rich basic character, and they both encode multiple binding sites for SH3 domain-containing binding partners. Nonetheless, the PRD of Dyn2 seems to be critically required for its function in fibroblasts. Replacing the Dyn2 PRD with that from Dyn1 significantly reduces the ability of Dyn2 to rescue CME in the Dyn1/Dyn2 KO cells. We previously showed that Dyn2 was more effectively targeted to CCPs than Dyn1 in fibroblasts (10), and indeed, the Dyn1-PRD2 chimera was more active than Dyn1. Live cell total internal reflection fluorescence microscopy in nonneuronal cells has shown that Dyn2-GFP exhibits a longer lifetime at CCPs than Dyn1-GFP (34). Together, these data suggest that Dyn2 binds either specifically or with higher affinity to an SH3 domain-containing binding partner(s) required to target it to CCPs. Whether enhanced interactions with other binding partners are also required for other stages in CME remains to be determined.
PH Domains and PRDs Regulate Dynamin Activity.
Limited proteolysis or deletion studies showed that removal of the PRD decreased, whereas removal of the PH domain increased dynamin's assembly-stimulated GTPase activity in low salt (27, 35). These findings suggested that the PH domain and PRD were negative and positive regulators of dynamin assembly, respectively. Dyn1 is phosphorylated at the synapse, and phosphorylation of the PRD has been shown to inhibit its assembly, membrane binding, and interactions with SH3 domain-containing partners (36, 37). Thus, we propose that Dyn1 might be negatively regulated (i.e., clamped) by phosphorylation at the synapse. This is parallel to the need to clamp components of the exocytic machinery on docked synaptic vesicles for rapid Ca2+-triggered release (38). Calcineurin-dependent dephosphorylation of Dyn1 (39) would release this negative inhibition, allowing it to rapidly capture, squeeze, and pinch off budding membranes. In contrast, in fibroblasts, Dyn2 is recruited to nascent CCPs and has been suggested to play a role in regulating early events in CCP stabilization and maturation (5). We propose that the curvature sensitivity of the PH domain coupled with PRD-mediated interactions of Dyn2 with coated pit components serve as components of an assembly switch that mediates fission only after the deeply invaginated CCP has fully matured and acquired a narrow neck. The differential localization and dynamics of Dyn1-GFP and Dyn2-GFP described above are consistent with this scenario. Interestingly, the SH3 domain-containing proteins that bind dynamin can also positively or negatively regulate its assembly activity (15, 26). Thus, interactions with the PRD might also contribute to differential regulation of these two dynamin isoforms.
A critical role for the PH domain in regulating dynamin function, both in vitro and in vivo, has recently been established by analysis of the biochemical properties of PH domain mutants in Dyn2 that are linked to centronuclear myopathy (CNM), a congenital muscle disease (40). These mutations map to the C-terminal α-helix, which is on the opposite side from the PH domain's PI4,5P2-binding pocket formed by VL1, VL2, and VL3 (41). This helix has been shown in other PH domains to be important for interactions with and regulation of small GTPases (42). The CNM mutations seemed to affect the intramolecular regulation of dynamin, resulting in enhanced basal GTPase activity and enhanced self-assembly without effecting PI4,5P2-binding affinity (41). Kenniston and Lemmon (41) suggested a role for the PH domain in intramolecular interactions that regulate dynamin activity, although the identity of other domain(s) engaged in these intramolecular interactions remains unknown. Because dynamin is the master regulator of endocytosis in mammalian cells, additional work is necessary to define the mechanisms, in turn, that regulate its tissue-specific activities.
Methods
Plasmids and Retroviruses.
Dyn1/2 chimeras were generated by seamless cloning (43) (Fig. S2_A_). For protein expression, dynamin constructs were cloned into pIEX6 (Novagen), resulting in N-terminal His-tagged proteins for expression in Sf9 cells. For reconstitution of Dyn1/2-conditional KO MEFs, N-terminal HA-tagged dynamin was cloned in pMIEG3 (derived from pMSCV; Novagen), resulting in the bicistronic expression of HA-Dyn and GFP (10). Retroviruses were prepared as previously described (10).
Purification and Storage of Dynamin.
Dynamin proteins were expressed in Sf9 cells transiently transfected with various constructs as previously described (19) and purified by affinity chromatography as described previously using GST-tagged amphiphysin-II SH3 domain as an affinity ligand (14). Purified dynamin was dialyzed overnight in 20 mM Hepes (pH 7.5), 150 mM KCl, 1 mM DTT, 1 mM EGTA, and 10% glycerol. Storage in 10% glycerol seemed to be important, because Dyn2 was more prone to aggregation when stored in 50% glycerol. Protein concentration was determined by A280 with εHis-Dyn1 = 56,185 and εHis-Dyn2 = 54,695. Proteins were stored at −80 °C and used within 1 mo.
Preparation of Lipid Templates.
For liposomes, lipid mixtures (DOPC:DOPS:PIP2 at 80:15:5) were dried, rehydrated in buffer [20 mM Hepes (pH 7.5), 150 mM KCl] to a final concentration of 1 mM, and subjected to a series of freeze–thaw cycles before extrusion through polycarbonate membranes (Whatman) with a pore size ranging from 50 to 1,000 nm using an Avanti Mini-Extruder. Lipid nanotubes composed of DOPC:DOPS:PIP2:GalCer (40:15:5:40) were generated according to procedures described previously (13, 14). SUPER templates were generated as previously reported (18, 23); all solutions for their preparation were filtered through a 0.22-μm filter, and incubations were performed in low-adhesion microcentrifuge tubes (USA Scientific). Briefly, 5 × 106 silica beads (silicon oxide microspheres, d = 4.97 μm; Corpuscular) were incubated with 20 nmol 100-nm liposomes (DOPC:DOPS:PIP2:RhoPE at 79:15:5:1) in 20 mM Hepes (pH 7.5) and 600 mM NaCl in a final volume of 100 μL. The mix was incubated for 30 min at room temperature. To wash the templates, 1 mL water was added, and tubes were centrifuged in a swing-out centrifuge rotor (Allegra 6R Centrifuge; Beckman); 1 mL supernatant was removed by pipetting, and the residual 100-μL reaction was mixed with 1 mL water by gentle vortexing (medium speed) and washing four times.
Fission Activity of Dynamin.
All experiments were performed in low-adhesion microcentrifuge tubes (USA Scientific). For fission activities, 5 × 105 SUPER templates were added to 100 μL buffer containing 20 mM Hepes (pH 7.5), 150 mM KCl, 1 mM MgCl2, 1 mM GTP, and indicated concentrations of dynamin for 30 min at room temperature. During this incubation, templates were allowed to settle without additional mixing. Templates were pelleted at 260 × g in a swing-out centrifuge rotor (Allegra 6R Centrifuge; Beckman), and 75 μL supernatant was mixed with 25 μL 0.4% Triton X-100 to dissolve vesicles. Total fluorescence of templates was determined in a separate reaction containing 5 × 105 SUPER templates in 0.1% Triton X-100. Fluorescence was measured in a 96-well fluorescent plate reader (Bio-Tek Instruments ), and fission activity is expressed as percentage of total fluorescence on SUPER templates.
Fluorescent Microscopy of Dynamin–SUPER Template Interactions.
Assays for fission activity on membrane tethers were performed in Lab-Tek chambers (Nunc) that had been preincubated for 20 min with 1% fatty acid-free BSA. These were washed, and 200 μL 20 mM Hepes (pH 7.5), 150 mM KCl, 1 mM MgCl2, and 1 mM GTP with an oxygen scavenger system, which was essential to prevent photo damage of dynamin and lipids and consisted of 50 mg/mL glucose oxidase (Sigma), 10 mg/mL catalase (Roche), 25 mM glucose (Sigma), and 1 mM DTT, was added. To this, we added 5 × 105 SUPER templates and allowed them to settle onto the coverslip. Tethers were generated by the addition of 20-μm glass beads, which were gently rolled over the surface of the SUPER templates through gentle agitation of the Lab-Tek chamber. Fluorescence imaging was carried out at room temperature on an inverted Olympus IX-70 microscope with a 100×, 1.35-NA oil-immersion objective equipped with an ORCA ER CCD camera (Hamamatsu) and 617/73-nm emission filters for RhPE-labeled membranes. Dynamin was added to a final concentration of 0.5 μM to the corner and allowed to diffuse across the chamber. Curvature generation experiments were executed as above except that SUPER templates were allowed to settle in a buffer containing 0.5 μM dynamin without GTP. Pictures were taken after an incubation of 10 min at room temperature.
Fluorescence-Based Assays for Membrane Binding and Self-Assembly.
To determine membrane binding, NBD-labeled reactive-Cys-less (RCL) -Dyn1 G532C and RCL-Dyn2 S532C were generated as previously reported (15). For measurements, 0.05 μM NBD-labeled dynamin was mixed with 0.45 μM unlabeled protein in buffer containing 20 mM Hepes (pH 7.5), 150 mM KCl, and 2 mM MgCl2. The solution was excited at 470 nm, and emission intensity was measured at 530 nm. After initial fluorescence (_F_0) was established, lipid template (50 μM) was added to the solution, and maximum fluorescence intensity was taken after incubation for 10 min at room temperature. Membrane binding is expressed as fold difference of fluorescence and normalized to values observed for lipid nanotubes.
For self-assembly, BODIPY (BODIPY-FL C1-IA) -labeled Dyn1 was generated as previously reported (15). Labeling preferentially occurs in the GED of dynamin at Cys-708; however, this residue is absent in Dyn2. Therefore, the corresponding residue in Dyn2 Ser702 was mutated to a Cysteine for subsequent labeling. Labeled dynamin (0.5 μM) was incubated with buffer containing 20 mM Hepes (pH 7.5), 150 mM KCl, and 2 mM MgCl2. BODIPY fluorescence was excited at 490 nm, and emission was measured at 510 nm to establish _F_0. For self-assembly, lipid template (50 μM) was added to the solution, and fluorescence intensity (F) was followed for 10 min at room temperature. Some quenching occurred in the absence of liposomes and was subtracted as background. Quenching is expressed as (_F_0 − F)/_F_0.
GTPase Activity.
GTP hydrolysis by dynamin was measured as a function of time using a colorimetric malachite green assay that detects the release of inorganic phosphate (13). Briefly, 0.5 μM dynamin or different concentrations of dynamin as indicated were incubated without (basal) or with (assembly-stimulated) lipid templates (150 μM) of different curvature in a buffer containing 20 mM Hepes (pH 7.5), 150 mM KCl, 1 mM GTP, and 2 mM MgCl2. Aliquots were taken at several time points, free phosphate was determined using malachite green, and rates of hydrolysis were calculated. To determine whether the two isoforms exhibited different cooperative behavior, we measured assembly-stimulated GTPase activity of varying concentrations of dynamin on lipid nanotubes or 100-nm liposomes. To determine significance, we plotted log(vol/volmax − v) vs. log[Dyn] to obtain a linear plot. The slopes for Dyn1 and Dyn2 were not significantly different (P > 0.5) when measured either on lipid nanotubes or 100-nm liposomes.
EM.
EM was done as previously described (19). Briefly, 1 μM Dyn1 or Dyn2 was incubated with 25 μM liposomes of 1,000 or 400 nm or lipid nanotubes in 20 mM Hepes (pH 7.5) and 150 mM KCl for 30 min, and subsequently, it was adsorbed to carbon-coated grids and stained with 1% uranyl acetate.
Reconstitution of Dyn1/2 KO cells and Tfn Uptake Assays.
Dyn1 and Dyn2 conditional KO mouse embryonic fibroblasts were provided by Pietro de Camilli (Yale University, New Haven, CT) (30). Cells were cultured in DMEM containing 10% FBS and 10 mM Hepes (pH 7.4). Cells were infected with retrovirus expressing the respective HA-dynamin protein and GFP from a bicistronic vector. Typically, 4 d after infection, cells were sorted by FACS for very low GFP signal to obtain cells expressing low levels of dynamin. KO of endogenous dynamin was induced by the addition of 3 μM 4-hydroxytamoxifen (4-OHT) to the culture medium for 2 d and subsequent incubation with 0.3 μM tamoxifen for an additional 2 d. Expression levels were determined by immunoblotting against the HA epitope using the 12CA5 antibody purified from hybridomas obtained from Ian Wilson (The Scripps Research Institute, La Jolla, CA) or against Dyn2 using sc-6400 from Santa Cruz Biotechnology. Tfn uptake experiments were performed as previously described (4). Briefly, ∼2 × 105 cells were incubated in 50 μL PBS4+ (PBS containing 0.2% BSA, 5 mM glucose, 1 mM MgCl2, 1 mM CaCl2) containing 4 μg/mL b-Tfn for 5 min at 37 °C. Extracellular b-Tfn was quenched by the addition of excess avidin and subsequently quenched by biocytin. Internalized b-Tfn was detected by ELISA using streptavidin-HRP. Data are normalized to total cell surface-bound b-Tfn, and averages are divided by protein expression levels to determine relative Tfn uptake activity.
Supplementary Material
Supporting Information
Acknowledgments
We thank Pietro De Camilli (Yale University) for generously providing the Dyn1/2 knockout mouse embryo fibroblasts, which were developed with the partial support of National Institutes of Health (NIH) Grant 2R37NS036251. We also thank Malcolm Wood, Director of The Scripps Research Institutes (TSRI) Core EM facility, for negative-stain EM sample preparation and imaging, and we thank members of the Schmid laboratory for providing helpful discussion and careful reading of the manuscript. This is TSRI manuscript number 20997. Y.-W.L. was supported by Muscular Dystrophy Association Grant MDA-114824, and S.N. was supported by Deutsche Forschungsgemeinschaft Grant NE-1552/1-1. R.R. is a special fellow and T.J.P. was a fellow of the Leukemia and Lymphoma Society. S.M.F. was supported by a Canadian Institutes of Health Research postdoctoral fellowship. This work was supported by NIH Grants R01GM42455 and R01MH61345 (to S.L.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 10381.
References
- 1.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Damke H, Baba T, Warnock DE, Schmid SL. Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol. 1994;127:915–934. doi: 10.1083/jcb.127.4.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ehrlich M, et al. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 4.Sever S, Damke H, Schmid SL. Dynamin:GTP controls the formation of constricted coated pits, the rate limiting step in clathrin-mediated endocytosis. J Cell Biol. 2000;150:1137–1148. doi: 10.1083/jcb.150.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loerke D, et al. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 7.Praefcke GJ, McMahon HT. The dynamin superfamily: Universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 8.Urrutia R, Henley JR, Cook T, McNiven MA. The dynamins: Redundant or distinct functions for an expanding family of related GTPases? Proc Natl Acad Sci USA. 1997;94:377–384. doi: 10.1073/pnas.94.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferguson SM, et al. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316:570–574. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- 10.Liu YW, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol Biol Cell. 2008;19:5347–5359. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschuler Y, et al. Redundant and distinct functions for dynamin-1 and dynamin-2 isoforms. J Cell Biol. 1998;143:1871–1881. doi: 10.1083/jcb.143.7.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran R. Vesicle scission: Dynamin. Semin Cell Dev Biol. 2011;22:10–17. doi: 10.1016/j.semcdb.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Leonard M, Song BD, Ramachandran R, Schmid SL. Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol. 2005;404:490–503. doi: 10.1016/S0076-6879(05)04043-7. [DOI] [PubMed] [Google Scholar]
- 14.Stowell MH, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: Evidence for a mechanochemical molecular spring. Nat Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- 15.Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramachandran R, et al. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 18.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramachandran R, et al. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell. 2009;20:4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binns DD, et al. The mechanism of GTP hydrolysis by dynamin II: A transient kinetic study. Biochemistry. 2000;39:7188–7196. doi: 10.1021/bi000033r. [DOI] [PubMed] [Google Scholar]
- 21.Lin HC, Barylko B, Achiriloaie M, Albanesi JP. Phosphatidylinositol (4,5)-bisphosphate-dependent activation of dynamins I and II lacking the proline/arginine-rich domains. J Biol Chem. 1997;272:25999–26004. doi: 10.1074/jbc.272.41.25999. [DOI] [PubMed] [Google Scholar]
- 22.Warnock DE, Baba T, Schmid SL. Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol Biol Cell. 1997;8:2553–2562. doi: 10.1091/mbc.8.12.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pucadyil TJ, Schmid SL. Supported bilayers with excess membrane reservoir: A template for reconstituting membrane budding and fission. Biophys J. 2010;99:517–525. doi: 10.1016/j.bpj.2010.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roux A, et al. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshida Y, et al. The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J. 2004;23:3483–3491. doi: 10.1038/sj.emboj.7600355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soulet F, Schmid SL, Damke H. Domain requirements for an endocytosis-independent, isoform-specific function of dynamin-2. Exp Cell Res. 2006;312:3539–3545. doi: 10.1016/j.yexcr.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Artalejo CR, Lemmon MA, Schlessinger J, Palfrey HC. Specific role for the PH domain of dynamin-1 in the regulation of rapid endocytosis in adrenal chromaffin cells. EMBO J. 1997;16:1565–1574. doi: 10.1093/emboj/16.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ferguson SM, et al. Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell. 2009;17:811–822. doi: 10.1016/j.devcel.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fish KN, Schmid SL, Damke H. Evidence that dynamin-2 functions as a signal-transducing GTPase. J Cell Biol. 2000;150:145–154. doi: 10.1083/jcb.150.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shpetner HS, Herskovits JS, Vallee RB. A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem. 1996;271:13–16. doi: 10.1074/jbc.271.1.13. [DOI] [PubMed] [Google Scholar]
- 33.Smith SM, Renden R, von Gersdorff H. Synaptic vesicle endocytosis: Fast and slow modes of membrane retrieval. Trends Neurosci. 2008;31:559–568. doi: 10.1016/j.tins.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rappoport JZ, Simon SM. Real-time analysis of clathrin-mediated endocytosis during cell migration. J Cell Sci. 2003;116:847–855. doi: 10.1242/jcs.00289. [DOI] [PubMed] [Google Scholar]
- 35.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr Biol. 1999;9:257–260. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 36.Slepnev VI, Ochoa GC, Butler MH, Grabs D, De Camilli P. Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science. 1998;281:821–824. doi: 10.1126/science.281.5378.821. [DOI] [PubMed] [Google Scholar]
- 37.Smillie KJ, Cousin MA. Dynamin I phosphorylation and the control of synaptic vesicle endocytosis. Biochem Soc Symp. 2005;72:87–97. doi: 10.1042/bss0720087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang ZP, Südhof TC. Cell biology of Ca2+-triggered exocytosis. Curr Opin Cell Biol. 2010;22:496–505. doi: 10.1016/j.ceb.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JP, Sim AT, Robinson PJ. Calcineurin inhibition of dynamin I GTPase activity coupled to nerve terminal depolarization. Science. 1994;265:970–973. doi: 10.1126/science.8052858. [DOI] [PubMed] [Google Scholar]
- 40.Durieux AC, Prudhon B, Guicheney P, Bitoun M. Dynamin 2 and human diseases. J Mol Med. 2010;88:339–350. doi: 10.1007/s00109-009-0587-4. [DOI] [PubMed] [Google Scholar]
- 41.Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DiNitto JP, et al. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol Cell. 2007;28:569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Q. Seamless cloning and gene fusion. Trends Biotechnol. 2005;23:199–207. doi: 10.1016/j.tibtech.2005.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cousin MA, Robinson PJ. The dephosphins: Dephosphorylation by calcineurin triggers synaptic vesicle endocytosis. Trends Neurosci. 2001;24:659–665. doi: 10.1016/s0166-2236(00)01930-5. [DOI] [PubMed] [Google Scholar]
Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10381–10382.
AUTHOR SUMMARY
The plasma membrane, the envelope that surrounds all living cells, serves as both a physical barrier and a platform for communication between cells and their environment. Thus, the transport of molecules in vesicles from the plasma membrane into the cell is highly orchestrated. The major process by which cell surface proteins such as signaling receptors and transporters are internalized is dubbed clathrin-mediated endocytosis, named after the protein clathrin, which decorates the vesicles that ferry molecules into the cell. Clathrin-mediated endocytosis is also involved in the uptake of nutrients and other macromolecular cargo, and it is made possible by the assembly of the clathrin coat and other curvature-generating proteins that deform the underlying plasma membrane into clathrin-coated pits, which concentrate cargo, invaginate, and pinch off to form clathrin-coated endocytic vesicles.
Dynamin (Dyn) is an atypical member of the GTPase family of enzymes that regulates a range of physiologically important cellular reactions. Dynamin self-assembles into rings to form a collar-like structure at the necks of deeply invaginated clathrin-coated pits, where it catalyzes membrane fission to release cargo-laden endocytic vesicles into the cell. Dynamin, whose protein sequence is highly conserved among higher eukaryotes, is not found in yeast. Moreover, whereas less complex organisms like Caenorhabditis elegans and Drosophila express only a single isoform, mammals express three dyn isoforms (Dyn1, Dyn2, and Dyn3) in a tissue-specific manner. Most studies have focused on Dyn1, the first identified and most abundant neuron-specific isoform. Dyn1 is required for the stimulus-dependent rapid recycling of the membrane components of synaptic vesicles at nerve terminals, a function crucial for sustained neurotransmission. Indeed, dynamin's role in clathrin-mediated endocytosis was first discovered in the context of synaptic vesicle recycling in the fruit fly, Drosophila. When shifted to a nonpermissive temperature, flies bearing a temperature-sensitive mutation in the dynamin counterpart, shibire, become rapidly paralyzed. Their nerve terminals are depleted of synaptic vesicles and accumulate collared pits, suggesting a role for dynamin in membrane fission. Other evidence derived from studies in nonneuronal cells indicates that Dyn2, the ubiquitously expressed isoform, plays a dual role in clathrin-mediated endocytosis. These studies suggest that, in its unassembled state, Dyn2 (like other GTPase family members) regulates early stages of clathrin-mediated endocytosis by monitoring cargo recruitment, coat assembly, and curvature generation before self-assembling into the rings that mediate fission (1).
Strikingly, despite being 79% identical in protein sequence, the two isoforms are not fully functionally redundant; Dyn2 cannot efficiently restore rapid synaptic vesicle recycling in neurons from mice lacking Dyn1 (2), whereas Dyn1 cannot efficiently restore clathrin-mediated endocytosis in mouse fibroblasts lacking Dyn2 (3). Here, we report a comprehensive side by side biochemical characterization of Dyn1 and Dyn2 aimed at understanding the functional differences between these two closely related isoforms. We have discovered that Dyn1 can generate highly curved membranes from flat templates, whereas Dyn2 is a curvature sensor that will only self-assemble onto highly curved membrane templates. After they are assembled on curved, neck-like membranes, both isoforms can catalyze membrane fission. These differences provide mechanistic insight into the tissue-specific regulation of clathrin-mediated endocytosis.
Dynamin consists of five functionally distinct domains: a highly conserved N-terminal GTPase domain involved in the binding and hydrolysis of GTP, which is an energy currency for some cellular processes, a middle domain and GTPase effector domain that are required for self-assembly, a pleckstrin homology domain that binds to and inserts into the plasma membrane to generate membrane curvature, and a C-terminal proline/arginine domain that binds to protein partners necessary for targeting to clathrin-coated pits (1, 4). Structure function analyses of a series of Dyn1 mutants have revealed that its interdependent abilities to bind and self-assemble onto membranes, generate high curvature on the membranes, and hydrolyze GTP help catalyze membrane fission (4). A number of assays have been developed in the past to individually follow each of these different activities (1, 4), enabling us to directly compare the biochemical properties of Dyn1 and Dyn2 on membrane templates of differing curvature. By measuring these activities on membrane templates of increasing size and hence, decreasing curvature, we discovered that the assembly-stimulated GTPase activity of Dyn2 is highly sensitive to curvature. Unlike Dyn1, Dyn2 is unable to self-assemble onto large, flat membranes deposited on glass beads or catalyze vesicle formation from them. We generated chimeric Dyn1/Dyn2 molecules by swapping individual domains from one isoform with the other. These experiments identified the pleckstrin homology domain as being responsible for the curvature-generating and -sensing abilities of Dyn1 and Dyn2, respectively. Remarkably, additional analyses identified a single amino acid residue at position 600 within the pleckstrin homology domain—a tyrosine in Dyn1 and a leucine in Dyn2—that conferred the isoform-specific curvature-sensitive behavior.
To determine whether these different properties assayed in vitro account for their differential in vivo activities, we reconstituted Dyn1/2 chimeras in mouse embryo fibroblast cells that lacked Dyn1 and Dyn2. Chimeras bearing the Dyn2 proline/arginine domain were more efficiently targeted to clathrin-coated pits, and consistent with the biochemical studies, the efficiency of endocytosis was greater in cells expressing chimeras bearing the Dyn1 pleckstrin homology domain. Thus, although Dyn1 was less efficient than Dyn2 at supporting clathrin-mediated endocytosis in these nonneuronal cells, its pleckstrin homology domain enhanced the ability of Dyn1/Dyn2 chimeras to support clathrin-mediated endocytosis.
All species of mammalian Dyn2 contain the amino acid leucine at position 600 within the pleckstrin homology domain, whereas all mammalian Dyn1 and Dyn3 contain the curvature-generating amino acid tyrosine at this position. The only isoforms expressed in C. elegans and Drosophila also a tyrosine at this position. Yeast lacks dynamin, suggesting that it might have first evolved for rapid endocytosis at the synapse, a nervous system structure specific to multicellular animals. Subsequently, vertebrates seem to have evolved a second, ubiquitously expressed isoform that is more efficiently targeted to clathrin-coated pits but exhibits greater curvature sensitivity and weakened ability to catalyze membrane fission from planar membranes compared with its evolutionary progenitor.
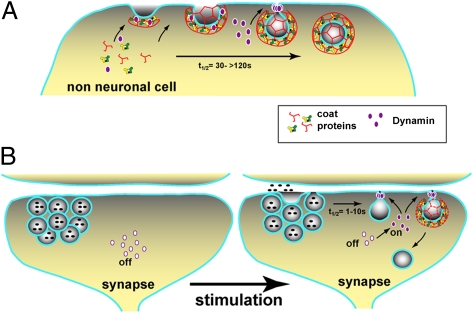
These results beg the question of why Dyn2 has evolved to be more sensitive to membrane curvature than Dyn1. We propose that, as a curvature sensor, Dyn2 is well-suited for its dual role in clathrin-mediated endocytosis in nonneuronal cells (1). Dyn2 is efficiently recruited to nascent clathrin-coated pits through its proline/arginine domain. However, given its biochemical properties, other factors, including coat assembly and recruitment of curvature-generating proteins, would be needed to form a narrow neck of sufficient curvature to trigger Dyn2 assembly. In this way, Dyn2 is positioned to regulate clathrin-mediated endocytosis by sensing when clathrin-coated pit maturation is complete and to mediate membrane fission only after other factors have triggered its assembly switch (Fig. P1_A_). In contrast, given its in vitro properties, we suggest that Dyn1 can support rapid endocytosis and recycling of synaptic vesicles, because it can efficiently bind, squeeze, and stabilize the necks of nascent pits, perhaps even without the help of accessory proteins or the clathrin coat (Fig. P1_B_). Indeed, Dyn1 activity may be inhibited at the resting synapse by phosphorylation and activated by dephosphorylation on neuronal stimulation (5) for rapid capture of the newly released synaptic vesicle membrane.
Fig. P1.
Differential regulation of dynamin isoforms in fibroblasts and at the synapse. (A) In fibroblasts, Dyn2 is recruited to nascent clathrin-coated pits through its proline/arginine domain, where it can regulate clathrin-coated pit maturation and the formation of a deeply invaginated pit by coat and accessory proteins. After the neck is sufficiently narrow, additional Dyn2 is recruited to the membrane through its curvature-sensitive pleckstrin homology domain to mediate fission. (B) In the resting synapse, the proline/arginine domain of Dyn1 is phosphorylated, inhibiting Dyn1 self-assembly and membrane binding. Dephosphorylation occurs rapidly on stimulation, allowing Dyn1 to rapidly self-assemble and mediate membrane fission and retrieval of synaptic vesicle membrane components, perhaps without the help of coat proteins (as illustrated) because of its powerful curvature-generating pleckstrin homology domain.
In summary, our findings present an example of how the divergence of two highly similar isoforms of a key enzyme can provide intrinsic elements of regulation to support tissue-specific functions. To gain additional understanding of the endocytic machinery, it will be necessary, in the future, to develop in vitro reconstitution systems for clathrin-mediated endocytosis using cell type-specific components.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See full research article on page E234 of www.pnas.org.
References
- 1.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferguson SM, et al. A selective activity-dependent requirement for dynamin 1 in synaptic vesicle endocytosis. Science. 2007;316:570–574. doi: 10.1126/science.1140621. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y-W, Surka MC, Schroeter T, Lukiyanchuk V, Schmid SL. Isoform and splice-variant specific functions of dynamin-2 revealed by analysis of conditional knock-out cells. Mol Biol Cell. 2008;19:5347–5359. doi: 10.1091/mbc.E08-08-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmid SL, Frolov VA. Dynamin: Functional design of a membrane fission catalyst. Annu Rev Cell Dev Biol. 2011;27:3.1–3.27. doi: 10.1146/annurev-cellbio-100109-104016. [DOI] [PubMed] [Google Scholar]
- 5.Cousin MA, Robinson PJ. The dephosphins: Dephosphorylation by calcineurin triggers synaptic vesicle endocytosis. Trends Neurosci. 2001;24:659–665. doi: 10.1016/s0166-2236(00)01930-5. [DOI] [PubMed] [Google Scholar]
Supplementary Materials
Supporting Information