Coilin forms the bridge between Cajal bodies and SMN, the Spinal Muscular Atrophy protein (original) (raw)
Abstract
Spinal muscular atrophy (SMA) is a genetic disorder caused by mutations in the human survival of motor neuron 1 gene, SMN1. SMN protein is part of a large complex that is required for biogenesis of various small nuclear ribonucleoproteins (snRNPs). Here, we report that SMN interacts directly with the Cajal body signature protein, coilin, and that this interaction mediates recruitment of the SMN complex to Cajal bodies. Mutation or deletion of specific RG dipeptide residues within coilin inhibits the interaction both in vivo and in vitro. Interestingly, GST-pulldown experiments show that coilin also binds directly to SmB′. Competition studies show that coilin competes with SmB′ for binding sites on SMN. Ectopic expression of SMN and coilin constructs in mouse embryonic fibroblasts lacking endogenous coilin confirms that recruitment of SMN and splicing snRNPs to Cajal bodies depends on the coilin C-terminal RG motif. A cardinal feature of SMA patient cells is a defect in the targeting of SMN to nuclear foci; our results uncover a role for coilin in this process.
Keywords: Coilin, SMN, SMA, snRNPs, arginine methylation, Cajal body
Eukaryotic nuclei are highly organized and contain numerous subdomains. These substructures are involved in various aspects of cellular metabolism including replication, transcription, and RNA processing (for reviews, see Lamond and Earnshaw 1998; Matera 1999; Cremer and Cremer 2001). One of the emerging principles of nuclear organization is that many individual domains are associated with specific genetic loci and can be regulated by chromosomal determinants. Another emerging principle is that associations between these various domains and loci are dynamic and can change in response to cellular signals (Misteli 2001). Thus, studies of the functional organization of the nucleus seek to understand this dynamic behavior and address how improper subcellular localization of macromolecules can lead to human disease. In particular, mislocalization of one important macromolecular complex has been etiologically linked to a neurodegenerative disorder called Spinal Muscular Atrophy (Frugier et al. 2000).
SMA is a genetic disease that is characterized by an autosomal recessive inheritance pattern. It is the most common hereditary cause of infant mortality (Pearn 1980). Although some SMA patients display decreased fetal movements, the clinical course of the disease typically begins postnatally, resulting in apoptosis of spinal motor neurons, followed by progressive paralysis in the limbs and trunk and, in its most severe form, infant death (Melki 1997; Talbot and Davies 2001 and references therein). Greater than 95% of the cases of SMA are caused by mutations in the survival of motor neurons (SMN) locus (Lefebvre et al. 1995).
There are typically two SMN genes, located within an inverted duplication on human chromosome 5q13 (Lefebvre et al. 1995). Deletion of the telomeric copy, SMN1, results in the disease, whereas similar mutations in the centromeric copy, SMN2, have no phenotype (for reviews, see Burghes 1997; Melki 1997). Thus, despite the fact that the two coding regions differ by only five silent point mutations, the genes produce two different isoforms of the SMN protein (Lorson et al. 1999). In mouse, Smn is an essential, single-copy locus and homozygous deletions result in early embryonic lethality (Schrank et al. 1997). This phenotype can be rescued in a dosage-dependent fashion by transgenic expression of the human SMN2 gene (Hsieh-Li et al. 2000; Monani et al. 2000).
SMN protein localizes to both the cytoplasm and the nucleus, but its nuclear staining is restricted to two different types of subdomains: gems and Cajal bodies (CBs; Liu and Dreyfuss 1996; Matera and Frey 1998; Carvalho et al. 1999; Young et al. 2000). Furthermore, SMN is part of a large complex of proteins that are involved in the biogenesis of snRNPs (Fischer et al. 1997; Liu et al. 1997; Pellizzoni et al. 1999; Meister et al. 2000). These proteins include Gemin2/SIP1, Gemin3/dp103, and Gemin4/GIP1 (Charroux et al. 1999, 2000; Grundhoff et al. 1999; Meister et al. 2000) as well as snRNP core components, collectively called Sm proteins (B/B′, D1–3, E-G). Indeed, the SMN complex is required for assembly of the Sm core domain of spliceosomal U snRNPs in vivo (Fischer et al. 1997; Meister et al. 2001). Interestingly, SMN contains a Tudor domain, which is common among many different RNP-binding proteins (Ponting 1997). The Tudor domain of SMN is known to directly interact with Sm proteins (Bühler et al. 1999; Selenko et al. 2001).
An important cellular feature of SMA is the failure of the SMN complex to localize within nuclear bodies (Coovert et al. 1997; Lefebvre et al. 1997; Frugier et al. 2000). Most SMA-causing mutations result in the expression of a truncated form of the protein (Lorson et al. 1999) that lacks sequences encoded by exon 7 (SMNΔEx7). The C-terminal region of SMN is required for self-oligomerization (Lorson et al. 1998), efficient binding of Sm proteins (Pellizzoni et al. 1999), and proper translocation to the nucleus (Frugier et al. 2000). Gangwani et al. (2001) recently showed that an essential zinc finger protein, called ZPR1, is also important for nuclear localization of SMN. However, the interaction between SMN and ZPR1 is likely to be indirect (for review, see Matera and Hebert 2001).
Factor(s) responsible for targeting SMN to CBs have not been described. The molecular links between CBs (which contain snRNPs) and gems (which do not) are also unclear. Recently, our laboratory showed that coilin, the CB signature protein, is required for recruitment of SMN and Sm proteins to CBs (Tucker et al. 2001). Targeted deletion of the C-terminal 487 amino acids of coilin resulted in formation of residual CBs that lack SMN and Sm snRNPs, but contain the nucleolar epitopes Nopp140 and fibrillarin (Tucker et al. 2001). Inspection of C-terminal coilin sequences revealed the existence of a conserved RG dipeptide motif similar to the C-terminal tails of Sm proteins. The RG-rich tails of Sm proteins D1, D3, and B/B′ have been shown to directly interact with SMN (Friesen and Dreyfuss 2000; Selenko et al. 2001).
In this report, we show that SMN and coilin coimmunoprecipitate from HeLa cell lysates. This interaction is direct, as shown by GST-pulldown experiments using recombinant proteins. Mutation of specific arginine residues within the coilin RG box inhibits the interaction both in vivo and in vitro. Mapping and competition experiments show that coilin and SmB′ share similar or overlapping binding sites on SMN and bind with approximately the same avidity. Finally, transfection of mouse coilin knockout cells with full-length coilin showed that SMN and Sm snRNPs were recruited to the CBs thus formed. However, transfection with coilin constructs lacking the RG motif failed to recruit Sm or SMN complex proteins. Thus, our results show that the coilin RG box is essential for localization of the SMN complex within CBs.
Results
The coilin RG box mediates interaction with the SMN complex
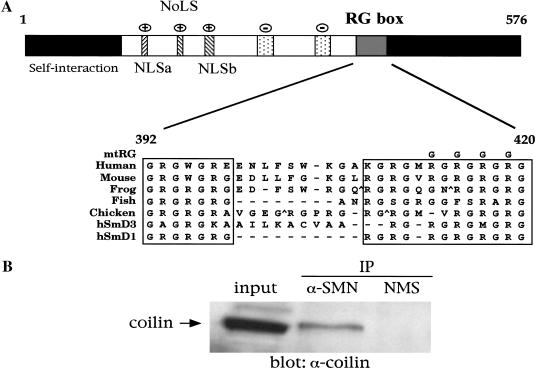
Vertebrate coilin proteins contain a stretch of arginine and glycine dipeptide (RG) residues in their C-terminal regions (Fig. 1A). This conserved domain is similar to the RG repeats present in metazoan Sm proteins (Salgado-Garrido et al. 1999; Brahms et al. 2000). The RG motifs present in human SmD1, D3, and B/B′ have been shown to mediate interaction with SMN (Friesen and Dreyfuss 2000; Selenko et al. 2001). Given that coilin and SMN colocalize within CBs, we wanted to test whether at least a fraction of these proteins might be part of the same macromolecular complex. Immunoprecipitation of HeLa lysate with anti-SMN antibodies showed that coilin interacts with SMN (Fig. 1B). The interaction was specific, because no coilin was recovered in control immunoprecipitations using normal mouse serum. To further define the interaction, we transfected HeLa cells with various GFP- or myc-tagged coilin constructs (Fig. 2A). Coimmunoprecipitation of cell lysates with anti-GFP or anti-myc antibodies revealed that the interaction is detectable in both directions (Fig. 2B, top; lanes 2, 4, and 6). No SMN was recovered from lysates transfected with empty GFP vector (data not shown).
Figure 1.
Coilin contains a conserved RG box and interacts with SMN. (A) Schematic representation of human coilin. The self-interaction domain and positively charged nuclear and nucleolar localization signals (NLS, NoLS) are indicated (Hebert and Matera 2000). The two stippled regions represent potentially negatively charged regions and reside upstream of the bipartite RG box, spanning residues 392–420 in human. For comparison, similar RG-box motifs from other vertebrate coilins as well as from the human SmD3 and SmD1 proteins are aligned. Carats (^) mark sites where two to three amino acids were excluded to facilitate the alignment. Gaps are denoted by a dash. The top line of the alignment shows the position of a mutation of the RG box (substitution of four arginines to glycines) used in this study (mtRG). (B) Coilin interacts with SMN in HeLa lysate. Immunoprecipitation of the lysate by either an anti-SMN antibody or normal mouse serum (NMS) was conducted as described in Materials and Methods, followed by SDS-PAGE and Western blotting with anticoilin antibodies. The input lane shows 8% of the lysate used in the immunoprecipitation reactions.
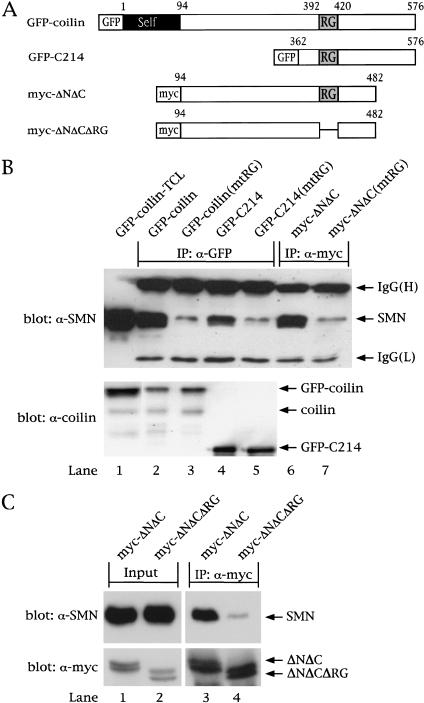
Figure 2.
The coilin RG box mediates interaction with SMN. (A) Schematic of coilin constructs. The RG box is indicated, as is the tag used for each construct. GFP–C214 contains the C-terminal 214 amino acids of coilin, residues 362–576. Myc-tagged coilinΔNΔC spans residues 94–482, as shown. The RG box of each construct was mutated (mtRG), as shown in Fig. 1A. Furthermore, the entire RG box (392–420) was deleted in myc–ΔNΔC (myc–ΔNΔCΔRG). (B) Coilin interaction with SMN is dependent on the RG box. HeLa cells were transfected with the various coilin constructs, and the lysates were immunoprecipitated with anti-GFP (lanes 2–5) or anti-myc (lanes 6,7) antibodies, as described in Materials and Methods. The beads were washed and subjected to SDS-PAGE and Western blotting with anti-SMN antibodies (top). The immunoglobulin (IgG) heavy (H) and light (L) chains are marked. The input lane shows 5% of the lysate used in the immunoprecipitation reactions. The same immunoprecipitates were electrophoresed and blotted with anticoilin antibodies to show that equal amounts of protein were pulled down (bottom). (C) Deletion of the RG box in coilin reduces SMN interaction. HeLa cells were transfected with myc–ΔNΔC or myc–ΔNΔCΔRG, and the lysates were immunoprecipitated with monoclonal anti-myc antibodies, followed by SDS-PAGE and Western blotting with anti-SMN antibodies (top). The same blot was reprobed with polyclonal anti-myc antibodies (bottom). The input lanes show 5% of the lysate used in the reactions.
To assess the role of the coilin RG box, we mutated four of the arginines in this motif to glycines (Fig. 1A). After transfection of these mutant (mtRG) constructs, coimmunoprecipitation showed that arginine substitution greatly reduced the amount of SMN recovered (Fig. 2B, top; lanes 3, 5, and 7). Furthermore, expression of the coilin C terminus was sufficient for the interaction. Control blots of the same immunoprecipitation reactions were run in parallel and probed with anticoilin antibodies, showing that equivalent amounts of protein were recovered for each set of constructs (Fig 2B, bottom; cf. lanes 2 and 3 or 4 and 5). Previous studies of SMN and Sm protein interactions could only infer the importance of the arginine residues, because only truncation mutants were studied (Friesen and Dreyfuss 2000; Selenko et al. 2001). Here, we show that arginine substitution rather than truncation is important for binding to SMN. On the basis of these observations, we conclude that arginines within the coilin RG box are important for interaction with SMN in vivo.
Interestingly, the interaction between coilin and SMN need not be restricted to CBs, as the myc-coilinΔNΔC protein is completely nucleoplasmic (Hebert and Matera 2000) and the GFP-C214 construct localizes diffusely throughout the entire cell, similar to GFP alone (data not shown). Thus, the coilin self-association domain (Hebert and Matera 2000) is not necessary for the interaction with SMN. We note that recovery of SMN with coilin-C214 was not quite as robust as it was with the longer constructs (Fig. 2B, cf. lanes 2 and 6 with lane 4), suggesting that coilin residues 94–362 may make additional contacts with SMN. Indeed, slight residual binding to SMN was observed even when the entire coilin RG domain was deleted (Fig. 2C). Alternatively, it is possible that the primarily cytoplasmic GFP-C214 protein does not compete as well for SMN binding sites as does nuclear coilin (see below). Additional experiments revealed that Gemin2 and Gemin3 coprecipitate with SMN and GFP-coilin (data not shown). Thus, in vivo, it is likely that coilin associates with the multisubunit SMN complex (Liu et al. 1997; Pellizzoni et al. 1999; Meister et al. 2000), raising questions as to the precise nature of the interaction.
Coilin interacts directly with SMN
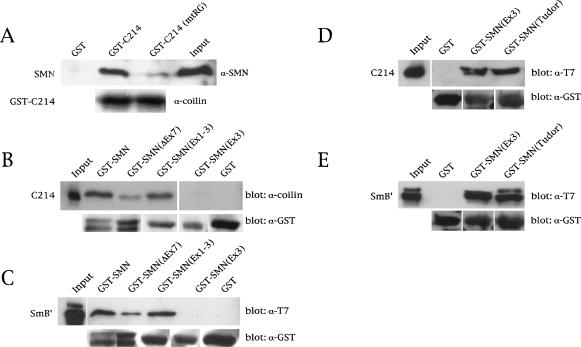
We therefore generated and purified His- and GST-tagged versions of both SMN and coilin and conducted in vitro pulldown assays. As shown in Figure 3A, a GST fusion of the coilin C-terminal 214 residues (GST–C214) interacts directly with His-tagged, full-length SMN. GST alone showed no detectable binding. As we found in vivo (Fig. 2B), mutation of the coilin RG box significantly reduced the interaction (Fig. 3A, top). Control blots with anticoilin (Fig. 3A, bottom) or anti-GST (Fig. 3B, bottom) antibodies showed that comparable amounts of the GST constructs were used in the assay. These results show that the interaction between coilin and SMN is direct and is mediated by the coilin RG box.
Figure 3.
Coilin directly interacts with SMN via the RG box. (A) Binding assays of His–SMN and GST–C214 or GST–C214(mtRG) were conducted as described in Materials and Methods, followed by SDS-PAGE and Western blotting with anti-SMN antibodies (top). The same blot was reprobed with anticoilin antibodies (bottom) to show that equal amounts of GST–C214 and GST–C214(mtRG) were used. (B) Mapping of the coilin interaction site on SMN. Various GST–SMN fusions were tested for their ability to bind His–C214. GST–SMNΔEx7 does not contain exon 7 sequences. GST–SMN(Ex1–3) and GST–SMN(Ex3) contain the first three or only the third exon of SMN, respectively. The Western blot was probed with an anticoilin antibody (top). The same blot was reprobed with anti-GST antibodies to verify that equal amounts of beads were used in the assay (bottom). (C) Control pulldowns with the same GST–SMN constructs used in B were assayed using His–T7–SmB′. Western blotting was performed with an anti-T7 antibody (top) or an anti-GST antibody (bottom). The input lanes for all reactions are equivalent to 10% of that used in the binding assay. (D,E) The Tudor domain of SMN mediates binding to coilin and SmB′. GST–SMN(Ex3), encompassing SMN residues 91–158, and GST–SMN(Tudor), spanning residues 83–173, were used in pulldown assays with purified coilin-C214 or SmB′. A less stringent buffer was used in these binding assays compared with the buffer used in A–C (see Materials and Methods). The blots were reprobed with an antibody to GST to verify that equal amounts of beads were used in the assay. The input lane for D was equivalent to 10% of the C214 used in the binding assay, whereas the input lane for E represents 20% of the SmB′ used in the binding reaction.
Given that SMN can self-oligomerize (Lorson et al. 1998; Pellizzoni et al. 1999), it was important not only to determine whether the coilin interaction required SMN oligomers, but to localize the coilin binding site on the SMN protein. We therefore reversed the direction of the GST pulldowns described above, tethering various GST–SMN constructs to the beads and testing with His-tagged coilin-C214. Figure 3B shows that coilin-C214 can interact with bead-bound SMN (i.e., SMN that has not oligomerized). Furthermore, deletion of SMN exon 7 (SMNΔEx7, the principal SMN isoform in SMA patients) reduced the binding activity, whereas that of SMN(Ex1–3) was relatively unaffected. The exon 3-only fusion, SMN(Ex3), showed an extremely faint band under relatively stringent wash conditions (see Materials and Methods), whereas GST alone did not bind at all (Fig. 3B). As a control, we tested the binding of His–SmB′ to the same GST–SMN constructs and obtained equivalent results (Fig. 3C).
There is some discrepancy in the literature regarding the precise Sm protein binding site(s) within SMN. Dreyfuss and colleagues have implicated exon 7 (Charroux et al. 1999; Pellizzoni et al. 1999), whereas the Fischer group identified the so-called Tudor domain of SMN (essentially exon 3) as the important region (Bühler et al. 1999; Selenko et al. 2001). Significantly, none of the previous domain-mapping studies used purified recombinant proteins for both reactants. Instead, they substituted in vitro translation products of the various SMN subfragments, which potentially could contain additional components of the SMN complex present in the reticulocyte lysate. Not only did we perform the binding reactions using purified recombinant proteins, but our wash conditions contained similar salt concentrations and more than 10 times the amount of detergent used previously.
Because neither coilin nor SmB′ bound tightly to the GST–SMN(Ex3) construct, we then asked whether more relaxed washing conditions (Charroux et al. 1999; see Materials and Methods for details) would still elicit specific binding. Indeed, we found that whereas GST alone did not bind under these conditions, GST–SMN(Ex3) and a slightly larger Tudor domain construct GST–SMN(Tudor) were efficiently bound by both His–C214 (Fig. 3D) and His–SmB′ (Fig. 3E). Together, these data clearly show that the RG dipeptide motifs present in coilin mimic those within the C-terminal tails of Sm proteins in their direct interaction with the SMN Tudor domain.
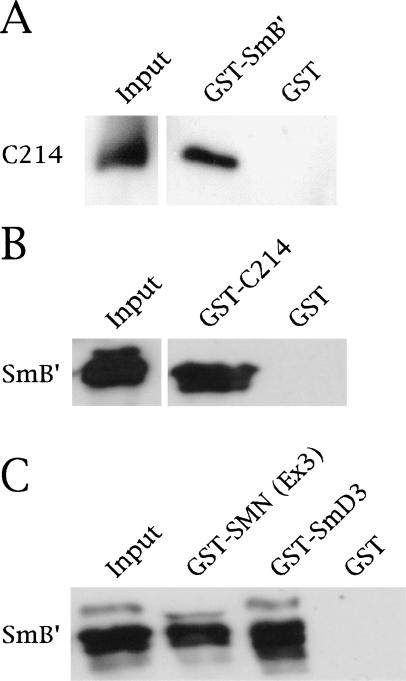
Coilin interacts directly with SmB′
Several lines of evidence suggest an interaction between coilin and Sm proteins. First, Bauer and Gall (1997) stated that coilin can be immunoprecipitated from Xenopus germinal vesicle extract by anti-Sm antibody Y12. We since have confirmed this observation in HeLa cell extracts (data not shown). Second, certain cell lines display CBs that are separable from gems (e.g., HeLa PV; Liu and Dreyfuss 1996). These CBs thus are enriched in snRNAs and Sm proteins but lack detectable concentrations of the SMN complex. It therefore is possible that coilin and at least some of the Sm proteins might interact directly. We tested this idea and found that, indeed, GST–SmB′ recovered His–C214 in a pulldown assay (Fig. 4A), whereas GST alone did not. The interaction proved faithful in the reverse direction as well, because GST–C214 bound to His–SmB′ (Fig. 4B). As a control for proper folding of His–SmB′, we showed that it could interact with both GST–SmD3 and GST–SMN(Ex3) (Fig. 4C). We currently are investigating in detail the regions within coilin and various Sm proteins that are required for this interaction. However, it seems likely that residues within the distal, highly conserved coilin C terminus (Fig. 1A) are responsible for the binding to SmB/B′, and perhaps with other Sm proteins as well.
Figure 4.
Coilin interacts with SmB′. (A) Direct interaction of His–C214 with GST–SmB′, but not with GST alone. His–C214 was detected by anticoilin antibodies. (B) His–T7–SmB′ interacts with GST–C214. The blot was probed with anti-T7 antibodies (top) to detect His–T7–SmB′. Reprobing with anticoilin antibodies showed equal levels of GST (data not shown). (C) The SmB′ used in this study is properly folded because it interacts with GST–SmD3 and GST–SMN(Ex3). The input lanes account for 20% of the His-tagged reagents in A and B and 33% of the amount in C.
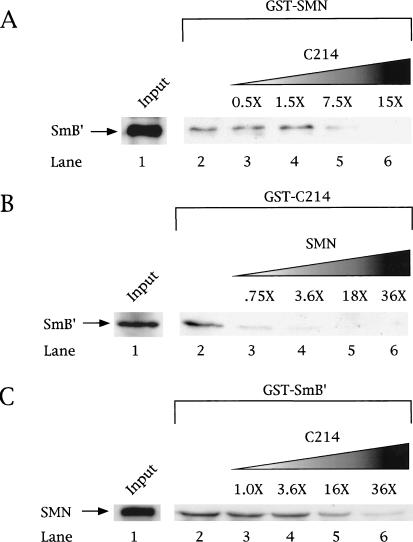
Coilin competes with SmB′ for binding to SMN
Because coilin and SmB′ both contain RG dipeptides and bind to the various GST–SMN constructs with similar affinities (Fig. 3), we wondered if they might occupy overlapping binding sites. We tested the competitive nature of the interaction in three different ways. Pulldown assays were performed using fixed amounts of His–T7–SmB′ and the GST constructs, either GST–SMN (Fig. 5A) or GST–C214 (Fig. 5B). Variable amounts of His–C214 (Fig. 5A) or His–SMN (Fig. 5B) then were added. In each case, increasing amounts of His–C214 or His–SMN competed with His–T7–SmB′ for binding to GST–SMN or GST–C214, respectively. Indeed, a 15-fold molar excess of His–C214 precludes any His–SmB′ binding to GST–SMN (Fig. 5A). Similarly, an excess of His–SMN is sufficient to abolish His–SmB′ binding to GST–C214 (Fig. 5B). Finally, we also tethered GST–SmB′ to the beads and found that increasing amounts of coilin-C214 also could compete with soluble (i.e., oligomerized) SMN (Fig. 5C). As expected, the oligomeric SMN bound more tightly to SmB′ than did the GST-bound SMN (Pellizzoni et al. 1999), thus requiring more coilin competitor (Fig. 5, cf. C and A). Regardless of which reactant was tethered to the beads, the other two proteins competed for its binding. Given that SmB′ interacts directly with both coilin and SMN, the reduction of SmB′ recovery in Figure 5A and B reflects not only a saturation of the GST component, but a sequestration of SmB′ through its interaction with the more abundant protein.
Figure 5.
Coilin competes with SmB′ and SMN for binding. Competition experiments were conducted using GST–SMN (A), GST–C214 (B), or GST–SmB′ (C) beads incubated with a constant amount of His–T7–SmB′ (A,B) or SMN (C). Increasing amounts of His–C214 (A,C) or SMN (B) were added in separate reactions at the indicated fold excess, based on protein level, relative to the amount of His–T7–SmB′ (or SMN) present (lanes 3–6). The beads were washed, as described in Materials and Methods, and subjected to SDS-PAGE and Western blotting with an anti-T7 antibody (for detection of SmB′ and C214) or an anti-SMN antibody. The input lane (lane 1) shows an amount of protein equivalent to 20% of that used in lanes 2 and 4. Lane 2 in all panels shows the amount of His–T7–SmB′ (A,B) or SMN (C) recovered in the absence of competitor. It is clear that addition of increasing amounts of competitor did not poison the GST beads because companion blots showed increasing amounts of either C214 (A,C) or SMN (B) were bound as binding to the other component decreased (data not shown).
The coilin RG box recruits the SMN complex to CBs
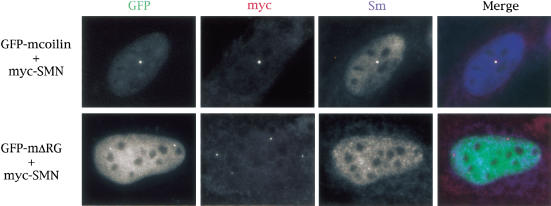
We have shown that coilin and SMN interact both in cell extracts and directly in vitro. Together with evidence that coilin and SMN interact genetically (Tucker et al. 2001), these results led us to hypothesize that coilin is the factor that recruits the SMN complex (and Sm snRNPs) to the CB. To test this idea, we took advantage of mouse embryonic fibroblast (MEF) cell lines derived from knockout mice that lack the C-terminal 487 amino acids of coilin, including the RG-box motif. These MEFs do not display normal CBs but instead form residual CBs that lack coilin, SMN, and snRNPs (Tucker et al. 2001). Transient expression of full-length mouse GFP-coilin restored formation of normal CBs (Tucker et al. 2001). As shown in Figure 6, knockout cells transfected with GFP–mcoilin and myc-SMN show prominent CBs that contain Sm snRNPs. However coexpression of a coilin RG-box deletion (GFP–mcoilinΔRG) tells a dramatically different story, generating distinct coilin and SMN foci (Fig. 6). In no case did we observe a cell (n > 400) with coincident GFP–mcoilinΔRG and myc–SMN dots. Intriguingly, we found that Sm proteins are not enriched in the GFP–mcoilinΔRG foci, suggesting that either the coilin RG box is important for proper folding of the C-terminal region or that Sm proteins bind to the RG box itself. Duplicate experiments on a second knockout MEF cell line resulted in the same phenotype. These results provide mechanistic evidence that the coilin RG-box motif is essential for proper CB formation.
Figure 6.
The RG box of coilin is required for recruitment of the SMN complex to the Cajal body. A mouse embryonic fibroblast cell line derived from coilin knockout mice (Tucker et al. 2001) was cotransfected with GFP-mouse coilin (GFP–mcoilin) and human myc-tagged SMN (top). SMN was visualized by an anti-myc antibody whereas Sm proteins were detected using the antibody Y12. A white signal in the merged image shows that all three signals are coincident. Additionally, mouse coilin deleted of the RG box (GFP–mΔRG) was cotransfected into the KO line with myc–SMN (bottom). The presence of both red and green signals, without overlap, can be observed in merged image. Cells were processed as described in Materials and Methods.
Discussion
We have shown that SMN, the spinal muscular atrophy gene product, interacts with coilin, the CB marker protein via the coilin C-terminal RG-box motif. The interaction is direct, because substitution or deletion of specific arginine residues abrogates binding both in vivo and in vitro. Furthermore, coilin competes with Sm proteins for binding to SMN. Transfection experiments in coilin knockout cell lines revealed that the RG box is required for recruitment of the SMN complex to CBs. We note that deletion of the RG box in mouse coilin resulted in an increased nucleoplasmic fraction of the protein (Fig. 6, cf. localization of GFP–mcoilin with GFP–mcoilinΔRG). Therefore, the coilin RG box also may play a role in the overall folding and/or posttranslational modification of the protein, which could affect its subsequent subcellular localization. Interestingly, the RG-box deletion described in this study resembles the CB/gem separation phenotype observed in certain strains of HeLa cells (Liu and Dreyfuss 1996; Matera and Frey 1998; Sleeman and Lamond 1999), with the important caveat that the GFP–mcoilinΔRG foci do not recruit snRNPs. Recently, Brahms et al. (2000) showed that RG dipeptides in human Sm proteins contain symmetrically dimethylated arginines, the presence of which forms the epitope for the well-known anti-Sm monoclonal antibody Y12. Unmethylated RG repeats are not recognized by this antibody. Plausibly, arginines within the coilin RG motif also may be methylated, and such modification could not only contribute to the Y12 staining pattern itself, but alter the binding of coilin to SMN. In fact, Friesen et al. (2001) recently have shown that arginine methylation substantially increases the binding affinity of Sm proteins to SMN. Thus, differences in expression of coilin modifiers in the various cell lines could account for the observed variability in gem versus CB expression. Indeed, the fact that gems and CBs tend to exist as separate entities in fetal tissues, whereas in adults the two structures are fused (Young et al. 2001), argues that this process is developmentally regulated. Arginine methylation therefore may serve as a molecular switch that controls coilin and Sm protein interactions with SMN. Certain coilin constructs like C214 might not have equal access to the proper arginine methyltransferases, perhaps accounting for why cytoplasmic GFP–coilin–C214 might be less effective at binding SMN than its nuclear counterparts (Fig. 2B).
Self-association and nuclear body formation
How does nuclear SMN play a role in snRNP biogenesis or recycling when it is concentrated in gems, which do not contain snRNPs (Liu and Dreyfuss 1996; Dietz 1998)? The finding that coilin is directly required for recruitment of the SMN complex to CBs solves this long-standing question in SMA biology. In other words, coilin is the bridge between CBs and gems. Thus, an important factor in gem formation simply may be the local concentrations of the various interactors. For example, considering that coilin is primarily nuclear, the relative concentrations of SMN and Sm proteins in the cytoplasm would favor that combination. However, when the SMN/snRNP complex enters the nucleus, it encounters high levels of coilin, especially within the CB. We have shown that coilin competes with SmB′ for binding sites on SMN (Fig. 5). In this scenario, self-oligomerized coilin proteins in the CB (Hebert and Matera 2000) conceivably could bind to both Sm and SMN, therefore dissociating the SMN/snRNP complex. SMN, liberated from its snRNP cargo, then would be free to leave the CB, possibly to regenerate splicing snRNPs (Pellizzoni et al. 1998) or shuttle back to the cytoplasm to help assemble new ones (Matera and Hebert 2001). Alternatively, a large fraction of coilin is localized diffusely in the nucleoplasm, perhaps reflecting a coilin/Sm or a coilin/SMN pool. The presence of gems therefore may reflect a local surplus of SMN complexes, resulting in the formation of gems by SMN self-association (Hebert and Matera 2000).
SMA: the coilin connection?
Cells derived from SMA patients and mouse models of the disease display defects in the nuclear targeting of SMNΔEx7 (Frugier et al. 2000). Genetic studies have shown that the severity of SMA is directly dependent on the expression of full-length SMN protein (Hsieh-Li et al. 2000; Monani et al. 2000). In fact, _Cre_-mediated deletion of Smn exon 7 in mouse neuronal cells results in an SMA-like phenotype as well as a pronounced defect in coilin localization, forming large paranucleolar aggregates (Frugier et al. 2000). Recent work from our laboratory (Tucker et al. 2001) provides additional evidence for a genetic interaction between coilin and SMN. Given the molecular results presented here, it will be interesting to see whether deletion of the coilin C terminus exacerbates, or possibly even alleviates, the phenotype of SMN heterozygotes (Jablonka et al. 2000) when animals bearing these mutations are crossed.
Materials and methods
Plasmid construction and mutagenesis
For GFP-tagged fusion proteins, we used the pEGFP vector series (Clontech). GFP-tagged coilin has been described previously (Hebert and Matera 2000). The GFP-C214 construct, in which the C-terminal 214 amino acids of coilin were cloned in frame with GFP, was generated by use of _Pst_I restriction sites in coilin and the GFP vector. To make myc-coilinΔNΔC (coilin residues 94–482), we introduced a stop codon at residue 482 in myc-coilin(94–576), by using the Quick Change Mutagenesis kit (Stratagene). To mutate the coilin RG box, we mutagenized templates by using mtRG forward primer: 5′-G GGA CGG GGC ATG GGA GGC GGA GGT GGA GGA GGA GGC CAT CCT GTT TCC-3′ and mtRG reverse primer 5′-GGA AAC AGG ATG GCC TCC TCC TCC ACC TCC GCC TCC CAT GCC CCG TCC C-3′ (R to G codon changes are underlined). The RG box in myc-coilinΔNΔC was deleted using partially overlapping primers: ΔRG forward 5′-ATC GTC GCA GGA TCC GGG CAT CCT GTT TCC TGT GTT GTA AAT AG-3′ and ΔRG reverse 5′-GGA TCC TGC GAC GAT TCC TAA ACT AGC AGG GAG AGA CAC ACT GGG-3′. The RG box in GFP-mouse coilin (Tucker et al. 2001) was deleted using partially overlapping primers: ΔRGm forward 5′-ATC GTC GCA GGA TCC GGG CAA GCG GTC TCC TGT GTC TTT AAT AG-3′ and ΔRGm reverse 5′-GGA TCC TGC GAC GAT TCC CAA ACT GGT GGG GAC AGG CGT ACT GGG-3′ (the underlined regions mark an overlapping, in-frame linker). By using these primers in a Quick Change reaction, we were able to generate a deletion of the entire RG box in both human and mouse backgrounds and include the five-residue linker. The GST fusions harboring SMN, SMN(ΔEx7), SMN(Ex1–3), and SMN(Ex3) were kind gifts of E. Androphy (Tufts University, Boston, MA) and C. Lorson (Arizona State University, Tempe). GST–SMN(Tudor) (residues 83–173) was a gift from P. Selenko (EMBL, Heidelberg, Germany). Human myc-tagged SMN was a kind gift of G. Dreyfuss (HHMI, University of Pennsylvania, Philadelphia). The SmB′ cDNA (a kind gift of T. Gray, Wadsworth Center, Albany, NY) was cloned in frame with GST in pGEX-2T (Pharmacia) and His-T7 into pET28a (Novagen) by PCR using specific primers. The C214 fragment of coilin was cloned into pGEX-3X and pET28a by using standard molecular biological techniques, as was the cloning of SMN into pET28a. Constructs were verified by sequencing and Western blotting.
GST-pulldown assays
GST- and His-tagged constructs were transformed into Escherichia coli BL21(DE3)pLysS cells, induced, and purified using either glutathione beads (Pharmacia) or Ni2+ agarose beads (QIAGEN), as per the manufacturer’s specifications. His-tagged proteins were dialyzed after elution against 20 mM Tris at pH 7.5, 300 mM NaCl, 2 mM EDTA, 40% glycerol, and one tablet per 50 mL of Complete EDTA-free protease inhibitor cocktail tablets (Roche). The Bradford assay reagent from Bio-Rad was used to quantify the concentrations of purified components. Recombinant proteins were checked by Coomassie staining and Western blotting to ensure purity and lack of degradation. Binding reactions typically contained 1 μg of the GST fusion protein (on beads) and ∼0.6 μg of the His-T7-tagged protein in 1 mL of a modified RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA), plus 2 mM dithiothreitol (DTT). The reactions were incubated with gentle inversion for 1 h at 4°C, followed by 5 washes (1 mL each) with RIPA plus DTT. The beads were resuspended in 15 μL 5 × SDS loading buffer, boiled, and subjected to SDS-PAGE as described (Hebert and Matera 2000). The assays shown in Figure 3, D and E, were conducted as above except that a buffer adapted from Charroux et al. (1999) was used. Namely, these reactions were incubated and washed in 50 mM Tris-HCl at pH 7.5, 100 mM NaCl, 0.05% NP-40, 2 mM EDTA, and 2 mM DTT. Primary antibodies used include anti-SMN 7B10 (1 : 1000; Meister et al. 2000), anti-SMN N-19 (1 : 100; Santa Cruz), anti-T7 (1 : 1000; Novagen), anti-GST (1 : 1000; Santa Cruz), and anticoilin R288 (1 : 500; Andrade et al. 1993). The competition experiments (Fig. 5) were performed by incubating a fixed amount of the GST fusion with a constant amount of His–T7–SmB′ (Fig. 5A,B) or His–T7–SMN (Fig. 5C). Separate reactions were conducted with increasing levels of the competitor protein. The beads were washed as described above and analyzed by SDS-PAGE and Western blotting.
Immunoprecipitation, transfection, and immunofluorescence
HeLa cells were grown in DMEM (GIBCO BRL), supplemented with 10% FBS (GIBCO BRL), and then harvested, washed in PBS, and snap-frozen in liquid nitrogen for 30 sec. One hundred microliters of modified RIPA buffer was added to the cell pellet. A suspension then was generated using a Teflon pestle (10–15 strokes), and an additional 900 μL of RIPA was added. The cell suspension then was incubated for 30 min at 4°C with gentle inversion, followed by centrifugation to pellet cellular debris. The lysate subsequently was incubated with the anti-SMN antibody 7B10 (Meister et al. 2000) or an equivalent amount of normal mouse serum. The reactions were incubated overnight at 4°C with gentle inversion, followed by the addition of 60 μL of 50% Protein G–Sepharose (Pharmacia) for 2 h. The beads were washed five times with 1 mL RIPA, resuspended in 30 μL of 5× SDS loading buffer, boiled, and subjected to SDS-PAGE as described (Hebert and Matera 2000), followed by detection of coilin using anticoilin antibody R288 (1 : 500; Andrade et al. 1993). Where indicated, HeLa cells were transfected for 24 h using SuperFect (QIAGEN) according to the manufacturer’s specifications. For coimmunoprecipitation of endogenous SMN with various coilin fragments, cells were transfected and lysed as described above. Lysates containing GFP-tagged proteins were incubated with anti-GFP antibodies (Clontech), followed by incubation with Protein G–Sepharose beads, and washed as described above. Myc-tagged coilin constructs were immunoprecipitated with anti-myc antibodies (clone 9E10; Santa Cruz) and incubated with Protein G–Sepharose. After SDS-PAGE and transfer, SMN was detected by use of anti-SMN antibody 7B10 (1 : 1000). MEF cells were transfected using LipofectAMINE (GIBCO BRL) for 24 h according to the manufacturer's specifications. Cells were grown on chambered slides (Nunc), fixed in paraformaldehyde, and permeabilized in Triton X-100 as described (Frey and Matera 1995). The cells were incubated with a polyclonal anti-myc antibody (1 : 40; Santa Cruz) to detect SMN and the anti-Sm monoclonal antibody Y12 (1 : 800; Lerner et al. 1981). Fluorochrome conjugated secondary antibodies anti-rabbit Texas Red (Molecular Probes) and anti-mouse Cy5 (Amersham) were used and images were acquired and processed as described (Hebert and Matera 2000).
Acknowledgments
We thank K. Tucker for helpful advice and insight regarding the coilin RG-box motif. We are indebted to E. Androphy, E. Chan, G. Dreyfuss, U. Fischer, T. Gray, M. Sattler, and J. Steitz for critical reagents. M.D.H. was supported in part by an NIH postdoctoral training fellowship (T32-HD07518). This work was supported by grants to A.G.M. from the NIH (GM53034 and NS41617) and the Muscular Dystrophy Association.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gxm26@po.cwru.edu; FAX (216) 368-3432.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.908401.
References
- Andrade LEC, Tan EM, Chan EKL. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DW, Gall JG. Coiled bodies without coilin. Mol Biol Cell. 1997;8:73–82. doi: 10.1091/mbc.8.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Lührmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem. 2000;275:17122–17129. doi: 10.1074/jbc.M000300200. [DOI] [PubMed] [Google Scholar]
- Bühler D, Raker V, Lührmann R, Fischer U. Essential role for the Tudor domain of SMN in spliceosomal U snRNP assembly: Implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- Burghes AHM. When is a deletion not a deletion? When it is converted. Am J Hum Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3. A novel dead box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1194. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Yong J, Shevchenko A, Mann M, Dreyfuss G. Gemin4: A novel component of the SMN complex that is found in both gems and nucleoli. J Cell Biol. 2000;148:1177–1186. doi: 10.1083/jcb.148.6.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert D, Le T, McAndrew P, Strasswimmer J, Crawford T, Mendell J, Coulson S, Androphy E, Prior T, Burghes A. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- Dietz H. Polishing the cutting edge of gems. Nat Genet. 1998;20:321–322. doi: 10.1038/3784. [DOI] [PubMed] [Google Scholar]
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase cells. Proc Natl Acad Sci. 1995;92:5915–5919. doi: 10.1073/pnas.92.13.5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- Frugier T, Tiziano FD, Cifuentes-Diaz C, Miniou P, Roblot N, Dierich A, Le Meur M, Melki J. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the Survival Motor Neurons protein. Nat Cell Biol. 2001;3:376–383. doi: 10.1038/35070059. [DOI] [PubMed] [Google Scholar]
- Grundhoff AT, Kremmer E, Tureci O, Glieden A, Gindorf C, Atz J, Mueller-Lantzsch N, Schubach WH, Grässer FA. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J Biol Chem. 1999;274:19136–19144. doi: 10.1074/jbc.274.27.19136. [DOI] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- Jablonka S, Schrank B, Kralewski M, Rossoll W, Sendtner M. Reduced survival motor neuron (Smn) gene dose in mice leads to motor neuron degeneration: An animal model for spinal muscular atrophy type III. Hum Mol Genet. 2000;9:341–346. doi: 10.1093/hmg/9.3.341. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Earnshaw WC. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc Natl Acad Sci. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: Multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR. Coiled bodies and gems: Janus or Gemini? Am J Hum Genet. 1998;63:317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Hebert MD. The survival motor neurons protein uses its ZPR for nuclear localization. Nat Cell Biol. 2001;3:E93–E94. doi: 10.1038/35070157. [DOI] [PubMed] [Google Scholar]
- Meister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum Mol Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- Meister, G., Bühler, D., Pillai, R., Lottspeich, F., and Fischer, U. 2001. A multiprotein complex mediates ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. (in press). [DOI] [PubMed]
- Melki J. Spinal muscular atrophy. Curr Opin Neurol. 1997;10:381–385. doi: 10.1097/00019052-199710000-00005. [DOI] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossol W, Prior TW, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- Pearn J. Classification of spinal muscular atrophies. Lancet. 1980;8174:919–922. doi: 10.1016/s0140-6736(80)90847-8. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP. Tudor domains in proteins that interact with RNA. Trends Biochem Sci. 1997;22:51–52. doi: 10.1016/s0968-0004(96)30049-2. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrank B, Gotz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selenko P, Sprangers R, Stier G, Buhler D, Fischer U, Sattler M. SMN Tudor domain structure and its interaction with the Sm proteins. Nat Struct Biol. 2001;8:27–31. doi: 10.1038/83014. [DOI] [PubMed] [Google Scholar]
- Sleeman JE, Lamond AI. Nuclear organization of pre-mRNA splicing factors. Curr Opin Cell Biol. 1999;11:372–377. doi: 10.1016/S0955-0674(99)80052-0. [DOI] [PubMed] [Google Scholar]
- Talbot K, Davies KE. Spinal muscular atrophy. Semin Neurol. 2001;21:189–197. doi: 10.1055/s-2001-15264. [DOI] [PubMed] [Google Scholar]
- Tucker KE, Berciano MT, Jacobs EY, LePage D, Shpargel KB, Rossire JJ, Chan EKL, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy determining gene product. J Cell Biol. 2001;154:293–307. doi: 10.1083/jcb.200104083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PJ, Le TT, thi Man N, Burghes AH, Morris GE. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- Young PJ, Le TT, Dunckley M, thi Man N, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: Nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]