Fusing a lasting relationship between ER tubules (original) (raw)
. Author manuscript; available in PMC: 2012 Jul 1.
Published in final edited form as: Trends Cell Biol. 2011 May 6;21(7):416–423. doi: 10.1016/j.tcb.2011.03.009
Abstract
Atlastin is an integral membrane GTPase localized to the endoplasmic reticulum (ER). In vitro and in vivo analyses indicate that atlastin is a membrane fusogen capable of driving membrane fusion, suggesting a role in ER structure and maintenance. Interestingly, mutations in the human atlastin-1 gene, SPG3A, cause a form of autosomal dominant hereditary spastic paraplegia (HSP). The etiology of HSP is unclear but two predominant forms of the disorder are caused by mutant proteins that affect ER structure, formation, and maintenance in motor neurons. In this review, we describe what is known about the molecular mechanism of atlastin function and its potential role in HSP. Greater understanding of the function of atlastin and associated proteins should lend significant insight into normal ER biogenesis and maintenance, as well as the pathology of disease.
Membrane fusion in organelle function
The biogenesis and maintenance of eukaryotic organelles is a complex and dynamic process that requires many protein and lipid components in order to generate the compartmentalization of function that is typical of the eukaryotic cell. In some cases, like the Golgi complex, proper organelle function depends upon communication between different compartments via vesicular transport. In other cases, such as mitochondria, organelle structure is highly dynamic with membranes undergoing regular fission and fusion events, a process that is required for normal mitochondrial function. Yet a third category of organelles, which includes the ER, lysosomes, and perhaps peroxisomes, employs both vesicular transport and fusion of parts of the organelle to carry out their normal biochemical functions. In all cases, proper organelle function requires membrane fusion events.
All known biological membrane fusion is driven by specific fusion proteins, or fusogens. The first membrane fusion proteins characterized in detail were viral fusion proteins from enveloped viruses1–6. Intracellular fusion in the secretory pathway mediated by a protein family collectively known as SNAREs 7,8 has received much attention over the last 15 years. Recently, the identification of fusogens responsible for organelle fusion, such as occurs in ER and mitochondria, have received closer attention. Mitochondria undergo constant fusion and fission events to form highly dynamic networks whose morphology results from a balance between these two processes9–14. Mitochondrial shape changes are important for proper function and inheritance15 and loss of these dynamics can result in disease13,16,17. Two large GTPases called Mitofusins (Fzo)9,18–21 and OPA1 (MGM1)22–26 are thought to fuse the outer and inner mitochondrial membrane, respectively.
The ER is also a highly dynamic organelle that exists as an interconnected network of tubes and sheets27–29. While ER sheets are mostly perinuclear and contiguous with the outer nuclear membrane, peripheral tubular extensions of the ER move along microtubule tracks, and frequently join together by membrane fusion. Regions of the peripheral ER maintain close contact with virtually all other cytoplasmic organelles, including mitochondria, peroxisomes, chloroplasts, and Golgi, as well as the plasma membrane. These diverse associations might allow non-vesicular transport of ER-synthesized lipids and sterols as well as interorganelle calcium homeostasis29. Membrane fusion allows the ER to maintain a dynamic network that can quickly change shape and preserve lumen continuity while adapting to the changing cytoplasmic environment.
Insight into how ER fusion occurs was recently revealed by the identification of a new membrane fusion protein called atlastin30. In vitro and in vivo evidence suggest that atlastin is likely responsible for generating and maintaining the dynamic nature of peripheral ER tubules 30,31. In addition, atlastin dysfunction results in a form of Hereditary Spastic Paraplegia (HSP, also called familial spastic paraparesis or Strümpell-Lorrain disease), a group of inherited neurological disorders characterized by progressive lower extremity weakness and spasticity32,33. This intriguing evidence suggests that ongoing shape changes and lumen continuity are important for normal ER function, and that loss of these could be implicated in a disease state. In this review, we will focus on the mechanistic basis of ER membrane fusion driven by atlastin and discuss its potential role in the etiology of Hereditary Spastic Paraplegia.
Atlastin genetics, domain architecture, and structure
Atlastin and HSP
Atlastin is the product of the SPG3A (Spastic Paraplegia Gene 3A) locus34 and mutation of this gene is responsible for a form of HSP. The neuropathological basis for compromised motor function in HSP is likely length-dependent axonopathy of the corticospinal tract33. Genetic analysis has identified more than 40 different loci involved in HSP (SPG1-45)32,33,35,36 and 20 HSP loci have been molecularly identified35. Over half of all autosomal dominant HSP (ADHSP) cases occur due to mutation in one of three proteins: Spastin (SPAST, SPG4,), a microtubule severing protein; Atlastin-1 (ATL1, SPG3A), a GTPase; and receptor expression enhancing protein -1 (REEP1, SPG3137), first identified for its role in trafficking receptors to the plasma membrane. Spastin mutations occur in 40–45% of ADHSP38, while Atl1 and REEP1 account for 10%39 and 3%40, respectively. Molecular analysis of patients with HSP has identified 44 mutations in the ATL1 gene. All of these lesions are dominant alleles and mutations have been identified in all domains of the protein.
Domain architecture
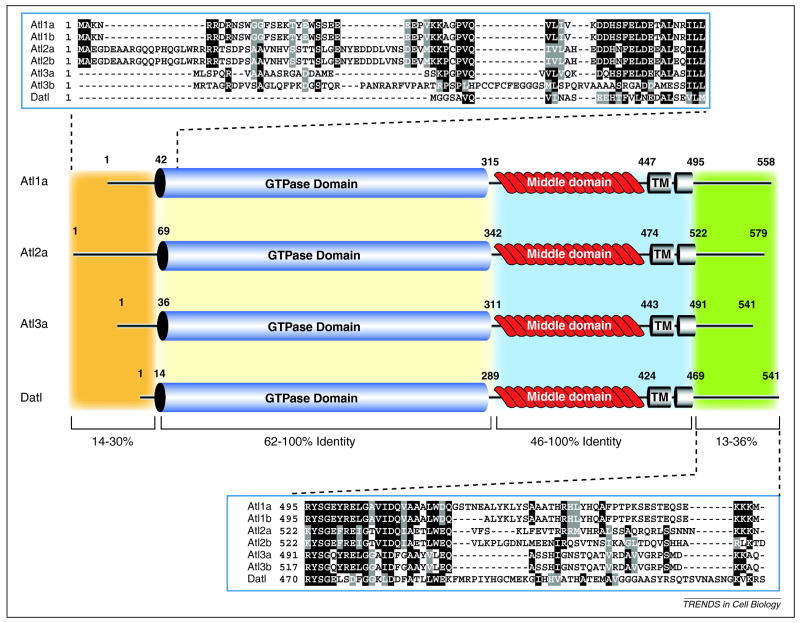
Human atlastin-1 (Atl1) is a 558 amino acid (63 kDa) multidomain protein (Figure 1). It has a short N-terminal domain, followed by a well-conserved GTPase domain, a middle domain with undefined function, two tandem transmembrane domains and a C-terminal cytoplasmic domain41. The original atlastin gene was renamed atlastin-1 following the identification of two additional paralogs (atlastin-2 and atlastin-3)41.
Figure 1. Domain architecture and protein similarity between Human and Drosophila atlastins.
Human Atl1a (NP_056999.2), Atl2a (NP_071769.2), Atl3a (NP_056274.3) and Datl (AAF56318.1) are shown. The number of amino acid residues and location of identified domains are indicated on the protein schematic. The three human paralogs and Drosophila ortholog are subdivided into four regions indicated by the colored boxes. The variable length N-terminus is colored orange, the highly conserved GTPase domain depicted as a blue cylinder, the INTERPRO domain IPR015894 (GBP-N-term)76 is colored tan, the “middle domain” containing the 2 tandem transmembrane domains is colored light blue and the variable length C-terminus is colored light green. Multiple sequence alignments (ClustalW2) of the extreme N- and C-terminus of all 6 human atlastin isoforms and Datl are shown (top and bottom) to highlight sequence divergence.
Atlastin homologs in other species share a very similar domain architecture, typified by Drosophila melanogaster atlastin (Datl). The Drosophila genome produces a single atlastin protein that is 541 amino acids in length (Figure 1). The Datl sequence is highly homologous with all three human isoforms, ranging between 44–49% identical (61–68% similar) over the entire length of the protein. Functional homologs of atlastin are also found in yeast (Sey1p) and plants (RHD3)42,43. These proteins share limited sequence similarity with human atlastin, yet they are GTPases and possess a similar domain structure31.
The atlastins are closest in sequence to the human guanylate binding proteins (GBPs). Together, they have been grouped with the dynamin family of GTPases and loosely termed “dynamin-like” family members44. The inclusion of atlastins and GBPs in this group is based entirely on homology within the N-terminal GTPase domain. The other conserved regions that define dynamins and are now known to provide membrane fission activity, including the GED, PH and PRD domains45, are absent in atlastin. Additionally, atlastin contains two tandem membrane spanning regions that are necessary for membrane fusion activity and are absent in dynamins. Distinct differences are even present within the conserved GTPase domain. Atlastins and GBPs share a unique RD motif in the G4 GTP-binding domain that interacts with the nucleotide base that is different from what is found in dynamins, dynamin-like proteins and even mitofusins44,46. Based on recent functional analysis of atlastin30 and the strong in vivo evidence that mitofusins are involved in mitochondrial membrane fusion47, we conclude that atlastins and mitofusins act as fusion GTPases, while dynamins and related fission molecules such as DRP1 act as fission GTPases.
All atlastins contain two hydrophobic regions that are predicted to span the membrane. Biochemical fractionation revealed that Atl1 is an integral membrane protein with both N- and C-termini exposed to the cytoplasm41. Most large GTPases form higher order oligomeric structures and Atl1 has been shown to self-associate. In vitro analysis of the N-terminal cytoplasmic domain of Atl1 revealed that it is a monomer that shifts to a size consistent with a dimer in the presence of nonhydrolyzable GTP48. However, immunoprecipitated full-length Atl1 migrated as an apparent homotetramer by gel filtration41, as did Atl2 and Atl349. Either the transmembrane segments and the C-terminal tail influence oligomerization, or the immunoprecipitated material from detergent extracts contained additional mass not attributable to atlastin. Attempts to identify heteroligomeric complexes has met with mixed results. Endogenous Atl1 and Atl2 do not coprecipitate other atlastins; however, mixed heteroligomeric complexes can be coprecipitated when atlastins are overexpressed49.
Atlastin structure
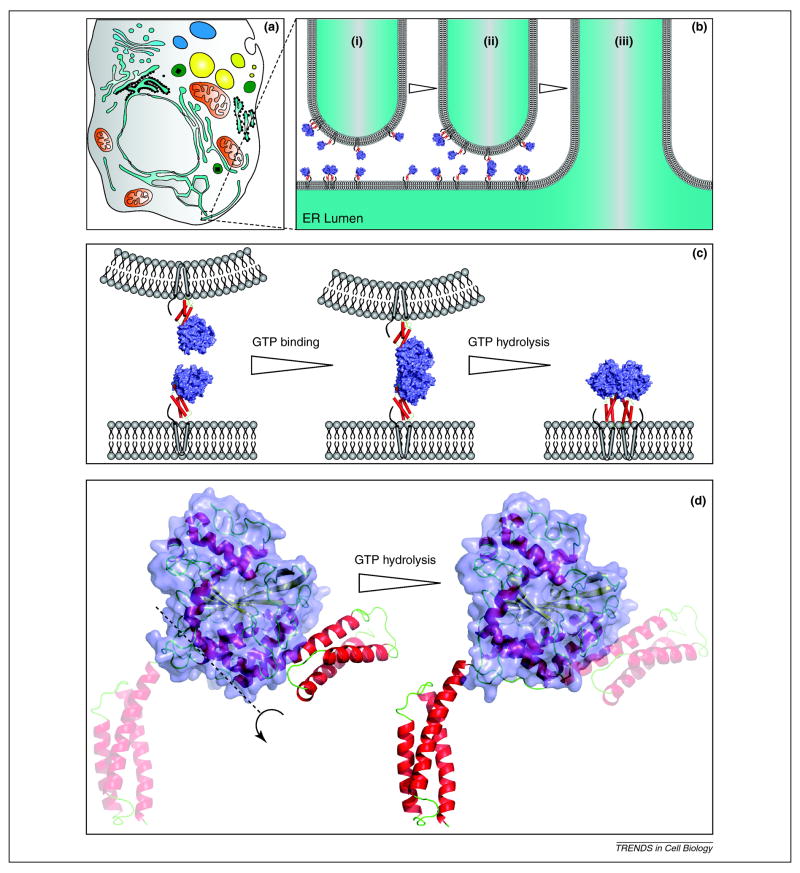
The structure of the N-terminal cytoplasmic domain (residues 1–446) of human Atl1 was solved by X-ray crystallography in two recent studies48,50 (Figure 2C). The globular GTPase domain bears a strong resemblance to the equivalent region of guanylate binding protein1 and dynamin. The middle domain folds into a three-helix bundle that connects to the GTPase domain by a flexible linker. Interestingly, two crystal forms were identified that differed in the relative arrangement of the three-helix bundle and the GTPase domain resulting from alterations in the linker (Figure 2C). Additionally, crystal packing contacts between adjacent monomers in the crystal suggested that dimerization could occur between GTPase domains, similar to GBPs48. Dimer models of both crystal forms suggest the intriguing possibility that form 2 is a dimer that could bridge membranes and that form 1 represents a post-fusion structure (Figure 2B and C) 51. While these dimer models are suggestive, they must be interpreted cautiously. Additionally, analysis of Atl1 monomers and dimers in solution by small angle X-ray scattering (SAXS) showed that the solution structure differs significantly from the static X-ray structures of the monomer and the hypothetical dimers48.
Figure 2. Proposed model for atlastin function in ER membrane fusion.
(A) A stylized eukaryotic cell depicting cytoplasmic organelles and the phycial relationship to the ER. A growing ER tubule approaching a peripheral ER tubule is highlighted in the dashed box. (B) A 50 nm diameter ER tubule is shown in proximity to a perpendicular ER tubule (i). Various atlastin molecules are shown to scale in both membranes. As the growing tube makes a close approach, form 2 atlastin monomers signal GTP binding and dimerization is favored (ii). GTP hydrolysis triggers the transition from form 2 to form 1 driving membrane fusion (iii). (C) Larger scale representations of the relevant interactions shown in panel B. (D) Structural models of Atl1crystal form 2 (left) and crystal form 1 (right). The proposed conformational changes that rotates the three-helix bundle out of the plane of the page about the axis marked by a dashed line results in form 2. The transparent three helix bundle on the left is the future location following GTP hydrolysis while the transparent three helix bundle on the right is the previous location after the conformational change. The GTPase domain is shown as a surface representation (blue) in all panels while the three-helix bundle segments are shown as red cylinders (B and C) or red helices (D). The location of the transmembrane domains is indicated by grey cylinders and the C-terminal tail as a thick black line in B and C. Atl1 crystal form 2 (3Q5E) and crystal form 1 (3Q5D) were rendered in pymol.
Atlastin function in cultured cells and model organisms
Localization
The subcellular distribution of atlastin has been examined in many cell types by a variety of techniques. Atl1 has been found in the ER52,53, Golgi41, and vesicular structures around the ER43. The Atl1 paralogs Atl2 and Atl3 have also both been localized to the ER49, as has Drosophila atlastin30. Recent analysis of Atl1 localization in zebrafish primary neurons found that Atl1 co-localized with endosomal markers54. All atlastins that have been examined contain an ER retention signal at their extreme C-terminus, are most commonly found in the ER, and therefore likely function in this compartment; however, additional sites of action cannot be excluded due to the observed localization to other compartments.
Phenotypic effects of up- and down-regulation of atlastin expression
HeLa cells normally express low levels of endogenous Atl1 and abundant amounts of Atl2 and Atl349. siRNA-mediated knockdown of Atl2 or Atl3 results in a normal ER morphology by fluorescence microscopy with a small subset of double knockdown cells showing more tubular, less reticular ER. The most pronounced effect of atlastin reduction is on Golgi morphology49. In this study, the Golgi in Atl2 or Atl3 knockdown cells was fragmented resembling “mini-stacks” or elongated tubules, however, protein trafficking was normal. The reason for the dramatic change in Golgi morphology and relatively minor effect on ER structure remains obscure. These morphological changes may be an indirect result of altered communication between the ER and subsequent secretory compartments that is not reflected in bulk protein traffic.
In this same study, overexpression of wildtype myc-Atl1 -2 or -3 did not noticeably affect ER morphology by light microscopy, but produced a fragmented Golgi49. However, overexpression of GTPase-deficient mutants of atlastin resulted in more elongated and tubular ER with less branching, as well as a fragmented Golgi. These effects may be due to a dominant negative effect of the overexpressed protein. Overexpression of either wildtype or mutant atlastins did not significantly affect protein trafficking. The effects of atlastin reduction were also examined in neurons, the primary site of action for HSP. Atl1 knockdown in primary rat cerebral cortical neurons showed reduced axonal length, an increased number of neurons without axons and a reduced number of dendrites per cell55. It is possible that specific ER functions such as appropriate calcium homeostasis are required for proper neurite outgrowth or axonal pathfinding and these functions are disrupted when atlastin-mediated fusion is lost.
Atlastin interactions
Atl1 has been shown to associate with spastin, a microtubule severing protein52,56,57, and recent work has also demonstrated that atlastin interacts with all of the ER tubule forming proteins in the Reticulon and REEP/Yop/DP1 family31,57. An interaction between Atl1 and spastin has been reported by several groups49,52,56,57, but there is conflicting evidence with regard to the interacting regions. In one case, the N-terminal 80 amino acids of spastin (which includes a putative membrane spanning domain) were required for an interaction with the N-terminal cytoplasmic domain of Atl1, a construct that lacks transmembrane segments52. In another series of experiments, the C-terminus of Atl1 (residues 408–558, which contains the transmembrane domains and the C-terminal tail) was required for an interaction with full-length spastin, suggesting a mutually exclusive region of Atl1 required for spastin interaction 56. Most recently, an association between three HSP-related proteins, atlastin, spastin and REEP1, was reported57. This work showed that the interaction between atlastin and REEP1 likely occurs through hydrophobic membrane spanning domains of each protein57, similar to atlastin s interaction with other reticulons31. This recent study also reported that the interaction between spastin and atlastin required the N-terminal transmembrane segment of the M1 isoform57 of spastin and the two tandem TMDs of atlastin. The disagreement in the literature on which portion of atlastin is responsible for the interaction with spastin suggests that this interaction should be interpreted cautiously. Additionally, spastin interaction with atlastin is limited to atlastin-1, since atlastin-2 and atlastin-3 do not appear to interact with spastin49. While an association between the ER tube forming proteins and atlastin may spatially restrict the location of atlastin within the ER tube where it is functionally required for fusion, a functional link between the microtubule severing protein spastin and atlastin could be utilized to organize ER tubules and the microtubule cytoskeleton into a regulated network.
Atlastin function in model organisms
The first phenotypic characterization of atlastin function in animals was done in Drosophila where a P-element insertion in the fly atlastin gene (_atl_1) was identified during a large scale behavioral screen58. _atl_1 mutants are temporarily paralyzed by mechanical shock and classified as “bang-sensitive”. More recently, another atlastin mutant (_atl_2) was generated by imprecise excision of the original _atl_1 P-element59. This ~1,600 bp deletion mutant is likely a null allele and is pupal lethal. Immunolocalization experiments reveal that Datl is broadly expressed in the larval CNS as well as body wall muscle, but is not enriched within synapses. Analysis of the _atl_2 mutant indicated that loss of atlastin produces subtle effects on the neuromuscular junctions such as slightly increased synaptic bouton number and marginally reduced body wall muscle size. Examination of microtubule organization in muscle suggests that Datl may regulate microtubule organization and dynamics, as has been suggested by association with spastin. Furthermore, these authors suggest that Datl may also functionally and physically interact with spastin.
Datl function has been examined in vivo by RNA interference. Reduction of Datl results in minor changes in ER structure when examined by fluorescence microscopy; however, transmission electron microscopy reveals substantial ER fragmentation30. Fluorescence loss in photobleaching (FLIP) experiments demonstrate that Datl depletion and ER fragmentation results in pervasive ER lumen discontinuity. This disorganization of the ER likely has profound effects on normal ER function. Moreover, overexpression of Datl in flies produces a grossly aberrant ER that also contains Golgi markers. When the ER was examined by electron microscopy, abnormally enlarged cisternae were also observed30. The cellular phenotypes of Datl reduction and overexpression strongly suggest that Datl plays an important role in ER maintenance and biogenesis. In the absence of Datl function, the ER fragments and continuity between ER subdomains is lost.
A recent study examined the consequences of altering Atl1 levels in the zebrafish Danio rerio by morpholino-mediated knockdown and overexpression54. Decreased levels of Atl1 resulted in aberrant spinal motor axons, which caused a reduction of larval movement. Molecular analysis indicated that bone morphogenic protein (BMP) signaling was upregulated in the knockdown and inhibited by Atl1 overexpression, suggesting a role in receptor trafficking. These authors also showed that Atl1 localizes to endosomes and that Atl1 deletion did not affect ER structure when examined by fluorescence microscopy. While it is possible that Atl1 functions differently in teleosts, the abnormalities seen in motor axons could also be explained by ER morphological changes not seen by light microscopy, although this remains to be tested. The presence of Atl1 in endosomes is a bit more difficult to reconcile with work from other species.
Atlastin is a fusion protein
The atlastin phenotypes observed in vivo in Drosophila are consistent with a role for Datl in the fusion of ER membranes. The protein(s) responsible for the fusion of ER membranes have been unclear until the recent functional characterization of Datl30. The role of atlastin as a membrane fusion protein was explored by reconstitution studies and enzymatic analysis30. Bacterially expressed Datl reconstituted into synthetic liposomes 60 was analyzed for membrane fusion activity utilizing a lipid mixing assay61 that has been used extensively for the analysis of membrane fusion by the SNARE proteins8,62–72. Atlastin promotes robust lipid mixing that is completely dependent on divalent metals and GTP. No fusion occurs when non-hydrolyzable GTP analogs are included in the reaction. Additionally, a mutant atlastin (K51A) that prevents nucleotide binding does not support membrane fusion. In fact, this mutant is a dominant inhibitor of membrane fusion when liposomes that contain wildtype atlastin are mixed with the liposomes containing the K51A mutant atlastin.
Additional support for the role of atlastin as an ER fusogen was recently provided by antibody inhibition studies of ER tubule formation en route to nuclear envelope regeneration in Xenopus egg extracts31. Reformation of ER tubules from fragmented ER vesicles is known to be a GTP-dependent process73, and antibodies against Xenopus atlastin inhibits tubule reformation similar to GTPγS31,57. This result suggests that atlastin plays a role in ER tubule formation in Xenopus.
The structure-function studies of Datl30 and the recent crystal structure of Atl148 have led us to develop the following working model of atlastin function in membrane fusion (Figure 2). ER tubules often form three-way junctions when the tip of a mobile tubule fuses with the side of an existing tubule (Figure 2A and B). Atlastin may localize along the length and tips of the tube, perhaps directly though an association with the reticulon/REEP tube forming proteins. We suggest that atlastin exists in multiple conformations, both monomer and cis-dimers (i.e. dimers within the same membrane). We also make the assumption that atlastin undergoes a monomer to dimer transition that is dependent on bound nucleotide. This assumption is supported by gel filtration data with Atl148 and our unpublished data. The observation that Atl1 crystalizes in two distinct forms suggests that conformational rearrangement of the middle domain three-helix relative to the GTPase domain may also be important for the fusion mechanism. Crystal form 2 (Figure 2C) is oriented in a manner that would allow dimerization between the GTPase domains and connect adjacent bilayers (Figure 2B and C). We model this interaction as a first point of contact. An interaction between catalytic surfaces of the GTPase domain is plausible based on crystal packing contacts and homology with GBP1; however, we favor the idea that additional associations between juxtamembrane regions are required for fusion, based on the mechanism of other known fusion proteins such as SNAREs and viral envelope protein. We suggest that GTP binding permits association between the GTPase domains, although this interaction is unlikely to be the major contacts that drive dimerization. Figure 2C (left) depicts the crystal form 2 structures interacting between the membrane of an approaching tubule tip and the side of an established tubule, promoted by GTP binding. Nucleotide hydrolysis would then drive a structural rearrangement of the linker region that repositions the middle domain three-helix bundle. The transition from crystal form 2 to crystal form 1 could be accomplished by rotating the three-helix bundle about the axis indicated in Figure 2D. Since the C-terminus of the mobile three-helix bundle is firmly anchored in the membrane by two transmembrane domains, the movement shown in the schematic would induce close proximity between the two opposing bilayers, perhaps close enough to exclude water in the hydration shell. Additionally, the mechanical motion generated by the three-helix bundle transition could distort and sufficiently perturb the phospholipid bilayer structure to induce lipid mixing and membrane fusion.
Concluding remarks
The ER provides a plethora of functions required for normal cell function ranging from calcium homoestasis, glycosylation, and lipid biosynthesis, to acting as the initial way station for all secreted proteins, as well as most transmembrane proteins. Given the fundamental importance of such ER functions, it’s not difficult to envision how disrupting ER function could cause dire cellular consequences. Less obvious is the need to maintain specific ER morphologies. Why does the ER need to be constructed as sheet and tubes? Perhaps form does follow function and the diversity of ER biochemistry requires its elaborate reticular structure. Analysis of the ER fusion protein atlastin further suggests that the dynamic relationship between tube and sheet structures, and the ability to generate new connections, is also paramount.
Considering our limited understanding of the functional consequences of ER morphological changes, it is difficult to precisely interpret all of the phenotypes of atlastin misexpression. Regardless, cogent arguments can be made to explain many of them in the context of ER dysfunction. The pathology of HSP suggests that long motor neurons may be one of the most susceptible tissues to alterations in ER structure. Perhaps ER tubules that provide luminal connections to the cell body are needed in the synapses of these very long axons. The inability of motor neurons with mutations in Atl1 to generate ER that spans the physical distance between synaptic boutons and the cell body may lead to inappropriate neuronal activity and eventually neurodegeneration.
The identification of atlastin as a novel GTP-dependent membrane fusion protein also expands our understanding of the ways biological membranes merge. Traditional membrane fusion proteins such as viral fusion proteins and SNAREs use energy derived from metastable protein folding intermediates to drive fusion74,75. The use of chemical energy in the form of nucleotide hydrolysis at the point of membrane fusion is unique to a new class of membrane fusion protein exemplified by atlastin and perhaps mitofusin. Detailed analysis of this type of fusion mechanism has yet to be explored. Future mechanistic analysis will determine how atlastin uses GTP hydrolysis to generate force and move lipids. The knowledge that atlastin controls ER fusion opens an exciting new area of possibilities for understanding ER morphogenesis and function in general, as well as a providing a potential mechanistic basis of the pathophysiology of Hereditary Spastic Paraplegia.
Acknowledgments
We would like to thank Joseph Faust and Avani Verma for comments on the manuscript. Work in the McNew lab is supported by funds from the National Institutes of Health (GM071832) and the G. Harold and Leila Y. Mathers Foundation. Work in the Daga lab is supported by grants from the Italian Ministry of Health, the Association Française contre les Myopathies and the Fondazione Telethon.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blumenthal R, Clague MJ, Durell SR, Epand RM. Membrane fusion. Chem Rev. 2003;103:53–69. doi: 10.1021/cr000036+. [DOI] [PubMed] [Google Scholar]
- 2.Zaitseva E, Mittal A, Griffin DE, Chernomordik LV. Class II fusion protein of alphaviruses drives membrane fusion through the same pathway as class I proteins. J Cell Biol. 2005;169:167–77. doi: 10.1083/jcb.200412059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kielian M, Rey FA. Virus membrane-fusion proteins: more than one way to make a hairpin. Nat Rev Microbiol. 2006;4:67–76. doi: 10.1038/nrmicro1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martens S, McMahon HT. Mechanisms of membrane fusion: disparate players and common principles. Nat Rev Mol Cell Biol. 2008;9:543–56. doi: 10.1038/nrm2417. [DOI] [PubMed] [Google Scholar]
- 5.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–8. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sollner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362:318–24. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 8.Weber T, et al. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–72. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 9.Hoppins S, Nunnari J. The molecular mechanism of mitochondrial fusion. Biochim Biophys Acta. 2009;1793:20–6. doi: 10.1016/j.bbamcr.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 10.Lackner LL, Nunnari JM. The molecular mechanism and cellular functions of mitochondrial division. Biochim Biophys Acta. 2009;1792:1138–44. doi: 10.1016/j.bbadis.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan DC. Mitochondrial fusion and fission in mammals. Annu Rev Cell Dev Biol. 2006;22:79–99. doi: 10.1146/annurev.cellbio.22.010305.104638. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto K, Shaw JM. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu Rev Genet. 2005;39:503–36. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 13.Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol. 2007;8:870–9. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 14.Herzig S, Martinou JC. Mitochondrial dynamics: to be in good shape to survive. Curr Mol Med. 2008;8:131–7. doi: 10.2174/156652408783769625. [DOI] [PubMed] [Google Scholar]
- 15.Hermann GJ, Shaw JM. Mitochondrial dynamics in yeast. Annu Rev Cell Dev Biol. 1998;14:265–303. doi: 10.1146/annurev.cellbio.14.1.265. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Chan DC. Mitochondrial dynamics--fusion, fission, movement, and mitophagy--in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–76. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 18.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu Rev Biochem. 2007;76:751–80. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 19.Hermann GJ, et al. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J Cell Biol. 1998;143:359–73. doi: 10.1083/jcb.143.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legros F, Lombes A, Frachon P, Rojo M. Mitochondrial fusion in human cells is efficient, requires the inner membrane potential, and is mediated by mitofusins. Mol Biol Cell. 2002;13:4343–54. doi: 10.1091/mbc.E02-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–74. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 22.Kubokawa K, Miyashita T, Kubo Y. Isolation of a cDNA for a novel 120-kDa GTP-binding protein expressed in motor neurons in the salmon brain. FEBS Lett. 1998;431:231–5. doi: 10.1016/s0014-5793(98)00762-5. [DOI] [PubMed] [Google Scholar]
- 23.Wong ED, et al. The dynamin-related GTPase, Mgm1p, is an intermembrane space protein required for maintenance of fusion competent mitochondria. J Cell Biol. 2000;151:341–52. doi: 10.1083/jcb.151.2.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delettre C, et al. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat Genet. 2000;26:207–10. doi: 10.1038/79936. [DOI] [PubMed] [Google Scholar]
- 25.Alexander C, et al. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat Genet. 2000;26:211–5. doi: 10.1038/79944. [DOI] [PubMed] [Google Scholar]
- 26.Cipolat S, Martins de Brito O, Dal Zilio B, Scorrano L. OPA1 requires mitofusin 1 to promote mitochondrial fusion. Proc Natl Acad Sci U S A. 2004;101:15927–32. doi: 10.1073/pnas.0407043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol. 2009;25:329–54. doi: 10.1146/annurev.cellbio.042308.113324. [DOI] [PubMed] [Google Scholar]
- 28.Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–9. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 29.English AR, Zurek N, Voeltz GK. Peripheral ER structure and function. Curr Opin Cell Biol. 2009;21:596–602. doi: 10.1016/j.ceb.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orso G, et al. Homotypic fusion of ER membranes requires the dynamin-like GTPase Atlastin. Nature. 2009;460:978–83. doi: 10.1038/nature08280. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, et al. A class of dynamin-like GTPases involved in the generation of the tubular ER network. Cell. 2009;138:549–61. doi: 10.1016/j.cell.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fink JK. Advances in the hereditary spastic paraplegias. Exp Neurol. 2003;184 (Suppl 1):S106–10. doi: 10.1016/j.expneurol.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Salinas S, Proukakis C, Crosby A, Warner TT. Hereditary spastic paraplegia: clinical features and pathogenetic mechanisms. Lancet Neurol. 2008;7:1127–38. doi: 10.1016/S1474-4422(08)70258-8. [DOI] [PubMed] [Google Scholar]
- 34.Zhao X, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29:326–31. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- 35.Blackstone C, O'Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2010;1:31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durr A. Genetic testing for the spastic paraplegias: drowning by numbers. Neurology. 2008;71:236–8. doi: 10.1212/01.wnl.0000320131.36091.a5. [DOI] [PubMed] [Google Scholar]
- 37.Zuchner S, et al. Mutations in the novel mitochondrial protein REEP1 cause hereditary spastic paraplegia type 31. Am J Hum Genet. 2006;79:365–9. doi: 10.1086/505361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hazan J, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 39.Namekawa M, et al. SPG3A is the most frequent cause of hereditary spastic paraplegia with onset before age 10 years. Neurology. 2006;66:112–4. doi: 10.1212/01.wnl.0000191390.20564.8e. [DOI] [PubMed] [Google Scholar]
- 40.Beetz C, et al. REEP1 mutation spectrum and genotype/phenotype correlation in hereditary spastic paraplegia type 31. Brain. 2008;131:1078–86. doi: 10.1093/brain/awn026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu PP, et al. Cellular localization, oligomerization, and membrane association of the hereditary spastic paraplegia 3A (SPG3A) protein atlastin. J Biol Chem. 2003;278:49063–71. doi: 10.1074/jbc.M306702200. [DOI] [PubMed] [Google Scholar]
- 42.Brands A, Ho TH. Function of a plant stress-induced gene, HVA22. Synthetic enhancement screen with its yeast homolog reveals its role in vesicular traffic. Plant Physiol. 2002;130:1121–31. doi: 10.1104/pp.007716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- 44.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–47. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 45.Mettlen M, Pucadyil T, Ramachandran R, Schmid SL. Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans. 2009;37:1022–6. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–4. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 47.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol. 2010;11:872–84. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 48.Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rismanchi N, Soderblom C, Stadler J, Zhu PP, Blackstone C. Atlastin GTPases are required for Golgi apparatus and ER morphogenesis. Hum Mol Genet. 2008;17:1591–604. doi: 10.1093/hmg/ddn046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bian X, et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc Natl Acad Sci U S A. 2011;108:3976–81. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daumke O, Praefcke GJ. Structural insights into membrane fusion at the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1019194108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sanderson CM, et al. Spastin and atlastin, two proteins mutated in autosomal-dominant hereditary spastic paraplegia, are binding partners. Hum Mol Genet. 2006;15:307–18. doi: 10.1093/hmg/ddi447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Namekawa M, et al. Mutations in the SPG3A gene encoding the GTPase atlastin interfere with vesicle trafficking in the ER/Golgi interface and Golgi morphogenesis. Mol Cell Neurosci. 2007;35:1–13. doi: 10.1016/j.mcn.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Fassier C, et al. Zebrafish atlastin controls motility and spinal motor axon architecture via inhibition of the BMP pathway. Nat Neurosci. 2010;13:1380–7. doi: 10.1038/nn.2662. [DOI] [PubMed] [Google Scholar]
- 55.Zhu PP, Soderblom C, Tao-Cheng JH, Stadler J, Blackstone C. SPG3A protein atlastin-1 is enriched in growth cones and promotes axon elongation during neuronal development. Hum Mol Genet. 2006;15:1343–53. doi: 10.1093/hmg/ddl054. [DOI] [PubMed] [Google Scholar]
- 56.Evans K, et al. Interaction of two hereditary spastic paraplegia gene products, spastin and atlastin, suggests a common pathway for axonal maintenance. Proc Natl Acad Sci U S A. 2006;103:10666–71. doi: 10.1073/pnas.0510863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Park SH, Zhu PP, Parker RL, Blackstone C. Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest. 2010 doi: 10.1172/JCI40979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee Y, et al. Loss of spastic paraplegia gene atlastin induces age-dependent death of dopaminergic neurons in Drosophila. Neurobiol Aging. 2008;29:84–94. doi: 10.1016/j.neurobiolaging.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 59.Lee M, et al. Drosophila Atlastin regulates the stability of muscle microtubules and is required for synapse development. Dev Biol. 2009;330:250–62. doi: 10.1016/j.ydbio.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 60.Rigaud JL, Levy D. Reconstitution of membrane proteins into liposomes. Methods Enzymol. 2003;372:65–86. doi: 10.1016/S0076-6879(03)72004-7. [DOI] [PubMed] [Google Scholar]
- 61.Struck DK, Hoekstra D, Pagano RE. Use of resonance energy transfer to monitor membrane fusion. Biochemistry. 1981;20:4093–9. doi: 10.1021/bi00517a023. [DOI] [PubMed] [Google Scholar]
- 62.McNew JA, et al. Compartmental specificity of cellular membrane fusion encoded in SNARE proteins. Nature. 2000;407:153–9. doi: 10.1038/35025000. [DOI] [PubMed] [Google Scholar]
- 63.McNew JA, Weber T, Engelman DM, Sollner TH, Rothman JE. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol Cell. 1999;4:415–21. doi: 10.1016/s1097-2765(00)80343-3. [DOI] [PubMed] [Google Scholar]
- 64.McNew JA, et al. Close is not enough: SNARE-dependent membrane fusion requires an active mechanism that transduces force to membrane anchors. J Cell Biol. 2000;150:105–17. doi: 10.1083/jcb.150.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parlati F, et al. Topological restriction of SNARE-dependent membrane fusion. Nature. 2000;407:194–8. doi: 10.1038/35025076. [DOI] [PubMed] [Google Scholar]
- 66.Parlati F, et al. Rapid and efficient fusion of phospholipid vesicles by the alpha- helical core of a SNARE complex in the absence of an N-terminal regulatory domain. Proc Natl Acad Sci U S A. 1999;96:12565–70. doi: 10.1073/pnas.96.22.12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paumet F, Rahimian V, Rothman JE. The specificity of SNARE-dependent fusion is encoded in the SNARE motif. Proc Natl Acad Sci U S A. 2004;101:3376–80. doi: 10.1073/pnas.0400271101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tucker WC, Weber T, Chapman ER. Reconstitution of Ca2+-regulated membrane fusion by synaptotagmin and SNAREs. Science. 2004;304:435–8. doi: 10.1126/science.1097196. [DOI] [PubMed] [Google Scholar]
- 69.Lynch KL, et al. Synaptotagmin-1 utilizes membrane bending and SNARE binding to drive fusion pore expansion. Mol Biol Cell. 2008;19:5093–103. doi: 10.1091/mbc.E08-03-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu X, Zhang F, McNew JA, Shin YK. Membrane fusion induced by neuronal SNAREs transits through hemifusion. J Biol Chem. 2005;280:30538–41. doi: 10.1074/jbc.M506862200. [DOI] [PubMed] [Google Scholar]
- 71.Schaub JR, Lu X, Doneske B, Shin YK, McNew JA. Hemifusion arrest by complexin is relieved by Ca2+-synaptotagmin I. Nat Struct Mol Biol. 2006;13:748–50. doi: 10.1038/nsmb1124. [DOI] [PubMed] [Google Scholar]
- 72.Vicogne J, et al. Asymmetric phospholipid distribution drives in vitro reconstituted SNARE-dependent membrane fusion. Proc Natl Acad Sci U S A. 2006;103:14761–6. doi: 10.1073/pnas.0606881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dreier L, Rapoport TA. In vitro formation of the endoplasmic reticulum occurs independently of microtubules by a controlled fusion reaction. J Cell Biol. 2000;148:883–98. doi: 10.1083/jcb.148.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McNew JA. Regulation of SNARE-Mediated Membrane Fusion during Exocytosis. Chem Rev. 2008;108:1669–1686. doi: 10.1021/cr0782325. [DOI] [PubMed] [Google Scholar]
- 75.Skehel JJ, Wiley DC. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–4. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 76.Hunter S, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]