Launching the T-Lineage Developmental Programme (original) (raw)
. Author manuscript; available in PMC: 2011 Jul 7.
Published in final edited form as: Nat Rev Immunol. 2008 Jan;8(1):9–21. doi: 10.1038/nri2232
Preface
Multipotent blood progenitor cells enter the thymus and begin a protracted differentiation process in which they gradually acquire T-cell characteristics while shedding their legacy of developmental plasticity. Notch signalling and basic helix-loop-helix E-protein transcription factors collaborate repeatedly to trigger and sustain this process throughout the period leading up to T-cell lineage commitment. Nevertheless, the process is discontinuous with separately regulated steps that demand roles for additional collaborating factors. This review discusses new evidence on the coordination of specification and commitment in the early T-cell pathway; effects of microenvironmental signals; the inheritance of stem-cell regulatory factors; and the ensemble of transcription factors that modulate the effects of Notch and E proteins, to distinguish individual stages and to polarize T-lineage fate determination.
Mammalian T cells of diverse functional types share a complex developmental history. They originate from pluripotent precursors in the bone marrow or fetal liver, which migrate to the thymus, the sole function of which is to initiate and sustain T-cell differentiation. Relatively few T-cell progenitors migrate into the thymus per day, but they respond to this new environment by proliferating extensively, while initiating the T-cell differentiation transcriptional programme1–4, and gradually turning off genes that allow differentiation to non-T-cell lineages. They then undergo T-cell receptor (TCR) gene rearrangements and assemble TCR complexes, upon which their future survival and functions will depend. These cells can mature into different T-cell lineages, including γδ T cells and αβ T cells. The αβ T cells diverge further into different sublineages such as CD4+ T cells, CD8+ T cells and NKT cells as well as regulatory T cells (Treg), each with greatly differing functions once they emigrate from the thymus to the periphery. However, these diverse T-cell lineages share their early developmental history, and many of the key genes required in later T-cell functions are first expressed prior to TCR rearrangement. As the regulatory mechanisms that guide stem cells into the T-cell lineage become increasingly well-defined, a sequence of events is revealed that may make early T-cell development a particularly illuminating paradigm for stem-cell-based modes of development in general.
The T-lineage commitment process consists of an irreversible progression of distinct developmental stages. Each of these stages has the potential to be sustained over multiple cell cycles. This implies that particular regulatory changes, perhaps environmentally triggered, must intervene to advance the cells from one state to the next. Clear changes in gene expression and developmental potential mark these transitions. Yet genetic evidence, from germline and conditional knockouts, emphasizes the requirements for a stable core group of transcription factors that act repeatedly at successive stages. In this Review, we explore how this image of regulatory persistence can be modified to explain an outcome of dramatic and irreversible developmental change.
Landmarks for early T-cell development
We first review what happens in T-cell specification, before considering why it happens. From fetal through adult life, T-cell precursors develop through distinct stages defined by multiparameter flow cytometry1,5–8. In mice, cell-transfer experiments in vivo, as well as clonal-cell- culture experiments in vitro, have revealed at which stages these cells lose the non-T-lineage developmental potential inherited from their stem-cell progenitors and when they gain specific T-lineage characteristics9–13. The exact types of precursor cells that can participate, the branch points, the alternative pathways, and the kinetics of T-cell development are increasingly well-defined in mice1,8,9,14,15 and humans7,16. This evidence, spanning contributions of many groups for at least two decades, is summarized for the murine case in Fig. 1, Table 1, and Box 1.
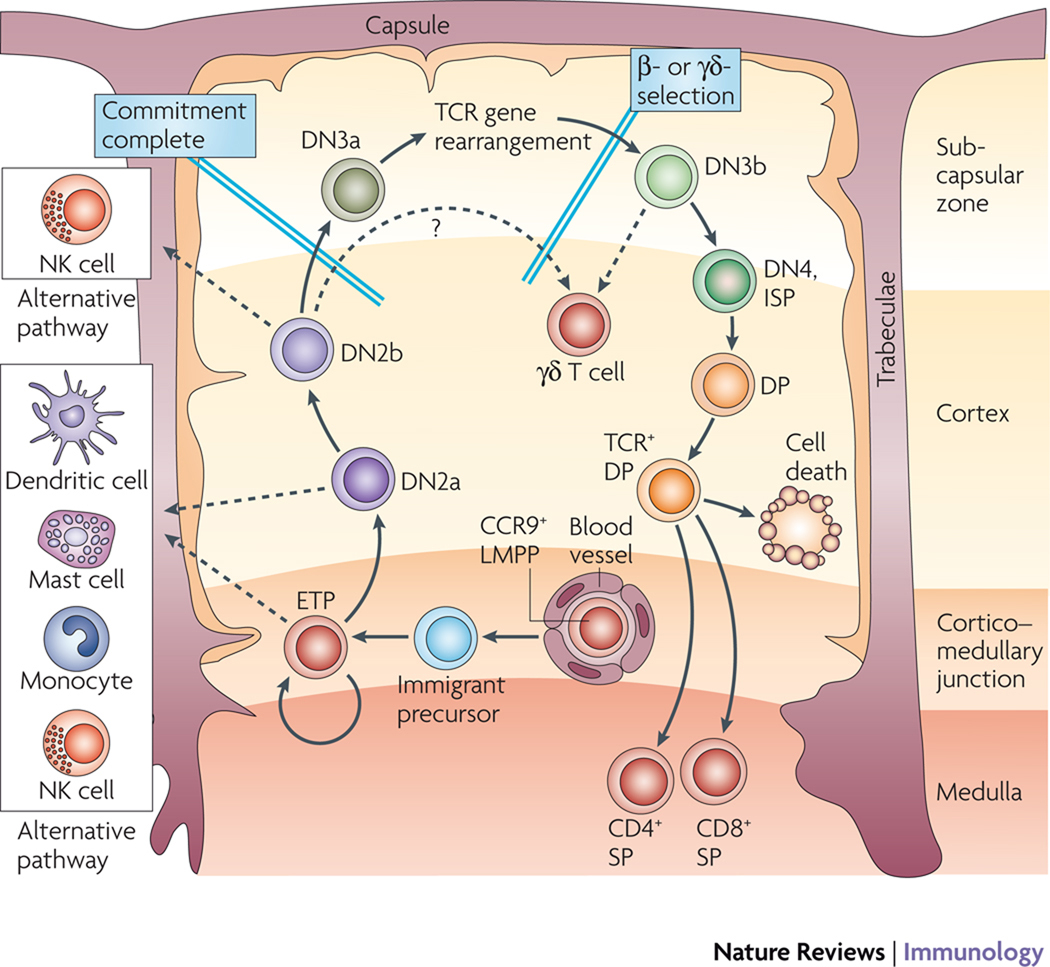
Figure 1. Stages in early T-cell development.
Top panel: Path of T-cell precursors through the thymus during development. A major pathway for adult murine thymocyte development is depicted, with cells shown from the point of entry into the thymus at the cortico–medullary junction through their migration to the outer rim of the cortex during the progression toward commitment, on the left-hand path. Early T-cell precursors (ETP), double negative (DN) 2 subsets, DN3 subsets, and DN4 cells are defined as described in Table 1. In the adult thymus, the immigrants initially enter the thymus through vessels near the cortical-medullary junction, and migrate outward through the thymus as shown as they pass through the indicated stages8. Note that at any given cell cycle, the ETPs appear to have the options either to continue their expansion, with minimal differentiation, in the cortical-medullary junction region or to differentiate into DN2 cells (presumably, DN2a cells as shown) and begin their migration outward through the cortex. For β-selection, they are situated in the extreme outer portion of the thymus (subscapsular zone). The reversal of migration for later stages of thymocyte development is shown on the right-hand path. Broken arrows depict alternative developmental pathways that are still open to ETP and different subsets of DN2 cells that are likely to correspond to DN2a and DN2b cells.
TABLE 1.
PHENOTYPIC MARKERS OF MURINE EARLY T-LINEAGE PRECURSORS
| ETP | DN2a* | DN2b* | DN3a | DN3b | DN4 | |
|---|---|---|---|---|---|---|
| c-KIT | Hi | Hi | Int | Lo | Lo | Lo |
| CD44 | Hi | Hi | Hi | Lo | Lo | Lo |
| CD25 | Neg | Hi | Hi | Hi | Hi/Int | Lo |
| CD24 | Neg/Int | Hi | Hi | Hi | Hi | Hi |
| CD27 | Hi | Hi | Int | Lo | Hi | Hi |
| Thy-1 | Lo | Hi | Hi | Hi | Hi | Hi |
| Proliferation | + | + | + | − | + | + |
| TCRβ rearrangement | GL > DJH | GL > DJH | GL > DJH | DJH,VDJH | VDJ+ | VDJ+ |
BOX 1: Purification of early thymic subsets.
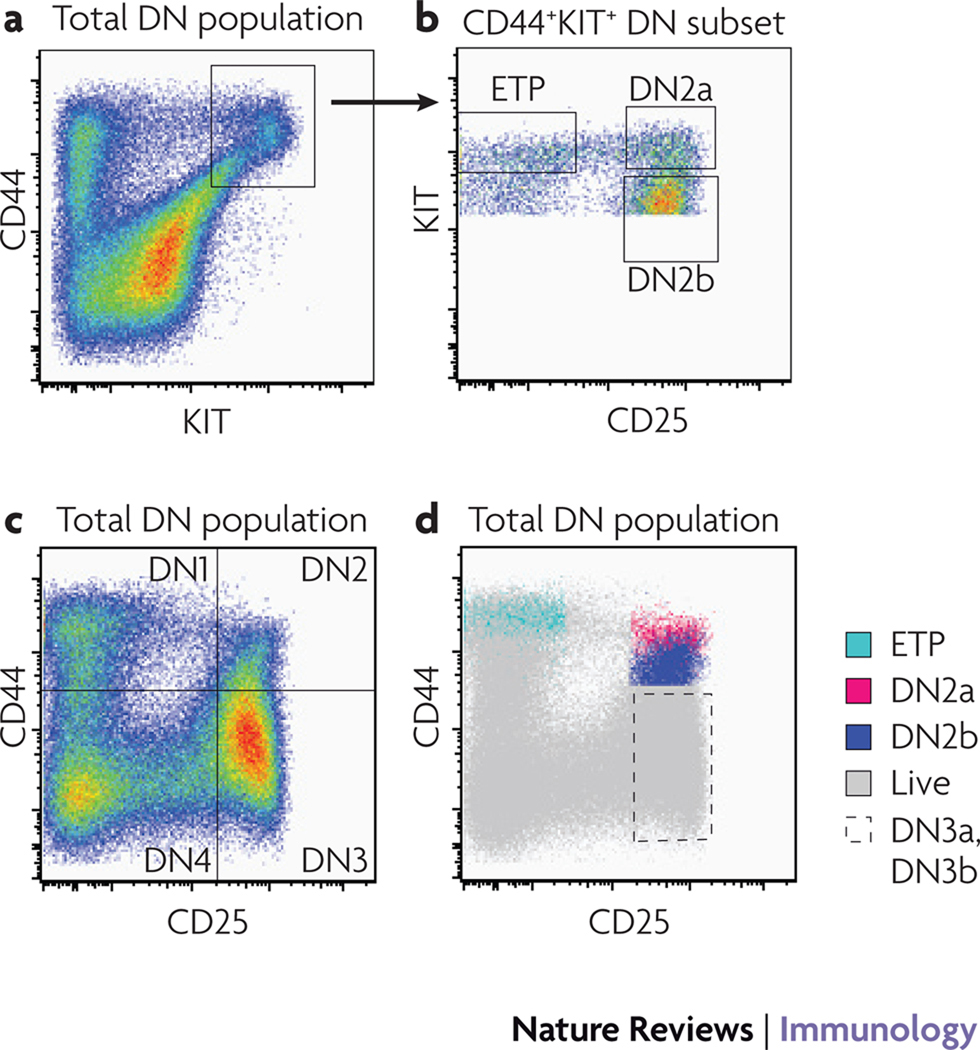
A high level of cell-subset purity is particularly important to allow gene expression to be analysed reproducibly and for the results to be correlated meaningfully with developmental assays. Although the CD4−CD8− double negative (DN) early T-cell subsets are usefully subdivided by the expression of CD25 and CD44, those parameters alone are insufficient for rigorous purification of DN subsets without contaminating cells6. In particular, the CD25−CD44+ so-called DN1 population is very heterogeneous and only a minority of thymocytes with this phenotype are actually early T-cell precursors25,131. In addition to unrelated haematopoietic cells, this population is often contaminated with mature natural killer (NK) cells, NKT cells and γδ T cells. Such cells often share high levels of CD44 expression with genuine T-cell precursors, and even when depletion protocols designed to remove cells expressing the T-cell receptor (TCR) and NK-cell markers as well as cells expressing CD4 and CD8, the low levels of expression of mature cell-surface receptors on these cell types cause inefficient depletion. Additional use of selection protocols based on surface expression of c-KIT allows for more efficient purification of the potent T-lineage precursors6,25,131, and in this Review we use the term ETP (early T-cell precursor)25 to refer to the c-KIT++ DN1 cells (Fig. 1, Fig. 3). Likewise, evidence is accumulating that DN2 cells are not a homogenous population. They can be subdivided by cell-surface expression levels of c-KIT into DN2a and DN2b populations (Fig. 3), and recent data show that this distinction may correspond to the loss of dendritic-cell potential33. DN3 cells can also be subdivided into those before and immediately after β (or γδ)-selection (DN3a and DN3b, respectively) based on size132 and CD27 expression27. These finer distinctions between sequential developmental stages give a sharper resolution to the changing gene regulatory events that take place during the course of T-cell differentiation.
Curiously, several distinct progenitor types can enter the T-cell pathway9,15. But one major progenitor source in adult mice consists of lympho-myeloid pluripotent precursors (LMPPs) in the bone marrow or blood, which can give rise to macrophages, dendritic cells (DCs), natural killer (NK) cells, B cells and T cells, but not erythrocytes or megakaryocytes17,18. The earliest human thymic precursors also have lymphoid and myeloid potential19,20. Within the murine LMPP population, cells with the capacity to migrate to the thymus are probably distinguished by expression of the CC chemokine receptor 9 (CCR9), in addition to the stem- and progenitor-cell markers cKIT (also known as CD117), stem-cell antigen 1 (SCA1), and the growth-factor-receptor tyrosine kinase FLT3 (also called FLK2), which define LMPPs9,21,22. Prethymic cells in the fetal blood and liver might have somewhat different properties from postnatal thymus-seeding cells23,24.
Development through the first “pro-T cell” stages, from early T precursor (ETP) to double negative 3 (DN3), is TCR independent and is coordinated with migration through distinct thymic microenvironments8 (Fig. 1). In ETP and DN2 stages, cells proliferate extensively while acquiring their first T-cell characteristics. Although precursors may actually initiate TCR (or immunoglobulin heavy chain) gene rearrangements abortively, some even before reaching the thymus25,26, none of these have the full V(D)J rearrangement required to permit expression of TCRβ, TCRγ, and/or TCRδ genes. As the cells reach the DN3 stage, they stop proliferating, greatly increase TCR gene rearrangement, and generate the first fully rearranged TCR loci. DN3 cells that succeed in making in-frame TCR gene rearrangements become activated by TCR-dependent selection (DN3b cells); this distinguishes them from DN3 cells that are not yet selected (DN3a cells)27. Expression of a TCRβ chain qualifies the cells to undergo β-selection, turning on expression of CD4 and CD8 to become double positive (DP) cells, and eventually acquiring cell-surface TCRαβ complexes. This prepares them for positive selection and negative selection to generate mature CD4+ or CD8+ TCRαβ+ T cells. Alternatively, DN3 cells that successfully rearrange TCRγ and δ chains instead of β chains are selected as γδ T cells.
The thymic epithelium provides a particularly potent combination of receptor ligands and growth factors to trigger and support pro-T-cell differentiation, proliferation and survival8,11,15,28. The most important of the environmental inputs for T-cell development are ligands for the Notch cell-surface receptors, specifically Delta-family ligands, Delta-like 1(DL1) and DL429. Notch signalling initiates and sustains the T-lineage programme throughout the pro-T-cell stages, and presentation of Notch ligands by stromal cells is the key element in successful tissue culture systems that support early T-cell development in vitro30. In addition, the thymic epithelium produces ligands for other signalling pathways and cytokines, such as the KIT ligand stem cell factor (SCF) and interleukin-7 (IL-7), both of which are critical for sustaining the proliferation that pro-T cells undergo at defined stages. Migration of the pro-T cells through different zones of the thymus may provide changing microenvironmental signals that help to drive T-cell differentiation programming8. Furthermore, more mature thymic T cells can themselves have direct or indirect feedback effects on the gene expression and differentiation of early T cells2,31.
Key transitions in programming for T-cell identity
Programming for T-lineage identity includes both specification, which confers T cell-specific functions, and commitment, or loss of the ability to adopt alternative developmental fates.
The cells gain T-cell characteristics not coordinately, but in a multistep cascade. Fig. 2a depicts these gene expression changes on a color-coded logarithmic scale (>5000-fold range), because, although relatively modest changes in gene expression can be functional, the T-cell specification process also includes changes of much greater magnitude. T-cell identity genes, as well as the transcription factors that regulate their expression, are turned on in a precise order. The T-cell identity genes encode products that are critical for TCR rearrangement [recombination activating gene 1 (RAG1) and RAG2], TCR complex assembly (such as the CD3 chains), and signalling components, including kinases, phosphatases and adaptor proteins such as LCK, ZAP70 (TCRζ-associated protein 70 kd), and LAT (linker of activated T cells). There is little activation of these genes in precursors before they enter the thymus (although there are some possible exceptions26,32), and within the thymus, ETPs express relatively few T-cell identity genes, mostly at low levels. Most of these genes begin to be upregulated in the DN2 stage, as Fig. 2a shows for Cd3g, Cd3e, Zap70 and Il7ra (CD127)27,33–38 (M.A.Y. unpublished observations). Finally, by the DN3a stage of T-cell development, T-lineage identity is set. Germline transcription of the TCRβ variable-region gene segments is also upregulated39 as VDJ rearrangement of TCRβ genes proceeds27. Even effector genes like IL2 become accessible for induction40. The cells are still far from mature T cells, but they have completed assembly of the whole TCR signalling apparatus that is necessary for β- or γδ-selection, as well as for positive and negative selection and mature cell functions thereafter.
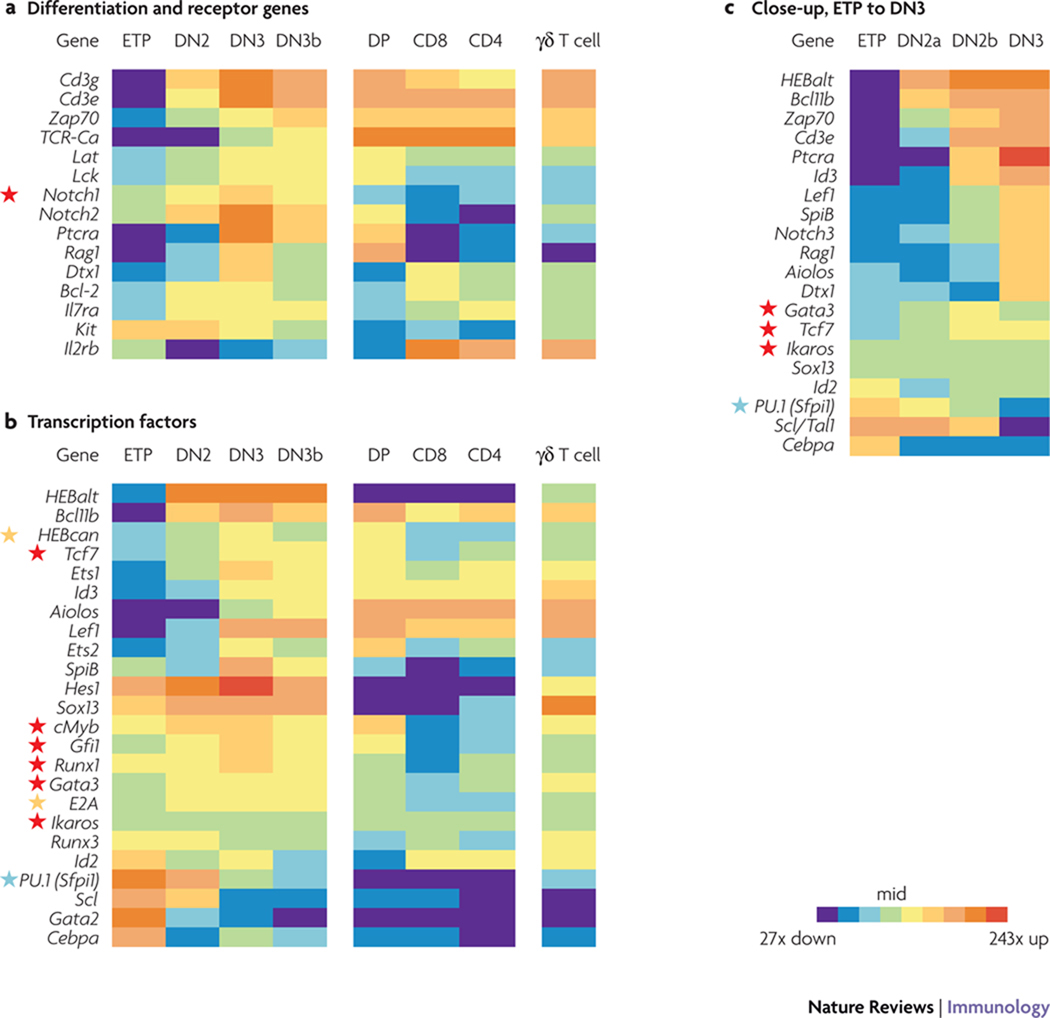
Figure 2. Expression levels of differentiation and regulatory genes in early T-cell development.
A comprehensive set of expression patterns is shown for 39 differentiation and regulatory genes in subsets of adult murine thymocytes. To summarize expression patterns of these genes for valid inter-gene comparisons, quantitative real-time RT-PCR results are collected here from a fixed set of 2–4 biological replicates of each of the populations indicated. All measurements were carried out as previously described27,34,35,38,50,52,53,68,96, and similar results are reported by others33,36,37,133. The amounts of mRNA expressed are normalized to β-actin at each developmental stage and the values depicted as “heat maps”, using a color code to indicate relative levels on a logarithmic scale as previously described53. Although small differences in gene expression levels can be meaningful, this method provides the dynamic range to depict changes of both smaller and larger magnitude. Briefly, the central value (“mid”) for each gene is the geometric mean of the maximum and minimum expression levels for that gene in the sample series, and the threefold range around this value is coded green. In (a) and (b), the mid value is for all cell subsets shown; in (c) it is the mid value for ETP, DN2a, DN2b, and DN3 cells only. The overall range of expression is then separated into discontinuous “bins” in which each shade redder represents a 3-fold increase in mRNA, while each shade bluer represents a 3-fold decrease. This analysis permits comparison between patterns of expression but does not indicate absolute RNA accumulations: poorly-expressed genes can give variability in the lower range as the central value itself is close to the detection threshold. Red stars indicate an essential member of the regulatory “core group”. Orange stars indicate partially redundant members of the “core group”. Blue stars mark PU.1, an early core group member that later switches to become an antagonist of T-cell development.
Concurrently, there are staged shifts in growth factor receptor expression (Fig. 2a). cKIT expression is essential in the earliest precursors41, then gradually downregulated through the DN3 stage37, whereas IL7Rα, which sustains most pro-T-cell proliferation42, is strongly expressed in DN2 and DN3 stages until after β-selection. Meanwhile, IL2Rβ, an important growth factor receptor for NK cells and mature T cells, is downregulated in a mirror opposite pattern to that of IL7Rα (Fig. 2a). This downregulation is functionally significant because IL2Rβ, although related to IL7Rα, transmits different signals that can divert immature thymocytes from the T-cell lineage to the myeloid lineage43.
Commitment also occurs in discrete steps. In adult mice, whereas some thymus-seeding progenitors initially retain B-lineage developmental potential, all rapidly lose this capability once inside the thymus, as FLT3 and CCR9 are downregulated44,45. T-cell precursors that populate the fetal thymus appear to have lost B-cell potential before entering the thymus11,12. Nevertheless, both adult and fetal thymic immigrants retain DC and NK-cell potentials until a later stage36,46–49, and at least some have macrophage and/or mast-cell potential as well36,50 (Fig. 1). Both ETPs and many DN2 cells show readily detectable NK-cell and DC potential if they are removed from the thymus and cultured in permissive conditions. However, by the time the cells reach the DN3 stage, they are committed to the T-cell lineage and can no longer develop into alternative cell types under any of these conditions (reviewed in ref.51).
Individual ETPs can embark on these alternative pathways somewhat more easily than DN2 cells can47,52. Until recently, the graded reduction in DC and NK-cell developmental potential at the DN2 stage could be interpreted stochastically, with DN2 cells simply becoming less likely to access their non-T-cell options than ETP. However, recent evidence33 shows a discrete transition in the midst of the DN2 stage, while the cells are still proliferating and before extensive TCRβ rearrangement. Activation of a green fluorescent protein transgene driven by the proximal Lck promoter proves to distinguish late from early DN2. At this point, cells specifically lose the ability to adopt the DC fate33 (Fig. 1). This transition seems likely to be correlated with the first slight reduction in expression of cKIT on nontransgenic DN2 cells that we will term the transition from DN2a to DN2b6 (Fig. 3). The discovery of this transition33 shows that the potential to develop into at least three different T-lineage alternatives – B-cell, DC, and NK-cell – may each be relinquished at successive stages. Commitment is not a single event, but a cascade of separate developmental renunciations that only ends at the DN3 stage when all non-T-cell options have been lost.
Figure 3. Phenotypic markers for the transitions through specification and commitment.
Left hand panels: flow cytometric profiles of CD4−CD8− double negative (DN) thymocytes from adult mice, gated for viability and absence of mature-cell lineage markers, using CD44 and c-KIT (top panel) and CD44 and CD25 (lower panel). Conventional DN1, DN2, DN3, and DN4 quadrants are marked on the lower left plot.
Upper right: profiles of c-KIT and CD25 expression in CD44+c-KIT+ DN thymocytes, distinguishing Early T-cell Precursors (ETP), DN2a, and DN2b subsets.
Lower right panel: location of the ETP, DN2a, DN2b, and DN3 cells within the CD44 and CD25 expression distribution of the whole DN population.
Loss of DC potential appears to be coupled with a discrete regulatory event that unleashes the full T-cell differentiation programme33, a coupling much tighter than was previously evident. Only a few of the T-cell-identity genes are upregulated by the DN2a stage (Fig. 2c). The earliest-responding genes are not differentiation genes but encode the transcription factors HEBalt (HeLa E-box binding factor, alternative form) and BCL11b (B-cell lymphoma 11b)53, discussed below. Some genes that play key roles in β-selection [Ptcra (preTCRα), and _Rag1_] are strongly upregulated only at the DN2b stage or at the DN2b to DN3a transition27,32,34,36,37 (Fig. 2c). The question is therefore what changes in transcription factor and signalling activities (jointly termed T-lineage regulatory factors) can explain these abrupt changes in gene expression and behavior.
T-lineage regulatory factors: core group
As shown in Fig. 2b,c, there are indeed major changes in transcription factor expression during the progression of T-cell precursors from ETP to DN3 stages. This is evident for the specific transcription factors chosen for presentation in Fig. 2b, which include most of the T-cell factors known to be essential (as indicated by the stars in Fig.2), as well as most factors implicated in lineage plasticity, those implicated in the regulation of TCR and other T-lineage gene expression, and their closest relatives.
Notch signalling is the best-studied T-lineage positive regulator. Upon triggering, the Notch intracellular domain (NotchIC) converts the RBPJ transcription factor (recombination site binding protein J, also known as CSL, core binding factor/suppressor of hairless/Lag-1) from a default repressor to an activator (reviewed in 29,54). Several T-lineage genes are directly activated by the Notch-RBPJ complex, including distinguishing markers for the DN2 and DN3 stages such as Ptcra and the DN2–DN3-specific marker Cd25 55–57. Another direct Notch target gene encodes the transcription factor HES1 (hairy, enhancer of split 1). NotchIC is the only regulator known to be able to drive ectopic T-cell development when overexpressed. However, Notch signalling alone is not sufficient to activate or sustain the T-cell programme.
Genetic evidence emphasizes the roles of a “core group” of diverse transcription factors or transcription factor families that must work with Notch in the T-cell specification process, either during or before the intrathymic T-cell precursor stages (Table 2)2,58–60. This core group of factors includes: GATA3 (GATA sequence-binding factor 3)61; MYB (myeloblastosis proto-oncogene); RUNX–CBFβ (runt domain or acute myelogenous leukemia factor – core binding factor β) complexes; basic helix-loop-helix E proteins62 such as E2A (Tcfe2a) and HEB (Tcf12); (TCF-1 (T-cell factor 1, encoded by Tcf7); the zinc-finger repressor Growth factor independence 1 (GFI1); and Ikaros family members (Table 2; Fig. 2, red and orange stars). These factors are discussed extensively in the following sections, as their roles are not only essential but are also recurrent in T-cell development. Most of these have expression patterns consistent with persistent roles throughout the ETP to DN3 stages (Fig. 2): they are already expressed relatively well at the ETP stage and are upregulated only modestly compared to many other pro-T cell genes. However, the continuous expression of these core genes cannot explain the discontinuities in differentiation gene expression.
TABLE 2.
TRANSCRIPTION FACTORS ESSENTIAL FOR EARLY T-CELL DEVELOPMENT
| FACTOR | FAMILY | TARGET SITE* | KNOCKOUT EFFECT | OVEREXPRESSIONEFFECT | SELECTEDREFERENCES† |
|---|---|---|---|---|---|
| RBPJ (CSL) (activated by Notch) | TIG | an(T/C)GTGG(G/a)AA(A/C)c(site for Drosophila orthologue only) | No T-cell development. Early B cell development unaffected. Later stage knockouts cause block to TCRβ rearrangement & β-selection with little effect on γδ cells | RBPJ overexpression not done. Overactivation by constitutive Notch causes T-ALL (can collaborate with loss of E proteins | 29,54 |
| GATA-3 | GATA (Two C4 zinc fingers) | (A/T)GATA(A/G) | No DN cells; if deleted later, block of β-selection and of CD4 SP generation | Loss of viability and diversion to mast cell fate | 50,61 |
| Myb | Myb | (C/T)AAC(G/T)G | Multiple blocks: defective progenitors, poor TCRβ rearrangement, poor DN3 to DP proliferation, poor DP survival, loss of CD4 cells, little effect on γδ | ? | 112–114 |
| TCF-1 (gene name Tcf7) | HMG | (A/C)A(A/C)AG | Severe block with loss of DN2 cells in adult, defective β-selection and loss of DP in young animals | ? | 86,87 |
| LEF-1 | HMG | (A/C)A(A/C)AG | No phenotype alone; but with TCF-1 mutant, β-selection in fetus completely blocked | ? | 134 |
| E2A | bHLH “E protein” | (A/G)CAG(G/C)TG | Defective DN2 generation, TCR rearrangement, DN3 checkpoint enforcement; leukemia (often in collaboration with activated Notch) | ? | 62,121 |
| HEB | bHLH “E protein” with diverse splice & promoter variants | CAG(G/C)TG | Weak phenotype alone. No β-selection if E2A also mutated; but γδ cells fine even in presence of HEB dominant negative | Early growth inhibition (HEBcan); acceleration followed by inhibition (HEBalt) | 62,64,121 |
| Gfi-1 | SNAG, 6 zinc finger | aAATC(A/t)c(A/T)G(C/t) | Inhibited generation of DN2 cells; also, overexpression of Id1, Id2 | ? | 84,85 |
| Ikaros (Ikzf1, Znfn1a1) | Ikaros, Ikzf | (T/C)(T/C)TGGGAG(A/G) | No fetal T cell development; highly defective T-lineage potential in adult prethymic cells. Dose effect: reduced activity causes T leukemia, may collaborate with activated Notch | ? | 18 |
| Runx1 (with CBFβ partner) | Runt | TG(T/c)GGT | Early stem cell defects; block to generation of DN3 cells; derepression of CD4 | ? | 77,115 |
| PU.1 | ETS | GAGGAA and diverse variants | No recognizable T cell development from earliest stages | Diversion to DC or monocyte lineage | 52,75,76,94,95 |
Stage-specific T-cell transcription factors
T-cell development also depends on factors that do not have sustained roles. Of these, the best-studied is the ETS (E26-transformation specific) family factor PU.1 (Fig. 2, blue star), another “core group” member, but with a difference. While it is vital for prethymic precursor development, PU.1 switches to a T-lineage antagonist once cells progress to the DN2a/b stages, and it must then be silenced. As described below, PU.1 is one of a cohort of prethymically inherited factors (Fig. 2, SCL [stem cell leukemia, or TAL1], GATA2, and C/EBPα [CCAAT-enhancer binding protein α]) that most likely sustain the lineage plasticity of cells in early stages of T-cell development. All of these factors are downregulated to essentially undetectable levels before β- or γδ-selection (Fig. 2b,c) and the downregulation of these non-T factors is strongly correlated with the activation of the T-cell gene expression programme.
Not all transcription factors inherited from haematopoietic stem cells are turned off during T-cell specification. Many of the core group factors have important prethymic roles as well. There is also a large set of transcription factor genes that continue to be expressed stably in multipotent progenitors all the way to cells undergoing β-selection53. But only a few transcription-factor genes are specifically turned on at a time that might contribute to the onset of T-lineage gene expression in the DN2 stage. These include the zinc finger transcription factor and tumor suppressor factor BCL11B63, a particular promoter use isoform of the E protein HEB called HEBalt53,64, and more transiently, a transcription factor involved in the Hedgehog signalling pathway, GLI265.
To explain the further increase in T-cell gene expression at the DN3 stage, several additional factors need to be considered, which have been implicated through direct studies of the regulatory elements of TCR and other T-cell genes (reviewed in 2). ETS-family and zinc-finger factors are prominent, especially ETS1, which works in an intimate collaboration with RUNX1-CBFβ 66,67. Both ETS1 and ETS2 are included in a group of transcription-factor genes that are strongly upregulated at the DN2 to DN3 transition (Fig. 2b), along with TCF-1 relative LEF-1 (lymphocyte enhancer factor 1)34. The DN3 stage is also distinguished by peak levels of expression of the PU.1-related factor SPIB and the Notch-RBPJ target HES1, which are shut off after β-selection. An additional high mobility group factor, SOX13, has recently been shown to have a specific, essential role for the generation of TCRγδ cells68; however, SOX13 is expressed throughout the pro-T cell stages, and is only downregulated in cells that take the αβ pathway.
Thus, although Notch signalling occurs throughout, the transcription factor profile of the cells through the ETP to DN3 stages consists of several distinct layers. On a background of factors inherited from prethymic progenitor cells53, there is maintenance or a gradual increase in expression of the core group factors; there is a staggered decline of factors that could otherwise drive the cells to alternative fates; and there are new sets of factors abruptly adding to the mixture at two precisely timed transitions, first to signal entry to the DN2 stage, and then as the cells reach the climax of cell-type specification and commitment in the DN3 stage. Although not well studied yet, these last groups of factors are not simply stage markers. Notch and the core group transcription factors have different effects on early T-cell precursors at different stages, because the ways transcription factors actually work in development (Box 2) depend on their dose, their precise splice isoforms, their position in a gene network, and their combinatorial interactions with other factors. It is likely that more dynamically regulated factors, collaborating with the core group, provide this stage specificity. The next sections focus in depth on how such interactions may operate at four pivotal transitions in T-cell specification.
BOX 2: Same transcription factor protein, different target gene effects.
Several general principles make it possible for the same transcription factor protein to have effects on widely different sets of target genes at different stages in a developmental process. First, the effect of a particular transcriptional regulator depends on its dose, due to DNA-binding biophysics. Transcription factors at high concentration can bind to a wider range of DNA sites than they can at low concentration. Second, the effect of any essential regulatory factor depends on combinatorial interaction with other factors and signalling pathways. These affect the binding of the essential factor to particular cis-regulatory elements, sometimes in an all-or-nothing fashion, and may change the net effect from activation to repression or vice versa. Finally, the effect also depends on the gene network within which the factor of interest operates. Among the target genes activated by a factor can be genes encoding inhibitors of the initial regulator itself. Such feedback negative regulation can make a significant difference between the impact of a factor when continuously expressed and the impact of the same factor when activated in a discontinuous or oscillating manner. Thus, it is likely to be significant that several Notch target genes expressed in pro-T cells encode inhibitors of Notch signalling itself93,133.
Competence to turn on the T-cell programme
Notch signalling is vital to initiate T-cell development, but it is not sufficient to activate T-lineage genes directly69. The immediate impacts of Notch signals have been greatly clarified by culture systems that present Notch ligands DL1 or DL4 to haematopoietic precursors on OP9 (or other) stromal cells, thus triggering a synchronized cascade of Notch responses35,70–74. Within several days of culture with OP9-DL1 cells and supportive cytokines, Thy1+ CD25+ DN2 cells and ultimately DP cells can be generated from various types of haematopoietic precursors, through a reproducible molecular pathway35,53,70,71,73. Although Notch signalling turns on tissue-nonspecific genes such as Hes1 and Nrarp in many cells within the population, expression of the key T-cell transcription factors GATA3 and TCF-1 is only induced within a subset of these precursors. A smaller subset of these then proceed to a DN2-like state, turning on CD25, Thy1, CD3ε, preTCRα, and BCL11b35,53. Because only select types of haematopoietic precursor activate these T-cell genes in response to Notch, it is clear that additional factors are needed35.
At least four transcription factor contributions now appear to be required to enable progenitors to undergo the T-lineage initiation response: PU.175,76; Ikaros18; RUNX family factors, working with CBFβ 77 (Y. Guo, I. Maillard, and N. A. Speck, personal communication); and E protein activity, from E2A and possibly its relatives such as HEB. E proteins are already expressed in haematopoietic precursors but are engaged to unknown extents in complexes with Id proteins or SCL (TAL1), which sequester them from activating T-lineage specific genes. In a multipotent haematopoietic progenitor cell line, E2A levels positively regulate some Notch target and T-lineage genes including Notch1 itself, with synergistic effects of E2A and Notch signalling together78. This is the first of multiple stages at which E proteins and Notch work together in early T-cell development (Fig. 4).
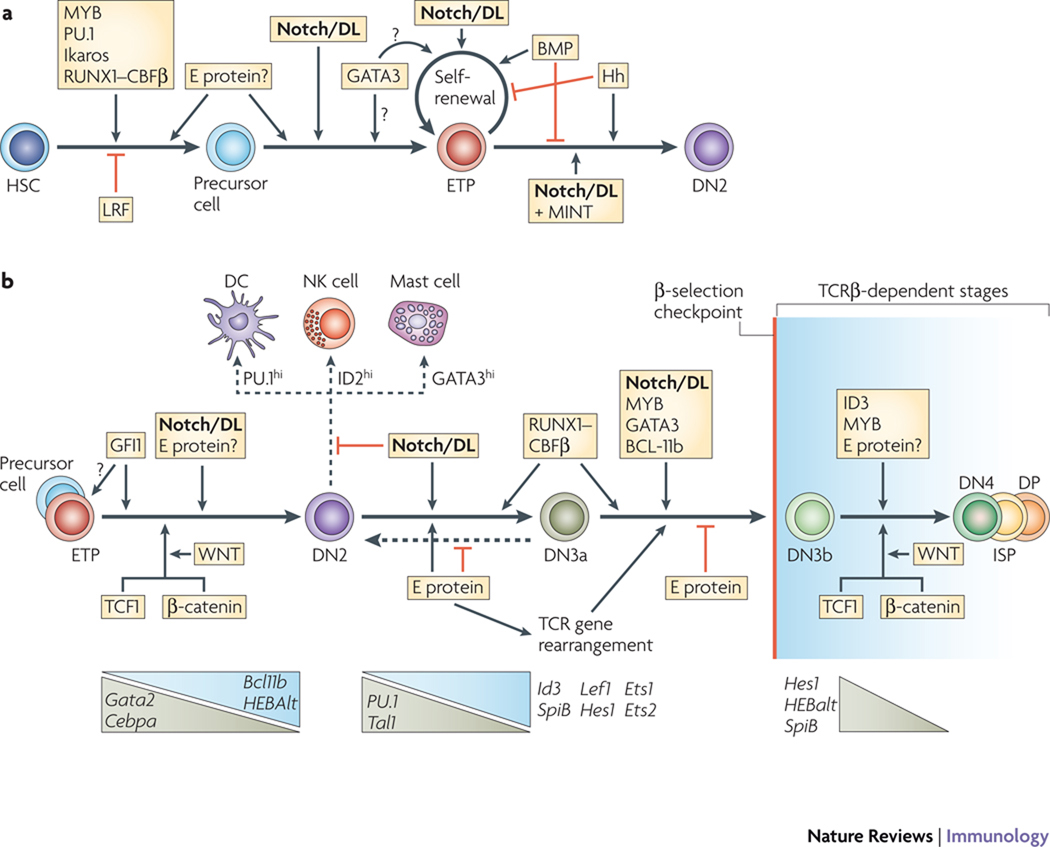
Figure 4. Genetic evidence for temporal roles of regulatory factors in early T-cell development.
The figure summarizes times in early T-cell development when the indicated genes are essential. Data are from germline and conditional knockout studies, with additional details from in vitro differentiation assays2,29,54,58–62,77,84,85,111–115. Note the repeated inputs from Notch signalling and E proteins at successive stages, and the importance of MYB, RUNX--CBFβ, and GATA3 both in the earliest stages and in the functions of double negative (DN)3 cells. DN2 stages have not yet been subdivided into DN2a and DN2b in any of the literature used as sources, so this distinction has not been included. Blue text and arrows: factors promoting T-cell differentiation at a particular transition (the order of arrows shown at any one transition does not necessarily indicate the order of transcription factor activity). Purple text: factors promoting alternatives to T-cell development. Panel (a): stages from haematopoietic stem cell (HSC) to the ETP-DN2 choice point. The stages at which cells become primed to respond to Notch signals by undergoing T-lineage specification are shown. Evidence for a possible early role of E protein is noted but its timing is uncertain. Prec. = any relevant prethymic precursor population (not specifically defined). Diamond: a choice point, discussed in the text, at which ETP cells may undergo further self-renewal or else proceed to the DN2 stages; however, both choices can enhance T-cell development although with different kinetics. Panel (b): recursive roles of E proteins and Notch/Delta (Notch/DL) signalling at stages from the ETP stage through β-selection. Developmental options still open at the DN2 stage(s), and the potential reversal of the DN2 to DN3 transition, are alternatives shown by broken arrows. The broken gray line depicts the β-selection checkpoint, that is the boundary between TCR-independent developmental stages on the left and the later TCR-dependent developmental stages in the shaded zone on the right. The later developmental changes from DN4 to ISP (immature single positive) to DP are not shown as discrete transitions. Below scheme: grey triangles show approximate timing of major increases (upward-pointing) or decreases (downward-pointing) in the expression of other indicated transcription factor genes (cf. Fig. 2) as discussed in the text.
Competence to undergo the T-lineage response to Notch signals is under negative as well as positive regulation in haematopoietic precursors. One newly defined mechanism is mediated by a transcriptional repressor, LRF (leukaemia/lymphoma related factor, also known as Zbtb7a or Pokémon). LRF makes B-cell development possible by counteracting Notch responsiveness in prethymic haematopoietic precursors79. It limits expression of Notch1 and Notch3 themselves, and, either by direct repression or as a result of reduced Notch expression, it keeps T-lineage genes silent. In the absence of LRF, T-cell specification genes are spontaneously activated in bone marrow progenitors79.
Thus, even before Notch signalling, T-cell precursor competence is defined by expression of PU.1, Ikaros, RUNX-CBFβ complexes, and readily mobilized E proteins. In each subset, LRF may also be important for establishing the threshold that the Notch signals must overcome in order to initiate the T-cell programme in those cells.
ETP to DN2: delay vs. progression
ETPs are clearly excellent T-cell precursors, but they present some paradoxes. ETPs profoundly depend on Notch-driven transcription for their maintenance45,80,81, and they already strongly express the Notch-promoted genes Hes1, Gata3, and Tcf7 at levels well above those of prethymic progenitors53,80. However, unlike cells responding to Notch in vitro, adult murine ETPs delay many days (~10 days) and many cell cycles before activating DN2-stage Notch target genes8. Although some ETPs already have DJβ TCR gene rearrangements25, their expression of RAG recombinases is barely detectable (Fig. 2a,c). The length of time spent proliferating in the ETP stage is also variable and subject to regulation: it is much shorter for fetal thymocytes, and the size of the ETP pool is adjusted dynamically based on niche availability and homeostatic feedback82,83. So, even when the requirements are met for initiating T-lineage development in response to Notch signals, presumably including MYB, RUNX–CBFβ, Ikaros, and E proteins, there must be additional regulatory inputs that control whether precursors will stay in a “holding pattern” for another cell cycle as ETPs or “graduate” onward as DN2 cells (Fig. 1; Fig. 4a).
Two regulators specifically needed to generate DN2 cells are TCF1 and the transcriptional repressor GFI184,85 (Fig. 4b). It is not clear yet whether GFI1 controls the ETP to DN2 transition itself or helps to make competent ETPs, and there is little increase in GFI1 from ETPs to DN2 (Fig. 2b). TCF1 is an attractive candidate for a rate-limiting positive regulator of the DN2 transition because of its increasing, Notch-dependent expression pattern34,53 and because many T-lineage differentiation genes, including Cd3g and Cd3e, have predicted TCF1 binding sites in their promoter regions (H. T. Petrie and M. Fallahi, personal communication). Analogously to the Notch pathway, microenvironmental WNT signalling can convert TCF1 to an activator through interaction with β-catenin86,87. However, indirect evidence suggests that WNT signalling may already be occurring in most ETPs87,88. Additional regulatory inputs that delay ETPs from proceeding to the DN2 stage must exist in the adult thymus, and notably, at least three microenvironmental signalling systems are implicated.
Progression of ETPs to the DN2 stage can be held back by the action of certain members of the bone morphogenetic protein (BMP) and fibroblast growth factor (FGF) families89–91, which appear to promote ETP self-renewal. The effects of these environmental signals seem to be counteracted by activation of the hedgehog signalling pathway, working through GLI2, which appears to enhance the generation of DN2 cells65 (Fig. 4a).
Notch itself may also play a two-sided role. There is a natural damper of Notch-dependent transcription, MSX2-interacting protein (MINT)92, which is normally expressed abundantly in ETPs, with levels declining into DN3 and after β-selection. MINT-deficiency should increase Notch-mediated transcriptional activation, and this might be expected to enhance T-cell specification. The observed phenotype, however, is the opposite: MINT mutants accumulate ETPs with reduced transit to the DN2 stage. Is too much Notch signalling a stalling factor that keeps cells in the ETP stage? Crosstalk with Hedgehog, BMP, or WNT pathway mediators could occur. Another pathway may be through direct Notch target gene NRARP, a cytoplasmic ankyrin-repeat protein that can act as a potent feedback inhibitor of Notch signalling, targeting a different aspect of the Notch signalling pathway than MINT93. When MINT is deleted, ETPs accumulate much higher levels of RNA encoding NRARP than normal ETPs, losing one kind of Notch modulator but gaining another92,93, and this could also bias MINT-deficient ETPs to expand without progressing. In either case, the role of MINT shows that a Notch-dependent gene regulatory network, not simply Notch signalling alone, may channel cells differentially between ETP self-renewal and transit to the DN2 stage.
Lineage plasticity in the early T-cell precursor stages
Notch signalling plays an important role in the “negotiation” over cell fate throughout the ETP stage and into the DN2 stage. Although all but the earliest CCR9+ FLT3+ immigrants have lost B cell potential, at least some of these cells retain myeloid, DC, NK-cell, and even (rarely) mast-cell50 potential, revealed under permissive environmental conditions. This lineage instability reflects the mixture of T-lineage and persisting stem-cell transcription factors in ETP and DN2 cells (reviewed in refs. 51,58,94) (Fig. 2). “Non-T factors” present in ETP and DN2a cells, such as ID2, SCL, C/EBPα, GATA2, and PU.1, are likely to play direct roles in maintaining non-T-cell developmental options. C/EBPα and PU.1 are each sufficient to impose a myeloid or DC programme on T-lineage precursors, if expressed ectopically after their endogenous expression is normally turned off52,95–97, and PU.1 is required for normal DC development from thymocytes97. ID2 and SCL work as modulators or antagonists of E-protein activity, and this antagonism is crucial for ID2 to promote NK-cell development98,99.
Not only do these non-T-cell factors promote specific alternative pathways, but they also make the regulatory state of ETPs and DN2 cells intrinsically unstable. In these stages, even GATA3, a transcription factor of the T-lineage core group, can drive alternative fates, only supporting the early stages of T-cell development within a highly limited dose-response range50. Increased concentrations of GATA3 drive ETPs and DN2 cells into the mast-cell lineage50, although GATA3 is clearly and repeatedly essential for T-cell development at multiple stages59,61. Early T-cell precursors are also sensitive to cytokine stimulation of ectopic receptors, such as the GM-CSF receptor or the IL-2Rβ–γc complex: cytokine signalling of these receptors is sufficient to activate a myeloid programme in ETPs or DN2 cells43,100. This can occur by triggering upregulation of C/EBPα and/or PU.1 activity101,102, or by precocious activation of protein kinase C or RAS, mediators normally activated by TCR signalling but which strongly enhance lineage diversion if PU.1 is present96. The cells only stabilize their T-cell identity when these non-T factors are finally silenced.
Uncommitted precursors are probably restrained from these developmental alternatives in vivo by interaction with intrathymic Notch ligands19,50,52,95. Notch signalling inhibits, though it does not eliminate, NK-cell differentiation potential47,103,104. Although Notch signalling does not repress PU.1 directly, it does protect newly induced T-lineage differentiation and regulatory genes from repression in the presence of high concentrations of PU.152, thus tilting the balance of a bistable regulatory switch toward T-lymphoid and against myeloid fate58. Through a different pathway, Notch–Delta interactions also restrain GATA3-overexpressing cells from diversion to the mast-cell lineage50. The biochemical mechanisms remain to be determined, but in each case, Notch signalling acts to preserve access to the T-lineage programme without extinguishing regulators for other developmental options.
Effects on E proteins may be at the crux of the balance between T-cell and non-T-cell developmental alternatives58 for ETPs and DN2 cells. Both the NK-cell pathway and the myeloid and DC alternatives to T-cell development involve repressing or neutralizing E proteins. When either PU.1 or C/EBPα is re-introduced to committed DN3 cells to divert them to a myeloid or DC fate, one of their earliest effects is to inhibit net E-protein activity52,95. HEBalt is a particularly sensitive repression target52,96. Correspondingly, Notch signalling maintains HEBalt and inhibits upregulation of Id2 in response to PU.152. The different roles of various E protein heterodimers make them challenging to study, but these results suggest that E-protein activity may be pivotal for the lineage fidelity of early T-cell precursors.
Why is broad lineage plasticity retained so long by early T-cell precursors? One possible answer is that until TCR gene rearrangement, pro-T cells regulate their proliferation through mechanisms shared with multipotent haematopoietic progenitors, and that the non-T factors are part of this mechanism. ID2 is directly involved in cell-cycle progression in many cell types105, and it may be indispensable to maintain proliferation in the presence of high levels of E-protein activity until another, lymphoid-specific ID factor, ID3, becomes induced (Fig. 2). Both PU.1 and SCL are important for maintenance of multipotent haematopoietic progenitors75,76,106,107, possibly through target genes such as cKIT108,109 and the cell-cycle activator CDK6110. Thus, throughout the pro-T cell stages, the T-cell-specific gene expression programme may gradually be assembled onto the “armature” of a proliferating multipotent progenitor state. If so, this “armature” can apparently be removed only when assembly is complete.
T-lineage commitment: transit to the DN3 stage
After the plasticity of the ETP and DN2 stages, the DN3 stage marks a break. Not only do most T-cell genes become fully activated, but also, the cells lose all non-T developmental options and stop proliferating. The transition from proliferative expansion to cell-cycle arrest enables the TCR gene rearrangement process to be fully activated. During in vitro differentiation, the DN2 population often expands abnormally at the expense of DN3 generation, implying that this transition also requires specific regulation72,111.
A phenotypically normal DN2 to DN3 transition depends on RUNX1–CBFβ complexes, and RUNX1–CBFβ as well as MYB, GATA3, and BCL11B are needed for full TCRβ gene rearrangement and competence to undergo β-selection77,111–116 (Fig. 4b). Cis-regulatory analysis of TCR and RAG1/2 indicates specific sites where most of these factors can be implicated in direct regulation (rev. in ref.2). Although none of these transcription factors increases much in expression from DN2 to DN3 (Fig. 2b,c), their functions are very likely modified by the induction of new participants. Of particular interest, ETS1 and ETS2 are sharply upregulated at this point (Fig. 2b,c). ETS1 collaborates with Runx in TCR gene expression and may enhance Runx factor activity profoundly67. Another ETS factor, SPIB, may contribute to checkpoint arrest97. PU.1 specifically inhibits ETS1, ETS2, and SPIB expression in thymocytes52, and so the timing of their induction may await the downregulation of PU.1.
As in all the previous stages, Notch and E proteins play major roles too. The DN3 stage is the peak period for expression of Notch/RBPJ and E protein target genes such as Deltex1, Rag1, Hes1, and Ptcra (Fig. 2). Murine DN3a pro-T cells, prior to β- or γδ-selection, are acutely dependent on Notch signalling to maintain viability through phosphoinositide 3-kinase (PI3K) pathway signals and activation of the proto-oncogene cMyc117–119. E2A and HEB activity are crucial not only for TCR gene rearrangement62, but also for the ability to undergo normal β-selection even when a rearranged TCR transgene is supplied120. E2A and HEB together are particularly implicated in maintaining the DN3-stage proliferative arrest121: in part they activate expression of growth inhibitory factors, such as suppressor of cytokine signalling (SOCS)1 and SOCS3 proteins, which uncouple growth-factor receptors like IL-7R from their signalling pathways; and in part they directly inhibit cell-cycle-activating genes such as Cdk6122. This cell-cycle-arresting role of E proteins is important for checkpoint control (Fig. 4b). When E2A is mutated, not only is TCR gene rearrangement defective, but also the penalty for failure is removed, and cells can proceed abnormally through the β-selection checkpoint123. Such E2A-deficient unchecked cells are prone to leukaemic transformation. However, note that if E2A and HEB were to exercise this function earlier, they would abort T-cell development, as is seen when these factors are experimentally overexpressed64. Finally, there is new evidence that E proteins are also responsible for making the DN2 to DN3 transition irreversible. Both E2A and HEB together have been conditionally deleted in DN3 thymocytes, and the loss of these E proteins not only prevents proliferative arrest at the DN3 stage but also enables DN3 cells to undergo reverse differentiation, to return to a DN2-like phenotype121. A key question is now whether such removal of E-protein activity may also be sufficient to restore expression of the progenitor-cell regulatory factors and broader developmental potentials of earlier states.
Directly or indirectly, the uniqueness of the DN3 state may depend on particularly high doses of both Notch and E-protein activity. The peak-level Notch target gene expression could reflect naturally decreased MINT expression in the DN3 cells92; or enhanced responsiveness to Delta-family Notch ligands through increased expression of the modifier Lunatic Fringe (Lfng)124; or migration into an intrathymic zone8 (subcapsular region, Fig. 1) that presents an increasingly potent Notch ligand, DL4125. In parallel, although E-protein (E2A and HEBcan) gene expression itself is relatively unchanged throughout development64 (Fig. 2b), the accumulation of E protein greatly increases at the DN3a stage126. Reduced SCL and ID2 expression could be responsible, although surprisingly, ID3 expression increases at this stage and might be expected to continue antagonizing E-protein activity34 (Fig. 2b,c). The high accumulation of HEBalt in DN3 cells may contribute, either as a highly stabilizing partner for full-length E proteins or as an ID-interaction decoy64. Adjustments in levels of Notch and E proteins can apparently compensate for each other78. Fetal murine pro-T cells express surprisingly high levels of Id2 which should inhibit E protein activity, but transcriptional evidence also suggests they experience unusually strong Notch activity which may compensate34.
Commitment depends not only on irreversibility but also on repression, and the main unresolved issue about the DN2 to DN3 transition is how the “non-T factors” become silenced in the DN2 and DN3 stages. The sweeping downregulations of factors like PU.1 and SCL/TAL1 from high stem-cell levels53 are among the most profound regulatory changes that occur during T-cell specification. However, very little is known yet about T-lineage-promoting repression. Notch signalling induces expression of a repressor, HES1, but this factor probably does not repress PU.1, as it is not required to block myeloid potential or confer T-cell potential in fetal progenitors23. It may be more important that RUNX, MYB, and TCF-1/LEF-1 factors, all expressed at peak levels in DN3 cells, also have the capacity to act as repressors, especially in certain isoforms. In this context, the repressor activity of BCL11B is also of particular interest. The key factors may be defined by their interaction with the corepressor NCOR1 (nuclear receptor co-repressor 1), which is reportedly essential to prepare cells for β-selection127. Cis-regulatory analysis of repression targets Scl/Tal1 and Sfpi1 (PU.1) could “bait the trap” to identify repressors mediating commitment.
From “haematopoietic” to “immunological” T-cell development
From β-selection or γδ-selection onwards, multiple regulatory changes are triggered by TCR signals, but T lineage identity is completely and stably established. The basic set of T-lineage effector genes continues to be expressed as the cells diverge to αβ, γδ and other specialized T-cell lineages (Fig. 2a), even though quite different combinations of transcription factors are used in γδ and different subsets of αβ-lineage cells27,59,68,128 (Fig. 2b). Most importantly, neither the thymic microenvironment nor the DN3 transcription factor set are needed to maintain the T-lymphoid identity of the cells. Notch signal-dependent transcription becomes dispensable129, E-protein activity is at least reduced, and many of the transcription factors that participated in the early T-cell precursor stages decrease or disappear37,38 (Fig. 2b).
This means that the specification mechanisms that turned on T-cell genes probably work in a ratchet-like fashion to make activation of genes like Cd3g and Cd3e virtually irreversible. One way this could occur is by substituting the roles of the transcription factors that initiate development with different members of the same transcription factor family that are expressed stably in mature cells. For example, the ETS-family transcription factor GABPα can drive expression of IL-7Rα in PU.1-negative cells through the same cis-element that PU.1 uses when it is present130. Another mechanism is probably the establishment of durable epigenetic changes in the accessibility of T-cell and non-T-cell genes, which could make continuing T-cell gene expression a default, no longer dependent on delicately titrated regulatory balances. Such mechanisms are probably essential to explain the completion of the specification process and may be even more important for establishing a strong barrier against reactivation of the non-T-cell regulators that were expressed at earlier stages.
Concluding remarks
T-cell specification is a segmented process. The completion of T-lineage commitment is far removed from the initial responses to Notch signalling, with distinct steps in between that are sensitive to environmental influence. Ongoing requirements for Notch and E proteins provide continuities throughout this process, but different sets of Notch and E protein target genes are activated at each step. As discussed here, this is because each step is distinguished by changes in other transcription factors and sensitivities to changing cytokines and other signals encountered during migration through the thymus that create a constantly shifting regulatory context. As suggested elsewhere7, some of the remarkable developmental stability of T-cell identity after β-selection may be due to the completeness with which the cells finally extinguish expression of the regulators for competing fates. The identity of the critical silencing functions is another remaining mystery. It will now be important to determine the exact mechanisms that control deployment of Notch/RBPJ, E proteins, and other “core group” factors that seem to be present throughout, to activate precisely shifting sets of T-lineage genes at each step.
Acknowledgements
The authors wish to thank T. Graf, H. Kawamoto, B. Kee, C. Murre, H. Petrie, and N. Speck for valuable discussions and generous sharing of unpublished results. We apologize to many authors whose work we could not adequately cite. We also thank members of the Rothenberg group for collegial interchange, advice, and permission to cite unpublished work, and N. Feng, R. Butler, and D. Perez for technical support. The authors were supported by grants from the NIH to E.V.R. (R01 CA90233 and R01 CA98925), M.A.Y. (R01 AI064590), and J.E.M. (F32 AI068366). E.V.R. gratefully acknowledges the Albert Billings Ruddock Professorship.
REFERENCES
- 1.Hayday AC, Pennington DJ. Key factors in the organized chaos of early T cell development. Nature Immunol. 2007;8:137–144. doi: 10.1038/ni1436. [DOI] [PubMed] [Google Scholar]
- 2.Anderson MK. At the crossroads: diverse roles of early thymocyte transcriptional regulators. Immunol. Rev. 2006;209:191–211. doi: 10.1111/j.0105-2896.2006.00352.x. [DOI] [PubMed] [Google Scholar]
- 3.Petrie HT. Cell migration and the control of post-natal T-cell lymphopoiesis in the thymus. Nature Rev. Immunol. 2003;3:859–866. doi: 10.1038/nri1223. [DOI] [PubMed] [Google Scholar]
- 4.Rothenberg EV. Stepwise specification of lymphocyte developmental lineages. Curr. Opin. Genet. Dev. 2000;10:370–379. doi: 10.1016/s0959-437x(00)00098-8. [DOI] [PubMed] [Google Scholar]
- 5.Shortman K, et al. The linkage between T-cell and dendritic cell development in the mouse thymus. Immunol. Rev. 1998;165:39–46. doi: 10.1111/j.1600-065x.1998.tb01228.x. [DOI] [PubMed] [Google Scholar]
- 6.Ceredig R, Rolink T. A positive look at double-negative thymocytes. Nature Rev. Immunol. 2002;2:888–897. doi: 10.1038/nri937. This review was first to emphasize the canonical importance of using high c-Kit expression to define T-cell precursors in the DN1 population as well as to characterize DN2 cells. DN1 cells isolated as recommended here largely correspond to the ETP and DN1a,b cells confirmed as precursors in refs.25 and 131.
- 7.Blom B, Spits H. Development of human lymphoid cells. Annu. Rev. Immunol. 2006;24:287–320. doi: 10.1146/annurev.immunol.24.021605.090612. [DOI] [PubMed] [Google Scholar]
- 8.Petrie HT, Zuniga-Pflucker JC. Zoned out: functional mapping of stromal signaling microenvironments in the thymus. Annu. Rev. Immunol. 2007;25:649–679. doi: 10.1146/annurev.immunol.23.021704.115715. [DOI] [PubMed] [Google Scholar]
- 9.Bhandoola A, von Boehmer H, Petrie HT, Zuniga-Pflucker JC. Commitment and developmental potential of extrathymic and intrathymic T cell precursors: plenty to choose from. Immunity. 2007;26:678–689. doi: 10.1016/j.immuni.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Wu L. T lineage progenitors: the earliest steps en route to T lymphocytes. Curr. Opin. Immunol. 2006;18:121–126. doi: 10.1016/j.coi.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Jenkinson EJ, Jenkinson WE, Rossi SW, Anderson G. The thymus and T-cell commitment: the right niche for Notch? Nature Rev. Immunol. 2006;6:551–555. doi: 10.1038/nri1883. [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto H. A close developmental relationship between the lymphoid and myeloid lineages. Trends Immunol. 2006;27:169–175. doi: 10.1016/j.it.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Pelayo R, et al. Lymphoid progenitors and primary routes to becoming cells of the immune system. Curr. Opin. Immunol. 2005;17:100–107. doi: 10.1016/j.coi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 14.Garbe AI, von Boehmer H. TCR and Notch synergize in αβ versus γδ lineage choice. Trends Immunol. 2007;28:124–131. doi: 10.1016/j.it.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Boehm T, Bleul CC. Thymus-homing precursors and the thymic microenvironment. Trends Immunol. 2006;27:477–484. doi: 10.1016/j.it.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Weerkamp F, Pike-Overzet K, Staal FJT. T-sing progenitors to commit. Trends Immunol. 2006;27:125–131. doi: 10.1016/j.it.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida T, Ng SY, Zuniga-Pflucker JC, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nature Immunol. 2006;7:382–391. doi: 10.1038/ni1314. This paper uses a sophisticated combination of gene expression analysis and in vitro development from defined precursors to establish the exact functions of Ikaros in distinct types of multipotent hematopoietic progenitors. It documents quantitatively the importance of Ikaros for T-cell development from lymphomyeloid precursors.
- 19.Garcia-Peydro M, de Yebenes VG, Toribio ML. Notch1 and IL-7 receptor interplay maintains proliferation of human thymic progenitors while suppressing non-T cell fates. J. Immunol. 2006;177:3711–3720. doi: 10.4049/jimmunol.177.6.3711. [DOI] [PubMed] [Google Scholar]
- 20.De Smedt M, Hoebeke I, Reynvoet K, Leclercq G, Plum J. Different thresholds of Notch signaling bias human precursor cells toward B-, NK-, monocytic/dendritic-, or T-cell lineage in thymus microenvironment. Blood. 2005;106:3498–3506. doi: 10.1182/blood-2005-02-0496. [DOI] [PubMed] [Google Scholar]
- 21.Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J. Immunol. 2007;178:858–868. doi: 10.4049/jimmunol.178.2.858. [DOI] [PubMed] [Google Scholar]
- 22.Schwarz BA, et al. Selective thymus settling regulated by cytokine and chemokine receptors. J. Immunol. 2007;178:2008–2017. doi: 10.4049/jimmunol.178.4.2008. [DOI] [PubMed] [Google Scholar]
- 23.Masuda K, et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–4060. doi: 10.1038/sj.emboj.7600878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yokota T, et al. Unique properties of fetal lymphoid progenitors identified according to RAG1 gene expression. Immunity. 2003;19:365–375. doi: 10.1016/s1074-7613(03)00231-0. [DOI] [PubMed] [Google Scholar]
- 25.Allman D, et al. Thymopoiesis independent of common lymphoid progenitors. Nature Immunol. 2003;4:168–174. doi: 10.1038/ni878. [DOI] [PubMed] [Google Scholar]
- 26.Igarashi H, Gregory SC, Yokota T, Sakaguchi N, Kincade PW. Transcription from the RAG1 locus marks the earliest lymphocyte progenitors in bone marrow. Immunity. 2002;17:117–130. doi: 10.1016/s1074-7613(02)00366-7. [DOI] [PubMed] [Google Scholar]
- 27.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV. Developmental and molecular characterization of emerging β- and γδ-selected pre-T cells in the adult mouse thymus. Immunity. 2006;24:53–64. doi: 10.1016/j.immuni.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 28.Schmitt TM, Zuniga-Pflucker JC. Thymus-derived signals regulate early T-cell development. Crit. Rev. Immunol. 2005;25:141–160. doi: 10.1615/critrevimmunol.v25.i2.40. [DOI] [PubMed] [Google Scholar]
- 29.Maillard I, Fang T, Pear WS. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 2005;23:945–974. doi: 10.1146/annurev.immunol.23.021704.115747. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt TM, Zúñiga-Pflücker JC. Induction of T cell development from hematopoietic progenitor cells by Delta-like-1 in vitro. Immunity. 2002;17:749–756. doi: 10.1016/s1074-7613(02)00474-0. By establishing a highly robust, accessible in vitro culture system for early T-cell development, this paper liberated T-cell development from the “black box” of the thymic microenvironment and profoundly transformed the field.
- 31.Silva-Santos B, Pennington DJ, Hayday AC. Lymphotoxin-mediated regulation of γδ cell differentiation by αβ T cell progenitors. Science. 2005;307:925–928. doi: 10.1126/science.1103978. [DOI] [PubMed] [Google Scholar]
- 32.Gounari F, et al. Tracing lymphopoiesis with the aid of a pTα-controlled reporter gene. Nature Immunol. 2002;3:489–496. doi: 10.1038/ni778. [DOI] [PubMed] [Google Scholar]
- 33.Masuda K, et al. T cell lineage determination precedes the initiation of TCRβ rearrangement. J. Immunol. 2007;179:3699–3706. doi: 10.4049/jimmunol.179.6.3699. Using a pLck—GFP transgene to mark a positive switchpoint in T-cell gene expression in living thymocytes, the authors discovered that the DN2 stage is composed of two distinct substages, and that dendritic cell developmental potential is lost by the time that the pLck-GFP transgene is activated. These authors are also pioneers of in vitro clonal analysis of T-lineage commitment.
- 34.David-Fung ES, et al. Progression of regulatory gene expression states in fetal and adult pro-T cell development. Immunol. Rev. 2006;209:212–236. doi: 10.1111/j.0105-2896.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghon TN, David E-S, Zúñiga-Pflücker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19:965–978. doi: 10.1101/gad.1298305. This paper dissects the timing and gene expression signatures correlated with lineage commitment in vitro in the presence or absence of Notch/Delta signals. It shows delayed, nonuniform kinetics of gene activation in both T-cell and B-cell conditions and a surprising degree of reversibility at the population level, both in gene expression and in developmental fate choice.
- 36.Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. doi: 10.1182/blood-2004-08-3087. A detailed phenotypic and behavioral analysis of primitive T-cell precursors in the adult mouse thymus, this study uses limiting dilution cultures and gene expression analysis to show that macrophage developmental potential is maintained by ETP and DN2 thymocytes whereas B-cell potential is not. Macrophages generated from ETP and DN2 cells are phagocytically active and yet may contain limited TCRβ gene rearrangements, confirming their T-lineage origin.
- 37.Tabrizifard S, et al. Analysis of transcription factor expression during discrete stages of postnatal thymocyte differentiation. J. Immunol. 2004;173:1094–1102. doi: 10.4049/jimmunol.173.2.1094. This paper has provided the benchmark for expression analysis of transcription factor genes in murine pro-T cells, based on hybridization to Affymetrix microarrays. The supplementary Tables provide a rich trove of data on hundreds of regulatory genes, and the body of the paper identifies genes that are particularly useful for defining specific transitions.
- 38.Yui MA, Rothenberg EV. Deranged early T cell development in immunodeficient strains of nonobese diabetic mice. J. Immunol. 2004;173:5381–5391. doi: 10.4049/jimmunol.173.9.5381. [DOI] [PubMed] [Google Scholar]
- 39.Chen F, Rowen L, Hood L, Rothenberg EV. Differential transcriptional regulation of individual TCR Vβ segments before gene rearrangement. J. Immunol. 2001;166:1771–1780. doi: 10.4049/jimmunol.166.3.1771. [DOI] [PubMed] [Google Scholar]
- 40.Rothenberg EV, Diamond RA, Pepper KA, Yang JA. Interleukin-2 gene inducibility in T cells prior to T-cell receptor expression: changes in signaling pathways and gene expression requirements during intrathymic maturation. J. Immunol. 1990;144:1614–1624. [PubMed] [Google Scholar]
- 41.Massa S, Balciunaite G, Ceredig R, Rolink AG. Critical role for c-kit (CD117) in T cell lineage commitment and early thymocyte development in vitro. Eur. J. Immunol. 2006;36:526–532. doi: 10.1002/eji.200535760. [DOI] [PubMed] [Google Scholar]
- 42.Kang J, Der SD. Cytokine functions in the formative stages of a lymphocyte's life. Curr. Opin. Immunol. 2004;16:180–190. doi: 10.1016/j.coi.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 43.King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4508–4513. doi: 10.1073/pnas.072087899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benz C, Bleul CC. A multipotent precursor in the thymus maps to the branching point of the T versus B lineage decision. J. Exp. Med. 2005;202:21–31. doi: 10.1084/jem.20050146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambandam A, et al. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nature Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- 46.Lu M, et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J. Immunol. 2005;175:5848–5856. doi: 10.4049/jimmunol.175.9.5848. [DOI] [PubMed] [Google Scholar]
- 47.Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J. Exp. Med. 2004;200:469–479. doi: 10.1084/jem.20040394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen HQ, et al. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J. Immunol. 2003;171:3401–3406. doi: 10.4049/jimmunol.171.7.3401. [DOI] [PubMed] [Google Scholar]
- 49.Wu L, Li C-L, Shortman K. Thymic dendritic cell precursors: relationship to the T lymphocyte lineage and phenotype of the dendritic cell progeny. J. Exp. Med. 1996;184:903–911. doi: 10.1084/jem.184.3.903. This is the original paper that showed persistence of a non-lymphoid developmental option into the DN2 stage of thymocyte development, which was shut off in the DN3 stage. Although clonal analysis was not yet possible, the authors’ findings have been repeatedly confirmed by more recent work.
- 50.Taghon T, Yui MA, Rothenberg EV. Mast cell lineage diversion of T lineage precursors by the essential T-cell transcription factor GATA-3. Nature Immunol. 2007;8:845–855. doi: 10.1038/ni1486. This paper shows that even the essential T-lineage transcription factor GATA-3 can act as a diversionary factor for ETP and DN2 T-cell precursors, if it is expressed at an elevated level. In this case the effect of Notch is reversed so as to become an antagonist of GATA-3 instead of a collaborator. More generally, these findings show an unexpectedly close relationship between the mast-cell developmental programme and the early stages of the T-lineage programme.
- 51.Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. doi: 10.1016/j.immuni.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Franco CB, et al. Notch/Delta signaling constrains re-engineering of pro-T cells by PU.1. Proc. Natl. Acad. Sci. U S A. 2006;103:11993–11998. doi: 10.1073/pnas.0601188103. This paper details the regulatory cascade induced by PU.1 as it diverts thymocytes from the T-cell program to the myeloid or dendritic-cell pathway. In DN2 and later thymocytes, Notch-Delta signaling precisely neutralizes a subset of PU.1 effects, restoring the T-lineage levels of particular target genes. These target genes are shown to be fundamental for T-lineage identity, as they are sufficient to preserve the T-lineage developmental programme even in face of supraphysiological PU.1 expression levels.
- 53.Tydell CC, et al. Molecular dissection of prethymic progenitor entry into the T lymphocyte developmental pathway. J. Immunol. 2007;179:421–438. doi: 10.4049/jimmunol.179.1.421. Using a global gene discovery approach based on de novo cloning, the authors report a panel of transcription factor and signalling genes that are upregulated in early T-lineage development more than in precursors for other lineages. Surprisingly, most of the “T-lineage” regulatory candidates are proven to be “legacy factors”, expressed at stable levels from stem or multipotent progenitors. However, Bcl11b is induced in a uniquely strong, specific way during the ETP to DN2 transition in vivo and at a corresponding point in vitro.
- 54.Tanigaki K, Honjo T. Regulation of lymphocyte development by Notch signaling. Nature Immunol. 2007;8:451–456. doi: 10.1038/ni1453. [DOI] [PubMed] [Google Scholar]
- 55.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deftos ML, Huang E, Ojala EW, Forbush KA, Bevan MJ. Notch1 signaling promotes the maturation of CD4 and CD8 SP thymocytes. Immunity. 2000;13:73–84. doi: 10.1016/s1074-7613(00)00009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maillard I, et al. The requirement for Notch signaling at the β-selection checkpoint in vivo is absolute and independent of the pre-T cell receptor. J. Exp. Med. 2006;203:2239–2245. doi: 10.1084/jem.20061020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rothenberg EV. Regulatory factors for initial T lymphocyte lineage specification. Curr. Opin. Hematol. 2007;14:322–329. doi: 10.1097/MOH.0b013e3281de72a8. [DOI] [PubMed] [Google Scholar]
- 59.Rothenberg EV, Taghon T. Molecular Genetics of T Cell Development. Annu. Rev. Immunol. 2005;23:601–649. doi: 10.1146/annurev.immunol.23.021704.115737. [DOI] [PubMed] [Google Scholar]
- 60.Staal FJT, Weerkamp F, Langerak AW, Hendriks RW, Clevers HC. Transcriptional control of T lymphocyte differentiation. Stem Cells. 2001;19:165–179. doi: 10.1634/stemcells.19-3-165. [DOI] [PubMed] [Google Scholar]
- 61.Ho IC, Pai SY. GATA-3 - not just for Th2 cells anymore. Cell. Mol. Immunol. 2007;4:15–29. [PubMed] [Google Scholar]
- 62.Murre C. Helix-loop-helix proteins and lymphocyte development. Nature Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- 63.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of αβ T lymphocytes. Nature Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 64.Wang D, et al. The basic helix-loop-helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J. Immunol. 2006;177:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- 65.El Andaloussi A, et al. Hedgehog signaling controls thymocyte progenitor homeostasis and differentiation in the thymus. Nature Immunol. 2006;7:418–426. doi: 10.1038/ni1313. [DOI] [PubMed] [Google Scholar]
- 66.Goetz TL, Gu TL, Speck NA, Graves BJ. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor α2. Mol. Cell. Biol. 2000;20:81–90. doi: 10.1128/mcb.20.1.81-90.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gu TL, Goetz TL, Graves BJ, Speck NA. Auto-inhibition and partner proteins, core-binding factor β (CBFβ) and Ets-1, modulate DNA binding by CBFα2 (AML1) Mol. Cell. Biol. 2000;20:91–103. doi: 10.1128/mcb.20.1.91-103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melichar HJ, et al. Regulation of γδ versus αβ T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 69.Weerkamp F, et al. Identification of Notch target genes in uncommitted T-cell progenitors: No direct induction of a T-cell specific gene program. Leukemia. 2006;20:1967–1977. doi: 10.1038/sj.leu.2404396. [DOI] [PubMed] [Google Scholar]
- 70.Höflinger S, et al. Analysis of Notch1 function by in vitro T cell differentiation of Pax5 mutant lymphoid progenitors. J. Immunol. 2004;173:3935–3944. doi: 10.4049/jimmunol.173.6.3935. [DOI] [PubMed] [Google Scholar]
- 71.Schmitt TM, et al. Induction of T cell development and establishment of T cell competence from embryonic stem cells differentiated in vitro. Nature Immunol. 2004;5:410–417. doi: 10.1038/ni1055. [DOI] [PubMed] [Google Scholar]
- 72.Huang J, et al. Propensity of adult lymphoid progenitors to progress to DN2/3 stage thymocytes with Notch receptor ligation. J. Immunol. 2005;175:4858–4865. doi: 10.4049/jimmunol.175.8.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Krueger A, Garbe AI, von Boehmer H. Phenotypic plasticity of T cell progenitors upon exposure to Notch ligands. J. Exp. Med. 2006;203:1977–1984. doi: 10.1084/jem.20060731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lefort N, et al. Short exposure to Notch ligand Delta-4 is sufficient to induce T-cell differentiation program and to increase the T cell potential of primary human CD34+ cells. Exp. Hematol. 2006;34:1720–1729. doi: 10.1016/j.exphem.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 75.Dakic A, et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J. Exp. Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Iwasaki H, et al. Distinctive and indispensable roles of PU.1 in maintenance of hematopoietic stem cells and their differentiation. Blood. 2005;106:1590–1600. doi: 10.1182/blood-2005-03-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Talebian L, et al. T-lymphoid, megakaryocyte, and granulocyte development are sensitive to decreases in CBFβ dosage. Blood. 2007;109:11–21. doi: 10.1182/blood-2006-05-021188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J. Exp. Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. The authors make use of a valuable E2A knockout hematopoietic progenitor cell line and an elegant tamoxifen induction strategy to identify probable direct targets of E2A that are accessible in a precursor-cell context. The results argue strongly against a binary opposition of Notch and E2A effects and support a synergistic and combinatorial interaction for activation of T-lineage genes.
- 79.Maeda T, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan JB, Visan I, Yuan JS, Guidos CJ. Requirement for Notch1 signals at sequential early stages of intrathymic T cell development. Nature Immunol. 2005;6:671–679. doi: 10.1038/ni1217. [DOI] [PubMed] [Google Scholar]
- 81.Krueger A, von Boehmer H. Identification of a T lineage-committed progenitor in adult blood. Immunity. 2007;26:105–116. doi: 10.1016/j.immuni.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Laurent J, Bosco N, Marche PN, Ceredig R. New insights into the proliferation and differentiation of early mouse thymocytes. Int. Immunol. 2004;16:1069–1080. doi: 10.1093/intimm/dxh108. [DOI] [PubMed] [Google Scholar]
- 83.Prockop SE, Petrie HT. Regulation of thymus size by competition for stromal niches among early T cell progenitors. J. Immunol. 2004;173:1604–1611. doi: 10.4049/jimmunol.173.3.1604. [DOI] [PubMed] [Google Scholar]
- 84.Yücel R, Karsunky H, Klein-Hitpass L, Möröy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J. Exp. Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hock H, et al. Intrinsic requirement for zinc finger transcription factor Gfi-1 in neutrophil differentiation. Immunity. 2003;18:109–120. doi: 10.1016/s1074-7613(02)00501-0. [DOI] [PubMed] [Google Scholar]
- 86.Schilham MW, et al. Critical involvement of Tcf-1 in expansion of thymocytes. J. Immunol. 1998;161:3984–3991. [PubMed] [Google Scholar]
- 87.Weerkamp F, et al. Wnt signaling in the thymus is regulated by differential expression of intracellular signaling molecules. Proc. Natl. Acad. Sci. U S A. 2006;103:3322–3326. doi: 10.1073/pnas.0511299103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rosenbauer F, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nature Genet. 2006;38:27–37. doi: 10.1038/ng1679. [DOI] [PubMed] [Google Scholar]
- 89.Hager-Theodorides AL, et al. Bone morphogenetic protein 2/4 signaling regulates early thymocyte differentiation. J. Immunol. 2002;169:5496–5504. doi: 10.4049/jimmunol.169.10.5496. [DOI] [PubMed] [Google Scholar]
- 90.Tsai PT, Lee RA, Wu H. BMP4 acts upstream of FGF in modulating thymic stroma and regulating thymopoiesis. Blood. 2003;102:3947–3953. doi: 10.1182/blood-2003-05-1657. [DOI] [PubMed] [Google Scholar]
- 91.Varas A, et al. The role of morphogens in T-cell development. Trends Immunol. 2003;24:197–206. doi: 10.1016/s1471-4906(03)00033-4. [DOI] [PubMed] [Google Scholar]
- 92.Tsuji M, Shinkura R, Kuroda K, Yabe D, Honjo T. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc. Natl. Acad. Sci. U S A. 2007;104:1610–1615. doi: 10.1073/pnas.0610520104. The unexpected phenotype of removing an inhibitor of Notch-dependent transcription reveals that Notch effects on early T cells depend on a gene regulatory network based on feedback negative regulation. The action of Mint and effects on target gene Nrarp distinguish Notch promotion of ETP self-renewal from Notch promotion of DN2 cell generation.
- 93.Yun TJ, Bevan MJ. Notch-regulated ankyrin-repeat protein inhibits Notch1 signaling: multiple Notch1 signaling pathways involved in T cell development. J. Immunol. 2003;170:5834–5841. doi: 10.4049/jimmunol.170.12.5834. [DOI] [PubMed] [Google Scholar]
- 94.Laiosa CV, Stadtfeld M, Graf T. Determinants of lymphoid-myeloid lineage diversification. Annu. Rev. Immunol. 2006;24:705–738. doi: 10.1146/annurev.immunol.24.021605.090742. [DOI] [PubMed] [Google Scholar]
- 95.Laiosa CV, Stadtfeld M, Xie H, Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. In an extensive and elegantly controlled study, the authors prove that re-expression of either C/EBPα or PU.1 in fully-committed DN3 cells is sufficient to reverse commitment and transform the thymocytes to myeloid or dendritic cells. The authors show some rescue of Thy-1+ cells by coexpression of NotchIC or GATA-3, but not by E2A. NotchIC is more effective at neutralizing the PU.1 effect than at neutralizing effects of C/EBPα, thus emphasizing the critical importance for thymocytes of extinguishing C/EBPα expression early.
- 96.Dionne CJ, et al. Subversion of T lineage commitment by PU.1 in a clonal cell line system. Dev. Biol. 2005;280:448–466. doi: 10.1016/j.ydbio.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 97.Lefebvre JM, et al. Enforced expression of Spi-B reverses T lineage commitment and blocks β-selection. J. Immunol. 2005;174:6184–6194. doi: 10.4049/jimmunol.174.10.6184. [DOI] [PubMed] [Google Scholar]
- 98.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. J. Exp. Med. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ikawa T, Fujimoto S, Kawamoto H, Katsura Y, Yokota Y. Commitment to natural killer cells requires the helix-loop-helix inhibitor Id2. Proc. Natl. Acad. Sci U.S.A. 2001;98:5164–5169. doi: 10.1073/pnas.091537598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K. Enforced Granulocyte/Macrophage Colony-Stimulating Factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J. Exp. Med. 2003;197:1311–1322. doi: 10.1084/jem.20021843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hsu CL, et al. Antagonistic effect of CCAAT enhancer-binding protein-α and Pax5 in myeloid or lymphoid lineage choice in common lymphoid progenitors. Proc. Natl. Acad. Sci. U S A. 2006;103:672–677. doi: 10.1073/pnas.0510304103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shibata Y, et al. GM-CSF regulates alveolar macrophage differentiation and innate immunity in the lung through PU.1. Immunity. 2001;15:557–567. doi: 10.1016/s1074-7613(01)00218-7. [DOI] [PubMed] [Google Scholar]
- 103.Carotta S, Brady J, Wu L, Nutt SL. Transient Notch signaling induces NK cell potential in Pax5-deficient pro-B cells. Eur. J. Immunol. 2006;36:3294–3304. doi: 10.1002/eji.200636325. [DOI] [PubMed] [Google Scholar]
- 104.Rolink AG, Balciunaite G, Demoliere C, Ceredig R. The potential involvement of Notch signaling in NK cell development. Immunol. Lett. 2006;107:50–57. doi: 10.1016/j.imlet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 105.Yokota Y, Mori S. Role of Id family proteins in growth control. J.Cell. Physiol. 2002;190:21–28. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- 106.Brunet de la Grange P, et al. Low SCL/TAL1 expression reveals its major role in adult hematopoietic myeloid progenitors and stem cells. Blood. 2006;108:2998–3004. doi: 10.1182/blood-2006-05-022988. [DOI] [PubMed] [Google Scholar]
- 107.Curtis DJ, et al. SCL is required for normal function of short-term repopulating hematopoietic stem cells. Blood. 2004;103:3342–3348. doi: 10.1182/blood-2003-09-3202. [DOI] [PubMed] [Google Scholar]
- 108.Fisher RC, Lovelock JD, Scott EW. A critical role for PU.1 in homing and long-term engraftment by hematopoietic stem cells in the bone marrow. Blood. 1999;94:1283–1290. [PubMed] [Google Scholar]
- 109.Krosl G, et al. Transcription factor SCL is required for c-kit expression and c-Kit function in hemopoietic cells. J. Exp. Med. 1998;188:439–450. doi: 10.1084/jem.188.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Matushansky I, Radparvar F, Skoultchi AI. CDK6 blocks differentiation: coupling cell proliferation to the block to differentiation in leukemic cells. Oncogene. 2003;22:4143–4149. doi: 10.1038/sj.onc.1206484. [DOI] [PubMed] [Google Scholar]
- 111.Kawazu M, et al. Functional domains of Runx1 are differentially required for CD4 repression, TCRβ expression, and CD4/8 double-negative to CD4/8 double-positive transition in thymocyte development. J. Immunol. 2005;174:3526–3533. doi: 10.4049/jimmunol.174.6.3526. [DOI] [PubMed] [Google Scholar]
- 112.Lieu YK, Kumar A, Pajerowski AG, Rogers TJ, Reddy EP. Requirement of c-myb in T cell development and in mature T cell function. Proc. Natl. Acad. Sci. U S A. 2004;101:14853–14858. doi: 10.1073/pnas.0405338101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bender TP, Kremer CS, Kraus M, Buch T, Rajewsky K. Critical functions for c-Myb at three checkpoints during thymocyte development. Nature Immunol. 2004;5:721–729. doi: 10.1038/ni1085. [DOI] [PubMed] [Google Scholar]
- 114.Emambokus N, et al. Progression through key stages of haemopoiesis is dependent on distinct threshold levels of c-Myb. EMBO J. 2003;22:4478–4488. doi: 10.1093/emboj/cdg434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Growney JD, et al. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 2005;106:494–504. doi: 10.1182/blood-2004-08-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Inoue J, et al. Expression of TCRαβ partly rescues developmental arrest and apoptosis of αβ T cells in Bcl11b−/− mice. J. Immunol. 2006;176:5871–5879. doi: 10.4049/jimmunol.176.10.5871. [DOI] [PubMed] [Google Scholar]
- 117.Ciofani M, Zúñiga-Pflücker JC. Notch promotes survival of pre-T cells at the β-selection checkpoint by regulating cellular metabolism. Nature Immunol. 2005;6:881–888. doi: 10.1038/ni1234. [DOI] [PubMed] [Google Scholar]
- 118.Weng AP, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol. Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wojciechowski J, Lai A, Kondo M, Zhuang Y. E2A and HEB are required to block thymocyte proliferation prior to pre-TCR expression. J. Immunol. 2007;178:5717–5726. doi: 10.4049/jimmunol.178.9.5717. Using a double conditional knockout mouse system, the authors show that cells which reach the DN3 stage before deleting E2A and HEB seem to reverse course after deleting them, and return to extended IL-7-dependent proliferation and a DN2-like phenotype. At this stage of development, E proteins appear to have a role specifically focused on establishing the unique features of DN3 cells, in spite of other evidence for E protein activity in earlier stages.
- 122.Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc. Natl. Acad. Sci. U S A. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Visan I, et al. Regulation of T lymphopoiesis by Notch1 and Lunatic fringe-mediated competition for intrathymic niches. Nature Immunol. 2006;7:634–643. doi: 10.1038/ni1345. [DOI] [PubMed] [Google Scholar]
- 125.Besseyrias V, et al. Hierarchy of Notch-Delta interactions promoting T cell lineage commitment and maturation. J. Exp. Med. 2007;204:331–343. doi: 10.1084/jem.20061442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Engel I, Johns C, Bain G, Rivera RR, Murre C. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 2001;194:733–746. doi: 10.1084/jem.194.6.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Jepsen K, et al. Combinatorial roles of the nuclear receptor corepressor in transcription and development. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 128.Pennington DJ, et al. The inter-relatedness and interdependence of mouse T cell receptor γδ+ and αβ+ cells. Nature Immunol. 2003;4:991–998. doi: 10.1038/ni979. [DOI] [PubMed] [Google Scholar]
- 129.Tanigaki K, et al. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity. 2004;20:611–622. doi: 10.1016/s1074-7613(04)00109-8. [DOI] [PubMed] [Google Scholar]
- 130.Xue H-H, et al. GA binding protein regulates interleukin 7 receptor α-chain gene expression in T cells. Nature Immunol. 2004;5:1036–1044. doi: 10.1038/ni1117. [DOI] [PubMed] [Google Scholar]
- 131.Porritt HE, et al. Heterogeneity among DN1 prothymocytes reveals multiple progenitors with different capacities to generate T cell and non-T cell lineages. Immunity. 2004;20:735–745. doi: 10.1016/j.immuni.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 132.Hoffman ES, et al. Productive T-cell receptor β-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev. 1996;10:948–962. doi: 10.1101/gad.10.8.948. [DOI] [PubMed] [Google Scholar]
- 133.Izon DJ, et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity. 2002;16:231–243. doi: 10.1016/s1074-7613(02)00271-6. [DOI] [PubMed] [Google Scholar]
- 134.Okamura RM, et al. Redundant regulation of T cell differentiation and TCRα gene expression by the transcription factors LEF-1 and TCF-1. Immunity. 1998;8:11–20. doi: 10.1016/s1074-7613(00)80454-9. [DOI] [PubMed] [Google Scholar]